Abstract
TGFβ plays key roles in fibrosis and cancer progression, and latency is conferred by covalent linkage to latent TGFβ binding proteins (LTBPs). LTBP1 is essential for TGFβ folding, secretion, matrix localization and activation but little is known about its structure due to its inherent size and flexibility. Here we show that LTBP1 adopts an extended conformation with stable matrix-binding N-terminus, extended central array of 11 calcium-binding EGF domains and flexible TGFβ-binding C-terminus. Moreover we demonstrate that LTBP1 forms short filament-like structures independent of other matrix components. The termini bind to each other to facilitate linear extension of the filament, while the N-terminal region can serve as a branch-point. Multimerization is enhanced in the presence of heparin and stabilized by the matrix cross-linking enzyme transglutaminase-2. These assemblies will extend the span of LTBP1 to potentially allow simultaneous N-terminal matrix and C-terminal fibrillin interactions providing tethering for TGFβ activation by mechanical force.
TGFβ is a potent cytokine with key roles in development and homeostasis. TGFβ is secreted in a latent form, non-covalently bound to its cleaved propeptide which is disulphide linked to LTBP family proteins which assist in folding and secretion1. However, there are also instances where LTBPs are secreted independent of TGFβ2. The TGFβ-binding LTBPs −1, −3 and −4 are critical for correct deposition and subsequent bioavailability of TGFβ in the extracellular matrix3. LTBP2 does not bind TGFβ and has TGFβ-independent functions4, while the other LTBPs are also involved in cell adhesion5, elastogenesis6 and stabilization of cell surface receptors7, independent of their TGFβ-binding role.
In addition to these roles, it has also been speculated that LTBPs may contribute to matrix architecture based on their similarity to the fibrillin family8. They are composed of three 8-cysteine domains, sixteen EGF-like repeats and a hybrid domain with proline-rich interdomain regions9. LTBP1 is known to be calcium binding and this interaction is thought to induce structural changes and protect the protein from proteolysis10. In TGFβ-binding members of the family, the latent complex associates with the second 8-cysteine domain11. In addition to localization of the growth factor, this association allows mechanical activation of TGFβ12 through interaction of the TGFβ prodomain with integrin αVβ63. Tensile force exerted through matrix-anchored LTBP1 and latent complex bound integrins alters the prodomain conformation releasing the growth factor13. The C-terminus of LTBP1 also associates with fibrillin8, while the N-terminus binds to heparin14,15, fibulin-516 and fibronectin17. The N-terminus has been shown to be a substrate for transglutaminase-2 (TG2)18 which promotes its covalent incorporation into the extracellular matrix19. LTBPs can undergo alternative splicing as described for LTBP120 (of which the best characterized are short-form (LTBP1S) and long-form21), LTBP2 and LTBP422.
For a protein with clear mechanical functions, study of the structure is critical to a full understanding of its effects. The TGFβ binding domain has been previously solved revealing a disulfide bond which can be removed without disrupting domain fold, flanked by charged residues thought to mediate complex formation23. Moreover NMR studies have demonstrated that the domain pairs cbEGF-TB3 and TB3-EGF (both towards the C-terminal end of LTBP1) had high flexibility24 which may increase its accessibility for matrix interaction. The central region is occupied by eleven calcium-binding EGF-like (cbEGF) repeats. This type of domain arrangement has been predicted to produce a rod-like structure stabilized by calcium binding between neighboring domains25. However, in the related protein fibrillin-1, it produces an extended non-linear conformation26. A hinge region has been postulated to exist in the N-terminus of LTBP1 based on accessibility of a protease sensitive sequence27 though it is not clear whether this is a flexible region or whether it adapts discrete conformations in the presence of binding partners.
While it has been recently established that the function of LTBPs extends beyond TGFβ regulation6,7, the role of LTBPs in matrix architecture remains unresolved8. Here we investigate the nanostructure of LTBP1 and its capacity to assemble higher order oligomers in the absence of other factors. Our study demonstrates that the N-terminal region is rigid while the central cbEGF region is flexible but held in a predominantly extended conformation by calcium. We show calcium dependent multimerization of LTBP1 into filament-like structures which is N-terminally driven, promoted in the presence of heparin and stabilized by the matrix cross-linking enzyme TG2.
Results
LTBP1 can oligomerize via the N-terminus in a calcium dependent manner
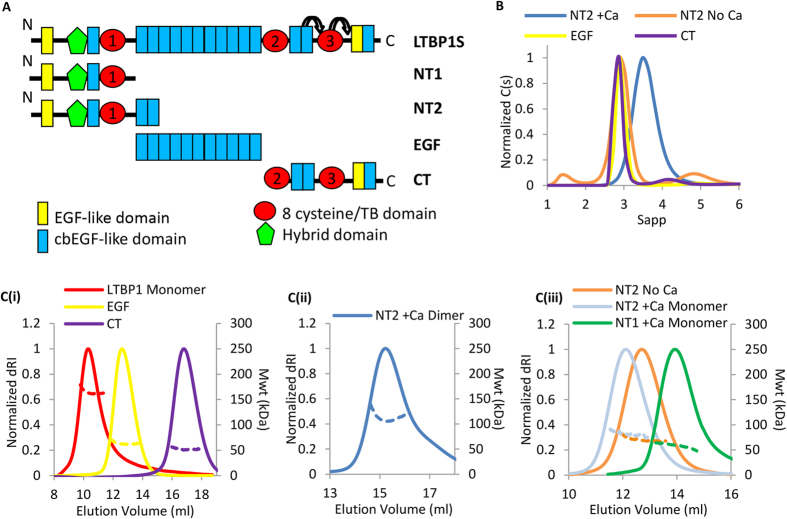
Schematic diagrams of LTBP1S and the truncated constructs used in this study are shown in Fig. 1A. These were expressed in HEK-293 EBNA cells and purified (Fig. S1). These constructs were characterized using size exclusion chromatography (SEC)-multiangle light scattering (MALS) and analytical ultracentrifugation (AUC)(Fig. 1B,C, respectively) to determine oligomeric state and hydrodynamic properties (Table 1). While the EGF and CT constructs were found to be monomers, constructs containing the N-terminal domains also contained dimers and/or larger species in addition to the monomeric species in the presence of 2 mM CaCl2. The dimers and oligomers could be dissociated in the presence of 2 mM EDTA or EGTA (Fig. 1B,C and S1B) suggesting that the interaction is calcium dependent. Monomeric LTBP1, NT1 and NT2 species (Fig. 1C(i) and (iii)) could be separated from dimer and higher order oligomers by SEC but reassociated with time in the presence of calcium so were therefore used rapidly following purification for the analyses described below. The hydrodynamic data obtained from AUC, including the hydrodynamic radii and frictional ratios indicate that all the LTBP1 constructs are elongated and non-globular (Table 1).
Figure 1. LTBP1 can oligomerize via the N-terminus in a calcium-dependent manner.
(A) Schematic diagram showing the domain organization of full-length LTBP1S and the constructs NT1, NT2, EGF and CT. The three TB domains are shown as red circles and numbered and flexible regions either side of TB3 defined in ref. 24 are indicated by arrows. (B) AUC analysis of the constructs, the NT2 region is analyzed with either 2 mM calcium (blue) or 2 mM EDTA (orange) to show a monomer-dimer shift. (C) SEC-MALS analysis showing differential refractive index (dRI) solid line and molecular mass (kDa) dashed line as a function of elution volume for (i) LTBP1, EGF and CT, (ii) NT2 dimer in the presence of calcium, (iii) NT2 monomer in the presence of calcium or 2 mM EGTA and NT1 monomer in the presence of calcium. For panels C(i) and C(iii) a Superdex 200 column was used whereas for panel C(ii) a Superose 6 column was used.
Table 1. Hydrodynamic parameters from different biophysical techniques.
| NT2 | EGF | CT | |||
|---|---|---|---|---|---|
| Hydrodynamic Properties | Monomer | Dimer | |||
| Molecular Weight (kDa) | Sequence | 66 | 132 | 54.5 | 45 |
| MALS | 70.9* | 135 | 62.5 | 52.5 | |
| AUC | 77* | 186 | 72 | 52 | |
| Dmax (nm) | SAXS | 20.0 | N.D. | 23.2 | 17.5 |
| Rg (nm) | SAXS | 6 | N.D. | 7.6 | 4.9 |
| Rh (nm) | MALS | 6.23* | 7.9 | 7.27 | 5.26 |
| AUC | 6* | 8.2 | 6.64 | 5.51 | |
| f/f0 | AUC | 1.95* | 2.44 | 2.87 | 2.34 |
| S20W | AUC | 3.08* | 3.9 | 2.7 | 2.75 |
N.D. = not determined.
*Data collected in the presence of 2 mM EDTA.
LTBP1 N-terminus has an elongated stable conformation
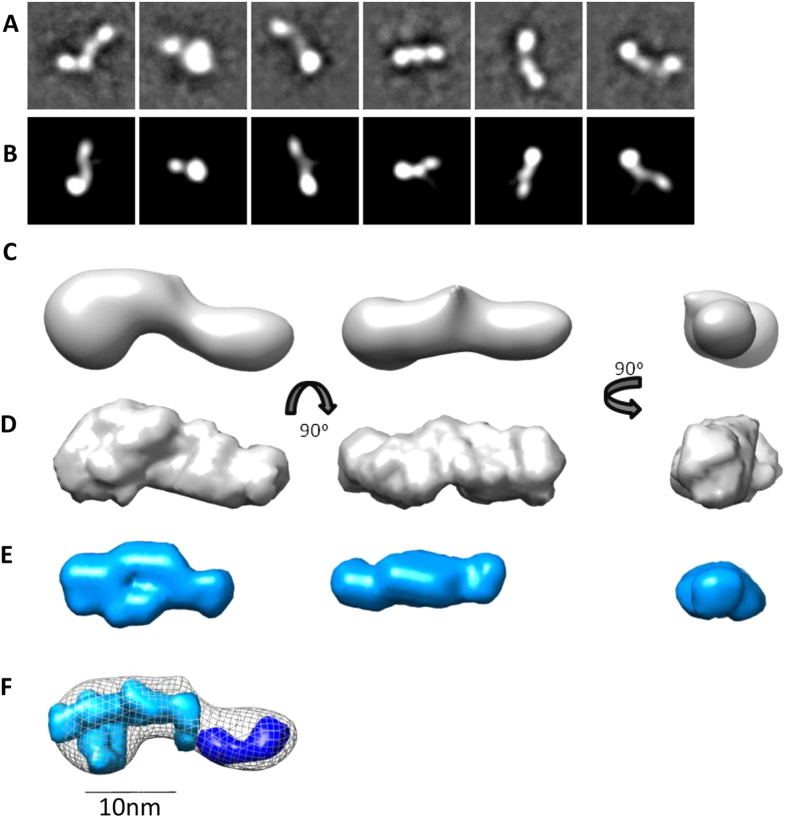
The N-terminus of LTBP1 has many interdomain linkers and a putative hinge region27 so may be expected to demonstrate flexibility. To determine if this region was flexible, monomeric NT2 was prepared by SEC in the presence of calcium to isolate a monomeric species and was imaged by negative staining EM. 10,000 particles were selected and classified (Fig. 2A) and a 3D model at 24 Å resolution calculated by angular reconstitution (Fig. 2B,C). The convergence to a single model suggests that the protein is relatively inflexible. The structure had an elongated shape with dense lobe at one end, with dimensions 22 nm × 9 nm. The structure was further assessed in solution using small angle X-ray scattering (SAXS). Data on NT1 and NT2 regions both in the presence of calcium were obtained using SEC-SAXS, in which scattering measurements are made directly on the eluant from SEC to collect data on monomeric species. Data quality were assessed using the Guinier region (Figs S2 and S3) and to determine the radius of gyration (Rg) shown in Table 1. Analysis of flexibility from the Porod-Debye plot (Figs S2E and S3E) indicated that the N-terminal regions adopt a single conformation, a finding which supports the EM data. The maximum dimension (Dmax) for the NT2 region was 20 nm which is also consistent with the EM 3D model and 15 nm for the NT1 region. Ab initio models for the NT2 region strongly correlate with the EM model (Fig. 2D) and as expected the NT1 region was smaller with a shorter elongated protrusion (Fig. 2E). The relative densities of the NT1 and NT2 regions were modelled with MONSA28(Fig. 2F). The additional density present in the NT2 construct is consistent with the presence of an additional two cbEGF domains and locates the N-terminus in the larger lobe.
Figure 2. The N-terminal region of LTBP1 has an elongated stable conformation.
(A). Negative staining EM of NT2, analyzing 10,000 particles from 150 micrographs. Representative class averages from 100 classes generated using EMAN2. (B) Matched projections from the 3D model (box size = 35.8 nm). (C) EM 3D reconstruction of NT2 shown in three orthogonal views. SAXS analysis of the NT2 and NT1 regions in solution. Ab initio models of NT2 (D) and NT1 (E) regions generated using DAMMIF software shown in three orthogonal views. (F) Modelling of the difference in density between the NT2 and NT1 regions was performed with MONSA. The NT1 density is shown in cyan and the additional density in the NT2 region is shown in blue and superimposed with the NT2 EM model. For panels (C–F) the scale bar = 10 nm.
LTBP1 N-terminus displays calcium dependent oligomerization enhanced by heparin binding
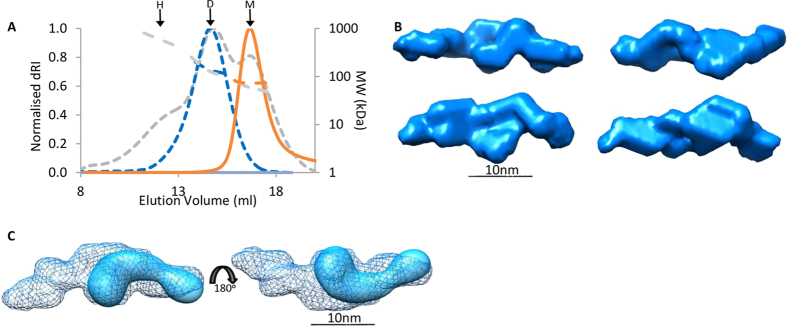
We then investigated whether the N-terminal oligomers are able to form spontaneously in the absence of endogenous matrix components. From SEC, a monomer of NT2 was isolated and then stored overnight at 4 °C in either 2 mM calcium or 2 mM EGTA. These samples were then analyzed by MALS and in the absence of calcium (2 mM EGTA) the NT2 region remained monomeric. However, in the presence of calcium a mixture of distinct peaks corresponding approximately to monomer, dimer and higher order assemblies were observed (Fig. 3A). The mass measurement in MALS is more accurate for a single species therefore to verify the size of the predominant species, the dimer fraction was rerun immediately, confirming the dimer to be the major species. However, if the dimer was stored for longer periods of time, it would oligomerize into a mixture of dimer and higher order species. These data demonstrate that oligomerization can occur spontaneously in vitro in the absence of other factors to form an equilibrium of monomer, dimer and higher order oligomers and suggests that multimerization is calcium-dependent. To determine the arrangement of the N-terminal oligomers, SAXS data for a dimer of the NT1 region was collected. Ab initio models of the dimer were calculated in the absence of symmetry constraints and a panel of representative shapes shown (Fig. 3B). These all indicate a head-to-head arrangement via the globular region, suggesting an interaction mediated via the very N-terminal domains. This is further illustrated with the superimposition of the EM map which fits in this orientation into the dimer (Fig. 3C).
Figure 3. LTBP1 N-terminus multimerizes in a head-to-head manner.
(A) SEC-MALS analysis of NT2 in 2 mM calcium or 2 mM EGTA on a Superose 6 column. Spontaneous oligomerization in the presence of calcium resulted in a mix of monomer (M = 54 kDa), dimer (D = 124 kDa) and higher order structures (H = 565 kDa) (gray trace) whereas in 2 mM EGTA only a monomer was observed (orange). To verify that the dominant species was a dimer, this fraction was run again (blue) and had Mwt of 135 kDa (Rh = 7.6 nm ± 1.017%). (B) SAXS data was collected on a dimer of the NT1 region and analyzed by ab initio modelling. A panel of four representative ab initio models is shown. (C) The NT1 monomer can be modelled into the dimer envelope suggesting an end-to-end arrangement.
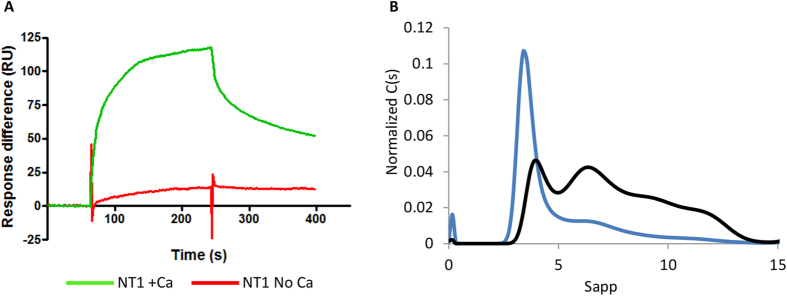
Previous studies have shown that full length LTBP1 binds to heparin29. In order to investigate which region in LTBP1 is responsible for this interaction, Surface Plasmon Resonance was used to test for binding of each of the fragments against immobilized heparin dp-20 in the presence of calcium or EGTA. While no binding was observed to the cbEGF or C-terminal regions (Fig. S4), the N-terminus showed binding in a calcium-dependent manner (Fig. 4A). Heparin has previously been demonstrated to affect the oligomeric assembly of related matrix proteins30. We therefore investigated the larger structures formed by the N-terminus and whether heparin has an effect on multimerization. Whereas NT1 predominantly formed a dimer, when pre-incubated with heparin a range of higher order species were observed indicating heparin enhanced multimerization (Fig. 4B).
Figure 4. LTBP1 N-terminus displays calcium-dependent binding to heparin which promotes oligomerization.
(A) Biacore analysis of 50 nM NT1 binding to immobilized heparin dp-20 in the presence of calcium (green) or 5 mM EGTA (red). (B) AUC analysis of NT1 in the absence (blue) and presence of heparin (black) showing increased propensity to form higher order oligomers.
The central cbEGF region of LTBP1 is held in an extended conformation by calcium
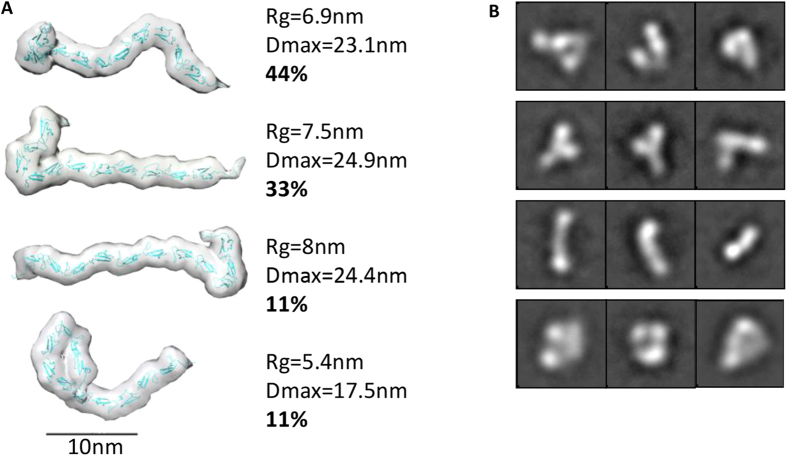
Arrays of cbEGF domains have been predicted to have a linear rod like arrangement based on the extrapolated structure of cbEGF domain pairs from fibrillin-125. However, solution studies on longer arrays of cbEGF domains have indicated that they may have a more compact arrangement26. To determine how the 11 contiguous cbEGF domains in LTBP1 are structured, we used SAXS to determine their shape in solution. Data were collected in the presence and absence of calcium to determine how this effects their flexibility and conformation. In the presence of calcium this region had a Rg of 7.6 nm and an average Dmax of 23.2 nm, compared to a rod of 11 cbEGF domains which would have a theoretical Dmax of 32 nm and Rg of 9.6 nm. However after treatment with EDTA the Rg decreased to 5.4 nm indicating a conformational collapse on removal of calcium (Fig. S5). In the presence and absence of calcium the cbEGF region demonstrated flexibility as assessed by q4 plot31 (Fig. S5). Therefore, the data were processed assuming this region adopted an ensemble of different conformers in solution. Using the ensemble optimization method (EOM), 10,000 models were generated which are then compared with the experimental SAXS data to calculate an ensemble that together account for the scattering data. In the presence of calcium the ensemble contained four different conformations in different proportions with Rg ranging from 5.4–8 nm (Fig. 5A). The Rh values obtained from MALS and AUC and frictional ratio (Table 1) also suggest that elongated conformations are common in solution. These conformations ranged from an elongated structure with a kink at one end to a collapsed conformation with half the domains folding back on themselves. To validate whether these conformations are observed experimentally, the cbEGF region in the presence of calcium was imaged by EM. A range of class averages were observed that correlated with the models predicted indicating that this region has flexibility even in the presence of calcium (Fig. 5B). The overall conformation of this region is elongated but not linear and supports the concept that contiguous arrays of cbEGF domains are not rod-like units.
Figure 5. The central region of 11 cbEGF repeats of LTBP1 is flexible but held in an extended conformation by calcium.
(A) Array of models produced using EOM from the EGF region X-ray scattering data in the presence of calcium. Extended conformations are more prevalent. (B) Class averages from negative stain EM showing a range of conformations reflected in the SAXS models, which are unable to resolve to a single 3D model (box size = 44.8 nm).
The C-terminal LTBP1 region is flexible
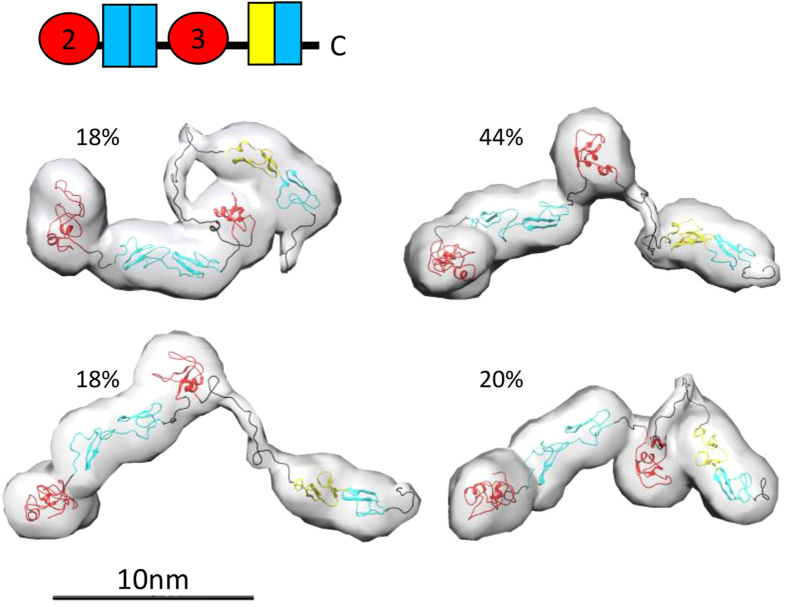
Recent NMR studies have shown that the three terminal domains of LTBP1 have considerable domain-domain flexibility24. In order to determine if the C-terminal region of LTBP1 encompassing these domains plus the upstream domain responsible for binding latent TGFβ is also flexible we performed SAXS. These data indeed show that this longer region is flexible (Fig. S6) and ensemble analysis provides four models that collectively give rise to the scattering data. These models range in Rg from 4.5–6.2 nm and Dmax from 14.6–19.8 nm, which shows quite considerable variation in size, supporting the premise that this region is indeed flexible (Fig. 6). The Rg/Rh and frictional ratios are higher than expected for a globular protein suggesting moderate elongation of this region (Table 1).
Figure 6. The C-terminal LTBP1 region is flexible.
Array of models produced using the ensemble method from CT SAXS data with their relative proportion indicated. The two pairs of EGF domains in the CT construct were each treated as a rigid body. Scale bar = 10 nm.
Assembly of LTBP1 via N-N or N-C terminal interactions
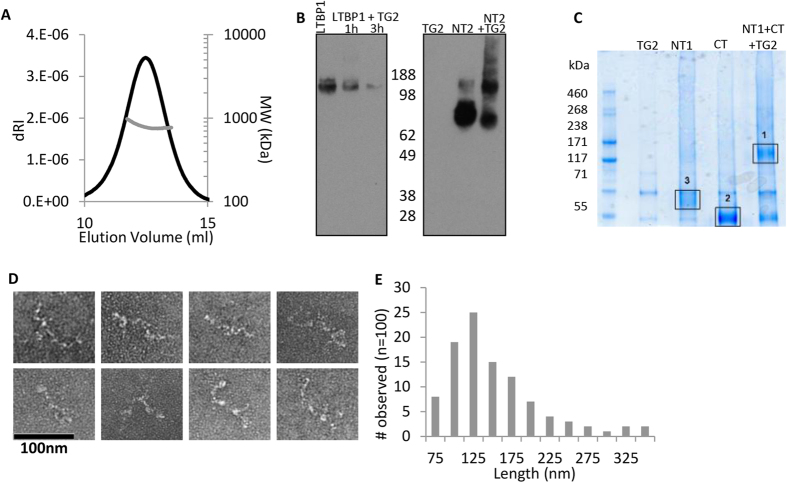
To determine the structure and oligomeric assembly of full-length LTBP1, it was purified and analyzed by MALS. Two main species were separated by SEC corresponding to a monomer and oligomer (Fig. S1B and Fig. 1C(i)). The molecular mass of the larger species was 790 kDa which is approximately 4–5 times the size of the monomer (Fig. 7A). In order to determine how this oligomer is formed, cross-linking experiments with TG2 were performed. It has already been demonstrated that LTBP1 is a substrate for TG218 and that the N-terminal region is involved in cross-linking but it is thought that LTBP1 is cross-linked to fibronectin or other matrix components32. To determine whether LTBP1 was able to form inter-molecular transglutaminase cross-links, full-length LTBP1 was cross-linked by TG2 as evidenced by the disappearance of this species on SDS-PAGE gel (Fig. 7B), presumably as the cross-linked form became too large to enter the gel. The NT, EGF and CT regions were then incubated in the presence of TG2. Only the NT region was able to be cross-linked to itself (Fig. 7B and Fig. S7), providing further support for self-association at the N-terminus. However when the N- and C-terminal regions were incubated together higher molecular weight species formed containing peptides from both the N- and C-terminal regions indicating that these regions interact and this could be stabilized by a cross-link (Fig. 7C and Fig. S7). Together these data suggest that LTBP1 can self-assemble, in the absence of other matrix molecules, via N-N or N-C terminal interactions. To determine what form the higher order LTBP1 oligomers took, they were imaged by EM which showed thin elongated structures (Fig. 7D) with average length of 130 nm (Fig. 7E). These length measurements are consistent with 4–5 LTBP1 molecules in an end-to-end arrangement.
Figure 7. LTBP1 oligomerization via N-N or N-C terminal interactions.
(A) SEC-MALS analysis of oligomeric LTBP1 indicates a molecular mass of 790 kDa. (B) Western blot showing cross-linking of LTBP1 using TG2, for full length LTBP1 a trace amount of dimer is visible after 1 hour. However after 3 hours the majority of cross-linked LTBP1 is too large to enter the gel. An equal amount of total protein was loaded into each well. The N-terminus alone forms monomer, dimer, trimer and tetramer with some higher order species. (C) TG2 cross-linking of the N- and C-terminal regions. Peptides from these bands were identified by MS with band 1 positive for both the N- and C-termini and bands 2 and 3 only positive for peptides from the C- or N-termini, respectively. (D) Oligomers were imaged using negative stain EM and analyzed using ImageJ. (E) Frequency of observation of the maximum straightened length of the oligomer (n = 100 from 32 images).
LTBP1 is elongated with flexible regions
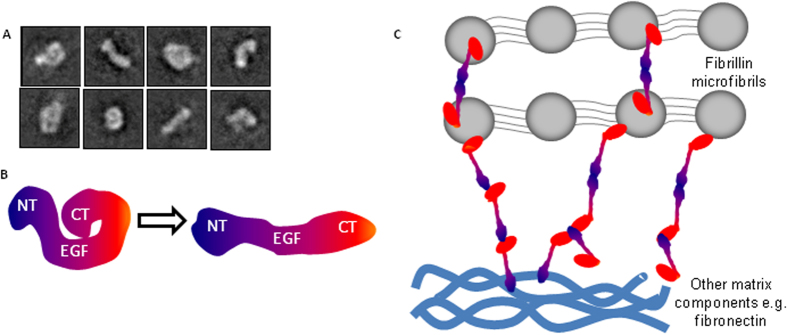
In order to determine how the shorter constructs are arranged in the full-length molecule, monomeric full length LTBP1 was applied to grids for EM analysis (Fig. 8A). A range of classes were observed including elongated shapes and others more compact with the termini folding inwards. The maximum contour length (55 nm) of the compact particles was measured using ImageJ and approximately corresponds to the combined maximum dimensions of the NT1 (15 nm), EGF (23.2 nm) and CT regions (17.5 nm). Some particles appear to adopt more extended conformations, however other class averages are shorter than expected, presumably due to flexibility around the C-terminus. A model is therefore proposed in Fig. 8B, whereby LTBP1 has a rigid N-terminal region, an extendable EGF region and flexible C-terminus with the potential to either extend to give rise to elongated shapes or fold back to give rise to the compact conformations. The oligomers have regions of density consistent with both elongated and compact LTBP1 conformations and so it is probable that both forms exist within these assemblies. This model indicates an end-to-end arrangement consistent with the assembled oligomeric filaments. In summary, we propose in Fig. 8C the arrangement for full length LTBP1 deposition in the matrix whereby linear strands can form in an N-N or N-C orientation with the N-terminus also able to act as a branching point.
Figure 8. LTBP1 has flexible regions and can form small filamentous structures.
(A) Selected class averages of full length LTBP1. Box size = 39.7 nm. (B) Cartoon schematic showing LTBP1 alternating between a compact and elongated conformation with flexibility toward the C-terminus. (C) Schematic diagram showing hypothesized arrangement of LTBP1 in the matrix with oligomers forming via N-N or N-C interactions cross-linked by TG2, with the N-terminus capable of acting as a branching point. In this way LTBP oligomers could span short distances between fibrillin microfibrils, and link fibrillin and fibronectin filaments.
Discussion
LTBP1 is essential for correct spatial and temporal regulation of TGFβ signaling and also important TGFβ-independent roles are emerging. However, to date studies on LTBP1 structure and assembly have been hampered by its large size and inherent flexibility24. Therefore we have determined the nanoscale structure of LTBP1 and higher order oligomers using combined structural and biochemical approaches. We show that the LTBP1 N-terminal region is not flexible, unlike the other regions of the molecule, and multimerizes into dimer and higher order oligomers in the presence of calcium. LTBP1 contains many calcium binding EGF domains and calcium levels in the matrix are typically many orders of magnitude higher than the intracellular concentration33. Our N-terminal constructs contain the putative “hinge” region, named because it contains protease-sensitive sequences flanked by ECM and TGFβ binding regions27, which suggests this region adopts a defined rather than flexible conformation perhaps able to adopt different conformations in the presence of binding partners. Full-length LTBP1 can adopt compact conformations with a flexible C-terminal region, these compact conformations may promote matrix diffusion and the partially buried C-terminus could serve to shield the growth factor from matrix proteases. Other conformations are more extended; the elongation of LTBP1 could contribute to mechanical force transduction from LTBP1 to the growth factor mediating integrin activation. Indeed tethering of the large latent TGFβ complex is required for force induced activation12.
It has been unclear whether LTBP1 is capable of forming its own fibrillar network or if it is dependent on scaffolds such as fibrillin or fibronectin for deposition34. LTBP1 and fibrillin have been shown to colocalize in early matrix deposition, yet in more mature cultures they occupy distinct networks35. Furthermore, fibroblasts lacking fibrillin1 did not deposit LTBP-136 and knockdown of fibrillin1 also disrupted LTBP1 deposition29, however, tissues lacking fibrillin1 stained positively for LTBP1 suggesting other molecules such as fibronectin or fibrillin2 can compensate37. Our data show that LTBP1 is able to self-associate in an N- to N- and N- to C-terminal manner, supporting the concept that LTBP1 can autonomously assemble. Our data suggests that LTBP1 elongates in an end-to-end manner while the N-terminus may act as a branching point. Small filaments seen to bridge fibrillin microfibrils in the ciliary zonules of the eye38, could represent short LTBP filaments connecting adjacent microfibrils, indeed LTBP2 is required for correct ciliary zonule structure and microfibril bundling4.
LTBP1 can be cross-linked to other ECM components by the matrix cross-linking enzyme TG218. We show that TG2 can also form cross-links between LTBP1 molecules, also confirming the specificity of self-association. The N-terminal region was able to cross-link to itself or to the C-terminus but no cross-links were detected between C-termini consistent with other studies18. In the absence of other matrix components, LTBP1 assembled preferentially into structures of approximately five monomer units as determined by mass and length. Interestingly, despite its similarity to fibrillin domain structure, LTBP1 forms elongated string-like structures whereas fibrillin forms bead-like globular structures39. However, LTBP1 and fibrillin also share structural features including an extended, non-linear conformation of cbEGF repeats26. While the independent LTBP1 structures are relatively short and without apparent bundling or periodicity, it is probable that LTBP1 assembly in vivo is heavily influenced by other matrix proteins.
Another candidate for influencing LTBP assembly is heparan sulfate (HS) and heparan sulfate proteoglycans (HSPGs). HS has previously been shown to bind to LTBP129, and this interaction is conserved in LTBP2 which, like LTBP1, interacts via the N-terminus15. This binding may disrupt elastogenesis by displacing elastin from cell surface HSPGs16. However LTBP4 promotes elastogenesis40 suggesting that the effect of LTBPs on this pathway may be type and/or tissue specific. Our data reveal a novel feature of the LTBP-HS interaction, as in the presence of heparin (a more-sulfated variant of HS), LTBP1 has an increased capacity to self-assemble. The function of LTBP1 oligomerization is not yet known. However, loss of LTBP2 which is not directly involved in growth factor activation causes ciliary zonule fragmentation due to tissue specific failure of microfibril function4. This links to the emerging role for the LTBP family in stabilization of matrix components either by tethering to the matrix or by transmitting mechanical force13. This must function through the covalent tethering of the N-terminus of LTBPs to one protein and the C-terminus to another. One intriguing possibility is that oligomerization of LTBP1 extends its reach, allowing it to span longer distances between binding partners and may provide additional covalently-linked tethering sites to facilitate mechanical activation.
In summary, we show that LTBP1 has the capacity to form autonomous oligomeric structures in a calcium dependent manner which is promoted by heparin and interactions can be stabilized by TG2 cross-linking. The assembly is N-terminally driven and we propose a model of combined N- to C- and N- to N- elongation with the N-terminus also functioning as a branching point. Our structural data demonstrate that LTBP1 has the ability to self-assemble in the absence of other components and the capacity to directly contribute to matrix architecture.
Materials and Methods
Protein Expression and Purification
Constructs LTBP1S, CT (residues 1002–1395)29, NT1 (residues 21–488), NT2 (residues 21–630), and EGF region (residues 526–1014) were expressed in HEK-293 EBNA cells and purified using C-terminal polyhistidine tags via nickel affinity chromatography followed by size exclusion chromatography (SEC). Purity of the constructs was verified using SDS-PAGE and the identity of proteins confirmed using Mass Spectrometry (MS) of excised bands.
Multi Angle Light Scattering
Samples (0.5 ml at ~0.5 mg ml−1) were injected onto a SEC column (Superdex 200 or Superose 6) at 0.75 ml min−1. Unless otherwise stated, buffer conditions were 10 mM Tris, 150 mM NaCl, 2 mM CaCl2, pH 7.4. Calcium concentrations reflect levels in the matrix and those used for previous studies on other members of the fibrillin superfamily26,41. Samples were passed through a Wyatt DAWN Heleod II EOS 18-angle laser photometer. This was coupled to a Wyatt Optilab rEX refractive index detector and data were analyzed using Astra 6 software.
Analytical Ultracentrifugation
Samples (~0.5 mg ml−1) were analyzed in the same buffers as for MALS. Studies were carried out using a Beckman XL-A ultracentrifuge as described previously42. To study changes in the presence of heparin, samples were pre-incubated for 12 hours with an approximately 10x molar excess of low molecular weight heparin (Iduron, 1800–7500 Da). Unbound heparin was removed using SEC.
Small Angle X-ray Scattering
Data were collected using inline SEC-SAXS on a Superdex 200 3.2/300 column at the ESRF on Beamline BM29 and at Diamond Light Source on beamline B21. Data were collected at 1 second intervals and the buffer blank was the eluent after one column volume. The scattering images obtained were spherically averaged using in-house software then protein scattering intensities were scaled and merged for each frame with a consistently similar Rg across the SEC elution peak and buffer subtracted using ScÅtter http://www.bioisis.net/tutorial/9. Ab initio modelling was performed using DAMMIF43 software in slow mode. The representative model had the lowest normalized spatial discrepancy from 20 repeats compared using DAMSEL44. Multiphase volumetric modelling was performed using MONSA28 to analyze the difference in density between NT1 and NT2 constructs. SwissModel was used to generate models of the individual domains with the exception of TB3 for which the structure is known23. As the EGF and CT regions were flexible they were analyzed as an ensemble, 10,000 models were generated using Ranch. Ranch generates 10,000 independent models using the amino acid sequence and high resolution structures of domains as the input. Within these constraints the models have random conformation and aim to cover all possible conformations. An ensemble of models representing the experimental SAXS data was generated by EOM from the pool of 10,000 models45.
Negative Stain EM
Full length LTBP1S, EGF and NT2 were negatively stained as in ref. 42. Data were recorded at 23000 × on a Tecnai Biotwin at 120 kV with a Gatan Orius CCD camera with a 1 s exposure at 0.5–1.5 μm defocus range at 2.8 Å/pixel for NT2 and 3.5 Å/pixel for EGF. Data were recorded on a FEI G2 Polara at 300 keV for LTBP1S at 3.1 Å/pixel. Using EMAN246, 10,000 particles were picked for EGF and NT2 and 2500 for full length LTBP1 using a combination of manual and semi-automated picking. Contrast function was corrected and each dataset subject to 2D classification. For NT2, these were used to generate an initial 3D model to seed six rounds of iterative refinement to produce a self-consistent 3D structure. Data was split into odd and even numbers and each refined independently to produce a resolution curve for each stage of refinement. With a Fourier shell correlation cut-off value of 0.143, the resolution estimate was 24 Å. Modelling was performed using UCSF Chimera47.
Cross-linking
Guinea pig liver derived commercial transglutaminase-2 (Sigma-Aldrich) was incubated with LTBP1 fragments in a ratio of 0.1:1 for 2 hours at 30 °C in 10 mM Hepes, 150 mM NaCl pH 7.4 (HBS) buffer containing 1 mM CaCl2. When cross-linking mixed samples, each component was included at equal molar ratios. Reactions were halted by heating to 90 °C and addition of detergent. Products were analyzed by western blotting using anti-histidine monoclonal antibody (R&D Systems).
Surface Plasmon Resonance
50 nM of recombinant fragments in HBS buffer with 0.005% (v/v) Tween-20 were injected over immobilized heparin saccharide dp-20 (Iduron) biotinylated using the cis-diol method on a streptavidin sensor chip (GE Healthcare). For calcium depleted conditions samples were prepared with 5mM of EGTA. All analyses were performed at least twice and traces were evaluated using BIAvaluation software (Biacore (GE Healthcare)).
Additional Information
How to cite this article: Troilo, H. et al. Independent multimerization of Latent TGFβ Binding Protein-1 stabilized by cross-linking and enhanced by heparan sulfate. Sci. Rep. 6, 34347; doi: 10.1038/srep34347 (2016).
Supplementary Material
Acknowledgments
We would like to thank Marge Howard in the Biomolecular analysis facility and staff in the EM facility; Diamond Light Source and ESRF (Midland BAG MX1580) for beamtime and Dr Mark Tully (beamline B21) and Dr Petra Pernot (beamline BM29) for assistance during SAXS data collection. In addition we would like to thank Rana Dajani and Hilda Diana Ruiz Nivia for technical assistance. HT was supported by BBSRC grant (Ref: BB/L00612X/1) to CB and CMK and RS by a Wellcome Trust studentship. The Wellcome Trust Centre for Cell-Matrix Research, University of Manchester is supported by core funding from the Wellcome Trust (088785/Z/09/Z).
Footnotes
The authors declare no competing financial interests.
Author Contributions H.T. and R.S. performed and analyzed experiments and contributed to the preparation of the figures. R.F.C. collected the data shown in Figure 8A. C.B. and C.M.K. conceived and coordinated the study. C.B. and H.T. wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
References
- Miyazono K., Olofsson A., Colosetti P. & Heldin C. H. A role of the latent TGF-beta 1-binding protein in the assembly and secretion of TGF-beta 1. EMBO J 10, 1091–1101 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantola A. K., Ryynanen M. J., Lhota F., Keski-Oja J. & Koli K. Independent regulation of short and long forms of latent TGF-beta binding protein (LTBP)-4 in cultured fibroblasts and human tissues. J Cell Physiol. 223, 727–736 (2010). [DOI] [PubMed] [Google Scholar]
- Annes J. P., Chen Y., Munger J. S. & Rifkin D. B. Integrin alphaVbeta6-mediated activation of latent TGF-beta requires the latent TGF-beta binding protein-1. J Cell Biol. 165, 723–734 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T. et al. Latent TGF-beta binding protein-2 is essential for the development of ciliary zonule microfibrils. Hum Mol Genet 23, 5672–5682 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vehvilainen P., Hyytiainen M. & Keski-Oja J. Latent transforming growth factor-beta-binding protein 2 is an adhesion protein for melanoma cells. J Biol Chem. 278, 24705–24713 (2003). [DOI] [PubMed] [Google Scholar]
- Dabovic B. et al. Function of latent TGFbeta binding protein 4 and fibulin 5 in elastogenesis and lung development. J Cell Physiol. 230, 226–236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C. T. et al. Latent transforming growth factor binding protein 4 regulates transforming growth factor beta receptor stability. Hum Mol Genet 24, 4024–4036 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai Z. et al. Latent transforming growth factor beta-binding protein 1 interacts with fibrillin and is a microfibril-associated protein.J Biol Chem. 278, 2750–2757 (2003). [DOI] [PubMed] [Google Scholar]
- Oklu R. & Hesketh R. The latent transforming growth factor beta binding protein (LTBP) family. Biochem J 352 Pt 3, 601–610 (2000). [PMC free article] [PubMed] [Google Scholar]
- Colosetti P., Hellman U., Heldin C. H. & Miyazono K. Ca2+ binding of latent transforming growth factor-beta 1 binding protein.FEBS Lett. 320, 140–144 (1993). [DOI] [PubMed] [Google Scholar]
- Gleizes P. E., Beavis R. C., Mazzieri R., Shen B. & Rifkin D. B. Identification and characterization of an eight-cysteine repeat of the latent transforming growth factor-beta binding protein-1 that mediates bonding to the latent transforming growth factor-beta1. J Biol Chem. 271, 29891–29896 (1996). [DOI] [PubMed] [Google Scholar]
- Buscemi L. et al. The single-molecule mechanics of the latent TGF-beta1 complex. Curr Biol 21, 2046–2054 (2011). [DOI] [PubMed] [Google Scholar]
- Shi M. et al. Latent TGF-beta structure and activation. Nature 474, 343–349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q. et al. Potential role for heparan sulfate proteoglycans in regulation of transforming growth factor-beta (TGF-beta) by modulating assembly of latent TGF-beta-binding protein-1. J Biol Chem. 282, 26418–26430 (2007). [DOI] [PubMed] [Google Scholar]
- Parsi M. K., Adams J. R., Whitelock J. & Gibson M. A. LTBP-2 has multiple heparin/heparan sulfate binding sites. Matrix Biol. 29, 393–401 (2010). [DOI] [PubMed] [Google Scholar]
- Sideek M. A., Menz C., Parsi M. K. & Gibson M. A. LTBP-2 competes with tropoelastin for binding to fibulin-5 and heparin, and is a negative modulator of elastinogenesis. Matrix Biol. 34, 114–123 (2014). [DOI] [PubMed] [Google Scholar]
- Kantola A. K., Keski-Oja J. & Koli K. Fibronectin and heparin binding domains of latent TGF-beta binding protein (LTBP)-4 mediate matrix targeting and cell adhesion. Exp Cell Res. 314, 2488–2500 (2008). [DOI] [PubMed] [Google Scholar]
- Nunes I., Gleizes P. E., Metz C. N. & Rifkin D. B. Latent transforming growth factor-beta binding protein domains involved in activation and transglutaminase-dependent cross-linking of latent transforming growth factor-beta. J Cell Biol. 136, 1151–1163 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verderio E. et al. Regulation of cell surface tissue transglutaminase: effects on matrix storage of latent transforming growth factor-beta binding protein-1. J Histochem Cytochem. 47, 1417–1432 (1999). [DOI] [PubMed] [Google Scholar]
- Oklu R., Hesketh T. R., Metcalfe J. C. & Kemp P. R. Expression of alternatively spliced human latent transforming growth factor beta binding protein-1. FEBS Lett. 435, 143–148 (1998). [DOI] [PubMed] [Google Scholar]
- Olofsson A. et al. Efficient association of an amino-terminally extended form of human latent transforming growth factor-beta binding protein with the extracellular matrix.J Biol Chem. 270, 31294–31297 (1995). [DOI] [PubMed] [Google Scholar]
- Michel K. et al. Analysis of the expression pattern of the latent transforming growth factor beta binding protein isoforms in normal and diseased human liver reveals a new splice variant missing the proteinase-sensitive hinge region. Hepatology 27, 1592–1599 (1998). [DOI] [PubMed] [Google Scholar]
- Lack J. et al. Solution structure of the third TB domain from LTBP1 provides insight into assembly of the large latent complex that sequesters latent TGF-beta. J Mol Biol. 334, 281–291 (2003). [DOI] [PubMed] [Google Scholar]
- Robertson I. B., Handford P. A. & Redfield C. NMR spectroscopic and bioinformatic analyses of the LTBP1 C-terminus reveal a highly dynamic domain organisation. Plos One 9, e87125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing A. K. et al. Solution structure of a pair of calcium-binding epidermal growth factor-like domains: implications for the Marfan syndrome and other genetic disorders. Cell 85, 597–605 (1996). [DOI] [PubMed] [Google Scholar]
- Baldock C. et al. Nanostructure of fibrillin-1 reveals compact conformation of EGF arrays and mechanism for extensibility. Proc Natl Acad Sci USA 103, 11922–11927 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J., Miyazono K., Heldin C. H. & Keski-Oja J. Latent transforming growth factor-beta 1 associates to fibroblast extracellular matrix via latent TGF-beta binding protein. J Cell Biol. 124, 171–181 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun D. I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 76, 2879–2886 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massam-Wu T. et al. Assembly of fibrillin microfibrils governs extracellular deposition of latent TGF beta. J Cell Sci. 123, 3006–3018 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedemann K., Batge B., Muller P. K. & Reinhardt D. P. Interactions of fibrillin-1 with heparin/heparan sulfate, implications for microfibrillar assembly. J Biol Chem. 276, 36035–36042 (2001). [DOI] [PubMed] [Google Scholar]
- Rambo R. P. & Tainer J. A. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers 95, 559–571 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J., Saharinen J., Hedman K. & Keski-Oja J. Latent transforming growth factor-beta 1 and its binding protein are components of extracellular matrix microfibrils. J Histochem Cytochem 44, 875–889 (1996). [DOI] [PubMed] [Google Scholar]
- Koch A. W., Engel J. & Maurer P. Calcium Binding to Extracellular Matrix Proteins, Functional and Pathological Effects. The Molecular Basis of Calcium Action in Biology and Medicine 1, 151–164 (2000). [Google Scholar]
- Unsold C., Hyytiainen M., Bruckner-Tuderman L. & Keski-Oja J. Latent TGF-beta binding protein LTBP-1 contains three potential extracellular matrix interacting domains. J Cell Sci. 114, 187–197 (2001). [DOI] [PubMed] [Google Scholar]
- Dallas S. L. et al. Role of the latent transforming growth factor beta binding protein 1 in fibrillin-containing microfibrils in bone cells in vitro and in vivo. J Bone Miner Res. 15, 68–81 (2000). [DOI] [PubMed] [Google Scholar]
- Ono R. N. et al. Latent transforming growth factor beta-binding proteins and fibulins compete for fibrillin-1 and exhibit exquisite specificities in binding sites. J Biol Chem. 284, 16872–16881 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberberg L. et al. Specificity of latent TGF-beta binding protein (LTBP) incorporation into matrix: role of fibrillins and fibronectin. J Cell Physiol. 227, 3828–3836 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E. C., Roth R. A., Heuser J. E. & Mecham R. P. Ultrastructural properties of ciliary zonule microfibrils. J Struct Biol. 139, 65–75 (2002). [DOI] [PubMed] [Google Scholar]
- Hubmacher D. et al. Biogenesis of extracellular microfibrils: Multimerization of the fibrillin-1 C terminus into bead-like structures enables self-assembly. Proc Natl Acad Sci USA 105, 6548–6553 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda K. et al. Latent TGF-beta binding protein 4 promotes elastic fiber assembly by interacting with fibulin-5. Proc Natl Acad Sci USA 110, 2852–2857 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock M. J. et al. Molecular basis of elastic fiber formation. Critical interactions and a tropoelastin-fibrillin-1 cross-link. J Biol Chem. 279, 23748–23758 (2004). [DOI] [PubMed] [Google Scholar]
- Troilo H. et al. Nanoscale structure of the BMP antagonist chordin supports cooperative BMP binding. Proc Natl Acad Sci USA 111, 13063–13068 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke D. & Svergun D. I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 42, 342–346 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozin M. & Svergun D. I. Automated matching of high- and low-resolution structural models. J. Appl. Cryst. 34, 33–41 (2001). [Google Scholar]
- Bernado P., Mylonas E., Petoukhov M. V., Blackledge M. & Svergun D. I. Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc. 129, 5656–5664 (2007). [DOI] [PubMed] [Google Scholar]
- Tang G. et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- Pettersen E. F. et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.