Abstract
Rad51 is a conserved eukaryotic protein that mediates the homologous recombination repair of DNA double-strand breaks that occur during mitosis and meiosis. In addition, Rad51 promotes mitochondrial DNA synthesis when replication stress is increased. Rad51 also regulates cell cycle progression by preserving the G2/M transition in embryonic stem cells. In this study, we report a novel function of Rad51 in regulating mitochondrial activity during in vitro maturation of mouse oocytes. Suppression of Rad51 by injection of Rad51 dsRNA into germinal vesicle-stage oocytes resulted in arrest of meiosis in metaphase I. Rad51-depleted oocytes showed chromosome misalignment and failures in spindle aggregation, affecting the completion of cytokinesis. We found that Rad51 depletion was accompanied by decreased ATP production and mitochondrial membrane potential and increased DNA degradation. We further demonstrated that the mitochondrial defect activated autophagy in Rad51-depleted oocytes. Taken together, we concluded that Rad51 functions to safeguard mitochondrial integrity during the meiotic maturation of oocytes.
Homologous recombination (HR) is the most prominent repair pathway for DNA double-strand breaks (DSBs). The central steps of HR involve the processing of DSB ends, a homology search and strand invasion. Efficient repair of DNA DSBs by HR depends on the formation of a Rad51 recombinase filament of broken single-stranded DNA (ssDNA) that facilitates strand invasion between the ssDNA and homologous double-strand DNA (dsDNA)1,2. If DNA DSBs are not properly repaired, they induce chromosome remodeling, cell cycle arrest or apoptosis. Because Rad51 performs an important function in HR, it protects genome integrity and enables cells to resist apoptosis triggered by DNA damage3,4,5. Additionally, Rad51 facilitates replication fork progression by preventing replication fork collapse, thereby ensuring cell proliferation in vertebrates6,7. In mouse embryonic stem cells, Rad51 regulates cell cycle progression by preserving the G2/M transition8.
Oocytes display a unique cell cycle progression during growth and meiotic maturation. In mammalian oocytes, meiosis generates haploid gametes after one round of chromosomal replication and two successive divisions, meiosis I and meiosis II. Oocytes have two points of arrest in the meiotic cell cycle, during the diplotene stage of prophase I and during metaphase II (MII), which both occur prior to fertilization with sperm. Several reports have shown that HR and Rad51 play important roles in ensuring genome stability in oocytes by repairing DNA damage that is either chemically induced or age related3,9,10,11,12,13. Rad51 expression shows an age-dependent decrease in mouse oocytes and is closely linked to increased aneuploidy rates11,12. Microinjection of Rad51 into oocytes of AKR/J mice also suppresses apoptosis and reduces the extent of DNA DSBs13.
Oocyte meiotic maturation involves cytoplasm maturation as well as nuclear maturation. Nuclear maturation includes chromosome segregation, and cytoplasm maturation is associated with the dynamics of various cytoplasmic components, including membranous organelles. In vitro maturation analysis revealed that DNA damage has differential effects on oocyte maturation depending on the degree of the damage14,15,16,17. Upon slight DNA DSBs, oocytes from fully grown mice can undergo meiosis resumption as indicated by germinal vesicle breakdown (GVBD)17. However, oocytes with severe DNA DSBs do not extrude the first polar body (PB) and cannot reach the MII stage during in vitro maturation17. A negative correlation between degree of DSBs and embryo developmental capacity has also been reported18. Another study focused on the role of Rad51 in the meiotic maturation of undamaged oocytes19. In a mouse model carrying a hypomorphic allele of Rad51, which overcomes the early embryonic lethality of the Rad51 null allele, oocytes displayed precocious separation of sister chromatids, aneuploidy and broken chromosomes at MII, possibly due to defects in sister chromatid cohesion at MII19. However, the molecular mechanisms underlying Rad51 activity during the meiotic maturation of oocytes remain to be elucidated.
In this study, we report for the first time that Rad51 regulates mitochondrial activity during in vitro maturation of mouse oocytes. Oocytes went into MI arrest following the suppression of Rad51 during in vitro oocyte maturation. We found that Rad51 depletion led to reductions in ATP production, mitochondrial potential, and mitochondrial quantity in MI-stage oocytes, supporting a novel role of Rad51 in the maintenance of mitochondrial dynamics and activity during the meiotic cell cycle in oocytes.
Results
Silencing of Rad51 in GV oocytes resulted in MI arrest
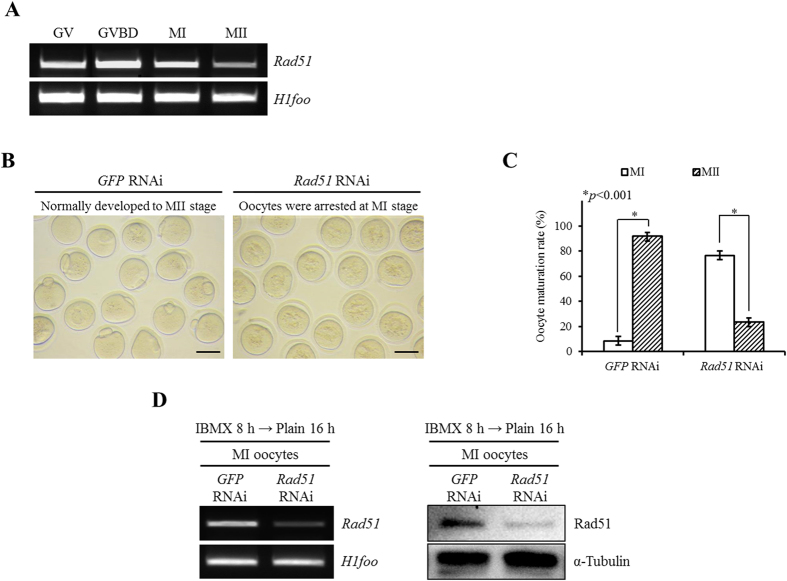
We first examined the expression level of Rad51 during oocyte maturation. As shown in Fig. 1A, Rad51 transcripts were expressed throughout meiosis. Specifically, the expression of Rad51 mRNA was maintained from the GV to the MI stage and reduced at the MII stage (Fig. 1A). In the control group, the GFP dsRNA-injected oocytes completed the meiosis process and developed normally to the MII stage with emission of first PBs (91.49%; Fig. 1B,C, Table 1, Video S1). In contrast, based on morphology, 76.48% of the Rad51-silenced oocytes were arrested at the MI stage, and 23.52% of the oocytes showed the first PB extrusion (Fig. 1B,C, Table 1, Video S2). We confirmed that there were reductions in Rad51 mRNA and protein levels in the MI-arrested oocytes after treatment with Rad51 RNAi (Fig. 1D).
Figure 1. Microinjection of Rad51 dsRNA into the cytoplasms of GV oocytes resulted in cell arrest at the MI stage.
(A) Typical pattern of Rad51 mRNA expression during oocyte maturation. For the PCR reaction, a single oocyte-equivalent mRNA was used as a template for the amplification of each gene. GV-, GVBD-, MI- and MII-stage oocytes were harvested after in vitro culture for 0, 2, 8 and 16 hours, respectively. H1foo was used as an internal control. (B) Micrographs of oocytes treated with GFP RNAi as a control or with Rad51 RNAi. The scale bars indicate 100 μm. GFP RNAi, GFP dsRNA-injected MII oocyte; Rad51 RNAi, Rad51 dsRNA-injected MI oocyte. (C) In vitro maturation rates of mouse oocytes after microinjection with Rad51 or GFP dsRNA into GV-stage oocytes. The asterisks represent statistical significance at p < 0.001. (D) Rad51 RNAi treatment resulted in specific suppression of Rad51 mRNA (left panel) and protein (right panel) expression. Protein lysate from 150 MI oocytes was loaded into each lane. H1foo and α-Tubulin were used as internal controls for mRNA and protein expression, respectively. IBMX 8 h → Plain 16 h, Rad51-injected oocytes cultured in IBMX supplement for 8 hours followed by 16 hours of culture in plain M16 medium.
Table 1. In vitro maturation of mouse oocytes after microinjection of Rad51 dsRNA into GV oocytes.
| Number of oocytes (%) |
|||
|---|---|---|---|
| Total | Metaphase I (MI) | Metaphase II (MII) | |
| GFP RNAi | 158 | 12 (8.51) | 146 (91.49) |
| Rad51 RNAi | 126 | 95 (76.48)* | 31 (23.52)* |
*Asterisks indicate significant differences compared to the value of the same stage of the GFP RNAi group (paired Student’s t-test, p < 0.001).
Silencing of Rad51 resulted in increased DNA damage during meiosis
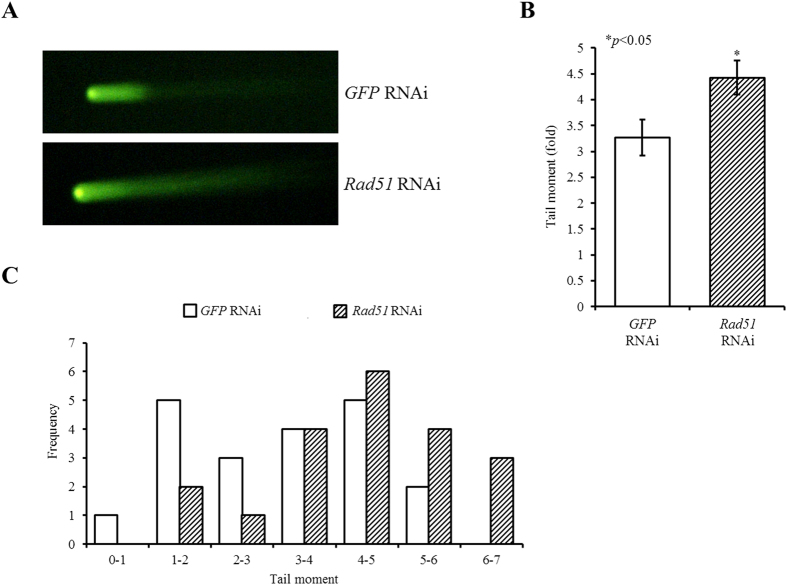
Rad51 plays an essential role in the homologous DNA strand exchange reaction that occurs during recombination and DNA repair. We examined whether depletion of Rad51 expression affected DNA integrity during oocyte maturation. The comet assay is a useful tool for the detection of single-strand breaks or DSBs in cells. We analyzed DNA damage in ZP-free Rad51-silenced oocytes using this assay. Inhibiting Rad51 expression in oocytes significantly increased DNA damage compared with the control oocytes (Fig. 2A,B). A typical histogram shows a rightward-shift in the tail moment of Rad51 RNAi (Fig. 2C), indicating that Rad51-silenced oocytes had more DNA damage. Thus, we concluded that Rad51-silencing damaged oocyte DNA and/or mitochondrial DNA (mtDNA), which may have been the main reason that these oocytes went into MI arrest.
Figure 2. Inhibition of Rad51 expression induced DNA damage in oocytes.
(A) Rad51-silenced MI oocytes were assessed for DNA damage using a comet assay. GFP dsRNA-injected MI oocytes exhibited slight DNA damage, whereas Rad51-silenced MI oocytes exhibited notable DNA damage. (B) Fold changes in tail moment following Rad51 RNAi are presented in a bar graph compared to that of the control group. The data are presented as the mean ± SEM. The asterisk represents statistical significance at p < 0.05. (C) Histogram of oocyte number (frequency) versus tail moment.
Rad51 depletion disrupted spindle and chromosomal configuration as well as the microtubule-organizing centers (MTOCs)
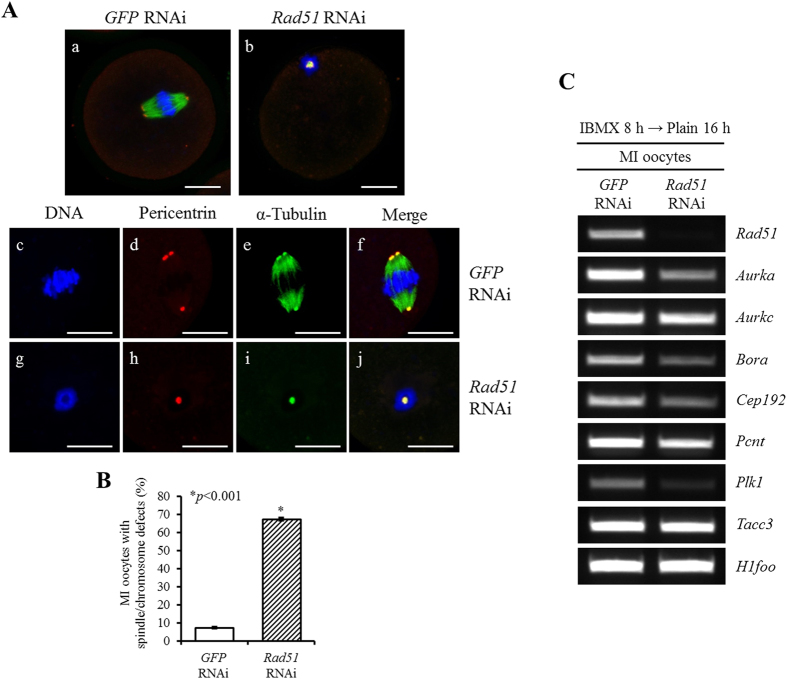
Although GVBD, the resumption of meiotic cell cycle, occurred in the Rad51-silenced oocytes, 76.48% of the oocytes could not complete meiosis and arrested at the MI stage. Because the Rad51-silenced oocytes failed to complete cytokinesis during oocyte nuclear maturation (Video S2), we evaluated the meiotic spindle structure, the chromosome alignment and the pericentrin distribution in the arrested oocytes using immunofluorescence staining (Fig. 3A). The GFP dsRNA-injected control MI oocytes exhibited meiotic spindles with a clear normal barrel shape (Fig. 3Aa,e,f) and well-aligned chromosomes at the metaphase plate (Fig. 3Aa,c,f). Indeed, pericentrin localized to the microtubule-organizing centers (MTOCs) at meiotic spindle poles in GFP dsRNA-injected control MI oocytes (Fig. 3Aa,d,f). In contrast, MI spindles were aggregated and intermingled with amassed chromosomes after Rad51 RNAi treatment (Fig. 3Ab,g–j). In addition, pericentrin localized as only one condensed spot with abnormally aggregated spindles and chromosomes in Rad51-depleted MI oocytes (Fig. 3Ab,h,j).
Figure 3. Reduction in Rad51 expression resulted in defective chromosome segregation and spindle assembly in MI oocytes.
(A) Immunofluorescence staining for the spindle and chromosomes in MI oocytes after injection with GFP- (a,c–f) or Rad51- (b,g–j) dsRNA in GV oocytes that were then allowed to mature in vitro. The oocytes were fixed in 4% paraformaldehyde and then stained with an antibody against Pericentrin (red) and α-Tubulin (green). DNA was counterstained with DAPI (blue). Lower panel shows the magnified region of the spindle-chromosome complex in another MI oocyte after GFP RNAi (c–f) and Rad51 RNAi (g–j). The scale bars indicate 20 μm. (B) Proportion of GFP or Rad51-dsRNA injected MI oocytes with defects of spindle assembly and chromosome configuration. (C) RT-PCR analysis of the expression of MTOC-related genes, including Aurka, Aurkc, Bora, Cep192, Pcnt, Plk1 and Tacc3, in Rad51-silenced MI oocytes. H1foo was used as an internal control.
Rad51 RNAi induced increased proportions of spindle assembly and chromosome configuration defects (Fig. 3B; 7.4% ± 0.5 in GFP RNAi vs. 67.5% ± 0.9 in Rad51 RNAi, p < 0.001). Mouse oocytes lack centrioles and possess MTOCs that are composed of pericentriolar material (PCM) proteins. These MTOCs are localized at the poles of the spindles and function as microtubule nucleating centers20,21,22. Because reduced Rad51 expression led to impairments in spindle assembly and pericentrin, we measured the expression levels of several essential PCM genes and found that six PCM genes (Aurka, Aurkc, Bora, Cep192, Pcnt and Plk1, but not Tacc3) were markedly reduced in Rad51-silenced MI-arrested oocytes (Fig. 3C).
Reduced Rad51 expression damaged mitochondrial dynamics and decreased ATP production
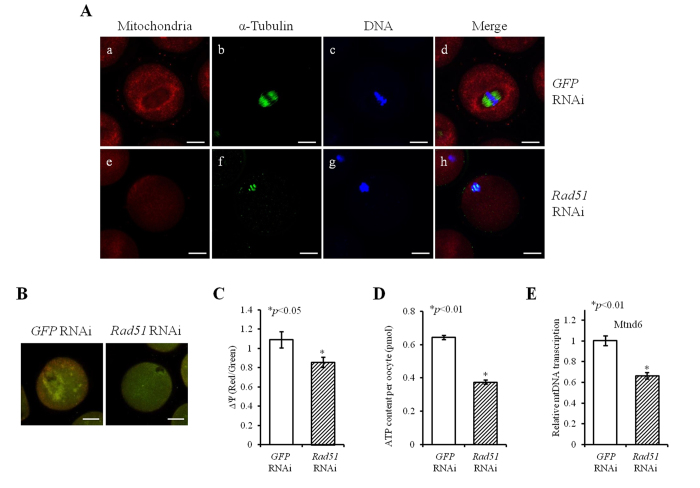
Because we observed DNA damage and spindle abnormalities, we next quantified the number and distribution of mitochondria in the ooplasm. Mitochondrial quantity was reduced in the ooplasm of Rad51-silenced MI oocytes compared to GFP-silenced MI oocytes, and the mitochondrial distribution pattern was also changed (Fig. 4A). In the control group, after treatment with GFP RNAi, mitochondria were mainly concentrated around the developing meiotic spindle and homogeneously distributed throughout the rest of the ooplasm (Fig. 4Aa–d). Conversely, there was no accumulation of mitochondria around the abnormally aggregated spindles in the Rad51-silenced MI oocytes (Fig. 4e–h).
Figure 4. Rad51 RNAi treatment caused a loss of mitochondrial membrane potential (ΔΨm) and impaired mitochondrial ATP production.
(A) Abnormal mitochondrial distribution was found in Rad51-silenced MI oocytes. GFP (a–d) or Rad51 (e–h) dsRNA was injected into MI oocytes that were precultured in M16 medium containing MitoTracker (mitochondria, red) for 30 minutes and then stained with an antibody against α-Tubulin (green). Cellular DNA was also stained (blue), and the cells were imaged by LSCM. The scale bars indicate 20 μm. (B) Effect of Rad51 on ΔΨm. ΔΨm in Rad51-silenced MI oocytes indicated by changes in RITC and FITC intensity, which were measured by LSCM as detailed in the Materials and Methods section. The scale bars indicate 20 μm. (C) Graphic representation of the results shown in (B). The data are presented as the mean ± SEM. The asterisk represents statistical significance at p < 0.05 as determined by a paired Student’s t-test. (D) Effects of Rad51 down-regulation on oocyte ATP levels. Reduction in Rad51 expression impaired mitochondrial ATP production. The data are presented as the mean ± SEM. The asterisk represents statistical significance at p < 0.01. (E) Rad51 RNAi treatment suppressed mitochondrial Mtnd6 mRNA expression. The data are presented as the mean ± SEM. The asterisk represents statistical significance at p < 0.01.
Next, the influence of Rad51 down-regulation in oocytes on mitochondrial membrane potential (ΔΨm) was measured to validate the abnormalities observed in the mitochondrial number and distribution pattern (Fig. 4B). The ratio between red and green fluorescence was significantly reduced in Rad51-silenced oocytes, indicating decreased mitochondrial membrane potential; the difference was statistically significant (Fig. 4C; 1.09 vs. 0.86, p < 0.05). We also evaluated the production of mitochondrial ATP in Rad51-silenced oocytes. The mean ATP concentrations in GFP RNAi oocytes and Rad51 RNAi oocytes were 0.64 ± 0.013 pmol and 0.38 ± 0.014, respectively, and this difference was statistically significant (Fig. 4D). In addition, the expression of mitochondria-encoded Mtnd6 transcripts, which are closely associated with mtDNA levels, was significantly decreased in the Rad51-silenced oocytes (Fig. 4E). Taken together, Rad51-silenced oocytes contain decreased levels of mtDNA, reduced mitochondrial membrane potential and reduced levels of mitochondrial ATP production, which collectively indicate that mitochondria in Rad51 RNAi oocytes are defective.
Rad51 depletion induced autophagy-mediated mitophagy of oocytes
Mitophagy is the selective segregation of damaged mitochondria and their subsequent degradation through autophagy23. Therefore, we assessed whether the mitochondrial reduction observed in Rad51-silenced oocytes was mitophagy-dependent. First, we observed the changes in six autophagy-related genes (Atg5, Atg7, Atg12, Becn1, Map1lc3a and Map1lc3b) via RT-PCR analyses. Among these genes, Atg7 was not detected in oocytes (Fig. 5A). In Rad51-silenced oocytes, the expression levels of three autophagy-related genes, Atg5, Atg12 and Becn1, were slightly decreased, while the expression levels of two autophagy-related genes, Map1lc3a and Map1lc3b, were markedly increased (Fig. 5A).
Figure 5. Rad51 RNAi treatment resulted in the removal of mitochondria by mitophagy.
(A) The mRNA expression profiles of autophagy-related genes were examined by RT-PCR. The expression levels of Atg5, Atg12 and Becn1 were slightly reduced, while the expression levels of Map1lc3a and Map1lc3b were increased in Rad51-silenced MI oocytes. H1foo was used as an internal control. IBMX 8 h → Plain 16 h, Rad51-silenced oocytes cultured in IBMX supplement for 8 hours followed by 16 hours of culture in plain M16 medium. (B) Immunofluorescence staining of Beclin1 in MI oocytes. Beclin1 is a marker protein for autophagy initiation. The oocytes were fixed in 4% paraformaldehyde and then stained with an antibody against Beclin1 (green). DNA was counterstained with DAPI (blue). Beclin1 expression is up-regulated in Rad51-silenced oocytes. The scale bars indicate 20 μm. (C) Rad51-silenced MI oocytes displayed increases in the size and number of Beclin1 puncta. The data are the numbers of Beclin1 puncta and are presented as the mean ± SEM. The asterisk represents statistical significance at p < 0.001 determined by a paired Student’s t-test. (D) Representative images showing the formation of Map1lc3b puncta. Map11c3b is a marker protein for mitophagy. Oocytes were fixed in 4% paraformaldehyde and then stained with DAPI (DNA, blue), MitoTracker (mitochondria, red) and anti-Map1lc3b antibody (green) and imaged using the LSCM. The 10 mM 3-MA was used to block autophagy. 3-MA-, oocytes were cultured without 3-MA addition; 3-MA+, oocytes were cultured with 3-MA. The scale bars indicate 20 μm. (E) Silencing of Rad51 expression resulted in the increased formation of Map1lc3b puncta. The average number of Map1lc3b puncta per oocyte was measured in MI oocytes after treatment of GFP (a–h) or Rad51 (i–p) RNAi and cultured in medium supplemented with (e–h, m–p) or without (a–d, i–l) 10 mM 3-MA. The data are presented as the mean ± SEM. Different letters (a~d) indicate significant differences at p < 0.001.
Beclin1 is a mammalian homolog of yeast autophagy-related gene 6 (Atg6) and is essential for autophagy. Induction or phosphorylation of Beclin1 is required to initiate autophagosome formation through the activation of class III phosphoinositide 3-kinase (PI3K)24. In GFP dsRNA-injected control oocyte, Beclin1 proteins were observed as small spots and were distributed evenly in the ooplasm (Fig. 5Ba,b). However, in Rad51-silenced MI oocytes, more and larger Beclin1 protein spots were detected in the ooplasm (Fig. 5Bc–d,C).
Another protein, Map1lc3b, a structural protein in the mitophagosomal membrane, is widely used as a marker of mitophagy25. After Rad51 RNAi treatment, MI-arrested oocytes were analyzed for Map1lc3 localization by counting the number of Map1lc3b puncta throughout the ooplasm. As expected, we found that Rad51 depletion increased the formation of Map1lc3b puncta compared to the control (Fig. 5Dc–d,k,l,E). When we tested the effects of the autophagy inhibitor 3-methyladenine (3-MA), an inhibitor of autophagy via the inhibition of PI3K activity, on Rad51 silencing-induced mitophagy, 3-MA selectively blocked autophagosome formation (Fig. 5Dk,l,o,p,E). Consistently, the amount of mitochondria, which was depleted in Rad51-silenced oocytes, was increased to levels comparable to those in the control by treatment with 3-MA (Fig. 5Db,j,f,n). These results suggest that Rad51 depletion induces autophagy-mediated mitophagy of oocytes.
Rad51 safeguards mitochondrial activity in oocytes
In Fig. 5, we noted that chromosomal alignment was not rescued by chemical inhibition of autophagy, despite the total number of rescued mitochondria (Fig. 5Da,i,e,m). To further examine whether defective spindle formation could be rescued using a mitophagy inhibitor, Rad51-silenced oocytes were treated with 3-MA and then loaded with MitoTracker and immunostained with α-Tubulin. GFP dsRNA-injected MI oocytes displayed normal meiotic spindles with aligned chromosomes incubated with (Fig. 6Ae,f) or without 3-MA (Fig. 6Aa,b). Consistent with the chromosome alignment results, disrupted meiotic spindle organization in Rad51-depleted oocytes (Fig. 6Ac,d) was not rescued by inhibition of mitophagy (Fig. 6Ag,h). To address whether the mitochondria accumulated in Rad51-depleted oocytes upon 3-MA treatment were functional, we examined the ΔΨm and mitochondrial ATP production in Rad51-silenced oocytes. We found that mitophagy inhibition did not rescue the decreases in ΔΨm caused by Rad51 depletion (Fig. 6B,C). Although ATP content was increased by 3-MA both in GFP RNAi- and Rad51 RNAi-treated oocytes, the amount of ATP in Rad51 RNAi oocytes was statistically lower than that of GFP RNAi oocyte (Fig. 6D). These results indicated that the mitochondria accumulated upon 3-MA treatment were not functional; furthermore, treatment also did not rescue the spindles and the chromosomal abnormalities.
Figure 6. Despite the accumulation of mitochondria by 3-MA, these mitochondria were malfunctional.
(A) Immunofluorescence staining for spindles and chromosomes after GFP (a,b,e,f) or Rad51 RNAi (c,d,g,h) and cultured without (a–d) or with 10 mM 3-MA (e–h). Oocytes were immunostained for spindles (α-Tubulin; green), mitochondria (MitoTracker; red) and chromosomes (DAPI; blue). The scale bars indicate 20 μm. (B) Effect of mitophagy inhibition on ΔΨm after Rad51 silencing. ΔΨm was measured using JC-1 staining. The ratio of RITC to FITC intensity was calculated. The scale bars indicate 20 μm. (C) Quantitative analysis of ΔΨm associated with Rad51 disruption and mitophagy inhibition is summarized. The data are presented as the mean ± SEM. Different letters (a~c) indicate significant differences at p < 0.05. (D) Effects of mitophagy inhibition on ATP levels in Rad51 down-regulated oocytes. ATP levels were measured as described in the Materials and Methods. The data are presented as the mean ± SEM. Different letters (a~c) indicate significant differences at p < 0.05.
Taken together, we concluded that Rad51 is involved in maintaining mitochondrial quantity and quality in mammalian oocytes. Rad51 depletion induced genome instability in nuclear DNA and/or mtDNA in oocytes, resulting in the dysfunction of mitochondria and the subsequent induction of autophagy-based mitophagy. Mitochondrial depletion caused abnormalities in the MTOCs and spindle formation and defective chromosome organization. These changes resulted in incomplete cytokinesis during meiotic division (Fig. 7).
Figure 7. Rad51 RNAi treatment induced mitochondrial malfunction and mitophagy and subsequently incomplete cytokinesis during oocyte meiosis.
Rad51 silencing-induced mitochondrial malfunction led the decreased ATP production and reduced ΔΨm in oocytes. Rad51-silenced oocytes had markedly fewer mitochondria as a result of the activation of mitophagy. In addition, abnormal spindle aggregation and chromosomal misalignment caused by low mitochondrial quality and quantity resulted in cytokinesis failure; as a result, Rad51-silenced oocytes were mainly arrested at MI.
Discussion
In the current study, we report a novel role for Rad51 associated with mitochondrial functioning during oocyte maturation in vitro. In Rad51-silenced MI-arrested oocytes, we observed a decreased quantity of mitochondria and a homogenous distribution of mitochondria throughout the cytoplasm. In contrast, in normal oocytes, mitochondria accumulated around developing meiotic spindles. Consistent with a previous report demonstrating that rearrangement of mitochondria during mouse oocyte maturation is associated with ATP production26, the ATP content in the Rad51-deficient MI-arrested oocytes was approximately 60% of that in the control oocytes. Furthermore, the mitochondrial membrane potential was also decreased in the Rad51-deficient oocytes, and the transcription of the mitochondrial gene Mtnd6 was approximately 70% of that in the control oocytes. These data suggest that Rad51 functions to maintain both the quantity and the quality of mitochondria in oocytes. It remains to be resolved how Rad51 depletion causes defects in mitochondrial activity and mitochondrial gene expression during oocyte maturation. Because large amounts of Rad51 are localized in the inner matrixes of mitochondria and because this protein functions to maintain mitochondrial gene copy number27,28,29, one possible mechanism for this process is the direct contribution of Rad51 to sustaining mitochondrial genome stability during oocyte maturation.
In mammalian oocytes, mitochondrial localization is microtubule-dependent30,31. Thus, the disruption in spindle assembly following Rad51 depletion, the abnormal distribution of mitochondria, and the reduced number of mitochondria could be all strongly interrelated, in agreement with previous findings that the anatomical association of spindles and mitochondria is linked to meiotic progression in oocytes30,32.
Recently, global trends have changed as a result of modern lifestyles—for example, women now regularly delay marriage and childbirth until they are in their 30s—which has led to growing problems in maternal reproductive senescence. One hallmark of oocyte aging is an accumulation of genetic mutations in genomes as well as in mtDNA3,12. Genomic instability and mitochondrial dysfunction in older women are strongly associated with poor reproductive performance and infertility11,31,33,34,35. Cytoplasmic transfer from young donors into older defective recipient oocytes was developed to treat infertility patients with some success, and the major beneficial cytoplasmic components in such cases are believed to be mitochondria36,37,38,39. The heteroplasmy found in children born from this cytoplasmic transfer technique has led to the ethical dilemma of whether such children have three parents and has resulted in moratoriums on ooplasm transfer in some countries33. However, a mitochondrial transfer technique developed for couples with mitochondrial diseases could be an exception to this ethical quandary. Nevertheless, we suggest using Rad51 microinjection to rescue reduced oocyte competency related to aging as an alternative to cytoplasmic transfer to avoid heteroplasmy. However, the optimal dose of Rad51 protein to rescue mitochondrial dysfunction in aged oocytes and the safety of this technique must still be resolved before it can be applied in human IVF programs or the somatic cell nuclear transfer technique.
Methods
Animals
Imprinting control region (ICR) mice (female and male), exclusively provided by Koatech (Pyeoungtack, Korea), were mated to produce embryos in the breeding facility at the CHA Research Institute of CHA University. All procedures described herein were reviewed and approved by the Institutional Animal Care and Use Committee of CHA University and were performed in accordance with Guiding Principles for the Care and Use of Laboratory Animals.
Reagents
Chemicals and reagents were obtained from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise noted.
Oocyte isolation
To isolate GV oocytes, three-week-old female ICR mice were injected with 5 IU pregnant mare’s serum gonadotropin (PMSG) and sacrificed 46 hours later. Isolated ovaries were punctured in M2 medium containing 0.2 mM 3-isobutyl-1-methyl-xanthine (IBMX), and cumulus-enclosed oocyte complexes (COCs) were collected. Cumulus cells were mechanically retrieved from oocytes by repeated extraction through a fine-bore pipette.
To obtain MII oocytes, we injected female mice with 5 IU eCG, followed by 5 IU hCG after 46 hours. MII oocytes were obtained from the oviduct 16 hours after hCG injection. The cumulus cells surrounding the MII oocytes were removed using hyaluronidase (300 U/ml). The isolated murine oocytes were snap frozen and stored at −70 °C prior to RNA isolation.
Messenger RNA isolation and RT-PCR
Oocyte mRNA was isolated using a Dynabeads mRNA DIRECT kit (Dynal Asa, Oslo, Norway) according to the manufacturer’s instructions. Briefly, oocytes were suspended with lysis/binding buffer and mixed with prewashed Dynabeads oligo dT25. After mRNA binding, the beads were washed with buffer A twice, followed by buffer B, and mRNA was eluted with Tris-HCl by incubation at 73 °C. Purified mRNA was used as a template for reverse transcription with an oligo (dT) primer according to the MMLV protocol. RT-PCR was performed with single oocyte-equivalent cDNA and primers (Table 2). Then, the RT-PCR products were separated by electrophoresis on a 1.5% agarose gel and analyzed using a Gel DocTM EZ Imager (Bio-Rad, Hercules, CA, USA). All experiments were repeated three times.
Table 2. Primer sequences and RT-PCR conditions.
| Gene symbol | Description | Accession numbers | Primer sequencesa | Annealing temperature | Product size |
|---|---|---|---|---|---|
| Rad51-Ab | RAD51 Recombinase | NM_011234.4 | For-AAGCTGCTTCAAGGTGGAAT | 60 °C | 507 bp |
| Rev-GAACATGGCTGCTCCATCTA | |||||
| Rad51-Bc | RAD51 Recombinase | NM_011234.4 | For- AGCTTTCAGCCAGGCAAAT | 60 °C | 265 bp |
| Rev-GCTTCAGCTTCAGGAAGACA | |||||
| Atg5 | Autophagy related 5 | NM_053069.5 | For- ATGTGCTTCGAGATGTGTGG | 60 °C | 215 bp |
| Rev- CAGGGGTGTGCCTTCATATT | |||||
| Atg7 | Autophagy related 7 | NM_001253717.1 | For-ATGCCAGGACACCCTGTGAACTTC | 60 °C | 351 bp |
| Rev-ACATCATTGCAGAAGTAGCAGCCA | |||||
| Atg12 | Autophagy related 12 | NM_026217.3 | For-TTCGGTTGCAGTTTCGCC | 63 °C | 311 bp |
| Rev-CCATGCCTGTGATTTGCAGTA | |||||
| Aurka | Aurora kinase A | NM_011497.4 | For-AGTTGGCAAACGCTCTGTCT | 60 °C | 160 bp |
| Rev-GTGCCACACATTGTGGTTCT | |||||
| Aurkc | Aurora kinase C | NM_001080965.1 | For-ACAACACCGGAACATCCTTC | 60 °C | 130 bp |
| Rev-TGCTGGTCCAACTTCTGATG | |||||
| Becn1 | Beclin1 | NM_019584.3 | For- TTTGACCATGCAATGGTAGC | 60 °C | 211 bp |
| Rev- TGGTCAGCATGAACTTGAGC | |||||
| Bora | Bora, aurora kinase A activator | NM_175265.4 | For-CCTGGTCCGCATACCTTTTA | 60 °C | 211 bp |
| Rev-CACGGTGTCATTTTCCCTCT | |||||
| Cep192 | Centrosomal protein 192kDa | NM_027556.1 | For-AGCTCAACTCGAGCCATCAT | 60 °C | 142 bp |
| Rev-GGGCTCAAAAACTTCAGTGC | |||||
| Map1lc3a | Microtubule-associated protein 1 light chain 3 alpha | NM_025735.3 | For-AGCTTCGCCGACCGCCGTAAG | 63 °C | 276 bp |
| Rev-CTTCTCCTGTTCATAGATGTCAGC | |||||
| Map1lc3b | Microtubule-associated protein 1 light chain 3 beta | NM_026160.4 | For-CGGAGCTTTGAACAAAGAGTG | 63 °C | 279 bp |
| Rev-TCTCTCACTCTCGTACACTTC | |||||
| Mtnd6 | Mitochondrially encoded NADH dehydrogenase 6 | NC_005089.1 | For-GAGGTTTAGGTTTAATTGTTAGTGGG | 60 °C | 175 bp |
| Rev- CAGTTAGATCCCCAAGTCTCTG | |||||
| Plk1 | Polo-like kinase 1 | NM_011121.3 | For-TGTAGTTTTGGAGCTCTGTCG | 60 °C | 148 bp |
| Rev-TCCCTGTGAATGACCTGATTG | |||||
| Pcnt | Pericentrin | NM_008787.3 | For- TGGAAAAGCTGTACCTGCAC | 60 °C | 192 bp |
| Rev- TGGTGAAGGGACGTGACATA | |||||
| Tacc3 | Transforming, acidic coiled-coil containing protein 3 | NM_001040435.3 | For-AAGTACAGCCAGAAAGACCTG | 60 °C | 299 bp |
| Rev-TCCTTCCGCTTCTCAAACC | |||||
| H1foo | H1 histone family, member O, oocyte-specific | NM_138311 | For- GCGAAACCGAAAGAGGTCAGAA | 60 °C | 378 bp |
| Rev- TGGAGGAGGTCTTGGGAAGTAA | |||||
| GFPb | Green fluorescent protein | KF111246.1 | For- TGTCCCAATTCTTGTTGAAT | 60 °C | 561 bp |
| Rev- TTGTCTGGTAAAAGGACAGG |
aFor, Forward; Rev, Reverse.
bPrimers were used for the preparation of Rad51 dsRNA and GFP dsRNA, respectively.
cDifferent set of primers was used to confirm the knockdown of Rad51 mRNA after RNAi treatment.
Quantitative real-time RT-PCR
Quantitative real-time RT-PCR was performed with single oocyte-equivalent cDNA and Mtnd6 primers (Table 2), as previously described, using an iCycler iQ™ Detection System (Bio-Rad)40. iQ SYBR Green Supermix PCR reagents (Bio-Rad) were used to monitor amplification, and the results were analyzed using iCycler iQ™ proprietary software. The melting curves were used to identify any nonspecific amplification products. The relative expression levels of the target genes were evaluated using the comparative CT method41,42, and all analytical procedures were repeated at least three times.
Preparation of Rad51 and GFP dsRNA
For RNAi experiments, we prepared Rad51 dsRNA. Rad51-A (Suppl. Fig. 1A) and GFP primers (Table 2) were used to amplify regions of Rad51 and GFP cDNA, respectively, which were then cloned into a pGEM-T Easy (Promega, Madison, WI, USA) vector. After linearization with SpeI, each RNA strand was transcribed in vitro, annealed at 75 °C and then cooled to room temperature (RT). dsRNA was purified with DNaseI and RNase digestion prior to dilution to 2.9 μg/μl. Purified dsRNA was verified by electrophoresis on a 1% agarose gel, which was used to compare the mobility of dsRNA with that of ssRNA (Suppl. Fig. 1B). GFP RNAi was used as an injection control.
Microinjection and in vitro culture
GV oocytes were microinjected with Rad51 dsRNA in M2 medium containing 0.2 mM IBMX. An injection pipette containing dsRNA solution was inserted into the cytoplasm of an oocyte, and 10 pl of dsRNA was microinjected using a constant flow system (Transjector; Eppendorf, Hamburg, Germany). To determine the rate of in vitro maturation, oocytes were cultured in M16 medium containing 0.2 mM IBMX for 8 hours, followed by culture in M16 alone for 16 hours in 5% CO2 at 37 °C. After the RNAi experiments were completed, the in vitro maturation rates and morphological changes were recorded as previously described40. 3-Methyladenine was used to inhibit autophagy. After RNAi, oocytes were cultured in M16 medium containing 10 mM 3-MA.
Time-lapse video microscopy
Time-lapse video observations were used to track phenotypic changes and the speed of oocyte maturation during in vitro culture as previously described43. A time-lapse microscope (JuLITM; Digital Bio, Seoul, Korea) was placed in an incubator under 5% CO2 at 37 °C, and a culture dish containing oocytes was placed on the microscope stage. Images were automatically captured every 5 minutes for 16 hours for GFP RNAi-treated oocytes or 36 hours for Rad51 RNAi-treated oocytes, and the sequential time-lapse images were converted into movie files using JuLI operation software.
Western blotting
Protein extracts (150 oocytes per lane) were separated using 12% SDS-PAGE. A PVDF membrane (Amersham Biosciences, Piscataway, NJ, USA) was used to transfer the separated proteins and then blocked for 1 hour in Tris-buffered saline-Tween (TBST) containing 5% non-fat milk. The blocked membranes were then incubated with rabbit anti-Rad51 antibody (1:1000; sc-8349, Santa Cruz Biotechnology, Dallas, TX, USA) or mouse anti-α-Tubulin antibody (1:1000; sc-8035, Santa Cruz Biotechnology) in TBST. After washing in TBST, membranes were incubated with HRP-conjugated anti-rabbit IgG (1:5000; #7074, Cell Signaling Technology, Danvers, MA, USA) or anti-mouse IgG (1:2000; A-2554) in TBST for 1 hour at RT. Bound antibodies were detected using an enhanced chemiluminescence detection system (Amersham Biosciences) according to the manufacturer’s instructions.
Comet assay
DNA damage was assessed using a neutral version of a comet assay to detect DSBs, as detailed in the manufacturer’s instructions (Trevigen, Gaithersburg, MD, USA). After the zona pellucida (ZP) was removed, oocytes were suspended in 1% low melting point agarose (LMA) at 37 °C, and then the mixture was pipetted onto a pre-coated slide (Trevigen). The slides were incubated at 4 °C in the dark for complete adherence and then immersed in 4 °C lysis solution from Trevigen for 4 hours. The slides were removed from the lysis solution and gently immersed in 4 °C 1X TAE buffer for 30 minutes before being electrophoresed for 40 minutes at 30 V. The slides were immersed in 1 M ammonium acetate at RT for 30 minutes and then immersed in 75% ethanol for fixation at RT for 30 minutes. After the fixation, the slides were incubated at 42 °C for at least 20 minutes until the LMA was fully dried and then stained with 1X SYBR® Green I Staining Solution for 5 minutes. The stained slides were observed under a fluorescence microscope. To quantify individual oocyte comets, analysis was performed using the CASP PC image analysis program according to a previously reported scoring method44. We scored each comet according to its tail moment (TM), which is the product of the tail length and the percent tail DNA, TM = TL × (%DNAT).
Immunofluorescence staining
Denuded oocytes were placed in PBS containing 0.1% polyvinyl alcohol (PBS-PVA), 4% paraformaldehyde, and 0.2% Triton X-100 and then fixed for 40 minutes at RT. After washing in PBS-PVA, fixed oocytes were blocked with 3% BSA-PBS-PVA for 1 hour and then incubated with mouse anti-α-Tubulin antibody (1:100), rabbit anti-Pericentrin antibody (1:100; PRB-432C, BioLegend, San Diego, CA, USA), rabbit anti-Beclin1 antibody (1:100; sc-11427, Santa Cruz Biotechnology), or rabbit anti-Map1lc3b antibody (1:100; ab48394, Abcam, Cambridge, MA, USA) at 4 °C overnight. After washing, the oocytes were incubated with an Alexa Fluor 488- or Alexa Fluor 555-conjugated secondary antibody (Molecular Probes, Eugene, OR, USA) for 1 hour at RT, and DNA was counterstained with DAPI (Promega).
Staining of mitochondria
To evaluate the distribution of mitochondria, oocytes were stained using MitoTracker Orange CMTMRos (Molecular Probes). MitoTracker, which fluoresces orange, was used at a concentration of 300 nM in M16 supplemented with 0.3% BSA for 30 minutes at 37 °C in the dark. After washing, the oocytes were fixed and immunofluorescently stained with an anti-α-Tubulin antibody and then counterstained with DAPI. The stained oocytes were stored below 4 °C until confocal microscopy was performed. The oocytes were examined using a laser scanning confocal microscope (LSCM; Leica, Wetzlar, Germany). A minimum of 30 oocytes per RNAi group was examined by LSCM.
Determination of mitochondrial membrane potential (ΔΨm) by JC-1
To measure oocyte mitochondrial activity, ΔΨm assays were performed using the ΔΨm-sensitive fluorescent probe JC-1 (Thermo Fisher Scientific, Waltham, MA, USA). Oocytes were cultured in the dark for 20 minutes in the presence of JC-1 at a concentration recommended by a manufacturer (1 μg/mL) and subsequently imaged on an LSCM using the fluorescein isothiocyanate (FITC, green) and rhodamine isothiocyanate (RITC, red) channels. Two fluorescent images (green and red fluorescence) were acquired in the largest diameter plane of the oocytes. Using confocal software, the signal intensities and the ratio of RITC to FITC for each oocyte were calculated and used as the endpoint for the ΔΨm.
Measurement of ATP content
The ATP content in 40 oocytes in each group was measured using a Bioluminescent Somatic Cell Assay Kit (FL-ASC) based on the luciferin-luciferase reaction as previously described, with minor changes45,46. The bioluminescence of each sample was measured using a luminometer (Centro XS3 LB 960; Berthold Technologies, Bad Wildbad, Germany), and an 11-point standard curve (containing 0 to 5.0 pmol ATP) was included in each assay. The ATP content in the oocytes was expressed as pmol/oocyte.
Statistical analysis
Each experiment was repeated at least three times, and statistical analyses were performed using Student’s t-test. Data were derived from at least three separate and independent experiments and expressed as the mean ± SEM. The p values were calculated based on a paired t-test of the means from the GFP RNAi group and the Rad51 RNAi group, and p < 0.05 was considered statistically significant.
Additional Information
How to cite this article: Kim, K.-H. et al. The role of Rad51 in safeguarding mitochondrial activity during the meiotic cell cycle in mammalian oocytes. Sci. Rep. 6, 34110; doi: 10.1038/srep34110 (2016).
Supplementary Material
Acknowledgments
This work was supported by the Korea Science and Engineering Foundation of the Korean government (MOST) (2015R1A2A2A01003498) and by the Priority Research Centers Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science, and Technology (2009–0093821). This work was also performed with the support of the Cooperative Research Program for Agriculture Science & Technology Development (PJ010033), Rural Development Administration, Republic of Korea.
Footnotes
Author Contributions K.-H.K., K.-S.P. and K.-A.L. conceived and designed the experiments; K.-H.K., J.-H.P. and E.-Y.K. performed the experiments; K.-H.K., J.-H.P., J.-J.K., K.-S.P. and K.-A.L. analyzed the data; K.-H.K., J.-H.P., K.-S.P. and K.-A.L. wrote the manuscript; J.-J.K., K.-S.P. and K.-A.L. acquired financial support.
References
- Sugawara N., Wang X. & Haber J. E. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell 12, 209–219 (2003). [DOI] [PubMed] [Google Scholar]
- White C. I. & Haber J. E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 9, 663–673 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujjo L. L. et al. Enhancing survival of mouse oocytes following chemotherapy or aging by targeting Bax and Rad51. PLoS One 5, e9204 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. Wrestling off RAD51: A novel role for RecQ helicases. Bioessays 30, 291–295 (2008). [DOI] [PubMed] [Google Scholar]
- Klein H. L. The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair 7, 686–693 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E. et al. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J . 17, 598–608 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki T. et al. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Sci. USA 93, 6236–6240 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S. W., Kim D. K., Kim K. P. & Park K. S. Rad51 regulates cell cycle progression by preserving G2/M transition in mouse embryonic stem cells. Stem Cells Dev. 23, 2700–2711 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarin J. J., Perez-Albala S. & Cano A. Cellular and morphological traits of oocytes retrieved from aging mice after exogenous ovarian stimulation. Biol. Reprod. 65, 141–150 (2001). [DOI] [PubMed] [Google Scholar]
- Tatone C. et al. Age-associated changes in mouse oocytes during postovulatory in vitro culture: Possible role for meiotic kinases and survival factor BCL2. Biol. Reprod. 74, 395–402 (2006). [DOI] [PubMed] [Google Scholar]
- Hamatani T. et al. Age-associated alteration of gene expression patterns in mouse oocytes. Hum. Mol. Genet. 13, 2263–2278 (2004). [DOI] [PubMed] [Google Scholar]
- Pan H., Ma P., Zhu W. & Schultz R. M. Age-associated increase in aneuploidy and changes in gene expression in mouse eggs. Dev. Biol. 316, 397–407 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez G. I. et al. Genetic variance modifies apoptosis susceptibility in mature oocytes via alterations in DNA repair capacity and mitochondrial ultrastructure. Cell Death Differ. 14, 524–533 (2007). [DOI] [PubMed] [Google Scholar]
- Ma J. Y. et al. The effects of DNA double-strand breaks on mouse oocyte meiotic maturation. Cell Cycle 12, 1233–1241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marangos P. & Carroll J. Oocytes progress beyond prophase in the presence of DNA damage. Curr. Biol. 22, 989–994 (2012). [DOI] [PubMed] [Google Scholar]
- Zhang T. et al. Effects of DNA damage and short-term spindle disruption on oocyte meiotic maturation. Histochem. Cell Biol. 142, 185–194 (2014). [DOI] [PubMed] [Google Scholar]
- Lin F. et al. Different fates of oocytes with DNA double-strand breaks in vitro and in vivo. Cell Cycle 13, 2674–2680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohrer R. C., Coutinho A. R., Duggavathi R. & Bordignon V. The incidence of DNA double-strand breaks is higher in late-cleaving and less developmentally competent porcine embryos. Biol. Reprod. 93, 59 (2015). [DOI] [PubMed] [Google Scholar]
- Kuznetsov S. et al. RAD51C deficiency in mice results in early prophase I arrest in males and sister chromatid separation at metaphase II in females. J. Cell Biol. 176, 581–592 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szollosi D., Calarco P. & Donahue R. P. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J. Cell Sci. 11, 521–541 (1972). [DOI] [PubMed] [Google Scholar]
- Calarco-Gillam P. D., Siebert M. C., Hubble R., Mitchison T. & Kirschner M. Centrosome development in early mouse embryos as defined by an autoantibody against pericentriolar material. Cell 35, 621–629 (1983). [DOI] [PubMed] [Google Scholar]
- Maro B., Howlett S. K. & Webb M. Non-spindle microtubule organizing centers in metaphase II-arrested mouse oocytes. J. Cell Biol. 101, 1665–1672 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi G. & Schwarz T. L. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 20, 31–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang S., Shin H., Song H., Suh C. S. & Lim H. J. Autophagic activation in vitrified-warmed mouse oocytes. Reproduction 148, 11–19 (2014). [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T. & Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 (2011). [DOI] [PubMed] [Google Scholar]
- Yu Y., Dumollard R., Rossbach A., Lai F. A. & Swann K. Redistribution of mitochondria leads to bursts of ATP production during spontaneous mouse oocyte maturation. J Cell Physiol 224, 672–680 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies A. A. et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell 7, 273–282 (2001). [DOI] [PubMed] [Google Scholar]
- Gildemeister O. S., Sage J. M. & Knight K. L. Cellular redistribution of Rad51 in response to DNA damage: Novel role for Rad51C. J. Biol. Chem. 284, 31945–31952 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson S. E. et al. Pir51, a Rad51-interacting protein with high expression in aggressive lymphoma, controls mitomycin C sensitivity and prevents chromosomal breaks. Mutat. Res. 601, 113–124 (2006). [DOI] [PubMed] [Google Scholar]
- Van Blerkom J. Microtubule mediation of cytoplasmic and nuclear maturation during the early stages of resumed meiosis in cultured mouse oocytes. Proc. Natl. Acad. Sci. USA 88, 5031–5035 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q. Y. et al. Translocation of active mitochondria during pig oocyte maturation, fertilization and early embryo development in vitro. Reproduction 122, 155–163 (2001). [PubMed] [Google Scholar]
- Dalton C. M. & Carroll J. Biased inheritance of mitochondria during asymmetric cell division in the mouse oocyte. J. Cell Sci. 126, 2955–2964 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentov Y. & Casper R. F. The aging oocyte–can mitochondrial function be improved? Fertil. Steril. 99, 18–22 (2013). [DOI] [PubMed] [Google Scholar]
- Grindler N. M. & Moley K. H. Maternal obesity, infertility and mitochondrial dysfunction: Potential mechanisms emerging from mouse model systems. Mol. Hum. Reprod. 19, 486–494 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatten H., Sun Q. Y. & Prather R. The impact of mitochondrial function/dysfunction on IVF and new treatment possibilities for infertility. Reprod. Biol. Endocrinol. 12, 111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. et al. Ooplasmic transfer in mature human oocytes. Mol. Hum. Reprod. 4, 269–280 (1998). [DOI] [PubMed] [Google Scholar]
- Muggleton-Harris A., Whittingham D. G. & Wilson L. Cytoplasmic control of preimplantation development in vitro in the mouse. Nature 299, 460–462 (1982). [DOI] [PubMed] [Google Scholar]
- Barritt J., Willadsen S., Brenner C. & Cohen J. Cytoplasmic transfer in assisted reproduction. Hum. Reprod. Update 7, 428–435 (2001). [DOI] [PubMed] [Google Scholar]
- Harvey A. J., Gibson T. C., Quebedeaux T. M. & Brenner C. A. Impact of assisted reproductive technologies: A mitochondrial perspective of cytoplasmic transplantation. Curr. Top. Dev. Biol. 77, 229–249 (2007). [DOI] [PubMed] [Google Scholar]
- Kim K. H. et al. Gas6 downregulation impaired cytoplasmic maturation and pronuclear formation independent to the MPF activity. PLoS One 6, e23304 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J. S., Reed A., Chen F. & Stewart C. N. Jr. Statistical analysis of real-time PCR data. BMC Bioinformatics 7, 85 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Lee S. Y. et al. Thioredoxin-interacting protein regulates glucose metabolism and affects cytoplasmic streaming in mouse oocytes. PLoS One 8, e70708 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konca K. et al. A cross-platform public domain PC image-analysis program for the comet assay. Mutat. Res. 534, 15–20 (2003). [DOI] [PubMed] [Google Scholar]
- Combelles C. M. & Albertini D. F. Assessment of oocyte quality following repeated gonadotropin stimulation in the mouse. Biol. Reprod. 68, 812–821 (2003). [DOI] [PubMed] [Google Scholar]
- Van Blerkom J., Davis P. W. & Lee, J. ATP content of human oocytes and developmental potential and outcome after in-vitro fertilization and embryo transfer. Hum. Reprod. 10, 415–424 (1995). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.