Abstract
The objective of this study was to identify hub genes and pathways associated with hepatocellular carcinoma (HCC) by centrality analysis of a co-expression network. A co-expression network based on differentially expressed (DE) genes of HCC was constructed using the Differentially Co-expressed Genes and Links (DCGL) package. Centrality analyses, for centrality of degree, clustering coefficient, closeness, stress and betweenness for the co-expression network were performed to identify hub genes, and the hub genes were combined together to overcome inconsistent results. Enrichment analyses were conducted using Gene Ontology and Kyoto Encyclopedia of Genes and Genomes databases. Finally, validation of hub genes was conducted utilizing reverse transcription-polymerase chain reaction (RT-PCR) analysis. In total, 260 DE genes between normal controls and HCC patients were obtained and a co-expression network with 154 nodes and 326 edges was constructed. From this, 13 hub genes were identified according to degree, clustering coefficient, closeness, stress and betweenness centrality analysis. It was found that reelin (RELN), potassium voltage-gated channel subfamily J member 10 (KCNJ10) and neural cell adhesion molecule 1 (NCAM1) were common hub genes across the five centralities, and the results of RT-PCR analysis for RELN, KCNJ10 and NCAM1 were consistent with the centrality analyses. Pathway enrichment analysis of DE genes showed that cell cycle, metabolism of xenobiotics by cytochrome P450 and p53 signaling pathway were the most significant pathways. This study may contribute to understanding the molecular pathogenesis of HCC and provide potential biomarkers for its early detection and effective therapies.
Keywords: hepatocellular carcinoma, co-expression network, centrality, hub gene, pathway, reverse transcription-polymerase chain reaction
Introduction
Hepatocellular carcinoma (HCC) is the major histological subtype of primary liver malignancies, accounting for ~80% of the total liver cancer burden (1). The majority of cases of HCC are associated with cirrhosis caused by infection with chronic hepatitis B virus (HBV) or hepatitis C virus (HCV), alcoholic injury, and to a lesser extent from genetically determined disorders such as hemochromatosis (2). However, there are few effective treatments and early diagnoses, partly because the cell- and molecular-based mechanisms that contribute to the pathogenesis of this tumor type are poorly understood (3).
With the advances made in high-throughput experimental technologies, such technologies have been applied to the exploration of diagnostic gene signatures and biological processes of human diseases (4), which provide novel insights into the underlying biological mechanisms of HCC. Studies of HCC based on microarray expression have revealed guiding principles of its molecular initiation and progression, and these may provide guidance for the investigation of potential molecular biomarkers for the early detection of HCC (5,6). For example, Jia et al (6) suggested that phospholipase C β 1 (PLCB1) was a critical driver gene with causal roles in carcinogenesis and might have an important role in the pathogenesis of HCC. The cytochrome P450 family 2 subfamily B member 6 (CYP2B6) gene has been found to be relevant to tumor angiogenesis or drug metabolism predisposed to the development of treatment-related toxicity in HCC (7).
However, the results obtained have been inconsistent for a variety of reasons, including small sample size, measurement error, and different statistical methods being used (8). The overlap is very low for the most significantly dysregulated genes across multiple studies (9). Network-based approaches, particularly co-expression networks, are an effective means of conducting a mechanistic analysis by identifying potential molecular markers for malignancy and connecting them together (10). Therefore, the present study used co-expression network-based centrality analysis to gain a clear insight into the significant and targetable tumorigenic genes of HCC and the integrated result of the five centralities (degree centrality, clustering coefficient, stress centrality, betweenness centrality and closeness centrality) to resolve the inconsistent outcomes obtained by different methods, which may be applicable to the early detection and treatment of HCC.
The objective of this study was to identify hub genes and pathways associated with HCC on the basis of network centrality analysis. Co-expression networks of differentially expressed (DE) genes between normal controls and patients with HCC were constructed using the Differentially Co-expressed Genes and Links (DCGL) package. Clusters in the network were obtained using the Molecular Complex Detection (MCODE) algorithm. Centrality analyses for the co-expression network were performed based on degree, clustering coefficient, closeness, stress and betweenness. Enrichment analyses for DE genes were performed using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. Finally, validation of hub genes was conducted utilizing reverse transcription-polymerase chain reaction (RT-PCR) analysis.
Materials and methods
Ethics statement
A total of 24 patients with HCC admitted to our hospital between June 2013 and December 2014 were enrolled in the present study. Written informed consent was obtained from all participants prior to tissue collection. Ethical approval was granted by the Institutional Ethical Committee.
Datasets
In the present study, three gene expression profiles [GSE6222 (11), GSE41804 (12) and GSE51401] of patients with HCC and normal controls were downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). A total of 78 HCC samples and 38 normal controls were collected from the three datasets. The characteristics of the datasets are shown in Table I.
Table I.
Characteristics of the datasets.
Dataset preprocessing
Prior to analysis, the quality of the gene microarray probe-level data was controlled by standard procedures, which comprising background correction (13), normalization (14), probe correction (15) and summarization (13). Background correction was carried out using the Robust Multi-array Average (RMA) algorithm to eliminate the influence of nonspecific hybridization (13). The perfect match (PM) probes were modeled as the sum of a normal noise component N (normal with mean µ and variance σ2) and an exponential signal component S (exponential with mean α). The normal was truncated at zero to avoid any possibility of negatives, and the observed intensity O was adjusted by the following equation:
where a=s-µ-σ2α and b=σ. It should be noted that ϕ and are the standard normal distribution density and distribution functions respectively, and mismatch (MM) probe intensities were not corrected by the above procedure.
Normalization was performed through a quantiles-based algorithm (14). The goal of the quantile method was to make the distribution of probe intensities for each array in a set of arrays the same. This method was a specific case of the transformation:
where G was estimated by the empirical distribution of each array and F using the empirical distribution of the averaged sample quantiles.
Probes of PM/MM value were corrected utilizing the MAS approach (15). An ideal MM was subtracted from PM and would always be less than the corresponding PM. Thus it could safely be subtracted without risk of negative values being obtained.
Summarization of probes was dependent upon medianpolishing (13). A multichip linear model was fit to data from each probe set. In particular for a probe set k with i=1, …, Ik probes and data from j=1, …, J arrays, were fitted into the following model:
where αi was a probe effect and βj was the log2 expression value.
In the next stage, the preprocessed probe-level dataset in CEL format was converted into expression measures, and then screened by the feature filter method of a gene filter package (16).
Integration of multiple datasets
For the purpose of integrating the three datasets into a single group and removing the batch effects caused by the use of different experimentation plans and methodologies, the GENENORM method was applied in order to increase the comparability of the datasets at score normalization, and the expression values were calculated (17). The modified gene expression value was given by the expression:
where Xij indicated each gene expression value in each study; stood for the mean gene expression value in the dataset; K represented the number of the studies and was the standard deviation of gene expression value.
The distribution of merged data was inspected according to the plotMDS qualitative validation method to observe visually whether the samples from all studies would cluster together or have a dataset-bias (18). Finally, the expression profile dataset containing 20,102 genes was obtained.
Identification of DE genes
Genes differently expressed between patients with HCC and normal subjects were identified using the empirical Bayes method of the Linear Models for Microarray Data package (19). The approach is applicable for the analysis of factorial data with high density oligonucleotide microarray data. The false discovery rate (FDR) was controlled by Benjamini-Hochberg test (20). Only the genes which met the criterion (P<0.05, |log2FoldChange|>2) were selected as DE genes in this study.
Co-expression network construction
Some significant genes may not be identifiable through their own behavior, but exhibit quantifiable changes when considered in conjunction with other genes (for example, as a co-expression network). In this study, co-expression networks were constructed using DCGL to identify differentially co-expressed (DC) genes and links (21). The DCGL package contains four modules: Gene filtration, link filtration, differential co-expression analysis (DCEA) and differential regulation analysis (DRA) modules. Differential co-expression profile (DCp) and differential co-expression enrichment (DCe) were involved in the DCEA module for extracting DC genes and DC links. DCp worked on the filtered set of gene co-expression value pairs, where each pair was composed of two co-expression values worked out in two different conditions separately. The subset of co-expression value pairs associated with a particular gene, in two groups for the two conditions separately, was written as two vectors: X=(xi1, xi2, …, xin) and Y=(yi1, yi2, …, yin) where n is the number of co-expression neighbors for a gene. A length-normalized Euclidean distance was used to measure the differential co-expression (dC) of this gene (22).
A permutation test was performed to assess the significance of dC. In this test, the disease samples and normal controls were randomly permuted, and Pearson's correlation coefficient (PCC) was calculated. The sample permutation was repeated N times, and a large number of permutation dC statistics formed an empirical null distribution. Non-informative correlation pairs were filtered out with the half-thresholding strategy and pairs with FDR-adjusted P<0.05 were retained (20).
DCe was also used to identify DC genes and DC links, which are based on the limit fold change (LFC) model. First, correlation pairs were divided into 3 parts according to the pairing of signs of co-expression values and the number of co-expression values: Pairs with the same signs (N1), pairs with different signs (N2) and pairs with differently-signed high co-expression values (N3). The first two parts were processed with the LFC model separately to produce two subsets of DC links (K1, K2), while the third part (N3) was added to the set of DC links directly. Therefore, a total of K=N3 + K1 + K2 DC links were determined from a total of N gene links. For a gene (gi), the total numbers of links (ni) and DC links (ki) associated with it were counted. A binomial probability model was used to estimate the significance of the gene being a DC gene.
Differentially co-expression summarization (DCsum) was implemented to combine the results from the DCp and DCe methods. After obtaining the DC genes and DC links, the co-expression network was visualized using the Cytoscape 2.1 software (www.cytoscape.org).
Cluster identification of the co-expression network
The clusters of the co-expression network were identified by MCODE, which is a theoretical cluster algorithm that selects densely connected regions (23). The MCODE algorithm includes three main stages: Vertex weighting, complex prediction and optionally post-processing. At the stage of vertex weighting, all vertices based on their local network density were weighted using the highest k-core of the vertex neighborhood. At the second stage, the vertex-weighted graph was taken as input. A complex with the highest weighted vertex was seeded, and moved outward from the seed vertex recursively. It owned vertices in the complex whose weight was above a given threshold, a given percentage away from the weight of the seed vertex. Complexes with a core <2 (graph of minimum degree 2) were filtered. In this study, node density cutoff = 0.1, node score cutoff = 0.2, K-core = 2 and maximum depth = 100 were set as the parameters in MCODE for the detection of clusters in the co-expression network. In addition, clusters with <10 nodes were discarded.
Centrality analysis of the co-expression network
In network analysis, the determination of the importance of a particular node or edge in a network is a fundamental challenge, and quantifying centrality and connectivity helps to identify portions of the network that may play important roles (24). In the present study, the biological importance of genes was characterized based on the co-expression network using indices of topological centrality, including local scale (degree and clustering coefficient) and global scale (stress centrality, betweenness centrality and closeness centrality). The genes at the ≥95% quantile distribution in the significantly perturbed networks were defined as hub genes.
For the graph G=(V, E), V is the set of vertices representing nodes in a network, and E is the set of edges representing relationships between the nodes. A path from node s to t is defined as a sequence of edges (ui, ui+1), 0≤i ≤l, where u0=s and ul=t. The length of a path is the sum of the weights of edges, and d (s, t) was used to denote the distance between s and t (the minimum length of any path connecting s and t in G). The total number of shortest paths between vertices s and t was denoted by σst, and the number passing through node v was denoted by σst (v).
Degree centrality
Degree quantifies the local topology of each gene by summing up the number of its adjacent genes and gives a simple count of the number of interactions of a given node (25). The degree CD (v) of a node v was determined using the following formula:
Clustering coefficient
The clustering coefficient of a node v is the proportion of its neighbors that are also neighbors of each other (26). An example is a situation in which node v is connected to nodes s, t and l, and only nodes s and t are also connected. This metric provides a measure of local cliques, where information processing/fold change is particularly segregated from the rest of the network. An edge eij connected node vi and vj, and the local clustering coefficient C (v) for node v was given as:
where ki was the number of nodes, Ni was the neighborhood of vi and defined as its immediate connections as follows:
Closeness centrality
Closeness centrality is a measure of the shortest paths to access all other proteins in the network (27). The larger the value, the more central is the protein. The closeness centrality, Cc (v) was defined as the reciprocal of the average shortest path length and was computed as follows:
Meanwhile, in the undirected graph, d (s, s)=0 and d (s, t)=d (t, s).
Betweenness centrality
Betweenness centrality is a topological metric in graphs for determining how the neighbors of a node are interconnected and is considered the frequency with which a node is on the shortest path between two other nodes (28). The betweenness centrality of a node v was calculated by the expression:
Therefore, the calculation might be rescaled by dividing through by the number of pairs of nodes not including v, so that CB (v) [0,1].
Stress centrality
Stress centrality is a metric based on the number of nodes in the shortest path between two other nodes (29). A stressed node was a node traversed by a high number of shortest paths. The stress, Cs (v) was calculated as follows:
Functional and pathway enrichment analysis
To further investigate the functions of DE genes, GO functional enrichment and KEGG pathway enrichment analysis were performed using the online tool Database for Annotation, Visualization and Integrated Discovery (DAVID) (30). GO terms and KEGG pathways with P<0.05 were selected based on an expression analysis systematic explored (EASE) test implemented in the DAVID (31). The formula used for the EASE test was as follows:
in which n=a'+b+c+d was the number of background genes; a' was the gene number of one gene set in the gene lists; a'+b was the number of genes in the gene list including at least one gene set; a'+c was the gene number of one gene list in the background genes; a' was replaced with a=a'-1 in EASE.
Validation of hub genes using RT-PCR analysis
In this study, RT-PCR was utilized to validate the hub genes that had the highest degree and the most importance in the co-expression network. Total RNA was obtained from 24 HCC tumor samples and 24 matched non-cancerous samples from adjacent tissues, respectively, using TRIzol regent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The data were normalized to β-actin reference. Three common hub genes, reelin (RELN), potassium voltage-gated channel subfamily J member 10 (KCNJ10) and neural cell adhesion molecule 1 (NCAM1), were subjected to validation analysis.
RT-PCR was performed in two steps. For the first, cDNA synthesis, RNA was mixed with oligo (dT)18 primers adjusted to 10 µl and incubated at 70°C for 5 min. Next, RNA/primer mix was used in 20-µl reactions containing 2 µl RNasin (40 U/µl), 8.0 µl 5X reverse transcriptase buffer, 8.0 µl dNTPs and 2 µl AMV reverse transcriptase (5 U/µl) (all reagents from New England Biolabs, Inc., Ipswich, MA, USA). The reactions were incubated for 1 h at 42°C, 15 min at 70°C, and adjusted to a final volume of 50 µl. For the second-strand synthesis, PCRs were conducted using specific primers (Table II), 0.2 mM dNTPs, 1 unit Taq DNA polymerase, 10X PCR buffer and 2 µl first-strand cDNA. Reactions were performed using the following program: 2 min at 94°C for predenaturation, followed by 35 cycles of 20 sec at 94°C, 15 sec at 60°C and 1 min at 68°C, and a final 7 min extension at 72°C. Next, 5 µl PCR product was loaded onto a 1.5% agarose gel containing ethidium bromide. To assess the limit of detection (LOD), serial 10-fold dilutions of total RNA were used as a template in 25-µl RT-PCRs. The PCR products were purified using the QIAquick PCR purifiation kit (Qiagen, Hilden, Germany) and were analyzed using Quantity One Software for gel imaging analysis (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Table II.
Primer sequences for the candidate genes.
| Primers (5′-3′) | |||
|---|---|---|---|
| Genes | Forward | Reverse | Length (bp) |
| RELN | ACCAGTGGGCAGTCGATGACATCAT | CTTCATTAGCCAACATCAACCACAC | 489 |
| KCNJ10 | CATGGGGTGAGGGTTAGGAG | GGGAGTGGAGGATGGGTG | 284 |
| NCAM1 | ATGGAAACTCTATTAAAGTGAACCTGA | TAGACCTCATACTCAGCATTCCAGT | 186 |
| β-actin | AAGTACTCCGTGTGGATCGG | TCAAGTTGGGGGACAAAAAG | 651 |
RELN, reelin; KCNJ10, potassium voltage-gated channel subfamily J member 10; NCAM1, neural cell adhesion molecule 1.
Statistical analysis
Data were presented as the mean ± standard deviation, and all statistical analyses were carried out using SPSS 19.0 software (SPSS, Inc., Chicago, IL, USA). Student's t-test was used to determine the statistical significance of differences between groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of DE genes
There were 20,102 genes after integrating the datasets GSE6222, GSE41804 and GSE51401 into the merged gene expression dataset used to detect DE genes in this study. In total, 260 DE genes were identified between patients with HCC and normal controls with the thresholds of P<0.05, |log2FoldChange|>2.
Co-expression network construction and cluster identification
Many genes together play important roles in the accomplishment of a biological function, and highly co-expressed genes participate in similar biological processes and pathways. Notably, functionally related genes are frequently co-expressed across samples. DCGL was applied, with the use of DCp and DCe methods in the DCEA module, in order to construct a co-expression network based on the 260 DE genes of HCC. A total of 326 co-expression gene pairs were identified; the two genes in each pair were DC genes. Finally, a co-expression network with 154 nodes and 326 edges was visualized using Cytoscape (Fig. 1).
Figure 1.
Co-expression network of 260 differentially expressed genes. There were 154 nodes and 326 edges in the network, which was constructed using the Differentially Co-expressed Genes and Links (DCGL) package. Genes (nodes) were connected by edges if their vectors were sufficiently similar. Nodes represent genes, and each edge is associated with a pair of co-expressed genes.
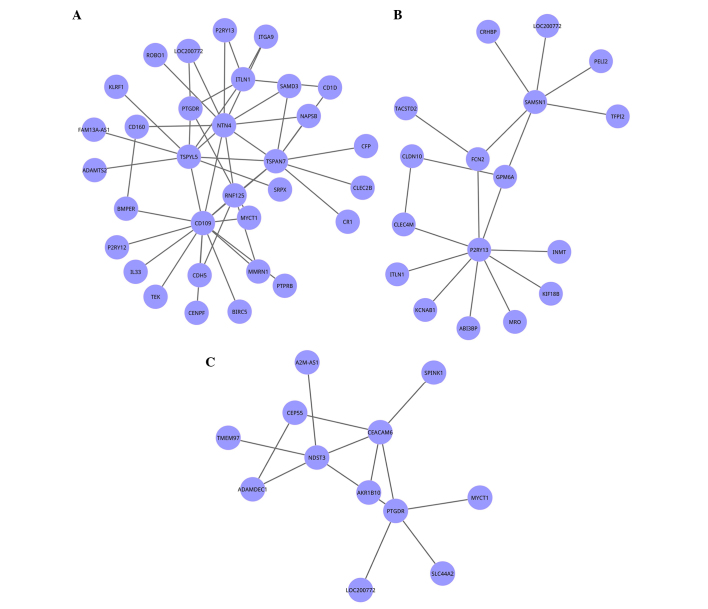
The MCODE algorithm was selected to mine subnetworks of the co-expression network. When a node density cutoff of 0.1, node score cutoff of 0.2, K-core of 2 and maximum depth of 100 were set, 3 clusters were identified having a gene number >10 (Fig. 2). In detail, cluster 1 possessed the most nodes (n=32), of which CD160 and CD109 connected with the greatest number of genes (n=16 and n=14, respectively) in the network. The total degree of cluster 1 was the highest (n=177).
Figure 2.
The three clusters identified in the co-expression network of the differentially expressed genes. (A) Cluster 1, (B) cluster 2 and (C) cluster 3. Cluster 1 possessed the greatest number of nodes (n=32), and the highest total degree (n=177). Nodes represent genes, and edges represent the interaction of genes.
Centrality analyses of the co-expression network
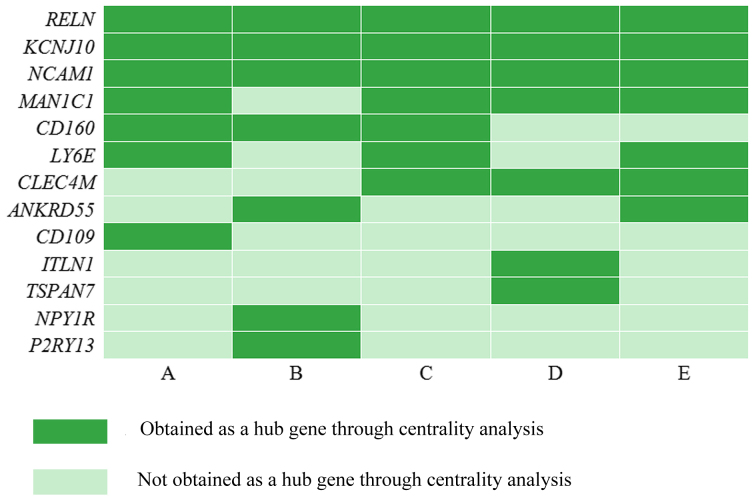
Centralities indicate the likelihood of a gene being functionally capable of holding communicating nodes together, for a node in a biological network. In this study, genes at the ≥95% quantile distribution in the co-expression network were defined as hub genes. Five types of centralities (degree, clustering coefficient, closeness, betweenness and stress) were calculated, based on the complex network, and it was found that hub genes or the top 5% of genes distributed in various centrality analyses of the same gene were not entirely consistent (Fig. 3). In total, 13 hub genes were obtained, of which RELN, KCNJ10 and NCAM1 were common hub genes across degree, clustering coefficient, closeness, betweenness and stress centrality analysis. In addition, mannosidase α class 1C member 1 (MAN1C1) was obtained by four methods (with clustering coefficient analysis being the exception), and CD160, lymphocyte antigen 6 complex, locus E (LY6E) and C-type lectin domain family 4 member M (CLEC4M) were detected using three of the five types.
Figure 3.
Distribution of hub genes identified by five types of centrality. (A) degree centrality; (B) clustering coefficient; (C) betweenness centrality; (D) closeness centrality and (E) stress centrality.
Functional and pathway enrichment analysis
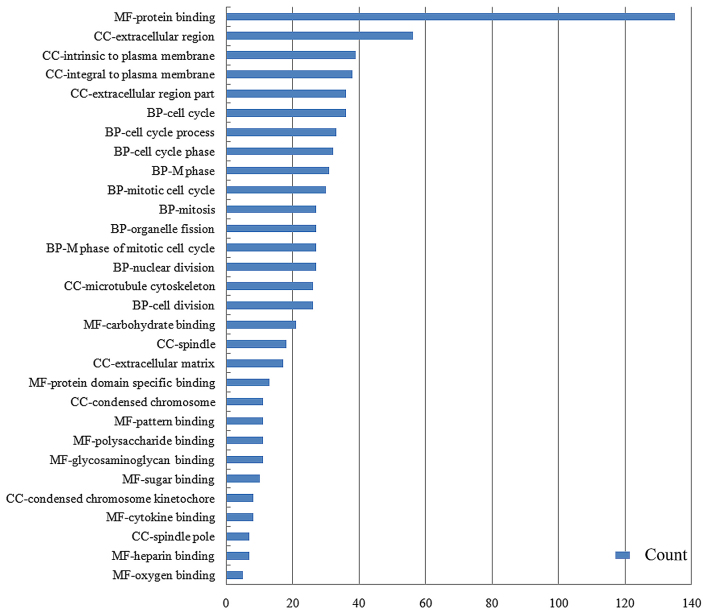
To identify the biological processes associated with gene expression changes in HCC, GO analysis was performed which covered three domains, namely molecular function (MF), biological process (BP) and cellular component (CC), for the 260 DE genes. The results showed that DE genes were enriched in 136 BP terms, 28 CC terms and 30 MF terms under the condition of P<0.05, and the top 10 terms in sequence of count value are shown in Fig. 4. Protein binding had the highest number of counts at 135. BP terms with a high count were associated with cell cycle and mitosis. Extracellular region and plasma membrane-related CC terms possessed high counts. When considering the terms according to P-value, the most significant terms of BP, CC and MF were mitosis with P=1.60E-16, spindle with P=4.44E-11 and carbohydrate binding with P=2.35E-07, respectively.
Figure 4.
Top 10 Gene Ontology terms in the biological process (BP), cellular component (CC) and molecular function (MF) domains in order of count value. Protein binding had the most counts (n=135). BP terms with high counts were associated with cell cycle and mitosis.
To further investigate the functions of DE genes, they were mapped to the KEGG database and 8 significant pathways with P<0.05 were identified (Table III). Cell cycle (P=1.21E-06), metabolism of xenobiotics by cytochrome P450 (P=5.35E-04) and p53 signaling pathway (P=5.67E-04) were the three most significant pathways. In addition to metabolism of xenobiotics by cytochrome P450, there were two other metabolic pathways, drug metabolism (P=4.13E-04) and linoleic acid metabolism (P=1.20E-02), which indicates that HCC has an association with metabolic biological processes.
Table III.
Significant enrichment pathways in hepatocellular carcinoma.
| Term | Count | P-value | Genes |
|---|---|---|---|
| Cell cycle | 13 | 1.21E-06 | CDK1, TTK, CDC20, PTTG1, SFN, MCM2, CCNB1, CCNE2, MAD2L1, CCNB2, BUB1, BUB1B, CCNA2 |
| Metabolism of xenobiotics by cytochrome P450 | 7 | 5.35E-04 | GSTA4, ADH4, CYP2C8, ADH1B, CYP2E1, CYP1A2, AKR1C1 |
| p53 signaling pathway | 7 | 5.67E-04 | CCNE2, CCNB1, CDK1, CCNB2, RRM2, SFN, THBS1 |
| Oocyte meiosis | 9 | 1.05E-03 | CCNE2, CCNB1, CDK1, MAD2L1, CCNB2, BUB1, CDC20, AURKA, PTTG1 |
| Drug metabolism | 6 | 4.13E-03 | GSTA4, ADH4, CYP2C8, ADH1B, CYP2E1, CYP1A2 |
| Linoleic acid metabolism | 4 | 1.20E-02 | AKR1B10, CYP2C8, CYP2E1, CYP1A2 |
| ECM-receptor interaction | 6 | 1.46E-02 | ITGA9, RELN, THBS1, COL5A2, HMMR, SPP1 |
| Progesterone-mediated oocyte maturation | 6 | 1.61E-02 | CCNB1, CDK1, MAD2L1, CCNB2, BUB1, CCNA2 |
ECM, extracellular matrix.
Validation of hub genes based on RT-PCR
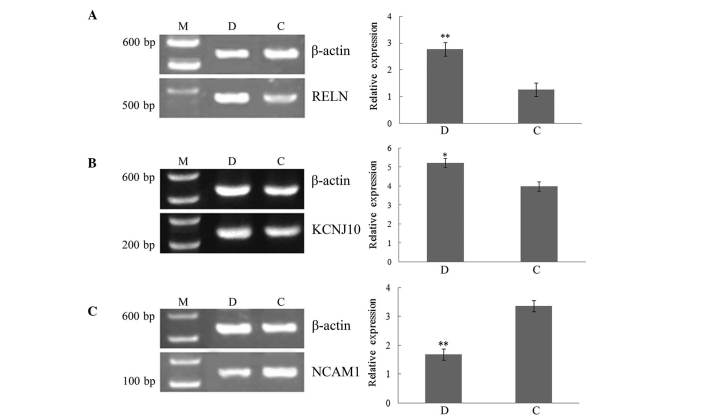
To confirm the hub genes identified on the basis of centrality analyses of the co-expression network and to investigate the key genes of HCC, RT-PCR analysis of three common hub genes (RELN, KCNJ10 and NCAM1) was conducted. RELN and KCNJ10 were upregulated DE genes, while NCAM1 was a downregulated DE gene. The RT-PCR results are shown in Fig. 5. The relative expression levels of RELN and KCNJ10 were increased, but that of NCAM1 was decreased in patients with HCC compared with healthy controls, which confirmed the DE gene analysis. Furthermore, the differences in these gene expression levels between normal controls and HCC patients were found to be statistically significant (for RELN and NCAM1, P<0.001; for KCNJ10, P<0.05). These results demonstrate that the common hub genes were significantly differentially expressed in patients with HCC.
Figure 5.
Reverse transcription-polymerase chain reaction results for the common hub genes (A) RELN, (B) KCNJ10 and (C) NCAM1 in patients with HCC and normal controls. Data are presented as the mean ± standard deviation *P<0.05 vs. the control group; **P<0.01 vs. the control group. M, marker; D, patients with HCC; C, normal control. HCC, hepatocellular carcinoma; RELN, reelin; KCNJ10, potassium voltage-gated channel subfamily J member 10; NCAM1, neural cell adhesion molecule 1.
Discussion
HCC is a highly prevalent malignancy worldwide with a heterogenetic molecular pathogenesis, that has not yet been fully clarified. Identifying the most significant genes and pathways associated with this disease contributes to understanding the molecular pathogenesis and providing potential biomarkers for effective therapies.
In the present study, hub genes were identified based on degree, clustering coefficient, closeness, betweenness and stress centrality analysis in a co-expression network of HCC and integrated the results of the five centralities to resolve the inconsistent outcomes provided by different methods. A total of 13 hub genes were identified and RELN, KCNJ10 and NCAM1 were common hub genes across the five centrality methods. In addition, the hub genes were validated utilizing RT-PCR analysis and the results were consistent with centrality analyses of the co-expression network. GO and pathway enrichment analysis of DE genes showed that cell cycle, mitosis and protein binding were the most relevant GO terms, while cell cycle, metabolism of xenobiotics by cytochrome P450 and p53 signaling pathway were the most significant pathways.
RELN is an extracellular 420-kDa glycoprotein that is involved in the regulation of neuronal migration during brain development (32). Varying levels of RELN expression had been reported in cancers. High expression levels of RELN have been reported in 87.5% of esophageal cancers (33) and 39% of prostate cancers (34). However, the expression of RELN is lost or highly reduced in gastric cancer (32) and in 72% of pancreatic cancers (35). Literature focused on RELN expression in HCC is lacking. In this study, it was discovered that RELN was an upregulated DE gene and significantly differently expressed based on RT-PCR assays as a hub gene in HCC. Moreover, it was found that DE genes of HCC were enriched in extracellular-related CC GO terms; notably, RELN encodes a large secreted extracellular matrix protein. These results indicate that RELN plays a significant role in HCC progression, which is consistent with previous studies. For instance, Okamura et al (36) revealed that RELN was a key regulatory gene associated with the recurrence of HCC. Furthermore RELN has been suggested to be involved in ECM-receptor interaction and focal adhesion, which might be the mechanism underlying the high metastasis rate of HCC with RELN mutations (37). ECM-receptor interaction containing RELN was found to be a significant pathway of HCC in the present study. Therefore, RELN appears to be significantly associated with HCC.
NCAM1, a downregulated DE gene, was identified as another common hub gene of HCC in the present study. NCAM1 encodes a cell adhesion protein which is a member of the immunoglobulin super family. NCAM has been found in cancer-initiating stem cells of the liver and is a marker of hepatic stem/progenitor cells (38). Balzarini et al (39) demonstrated a significant alteration of NCAM expression in HCC biopsies and underlined the importance of NCAM in the induction of abnormal neovascular formations in HCC vascular morphogenesis. As a member of the NCAM family, NCAM1 is a known hepatic stem/progenitor cell marker and has been experimentally demonstrated to be a direct target of miR-200c, which indicates that HCC has stem-like molecular characteristics and a poor prognosis (40). Furthermore, HCC tumor cells have been shown to be positive for NCAM1/CD56 immunohistochemically (41). In the present study, NCAM1 expression was confirmed in HCC by RT-PCR assays. Hence, it appears that NCAM1 is associated with HCC.
Functional enrichment analysis in the present study suggested that DE genes were enriched in cell cycle, mitosis and protein binding terms significantly, which was consistent with the functions of hub genes and significant pathways generally. Taking the cell cycle biological process as an example, cell cycle is the series of events that takes place in a cell leading to its division and duplication, and mitosis is a part of the cell cycle process. Dysregulation of cell cycle components may lead to tumor formation, and the roles that the cell cycle plays in HCC have been reported (42,43). When genes such as the cell cycle inhibitors cyclin-dependent kinases (CDK) and p53 mutate, they may cause cells to multiply uncontrollably, forming a tumor. Among the 260 DE genes of HCC identified in the present study, there were certain genes deeply associated with the cell cycle, such as CDK1, cell division cycle 20 (CDC20) and cyclin E2 (CCNE2). Furthermore, the p53 signaling pathway has a close association with the cell cycle (44).
It has been estimated that at least a third of all serious health problems are caused by metabolic disorders (45). The present study identified that HCC was associated with several metabolic pathways, such as metabolism of xenobiotics by cytochrome P450, drug metabolism and linoleic acid metabolism. Metabolism of xenobiotics by cytochrome P450 is a typical liver-function-specific pathway and is of importance in HCC (46). The members of cytochrome P450 (CYP) family, involved in a myriad of biological processes, is frequently dysregulated in liver cancer (47). In this pathway, three members of the CYP family were involved: CYP2C8, CYP2E8 and CYP1A2. Zhang et al (48) suggested that CYP2C8 was post-transcriptionally regulated by microRNAs in the human liver. Therefore, metabolism of xenobiotics by cytochrome P450 appears to be a significant pathway in HCC.
In conclusion, the present study identified hub genes (such as RELN, KCNJ10 and NCAM1) and pathways (for instance, cell cycle, metabolism of xenobiotics by cytochrome P450 and p53 signaling pathway) associated with HCC based on centrality analysis of a co-expression network and RT-PCR assays. This study may contribute to understanding the molecular pathogenesis of HCC and provide potential biomarkers for effective therapies of this disease.
Acknowledgements
This study was supported as a medical technology joint key project of Wuxi Medical Management Center (grant no. YGZX1202).
References
- 1.Bruix J, Sherman M. American Association for the Study of Liver Diseases: Management of hepatocellular carcinoma: An update. Hepatology. 2011;53:1020–1022. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki T, Kokudo N, Matsuyama Y, Izumi N, Ichida T, Kudo M, Ku Y, Sakamoto M, Nakashima O, Matsui O, et al. Prognostic impact of spontaneous tumor rupture in patients with hepatocellular carcinoma: An analysis of 1160 cases from a nationwide survey. Ann Surg. 2014;259:532–542. doi: 10.1097/SLA.0b013e31828846de. [DOI] [PubMed] [Google Scholar]
- 3.Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV-and HCV-associated hepatocellular carcinoma. Nat Rev Cancer. 2013;13:123–135. doi: 10.1038/nrc3449. [DOI] [PubMed] [Google Scholar]
- 4.Jordán F, Nguyen TP, Liu WC. Studying protein-protein interaction networks: A systems view on diseases. Brief Funct Genomics. 2012;11:497–504. doi: 10.1093/bfgp/els035. [DOI] [PubMed] [Google Scholar]
- 5.Wang L, Zang W, Xie D, Ji W, Pan Y, Li Z, Shen J, Shi Y. Comparison of hepatocellular carcinoma (HCC), cholangiocarcinoma (CC) and combined HCC-CC (CHC) with each other based on microarray dataset. Tumor Biol. 2013;34:1679–1684. doi: 10.1007/s13277-013-0702-6. [DOI] [PubMed] [Google Scholar]
- 6.Jia D, Wei L, Guo W, Zha R, Bao M, Chen Z, Zhao Y, Ge C, Zhao F, Chen T, et al. Genome-wide copy number analyses identified novel cancer genes in hepatocellular carcinoma. Hepatology. 2011;54:1227–1236. doi: 10.1002/hep.24495. [DOI] [PubMed] [Google Scholar]
- 7.Lee JH, Chung YH, Lee HC. Free Paper Session: HCC; Genetic polymorphisms associated with treatment toxicity after sorafenib combination therapy in Korean patients with hepatocellular carcinoma. Clin Mol Hepatol (Suppl) 2011;17:S36. [Google Scholar]
- 8.Reis AH, Vargas FR, Lemos B. More epigenetic hits than meets the eye: MicroRNAs and genes associated with the tumorigenesis of retinoblastoma. Front Genet. 2012;3:284. doi: 10.3389/fgene.2012.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang D, Han G, Feng X, Sun J, Duan Y, Lei H. Concerted perturbation observed in a hub network in Alzheimer's disease. PLoS One. 2012;7:e40498. doi: 10.1371/journal.pone.0040498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thériault BL, Dimaras H, Gallie BL, Corson TW. The genomic landscape of retinoblastoma: A review. Clin Experiment Ophthalmol. 2014;42:33–52. doi: 10.1111/ceo.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao YL, Sun YM, Chau GY, Chau YP, Lai TC, Wang JL, Horng JT, Hsiao M, Tsou AP. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008;27:5578–5589. doi: 10.1038/onc.2008.168. [DOI] [PubMed] [Google Scholar]
- 12.Hodo Y, Honda M, Tanaka A, Nomura Y, Arai K, Yamashita T, Sakai Y, Yamashita T, Mizukoshi E, Sakai A, et al. Association of interleukin-28B genotype and hepatocellular carcinoma recurrence in patients with chronic hepatitis C. Clin Cancer Res. 2013;19:1827–1837. doi: 10.1158/1078-0432.CCR-12-1641. [DOI] [PubMed] [Google Scholar]
- 13.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 15.Bolstad B. bioconductor.org/packages/devel/bioc/vignettes/affy/inst/doc/builtinMethods.pdf. affy: Built-in Processing Methods. 2013 Accessed December 20, 2014.
- 16.Lee J, Kim DW. Efficient multivariate feature filter using conditional mutual information. Electron Lett. 2012;48:161–162. doi: 10.1049/el.2011.3063. [DOI] [Google Scholar]
- 17.Taminau J. bioconductor.org/packages//2.11/bioc/vignettes/inSilicoMerging/inst/doc/inSilicoMerging.pdf. Using the inSilicoMerging package. 2014 Dec 20; Accessed.
- 18.Taminau J, Meganck S, Lazar C, Steenhoff D, Coletta A, Molter C, Duque R, de Schaetzen V, Solís DY Weiss, Bersini H, Nowé A. Unlocking the potential of publicly available microarray data using inSilicoDb and inSilicoMerging R/Bioconductor packages. BMC Bioinformatics. 2012;13:335. doi: 10.1186/1471-2105-13-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995:289–300. [Google Scholar]
- 21.Yang J, Yu H, Liu BH. cran.r-project.org/web/packages/DCGL/DCGL.pdf. Using the DCGL 2.0 Package. 2013 Accessed December 24, 2014.
- 22.Dokmanic I, Parhizkar R, Ranieri J, Vetterli M. Euclidean distance matrices: Essential theory, algorithms, and applications. IEEE Signal Process Mag. 2015;32:12–30. doi: 10.1109/MSP.2015.2398954. [DOI] [Google Scholar]
- 23.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bader DA, Madduri K. Parallel algorithms for evaluating centrality indices in real-world networks; 2006 International Conference on Parallel Processing (ICPP'06); Los Alamitos, CA. 2006. pp. 539–550. [DOI] [Google Scholar]
- 25.Haythornthwaite C. Social network analysis: An approach and technique for the study of information exchange. Libr Inform Sci Res. 1996;18:323–342. doi: 10.1016/S0740-8188(96)90003-1. [DOI] [Google Scholar]
- 26.Coman D, Rütimann P, Gruissem W. A flexible protocol for targeted gene co-expression network analysis. In: Concepción MR, editor. Plant Isoprenoids: Methods and Protocols. Springer; NY: 2014. pp. 285–299. [DOI] [PubMed] [Google Scholar]
- 27.Wasserman S, Faust K. Social Network Analysis: Methods and Applications. Cambridge University Press; Cambridge: 1994. [DOI] [Google Scholar]
- 28.Barthelemy M. Betweenness centrality in large complex networks. Eur Phys J B. 2004;38:163–168. doi: 10.1140/epjb/e2004-00111-4. [DOI] [Google Scholar]
- 29.Fekete SP, Kaufmann M, Kröller A, Lehmann K. A new approach for boundary recognition in geometric sensor networks. arXiv: cs/0508006. 2005 [Google Scholar]
- 30.Huangda W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Simon R. Microarray-based cancer prediction using single genes. BMC Bioinformatics. 2011;12:391. doi: 10.1186/1471-2105-12-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dohi O, Takada H, Wakabayashi N, Yasui K, Sakakura C, Mitsufuji S, Naito Y, Taniwaki M, Yoshikawa T. Epigenetic silencing of RELN in gastric cancer. Int J Oncol. 2010;36:85–92. [PubMed] [Google Scholar]
- 33.Wang Q, Lu J, Yang C, Wang X, Cheng L, Hu G, Sun Y, Zhang X, Wu M, Liu Z. CASK and its target gene Reelin were co-upregulated in human esophageal carcinoma. Cancer Lett. 2002;179:71–77. doi: 10.1016/S0304-3835(01)00846-1. [DOI] [PubMed] [Google Scholar]
- 34.Perrone G, Vincenzi B, Zagami M, Santini D, Panteri R, Flammia G, Verzì A, Lepanto D, Morini S, Russo A, et al. Reelin expression in human prostate cancer: A marker of tumor aggressiveness based on correlation with grade. Modern Pathol. 2007;20:344–351. doi: 10.1038/modpathol.3800743. [DOI] [PubMed] [Google Scholar]
- 35.Sato N, Fukushima N, Chang R, Matsubayashi H, Goggins M. Differential and epigenetic gene expression profiling identifies frequent disruption of the RELN pathway in pancreatic cancers. Gastroenterology. 2006;130:548–565. doi: 10.1053/j.gastro.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 36.Okamura Y, Nomoto S, Kanda M, Hayashi M, Nishikawa Y, Fujii T, Sugimoto H, Takeda S, Nakao A. Reduced expression of reelin (RELN) gene is associated with high recurrence rate of hepatocellular carcinoma. Ann Surg Oncol. 2011;18:572–579. doi: 10.1245/s10434-010-1273-z. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Qiu Z, Wei L, Tang R, Lian B, Zhao Y, He X, Xie L. Integrated analysis of mutation data from various sources identifies key genes and signaling pathways in hepatocellular carcinoma. PloS One. 2014;9:e100854. doi: 10.1371/journal.pone.0100854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuchiya A, Kamimura H, Tamura Y, Takamura M, Yamagiwa S, Suda T, Nomoto M, Aoyagi Y. Hepatocellular carcinoma with progenitor cell features distinguishable by the hepatic stem/progenitor cell marker NCAM. Cancer Lett. 2011;309:95–103. doi: 10.1016/j.canlet.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 39.Balzarini P, Benetti A, Invernici G, Cristini S, Zicari S, Caruso A, Gatta LB, Berenzi A, Imberti L, Zanotti C, et al. Transforming growth factor-beta1 induces microvascular abnormalities through a down-modulation of neural cell adhesion molecule in human hepatocellular carcinoma. Lab Invest. 2012;92:1297–1309. doi: 10.1038/labinvest.2012.94. [DOI] [PubMed] [Google Scholar]
- 40.Oishi N, Kumar MR, Roessler S, Ji J, Forgues M, Budhu A, Zhao X, Andersen JB, Ye QH, Jia HL, et al. Transcriptomic profiling reveals hepatic stem-like gene signatures and interplay of miR-200c and epithelial-mesenchymal transition in intrahepatic cholangiocarcinoma. Hepatology. 2012;56:1792–1803. doi: 10.1002/hep.25890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Connor K, Walsh JC, Schaeffer DF. Combined hepatocellular-cholangiocarcinoma (cHCC-CC): A distinct entity. Ann Hepatol. 2014;13:317–322. [PubMed] [Google Scholar]
- 42.Furuta M, Kozaki K, Tanimoto K, Tanaka S, Arii S, Shimamura T, Niida A, Miyano S, Inazawa J. The tumor-suppressive miR-497-195 cluster targets multiple cell-cycle regulators in hepatocellular carcinoma. PloS One. 2013;8:e60155. doi: 10.1371/journal.pone.0060155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim H, Lee K, Bae H, Eun JW, Shen Q, Park SJ, Shin WC, Yang HD, Park M, Park WS, et al. MicroRNA-31 functions as a tumor suppressor by regulating cell cycle and epithelial-mesenchymal transition regulatory proteins in liver cancer. Oncotarget. 2015;10:8089–8102. doi: 10.18632/oncotarget.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hahnvajanawong C, Ketnimit S, Pattanapanyasat K, Anantachoke N, Sripa B, Pinmai K, Seubwai W, Reutrakul V. Involvement of p53 and nuclear factor-kappaB signaling pathway for the induction of G1-phase cell cycle arrest of cholangiocarcinoma cell lines by isomorellin. Biol Pharm Bull. 2012;35:1914–1925. doi: 10.1248/bpb.b12-00118. [DOI] [PubMed] [Google Scholar]
- 45.Ford ES, Giles WH, Dietz WH. Prevalence of the metabolic syndrome among US adults: Findings from the third National Health and Nutrition Examination Survey. JAMA. 2002;287:356–359. doi: 10.1001/jama.287.3.356. [DOI] [PubMed] [Google Scholar]
- 46.Cheng S, Prot JM, Leclerc E, Bois FY. Zonation related function and ubiquitination regulation in human hepatocellular carcinoma cells in dynamic vs. Static culture conditions. BMC Genomics. 2012;13:54. doi: 10.1186/1471-2164-13-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu Y, Ping J, Chen H, Jiao L, Zheng S, Han ZG, Hao P, Huang J. A comparative analysis of liver transcriptome suggests divergent liver function among human, mouse and rat. Genomics. 2010;96:281–289. doi: 10.1016/j.ygeno.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 48.Zhang SY, Surapureddi S, Coulter S, Ferguson SS, Goldstein JA. Human CYP2C8 is post-transcriptionally regulated by microRNAs 103 and 107 in human liver. Mol Pharmacol. 2012;82:529–540. doi: 10.1124/mol.112.078386. [DOI] [PMC free article] [PubMed] [Google Scholar]