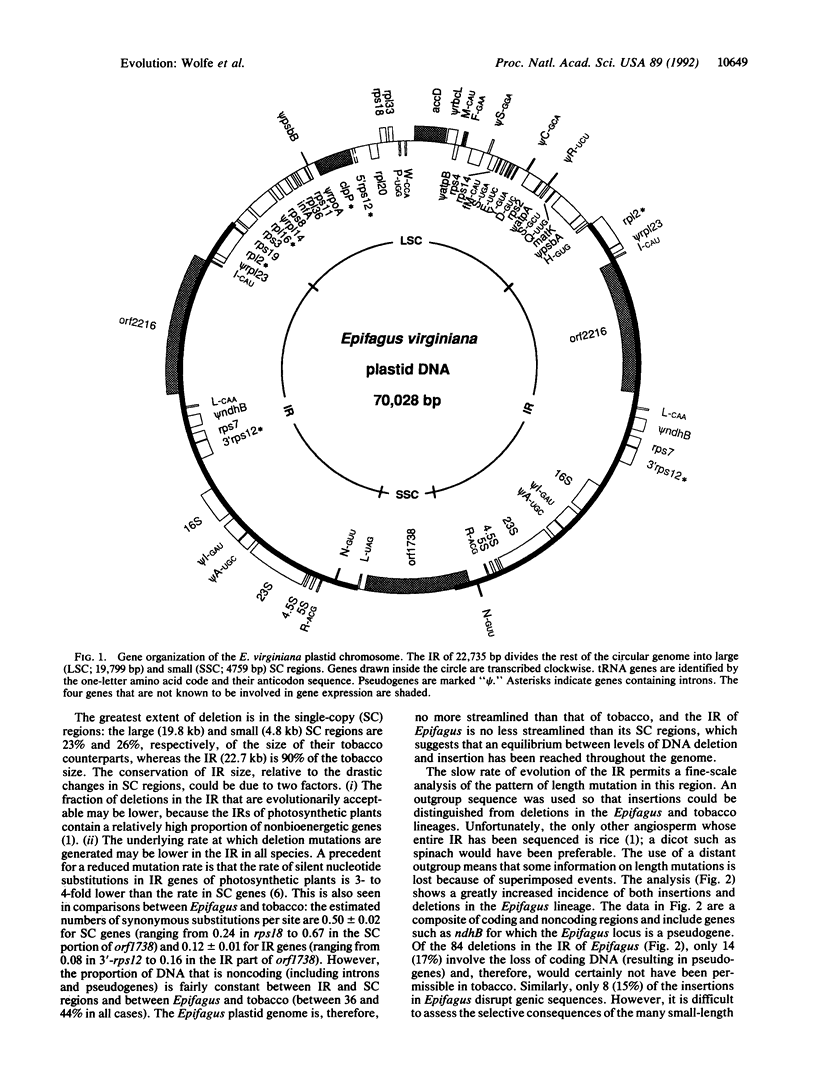
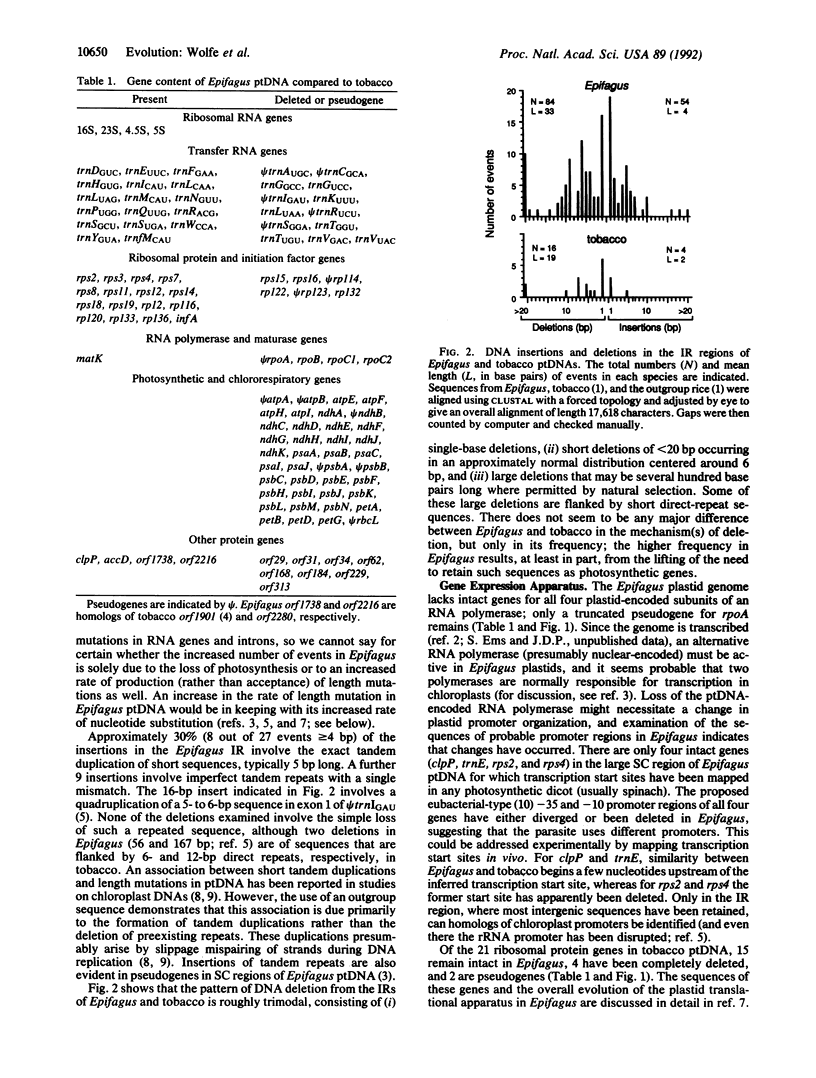
Abstract
Complete nucleotide sequencing shows that the plastid genome of Epifagus virginiana, a nonphotosynthetic parasitic flowering plant, lacks all genes for photosynthesis and chlororespiration found in chloroplast genomes of green plants. The 70,028-base-pair genome contains only 42 genes, at least 38 of which specify components of the gene-expression apparatus of the plastid. Moreover, all chloroplast-encoded RNA polymerase genes and many tRNA and ribosomal protein genes have been lost. Since the genome is functional, nuclear gene products must compensate for some gene losses by means of previously unsuspected import mechanisms that may operate in all plastids. At least one of the four unassigned protein genes in Epifagus plastid DNA must have a nongenetic and nonbioenergetic function and, thereby, serve as the reason for the maintenance of an active genome. Many small insertions in the Epifagus plastid genome create tandem duplications and presumably arose by slippage mispairing during DNA replication. The extensive reduction in genome size in Epifagus reflects an intensification of the same processes of length mutation that govern the amount of noncoding DNA in chloroplast genomes. Remarkably, this massive pruning occurred with a virtual absence of gene order change.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Basu M. K., Wilson H. J. Mercury risk from teeth. Nature. 1991 Jan 10;349(6305):109–109. doi: 10.1038/349109a0. [DOI] [PubMed] [Google Scholar]
- Goldberg A. L. The mechanism and functions of ATP-dependent proteases in bacterial and animal cells. Eur J Biochem. 1992 Jan 15;203(1-2):9–23. doi: 10.1111/j.1432-1033.1992.tb19822.x. [DOI] [PubMed] [Google Scholar]
- Goldring E. S., Grossman L. I., Krupnick D., Cryer D. R., Marmur J. The petite mutation in yeast. Loss of mitochondrial deoxyribonucleic acid during induction of petites with ethidium bromide. J Mol Biol. 1970 Sep 14;52(2):323–335. doi: 10.1016/0022-2836(70)90033-1. [DOI] [PubMed] [Google Scholar]
- Harada T., Ishikawa R., Niizeki M., Saito K. Pollen-derived rice calli that have large deletions in plastid DNA do not require protein synthesis in plastids for growth. Mol Gen Genet. 1992 May;233(1-2):145–150. doi: 10.1007/BF00587572. [DOI] [PubMed] [Google Scholar]
- Li S. J., Cronan J. E., Jr The genes encoding the two carboxyltransferase subunits of Escherichia coli acetyl-CoA carboxylase. J Biol Chem. 1992 Aug 25;267(24):16841–16847. [PubMed] [Google Scholar]
- Morden C. W., Wolfe K. H., dePamphilis C. W., Palmer J. D. Plastid translation and transcription genes in a non-photosynthetic plant: intact, missing and pseudo genes. EMBO J. 1991 Nov;10(11):3281–3288. doi: 10.1002/j.1460-2075.1991.tb04892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. D. Green ancestry of malarial parasites? Curr Biol. 1992 Jun;2(6):318–320. doi: 10.1016/0960-9822(92)90887-g. [DOI] [PubMed] [Google Scholar]
- Pfitzinger H., Weil J. H., Pillay D. T., Guillemaut P. Codon recognition mechanisms in plant chloroplasts. Plant Mol Biol. 1990 May;14(5):805–814. doi: 10.1007/BF00016513. [DOI] [PubMed] [Google Scholar]
- Richards C. M., Hinman S. B., Boyer C. D., Hardison R. C. Survey of plastid RNA abundance during tomato fruit ripening: the amounts of RNA from the ORF 2280 region increase in chromoplasts. Plant Mol Biol. 1991 Dec;17(6):1179–1188. doi: 10.1007/BF00028734. [DOI] [PubMed] [Google Scholar]
- Shimada H., Sugiura M. Fine structural features of the chloroplast genome: comparison of the sequenced chloroplast genomes. Nucleic Acids Res. 1991 Mar 11;19(5):983–995. doi: 10.1093/nar/19.5.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemeister G., Buchholz C., Hachtel W. Genes for the plastid elongation factor Tu and ribosomal protein S7 and six tRNA genes on the 73 kb DNA from Astasia longa that resembles the chloroplast DNA of Euglena. Mol Gen Genet. 1990 Feb;220(3):425–432. doi: 10.1007/BF00391749. [DOI] [PubMed] [Google Scholar]
- Siemeister G., Hachtel W. Organization and nucleotide sequence of ribosomal RNA genes on a circular 73 kbp DNA from the colourless flagellate Astasia longa. Curr Genet. 1990 May;17(5):433–438. doi: 10.1007/BF00334524. [DOI] [PubMed] [Google Scholar]
- Siemeister G., Hachtel W. Structure and expression of a gene encoding the large subunit of ribulose-1,5-bisphosphate carboxylase (rbcL) in the colourless euglenoid flagellate Astasia longa. Plant Mol Biol. 1990 May;14(5):825–833. doi: 10.1007/BF00016515. [DOI] [PubMed] [Google Scholar]
- Smith A. G., Wilson R. M., Kaethner T. M., Willey D. L., Gray J. C. Pea chloroplast genes encoding a 4 kDa polypeptide of photosystem I and a putative enzyme of C1 metabolism. Curr Genet. 1991 May;19(5):403–410. doi: 10.1007/BF00309603. [DOI] [PubMed] [Google Scholar]
- Weeden N. F. Genetic and biochemical implications of the endosymbiotic origin of the chloroplast. J Mol Evol. 1981;17(3):133–139. doi: 10.1007/BF01733906. [DOI] [PubMed] [Google Scholar]
- Wimpee C. F., Morgan R., Wrobel R. An aberrant plastid ribosomal RNA gene cluster in the root parasite Conopholis americana. Plant Mol Biol. 1992 Jan;18(2):275–285. doi: 10.1007/BF00034955. [DOI] [PubMed] [Google Scholar]
- Wimpee C. F., Wrobel R. L., Garvin D. K. A divergent plastid genome in Conopholis americana, an achlorophyllous parasitic plant. Plant Mol Biol. 1991 Jul;17(1):161–166. doi: 10.1007/BF00036822. [DOI] [PubMed] [Google Scholar]
- Wolfe K. H., Katz-Downie D. S., Morden C. W., Palmer J. D. Evolution of the plastid ribosomal RNA operon in a nongreen parasitic plant: accelerated sequence evolution, altered promoter structure, and tRNA pseudogenes. Plant Mol Biol. 1992 Apr;18(6):1037–1048. doi: 10.1007/BF00047707. [DOI] [PubMed] [Google Scholar]
- Wolfe K. H., Li W. H., Sharp P. M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A. 1987 Dec;84(24):9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K. H., Morden C. W., Ems S. C., Palmer J. D. Rapid evolution of the plastid translational apparatus in a nonphotosynthetic plant: loss or accelerated sequence evolution of tRNA and ribosomal protein genes. J Mol Evol. 1992 Oct;35(4):304–317. doi: 10.1007/BF00161168. [DOI] [PubMed] [Google Scholar]
- Wolfe K. H., Morden C. W., Palmer J. D. Ins and outs of plastid genome evolution. Curr Opin Genet Dev. 1991 Dec;1(4):523–529. doi: 10.1016/s0959-437x(05)80202-3. [DOI] [PubMed] [Google Scholar]
- Wolfe K. H., Morden C. W., Palmer J. D. Small single-copy region of plastid DNA in the non-photosynthetic angiosperm Epifagus virginiana contains only two genes. Differences among dicots, monocots and bryophytes in gene organization at a non-bioenergetic locus. J Mol Biol. 1992 Jan 5;223(1):95–104. doi: 10.1016/0022-2836(92)90718-y. [DOI] [PubMed] [Google Scholar]
- Wolfson R., Higgins K. G., Sears B. B. Evidence for replication slippage in the evolution of Oenothera chloroplast DNA. Mol Biol Evol. 1991 Sep;8(5):709–720. doi: 10.1093/oxfordjournals.molbev.a040680. [DOI] [PubMed] [Google Scholar]
- Zurawski G., Clegg M. T., Brown A. H. The Nature of Nucleotide Sequence Divergence between Barley and Maize Chloroplast DNA. Genetics. 1984 Apr;106(4):735–749. doi: 10.1093/genetics/106.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dePamphilis C. W., Palmer J. D. Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant. Nature. 1990 Nov 22;348(6299):337–339. doi: 10.1038/348337a0. [DOI] [PubMed] [Google Scholar]