Abstract
Purpose
The United States Immunodeficiency Network (USIDNET) patient registry was used to characterize the presentation, genetics, phenotypes, and treatment of patients with Hyper IgM Syndrome (HIGM).
Methods
The USIDNET Registry was queried for HIGM patient data collected from October 1992 to July 2015. Data fields included demographics, criteria for diagnosis, pedigree analysis, mutations, clinical features, treatment and transplant records, laboratory findings, and mortality.
Results
Fifty-two physicians entered data from 145 patients of ages 2 months to 62 years (median 12 years); 131 were males. Using patients’ age at last entry, data from 2072 patient years are included. Mutations were recorded for 85 subjects; 82 were in CD40LG. Eighteen subjects had non-X-linked HIGM. 40 % had a normal serum IgM and 15 %, normal IgA. Infections were reported for 91 %, with pulmonary, ear, and sinus infections being the most common. 42 % had Pneumocystis jirovecii pneumonia; 6 % had Cryptosporidium. 41 % had neutropenia. 78 % experienced non-infectious complications: chronic diarrhea (n = 22), aphthous ulcers (n = 28), and neoplasms (n = 8) including colon cancer, adrenal adenoma, liver adenocarcinoma, pancreatic carcinoid, acute myeloid leukemia, hepatoma, and, in a female with an autosomal dominant gain of function mutation in PIK3CD, an ovarian dysgerminoma. Thirteen patients had a hematopoietic marrow or stem cell transplant; three had solid organ transplants. Thirteen were known to have died (median age = 14 years).
Conclusions
Analysis of the USIDNET Registry provides data on the common clinical features of this rare syndrome, and in contrast with previously published data, demonstrates longer survival times and reduced gastrointestinal manifestations.
Keywords: Hyper IgM Syndrome, Primary immune deficiency, USIDNET, CD40/CD40L
Introduction
The Hyper IgM (HIGM) Syndromes are a group of rare genetic disorders leading to loss of T cell driven immunoglobulin class switch recombination (CSR) and/or defective somatic hypermutation as well as impaired T cell activation [1–4]. As is true for other primary immune deficiencies, serious infections, autoimmunity, inflammatory complications, and malignancy may develop [1, 5–8]. The most common causes are mutations in the CD40 Ligand (CD40LG) gene located at Xq26.3-27 leading to X-linked HIGM (XHIGM) in males [9, 10]. The CD40LG gene product is the CD40L transmembrane glycoprotein normally expressed on activated CD4+ T lymphocytes. Under tight regulation, CD40L interacts with CD40 that is constitutively expressed on B cells, macrophages, monocytes, and dendritic cells [10]. This interaction is a critical first step in B cell stimulation for growth, proliferation, CSR, and SHM resulting in the generation of both functional antibodies and memory B cells [1, 2]. Less frequent causes of the HIGM syndrome are autosomal recessive mutations (ARHIGM) in CD40 which also lead to loss of these B cell functions [11]. Other forms of ARHIGM result from mutations in activation-induced cytidine deaminase (AICDA) and uracil-DNA glycosylase (UNG) affecting males and females equally and leading to different clinical phenotypes, including significant lymphoid hypertrophy (due to large germinal centers) and a higher propensity for autoimmunity [12, 13]. AICDA codes for activation-induced cytidine deaminase (AID) which is active in the initiation of SHM and in the preferential selection of higher affinity B cells to proliferate in germinal centers for a full antibody and memory B cell response [1, 2, 12]. Both AID and UNG act downstream of CD40/CD40L in the CSR pathway and mutations in these genes lead to defects in CSR and SHM, and subsequently low serum levels of IgG, IgA, and IgE [12, 14]. Though bacterial infections are commonly seen in ARHIGM due to mutations in the AICDA/UNG genes, opportunistic pathogens are much more likely to occur in subjects with XHIGM and CD40 defects, as these defects involve impaired macrophage and T cell activation [9, 10, 15]. More complex and involving different mechanisms are the recently described autosomal dominant gain of function mutations in phosphoinositide 3-kinase catalytic delta component (PIK3CD) which may lead to hypogammaglobulinemia with increased serum IgM and increased susceptibility to infections, autoimmunity, lymphoid hyperplasia, and potentially lymphoma [16–19]. As males with mutations in NF kappa B essential modulator (IKBKG/NEMO) may have increased serum IgM levels as well as anhidrotic (or hypohidrotic) ectodermal dysplasia, they have been sometimes included in the HIGM designation [20].
While a number of reviews have examined the immunologic features, molecular causes and potential treatment options in HIGM syndrome, clinical reviews of large data sets are limited. Levy et al. [6] reported clinical and immunologic data on 56 patients with XHIGM, and Winkelstein et al. [5] examined these features in 79 subjects using a physician survey performed by the Immune Deficiency Foundation (IDF) from 1997 to 2002. As the IDF collection of data has continued to the present time utilizing a web-based entry system from the United States Immunodeficiency Network (USIDNET) [21], we now report an update of the clinical presentation, genetic mutations, immunologic phenotypes, and treatment practices for 145 HIGM patients from North America (including 75 patients not reported in the prior report of IDF data by Winkelstein et al.).
Methods
Since 1992, the IDF has collected survey data on subjects with primary immune deficiency diseases. The current online patient-consented USIDNET Registry is funded by the NIH and maintains clinical, laboratory, molecular, treatment, and quality of life data for patients with a number of primary immune defects, including those with an unclear final diagnosis. Data are entered from enrolling institutions in the USA and Canada as well as via a new link allowing patients to self-enroll and transfer medical records for entry into the Registry [21]. De-identified data available for research purposes were used for the current study, for which a query was submitted to the USIDNET requesting the most recent data for all HIGM patients through July 2015. The data fields surveyed included demographics, criteria used to render diagnosis, pedigree evaluation, gene mutation analysis, clinical features, treatment and transplant records, laboratory findings, and mortality.
Patients were considered to have HIGM if they had serum IgG concentrations at least 2 standard deviations below normal for their age and a confirmed mutation in any of the following genes: CD40LG (XHIGM) or AICDA, CD40, UNG (all ARHIGM), or PIK3CD (ADHIGM). Specific mutations are listed in Table 2 and reference the coding sequence of canonical transcripts: CD40LG (NM_000074.2), PIK3CD (NM_005026.3), AICDA (NM_020661.2). The mutations listed are based on information provided by the entering physician. For mutations that were ambiguous we have provided the most likely variation given the information provided and any published information cited by the enrolling physician. Subjects with the appropriate phenotype were also included if there were maternal cousins, uncles, or nephews with confirmed diagnosis of XHIGM, or if there were siblings with confirmed diagnosis of XHIGM or ARHIGM. Criteria for these phenotypically-diagnosed patients also included a serum IgG concentration at least 2 standard deviations below the normal for age with normal T and B cells numbers and at least two of the following: serum IgM concentration at least 2 standard deviations above normal for age; no antigen specific IgG antibody; recurrent bacterial infections in the first 5 years of life; Pneumocystis jirovecii (carinii) infection in the first year of life; neutropenia; Cryptosporidium-related diarrhea; sclerosing cholangitis; parvovirus induced aplastic anemia; or lymphoid hyperplasia (AICDA or UNG defect).
Table 2.
Recorded mutations
| Patient | Gene | Nucleotide changea | Protein change | Reference |
|---|---|---|---|---|
| Splice site mutations | ||||
| 1 | CD40LG | c.156+1G>T | [22, 32] | |
| 2 | CD40LG | c.156+1G>T | [22, 33] | |
| 3 | CD40LG | c.156+1G>T | [22] | |
| 4 | CD40LG | c.156+1G>T | [22] | |
| 5 | CD40LG | c.156+1G>T | [22, 34] | |
| 6 | CD40LG | c.156+1G>T | [22, 34] | |
| 7 | CD40LG | c.288+2T>A | Loss of exon 2 | [22] |
| 8 | CD40LG | c.346+1delG | Loss of exon 3 | [22] |
| 9 | CD40LG | c.346+1G>A | Loss of exon 3 | [22] |
| 10 | CD40LG | c.346+1G>A | Loss of exon 3 | [22] |
| 11 | CD40LG | c.346+2T>C | Loss of exon 3 | [22, 35] |
| 12 | CD40LG | c.346+5G>A | ||
| 13 | CD40LG | c.347−1G>A | Loss of exon 4 | [22] |
| 14 | CD40LG | c.347−1G>A | Loss of exon 4 | [22, 36] |
| 15 | CD40LG | c.409+1G>C | Loss of exon 4 | [22, 32] |
| 16 | CD40LG | c.409+1G>C | Loss of exon 4 | [22, 32, 37] |
| 17 | CD40LG | c.409+1G>C | Loss of exon 4 | [22, 32] |
| 18 | CD40LG | c.409+1G>C | Loss of exon 4 | [22, 32] |
| 19 | CD40LG | c.409+1G>C | Loss of exon 4 | [22] |
| 20 | CD40LG | – | Unknown splice defect | |
| Nonsense mutations | ||||
| 21 | CD40LG | c.78T>G | p.Y26* | [22] |
| 22 | CD40LG | c.208C>T | p.Q70* | [22] |
| 23 | CD40LG | c.216C>A | p.C72* | |
| 24 | CD40LG | c.419G>A | p.W140* | [22] |
| 25 | CD40LG | c.496C>T | p.Q166* | [22] |
| 26 | CD40LG | c.520C>T | p.Q174* | [22] |
| 27 | CD40LG | c.520C>T | p.Q174* | [22] |
| 28 | CD40LG | c.556C>T | p.Q186* | [22] |
| 29 | CD40LG | c.658C>T | p.Q220* | [22] |
| 30 | CD40LG | c.658C>T | p.Q220* | [22] |
| 31 | CD40LG | c.661C>T | p.Q221* | [22] |
| Missense mutations | ||||
| 32 | CD40LG | c.346G>A | p.G116S; Loss of exon 3 | [22] |
| 33 | CD40LG | c.384T>A; c.386A>G | p.S128R; p.E129G | |
| 34 | CD40LG | c.440C>A | p.T147N | |
| 35 | CD40LG | c.464T>C | p.L155P | |
| 36 | CD40LG | c.464T>C | p.L155P | |
| 37 | CD40LG | c.505T>G | p.Y169D | [22] |
| 38 | CD40LG | c.505T>G | p.Y169D | [22] |
| 39 | CD40LG | c.509A>G | p.Y170C | [22] |
| 40 | CD40LG | c.509A>G | p.Y170C | [22] |
| 41 | CD40LG | c.509A>G | p.Y170C | [22] |
| 42 | CD40LG | c.608G>T | p.R203I | [22] |
| 43 | CD40LG | c.623C>A | p.A208D | [33] |
| 44 | CD40LG | – | p.G219R | |
| 45 | CD40LG | c.680G>T | p.G227V | [22, 38] |
| 46 | CD40LG | c.680G>T | p.G227V | [22] |
| 47 | CD40LG | c.703G>C | p.A235P | [22] |
| 48 | CD40LG | c.761C>T | p.T254M | [22] |
| 49 | CD40LG | c.761C>T | p.T254M | [22] |
| 50 | CD40LG | c.773T>C | p.L258S | [22] |
| 51 | CD40LG | c.773T>C | p.L258S | [22] |
| 52 | PIK3CD | c.3061G>A (heterozygous) | p.E1021K | |
| Insertions and duplications | ||||
| 53 | CD40LG | c.380dupT | p.I127Ifs*3 | |
| 54 | CD40LG | c.383dupG | p.S128Rfs*2 | [22] |
| 55 | CD40LG | c.383dupG | p.S128Rfs*2 | [22, 32] |
| 56 | CD40LG | c.391dupA | p.S131Kfs*3 | [22] |
| 57 | CD40LG | c.391dupA | p.S131Kfs*3 | [22] |
| 58 | CD40LG | c.410-1_410ins9 | Ins 3aa btwn exons 4 & 5 | [22] |
| 59 | CD40LG | c.429dupA | p.G144Rfs*21 | [22, 34] |
| Deletions | ||||
| 60 | CD40LG | Large deletion (56 kb) including exons 1 through 5 | Undetectable cDNA | [22] |
| 61 | CD40LG | 1.5 kb deletion including exon 1 | – | [32] |
| 62 | CD40LG | 1.5 kb deletion including exon 1 | – | [32] |
| 63 | CD40LG | 1.5 kb deletion including exon 1 | – | [32] |
| 64 | CD40LG | c.157_160delATAG | p.I53Kfs*13 | [22] |
| 65 | CD40LG | – | Loss of exon 3b | [39] |
| 66 | CD40LG | – | Loss of exon 3b | |
| 67 | CD40LG | – | Loss of exon 3b | |
| 68 | CD40LG | c.289–25_32delAAAATGAC | – | |
| 69 | CD40LG | c.374_375delAT | p.H125Rfs*4 | |
| 70 | CD40LG | c.374_375delAT | p.H125Rfs*4 | |
| 71 | CD40LG | c.374_375delAT | p.H125Rfs*4 | |
| 72 | CD40LG | 10 kb deletion with breakpoint upstream of Exon 4 | Undetectable cDNA | [34] |
| 73 | CD40LG | 10 kb deletion with breakpoint upstream of Exon 4 | Undetectable cDNA | [34] |
| 74 | CD40LG | c.424_429delGAAAAA | p.E142_K143del | [22, 40] |
| 75 | CD40LG | c.559delG | p.A187Lfs*4 | |
| 76 | CD40LG | c.559delG | p.A187Lfs*4 | |
| 77 | CD40LG | c.590C>A; c.591delC | p.[S197Y;P198Pfs*44] | |
| 78 | CD40LG | c.590C>A; c.591delC | p.[S197Y;P198Pfs*44] | |
| 79 | CD40LG | c.736_740delCAAGT | p.Q246Efs*38 | |
| 80 | CD40LG | c.736_740delCAAGT | p.Q246Efs*38 | |
| 81 | CD40LG | Deletion of 8 bp in Exon 5 | – | |
| 82 | CD40LG | Deletion of more than 10 kb | – | |
| 83 | CD40LG | Deletion of more than 10 kb | – | |
| 84 | AICDA | – | Loss of exon 1 | |
Nucleotide changes reference coding sequence of canonical transcripts: CD40LG (NM_000074.2), PIK3CD (NM_005026.3), AICDA (NM_020661.2)
Patients were reported with a deletion of exon 3, likely due to an essential splicing mutation. However, the specific mutation was never delineated at the genomic level in these patients
Laboratory Studies
The laboratory values were recorded from local sites; only values entered during initial visits are used here. Age-adjusted standard reference norms were used for immunoglobulin levels. For some patients, the exact age was not recorded for the time of blood draw. In the analysis of these laboratory data, these patients were reported according to the largest possible normal range for approximate age. Blood counts were reported only if >50 patients had these data reported.
Clinical Data
Types and locations of infections were recorded in terms of “observed” (if a patient had experienced an infection at least once), “present” (if the patient was experiencing the infection at the time of report), or “prominent” (if the infection was chronic or frequently recurring). Types of infection (e.g., otitis) were recorded independently from infectious organisms (e.g., Streptococcus pneumoniae). For combined infectious types (e.g., skin/soft tissue infections) the number of patients who experienced at least one (but possibly more) of these conditions, are reported. Non-infectious complications are reported here in combined categories. “Gastrointestinal complications” included any patient reporting at least one of the following: non-infectious diarrhea (chronic or intermittent), inflammatory bowel disease, enteritis, and proctocolitis, gastroesophageal reflux, abdominal pain, constipation, vomiting, and/or protein-losing gastroenteropathy. “Sinopulmonary” complications included any patient reporting at least one of the following: sinus disease, allergic rhinitis, rhinorrhea, cough, rhinitis/occasional cough/nasal drip/nasal congestion, chest congestion, sinopulmonary facial swelling, dry cough, asthma, bronchiectasis, and phlegm production. “Dental/Oral” complications included periodontal disease, stomatitis, blunted and shortened roots of mandibular primary molars, gingival hypertrophy, adenoid hypertrophy, and/or abnormal tooth morphology. “Skin/Soft Tissue” complications included abscess, cellulitis, fungal dermatitis, impetigo, furuncles, diaper rash, nail infection, skin infections, rash, thrush, and pyoderma. “Neoplasm” included all forms of neoplasm. “Constitutional” symptoms included fatigue, fever, malaise, loss of appetite, night sweats, and weight loss.
Data Management and Statistical Analysis
For laboratory studies we report median and quartile ranges of the subjects for whom a value was determined. T tests, Chi-squared analysis, and odds ratios were performed using Excel (Microsoft, Inc.) and Prism (Graphpad Software, Inc.).
Results
Demographics and Diagnostic Criteria
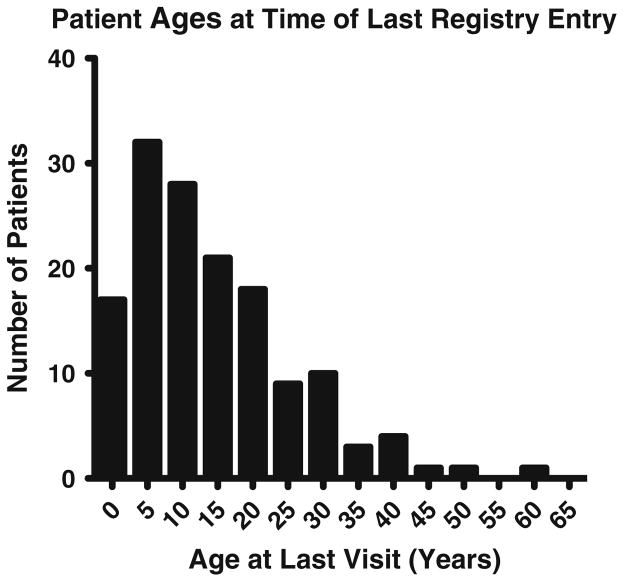
As of July 2015, the USIDNET Registry contained data for 145 patients (131 males and 14 females) who met the criteria for HIGM diagnosis entered by 52 physicians. Data are reported as they were entered, with varying degrees of completeness and specificity for each patient. For subjects with incomplete entries, only the data available, as outlined here, were included. Subjects were between the ages of 2 months and 62 years of age at entry, with a median age of 12 years (25th to 75th quartiles: 11–20 years) (Fig. 1). Their birth years were between 1947 and 2012 (Table 1). For 121 subjects, racial and ethnic characteristics were entered: the majority of patients were Caucasian (n = 85), followed by African American (n = 15), Pacific/Asian (n = 10), Hispanic subjects (n = 5) and mixed (n = 6). Considering the total number of years from the patients’ ages at the time of last data entry, there are 2072 patient years included in this report.
Fig. 1.
Patient ages
Table 1.
Demographic characteristics
| Characteristic | N (% total population) |
|---|---|
| Male | 131 (90 %) |
| Female | 14 (10 %) |
| Race/ethnicity | |
| Caucasian | 85 (59 %) |
| African American | 15 (10 %) |
| Asian or Pacific Islander | 10 (7 %) |
| Hispanic | 5 (3 %) |
| Mixed | 6 (4 %) |
| Unknown | 24 (17 %) |
| Year of birth (range) | 1947–2012 |
| Age at last entry | 2 months–62 years |
| Clinical diagnosis* | |
| XHIGM | 127 (88 %) |
| Specified CD40LG mutations | 82 (57 %) |
| HIGM, non-X-linked | 18 (12 %) |
| Females (mutation undocumented) | 12 (8 %) |
| AICDA (three males) | 4 (3 %) |
| PIK3CD (female) | 1 (<1 %) |
| CD40 (male) | 1 (<1 %) |
| Patients with known affected family members | 62 (43 %) |
Clinical diagnoses were based on sex, phenotype, family history and, when available, molecular data. For some patients, physicians reported a genetic diagnosis (such as AID or CD40 deficiency) without reporting the specific causative mutation(s). These patients are reported in the diagnostic summary here, but are not included in Table 2
For 127 patients the inheritance patterns, clinical phenotype and/or genetic studies supported the diagnosis of XHIGM. Sixty-two of these (49 %) had a family history of HIGM, including 41 with an affected brother. Other affected relatives were 15 maternal cousins, 6 maternal uncles, 2 great uncles, 2 nephews, and a grandfather. Fourteen female subjects were diagnosed as having HIGM; one was known to have a defect in AID and another female with the clinical phenotype of HIGM had an autosomal dominant gain of function mutation in PIK3CD. All together, four subjects (3 %), including three males, had mutations in AICDA. One male patient was diagnosed with mutations in CD40. For 84 subjects, the nucleotide locations affected and predicted effects were provided (Table 2). As related subjects may have been entered at different centers, presumed relatives are clustered together. These mutations included an array of missense, nonsense, deletion, insertion, splice site, and frame shift mutations, as noted in other comprehensive reports [22, 23].
Immunoglobulin Values
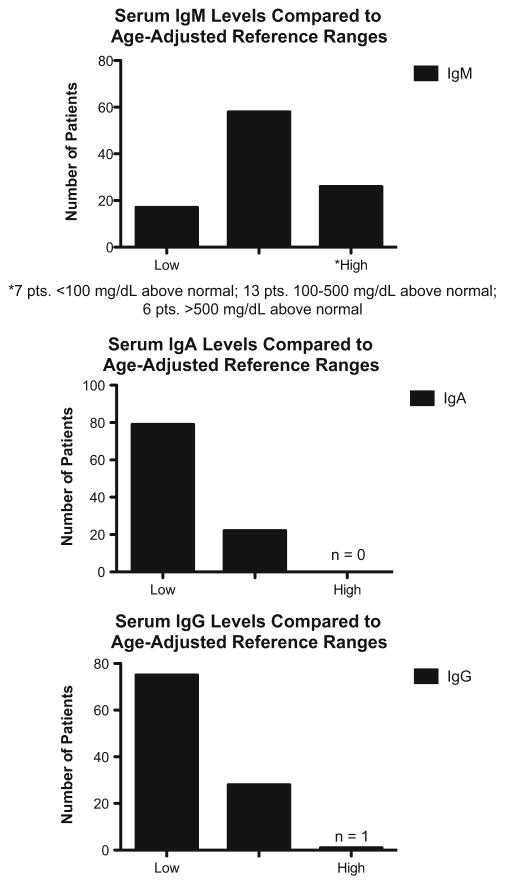
For 110 patients (76 % of total cohort) the range for serum IgM was 13 to 3,484 mg/dL, with a median of 103 mg/dL at first entry. Fifty-eight of these subjects had IgM levels within normal limits for age at first recorded blood draw, while 26 were above normal limits and 17 were below (Fig. 2). For serum IgA, the levels in 111 patients were 0 to 348 mg/dl with a median of 7 mg/dL; 22 of these subjects had a serum IgA within normal limits for age at first recorded blood draw, while 79 were below normal. For IgG (112 patients, most receiving IVIG) the levels recorded were 0 to 1,890 mg/dL, with a median 84 mg/dL. Serum IgE had been tested in 80 subjects; these levels had a median of 2 IU/mL; 78 subjects had levels under 15 IU, but one subject had a level of 31 IU and one with a known CD40LG defect had an elevated IgE level of 720 IU.
Fig. 2.
Immunoglobulin levels at time of diagnosis compared to age-adjusted reference ranges. Patients whose age was unknown at time of blood draw were considered according to the largest possible range of normal values
Hematologic and Lymphocyte Profiles
Sixty XIGM patients (41 %) had a history of chronic, cyclic or intermittent neutropenia, and 17 of these subjects had been treated with granulocyte-colony stimulating factor (G-CSF) for this. Patients who had experienced neutropenia had more infections than those who had not (odds ratio = 12.04, 95 % CI [2.74–52.88], p < 0.01). However, no significant difference in the maximum chronicity of documented infections existed between patients with versus those without neutropenia. Autoimmune/hematologic complications may occur in HIGM but the subjects entered in the Registry had only a few such events. Anemia was reported in 12 % (n = 18), thrombocytopenia in 3 % (n = 5), and Evan’s syndrome (auto-immune thrombocytopenia with autoimmune hemolytic anemia) in one subject. Lymphocyte numbers and/or subsets were reported for 71 subjects (Table 3). These data fell within the normal values for children ages 2–6 and 6–12 years of age for almost all subjects [24]. Data on CD27+ memory or isotype switched memory B cells were not collected.
Table 3.
Lymphocytes and subsets*
| Lymphocytes (cells/uL) | CD3 T (cells/uL) | CD4 helper T (cells/uL) | CD8 cytotoxic T (cells/uL) | CD19 B (cells/uL) | |
|---|---|---|---|---|---|
| Min | 2.4 | 142 | 95 | 133 | 52 |
| Q1 | 2.17 | 1.69 | 0.93 | 0.47 | 0.37 |
| Median | 4.29 | 2.69 | 1.93 | 0.82 | 0.90 |
| Q3 | 8.35 | 5.49 | 4.09 | 1.18 | 2.07 |
| Max | 19.19 | 9.89 | 8.39 | 4.06 | 9.222 |
| N | 71 | 57 | 62 | 62 | 57 |
Age 2–6: lymphocytes (cells/uL) = 3.60 (2.30–5.40); CD3 (cells/uL) = 2.39 (1.40–3.70); CD4 (cells/uL) = 1.38 (0.70–2.20); CD8 (cells/uL) = 0.84 (0.49–1.30); CD19 (cells/uL) = 0.75 (0.39–1.40). Age 6–12: Lymphocytes (cells/uL) = 2.70 (1.90–3.70) CD3 (cells/uL) = 1.82 (1.20–2); CD4 (cells/ uL) = 0.98 (0.65–1.5); CD8 (cells/uL) = 0.68 (0.37–1.10); CD19 (cells/uL) = 0.48 (0.27–0.86) [24]
Infections
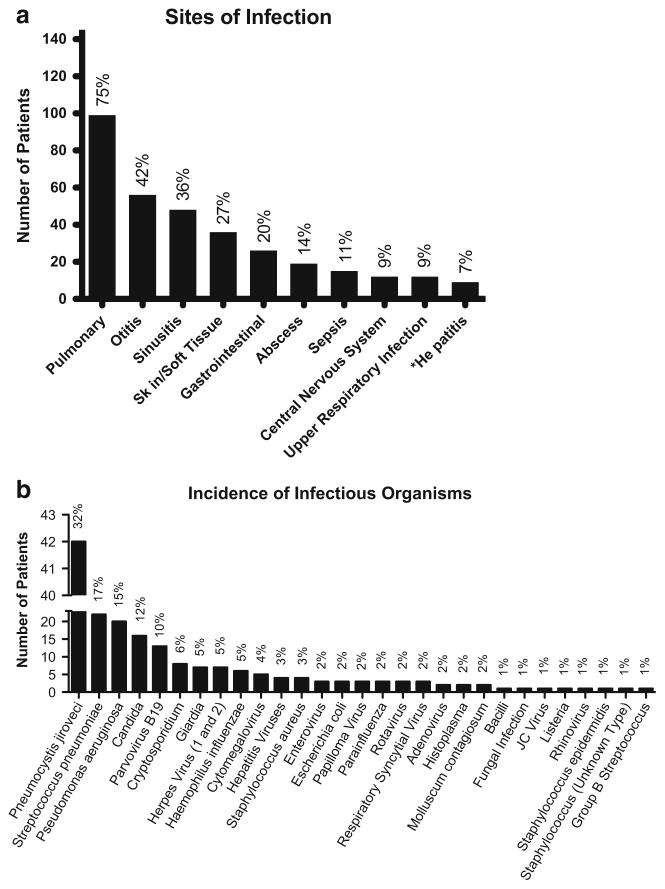
Clinical data were entered on 132 subjects. Of these, 185 specific infections were reported in 91 % of these patients (n = 120) with 52 % (n = 69) experiencing at least one prominent infection (Fig. 3a). Pulmonary infections were most commonly reported (99 subjects, 75 %). Ear infections were reported in 56 (42 %), sinus infections in 48 (36 %), and gastroenteritis in 26 (20 %). Other common infections were skin and soft tissue infections in 36, infectious diarrhea in 20, hepatitis in 9 (including sclerosing cholangitis in 4), and encephalitis in 9. The most common cause of infection was Pneumocystis jirovecii, which occurred in 42 subjects (32 %), followed by Streptococcus pneumoniae (n = 22, 17 %), and Pseudomonas aeruginosa (n = 20, 15 %). Most subjects with Pneumocystis jirovecii had XHIGM, but one was a female patient with a molecularly undefined HIGM syndrome. Other commoner infectious organisms were Candida spp. in 16 (12 %), Parvovirus B19 in 13 (10 %), and Cryptosporidium in 8 (6 %) (Table 4; Fig. 3b). Nine patients (7 %) had reported hepatitis (unknown cause), one of whom had documented cirrhosis and ultimately died of adenocarcinoma of the liver. 7 % had reported hepatitis infections (unknown cause), one of whom had documented cirrhosis and ultimately died of adenocarcinoma of the liver. A number of other viral infections were reported: herpes spp. (n = 7), cytomegalovirus (n = 5), enterovirus (n = 4), rotavirus (n = 3), respiratory syncytial virus (n = 3), parainfluenza (n = 3), molluscum (n = 2), and JC virus (n = 1).
Fig. 3.
a. Reported sites of infection in all patients (n = 145). The asterisk indicates causal organisms of Hepatitis unknown. b. Incidence of infectious organisms in only those patients for whom clinical data for infection was available (n = 132; remaining 12 patients did not have data on infection)
Table 4.
Infectious organisms
| Organisms recorded (188 infections) | Number |
|---|---|
| Pneumocystis jirovecii (carinii) | 42 |
| Streptococcus pneumoniae | 22 |
| Pseudomonas aeruginosa | 20 |
| Candida | 16 |
| Parvovirus B19 | 13 |
| Cryptosporidium | 8 |
| Giardia | 7 |
| Herpes virus (non-specified) | 7 |
| Haemophilus influenzae | 6 |
| Cytomegalovirus | 5 |
| Enterovirus | 3 |
| Escherichia coli | 3 |
| Rotavirus | 3 |
| Respiratory syncytial virus | 3 |
| Papillomavirus | 3 |
| Parainfluenza | 3 |
| Histoplasmosis | 2 |
| Molluscum | 2 |
| JC virus | 1 |
| Listeria | 1 |
| Hepatitis C | 1 |
| Hepatitis unknown | 1 |
| Herpes type 1 virus | 1 |
| Herpes type 2 virus | 1 |
Non-Infectious and Other Complications
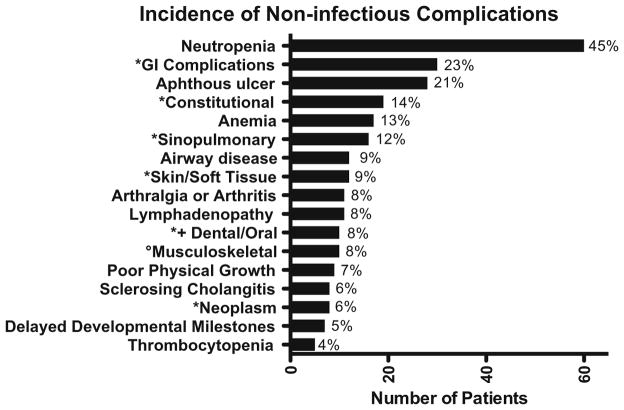
One hundred three patients (78 %) experienced non-infectious complications (Fig. 4). Gastrointestinal symptoms were the most prevalent (non-infectious diarrhea n = 22, other gastrointestinal symptoms without diarrhea n = 8). Four of the 22 patients with chronic or intermittent non-infectious diarrhea also reported poor growth. Aphthous ulcers were reported separately and affected 21 % of patients. Sixteen patients (12 %) experienced other sinopulmonary conditions. Eight patients had been diagnosed with sclerosing cholangitis, four of whom did not have documented accompanying Cryptosporidium infections. One of the five patients with cryptosporidiosis died of a hepatoma and two others received liver transplants; one patient died of sclerosing cholangitis secondary to cryptosporidiosis after solid organ transplant. Neoplasms (n = 7) were adrenal adenoma (with Cushing’s disease), liver adenocarcinoma, pancreatic carcinoid, acute myeloid leukemia, and hepatoma in subjects with XHIGM; one male with CD40 mutations had colon cancer; one female with a gain of function mutation in PIK3CD had resection of an ovarian dysgerminoma. Neurologic disease was reported in seven subjects with XHIGM: aseptic meningitis, ataxia with choreoathetosis, dysarthria, retinitis, and mental deterioration; ataxia with hypotonia and progressive multifocal encephalopathy; cerebral and optic atrophy; cortical infarcts due to meningitis; deafness; sensorineural deafness due to aminoglycosides; and mental retardation. Arthritis/arthralgia was noted in 8 % (n = 11), and irritable bowel disorder (IBD) in 2 % (n = 2). Other rarer conditions noted were growth hormone deficiency (n = 1), abnormal tooth morphology (n = 2), osteoporosis (n = 2) and cerebral cortical and optic atrophy (n = 1).
Fig. 4.
Incidence of non-infectious complications. The asterisk indicates combined variable (see “clinical data” in Methods). The degree sign indicates that musculoskeletal does not include arthritis/ arthralgia. The plus sign indicates that dental/oral does not include aphthous ulcers
Antimicrobials and Immunoglobulin Therapy
Sixty-one patients (46 % of patients with clinical data) were known to have been treated with oral trimethoprim/ sulfamethoxazole continuously or intermittently; for the other subjects this was not stated. Three patients, two with herpes simplex type 1 virus infections, used acyclovir continuously, and one patient was on a continuous anti-fungal (itraconazole) regimen. Dapsone, pentamidine, atovaquone, azithromycin, amoxicillin, cephalexin, and amoxicillin/clavulanic acid were also used on a continuous basis, but by less than 1 % of patients each. Five subjects had been treated with intravenous (IV) or oral steroids, and three subjects had been given thalidomide for unspecified reasons. Over 86 % of patients (n = 113) were known to have received immunoglobulin replacement therapy. Administration was primarily IV, though 28 patients were treated by intramuscular (prior to the 1980s) or subcutaneous administration routes (within the last decade).
Hematopoietic Stem Cell and Solid Organ Transplants
Sixteen patients (12 % of total cohort) were known to have received a transplant: 13 patients were given a hematopoietic stem cell transplant (HSCT) using matched sibling bone marrow (n = 5; ages 7, 8, 9 months, 6, and 10 years), cord blood (n = 2; ages 6 months and 3 years) parental bone marrow (n = 2; ages 8 months and 1 year), matched unrelated donors (n= 1, age 10 years) or stem cells, source not stated (n = 2, ages 6 months and 24 years, for AML). Three XHIGM patients received liver transplants (at ages 16, 25, and 38 years). Two of the four patients with documented cryptosporidiosis required a liver transplant. The only documented post-transplant outcomes were two deaths, both after liver transplant.
Mortality
Thirteen deaths were recorded in the registry (median age at time of death was 14 years, range = 1–39 years). Four patients died from liver complications (two after transplant), one from hemolytic uremic syndrome, two from pneumonias, and two from viral central nervous system (CNS) complications (progressive multifocal leukoencephalopathy from JC virus and encephalitis from Echovirus) (Table 5).
Table 5.
Mortality
| Cause of death | Age at death | Year of birth |
|---|---|---|
| Progressive multifocal leukoencephalopathy: JC virus | Unknown | 1989 |
| Liver adenocarcinoma | 17 | 1977 |
| PCP pneumonia | 9 months | 1993 |
| Encephalitis secondary to echovirus | 3 | 1988 |
| Hepatoma | 24 | 1967 |
| Hemolytic uremic syndrome/stroke | 26 | 1967 |
| Unknown | 2 | 2009 |
| Unknown | Unknown | 2005 |
| Unknown | Unknown | 1974 |
| Cryptosporidiosis with sclerosing cholangitisa | 39 | 1956 |
| CMV pneumoniab | 10 | 1988 |
| Liver failureb | 20 | 1985 |
| Unknownc | Unknown | 1977 |
Solid organ recipient
Bone marrow recipient
Stem cell recipient
Discussion
HIGM syndromes are rare immune defects with characteristic clinical and immunologic phenotypes. As previously noted, not all patients with gene defects associated with HIGM presented with elevated serum IgM; about half of the 110 subjects with IgM levels recorded had normal IgM levels for age and 20 % had significantly elevated levels. Higher serum IgM levels have previously been associated with chronic infections and increasing age, particularly when immunoglobulin replacement had not yet been initiated [6, 25]; however, we found no relationship between age at entry and serum IgM levels. Interestingly, even when considering loss of isotype switching as a main immunologic mechanism leading to HIGM, the serum IgA was normal in 22 subjects, and one subject had increased levels of serum IgE, suggesting alternative mechanisms for isotype switching, such as microbial activation of toll-like receptors, or B cell activating factor (BAFF) and its receptors [26].
The patients discussed here had many of the same infections as those in previous reports [5, 6, 11, 23]. Pneumonia was reported for the majority of subjects (63 %), followed by other upper and lower respiratory tract infections. Pneumocystis jirovecii pneumonia (PCP) was noted in 42 subjects (32 % of those with clinical data), but less than the 59 % noted in the previous US report [5] or in the European study where it was found to be the cause of almost all interstitial pneumonias [6]. While 80 % of the 35 XHIGM subjects in the Latin American report had pneumonia, only 26 % were related to Pneumocystis jirovecii as other organisms were identified. In addition, Aspergillus spp, Mycoplasma pneumoniae, Serratia marcescens, Klebsiella pneumoniae Acinetobacter, Microsporidium spp, Isospora belli, and Paracoccidioides brasiliensis, found in the Latin American cohort [23], were not reported in the USIDNET Registry.
In the current study, gastrointestinal tract disease leading to diarrhea (including both infectious and non-infectious types) occurred in 32 % (n = 42), similar to the 24 % reported by Winkelstein et al. [5] but not as frequent or severe as noted by Levy et al. [6] (Fisher’s exact p = 0.03), who reported 64 % of XHIGM patients with diarrhea leading to failure-to-thrive and requirement for parental nutrition in 13 subjects (23 %). Sclerosing cholangitis was reported in 19.6 % of subjects in the European report, in whom 55 % had Cryptosporidium as a cause [6]. The previous report from the older IDF data [5] noted sclerosing cholangitis and Cryptosporidium in five subjects (6 %). These previous numbers are similar to the current group, in which eight subjects (6 %) had sclerosing cholangitis, half of whom had Cryptosporidium, a known cause of this complication. While subclinical infections with Cryptosporidium detectable only by molecular means may have been present in either cohort [1, 27], the recorded data for the current cohort suggests a lower incidence of both sclerosing cholangitis and Cryptosporidium infection in the USIDNET Registry as compared to the European study (Fisher’s exact p value = 0.016 and <0.01, respectively), possibly due to more recent increased emphasis on Cryptosporidium prevention measures such as the use of bottled water. CD40L defects are thought to increase the risk of chronic Cryptosporidium infection and resultant in this complication since CD40 is expressed on bile duct epithelium, allowing for attachment of activated T cells and elimination of the parasite [28]. Sequelae of chronic cryptosporidiosis (cholangiopathy, sclerosing cholangitis, cirrhosis, cholangiocarcinoma, and other gastrointestinal malignancies) may necessitate liver transplantation [1, 8]. Three subjects in this Registry had a liver transplant for this complication and two of these patients died after transplant.
Chronic or intermittent neutropenia is often accompanied by oral and rectal ulcers, gingivitis, and proctitis, and is particularly common in males with XHIGM [1, 8]. In this study, 60 subjects (41 %) had neutropenia and 17 of these were known to have been treated with G-CSF, which is very similar to previous reports—63 % of subjects in older IDF report [5] and in 68 % in the European study [6]. Anemia and the auto-immune conditions previously noted in the HIGM reports were also found in a minority of subjects entered in this Registry [1, 8].
Most subjects were receiving immune globulin therapy, and more than half were receiving trimethoprim/ sulfamethoxazole on a continuous basis. While the only corrective treatment for HIGM is HSCT, long term data remain limited [8, 29, 30]. The USIDNET Registry currently contains limited information on 13 transplanted subjects. From a recent report of 175 XHIGM patients collected from both US and European data, 56 of 67 subjects who underwent HSCT had survived. The median survival time was similar for transplanted and non-transplanted subjects in this cohort, although median Lansky/Karnofsky scores were higher in those surviving this procedure (rank-sum p < 0.001) [29]. A report of 56 XHIGM patients in Japan showed greater overall survival rates in HSCT recipients, with significantly increased event-free and disease-free survival rates in patients younger than 5 years at the time of transplant [31]. As USIDNET continues data collection on HIGM, further information on such subjects will become available.
Conclusions
Given the rarity of HIGM, the large cohort of patients from the North America entered into the USIDNET Registry provides clinical, immunologic, and genetic features of this syndrome. While the USIDNET data reflect predominantly one time point, the data include over 2000 patient years of clinical history and thus provide both confirmation of previous reports and informative contrasts to these publications. In comparison to previous reports from Europe, reduced numbers of severe gastrointestinal manifestations and sclerosing cholangitis were noted. Data collected some 20 years ago suggest a mortality of 23.2 %, with the oldest subject being 23 years old [6]; in contrast, the patients registered here were between 2 months and 62 years of age with median of 12 years and 75 % quartile extending to age 20. The USIDNET data thus supplies a valuable perspective on the overall general status and health of subjects with HIGM.
Acknowledgments
This work was supported by the National Institutes of Health, AI 101093, AI-086037, AI-48693, T32-GM007280, The Jeffrey Modell Foundation, and the David S Gottesman Immunology Chair. We gratefully acknowledge the enthusiasm, expertise, and dedication of Tara Caulder, USIDNET Project Director, and Marla Goldsmith, USIDNET Registry Manager.
References
- 1.Davies EG, Thrasher AJ. Update on the hyper immunoglobulin M syndromes. Br J Haematol. 2010;149(2):167–80. doi: 10.1111/j.1365-2141.2010.08077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Durandy A, Kracker S. Immunoglobulin class-switch recombination deficiencies. Arthritis Res Ther. 2012;14(4):218. doi: 10.1186/ar3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qamar N, Fuleihan RL. The hyper IgM syndromes. Clin Rev Allergy Immunol. 2014;46(2):120–30. doi: 10.1007/s12016-013-8378-7. [DOI] [PubMed] [Google Scholar]
- 4.Picard C, Al-Herz W, Bousfiha A, Casanova JL, Chatila T, Conley ME, et al. Primary immunodeficiency diseases: an update on the classification from the International Union of Immunological Societies Expert committee for Primary Immunodeficiency 2015. J Clin Immunol. 2015 doi: 10.1007/s10875-015-0201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winkelstein JA, Marino MC, Ochs H, Fuleihan R, Scholl PR, Geha R, et al. The X-linked hyper-IgM syndrome: clinical and immunologic features of 79 patients. Medicine (Baltimore) 2003;82(6):373–84. doi: 10.1097/01.md.0000100046.06009.b0. [DOI] [PubMed] [Google Scholar]
- 6.Levy J, Espanol-Boren T, Thomas C, Fischer A, Tovo P, Bordigoni P, et al. Clinical spectrum of X-linked hyper-IgM syndrome. J Pediatr. 1997;131(1 Pt 1):47–54. doi: 10.1016/s0022-3476(97)70123-9. [DOI] [PubMed] [Google Scholar]
- 7.Jesus AA, Duarte AJ, Oliveira JB. Autoimmunity in hyper-IgM syndrome. J Clin Immunol. 2008;28(Suppl 1):S62–6. doi: 10.1007/s10875-008-9171-x. [DOI] [PubMed] [Google Scholar]
- 8.Hirbod-Mobarakeh A, Aghamohammadi A, Rezaei N. Immunoglobulin class switch recombination deficiency type 1 or CD40 ligand deficiency: from bedside to bench and back again. Expert Rev Clin Immunol. 2014;10(1):91–105. doi: 10.1586/1744666x.2014.864554. [DOI] [PubMed] [Google Scholar]
- 9.Fuleihan R, Ramesh N, Loh R, Jabara H, Rosen RS, Chatila T, et al. Defective expression of the CD40 ligand in X chromosome-linked immunoglobulin deficiency with normal or elevated IgM. Proc Natl Acad Sci U S A. 1993;90(6):2170–3. doi: 10.1073/pnas.90.6.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aruffo A, Farrington M, Hollenbaugh D, Li X, Milatovich A, Nonoyama S, et al. The CD40 ligand, gp39, is defective in activated T cells from patients with X-linked hyper-IgM syndrome. Cell. 1993;72(2):291–300. doi: 10.1016/0092-8674(93)90668-g. [DOI] [PubMed] [Google Scholar]
- 11.Lougaris V, Badolato R, Ferrari S, Plebani A. Hyper immunoglob-ulin M syndrome due to CD40 deficiency: clinical, molecular, and immunological features. Immunol Rev. 2005;203:48–66. doi: 10.1111/j.0105-2896.2005.00229.x. [DOI] [PubMed] [Google Scholar]
- 12.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102(5):565–75. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 13.Imai K, Slupphaug G, Lee WI, Revy P, Nonoyama S, Catalan N, et al. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat Immunol. 2003;4(10):1023–8. doi: 10.1038/ni974. [DOI] [PubMed] [Google Scholar]
- 14.Minegishi Y, Lavoie A, Cunningham-Rundles C, Bédard P-M, Hébert J, Côté L, et al. Mutations in activation-induced cytidine deaminase in patients with hyper IgM syndrome. Clin Immunol. 2000;97(3):203–10. doi: 10.1006/clim.2000.4956. [DOI] [PubMed] [Google Scholar]
- 15.Cabral-Marques O, Arslanian C, Ramos RN, Morato M, Schimke L, Soeiro Pereira PV, et al. Dendritic cells from X-linked hyper-IgM patients present impaired responses to Candida albicans and Paracoccidioides brasiliensis. J Allergy Clin Immunol. 2012;129(3):778–86. doi: 10.1016/j.jaci.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 16.Angulo I, Vadas O, Garçon F, Banham-Hall E, Plagnol V, Leahy TR, et al. Phosphoinositide 3-kinase δ gene mutation predisposes to respiratory infection and airway damage. Science. 2013;342(6160):866–71. doi: 10.1126/science.1243292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucas CL, Kuehn HS, Zhao F, Niemela JE, Deenick EK, Palendira U, et al. Dominant-activating germline mutations in the gene encoding the PI(3)K catalytic subunit p110delta result in T cell senescence and human immunodeficiency. Nat Immunol. 2014;15(1):88–97. doi: 10.1038/ni.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crank MC, Grossman JK, Moir S, Pittaluga S, Buckner CM, Kardava L, et al. Mutations in PIK3CD can cause hyper IgM syndrome (HIGM) associated with increased cancer susceptibility. J Clin Immunol. 2014;34(3):272–6. doi: 10.1007/s10875-014-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kracker S, Curtis J, Ibrahim MA, Sediva A, Salisbury J, Campr V, et al. Occurrence of B-cell lymphomas in patients with activated phosphoinositide 3-kinase delta syndrome. J Allergy Clin Immunol. 2014;134(1):233–6. doi: 10.1016/j.jaci.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zonana J, Elder ME, Schneider LC, Orlow SJ, Moss C, Golabi M, et al. A novel X-linked disorder of immune deficiency and hypohidrotic ectodermal dysplasia is allelic to incontinentia pigmenti and due to mutations in IKK-gamma (NEMO) Am J Hum Genet. 2000;67(6):1555–62. doi: 10.1086/316914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sullivan KE, Puck JM, Notarangelo LD, Fuleihan R, Caulder T, Wang C, et al. USIDNET: a strategy to build a community of clinical immunologists. J Clin Immunol. 2014;34(4):428–35. doi: 10.1007/s10875-014-0028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee WI, Torgerson TR, Schumacher MJ, Yel L, Zhu Q, Ochs HD. Molecular analysis of a large cohort of patients with the hyper immunoglobulin M (IgM) syndrome. Blood. 2005;105(5):1881–90. doi: 10.1182/blood-2003-12-4420. [DOI] [PubMed] [Google Scholar]
- 23.Cabral-Marques O, Klaver S, Schimke LF, Ascendino EH, Khan TA, Pereira PV, et al. First report of the hyper-IgM syndrome Registry of the Latin American Society for Immunodeficiencies: novel mutations, unique infections, and outcomes. J Clin Immunol. 2014;34(2):146–56. doi: 10.1007/s10875-013-9980-4. [DOI] [PubMed] [Google Scholar]
- 24.Shearer WT, Rosenblatt HM, Gelman RS, Oyomopito R, Plaeger S, Stiehm ER, et al. Lymphocyte subsets in healthy children from birth through 18 years of age: the pediatric AIDS Clinical Trials Group P1009 study. J Allergy Clin Immunol. 2003;112(5):973–80. doi: 10.1016/j.jaci.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Etzioni A, Ochs HD. The hyper IgM syndrome—an evolving story. Pediatr Res. 2004;56(4):519–25. doi: 10.1203/01.PDR.0000139318.65842.4A. [DOI] [PubMed] [Google Scholar]
- 26.He B, Santamaria R, Xu W, Cols M, Chen K, Puga I, et al. The transmembrane activator TACI triggers immunoglobulin class switching by activating B cells through the adaptor MyD88. Nat Immunol. 2010;11(9):836–45. doi: 10.1038/ni.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLauchlin J, Amar CF, Pedraza-Diaz S, Mieli-Vergani G, Hadzic N, Davies EG. Polymerase chain reaction-based diagnosis of infection with Cryptosporidium in children with primary immunodefi-ciencies. Pediatr Infect Dis J. 2003;22(4):329–35. doi: 10.1097/01.inf.0000059402.81025.cd. [DOI] [PubMed] [Google Scholar]
- 28.Hayward AR, Levy J, Facchetti F, Notarangelo L, Ochs HD, Etzioni A, et al. Cholangiopathy and tumors of the pancreas, liver, and biliary tree in boys with X-linked immunodeficiency with hyper-IgM. J Immunol. 1997;158(2):977–83. [PubMed] [Google Scholar]
- 29.De la Morena M, Leonard D. Long term clinical outcomes of patients with X-Linked Hyper IgM Syndrome, an international collaborative study. 16th Biennial Meeting of the European Society for Immunodeficiencies; October 31, 2014; Prague, Czech Republic: European Society for Immunodeficiencies; 2014. [Google Scholar]
- 30.Allewelt H, Martin PL, Szabolcs P, Chao N, Buckley R, Parikh S. Hematopoietic stem cell transplantation for CD40 ligand deficiency: single institution experience. Pediatr Blood Cancer. 2015 doi: 10.1002/pbc.25711. [DOI] [PubMed] [Google Scholar]
- 31.Mitsui-Sekinaka K, Imai K, Sato H, Tomizawa D, Kajiwara M, Nagasawa M, et al. Clinical features and hematopoietic stem cell transplantations for CD40 ligand deficiency in Japan. J Allergy Clin Immunol. 2015;136(4):1018–24. doi: 10.1016/j.jaci.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 32.Lin Q, Rohrer J, Allen RC, Larch M. A single strand conformation polymorphism study of CD40 ligand. Efficient mutation analysis and carrier detection for X-linked hyper IgM syndrome. J Clin Investig. 1996;97(1):196–201. doi: 10.1172/JCI118389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jain A, Atkinson TP, Lipsky PE, Slater JE, Nelson DL, Strober W. Defects of T-cell effector function and post-thymic maturation in X-linked hyper-IgM syndrome. J Clin Invest. 1999;103(8):1151–8. doi: 10.1172/jci5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seyama K, Nonoyama S, Gangsaas I, Hollenbaugh D, Pabst HF, Aruffo A, et al. Mutations of the CD40 ligand gene and its effect on CD40 ligand expression in patients with X-linked hyper IgM syndrome. Blood. 1998;92(7):2421–34. [PubMed] [Google Scholar]
- 35.Duplantier JE, Seyama K, Day NK, Hitchcock R, Nelson RP, Jr, Ochs HD, et al. Immunologic reconstitution following bone marrow transplantation for X-linked hyper IgM syndrome. Clin Immunol (Orlando, Fla) 2001;98(3):313–8. doi: 10.1006/clim.2000.4994. [DOI] [PubMed] [Google Scholar]
- 36.Scholl PR, O’Gorman MR, Pachman LM, Pachman LM, Haut P, Haut P, et al. Correction of neutropenia and hypogammaglobulinemia in X-linked hyper-IgM syndrome by allogeneic bone marrow transplantation. Bone Marrow Transplant. 1998;22(12):1215–8. doi: 10.1038/sj.bmt.1701512. [DOI] [PubMed] [Google Scholar]
- 37.Hollenbaugh D, Wu LH, Ochs HD, Ochs HD, Nonoyama S, Nonoyama S, et al. The random inactivation of the X chromosome carrying the defective gene responsible for X-linked hyper IgM syndrome (X-HIM) in female carriers of HIGM1. J Clin Investig. 1994;94(2):616–22. doi: 10.1172/JCI117377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allen RC, Armitage RJ, Conley ME, Rosenblatt H, Jenkins NA, Copeland NG, et al. CD40 ligand gene defects responsible for X-linked hyper-IgM syndrome. Science. 1993;259(5097):990–3. doi: 10.1126/science.7679801. [DOI] [PubMed] [Google Scholar]
- 39.Ramesh N, Fuleihan R, Ramesh V, Lederman S, Yellin MJ, Sharma S, et al. Deletions in the ligand for CD40 in X-linked immunoglobulin deficiency with normal or elevated IgM (HIGMX-1) Int Immunol. 1993;5(7):769–73. doi: 10.1093/intimm/5.7.769. [DOI] [PubMed] [Google Scholar]
- 40.Chu YW, Marin E, Fuleihan R, Ramesh N, Rosen FS, Geha RS, et al. Somatic mutation of human immunoglobulin V genes in the X-linked HyperIgM syndrome. J Clin Investig. 1995;95(3):1389–93. doi: 10.1172/JCI117791. [DOI] [PMC free article] [PubMed] [Google Scholar]