Abstract
Background:
Staphylococcus aureus is one of the predominant causes of skin and soft tissue infections (SSTIs), but limited data were available regarding the characterization of S. aureus from SSTIs patients in Jiangsu Province in China. We aimed to investigate the molecular epidemiology of S. aureus among SSTIs patients in two hospitals of Jiangsu Province.
Methods:
Sixty-two patients with SSTIs from two Chinese hospitals in Jiangsu Province were enrolled in this study, and 62 S. aureus isolates were collected from February 2014 to January 2015. S. aureus isolates were characterized by antimicrobial susceptibility testing, toxin gene detection, and molecular typing with sequence type, Staphylococcus protein A gene type, accessory gene regulator (agr) group, and Staphylococcal cassette chromosome mec type.
Results:
Sixteen (25.8%) methicillin-resistant S. aureus (MRSA) isolates were detected, and there was no isolate found resistant to vancomycin, teicoplanin, sulfamethoxazole-trimethoprim, and linezolid. The sei was the toxin gene most frequently found, and no lukS/F-PV-positive isolates were detected among the SSTIs’ patients. Molecular analysis revealed that ST398 (10/62, 16.1%; 2 MRSA and 8 methicillin-susceptible S. aureus) to be the dominant clone, followed by ST5 (8/62, 12.9%) and ST7 (8/62, 12.9%).
Conclusions:
The livestock ST398 was the most common clone among patients with S. aureus SSTIs in Jiangsu Province, China. Surveillance and further studies on the important livestock ST398 clone in human infections are necessarily requested.
Keywords: Livestock, Molecular Epidemiology, Skin and Soft Tissue Infections, ST398, Staphylococcus aureus
Introduction
Skin and soft tissue infections (SSTIs) range from relatively common superficial skin infections to rare but life-threatening infections such as necrotizing fasciitis or gas gangrene.[1] Staphylococcus aureus is the most common pathogen in SSTIs across all continents, causing a variety of SSTIs ranging from the benign (e.g., impetigo and uncomplicated cellulitis) to the immediately life-threatening.[2] While S. aureus has traditionally been the leading cause of SSTIs, its importance has ballooned in the past 15 years with the emergence of a worldwide epidemic of community-associated methicillin-resistant S. aureus (MRSA) SSTIs.[3] The developing resistance to antibiotics and the presence of virulence factors playing roles in SSTIs such as Panton-Valentine leukocidin (PVL) and toxic shock syndrome toxin (TSST) lead to major challenges in preventing and treating patients with S. aureus SSTIs.
In China, CC59 was the dominant clonal complex among S. aureus, especially MRSA among the patients with SSTIs, and ST7 has become the common clone recently in methicillin-susceptible S. aureus (MSSA) SSTIs.[4,5] The livestock-associated clone ST398, which was always associated with animals and farm workers,[6] was currently found frequently with a high positive rate of lukS/F-PV among S. aureus in SSTIs patients in China.[4,7] CC8 was the most common clone among SSTIs-associated S. aureus, especially MRSA in the United States and Japan.[8,9] Moreover, the prevalence of S. aureus colonization among SSTIs patients was high in the United States and USA300 (CC8) MRSA was often involved as reported.[10]
Jiangsu Province lies in the east of China and is adjacent to Shanghai. The livestock husbandry of the two cities enrolled in this study in Jiangsu Province was well developed. To the best of our knowledge, there were no published data regarding the molecular epidemiology of S. aureus among patients with SSTIs in Jiangsu Province in China. Thus, the aim of the current study was to investigate the genetic diversity of S. aureus from SSTIs patients in two Chinese hospitals in Jiangsu Province, China.
Methods
Study design and setting
A total of 62 patients with SSTIs from two Chinese hospitals in two cities (Affiliated Hospital of Nantong University and Jiangsu Taizhou People's Hospital, 41 and 21 patients, respectively) of Jiangsu Province were enrolled in this study, and 62 S. aureus isolates were collected during February 2014 to January 2015. The experiments in this study were performed in Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai.
This study was approved by the Ethics Committee of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, and the Review Board exempted the need for informed consent for this retrospective study mainly focused on bacteria without interventions involving patients.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed according to the guidelines of Clinical and Laboratory Standards Institute.[11] Antibiotics including penicillin (10 units), cefoxitin (30 μg), gentamicin (10 μg), kanamycin (30 μg), tobramycin (10 μg), erythromycin (15 μg), tetracycline (30 μg), teicoplanin (30 μg), minocycline (30 μg), ciprofloxacin (5 μg), clindamycin (2 μg), sulfamethoxazole-trimethoprim (25 μg), chloramphenicol (30 μg), rifampicin (5 μg), quinupristin-dalfopristin (15 μg), linezolid (30 μg), fusidic acid (10 μg), and mupirocin (5 μg and 200 μg) were tested by the disk diffusion method, and all the disks were Oxoid paper disks (Oxoid, UK). The minimum inhibitory concentration of vancomycin was detected by E-test (bioMerieux, France). The penicillin disk diffusion zone edge test was performed for β-lactamase detection, and inducible clindamycin resistance was determined by the D-test. S. aureus ATCC25923 and ATCC29213 were used as quality controls for the disk diffusion test and E-test, respectively.
Detection of toxin genes
A variety of clinically significant toxin genes were detected by polymerase chain reaction,[12] including lukS/F-PV (encoding Panton-Valentine leukocidin); tst (encoding toxic shock syndrome toxin 1); eta and etb (encoding exfoliative toxin A and B); sea-see and seg-sej (encoding staphylococcal enterotoxins SEA-SEE and SEG-SEJ); and sasX (encoded in a mobile genetic element), which act as a virulence determinant and play a key role in MRSA colonization and pathogenesis.
Molecular typing
Molecular typing including multilocus sequence typing (MLST) and Staphylococcus protein A gene (spa) typing was performed on all S. aureus.[13] For confirming the presence of MRSA, mecA detection was performed on all S. aureus collected. Staphylococcal cassette chromosome mec (SCCmec) types of MRSA were determined by the method described previously.[14]
Statistical analysis
The Chi-square or Fisher's exact test was used where appropriate using the SPSS software package (SPSS19.0, IBM-SPSS Inc, Armonk, NY, USA). A two-sided P < 0.05 was considered statistically significant.
Results
Clinical data
The median age of patients with SSTIs in this study was 50 years (range: 6 months to 95 years), and the gender distribution (male/female) was 40/22 (64.5%/35.5%). Forty of 62 patients were considered outpatients, and the rest 20 patients were inpatients. In terms of the infection types, abscesses (69.4%) was the most common infection type. Clinical data on any possible patient contact with animals were lack in this study.
Antimicrobial susceptibility testing
Sixteen (25.8%) isolates were confirmed as MRSA in this study. We did not find any isolated resistant to vancomycin, teicoplanin, sulfamethoxazole-trimethoprim, and linezolid. The resistance rates of other antibiotics tested are shown in Table 1. A total of 35 isolates (27 MSSA and 8 MRSA) were inducible resistance to clindamycin based on D-test results. One MRSA isolate (ST5-SCCmecI-t311) was observed showing high-level mupirocin resistance.
Table 1.
The antibiotic resistance rates of Staphylococcus aureus isolated from patients with skin and soft tissue infections in two Chinese hospitals
| Antibiotics | Resistance rate (%) | ||
|---|---|---|---|
| Total (n = 62) | MSSA (n = 46) | MRSA (n = 16) | |
| Penicillin | 90.3 | 87.0 | 100 |
| Gentamicin | 12.9 | 4.3 | 37.5 |
| Kanamycin | 22.6 | 8.7 | 62.5 |
| Tobramycin | 17.7 | 10.9 | 37.5 |
| Erythromycin | 69.4 | 65.2 | 81.3 |
| Tetracycline | 22.6 | 15.2 | 43.8 |
| Minocycline | 1.6 | 0 | 6.3 |
| Ciprofloxacin | 14.5 | 8.7 | 31.3 |
| Clindamycin* | 14.5 | 8.7 | 31.3 |
| Chloramphenicol | 4.8 | 4.3 | 6.3 |
| Rifampicin | 1.6 | 0 | 6.3 |
| Quinupristin-dalfopristin | 1.6 | 0 | 6.3 |
| Fusidic acid | 4.8 | 0 | 6.3 |
| Mupirocin† | 1.6 | 0 | 6.3 |
*35 isolates (27 MSSA and 8 MRSA) were D-test positive, indicating inducible clindamycin resistance; †One MRSA isolate presented high-level mupirocin resistance. MSSA: Methicillin-susceptible Staphylococcus aureus; MRSA: Methicillin-resistant Staphylococcus aureus.
Virulence factors
The toxin genes lukS/F-PV, etb, see, and sasX were not detected among all S. aureus isolates in this study. The sei was most frequently found among the toxin genes we screened for, occurring in 16 isolates (25.8%) as shown in Table 2. The tst was found in 3 isolates (1 MSSA and 2 MRSA) and all the 3 isolates were belonging to ST5. There was no significant difference observed in prevalence of the toxin genes between the MSSA and MRSA groups.
Table 2.
Prevalence of toxin genes among Staphylococcus aureus isolated from patients with skin and soft tissue infections in two Chinese hospitals
| Toxin genes | Positive rate (%) | χ2 | P | ||
|---|---|---|---|---|---|
| Total (n = 62) | MSSA (n = 46) | MRSA (n = 16) | |||
| lukS/F-PV | 0 | 0 | 0 | – | – |
| tst | 4.8 | 2.2 | 12.5 | 0.964 | 0.326 |
| eta | 3.2 | 4.3 | 0 | 0.001 | 0.979 |
| etb | 0 | 0 | 0 | – | – |
| sea | 8.1 | 4.3 | 18.8 | 1.663 | 0.197 |
| seb | 4.8 | 4.3 | 6.3 | <0.001 | 1.000 |
| sec | 19.4 | 13.0 | 37.5 | 0.873 | 0.350 |
| sed | 8.1 | 8.7 | 6.3 | <0.001 | 1.000 |
| see | 0 | 0 | 0 | – | – |
| seg | 12.9 | 10.9 | 18.8 | 0.142 | 0.706 |
| seh | 3.2 | 2.2 | 6.3 | – | 0.453 |
| sei | 25.8 | 23.9 | 31.3 | 0.061 | 0.806 |
| sej | 6.5 | 6.5 | 6.3 | <0.001 | 1.000 |
| sasX | 0 | 0 | 0 | – | – |
lukS/F-PV: Gene encoding Panton-Valentine leukocidin; tst: Gene encoding toxic shock syndrome toxin 1; eta and etb: Gene encoding exfoliative toxin A and B; sea-see and seg-sej: Gene encoding staphylococcal enterotoxins SEA-SEE and SEG-SEJ; sasX: Gene encoding mobile genetic element; Two-sided P value calculated by the Chi-square or Fisher’s exact test as appropriate. –: Not applicable; MSSA: Methicillin-susceptible Staphylococcus aureus; MRSA: Methicillin-resistant Staphylococcus aureus.
Molecular types
ST398 was the most common clone which detected in 10 isolates (2 MRSA and 8 MSSA) accounting for 16.1% among SSTIs patients. Both ST5 and ST7 were the second common clones (both 8/62, 12.9%), and 4 ST5 MRSA isolates and 1 ST7 MRSA isolate were identified as shown in Table 3. The relationships of S. aureus isolates are shown in a rough sketch produced by eBURST based on the MLST data of this study [Figure 1]. The spa type was identified in 38 types and it expressed a great diversity in this study. There was not an outstanding spa type that found in more than 5 isolates, and t4549 and t571 were the most common spa type relatively which were both detected in 4 isolates (6.5%). In total, we detected 5 SCCmec Type I, 3 SCCmec Type II, and 8 SCCmec Type V isolates. The agrI (32/62, 51.6%) was the most frequent agr group, followed by agrII (21/62, 33.9%), agrIII (5/62, 8.1%), and agrIV (4/62, 6.4%).
Table 3.
Molecular characteristics of Staphylococcus aureus isolated from patients with skin and soft tissue infections in two Chinese hospitals
| CC | ST | Isolates, n | SCCmec type | spa type (n) | Virulence factors (n) |
|---|---|---|---|---|---|
| 1 | 573 | 2 | t8915 (1), t4938 (1) | sec (1), sei (1) | |
| 2315 | 1 | t11687 (1) | sec (1), seg (1), sei (1) | ||
| 5 | 5 | 8 | I | t311 (1), t2460 (1) | tst (1), sec (1), seg (2), sei (2) |
| II | t311 (2) | tst (1), sea (2), sec (1), seg (1), sei (2) | |||
| t954 (2), t548 (1), t5734 (1) | tst (1), sed (3), seg (2), sei (4), sej (2) | ||||
| 965 | 2 | I | t15075 (1) | sed (1), sei (1), sej (1) | |
| t062 (1) | sea (1), seg (1), sei (1) | ||||
| 764 | 1 | II | t002 (1) | None | |
| 6 | 6 | 2 | t701 (2) | eta (1), sea (1) | |
| 7 | 7 | 8 | V | t796 (1) | None |
| t091 (3), t796 (2), t1685 (1), t3714 (1) | sec (1) | ||||
| 943 | 1 | t1867 (1) | seh (1) | ||
| 8 | 8 | 1 | t9101 (1) | sed (1), sej (1) | |
| 239 | 1 | V | t030 (1) | sea (1) | |
| 630 | 4 | V | t4549 (4) | None | |
| 1 | t5554 | None | |||
| 15 | 15 | 5 | t084 (3), t085 (1), t14014 (1) | None | |
| 468 | 1 | t774 (1) | None | ||
| 25 | 25 | 2 | t078 (2) | seb (2), sei (2) | |
| 59 | 59 | 1 | I | t437 (1) | seb (1), seh (1) |
| 88 | 88 | 1 | I | t15076 (1) | None |
| 4 | t3155 (2), t1376 (1), t15074 (1) | None | |||
| 121 | 120 | 1 | t435 (1) | sei (1) | |
| 2155 | 2 | t1425 (2) | eta (1), seg (1), sei (1) | ||
| 188 | 188 | 3 | t189 (3) | None | |
| 398 | 398 | 10 | V | t034 (1), t1928 (1) | None |
| t571 (4), t034 (2), t1451 (2) | None |
CC: Clonal complex; ST: Sequence type by multi-locus sequence typing; SCCmec: Staphylococcal cassette chromosome mec; spa: Staphylococcus protein A gene; None: No toxin gene detected.
Figure 1.
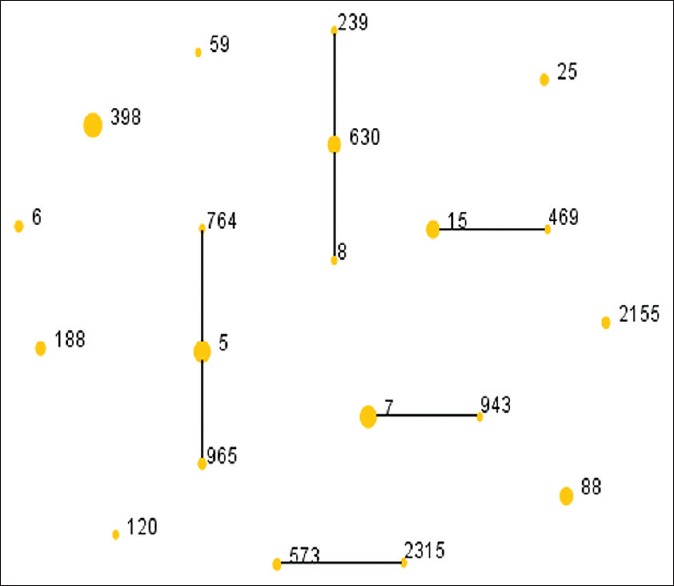
The diagram produced by eBURST with the stringent (default) group definition. Each number represents an MLST ST and the area of each circle indicates the prevalence of the ST in the MLST data of this study. MLST: Multilocus sequence typing; ST: Sequence type.
Discussion
In this study, 16 MRSA isolates were detected accounting for 25.8%, and it was higher than the occurrence of MRSA among S. aureus from SSTIs as we studied in Shanghai previously.[4] However, it was lower than the occurrence of MRSA among S. aureus SSTIs as reported in Canada.[15] Considering the importance of MRSA in SSTIs and its involvement associated with poor patient outcomes, vancomycin, teicoplanin, linezolid, daptomycin, tigecycline, and ceftaroline were recommended by European guidelines for the treatment of MRSA complicated SSTIs (cSSTIs).[16] According to the study which conducted more than 10 years in Europe, clinical efficacy and favorable outcomes of linezolid have been demonstrated for the treatment of MRSA cSSTIs, including the treatment of lower extremity infections.[16]
ST398 was found to be the most common clone within all the S. aureus isolates studied. ST398 was a livestock-associated clone and has been reported as a common but not the most common clone among patients with S. aureus SSTIs in Shanghai and Beijing, China before.[4,7] It has also been reported unexpectedly high among bone and joint infections and nasal-colonizing isolates in France.[17] The spa type t034 which was found among all ST398 S. aureus isolates was reported as a dominant type in pigs and pig farmers in Canada.[18] According to the documents in 2014, in the two cities in Jiangsu Province involved in this study, a total of 11.5 million pigs, 5.5 million sheep, and 208.6 million fowls were breed, of which 7.0 million pigs, 3.0 million sheep, and 142.6 million fowls were sold to nearby cities like Shanghai. The livestock husbandry in the two cities is well developed and a great number of people in these two cities are engaged in livestock husbandry or other related works. The high prevalence of ST398 S. aureus we observed in this study suggested that ST398 might be related to livestock or farm working. It was the limitation that we lack any clinical data on any possible patient contact with animals. Nevertheless, it was still suggested that CC398 S. aureus is mostly disseminated through direct contact to livestock, and a substantial proportion of patients seem to acquire MRSA CC398 through other pathways.[19] Epidemiological and genetic analyses revealed that human MRSA of unknown origin CC398 carriers carried MRSA from livestock origin, suggestive of indirect transmission. Although the exact transmission route remains unknown, direct human-to-human transmission remains a possibility as well.[20] In addition to the region or the transmission of CC398 S. aureus, the question how CC398 S. aureus could become the predominant clone in human infections so fast in China even in other countries during these years should be a matter of concern that need further studies.
Both ST5 and ST7 were the second common clones in this study. ST5 was a pandemic HA-MRSA clone disseminated internationally in Asia.[21] It was noteworthy that the toxin gene tst encoding TSST-1 was found in 3 isolates and all the 3 isolates were belonging to ST5. Moreover, the TSST-1 ST5 has been described in China and France before.[22,23] Besides frequently discovered among SSTIs patients in China currently, ST7 was also found to be the most common genotype of MSSA in invasive community-acquired S. aureus infection in Chinese children.[24]
CC8 (ST8, ST239, and ST630) was totally observed in 7 isolates as shown in Table 3. ST8 (USA300) has led to a high burden of SSTIs globally ever since its emergence in 2000 in the United States, as witnessed by S. aureus clone outbreak among SSTIs in many countries;[4] however, we only found one ST8 MSSA in SSTIs patients in this study. ST239 is recognized as a common epidemic clone in bloodstream infections[13] and ST630 was recently reported to cause severe infective endocarditis with systemic embolism in China.[25] CC59 which is always the most clonal complex among patients with SSTIs in China was found only in one MRSA isolate. PVL production by S. aureus may play a key role in the pathogenesis of S. aureus SSTIs.[26] However, we have not detected any lukS/F-PV-positive isolate among patients with SSTIs in this study.
In conclusion, ST398 was the most common clone among patients with S. aureus SSTIs in Jiangsu Province, China. The livestock ST398 might be the predominant clone in S. aureus, causing SSTIs in the region where livestock is well developed. Surveillance and further studies on the important livestock ST398 clone in human infections are necessarily requested.
Financial support and sponsorship
This work was supported by a grant from the National Natural Science Foundation of China (No. 81472010).
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
We thank Shao-Peng Chu (Laboratory, Affiliated Hospital of Nantong University, Nantong, Jiangsu Province) and Qing-Fang Zhang (Laboratory, Jiangsu Taizhou Peopleʼs Hospital, Taizhou, Jiangsu Province) for their kind help and cooperation in collecting isolates.
Footnotes
Edited by: Qiang Shi
References
- 1.Török ME, Conlon CP. Skin and soft tissue infections. Medicine. 2013;41:709–15. doi: 10.1016/j.mpmed.2013.10.001. [Google Scholar]
- 2.Cardona AF, Wilson SE. Skin and soft-tissue infections: A critical review and the role of telavancin in their treatment. Clin Infect Dis. 2015;61(Suppl 2):S69–78. doi: 10.1093/cid/civ528. doi: 10.1093/cid/civ528. [DOI] [PubMed] [Google Scholar]
- 3.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG., Jr Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28:603–61. doi: 10.1128/CMR.00134-14. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu FF, Hou Q, Yang HH, Zhu YQ, Guo XK, Ni YX, et al. Characterization of Staphylococcus aureus isolated from non-native patients with skin and soft tissue infections in Shanghai. PLoS One. 2015;10:e0123557. doi: 10.1371/journal.pone.0123557. doi: 10.1371/journal.pone.0123557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu F, Liu Y, Lv J, Qi X, Lu C, Ding Y, et al. Antimicrobial susceptibility, virulence determinant carriage and molecular characteristics of Staphylococcus aureus isolates associated with skin and soft tissue infections. Braz J Infect Dis. 2015;19:614–22. doi: 10.1016/j.bjid.2015.08.006. doi: 10.1016/j.bjid.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vandendriessche S, Vanderhaeghen W, Larsen J, de Mendonça R, Hallin M, Butaye P, et al. High genetic diversity of methicillin-susceptible Staphylococcus aureus (MSSA) from humans and animals on livestock farms and presence of SCCmec remnant DNA in MSSA CC398. J Antimicrob Chemother. 2014;69:355–62. doi: 10.1093/jac/dkt366. doi: 10.1093/jac/dkt366. [DOI] [PubMed] [Google Scholar]
- 7.Zhao C, Liu Y, Zhao M, Liu Y, Yu Y, Chen H, et al. Characterization of community acquired Staphylococcus aureus associated with skin and soft tissue infection in Beijing: High prevalence of PVL ST398. PLoS One. 2012;7:e38577. doi: 10.1371/journal.pone.0038577. doi: 10.1371/journal.pone.0038577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaltsas A, Guh A, Mediavilla JR, Varshney AK, Robiou N, Gialanellia P, et al. Frequency of panton-valentine leukocidin-producing methicillin-sensitive Staphylococcus strains in patients with complicated skin and soft tissue infection in bronx, new york. J Clin Microbiol. 2011;49:2992–5. doi: 10.1128/JCM.00704-11. doi: 10.1128/JCM.00704-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maeda T, Saga T, Miyazaki T, Kouyama Y, Harada S, Iwata M, et al. Genotyping of skin and soft tissue infection (SSTI)-associated methicillin-resistant Staphylococcus aureus (MRSA) strains among outpatients in a teaching hospital in Japan: Application of a phage-open reading frame typing (POT) kit. J Infect Chemother. 2012;18:906–14. doi: 10.1007/s10156-012-0506-4. doi: 10.1007/s10156-012-0506-4. [DOI] [PubMed] [Google Scholar]
- 10.Kumar N, David MZ, Boyle-Vavra S, Sieth J, Daum RS. High Staphylococcus aureus colonization prevalence among patients with skin and soft tissue infections and controls in an urban emergency department. J Clin Microbiol. 2015;53:810–5. doi: 10.1128/JCM.03221-14. doi: 10.1128/JCM.03221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.CLSI Document M100-S24. Vol. 34. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2014. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing;Twenty-Fourth Informational Supplement. [Google Scholar]
- 12.Gu FF, Han LZ, Chen X, Wang YC, Shen H, Wang JQ, et al. Molecular characterization of Staphylococcus aureus from surgical site infections in orthopedic patients in an orthopedic trauma clinical medical center in Shanghai. Surg Infect (Larchmt) 2015;16:97–104. doi: 10.1089/sur.2014.027. doi: 10.1089/sur.2014.027. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Wang WK, Han LZ, Liu Y, Zhang H, Tang J, et al. Epidemiological and genetic diversity of Staphylococcus aureus causing bloodstream infection in Shanghai 2009-2011. PLoS One. 2013;8:e72811. doi: 10.1371/journal.pone.0072811. doi: 10.1371/journal.pone.0072811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Yang HH, Huangfu YC, Wang WK, Liu Y, Ni YX, et al. Molecular epidemiologic analysis of Staphylococcus aureus isolated from four burn centers. Burns. 2012;38:738–42. doi: 10.1016/j.burns.2011.12.023. doi: 10.1016/j.burns. 271.12.023. [DOI] [PubMed] [Google Scholar]
- 15.Borgundvaag B, Ng W, Rowe B, Katz K EMERGency Department Emerging Infectious Disease Surveillance NeTwork (EMERGENT) Working Group. Prevalence of methicillin-resistant Staphylococcus aureus in skin and soft tissue infections in patients presenting to Canadian emergency departments. CJEM. 2013;15:141–60. doi: 10.2310/8000.2013.130798. [PubMed] [Google Scholar]
- 16.Bassetti M, Baguneid M, Bouza E, Dryden M, Nathwani D, Wilcox M. European perspective and update on the management of complicated skin and soft tissue infections due to methicillin-resistant Staphylococcus aureus after more than 10 years of experience with linezolid. Clin Microbiol Infect. 2014;20(Suppl 4):3–18. doi: 10.1111/1469-0691.12463. doi: 10.1111/1469-0691.12463. [DOI] [PubMed] [Google Scholar]
- 17.Valour F, Tasse J, Trouillet-Assant S, Rasigade JP, Lamy B, Chanard E, et al. Methicillin-susceptible Staphylococcus aureus clonal complex 398: High prevalence and geographical heterogeneity in bone and joint infection and nasal carriage. Clin Microbiol Infect. 2014;20:O772–5. doi: 10.1111/1469-0691.12567. doi: 10.1111/1469-0691.12567. [DOI] [PubMed] [Google Scholar]
- 18.Khanna T, Friendship R, Dewey C, Weese JS. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet Microbiol. 2008;128:298–303. doi: 10.1016/j.vetmic.2007.10.006. doi: 10.1016/j.vetmic.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Deiters C, Günnewig V, Friedrich AW, Mellmann A, Köck R. Are cases of Methicillin-resistant Staphylococcus aureus clonal complex (CC) 398 among humans still livestock-associated? Int J Med Microbiol. 2015;305:110–3. doi: 10.1016/j.ijmm.2014.11.007. doi: 10.1016/j.ijmm.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Lekkerkerk WS, van Wamel WJ, Snijders SV, Willems RJ, van Duijkeren E, Broens EM, et al. What is the origin of livestock-associated methicillin-resistant Staphylococcus aureus clonal complex 398 isolates from humans without livestock contact? An epidemiological and genetic analysis. J Clin Microbiol. 2015;53:1836–41. doi: 10.1128/JCM.02702-14. doi: 10.1128/JCM.02702-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen CJ, Huang YC. New epidemiology of Staphylococcus aureus infection in Asia. Clin Microbiol Infect. 2014;20:605–23. doi: 10.1111/1469-0691.12705. doi: 10.1111/1469-0691.12705. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Gu FF, Zhao SY, Xiao SZ, Wang YC, Guo XK, et al. Prevalence and molecular epidemiology of Staphylococcus aureus among residents of seven nursing homes in Shanghai. PLoS One. 2015;10:e0137593. doi: 10.1371/journal.pone.0137593. doi: 10.1371/journal.pone.0137593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robert J, Tristan A, Cavalié L, Decousser JW, Bes M, Etienne J, et al. Panton-valentine leukocidin-positive and toxic shock syndrome toxin 1-positive methicillin-resistant Staphylococcus aureus: A French multicenter prospective study in 2008. Antimicrob Agents Chemother. 2011;55:1734–9. doi: 10.1128/AAC.01221-10. doi: 10.1128/AAC.01221-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiao Y, Ning X, Chen Q, Zhao R, Song W, Zheng Y, et al. Clinical and molecular characteristics of invasive community-acquired Staphylococcus aureus infections in Chinese children. BMC Infect Dis. 2014;14:582. doi: 10.1186/s12879-014-0582-4. doi: 10.1186/s12879-014-0582-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng B, Jiang S, Xu Z, Xiao Y, Li L. Severe infective endocarditis with systemic embolism due to community associated methicillin-resistant Staphylococcus aureus ST630. Braz J Infect Dis. 2014;19:85–9. doi: 10.1016/j.bjid.2014.07.006. doi: 10.1016/j.bjid.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gould IM. Antibiotics, skin and soft tissue infection and meticillin-resistant Staphylococcus aureus: Cause and effect. Int J Antimicrob Agents. 2009;34(Suppl 1):S8–11. doi: 10.1016/S0924-8579(09)70542-4. doi: 10.1016/s0924-8579(09)70542-4. [DOI] [PubMed] [Google Scholar]


