Abstract
The Zika virus (ZIKV) is a flavivirus of the family Flaviviridae, which is similar to dengue virus, yellow fever and West Nile virus. Recent outbreaks in South America, Latin America, the Caribbean and in particular Brazil have led to concern for the spread of the disease and potential to cause Guillain-Barré syndrome and microcephaly. Although ZIKV has been known of for over 60 years there is very little in the way of knowledge of the virus with few publications and no crystal structures. No antivirals have been tested against it either in vitro or in vivo. ZIKV therefore epitomizes a neglected disease. Several suggested steps have been proposed which could be taken to initiate ZIKV antiviral drug discovery using both high throughput screens as well as structure-based design based on homology models for the key proteins. We now describe preliminary homology models created for NS5, FtsJ, NS4B, NS4A, HELICc, DEXDc, peptidase S7, NS2B, NS2A, NS1, E stem, glycoprotein M, propeptide, capsid and glycoprotein E using SWISS-MODEL. Eleven out of 15 models pass our criteria for selection. While a ZIKV glycoprotein E homology model was initially described in the immature conformation as a trimer, we now describe the mature dimer conformer which allowed the construction of an illustration of the complete virion. By comparing illustrations of ZIKV based on this new homology model and the dengue virus crystal structure we propose potential differences that could be exploited for antiviral and vaccine design. The prediction of sites for glycosylation on this protein may also be useful in this regard. While we await a cryo-EM structure of ZIKV and eventual crystal structures of the individual proteins, these homology models provide the community with a starting point for structure-based design of drugs and vaccines as well as a for computational virtual screening.
Keywords: Aedes mosquito, dengue virus, drug discovery, ebola virus, flavivirus, microcephaly, yellow fever, Zika virus
Introduction
All flaviviruses are spherical and contain a genome of approximately 11kb that functions as mRNA and encodes a polyprotein that leads to 10 proteins 1. Examples include dengue virus, yellow fever and West Nile virus 2. The recent pandemic of ZIKV occurring in South America, Latin America, the Caribbean and in particular Brazil spread by the Aedes mosquito has awakened dormant interest in this flavivirus which is a mild dengue-like disease 3. However several documented cases of Guillain-Barré syndrome and other neurologic conditions represent important complications of the disease. In recent weeks the extent of the disease has also become apparent as new discoveries and announcements are made almost daily. Though clearly we have a considerable number of significant gaps in our knowledge which need addressing 4.
The most concerning issue however is microcephaly observed in women who had ZIKV during pregnancy. There have been multiple cases of ZIKV found in fetal or newborn brain tissue that had signs of prenatal damage. The virus seems to have neurotropism in fetal brains, which may account for the presumed association between the infection and microcephaly 5, 6. The fetus in the recent case study had microcephaly with calcifications and ZIKV was found in the brain 6. The ZIKV strain was identified as from French Polynesia (GenBank accession number KJ776791) and several polymorphisms were noted in the NS1, NS4B and FtsJ like methyltransferase regions. While the findings are not absolute proof that ZIKV causes microcephaly, the evidence from this case report strengthens the linkage 7. Experts involved in the decision on the World Health Organization determined Public Health Emergency of International Concern (PHEIC) recommended the need for more research into the microcephaly link and need for an animal model to be developed. This group also interestingly called for open data sharing 8. Early work 45 years ago in inoculated newborn mice showed that ZIKV had neurological effects, enlarging astroglial cells and destroying pyriform cells. At the same time virus formation within the endoplasmic reticulum was also visualized 9. We are not aware of any studies of effects of ZIKV on human brain or brain cells. Localization of such viruses to the brain is not unusual for flaviviruses i.e. West Nile virus and this tropism may arise from viral binding to glycosaminoglycans, as has been observed for dengue virus in human microvascular endothelial cells 10. Heparan sulfate and the C-type lectin DC-SIGN (dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin) are well characterized attachment structures for flaviviruses on cells. Interfering with glycan binding is one potential approach to preventing virus entry. Another is to acidify the endosome as has been demonstrated in vitro with chloroquine for dengue virus infection 2. Several entry and adhesion factors, including DC-SIGN, Tyro3, and AXL as well as others, have been shown to permit ZIKV entry in human skin cells 11.
The routes for transmission of ZIKV besides mosquito are of some concern. Recent US CDC guidance to pregnant women describes precautions against sexual transmission of ZIKV 12 and that the virus can persist for up to 12 weeks 13. Possible ZIKV transmission through blood transfusion in French Polynesia was described by detecting the virus in 3% of asymptomatic blood donors 14. Given how widespread ZIKV has become, there is a risk of depleting the blood supply, if donation after potential virus exposure is deferred. Methods have also been developed to inactivate ZIKV in plasma using amotosalen and UVA illumination 15. There are issues with detection of ZIKV as a false positive dengue NS1 antigen test in a traveler to Switzerland was found to have the virus later. Therefore, cross-reactivity appears to be an issue in detection 16 and this also suggests the need for better diagnostics to be developed.
Structural knowledge of the ZIKV proteins may allow us to understand exposed epitopes which will facilitate the development of specific diagnostic reagents that differentiate it from dengue and other flaviviruses. Furthermore, open sharing of the three-dimensional arrangement of viral surface proteins could allow the mapping of potential neutralizing epitopes, guiding efforts to rationally design effective vaccines. We recently developed a preliminary model for ZIKV glycoprotein E based on the dengue virus glycoprotein E late stage fusion intermediate as a trimer 4. We now provide homology models of the glycoprotein E based on a dimer structure as well as attempts at modeling the other proteins in ZIKV. We have also investigated the likely glycosylation sites of the ZIKV envelope glycoprotein. Glycosylation may obstruct the binding of antibodies, or block access to potential underlying peptide antigens, so glycans may be an important consideration in diagnostic and vaccine development. Previously, glycosylation analysis using several computational tools predicted mammalian N-linked glycosylation at Asn-154 in most ZIKV strains 17, a site known to be important in other flaviviruses. N-glycosylation of the dengue envelope glycoprotein at two sites has been shown to mediate interactions with DC-SIGN 18. Insect cell expression of dengue shows both high-mannose and complex glycans 19. However, flavivirus glycosylation in model systems such as insect cell culture and mammalian tumor cell lines may not represent the true infective insect and mammalian glycoprotein. Mammalian O-linked glycosylation was predicted at Thr-245 and Thr-381 in some isolates 17. Yet, reliable prediction is hampered by a lack of O-glycosylation consensus sequences and preferences for O-glycosylation driven by structural characteristics 20.
Understanding the three-dimensional structure of antigenic ZIKV proteins may help accelerate the development of antibodies for diagnostics and rationally designed vaccines. In addition, the comparison of the assembled surface glycoprotein of ZIKV with that of dengue virus may help understand the accessible epitopes for the development of anti-flaviral vaccines in general. There is considerable prior work including structure-based design and virtual screening for dengue, yellow fever and other related flaviviruses to develop antivirals to target envelope glycoproteins 21– 26, guanylyltransferase 27, capsid protein NS3 helicase, NS2B-NS3 protease and NS5 polymerase 28, 29, as well as whole cell screens 30 which have produced many molecules potentially useful against ZIKV in vitro. Early work has only tested a small number of FDA-approved drugs against ZIKV including (EC 50 in parenthesis) interferon (34.3 IU/ml), ribavirin (143 ug/ml), 6-azauridine (1.5 ug/ml) and glycyrrhizin (384 ug/ml) 31. A recent paper by Hamel et al., from 2015 also showed interferon inhibited ZIKV replication in primary skin fibroblasts 11.
However, the use of compounds against ZIKV should take into account the treatment of pregnant women, and many of the potential options are unsuitable for use in pregnancy because of toxicity and/or teratogenicity. Despite limited human data, the available data in animal models suggests caution. Azauridine is highly toxic to the fetus in model systems (for example 32). Ribavirin is not recommended for use in pregnancy due to embryotoxic and teratogenic effects 33. Interferon is a potential abortifacient 34. We also need to consider the treatment of fetus as well as children (that might become infected after birth) and the relatively small subsection of FDA approved drugs that are approved for pediatric use 35. Therefore, alternative potential drugs for ZIKV are needed. The risks of medication use in pregnancy are notable. In particular, these include teratogenicity concerns. There is also the issue that the disease does not usually pose a direct risk to pregnant women themselves, so it’s important any drug, which will not be improving their own health, does not damage their health. Pregnancy can also create increased risks and liver problems, a particular concern with any new drug as it can also affect drug distribution. In pregnancy, a ZIKV infection at 13 weeks gestation was coupled with persistent virus in a fetus at 32 weeks 6. Treatment of symptomatic pregnant women may reduce risks of transmission to the fetus. A potential drug for ZIKV could also protect fetuses from damage by reducing transmission in the general population. If the drug appeared to reduce the duration of symptoms (which though mild can be annoying) and in turn reduced viral load, reducing the chance of transmission, this could benefit them. For example, cholera patients are often given antibiotics to reduce transmission. Also those with the flu are prescribed Tamiflu/Oseltamivir to reduce the duration of symptoms, minimize the severity of symptoms, but its use might also reduce transmission of flu in the general population. Since ZIKV has been found in semen many weeks after symptoms resolve 12, treatment of male partners may reduce viral load and reduce the long term risk of transmission.
To help accelerate drug discovery through computational analysis, we have now developed homology models of ZIKV proteins that may serve as potential drug and vaccine targets. To complement high-throughput screening efforts, we could perform virtual screening against the proteins in ZIKV. While there are crystal structures for proteins from dengue 36, 37, yellow fever, West Nile virus and other flaviviruses 38– 44 there are (to date) none for ZIKV. Therefore we are limited to generating homology models, although the close evolutionary relationships between flavivirus and their component proteins and genomes represents a valid approach 45.
Methods
Protein sequence alignment
As a prelude to modeling we assessed what 3D structural data had significant identity scores to a representative ZIKV polyprotein. For this we chose UniProtKB Q32ZE1_9FLAV. While we have requested the promotion of this entry to the Swiss-Prot expert review level it has been selected as the representative sequence for the UniRef90_Q32ZE1 entry that currently clusters 108 ZIKV individual sequence entries at 90% (or above) amino acid identity. We then performed a BLAST search of this against Protein Data Bank (PDB) sequence entries. Protein BLAST analysis was also performed for each ZIKV protein sequence 46 to identify the closest proteins 47 and understand potential evolution.
Homology modeling
The amino acid sequences of ZIKV strain (GenBank accession number KJ776791 48) were retrieved from the GenBank database 49 and used as targets for homology modelling using the SWISS-MODEL server 50, 51. The latter performed the target-template sequence alignment after searching the putative X-ray template proteins in PDB for generating the 3D models for all target sequences. The best homology models were selected according to Global Model Quality Estimation (GMQE) and QMEAN statistical parameters. GMQE is a quality estimation which combines properties from the target-template alignment. The quality estimate ranges between 0 and 1 with higher values for better models. QMEAN4 scoring function consisting of a linear combination of four structural descriptors as described elsewhere in more detail 52, 53. The pseudo energies returned from the four descriptors are related to what we would expect from high resolution X-ray structures of similar size using a Z-score scheme. Further, built models were exported to the SAVES server Version 4 54 and their overall stereochemical quality, including backbone torsional angles through the Ramachandran plot, was checked according to PROCHECK 55. Lastly, each model was refined by an energy minimization protocol, using the Smart Minimizer algorithm in Discovery Studio version 4.1 (Biovia, San Diego, CA).
Site of glycosylation prediction
Mammalian N-glycosylation sites were predicted for glycoprotein E by submitting the sequence to web-based tools namely N-GlycoSite 56, GlycoEP 57, 58 and NetNGlyc Version 1.0 59.
Illustration for Zika virion and animation
The Zika virion illustrations were created by combining the homology model of the envelope ZIKV glycoprotein E with the symmetry data from the dengue virus envelope. PDB ID:1K4R 60 contains the coordinates for three copies of the protein subunit of the dengue virus envelope, along with the symmetry data necessary to create the 180-subunit icosahedral structure of the complete viral envelope. The PyMOL Molecular Graphics System, Version 1.7.6.0. Schrödinger, LLC. was used to export the surface models of the three proteins in .obj format. Then they were imported into Lightwave 3D (NewTek, San Antonio, TX) where the symmetry data was used to instance copies of the model into the icosahedral envelope. The entire structure was copied several times and lighting applied as a surfacing effect to create a visually pleasing composition, and the image rendered out.
The next step was to import into Pymol the homology model of the ZIKV envelope protein which was homology modeled using PDB ID: 3P54 (from Japanese Encephalitis Virus) as a template. A surface model of this protein was exported from Pymol as an .obj, and imported into Lightwave in place of the dengue model, using the same symmetry operators to create the envelope array. Everything else about the picture was left the same (color, composition, lighting, etc...) to allow the structural differences to be more apparent, and that image rendered out as well.
The last step was to overlay the detailed area of the two images and create an animated gif to flip back and forth between the two images of ZIKV and dengue, again to allow the differences to be more clearly seen. The structure of the Zika virion could be explored in a similar manner, using known data from other flaviviruses as a guide.
ZIKV glycoprotein E homology model conformation comparison
The immature 4 and mature (this study) homology models for glycoprotein E were compared using the ‘align and superimpose’ proteins protocol in Discovery Studio Version 4.1 (Biovia, San Diego, CA).
Results
Sequence alignment across flaviviruses
A BLAST search of ZIKV polyprotein against PDB sequence entries shows the highest scoring matches with 55–70% sequence identity ( Supplementary material S1). Protein BLAST analysis of the individual ZIKV protein sequences show that many of the proteins are similar to the same protein from Spondweni virus in 12 out of 15 cases ( Table 1). Exceptions were: FtsJ which was closer to the Murray Valley encephalitis virus, NS1 which was more similar to dengue virus 3, and glycoprotein E which was closest to the dengue virus 1 protein. These results are in general accordance with whole sequence analyses 45.
Table 1. Protein BLAST search results - closest non-ZIKV proteins.
Homology modeling
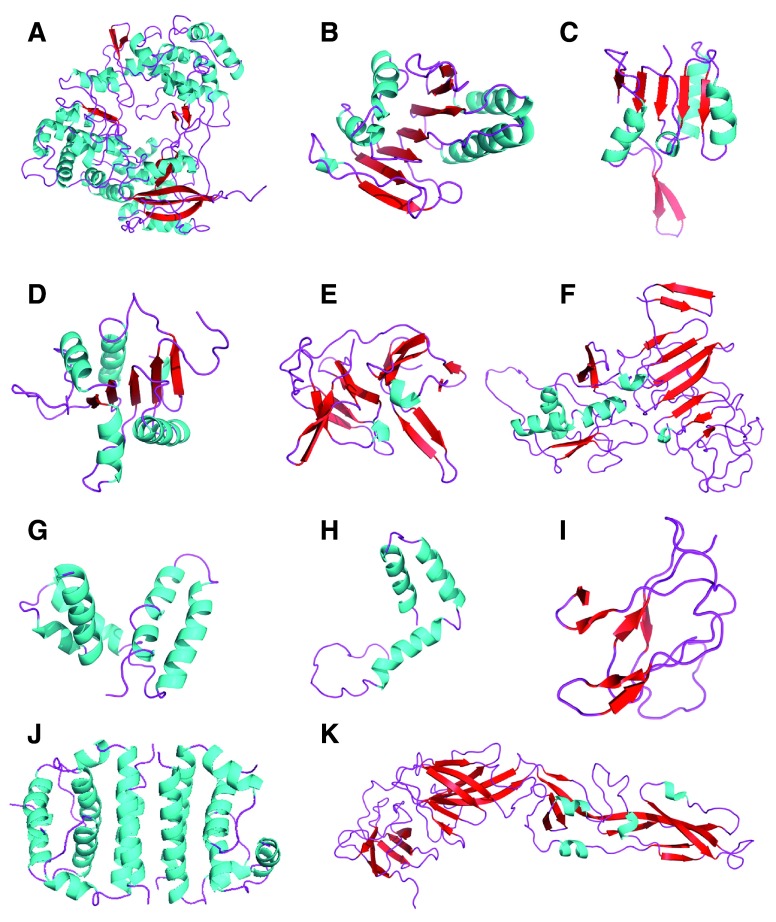
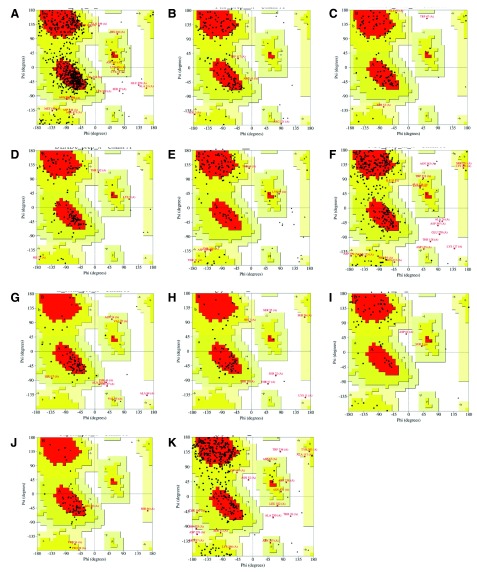
The SWISS-MODEL server was used to generate alignments ( Supplementary material S2) and homology models for all Zika proteins ( Table 2, Figure 1, Supplementary material S3). First, we selected suitable template protein structures in PDB, observing the following criteria: the template should have a high coverage (i.e., > 65% of target aligned to template) and sequence identity >30%. Then, we used GMQE and QMEAN4 scoring function as an initial criteria to discriminate good from bad models. Acceptable alignment values and higher GMQE and QMEAN4 scores were obtained during modeling, suggesting statistically acceptable homology models were generated for 11 proteins: NS5, FtsJ, HELICc, DEXDc, peptidase S7, NS1, E stem, glycoprotein M, propeptide, capsid, and glycoprotein E ( Table 2, Figure 1 and Figure 2). The Ramachandran plots for these 11 proteins provide further evidence of their acceptability ( Figure 2). On the other hand, because of low GMQE scores and of low coverage observed in X-ray template proteins available in the PDB, homology models for NS4B, NS4A, NS2B, and NS2A proteins appeared to have limitations regarding active sites and epitopes and they could not be validated.
Figure 1.
Selected ZIKV NS5 ( A), FtsJ ( B), HELICc ( C), DEXDc ( D), Peptidase S7 ( E), NS1 ( F), E Stem ( G), Glycoprotein M ( H), Propeptide ( I), Capsid ( J), and Glycoprotein E ( K) homology models (minimized proteins) that had good sequence coverage with template proteins developed with SWISS-MODEL.
Figure 2.
Ramachandran plots for ZIKV NS5 ( A), FtsJ ( B), HELICc ( C), DEXDc ( D), Peptidase S7 ( E), NS1 ( F), E Stem ( G), Glycoprotein M ( H), Propeptide ( I), Capsid ( J), and Glycoprotein E ( K) obtained by PROCHECK, showing the dihedral angles Psi and Phi of amino acid residues. Red represents most favored regions; yellow represents additional allowed regions; beige represents generously allowed regions; and white areas are disallowed regions.
Table 2. Summary of ZIKV homology model statistics.
The global and per-residue model quality has been assessed using the QMEAN scoring function 53. For improved performance, weights of the individual QMEAN terms have been trained specifically for SWISS-MODEL 50, 51, 71– 73. GMQE = Global Model Quality Estimation, QMEAN4 is a scoring function consisting of a linear combination of four structural descriptors as described elsewhere in more detail 52, 53.
| Protein | Coverage | Sequence
Identity |
GMQE | MEAN4 | PROCHECK analysis | |||
|---|---|---|---|---|---|---|---|---|
| Most
favored regions |
Additional
allowed regions |
Generously
allowed regions |
Disallowed
regions |
|||||
| NS5 | 100% | 53% | 0.89 | -2.80 | 66.3% | 30.9% | 2.3% | 0.9% |
| FtsJ | 99% | 54% | 0.94 | -1.23 | 69.5% | 27.7% | 2.1% | 0.7% |
| HELICc | 100% | 55% | 0.94 | -1.60 | 69.3% | 27.7% | 2.0% | 1.0% |
| DEXDc | 100% | 54% | 0.93 | -0.92 | 70.3% | 27.3% | 1.6% | 0.8% |
| Peptidase S7 | 100% | 53% | 0.92 | -0.25 | 67.5% | 28.3% | 4.2% | 0.0% |
| NS1 | 99% | 48% | 0.77 | -4.09 | 58.4% | 36.3% | 3.3% | 2.0% |
| E Stem | 100% | 46% | 0.74 | -8.02 | 62.5% | 27.5% | 6.3% | 3.8% |
| Glycoprotein M | 100% | 40% | 0.73 | -6.90 | 60.6% | 28.8% | 4.5% | 6.1% |
| Propeptide | 87% | 47% | 0.74 | -0.70 | 53.7% | 43.3% | 1.5% | 1.5% |
| Capsid | 65% | 42% | 0.50 | -3.54 | 69.5% | 27.1% | 3.4% | 0.0% |
| Glycoprotein E | 99% | 47% | 0.81 | -3.76 | 63.5% | 31.3% | 3.7% | 1.4% |
| NS4A | 20% | 29% | 0.07 | -1.75 | ─ | ─ | ─ | ─ |
| NS2B | 37% | 48% | 0.21 | -0.49 | ─ | ─ | ─ | ─ |
| NS2A | 16% | 31% | 0.04 | -3.10 | ─ | ─ | ─ | ─ |
| NS4B | 14% | 30% | 0.03 | -2.81 | ─ | ─ | ─ | ─ |
The best NS5 homology model was built using the full-length Japanese encephalitis virus NS5 as a template (PDB ID: 4K6M) 61. The homology model generated for the FtsJ protein was built using the crystal structure of the West Nile virus methyltransferase (PDB ID: 2OY0) 62. The best HELICc model was built using dengue virus helicase/nucleoside triphosphatase catalytic domain (PDB ID: 2BHR) 63 whereas the DEXDc model was built using the structure of the Murray Valley encephalitis virus RNA helicase (PDB ID: 2V8O) 64. The peptidase S7 model was built using the West Nile virus Ns2B-Ns3 protease (PDB ID: 2YOL) 65 and the NS1 model was built using West Nile virus non-structural protein 1 as template (PDB ID: 4O6D) 66. In addition, the E stem model was built using the cryo-electron microscopy (cryo-EM) structure of dengue virus capsid protein heterotetramer (PDB ID: 3J2P) 67 whereas the glycoprotein M model was built using the cryo-EM structure of dengue virus as a template (PDB ID: 3J27) 67. The propeptide model was built using the crystal structure of the precursor membrane protein-envelope protein heterodimer from the dengue 2 virus at low pH (PDB ID: 3C5X) 68 and the capsid model was built using the core (C) protein from West Nile virus, subtype Kunjin (PDB ID: 1SFK) 69. Finally, the best glycoprotein E model for the mature protein was built using the following sequence taken from the polyprotein, where the part corresponding to E is from residues 291-592, while the IG-like domain III is from residues 601-693. The model used the crystal structure of the Japanese encephalitis virus envelope protein, strain SA-14-14-2 as a template (PDB ID: 3P54) 70. After building of homology models, we performed an additional validation in order to explore stereochemical quality of dihedral angles phi against psi of amino acid residues in modeled structures and identify sterically allowed regions for these angles using PROCHECK analysis. The results shown in Table 2 and Figure 2 reveal that 58.4─70.3% residues of the modeled proteins are within the most favored regions (red), 27.1─43.3% residues of modeled proteins are within the additional allowed regions (yellow), 1.5─6.3% residues of modeled proteins are within the generously allowed regions (beige), and only 0.0─6.1% residues of modeled proteins are within the disallowed regions (white). These results showed that the overall stereochemical properties of the generated models were highly reliable and the models could be useful to future molecular modeling studies.
Site of glycosylation prediction
Several web-based tools were used for N-glycosylation site predictions as it provides a more thorough approach. N-GlycoSite 56 suggested N154 as a single N-glycosylation site matching the N-X-S/T/C consensus sequence. The same site was identified by GlycoEP using BPP settings (binary profile of patterns) 57, 58 giving a score of 0.65/1.00. NetNGlyc 59 also gave the same predicted site, with a jury agreement of 6/9.
Zika virion compared to dengue virion
A qualitative analysis of the Zika virion (which was constructed based on the dengue virion) can be compared to the dengue cryo-EM virion ( Figure 3) and indicates that Zika appears to have slightly more raised ‘pimples’ on the surface. The glycoprotein E dimer in ZIKV also has a narrow ‘letter-box’ groove while the dengue virion has a bigger ‘pore‘ between the intersection of 5 dimers (5 fold axis). These differences are considerably more apparent in the animation ( Supplementary material S4). It is important to note that the differences may also be artefacts of the homology modeling approach and template used for modeling ZIKV glycoprotein E.
Figure 3. Comparison of Zika and dengue virion illustrations.
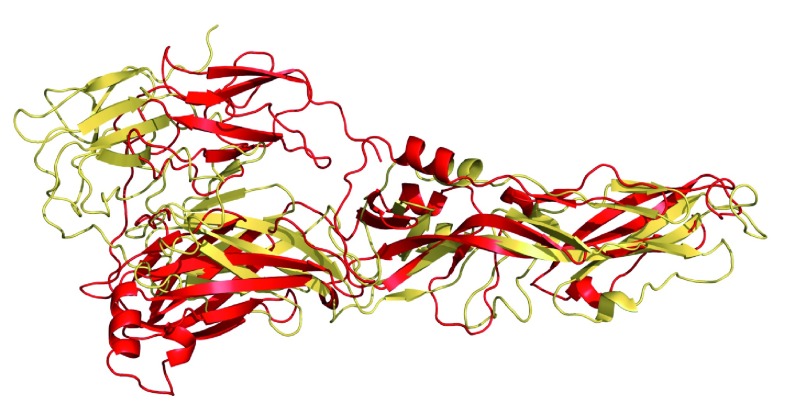
ZIKV glycoprotein E homology model conformation comparison
The homology models developed using two different templates namely the immature protein which was based on the dengue crystal structure 4gsx as a template 50, 71– 73 and the mature protein which was based on PDB ID:3P54 from Japanese encephalitis virus showed a large difference (RMSD 13.47Å) ( Figure 4). These proteins also demonstrate differences around the pocket used centered on the residues 270-277.
Figure 4. Overlap of ZIKV homology models for glycoprotein E, Yellow = mature conformation (this study) compared with the immature conformation (red) 4.
Discussion
The genus Flavivirus consists of 70 viruses many of which can cause severe human disease. There have been few sequence analyses of ZIKV previously in comparison to other flaviviruses. The genus Flavivirus produces a monophyletic tree with ZIKV being closest to Spondweni virus 74 while mosquito borne, tick borne and no-vector viruses cluster separately 45. A BLAST analysis of all the ZIKV proteins in this study suggests for 12 of 15, their closest protein is in Spondweni virus ( Table 1). More often strain sequences are compared within ZIKV and these showed variations in the NS5 gene 75 and glycoprotein E 17. This is important as it would suggest perhaps targeting other proteins would have less issue with resistance or variability due to the strain of ZIKV.
If we are to address ZIKV in the short term while we await a vaccine we need to rapidly identify an antiviral, and preferably one that can be used against other related flaviviruses. Ideally we would need to treat pregnant women and provide them with prophylaxis that was safe to them and their fetus. Such an antiviral could also be used to reduce transmission in the population in general (by reducing viral load and symptoms and/or duration). As noted a decade ago and is still is true today, no antiviral drug is approved for any flavivirus to date 76. It has been suggested that one of the ways to target these viruses is to interfere with the NS2B/NS3 protease complex 76. Understanding of flavivirus proteins and other RNA viruses has benefited from the EU funded project VIZIER 77, in particular several West Nile virus, dengue virus and other flavivirus structures of NS3 or NS5 were solved during this project and allosteric inhibitor sites were identified on NS5 78. Multiple pharmaceutical companies have worked on this target for HCV leading to clinical candidates like IDX320 79, danoprevir (ITMN-191/R7227) 80, GS-9256 81 and others 82, 83. The only HCV protease targeting FDA approved drug is simeprevir, TMC435 84, 85 and its use is avoided in pregnancy. Other HCV protease compounds are in clinical trials or submitted for FDA approval including Ledipasvir (formerly GS-5885) 86. Testing these molecules against ZIKV in vitro would be useful.
We recently described 6 steps which could be taken to kick start research on ZIKV 4, one of which was to develop homology models for ZIKV proteins that are similar to those targeted by molecules that are also active against the dengue virus. Such an approach would then enable docking of compound libraries of known antivirals, FDA approved drugs or other compounds 4. Ideally generating homology models with a single tool may not be enough. In particular, for those proteins with low sequence identity the use of servers and methods that use threading may be worthwhile (e.g. I-TASSER 87– 89). However these methods are generally only accessible to academics while others are required to license the technologies. This is ironic as these technologies were developed in most cases with NIH and NSF funds. An alternative commercial homology modeling approach (MODELLER) was also used and generated a NS5 homology model and the top hit was also the Japanese encephalitis virus RdRp domain (PDB ID: 4HDH) compared with PDB ID: 4K6M from SWISS-MODEL 61. 4HDH also includes the ATP and zinc metal where the catalytic centers are. The dengue virus 3 polymerase (PDB ID: 4HHJ) 90 has very high sequence homology and comes up as a potential target in MODELLER, which illustrates that all these viral RNA dependent polymerases are very similar.
While it is likely that the eventual availability of crystal structures of ZIKV proteins would improve the results of docking, the homology models described here ( Figure 1, Figure 2, Supplementary material S2) represent a starting point that can be used to help prioritize compounds for testing as described previously 4. Proteins with templates above 25–40% sequence identity might suggest the proteins are related while below this is a twilight zone. Homology modeling is thought to fill in the gaps between proteins with x-ray structures and those with none 91. Experimental testing of homology models and crystal structures indicate that a similar enrichment rate can be achieved when identifying active compounds in a set decoys 92. Others have also described homology models that may be an excellent alternative when crystal structures are unavailable for human GPCRs 93, 94, and have led to the first identification of inhibitors of the Mycobacterium tuberculosis Topoisomerase I after virtual screening 95, 96 prior to the crystal structure becoming available 97. Certainly there are still considerable challenges using homology models such as prediction of the correct binding pose 98 but there are plenty of success stories 98– 100. While databases of homology models exist like MODBASE 101 and SWISS-MODEL 50, 51, 71– 73 neither of these have any ZIKV protein homology models at the time of writing. There are many structural genomics initiatives and yet it would seem there are few if any continuing the work of VIZIER working on flaviviruses or emerging viruses.
Availability of structures are important as the structure of the ZIKV glycoprotein could be useful for design of antibodies selective for the virus which will be critical for the development of diagnostics, and understanding antibody binding also for the use of IV immunoglobulin in pregnancy and the organization of the epitopes on viral proteins may facilitate early work in vaccine development. There are further implications for understanding the antibody binding epitopes, which are sometimes shared between different flaviviruses. Broadly protective vaccines for flaviviruses may allow the simultaneous targeting of ZIKV and related viruses such as dengue 102. Understanding glycosylation is therefore important. To date Asn-154 is mentioned in Faye et al., 17 as a glycosylation site, as are Thr-170 (mucin type O-linked glycosylation) and other mucin sites at Thr-245 and Thr-381. Other probably O-GlcNAC attachment sites (Ser-142, Ser-227, Thr-231, Ser-304, Thr-366, and Thr-381) were also predicted. Our analysis of N-glycosylation with 3 different websites suggest Asn-154 also as a likely site of N-glycosylation in agreement with dengue virus 18.
Cryo-EM has been used to show how glycoprotein E dimers arrange on the surface of virions for flaviviruses including tick-borne encephalitis virus 103, West Nile virus 104, dengue virus 1 105 and dengue virus 4 106 ( Supplementary material S3). The early work on the tick-borne encephalitis virus 103 suggested 30 dimers on the surface and also pointed to how the glycoprotein E dimers can reorganize under low pH to form a trimer. The packing of the dimers in the dengue virion is different to tick-borne encephalitis with the glycoprotein E dimers showing 30 ‘herringbone rafts’ each containing three dimers to result in 180 copies of the protein 67, 107. West Nile virus again has a different arrangement with 60 trimers shown in the structure of the immature virus 104 ( Supplementary material S4). Even between the dengue serotypes 1, 2 and 4 for which there are cryo-EM structures 105, 106 it is apparent while the rafts are very similar (as are the sequence identities [60%]) there is a different charge distribution of the surface of each. Dengue serotype 2 had larger continuous patches of positive charges which was proposed to enable improved binding to heparan sulfate. This might also be the case for ZIKV in that the charge pattern is again different and could be key for vaccine development. The availability of virion structures makes it feasible to understand structure function of the complete virus such as assessment of membrane curvature and how organization of membrane proteins affects this 43.
A model of the Zika virion was constructed as an illustration using the homology model of the glycoprotein E dimer ( Figure 3). While the combined protein sequence of glycoprotein E and the immunoglobulin like domain is closest to dengue virus 1 (57 percent identity, Table 1) the closest template was for the crystal structure of the Japanese encephalitis virus envelope protein (53.12 percent identity, Table 2). This would suggest the virion should more closely resemble that of dengue virus 1, while producing a homology model based on a more distant virus might not be ideal. The homology model of glycoprotein E developed for the mature conformation in this study is significantly different from that developed previously for the immature conformation ( Figure 4). The proposed binding site centered around residues 270-277 appears shallower in the mature conformation and this would certainly affect the kinds of molecules that it could interact with. It might also point to the need to interfere with the immature conformation as preferable versus the mature conformation. Ultimately perhaps this model of the Zika virion could help us understand how drugs could access the virus. Viruses affecting pregnancy, like say Varicella which causes microcephaly and other developmental problems 106, 107, are often treated with IV immunoglobulin, i.e. antibodies, as well as antivirals to reduce the effect of the virus (or to avoid infection if given soon after exposure). The models could help us design combination approaches possibly targeting multiple proteins that might prevent drug resistance from occurring also.
Does having the homology models and the virion illustration help understand function? Well, the surface charge pattern might be inferred from the homology model and could be compared with dengue and other filoviruses for which there are cryo-EM structures. This may in turn present opportunities for vaccine design by indicating accessible surfaces and properties, allowing mapping of epitopes, design of accessible fragments and peptides for vaccine/diagnostic design. Vaccines themselves might be the only way to avoid the inevitable, otherwise, simply reducing the spread of ZIKV would just delay it. Women ultimately may just want to ‘get it over with’ and have ZIKV before they get pregnant and hope there is lasting immunity.
In summary, in the absence of crystal structures for any of the proteins comprising the ZIKV, we are left to attempt to construct homology models which we have done using the freely available SWISS-MODEL server. Further preparation of these models required freely available and commercial tools. In the case of the ZIKV glycoprotein E homology model, this has the added benefit of enabling the construction of a full virion. By comparing the Zika virion to the existing structures for other flaviviruses we can see similarities and differences on the surface ( Supplementary material S4, Supplementary material S5). This relatively crude approach could help to understand how we might develop antivirals and vaccines against it. In addition we now provide homology models as a starting point for (small and large scale) docking studies and further evaluation which may complement other modeling efforts for ZIKV 110. Ultimately the results of their use can be compared with using ZIKV crystal structures once generated.
Acknowledgments
Dr’s Priscilla Yang, Andrew Marsh, Derek Gatherer, Lucio Freitas-Junior, Daniel Mietchen, Joel S Freundlich, Jair Siqueira-Neto, Antony J. Williams, Alex Perryman and Mr. Tom Stratton are thanked for their helpful discussions and tweets. Biovia is kindly acknowledged for providing Discovery Studio to SE.
Funding Statement
CS was supported by Wellcome Trust Grant (to the IUPHAR/BPS Guide to PHARMACOLOGY) Number 099156/Z/12/Z.
I confirm that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 2 approved with reservations]
Supplementary material
.
.
References
- 1. Mlera L, Melik W, Bloom ME: The role of viral persistence in flavivirus biology. Pathog Dis. 2014;71(2):137–63. 10.1111/2049-632X.12178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pierson TC, Kielian M: Flaviviruses: braking the entering. Curr Opin Virol. 2013;3(1):3–12. 10.1016/j.coviro.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fauci AS, Morens DM: Zika Virus in the Americas--Yet Another Arbovirus Threat. N Engl J Med. 2016;374(7):601–4. 10.1056/NEJMp1600297 [DOI] [PubMed] [Google Scholar]
- 4. Ekins S, Mietchen D, Coffee M, et al. : Open drug discovery for the Zika virus [version 1; referees: awaiting peer review]. F1000Res. 2016;5:150 10.12688/f1000research.8013.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Martines RB, Bhatnagar J, Keating MK, et al. : Notes from the Field: Evidence of Zika Virus Infection in Brain and Placental Tissues from Two Congenitally Infected Newborns and Two Fetal Losses - Brazil, 2015. MMWR Morb Mortal Wkly Rep. 2016;65(6):159–60. 10.15585/mmwr.mm6506e1 [DOI] [PubMed] [Google Scholar]
- 6. Mlakar J, Korva M, Tul N, et al. : Zika Virus Associated with Microcephaly. N Engl J Med. 2016. 10.1056/NEJMoa1600651 [DOI] [PubMed] [Google Scholar]
- 7. Rubin EJ, Greene MF, Baden LR: Zika Virus and Microcephaly. N Engl J Med. 2016. 10.1056/NEJMe1601862 [DOI] [PubMed] [Google Scholar]
- 8. Heymann DL, Hodgson A, Sall AA, et al. : Zika virus and microcephaly: why is this situation a PHEIC? Lancet. 2016;387(10020):719–21. 10.1016/S0140-6736(16)00320-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bell TM, Field EJ, Narang HK: Zika virus infection of the central nervous system of mice. Arch Gesamte Virusforsch. 1971;35(2):183–93. 10.1007/BF01249709 [DOI] [PubMed] [Google Scholar]
- 10. Vervaeke P, Alen M, Noppen S, et al. : Sulfated Escherichia coli K5 polysaccharide derivatives inhibit dengue virus infection of human microvascular endothelial cells by interacting with the viral envelope protein E domain III. PLoS One. 2013;8(8):e74035. 10.1371/journal.pone.0074035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hamel R, Dejarnac O, Wichit S, et al. : Biology of Zika Virus Infection in Human Skin Cells. J Virol. 2015;89(17):8880–96. 10.1128/JVI.00354-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anon. Zika and Sexual Transmission.2016. Reference Source [Google Scholar]
- 13. Oduyebo T, Petersen EE, Rasmussen SA, et al. : Update: Interim Guidelines for Health Care Providers Caring for Pregnant Women and Women of Reproductive Age with Possible Zika Virus Exposure - United States, 2016. MMWR Morb Mortal Wkly Rep. 2016;65(5):122–7. 10.15585/mmwr.mm6505e2 [DOI] [PubMed] [Google Scholar]
- 14. Musso D, Nhan T, Robin E, et al. : Potential for Zika virus transmission through blood transfusion demonstrated during an outbreak in French Polynesia, November 2013 to February 2014. Euro Surveill. 2014;19(14). pii: 20761. 10.2807/1560-7917.ES2014.19.14.20761 [DOI] [PubMed] [Google Scholar]
- 15. Aubry M, Richard V, Green J, et al. : Inactivation of Zika virus in plasma with amotosalen and ultraviolet A illumination. Transfusion. 2016;56(1):33–40. 10.1111/trf.13271 [DOI] [PubMed] [Google Scholar]
- 16. Gyurech D, Schilling J, Schmidt-Chanasit J, et al. : False positive dengue NS1 antigen test in a traveller with an acute Zika virus infection imported into Switzerland. Swiss Med Wkly. 2016;146:w14296. [DOI] [PubMed] [Google Scholar]
- 17. Faye O, Freire CC, Iamarino A, et al. : Molecular evolution of Zika virus during its emergence in the 20 th century. PLoS Negl Trop Dis. 2014;8(1):e2636. 10.1371/journal.pntd.0002636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alen MM, Kaptein SJ, De Burghgraeve T, et al. : Antiviral activity of carbohydrate-binding agents and the role of DC-SIGN in dengue virus infection. Virology. 2009;387(1):67–75. 10.1016/j.virol.2009.01.043 [DOI] [PubMed] [Google Scholar]
- 19. Lei Y, Yu H, Dong Y, et al. : Characterization of N-Glycan Structures on the Surface of Mature Dengue 2 Virus Derived from Insect Cells. PLoS One. 2015;10(7):e0132122. 10.1371/journal.pone.0132122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nishikawa I, Nakajima Y, Ito M, et al. : Computational prediction of O-linked glycosylation sites that preferentially map on intrinsically disordered regions of extracellular proteins. Int J Mol Sci. 2010;11(12):4991–5008. 10.3390/ijms11124991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Z, Khaliq M, Zhou Z, et al. : Design, synthesis, and biological evaluation of antiviral agents targeting flavivirus envelope proteins. J Med Chem. 2008;51(15):4660–71. 10.1021/jm800412d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Poh MK, Yip A, Zhang S, et al. : A small molecule fusion inhibitor of dengue virus. Antiviral Res. 2009;84(3):260–6. 10.1016/j.antiviral.2009.09.011 [DOI] [PubMed] [Google Scholar]
- 23. Wang QY, Patel SJ, Vangrevelinghe E, et al. : A small-molecule dengue virus entry inhibitor. Antimicrob Agents Chemother. 2009;53(5):1823–31. 10.1128/AAC.01148-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mayhoub AS, Khaliq M, Kuhn RJ, et al. : Design, synthesis, and biological evaluation of thiazoles targeting flavivirus envelope proteins. J Med Chem. 2011;54(6):1704–14. 10.1021/jm1013538 [DOI] [PubMed] [Google Scholar]
- 25. Schmidt AG, Lee K, Yang PL, et al. : Small-molecule inhibitors of dengue-virus entry. PLoS Pathog. 2012;8(4):e1002627. 10.1371/journal.ppat.1002627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou Z, Khaliq M, Suk JE, et al. : Antiviral compounds discovered by virtual screening of small-molecule libraries against dengue virus E protein. ACS Chem Biol. 2008;3(12):765–75. 10.1021/cb800176t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stahla-Beek HJ, April DG, Saeedi BJ, et al. : Identification of a novel antiviral inhibitor of the flavivirus guanylyltransferase enzyme. J Virol. 2012;86(16):8730–9. 10.1128/JVI.00384-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Behnam MA, Nitsche C, Boldescu V, et al. : The Medicinal Chemistry of Dengue Virus. J Med Chem. 2016. 10.1021/acs.jmedchem.5b01653 [DOI] [PubMed] [Google Scholar]
- 29. Vincetti P, Caporuscio F, Kaptein S, et al. : Discovery of Multitarget Antivirals Acting on Both the Dengue Virus NS5-NS3 Interaction and the Host Src/Fyn Kinases. J Med Chem. 2015;58(12):4964–75. 10.1021/acs.jmedchem.5b00108 [DOI] [PubMed] [Google Scholar]
- 30. Shum D, Smith JL, Hirsch AJ, et al. : High-content assay to identify inhibitors of dengue virus infection. Assay Drug Dev Technol. 2010;8(5):553–70. 10.1089/adt.2010.0321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Crance JM, Scaramozzino N, Jouan A, et al. : Interferon, ribavirin, 6-azauridine and glycyrrhizin: antiviral compounds active against pathogenic flaviviruses. Antiviral Res. 2003;58(1):73–9. 10.1016/S0166-3542(02)00185-7 [DOI] [PubMed] [Google Scholar]
- 32. Raska K, Jr, Zedeck MS, Welch AD: Relationship between the metabolic effects and the pregnancy-interrupting property of 6-azauridine in mice. Biochem Pharmacol. 1966;15(12):2136–8. 10.1016/0006-2952(66)90250-4 [DOI] [PubMed] [Google Scholar]
- 33. Kochhar DM, Penner JD, Knudsen TB: Embryotoxic, teratogenic, and metabolic effects of ribavirin in mice. Toxicol Appl Pharmacol. 1980;52(1):99–112. 10.1016/0041-008X(80)90252-5 [DOI] [PubMed] [Google Scholar]
- 34. Entrican G: Immune regulation during pregnancy and host-pathogen interactions in infectious abortion. J Comp Pathol. 2002;126(2–3):79–94. 10.1053/jcpa.2001.0539 [DOI] [PubMed] [Google Scholar]
- 35. Blatt J, Farag S, Corey SJ, et al. : Expanding the scope of drug repurposing in pediatrics: the Children's Pharmacy Collaborative. Drug Discov Today. 2014;19(11):1696–8. 10.1016/j.drudis.2014.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klein DE, Choi JL, Harrison SC: Structure of a dengue virus envelope protein late-stage fusion intermediate. J Virol. 2013;87(4):2287–93. 10.1128/JVI.02957-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Modis Y, Ogata S, Clements D, et al. : A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci U S A. 2003;100(12):6986–91. 10.1073/pnas.0832193100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu J, Bera AK, Kuhn RJ, et al. : Structure of the Flavivirus helicase: implications for catalytic activity, protein interactions, and proteolytic processing. J Virol. 2005;79(16):10268–77. 10.1128/JVI.79.16.10268-10277.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Geiss BJ, Thompson AA, Andrews AJ, et al. : Analysis of flavivirus NS5 methyltransferase cap binding. J Mol Biol. 2009;385(5):1643–54. 10.1016/j.jmb.2008.11.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Y, Corver J, Chipman PR, et al. : Structures of immature flavivirus particles. EMBO J. 2003;22(11):2604–13. 10.1093/emboj/cdg270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brecher M, Chen H, Li Z, et al. : Identification and Characterization of Novel Broad-Spectrum Inhibitors of the Flavivirus Methyltransferase. ACS Infect Dis. 2015;1(8):340–349. 10.1021/acsinfecdis.5b00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Akey DL, Brown WC, Konwerski JR, et al. : Use of massively multiple merged data for low-resolution S-SAD phasing and refinement of flavivirus NS1. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 10):2719–29. 10.1107/S1399004714017556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang W, Kaufmann B, Chipman PR, et al. : Membrane curvature in flaviviruses. J Struct Biol. 2013;183(1):86–94. 10.1016/j.jsb.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Aleshin AE, Shiryaev SA, Strongin AY, et al. : Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold. Protein Sci. 2007;16(5):795–806. 10.1110/ps.072753207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kuno G, Chang GJ, Tsuchiya KR, et al. : Phylogeny of the genus Flavivirus. J Virol. 1998;72(1):73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anon. BLAST. Reference Source [Google Scholar]
- 47. Altschul SF, Gish W, Miller W, et al. : Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 48. Baronti C, Piorkowski G, Charrel RN, et al. : Complete coding sequence of Zika virus from a French polynesia outbreak in 2013. Genome Announc. 2014;2(3): pii: e00500-14. 10.1128/genomeA.00500-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Benson DA, Cavanaugh M, Clark K, et al. : GenBank. Nucleic Acids Res. 2013;41(Database issue):D36–42. 10.1093/nar/gks1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Biasini M, Bienert S, Waterhouse A, et al. : SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(Web Server issue):W252–8. 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bordoli L, Kiefer F, Arnold K, et al. : Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4(1):1–13. 10.1038/nprot.2008.197 [DOI] [PubMed] [Google Scholar]
- 52. Benkert P, Tosatto SC, Schomburg D: QMEAN: A comprehensive scoring function for model quality assessment. Proteins. 2008;71(1):261–77. 10.1002/prot.21715 [DOI] [PubMed] [Google Scholar]
- 53. Benkert P, Künzli M, Schwede T: QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009;37(Web Server issue):W510–4. 10.1093/nar/gkp322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anon. The Structure Analysis and Verification Server. Reference Source [Google Scholar]
- 55.Anon. PROCHECK and PROCHECK-NMR. Reference Source [Google Scholar]
- 56.Anon. N-GlycoSite. Reference Source [Google Scholar]
- 57.Anon. GlycoEP. Reference Source [Google Scholar]
- 58. Chauhan JS, Rao A, Raghava GP: In silico platform for prediction of N-, O- and C-glycosites in eukaryotic protein sequences. PLoS One. 2013;8(6):e67008. 10.1371/journal.pone.0067008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anon. NetNGlyc. Reference Source [Google Scholar]
- 60. Kuhn RJ, Zhang W, Rossmann MG, et al. : Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108(5):717–725. 10.1016/S0092-8674(02)00660-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lu G, Gong P: Crystal Structure of the full-length Japanese encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface. PLoS Pathog. 2013;9(8):e1003549. 10.1371/journal.ppat.1003549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhou Y, Ray D, Zhao Y, et al. : Structure and function of flavivirus NS5 methyltransferase. J Virol. 2007;81(8):3891–903. 10.1128/JVI.02704-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xu T, Sampath A, Chao A, et al. : Structure of the Dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 Å. J Virol. 2005;79(16):10278–88. 10.1128/JVI.79.16.10278-10288.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mancini EJ, Assenberg R, Verma A, et al. : Structure of the Murray Valley encephalitis virus RNA helicase at 1.9 Angstrom resolution. Protein Sci. 2007;16(10):2294–300. 10.1110/ps.072843107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hammamy MZ, Haase C, Hammami M, et al. : Development and characterization of new peptidomimetic inhibitors of the West Nile virus NS2B-NS3 protease. ChemMedChem. 2013;8(2):231–41. 10.1002/cmdc.201200497 [DOI] [PubMed] [Google Scholar]
- 66. Akey DL, Brown WC, Dutta S, et al. : Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science. 2014;343(6173):881–5. 10.1126/science.1247749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang X, Ge P, Yu X, et al. : Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat Struct Mol Biol. 2013;20(1):105–10. 10.1038/nsmb.2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li L, Lok SM, Yu IM, et al. : The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science. 2008;319(5871):1830–4. 10.1126/science.1153263 [DOI] [PubMed] [Google Scholar]
- 69. Dokland T, Walsh M, Mackenzie JM, et al. : West Nile virus core protein; tetramer structure and ribbon formation. Structure. 2004;12(7):1157–63. 10.1016/j.str.2004.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Luca VC, AbiMansour J, Nelson CA, et al. : Crystal structure of the Japanese encephalitis virus envelope protein. J Virol. 2012;86(4):2337–46. 10.1128/JVI.06072-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Arnold K, Bordoli L, Kopp J, et al. : The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. 10.1093/bioinformatics/bti770 [DOI] [PubMed] [Google Scholar]
- 72. Kiefer F, Arnold K, Künzli M, et al. : The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37(Database issue):D387–92. 10.1093/nar/gkn750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Guex N, Peitsch MC, Schwede T: Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. 2009;30(Suppl 1):S162–73. 10.1002/elps.200900140 [DOI] [PubMed] [Google Scholar]
- 74. Kokernot RH, Smithburn KC, Muspratt J, et al. : Studies on arthropod-borne viruses of Tongaland. VIII. Spondweni virus, an agent previously unknown, isolated from Taeniorhynchus (Mansonioides) uniformis. S Afr J Med Sci. 1957;22(2–3):103–12. [PubMed] [Google Scholar]
- 75. Tognarelli J, Ulloa S, Villagra E, et al. : A report on the outbreak of Zika virus on Easter Island, South Pacific, 2014. Arch Virol. 2016;161(3):665–8. 10.1007/s00705-015-2695-5 [DOI] [PubMed] [Google Scholar]
- 76. Bessaud M, Pastorino BA, Peyrefitte CN, et al. : Functional characterization of the NS2B/NS3 protease complex from seven viruses belonging to different groups inside the genus Flavivirus. Virus Res. 2006;120(1–2):79–90. 10.1016/j.virusres.2006.01.021 [DOI] [PubMed] [Google Scholar]
- 77.Anon. VIZIER. Reference Source [Google Scholar]
- 78. Bollati M, Alvarez K, Assenberg R, et al. : Structure and functionality in flavivirus NS-proteins: perspectives for drug design. Antiviral Res. 2010;87(2):125–48. 10.1016/j.antiviral.2009.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Parsy CC, Alexandre FR, Bidau V, et al. : Discovery and structural diversity of the hepatitis C virus NS3/4A serine protease inhibitor series leading to clinical candidate IDX320. Bioorg Med Chem Lett. 2015;25(22):5427–36. 10.1016/j.bmcl.2015.09.009 [DOI] [PubMed] [Google Scholar]
- 80. Jiang Y, Andrews SW, Condroski KR, et al. : Discovery of danoprevir (ITMN-191/R7227), a highly selective and potent inhibitor of hepatitis C virus (HCV) NS3/4A protease. J Med Chem. 2014;57(5):1753–69. 10.1021/jm400164c [DOI] [PubMed] [Google Scholar]
- 81. Sheng XC, Casarez A, Cai R, et al. : Discovery of GS-9256: a novel phosphinic acid derived inhibitor of the hepatitis C virus NS3/4A protease with potent clinical activity. Bioorg Med Chem Lett. 2012;22(3):1394–6. 10.1016/j.bmcl.2011.12.038 [DOI] [PubMed] [Google Scholar]
- 82. Schoenfeld RC, Bourdet DL, Brameld KA, et al. : Discovery of a novel series of potent non-nucleoside inhibitors of hepatitis C virus NS5B. J Med Chem. 2013;56(20):8163–82. 10.1021/jm401266k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kazmierski WM, Hamatake R, Duan M, et al. : Discovery of novel urea-based hepatitis C protease inhibitors with high potency against protease-inhibitor-resistant mutants. J Med Chem. 2012;55(7):3021–6. 10.1021/jm201278q [DOI] [PubMed] [Google Scholar]
- 84. Rosenquist Å, Samuelsson B, Johansson PO, et al. : Discovery and development of simeprevir (TMC435), a HCV NS3/4A protease inhibitor. J Med Chem. 2014;57(5):1673–93. 10.1021/jm401507s [DOI] [PubMed] [Google Scholar]
- 85. Cummings MD, Lin TI, Hu L, et al. : Discovery and early development of TMC647055, a non-nucleoside inhibitor of the hepatitis C virus NS5B polymerase. J Med Chem. 2014;57(5):1880–92. 10.1021/jm401396p [DOI] [PubMed] [Google Scholar]
- 86. Link JO, Taylor JG, Xu L, et al. : Discovery of ledipasvir (GS-5885): a potent, once-daily oral NS5A inhibitor for the treatment of hepatitis C virus infection. J Med Chem. 2014;57(5):2033–46. 10.1021/jm401499g [DOI] [PubMed] [Google Scholar]
- 87. Zhang Y: I-TASSER. Reference Source [Google Scholar]
- 88. Zhang Y: I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. 10.1186/1471-2105-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yang J, Yan R, Roy A, et al. : The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12(1):7–8. 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Noble CG, Lim SP, Chen YL, et al. : Conformational Flexibility of the Dengue Virus RNA-Dependent RNA Polymerase Revealed by a Complex with an Inhibitor. J Virol. 2013;87(9):5291–5295. 10.1128/JVI.00045-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Flower DR: Bioinformatics for vaccinology. Chichester: Wiley-Blackwell,2008. 10.1002/9780470699836 [DOI] [Google Scholar]
- 92. Du H, Brender JR, Zhang J, et al. : Protein structure prediction provides comparable performance to crystallographic structures in docking-based virtual screening. Methods. 2015;71:77–84. 10.1016/j.ymeth.2014.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Levoin N, Calmels T, Krief S, et al. : Homology Model Versus X-ray Structure in Receptor-based Drug Design: A Retrospective Analysis with the Dopamine D3 Receptor. ACS Med Chem Lett. 2011;2(4):293–7. 10.1021/ml100288q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Carlsson J, Coleman RG, Setola V, et al. : Ligand discovery from a dopamine D 3 receptor homology model and crystal structure. Nat Chem Biol. 2011;7(11):769–78. 10.1038/nchembio.662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Godbole AA, Ahmed W, Bhat RS, et al. : Targeting Mycobacterium tuberculosis topoisomerase I by small-molecule inhibitors. Antimicrob Agents Chemother. 2015;59(3):1549–57. 10.1128/AAC.04516-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Godbole AA, Ahmed W, Bhat RS, et al. : Inhibition of Mycobacterium tuberculosis topoisomerase I by m-AMSA, a eukaryotic type II topoisomerase poison. Biochem Biophys Res Commun. 2014;446(4):916–20. 10.1016/j.bbrc.2014.03.029 [DOI] [PubMed] [Google Scholar]
- 97. Tan K, Cao N, Cheng B, et al. : Insights from the Structure of Mycobacterium tuberculosis Topoisomerase I with a Novel Protein Fold. J Mol Biol. 2016;428(1):182–93. 10.1016/j.jmb.2015.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Nguyen ED, Norn C, Frimurer TM, et al. : Assessment and challenges of ligand docking into comparative models of G-protein coupled receptors. PLoS One. 2013;8(7):e67302. 10.1371/journal.pone.0067302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. da Silveira NJ, Arcuri HA, Bonalumi CE, et al. : Molecular models of NS3 protease variants of the Hepatitis C virus. BMC Struct Biol. 2005;5:1. 10.1186/1472-6807-5-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lee TV, Johnson RD, Arcus VL, et al. : Prediction of the substrate for nonribosomal peptide synthetase (NRPS) adenylation domains by virtual screening. Proteins. 2015;83(11):2052–66. 10.1002/prot.24922 [DOI] [PubMed] [Google Scholar]
- 101.Anon. PEPI-TiDP23-C103: First-in-Human Study to Examine the Safety, Tolerability, and Plasma Pharmacokinetics of Increasing Single and Repeated Oral Doses of TMC558445 and of a Combined Single Day Dosing of Oral TMC558445 and Oral TMC310911 and Also Oral Darunavir.2009. Reference Source [Google Scholar]
- 102. Keck ZY, Enterlein SG, Howell KA, et al. : Macaque Monoclonal Antibodies Targeting Novel Conserved Epitopes within Filovirus Glycoprotein. J Virol. 2015;90(1):279–91. 10.1128/JVI.02172-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ferlenghi I, Clarke M, Ruttan T, et al. : Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol Cell. 2001;7(3):593–602. 10.1016/S1097-2765(01)00206-4 [DOI] [PubMed] [Google Scholar]
- 104. Zhang Y, Kaufmann B, Chipman PR, et al. : Structure of immature West Nile virus. J Virol. 2007;81(11):6141–5. 10.1128/JVI.00037-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kostyuchenko VA, Zhang Q, Tan JL, et al. : Immature and mature dengue serotype 1 virus structures provide insight into the maturation process. J Virol. 2013;87(13):7700–7. 10.1128/JVI.00197-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kostyuchenko VA, Chew PL, Ng TS, et al. : Near-atomic resolution cryo-electron microscopic structure of dengue serotype 4 virus. J Virol. 2014;88(1):477–82. 10.1128/JVI.02641-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhang Y, Zhang W, Ogata S, et al. : Conformational changes of the flavivirus E glycoprotein. Structure. 2004;12(9):1607–18. 10.1016/j.str.2004.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Scheffer IE, Baraitser M, Brett EM: Severe microcephaly associated with congenital varicella infection. Dev Med Child Neurol. 1991;33(10):916–20. 10.1111/j.1469-8749.1991.tb14803.x [DOI] [PubMed] [Google Scholar]
- 109. Deasy NP, Jarosz JM, Cox TC, et al. : Congenital varicella syndrome: cranial MRI in a long-term survivor. Neuroradiology. 1999;41(3):205–7. 10.1007/s002340050736 [DOI] [PubMed] [Google Scholar]
- 110. Gatherer D: Zika virus protein structure homology modelling.2016. Reference Source [Google Scholar]
- 111.Anon. EMDataBank. Reference Source [Google Scholar]