Abstract
Sperm Lysozyme-Like Protein 1 (SLLP1) is one of the lysozyme-like proteins predominantly expressed in mammalian testes that lacks bacteriolytic activity, localizes in the sperm acrosome, and exhibits high affinity for an oolemmal receptor, SAS1B. The crystal structure of mouse SLLP1 (mSLLP1) was determined at 2.15Å resolution. mSLLP1 monomer adopts a structural fold similar to that of chicken/mouse lysozymes retaining all four canonical disulfide bonds. mSLLP1 is distinct from c-lysozyme by substituting two essential catalytic residues (E35T/D52N), exhibiting different surface charge distribution, and by forming helical filaments approximately 75Å in diameter with a 25Å central pore comprised of six monomers per helix turn repeating every 33Å. Cross-species alignment of all reported SLLP1 sequences revealed a set of invariant surface regions comprising a characteristic fingerprint uniquely identifying SLLP1 from other c-lysozyme family members. The fingerprint surface regions reside around the lips of the putative glycan binding groove including three polar residues (Y33/E46/H113). A flexible salt bridge (E46-R61) was observed covering the glycan binding groove. The conservation of these regions may be linked to their involvement in oolemmal protein binding. Interaction between SLLP1 monomer and its oolemmal receptor SAS1B was modeled using protein-protein docking algorithms, utilizing the SLLP1 fingerprint regions along with the SAS1B conserved surface regions. This computational model revealed complementarity between the conserved SLLP1/SAS1B interacting surfaces supporting the experimentally-observed SLLP1/SAS1B interaction involved in fertilization.
Keywords: SLLP1, crystal structure, refolding, SAS1B, protein-protein interaction, fertilization
Introduction
Mammalian fertilization provides an example of several inter-cellular processes, including cell specific sperm-egg recognition, inter-cellular adhesion, signaling, regulated exocytosis, fusion, phagocytosis, and sperm internalization (Primakoff & Myles, 2002), which ultimately initiate downstream intra-cellular processes of nuclear fusion and syngamy. Structural analyses of sperm specific proteins that exhibit high affinity interactions with egg specific surface proteins may reveal key information to elucidate molecular interactions before and during the fertilization process. Testicular proteins are also gaining considerable interest as cancer-testis antigens for immunotherapy in tumor treatment (Atanackovic et al., 2006) including Sperm Lysozyme-Like Protein 1 (SLLP1) (Wang et al., 2004). Although several sperm proteins have been implicated as binding partner pairs in various stages of the fertilization process (Vjugina & Evans, 2008), study of the 3-D structures engaged in these interactions is rudimentary. Only a few structures of sperm and egg proteins have been reported to date including spermadhesin (Romero et al., 1997), β-acrosin (Tranter et al., 2000), Sp18 (Kresge et al., 2001), MSP (Miao et al., 2008), MFP2 (Grant et al., 2005), A1BP (Jha et al., 2008), GAPDH (Frayne et al., 2009) and ZP3 (Han et al., 2010). In particular, no complex structure has been determined for a sperm protein implicated in sperm-oolemma binding, nor have 3-D models been advanced for a sperm ligand complexed with its oocyte binding partner.
SLLP1 was localized in the sperm acrosome and named after its high sequence homology with c-lysozyme (~50% sequence identity and >60% sequence similarity) (Mandal et al., 2003). Evolutionarily, SLLP1 is present exclusively in mammals, with orthologues identified in 20 species including those from Proboscidea and Primates, and across mammalian species SLLP1 shows very high sequence identity (≥73%). The 5.0–5.2 isoelectric point of SLLP1 is significantly more acidic than the pI of c-lysozyme (10.7) due to the prevalence of acidic residues on the protein surface. In contrast to c-lysozyme SLLP1 lacks bacteriolytic activity (Mandal et al., 2003) but binds to both unfertilized and fertilized eggs (Herrero et al., 2005). As revealed by fluorescence microscopy, SLLP1 binds predominantly to the oocyte perivitelline matrix and plasma membrane, showing particularly strong binding with the microvillar region of the ovulated mouse oocyte, the domain where fertilization occurs (Herrero et al., 2005). Both recombinant SLLP1 and its antiserum block sperm-egg interaction and fusion, indicating the possible role of SLLP1 in sperm/egg adhesion (Herrero et al., 2005). Thus the interaction of SLLP1 with candidate binding partners on the oocyte surface is believed to be important in mediating fertilization (Sachdev et al., 2012).
Several lines of evidence support the interaction of SLLP1 with the oocyte specific astacin-like metalloprotease SAS1B (Sperm Acrosomal SLLP1 Binding; a.k.a. ovastacin) (Quesada et al., 2004; Sachdev et al., 2012). These include anatomical co-localization, co-immunoprecipitation, yeast-two-hybrid and direct measurements of affinity (Sachdev et al., 2012). The SLLP1-SAS1B interaction has been reported to have a KD of 0.32 nM (Sachdev et al., 2012). Knockout (Burkart et al., 2012; Sachdev et al., 2012) of the SLLP1 oolemmal binding partner protein SAS1B in mice resulted in subfertility. In addition, SAS1B protein (ovastacin) has also been demonstrated localize in cortical granules in the sub-oolemmal cytoplasm (Pires et al., 2013) with ZP2-cleavage activity (Avella et al., 2013; Burkart et al., 2012) that could be inhibited by fetuin-B (Dietzel et al., 2013; Stocker et al., 2014), which reflects its role in the block to polyspermy following fertilization. Moreover, our recent study shows the presence of six alternative splice variants for SAS1B (Pires et al., 2013), which may relate to the versatile roles this protein plays during fertilization.
In the current study, the crystal structure of mouse SLLP1 (mSLLP1) was determined at 2.15Å resolution. In the crystal, mSLLP1 molecules formed helical filaments approximately 75Å in diameter with a 25Å central pore, a 33Å pitch, and ~15° helix angle. The helical filament contained six mSLLP1 monomers per helical turn. mSLLP1 monomer adopts a fold similar to that of c-lysozyme but with a modified groove corresponding to the substrate-binding site in c-lysozyme. In addition, the surface residues as well as surface charge differ significantly between c-lysozyme and mSLLP1. The structure of the mSLLP1 monomer was utilized to build a computation model of its complex with mouse SAS1B (mSAS1B) to predict and understand interacting surfaces mediating SLLP1 and SAS1B binding.
Materials and Methods
Cloning, Expression, and Protein Purification
The Sperm acrosome associated 3 (SPACA3) gene from Mus musculus that encodes the mSLLP1 protein was amplified from a cDNA library by PCR and cloned into the pET28b+ vector (Novagen Inc., Madison, WI) using NcoI and XhoI sites as described previously (Herrero et al., 2005). This construct possessed two additional N-terminal residues (MA) and eight additional C-terminal residues (LEHHHHHH) including 6-His to facilitate nickel affinity purification. The resulting plasmid was transformed into E. coli BL21(DE3). Colonies of the fresh transformants were inoculated into 50 mL Luria broth (LB) culture with a kanamycin concentration of 50µg/mL, followed by inoculation into four liters of LB media in baffled-flasks at 37°C. When cell growth reached an optical density A600 of ~0.8, mSLLP1 expression was induced by addition of 1mM isopropyl-1-thio-D-galactopyranoside (IPTG) to the culture and cultures were grown at 37°C for another 4 hours. Cells were harvested by centrifugation and the cell pellets were sonicated in lysis buffer (200 mM NaCl, 50 mM Tris pH 7.5) containing 2 protease inhibitors (1mM phenylmethylsulfonyl fluoride (PMSF) and 1 mM benzamidine). The lysis mixture was subjected to centrifugation and the pellets containing inclusion bodies were collected. The inclusion bodies were solubilized in 200 mM NaCl, 50 mM Tris pH 7.5, 8M Urea, 5 mM imidazole for 4 hours at 4°C and cleared by centrifugation. The supernatant was applied to Ni–NTA affinity resin (Qiagen, USA). The column was washed thoroughly to remove nonspecifically bound contaminants with 200 mM NaCl, 50 mM Tris pH 7.5, 8M Urea, 30 mM imidazole. The bound His-tagged mSLLP1 protein was eluted with 200 mM NaCl, 50 mM Tris pH 7.5, 8M Urea, 250 mM imidazole (15 mL). The protein concentration in the eluate was quantified with the Bradford assay (Bio-Rad Protein Assay).
Refolding and further purification
The purified mSLLP1 (~1 mg/mL) was refolded by fast dilution with refolding buffer containing 200 mM NaCl, 50 mM Tris pH 8.5, 5mM EDTA, 0.4mM glutathione, and 2mM oxidized glutathione. 15 ml of a 1 mg/mL solution of mSLLP1 containing 8 M urea was titrated into an equal volume of refolding buffer dropwise and stirred for 8 hours under reduced pressure (non-aerated conditions, low oxygen) at room temperature. The same dilution-titration was repeated 3 more times to reach a final volume of 240 mL with a urea concentration of 0.5 M. The protein solution was then concentrated and buffer-exchanged to a final concentration of ~15 mg/mL in 700 µL buffer (50mM NaCl, 10 mM HEPES pH7.5) with a Biomax concentrator (Millipore Inc., Bedford, MA) and purified in the same buffer by size-exclusion chromatography using a Superdex-200 gel filtration column 10/60 with 1 mL/min flow rate (Amersham Pharmacia Biotech Inc., Piscataway, NJ). Two fractions were collected from size exclusion chromatography. The major fraction was discarded and the minor monomeric fraction (retention time ~98.0 minutes) was collected and concentrated (Supplemental Fig. 1). Various attempts to crystallize protein from this preparation failed.
The same mSLLP1 preparation was further subjected to unfolding, followed by incubation with SulfoLink iodine resin (Pierce, Rockford, IL) for 24 hours. Improperly folded mSLLP1 molecules that possessed free thiol groups from cysteine residues were immobilized on SulfoLink resin. Properly folded mSLLP1 molecules that had all eight cysteine residues engaged in disulfide bridges did not interact with SulfoLink and remained in solution. This supernatant was then subjected to a second size-exclusion chromatography using Superdex-200 gel filtration column following same process described above. A monomeric peak of mSLLP1 (Retention time 99.8 minutes) was collected, concentrated and diffraction quality crystals were successfully obtained by crystallization (Supplemental Fig. 1).
Egg plasma membrane binding using recombinant refolded mSLLP1
An oocyte binding assay was performed to check by fluorescence microscopy the biological activity of mSLLP1 samples undergoing refolding (Herrero et al., 2005). ICR mice were used in all experiments. All animals were handled according to both the institutional guidelines approved by the animal care and use committee of the University of Virginia and the Guide for the Care and Use of Laboratory Animals (8th Edition) available from the Office of Laboratory Animal Welfare (OLAW) website (http://grants.nih.gov/grants/olaw/). 10–12 week old female mice were superovulated with 10 IU pregnant mare’s serum gonadotropin (PMSG), followed by intraperitoneal injection of 10 IU human chorionic gonadotrophin (hCG) after 48 hours. Mice were anesthetized by isoflurane and sacrificed 14 hours post hCG injection. Both oviducts were removed and overlaid immediately in 5% CO2 pre-equilibrated mineral oil. The oviducts were teased apart and the cumulus masses were collected. The cumulus cells were removed by hyaluronidase treatment to produce cumulus-free zona-intact oocytes. Half of the oocytes were treated by acid Tyrode’s solution (pH 2.5, Sigma, USA) for 25–30 seconds to remove the zona pellucida (ZP) and produce zona-free oocytes. Both zona-intact (+ZP) and zona-free (−ZP) oocytes were washed 5–7 times in 50 µL TYH medium and incubated in TYH medium at 5% CO2 to recover for 3 hours at 37°C. Both sets of oocytes were then incubated with 50 µg/mL refolded mSLLP1 at 5% CO2 for 1 h at 37°C. The resulting oocytes were further washed 5–7 times in medium with 5% normal goat serum (NGS) and incubated in the same media for 30 min followed by 5–7 washes with TYH medium followed by incubation with guinea pig anti-recombinant-mSLLP1 polyclonal antibody (1:50 in 5% NGS) at 5% CO2 for an hour at 37°C. The oocytes were then washed 5–7 times with 5% NGS media and incubated with goat anti-guinea pig/FITC antisera (1:200 in 5% NGS) at 5% CO2 for an additional hour at 37°C. Oocyte were washed 5 times with 5% NGS media and oocyte nuclei were visualized under a Zeiss Standard 18 ultraviolet microscope after staining with 1µg/mL Hoechst for 10 minutes and washing with NGS media. Fluorescent images of oocytes were captured with MrGrab 1.0 (Carl Zeiss Vision GmbH, Germany).
Crystallization, Data Collection and Processing
Crystallization was performed using the hanging drop vapor diffusion method at 21°C. Drops were composed of 2µL of the reservoir solution containing 100 mM Bis-Tris pH 6.5, 20% w/v PEG MME 5000, and 20% ethanol with equal volume of the concentrated refolded mSLLP1 (12 mg/mL) in crystallization buffer (200 mM NaCl, 10 mM HEPES pH 7.5). Needle-shaped crystals formed within 36 hours and grew to the final maximal size at around 72 hours (Supplemental Fig. 2). mSLLP1 crystals were flash cooled in liquid nitrogen immediately after harvesting. Diffraction images were collected at 100K on the 19-ID beamline operated by Structural Biology Center (SBC) (Rosenbaum et al., 2006) at the Advanced Photon Source (Argonne National Laboratory). Data collection was controlled and diffraction data were processed with HKL-2000 (Otwinowski & Minor, 1997).
Structure Determination, Refinement and Validation
Structure was determined by the molecular replacement (MR) technique. The MR search model was an mSLLP1 homology model constructed by MODELLER (Sali & Blundell, 1993) using hen egg white lysozyme (PDB code: 1LSG) as a template (Donahue et al., 1994). mSLLP1 crystallized in the space group P65 and contained three monomers in the asymmetric unit. The rotation matrix and translational vector for the first chain was found using EPMR (Kissinger et al., 2001) which utilizes an evolutionary search approach, resulting in a partial model that was used to search for additional copies of mSLLP1 molecule. The rotation matrices and translational vectors for both the second and third copies were found using MOLREP by supplying the best partial output model from previous MR run (Vagin & Teplyakov, 1997). Manual model optimization and refinement was performed in COOT (Emsley et al., 2010) with subsequent global refinement in REFMAC5 (Murshudov et al., 1997) and validation using Molprobity (Chen et al., 2010) as integrated in the HKL-3000 (Minor et al., 2006). All model quality indicators were either within the normal range or better. Data collection, structure determination, and refinement statistics are summarized in Table 1. The coordinates and experimental structure factors were deposited to the PDB with the accession code 4YF2.
Table 1.
Data collection, structure determination, and refinement statistics.
| PDB accession code | 4YF2 |
| Data Collection | |
| Wavelength (Å) | 0.9795 |
| Space group | P65 |
| Unit cell dimensions (Å, °) | a=136.6, b=136.6, c=33.3, 90.0, 90.0, 120.0 |
| Resolution Range (Å) a | 50.00–2.15 (2.19–2.15) |
| Rmerge b | 0.110 (0.918) |
| Mean I/σ | 7.0 (1.9) |
| Redundancy | 4.8 (4.7) |
| No. of unique reflections | 19,794 |
| Completeness (%) | 99.9 (100.0) |
| Structure Refinement | |
| Rwork/Rfree (%) | 18.8 / 21.7 |
| No. of protein atoms | 3046 |
| No. of solvent atoms | 132 |
| Average B-factor (Å2) | 45 |
| Geometrical RMS deviations | |
| bond length (Å) bond angle (°) |
0.017 1.7 |
| Ramachandran Plot (%) c | |
| Most favoured Allowed |
97.4 2.6 |
Data for the highest resolution shell are shown in parentheses.
Rmerge=Σ(|Ij-<I>|)/Σ<I>, where Ij is the observed intensity of reflection j and <I> is the average intensity of multiple observations. The calculation is performed for merged Bijovet pairs.
Ramachandran Plot statistics were generated by Molprobity.
Structure Analysis and Visualization
Molecular figures were prepared using PyMOL (Schrödinger, 2010). Surface Poisson-Boltzmann electrostatic potentials were generated by PDB2PQR (Dolinsky et al., 2007) and APBS tools (Baker et al., 2001). The sequence conservation score calculation was performed by the ConSurf server (Ashkenazy et al., 2010) using the empirical Bayesian method (Mayrose et al., 2004). Similar sequences were retrieved from UniProt with maximal sequence identity of 98% and an E-value cutoff of 0.0001. Since the current study involves proteins of different functions within the same c-lysozyme superfamily, the sequences used to calculate the conservation score were selected manually to ensure the correct functional family assignments. All SLLP1 orthologues used for multiple sequence alignment and invariant residues derivation were identified using HomoloGene (Feolo et al., 2000) with additional PSI-BLAST (Altschul & Koonin, 1998) using the gene name SPACA3 (Strausberg et al., 2002). SLLP2 orthologues were identified in a similar manner using its corresponding gene name SPACA5. Human SLLP2 homologous models were constructed by MODELLER (Sali & Blundell, 1993) using mSLLP1 structure as the template.
Defining and comparing invariant residues for highly conserved proteins
SLLP1 is highly conserved amongst mammals with pairwise sequence identity more than 73%. A method similar to Consurf (Ashkenazy et al., 2010) was used for analysis of both SLLP1 and lysozyme genes for identifying invariant residues of gene products and was performed independently for each gene across species. The consensus outcome profiles of these two genes across species were then compared to identify non-invariant residues. This protocol was further employed to characterize the most relevant residues that may be responsible for oocyte recognition from less significant ones. In this adapted procedure, all invariant residues were grouped into three categories compared to lysozyme (Supplemental Fig. 3) and mapped on the surface of the mSLLP1 structure (Supplemental Fig. 4). Invariant surface residues in SLLP1 proteins across species were identified by custom queries to the NEIGHBORHOOD database (Zheng et al., 2008). These invariant residues were further compared with a representative set of five lysozymes from different species (chicken, human, mouse, bovine stomach, and dog milk) to identify unique residues in SLLP1s across species. Invariant residues that were present in SLLP1 proteins across species but either absent or rarely observed in lysozymes were considered SLLP1 specific and potentially functionally relevant (colored in red). Other remaining invariant residues common to both SLLP1s and lysozyme were considered less relevant (colored in yellow). In addition, other conserved residues in lysozymes were considered important to maintain the enzymatic fold and were mainly present at the bottom of the putative glycan binding groove (colored in green). Invariant residues of lysozymes that were not observed in SLLP1s were employed as a positive control. The same analysis was also performed for SLLP2 (gene product of SPACA5) due to the high pairwise sequence identity (>79%) within the SLLP2 proteins across species.
Chitin binding assay
Recombinant chicken lysozyme and human SLLP1 (hSLLP1) were prepared by the same protocol of protein production and purification for mSLLP1 as described above. The bindings of recombinant mSLLP1, recombinant hSLLP1 and recombinant chicken lysozyme to chitin were compared (in a qualitative and not quantitative manner) by applying each protein to a column hand-packed with crab chitin powder (Sigma) (Fukamizo, 2000). The column was attached to an AKTA FPLC system and pre-equilibrated with 200 mM NaCl, 50 mM Tris pH 8.5. Following protein application, column was washed with pre-equilibration buffer and then eluted with 200 mM NaCl, 100 mM acetic acid. Chicken lysozyme was used as a positive control (Supplemental Fig. 5).
Glycoarray binding assay
Both purified recombinant mSLLP1 and hSLLP1 were diluted to 0.5 mg/mL in 200 mM NaCl, 10 mM HEPES pH 7.5 and sent to the Consortium for Functional Glycomics (CFG) for assessment of oligosaccharide binding (Drickamer & Taylor, 2002) using the following protocol. SLLP1 samples were diluted to 200 µg/mL in TSM binding buffer (20 mM Tris pH 7.4, 90 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, 0.05% Tween-20 and 1% BSA). Six independent assays were performed using a printed array (version 3.0) (http://www.functionalglycomics.org/static/consortium/resources/resourcecoreh10.shtml) that comprised 320 different glycans. In each assay, 70 µL of the diluted sample was applied to the printed microarray, covered with a coverslip, and incubated in a humidified chamber for one hour at room temperature, followed by removal of the coverslip and 4 washes with TSM washing buffer (20 mM Tris pH7.4, 90 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, 0.05% Tween-20). The microarray was then incubated with a fluorescently labeled anti-penta His IgG Alexa 488 diluted to 5µg/mL in TSM buffer (20 mM Tris pH7.4, 90 mM NaCl, 2 mM CaCl2, 2 mM MgCl2) for another hour, followed by four times wash in TSM washing buffer, once in TSM buffer and once in deionized water. The microarray was then spun dry and scanned using a Perkin Elmer Microscanarray XL4000 scanner. The signals were quantified by integration of the fluorescence intensity of each peak using Imagene (V.6) image analysis software. A C-type lectin influenza A virus protein H5N1 hemagglutinin was used as a positive control.
In gel bacteriolytic assay of recombinant hSLLP1, wild type and mutant forms
The mature hSLLP1 (128 residues: KLY --- CDF) (Mandal et al., 2003) protein encoding plasmid (vector pET28b+) was PCR amplified for 31 cycles in 25 µL reaction along with mutagenic oligonucleotide primers, buffer, dNTP and QuickChange multi enzyme blend as per manufacturer’s protocol (QuikChange multi-site-directed mutagenesis kit, Stratagene, La Jolla, CA) to rescue four conserved residues of chicken lysozyme including both catalytic residues (T35E, N52D, N53Y, M95K). The reaction product was then treated with DpnI and used to transform competent cells. Plasmids were isolated and sequenced to confirm the four site-directed mutations in hSLLP1 sequence.
BL21(DE3) E. coli cells were transformed with wild type hSLLP1, mutant hSLLP1, wild type human CABYR (negative control) (Naaby-Hansen et al., 2002) and human lysozyme (positive control, accession P61626) expression plasmids. Cells were grown in low salt LB media with MgSO4 (0.1 g/L) and glycerol (2.5 ml/L) in phosphate buffer (pH 7.0) and induced with 0.5 mM IPTG for 3 hours. All uninduced and induced E. coli cell extracts were suspended in Laemmli sample buffer containing 1% SDS without β-mercaptoethanol and heated at 60°C for 3 min. Treated cell extracts were resolved by SDS-PAGE analysis (15% resolving gels) containing inactivated bacteriolytic substrate, Micrococcus luteus (0.2%, 121°C, 30 min, 17 psi). The opaque gels were incubated overnight in 40 mM phosphate buffer (pH 7.4) or phosphate-citrate buffer (pH 5.2) with 1% Triton X-100 at 37°C. In the scanned gel lysozyme activity appeared as clear lysis zones (Audy et al., 1989).
Protein-protein docking in silico
A compact mouse SAS1B (mSAS1B) atomic structure was computationally modeled using a fully-automated procedure on the Robetta server (Kim et al., 2004). The Ginzu method as implemented on the Robetta server was used for secondary structure, signal peptide and domain prediction. Comparative models of individual domains were built from structures detected by PSI-BLAST. Two structures were used as the parent structures for comparative modeling (PDB code: 4GWM, 1K4N). Loop regions were assembled from fragments and optimized to fit the aligned template structure using the ROSETTA fragment insertion method.
Protein-protein docking between mSLLP1 and the computationally model of mSAS1B was performed using LZerD (Li & Kihara, 2012). Results were ranked according to LZerD score and the top 30 docking conformations were visually inspected based on surface conservation and surface charge complementarity. An α-lactalbumin / β-1,4-galactosyltransferase complex crystal structure (PDB code: 1NWG) (Ramakrishnan et al., 2001) was used as positive control to validate the screening criteria of the mSLLP1-mSAS1B complex. Surface Poisson-Boltzmann electrostatic potentials for both SAS1B and β-1,4-galactosyltransferase were generated by PDB2PQR (Dolinsky et al., 2007) and APBS tools (Baker et al., 2001). The sequence conservation scores for both were generated on the ConSurf server (Ashkenazy et al., 2010) choosing only mammalian sequences. The area of interface formed by buried surfaces upon complex formation was estimated by PISA (Krissinel & Henrick, 2007). A plausible complex model that satisfied available experimental evidences was derived.
Results
Refolding and crystallization of recombinant mSLLP1 expressed in bacteria
The purification and crystallization of recombinant mSLLP1 expressed in E. coli proved challenging as the desired protein was insoluble and accumulated in inclusion bodies. This was most likely due to inability of mSLLP1 to form four disulfide bridges required for the proper folding in the reductive environment of bacterial cytoplasm. In order to promote the formation of disulfide bridges, both oxidized and reduced forms of glutathione were present during refolding of mSLLP1 solubilized in the presence of urea. Nevertheless, the attempts to crystallize refolded monomeric recombinant mSLLP1 failed to produce crystals. The failure to crystallize refolded mSLLP1 was assumed to be related to the presence of a minor improperly folded fraction of the protein. The improperly folded fraction was likely to contain erroneously formed S-S bridges and non-oxidized cysteine residues that contained SH groups. In order to remove the mSLLP1 fraction that contained non-oxidized cysteine residues, the monomeric purified protein was denatured in non-reducing conditions and treated with SulfoLink resin that covalently modifies thiol groups. A second refolding followed by size exclusion chromatography produced a mSLLP1 preparation that yielded diffraction quality crystals. The protocol for the production of mSLLP1 protein used in the crystallization study employed two refolding steps. Whether mSLLP1 protein produced without the first refolding step may yield a crystallizable fraction was not tested. The covalent modification of thiol groups deserves consideration as a general strategy for improving crystallization of refractory recombinant proteins.
Functional competence of refolded recombinant mSLLP1
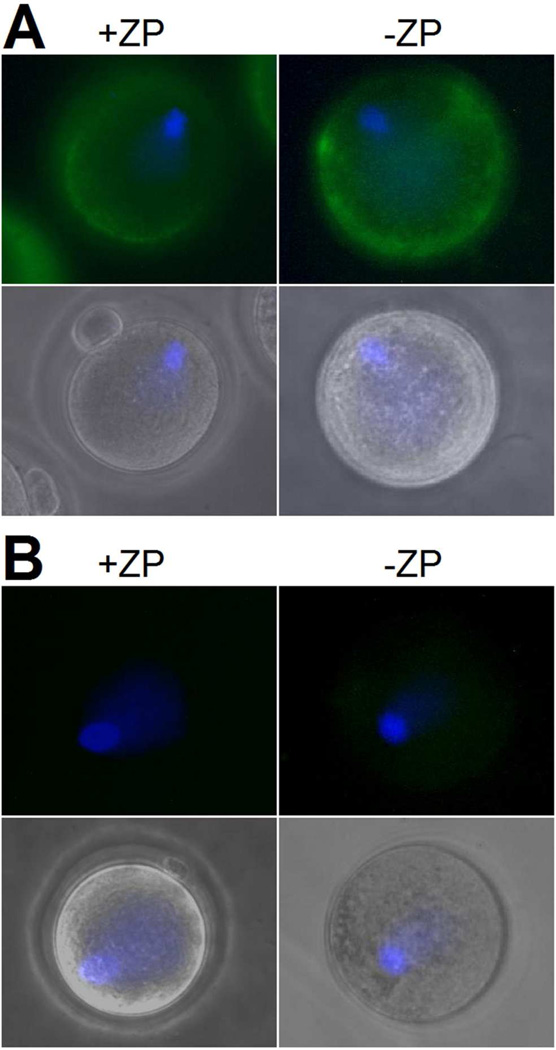
The mSLLP1 fraction undergoing refolding for crystallography was tested in a functional biological assay, oolemmal binding, as reported previously (Herrero et al., 2005). Following incubation of oocytes with refolded recombinant mSLLP1, both unfertilized zona-intact (ZI) and zona-free (ZF) oocytes exhibited fluorescent signals on the oocyte surface and within the perivitelline space (Fig. 1). More intense fluorescent signal was observed antipodal to the eccentrically position Hoechst stained MII arrested nucleus, suggesting denser complementary mSLLP1 binding sites in the microvillar domain in ovulated oocytes before fertilization. In control studies without mSLLP1, no fluorescent signal was detected.
Figure 1.
Indirect immunofluorescence of mSLLP1 complementary binding sites on the oocyte surface with (+ZP) and without Zona Pelllucida (−ZP). (A) mSLLP1 was fluorescently-labeled and shown in green, the nucleus was Hoechst stained and shown in blue. (B) Control experiments were performed without incubation of mSLLP1.
mSLLP1 structure
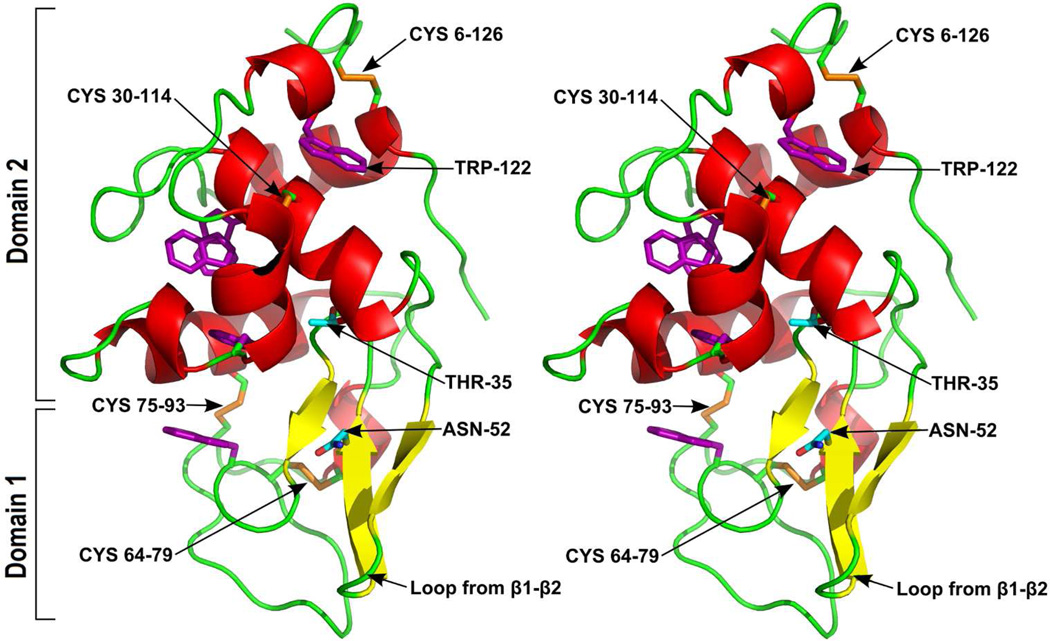
mSLLP1 was crystallized in the P65 space group with three mSLLP1 molecules in the asymmetric unit. The diffraction data was collected to a resolution of 2.15 Å, the structure was solved by molecular replacement, and refined to an R factor 18.8% and Rfree 21.7% (Table 1). The 128 amino acids of mSLLP1 (Fig. 2) form a compact α/β fold characteristic of the homologous c-lysozyme superfamily. Currently the structure most similar to mSLLP1 in the PDB is hen egg white lysozyme (HEWL, PDB code: 2VB1) which exhibits an almost identical main chain backbone tracing (DALI Z-score 21.8–24.9). Because of this similarity, HEWL crystal structure was chosen to compare with mSLLP1 instead of mouse lysozyme for which only a solution NMR structure is available (Obita et al., 2003). The root mean square deviation (RMSD) of mSLLP1 and the reported HEWL crystal structures ranges from 0.7 to 1.5Å for all 128 aligned Cα atoms present in mSLLP1. mSLLP1 retained all four characteristic disulfide bridges in c-lysozymes (Cys6-Cys126, Cys30-Cys114, Cys64-Cys79, and Cys75-Cys93). The N- and C- termini of mSLLP1 are positioned near each other due to the formation of the disulfide bond Cys6-Cys126. The C-terminal affinity purification tag (LEHHHHHH) were not observed in the final electron density map (Fig. 2).
Figure 2.
Stereo views of the mSLLP1 structure. The cartoon of the SLLP1 protein backbone is colored according to secondary structure with α-helices in red, β-strands in yellow and loops in green. Domain 1 consists principally of β-strands and domain 2 mainly of α-helices as indicated. The four disulfide bonds are shown as orange sticks. The five structurally important tryptophan residues are shown in purple. Unique SLLP1 residues Thr35 and Asn52 are shown in cyan with oxygen in red and nitrogen in blue (equivalent residues Glu35 and Asp52 in c-lysozyme are critical for catalysis).
A DALI search (Holm & Sander, 1998) revealed that in addition to c-type lysozymes, several additional proteins display structural similarity to mSLLP1. Among these proteins, α-lactalbumin (Pike et al., 1996) has the highest similarity to SLLP1 with a Z-score around 19.0 and Cα RMSD range between 1.4 and 1.8 Å for 122 aligned residues. The other proteins structurally similar to mSLLP1 fall into the broad lysozyme category including lytic transglycosylase (Fokine et al., 2008), invertebrate-type lysozyme (Goto et al., 2007), goose-type lysozyme (Weaver et al., 1995), and phage-type lysozyme (Xu et al., 2005), all of which are enzymes with peptidoglycan digestion activity capable of cleaving bacterial cell walls (E.C. 3.2.1.17). The proteins of the broad lysozyme category exhibit a range of Z-scores between 4.6 and 7.6, and the RMSD on Cα atoms range between 2.7 Å and 2.9 Å for 80 ~ 110 aligned residues despite of their relatively low sequence identity (9% ~ 13%) with mSLLP1.
The c-Lysozyme superfamily of proteins, including mSLLP1, contains a conserved hydrophobic core surrounded by highly variant hydrophilic residues, a feature common in soluble globular proteins. In addition to the four invariant tryptophan residues, Trp28, Trp63, Trp97, and Trp100, that contribute significantly to the formation of the hydrophobic core, mSLLP1 contains one additional tryptophan Trp122, that is also present in HEWL but absent in both mouse and human lysozymes. Conservation of this hydrophobic core is important for maintaining the overall architecture of the SLLP1 structure.
Characteristic hydrophilic groove in mSLLP1 monomer and the SLLP1 surface fingerprint
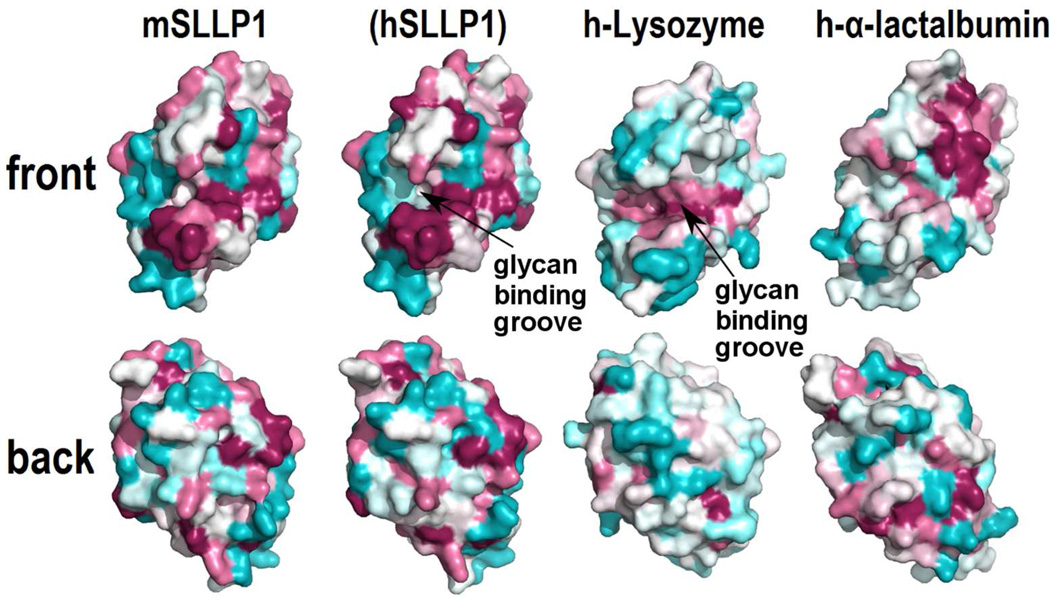
The structure of mSLLP1 reveals a deep cleft that divides the molecule into two domains linked by helix Leu87-Val98. Domain 1 consists of a β-sheet (Asn39-Tyr78) and domain 2 is composed of α-helices (Lys1-Phe38, Gln99-Asp127) (Fig. 2). The cleft consists of a hydrophilic groove similar to the glycan binding groove that is highly conserved within the lysozyme proteins across species (Fig. 3, h-lysozyme). In mSLLP1 as well as in lysozymes, the central region of the groove contains characteristic acidic residues. Comparison of surface electrostatic potential (Fig. 4) and invariant residues reveal this common core.
Figure 3.
ConSurf sequence conservation score plotted on the structure surface for mSLLP1/hSLLP1 compared with human lysozyme and human α-lactalbumin. Lysozymes from a wide variety of vertebrates were used for sequence conservation calculation. SLLP1 family of proteins and α-lactalbumin only exist in mammals and only the relevant mammalian sequences were used for ConSurf calculation. Conserved residues are shown in deep salmon and the most variable regions are shown in cyan. The back view is generated by a 180° rotation on the Y-axis from the front view. The hSLLP1 homologous model was not determined experimentally as indicated in parenthesis. Data for other members of the SLLP family are shown in Supplemental Fig. 9.
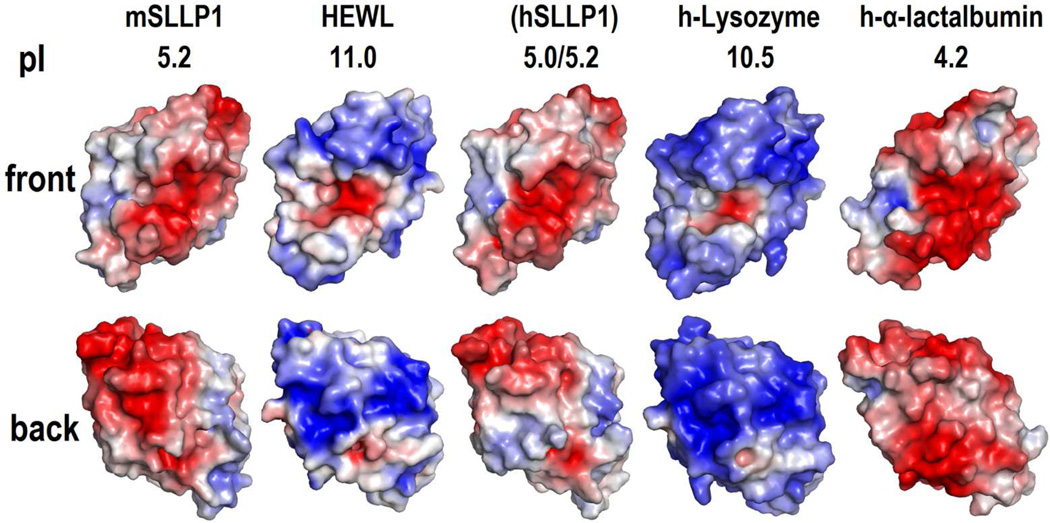
Figure 4.
Poisson-Boltzmann electrostatic potential on the solvent accessible surface of mSLLP1 and hSLLP1 compared with HEWL, human lysozyme, and human α-lactalbumin. The electrostatic surface is indicated by a continuous color scale between −5 kT/e (red) and 5 kT/e (blue). The back view is generated by a 180° rotation on Y-axis from front view. The hSLLP1 homology model was determined computationally [not experimentally] as indicated in parenthesis. Homology models for other members of the human SLLP family are shown in Supplemental Fig. 8.
In contrast, comparison of the peripheral part of the grooves in mSLLP1 and lysozymes reveal dissimilar residues or identical residues adopting a different side-chain conformation. As a result, the shape and the size of the grooves in SLLP1 and c-lysozyme differ markedly (Fig. 5).
Figure 5.
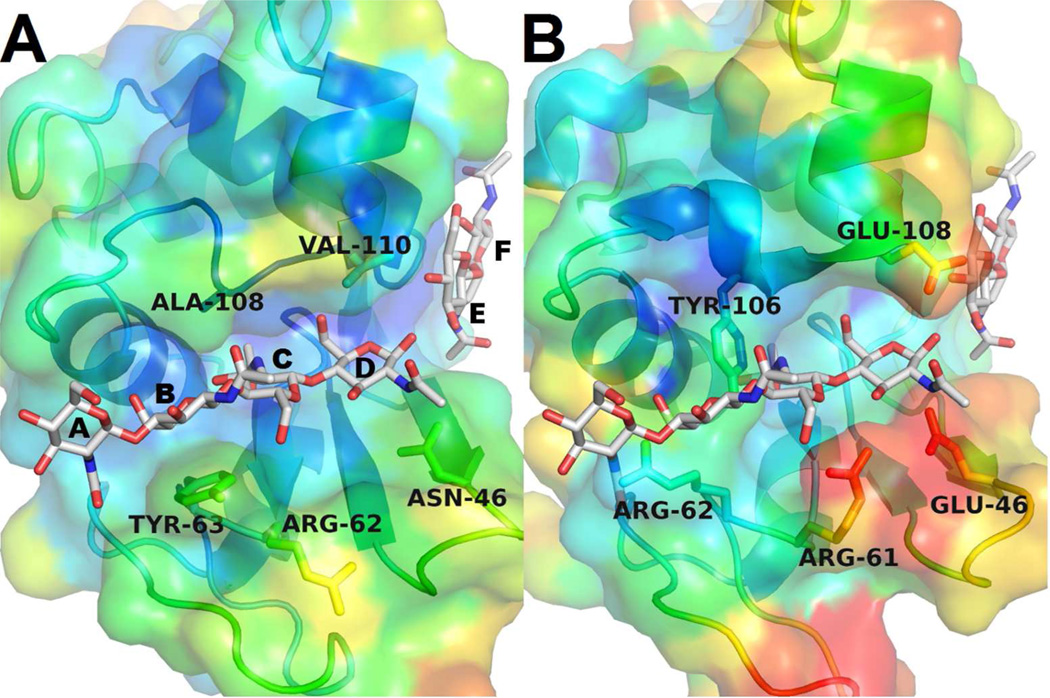
The mSLLP1 groove compared with the c-lysozyme glycan binding groove. Proteins are colored according to the B-factor for each atom, with red indicating flexible regions (high B-factor) and blue indicating more stable regions (low B-factor). (A) The crystal structure of (NAG)4 + (NAG)2 bound to HEWL (PDB code: 1LZS). The six glycan binding sites in the HEWL glycan binding groove are sequentially labeled by the A–F convention. (B) The SLLP1 groove differs strikingly from HEWL. When the NAG glycan is placed within the SLLP1 structure, positions A–D are excluded from access and docking of the glycan due to the interference by the side chains of five peripheral residues (Glu46, Arg61, Arg62, Tyr106, Glu108) displayed as sticks.
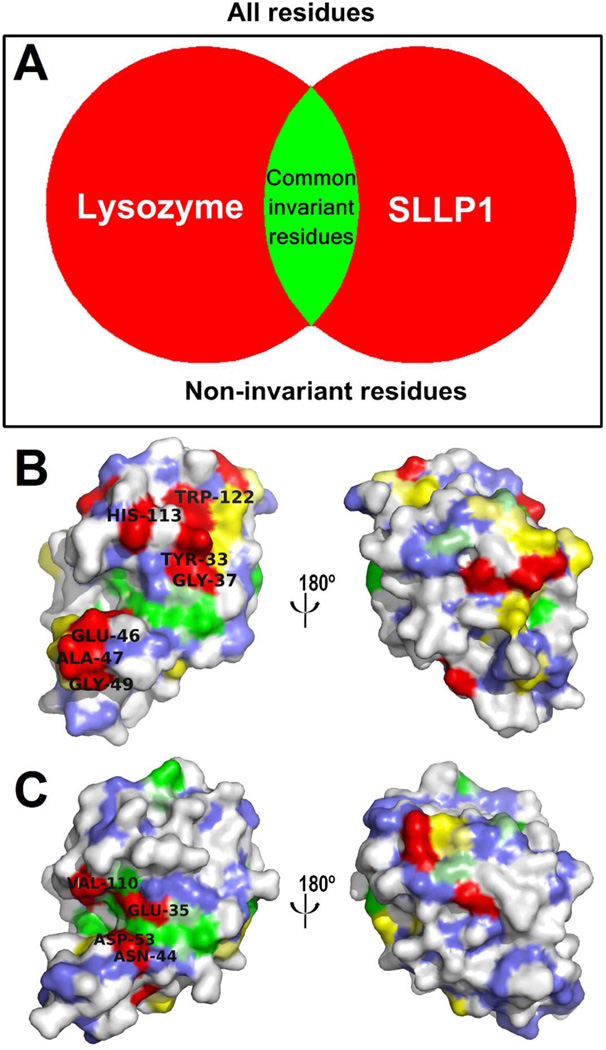
Conserved invariant residues present within various SLLP1s across species (Supplemental Fig. 3A) but not invariant within c-lysozymes (Fig. 6A) were analyzed. A unique SLLP1 fingerprint can be discerned among SLLP1 proteins present in mammalian species. Examination of 20 different SLLP1 proteins (Supplemental Fig. 3A) with amino acid identities above 75% identifies 57 invariant residues including eight cysteine residues involved in the formation of four disulfide bridges. Of the remaining 49 invariant SLLP1 residues, 35 are common to the c-lysozyme family (Fig. 6B, colored green and yellow and remaining 14 residues were defined as forming the distinct SLLP1 fingerprint due to their presence on the molecular surface (Fig. 6B, colored red).
Figure 6.
(A) Venn diagram of invariant residues in SLLP1 and lysozyme. (B) SLLP1 invariant residues derived from multiple sequence alignment mapped on the structure surface color coded by their agreement with corresponding residues in lysozyme shown in panel C. SLLP1 invariant residues that are not found or rare in c-lysozymes are colored in red. Invariant residues that are also common or conserved in c-lysozymes are colored in yellow and green, respectively. Other surface-exposed main-chain atoms are colored in blue and variable side-chain atoms remain in white. The residues in the extended invariant region near the putative glycan binding groove are labeled. Multiple sequence alignments are shown in Supplemental Fig. 3A with similar coloring convention. (C) Lysozyme color coded by its agreement with corresponding residues in SLLP1. The glycan binding groove residues are labeled. Data for SLLP2 are shown in Supplemental Fig. 3B and Supplemental Fig. 4.
The SLLP1 fingerprint region includes a cluster of seven SLLP1-specific residues distributed around the putative glycan binding groove with Glu46 / Ala47 / Gly49 on one side and Tyr33 / Gly37 / His113 / Trp122 on the other side (Fig. 6B, colored red). The surface region that includes these seven residues along with the adjacent conserved residues (Fig. 6B, colored green and blue) is the largest invariant surface area of the mSLLP1 structure. In addition there is also a smaller cluster of invariant residues antipodal to the hydrophilic groove but the contribution of SLLP1-specific residues to this cluster is small. Lysozymes, by contrast, do not possess additional conserved regions beyond their glycan binding groove (Fig. 6C).
SLLP1 lacks glycan binding and hydrolase activity
The potential involvement of the lysozyme-like groove of SLLP1 in glycan binding was tested using chitin affinity binding. Recombinant chicken lysozyme tightly bound to the chitin column at neutral pH and was eluted in acidic conditions as described previously (Fukamizo, 2000). In contrast, neither mSLLP1 nor hSLLP1 demonstrated detectable binding to the chitin column (Supplemental Fig. 5), although oolemmal surface binding activity of these preparations had been demonstrated (Fig. 1).
The SLLP1 preparations with demonstrated oolemmal surface binding activity were also tested for their affinity toward a diverse set of glycans using a glycoarray containing 320 different glycans in Consortium for Functional Glycomics (CFG) facility. However, no significant array binding was detected for either mSLLP1 or hSLLP1 (Supplemental Fig. 6). The service provided by CFG facility is well-validated and, using the same protocol as used in the case of SLLP1, CFG has conclusively identified the specific glycan substrates for many glycan-binding proteins, including a C-type lectin Influenza A virus protein H5N1 hemagglutinin (bottom panel, Supplemental Fig. 6).
Unlike lysozyme, both mSLLP1 and hSLLP1 demonstrated a lack of bacteriolytic activity due to the absence of both critical catalytic residues of chicken lysozyme (E35 and D52). Wild type SLLP1 also revealed non-bacteriolytic properties when secreted by yeast, Pichia pastoris (Mandal et al., 2003). To recover the bacteriolytic activity of SLLP1, both the non-catalytic residues of hSLLP1 were rescued by multiple site-directed mutagenesis process along with two conserved residues of chicken-type lysozymes (Mandal et al., 2003) (T35E, N52D, N53Y, M95K). Yet in gel bacteriolytic assay of mutated hSLLP1 using 0.2% Micrococcus luteus in non-reducing SDS-PAGE analyses continued to show a lack of any significant bacteriolytic activity, indicating that the differences between lysozyme and SLLP1 responsible for the bacteriolytic activity involve far more than the few invariant residues around lysozyme’s catalytic site. Positive control recombinant lysozyme demonstrated excellent bacteriolysis as clear zone of lysis (Supplemental Fig. 7) when expressed using identical vector and host cells, while negative control with the CABYR construct showed no bacteriolysis.
Glycan binding groove modified in SLLP1
The classical lysozyme substrate tetrasaccharide N-acetyl-glucosamine (NAG)4, deposited as a HEWL-NAG complex (PDB code: 1LSG), was placed in the mSLLP1 crystal structure as a reference glycan to analyze groove geometry and visualize substrate-enzyme interactions. In addition to the SLLP1 Thr35 and Asn52 residues that substitute for the critical catalytic Glu35 and Asp52 residues in lysozyme (Mandal et al., 2003), five additional residues, Glu46, Arg61, Arg62, Tyr106, Glu108, from the SLLP1 groove were found to interfere significantly with sugar rings A–D of the NAG glycan (Fig. 5). The Arg62 side chain prevents alignment of the sugar backbone within the groove by blocking the alignment of sugar rings A and B, while the aromatic side chain of Tyr106 protrudes directly into rings B and C. The steric hindrances conferred by Arg62 and Tyr106 would likely prevent the binding of an oligosaccharide substrate with a conformation similar to that of the lysozyme-NAG complex. Further, the SLLP1 Glu46 side chain interacts with the side chain of Arg61 and forms a salt bridge that covers the entrance of the glycan binding site. Moreover, the side chain of Glu108 appears to protrude out and partially block the catalytic groove. Due to the striking difference in the residues surrounding the putative glycan binding groove, in silico docking failed to allow the (NAG)4 conformation to fit satisfactorily into the mSLLP1 groove. However, a weaker glycan binding site in the SLLP1 groove is likely preserved for the other NAG fragment (NAG)2 sugar rings in positions E and F. This domain is preserved without evidence of steric hindrance in both HEWL (Fig. 6C) and SLLP1 (Fig. 6B).
Conserved surface residues of the mSLLP1 monomer
An oolemma receptor for SLLP1, SAS1B (Sperm Acrosomal SLLP1 Binding protein), also known as ovastacin (Quesada et al., 2004), is an astacin-like metalloproteinase associated with the oocyte cortical granules (Burkart et al., 2012; Pires et al., 2013), oocyte membrane and microvillar region (Sachdev et al., 2012). Non-glycosylated recombinant mSAS1B was reported to bind to mSLLP1 (Sachdev et al., 2012), indicating the importance of protein-protein interaction irrespective of the presence of glycans. In order to gain further insight into the SLLP1 and SAS1B interaction, computational modeling of the interaction was undertaken. Analysis of both SLLP1 and SAS1B surfaces were performed to determine the patches that are conserved in these proteins through analysis of the residues of both molecules from various different organisms. SLLP1 conservation was analyzed using SPACA3 gene product from 20 different mammalian species (Supplemental Fig. 3A) and mapped on the surface of mSLLP1 crystal structure (Fig. 3). SAS1B conservation was analyzed using astacin-like proteins with high sequence identities (>61%) to mSAS1B from 15 mammalian species (see Supplemental data) and mapped on a computational model (see section titled “Protein-protein docking in silico” in methods for rationale of the modeling) (Fig. 7B). The underlying assumption was that conservation of surface regions correlates with their functional importance for protein-protein interaction. Despite the high similarity of the protein fold and several common invariant residues in part of the grove (Ser34, Asn37, Ala40, Gly53, Ile54, Glu56, Figure 6, colored green), mSLLP1 exhibits distinctly different surface residues than does HEWL. The conserved surface of lysozymes is only within the catalytic groove, including residues not conserved in the SLLP1 proteins across species (Glu35, Asn44, Asp53, Val110) (Fig. 6C, colored red). Unlike lysozymes, the characteristic conserved surface of SLLP1 proteins is outside the groove (Fig. 3) and is discussed as SLLP1 surface fingerprints below. Notably, a significant change in the isoelectric points of mSLLP1 (pI 5.2) and HEWL (pI 11.0) is reflected in opposite surface charges at physiological pH. The Poisson-Boltzmann electrostatic potential distribution on the protein surface demonstrates that the surface of mSLLP1 is largely acidic (Fig. 4, colored red) contrary to very basic nature of the c-lysozyme surface (Fig. 4, colored blue). The hSLLP1 homology model constructed using the mSLLP1 structure as template expectedly exhibited an electrostatic potential distribution similar to that of mSLLP1 (Fig. 4, h-lysozyme pI 10.5 / hSLLP1 pI 5.2).
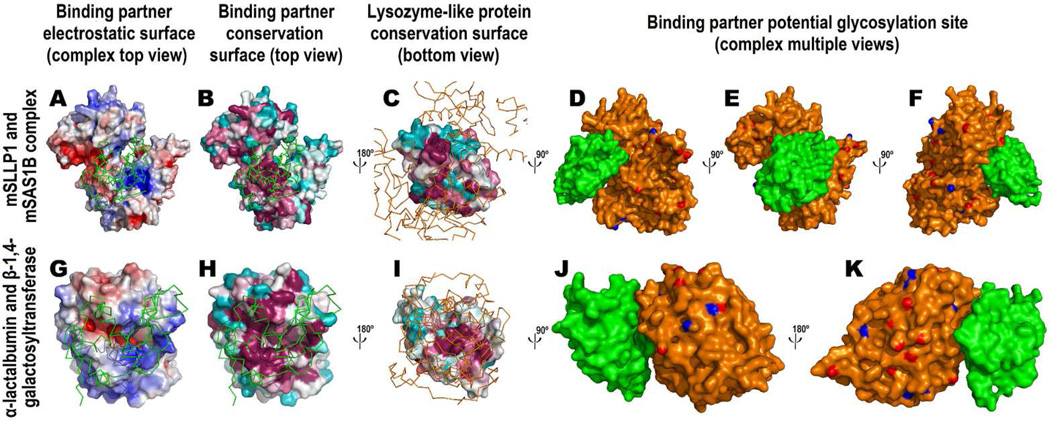
Figure 7.
Construction of mSLLP1/mSAS1B complex model (A–F) using α-lactalbumin / β-1,4-galactosyltransferase complex crystal structure (PDB code: 1NWG) as a positive control (G–K). The top view of the complex looks down the protein-protein axis of the binary complex with the lysozyme-like protein on the top (A,B,G,H), while the bottom view is the opposite direction with the binding partner on the top (C,I). The molecules on the top are represented by Cα ribbon (lysozyme-like protein in green and its binding partner in orange) to disclose the binding interface for the bottom molecule (A,B,C,G,H,I). For the molecules on the bottom, surface Poisson-Boltzmann electrostatic potentials for the (candidate) binding partner are shown with the same convention in Figure 4 (A,G) and surface conservation scores are shown with the same convention in Figure 5 (B,C,H,I). The complexes are also shown from multiple viewpoints with the lysozyme-like protein in green and its binding partner in orange. Hydroxyl oxygen atoms for serine/threonine residues are colored in red and amide nitrogen atoms in asparagine residues are colored in blue on the surface of the binding partner as potential glycosylation sites (D,E,F,J,K). A high resolution SAS1B computational model is shown with surface Poisson-Boltzmann electrostatic potentials in Supplemental Fig. 10.
Another protein that belongs to c-lysozyme superfamily and also participates in protein-protein binding is α-lactalbumin which interacts with β-1,4-galactosyltransferase (Pike et al., 1996). Interestingly, the conserved surface regions of α-lactalbumin are completely different from the characteristic conserved surface of SLLP1 (Fig. 3, h-α-lactalbumin), indicating that the critical set of α-lactalbumin residues involved in α-lactalbumin-β-1,4-galactosyltransferase interaction is distinctly different from the conserved SLLP1 residues used in SLLP1-SAS1B interaction. α-Lactalbumin is even more acidic than SLLP1 as characterized by its isoelectric point (pI) and Poisson-Boltzmann electrostatic potential distribution (Fig. 4, h-α-lactalbumin).
Computational model of mSLLP1 and SAS1B interaction
SAS1B protein (ovastacin) has been reported to spatially localize to both the oolemma (Sachdev et al., 2012) and cortical granules (Burkart et al., 2012; Pires et al., 2013) with different possible functions ascribed to the proteins in these locations. Before the cortical reaction, the enzymatic activity of SAS1B has been shown to be mediated by fetuin-B (Dietzel et al., 2013; Stocker et al., 2014). Eight lines of evidence reveal the strong binding between SLLP1 and SAS1B, which in turn support the hypothesis that SLLP1 binding to the oocyte surface is mediated by SAS1B. This evidence includes protein co-immunoprecipitations, yeast-two-hybrid analyses, anatomical co-localization, inhibition of sperm binding and fusion (Sachdev et al., 2012), microvillar domain binding, reduced fertility in mice bearing targeted deletion (Burkart et al., 2012; Sachdev et al., 2012), and Biacore affinity calculations (Sachdev et al., 2012). Since a binding affinity of SLLP1 for polysaccharides was not detected in glycan array studies, computational studies were undertaken to probe the protein-protein interaction between SLLP1 and SAS1B. The binding interactions were presumed to involve the conserved fingerprint surface regions of SLLP1 and conserved surfaces of SAS1B. The 3-D crystal structure of the SAS1B protein is yet unknown and a computation model of full-length SAS1B was built using a protocol combining both ab initio technique and comparative modeling of a known structure of a zinc-dependent metalloprotease domain as implemented on Robetta server (Kim et al., 2004).
The modeled structure of SAS1B consists of two domains connected by a short α-helix (160–167) and overall exhibits a palm-shape outline (Fig. 7A). The smaller N-terminal domain (1–155) of SAS1B consists principally of a β-sheet with few α-helices, while the larger C-terminal domain (170–414) is mainly α-helical. The highly conserved surfaces of both SLLP1 and SAS1B were assumed to be involved in the protein-protein interaction previously demonstrated (Sachdev et al., 2012) and hence subjected for complementary recognition by computation. Multiple SLLP1-SAS1B complex models with surface geometry and charge complementarities were generated but only one of these models exhibited a binding interface between SLLP1 and SAS1B that involved conserved surfaces from both proteins. In this model, the mSLLP1 structure was embedded between the two domains of the mSAS1B structure in agreement with the experimental data (Sachdev et al., 2012) wherein both C- and N- terminal domains exhibited independent affinity for SLLP1. The model exhibits a buried interface area of 1194 Å2 and the contribution from the N-terminal domain of SAS1B is higher than from its C-terminal domain in accordance with previously published data (Sachdev et al., 2012). Since the modeling of mSLLP1-mSAS1B interface is based on surface topology (Li & Kihara, 2012) and not atomic interactions, the binding free energy of dissociation estimated by PISA (Krissinel & Henrick, 2007) may not be an accurate measure to deduce the binding strength and thus not used. The computation modeling method was validated by building a protein-protein complex of human α-lactalbumin and its interacting protein, β-1,4-galactosyltransferase for which crystal structure of the complex is reported, considering that the α-lactalbumin is structurally similar to SLLP1. Even though their binding partners are vastly different, both of their surface conserved regions formed protein-protein interactions that were close to the major groove.
The power of this computational model of the SAS1B–SLLP1 interaction is that it now permits docking of putative inhibitory small molecules which can be tested in the various bioassays of SLLP1-SAS1B interaction (Sachdev et al., 2012). It must be noted that the structure of SAS1B is not experimentally derived. A theoretical model has its limitations with high uncertainty and inaccuracy at atomic level, which could render the modeling of the atomic interactions between SLLP1 and SAS1B disputable. However the computational modeling of the complex is based on the tertiary fold and the surface topology and not at residue or atomic level interactions and hence to support the model experimental data must be obtained. The theoretical model of SAS1B is therefore an excellent source towards generating various testable hypotheses via experimentation to draw any structural/functional conclusion.
mSLLP1 crystal packing
Examination of the arrangement of mSLLP1 molecule in the crystal lattice revealed the filamentous organization of mSLLP1 which is the major contributor to the crystal packing (Fig. 8A) and is also in line with the observed needle-shape of the mSLLP1 crystals (Supplemental Fig. 2). Additional investigation of the crystal packing of all lysozyme-like protein structures in the PDB revealed that no other protein with sequence similarity higher than 30% to mSLLP1 was reported with filamentous quaternary structure. Despite of the presence of more than 700 lysozyme-like structures in the PDB, mSLLP1 crystal structure uniquely revealed a filamentous crystal packing (Fig. 8C).
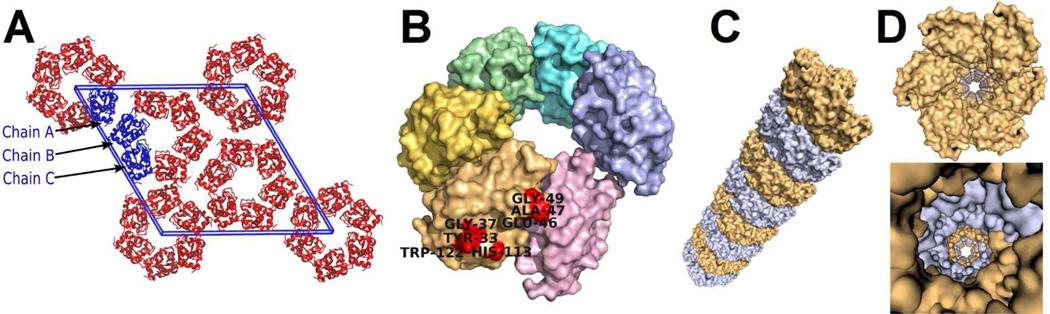
Figure 8.
The filamentous quaternary structure of mSLLP1 observed in the crystal. (A) The cross-section of filaments in the crystal lattice. The crystal cell is shown in blue. The three SLLP1 monomers within the asymmetric unit are also shown in blue. The SLLP1 monomers within the crystal cell generated by crystal symmetry operations are shown in red. Six monomers needed to complete the filament within the crystal cell are also in red. (B) Side view of six SLLP1 monomers that comprise one helix turn in the SLLP1 filament. The six monomers needed to complete a turn of the helix are shown in six different colors. The seven SLLP1 fingerprint residues (Fig. 6B) predicted to interact with SAS1B are colored in red. (C) Angled, exterior view of a SLLP1 filament segment consisting eight helical turns. Helical turns are colored in alternate colors. (D) Cross section view of the SLLP1 filament viewing the central pore.
The internal symmetry of the SLLP1 filaments resulted in six molecules comprising a helical turn corresponding to the crystal symmetry of six-fold screw axis in space group P65, with a 33 Ǻ pitch and a helix angle of 15° (Fig. 8B). The filament exhibits an outer-diameter of ~75Ǻ with a 25Å central pore. All three SLLP1 chains (Fig. 8A) observed in the asymmetric unit of the crystal are involved in the formation of SLLP1 filaments. Chains A related by crystallographic symmetry operation comprise one filament, while the alternating chains B and C form two other filaments in the crystal. When viewing along the c-axis of the crystal, the circular cross-sections of three SLLP1 filaments pack to fill a two-dimensional lattice (Fig. 8A).
None of the seven SLLP1 fingerprint residues (Fig. 6B) is involved in any filament formation. Four of the seven SLLP1 fingerprint residues (Tyr33, Gly37, His113, and Trp122) are exposed on the external surface of the filament, while the other three (Glu46, Ala47, Gly49) are exposed on the end of the filament not blocked by lateral filament formation but may be partially blocked by stacking filament formation (Fig. 8B). Out of the six putative glycan binding sites in mSLLP1 structure (Fig. 5A, sites A–F), two sites (sites A–B) are involved in the lateral formation of the filament, while the remainder four sites (sites C–F) are exposed on the external surface of the filament.
Discussion
SLLP1 is by far the best characterized c-type lysozyme-like protein in mammalian sperm with its defined intra-acrosomal localization (Herrero et al., 2005; Mandal et al., 2003) and role in fertilization as a binding partner for the oolemmal protein SAS1B (Sachdev et al., 2012). The availability of the SLLP1 crystal structure permits structural analysis of its biological activities (Herrero et al., 2005), opens opportunities to study molecular interactions that involve other sperm lysozyme-like proteins, suggests molecular surfaces for docking inhibitors that may block conception, and suggests intriguing new hypotheses regarding the functional role of the SLLP1 filament in vivo.
SLLP1 structure showed a non-conserved groove unfavorable for glycan binding
The mSLLP1 structure contains a groove similar to the glycan-binding groove in lysozyme. The presence of this groove suggested that SLLP1 may possess an affinity for oligosaccharides. However, SLLP1 lacked binding to an array of glycans, the classical substrate of lysozymes. SLLP1 also failed to bind to chitin although lysozyme tightly bound to the chitin column (Supplemental Fig. 5). SLLP1 lacked bacteriolytic activity due to the substitution of the key catalytic residues, Glu35HEWL and Asp52HEWL with Thr35 and Asn52 (Mandal et al., 2003). Moreover, the bacteriolytic activity of SLLP1 could not be rescued by the re-introduction of the lysozyme’s catalytic residues Glu35 and Asp52 (Supplemental Fig. 7).
The structural comparison between SLLP1 and c-type lysozymes revealed that the differences are not limited to just the two catalytic residues (Fig. 5). When compared with c-type lysozymes, the SLLP1 groove is only partially conserved at the center (Fig. 6B, colored green) and most importantly the side chains of Arg62 and Tyr106 protrude into the middle of the groove thus creating a steric hindrance that would significantly restrain the binding of the A–D ring of glycan in a conformation similar to that observed in lysozyme (Fig. 5). In addition side chains of another three residues (Arg61, Glu46, Glu108) are likely to impede the glycan recognition by partially blocking the entrance of the groove. A highly mobile salt bridge (Glu46-Arg61) was also found with partially disordered side chains (B factors in the range of 55 Å2 – 80 Å2), which may act as a switch to turn on/off the accessibility to the putative glycan binding groove under certain circumstances. In addition to differences in the inner groove surface, the surface charge characteristics of the outer regions of the groove were strikingly different (Fig. 4), which further provided evidence that SLLP1 and lysozyme differ in glycan recognition and binding affinity.
Analyses of solid state SLLP1 crystal structure do not rule out its glycan binding capability in solution. Glycan binding affinity experiments were carried out in solution using a diversified set of 320 different glycans microprinted in the glycoarray (Supplemental Fig. 6). No binding was detected, suggesting that the lysozyme-like groove in SLLP1 is unfavorable for glycan binding.
SLLP1 fingerprint identifies its competent functional surfaces
Even though attempts to assign the functional role of the SLLP1 groove to glycan recognition failed, structural analysis revealed the presence of unique fingerprint regions adjoining the groove in the mSLLP1 crystal structure. The delineation of the fingerprint regions was based on conserved invariant residues present within various SLLP1s across species (Supplemental Fig. 3A) but not invariant within c-lysozymes (Fig. 6A). With the crystal structure data for SLLP1 available, the unique presence of a characteristic SLLP1 fingerprint make its identification a powerful instrument to pinpoint competent surface regions on the SLLP1 structure that uniquely contribute to SLLP1’s function.
SLLP1’s biological function as an intra-acrosomal binding partner for the oolemmal surface protein SAS1B has been supported by eight lines of evidence previously (Sachdev et al., 2012). Both native SAS1B (which may be glycosylated) and non-glycosylated recombinant SAS1B (rSAS1B) demonstrated ability to bind to SLLP1 (Sachdev et al., 2012). The relative binding affinity between rSAS1B and SLLP1 was found to be in the nanomolar range (KD = 0.32 nM), and it was concluded that protein-protein interactions are likely to play a critical role for SLLP1 during mammalian fertilization (Sachdev et al., 2012). The fingerprint-defined competent functional surface of SLLP1 identified in the present study may provide candidate domains for the interaction between SLLP1 and SAS1B. To shed light on the details of this molecular interaction, computational modeling of SLLP1 with SAS1B was employed to study the binding interface by delineation of the binding characteristics between these two proteins.
Computational model of mSLLP1-mSAS1B complex revealed complementary binding surface
The mSLLP1-mSAS1B computational model was built using an evolutionary premise that functionally critical regions on a protein are conserved in order for the same mechanism of action to be inherited in different organisms during protein evolution. In the case of c-lysozyme, functionally critical regions required for the substrate binding in the catalytic active site are conserved (Fig. 3, h-Lysozyme). However, for a protein that binds to another macromolecule, for example α-lactalbumin, functionally critical regions required for protein binding are conserved (Fig. 3, h-α-lactalbumin). This evolutionary assumption is supported by the binding interface observed in an α-lactalbumin / β-1,4-galactosyltransferase complex crystal structure (PDB code: 1NWG) (Ramakrishnan et al., 2001). In this complex crystal structure, both α-lactalbumin and β-1,4-galactosyltransferase indeed present their largest conserve surface area complementary to each other (Fig. 7H, 7I).
Based upon this evolutionary premise that residues involved in the binding interface should be conserved across different species, characteristic regions for both mSLLP1 and mSAS1B were analyzed to form such an interface. mSLLP1 indeed exhibits a single prominent conserved region (Fig. 3, mSLLP1; Fig. 6B, all non-white colors; Fig. 7C). Similar to that of human α-lactalbumin, the conserved region in SLLP1 comprised the largest invariant surface including both the groove center (Fig. 6B, colored green) and the fingerprint regions adjoining the groove (Fig. 6B, colored red). Additionally invariant residues antipodal to the hydrophilic groove in SLLP1 are rare and presumably less critical for the recognition of SLLP1’s partner protein SAS1B (Fig. 6B). The conserved surface for SAS1B was also characterized and modeled (Fig. 7B). Conserved surfaces on both SLLP1 and SAS1B were then considered for the computational modeling of the binding interface in the mSLLP1-mSAS1B complex.
The satisfactory mSLLP1/mSAS1B complex model was built by considering the complementarity in surface geometry and electrostatic potential as essential criteria. The complex was built by using 3-D zernike descriptor implemented in the program LZerD (Li & Kihara, 2012), followed by selection of the complex that is having the highest complementarity, for example a positively charged region on mSAS1B surface (Fig. 7A) and a negatively charge region on mSLLP1 surface (Fig. 4). The final complex was selected by ensuring that the most conserved regions for both proteins assemble with maximum overlap to form the binding interface (Fig. 7B, 7C). The similar charge complementarity was also observed in the α-lactalbumin / β-1,4-galactosyltransferase complex. α-Lactalbumin exhibited strong negative charge all over the surface (Fig. 4, h-α-lactalbumin) while its binding partner exhibited a positively charged surface patch around the binding interface (Fig. 7G). As illustrated on three different orientations, mSLLP1 indeed docked onto the mSAS1B structure with larger buried interface (1194 Å2) (Fig. 7D–F) compared to α-lactalbumin/β-1,4-galactosyltransferase complex crystal structure (buried interface 570 Å2) (Fig. 7J, 7K). This high affinity protein-protein interaction between sperm and egg may be essential for fertilization.
The SLLP1-SAS1B computational model permits guidance on the design of site-specific mutagenesis studies of key residues involved in the binding to disrupt the complex formation, which may yield confirmatory insights into the binding mechanism at the molecular level. The model also allows virtual screening and docking of small molecule inhibitors into the surface interface for the disruption of this protein-protein interaction which can be further tested experimentally with particular attention to effects on conception (Sachdev et al., 2012). It must be noted that the structure of the mSLLP1-mSAS1B complex is not experimentally determined, but is a theoretical model where one of the two structures (mSAS1B) is itself an in silico model. Therefore, although plausible at the level of quaternary assembly, the model of the mSLLP1-mSAS1B complex may be very inaccurate at atomic level. Thus, the model must be experimentally validated, for instance through site-directed mutagenesis, before drawing any structural and functional analysis.
mSLLP1 crystals are composed of helical filaments
The mSLLP1 filaments observed in crystals reflect that, besides the soluble monomeric form, SLLP1 may exhibit a filamentous form at high protein concentrations, possibly in response to chemical change in the microenvironment. Dynamic Light Scattering (DLS) data suggests that the oligomeric state of mSLLP1 may be concentration dependent during crystallization (see Supplemental data). Undoubtedly, there is a dynamic equilibrium of SLLP1 between monomeric and filamentous states during in vitro crystallization. Noted examples of filamentous protein structures are cytoskeletal proteins including microfilaments (MF), intermediate filaments (IF) and microtubules (MT) (Frixione, 2000). The dimension of the SLLP1 filament (~75Å diameter) is between those of MFs (~55Å) and IFs (~100Å). However, the presence of a ~25Å central channel renders the SLLP1 filament structurally more similar to MTs, yet MTs are much larger in dimension (~240Å diameter with ~120Å central channel). Due to the similarities between cytoskeletal proteins and the 200µm needle-shaped crystal observed, the length of potential SLLP1 filament in vivo might reach micrometer range. However, further in vivo studies must be undertaken to determine whether an environment suitable for filament growth exists in vivo during acrosomal biogenesis, acrosomal matrix condensation and/or during the fertilization cascade at any of the steps of acrosome reaction, zona penetration, and sperm-egg binding. The presence of the 25Å central channel in the filament core leads to a speculation that if these filaments are detected in vivo, this channel may be a means to rapidly allow hydration and disassembly of the filament or the channel may admit and transmit molecules if the filament were to retain an intact rigid structure. The observation of the SLLP1 filament in crystallo opens opportunities to explore the presence of SLLP1 filaments in vivo; and suggests new models of events mediating sperm-egg interaction, given the role SLLP1 plays in the recognition of an oocyte-specific receptor SAS1B. For example, intra-acrosomal SLLP1 filaments could be an effective way to store high concentrations of SLLP1 molecules in the acrosome during acrosomal matrix condensation or filament growth could be a way to deposit soluble SLLP1 molecules (128 amino acids) onto membrane-bound SLLP1 precursor (163 amino acids). Based on these in crystallo observations investigators studying the ultrastructure of sperm-egg interaction should be on the lookout for 75Å filaments within the acrosome or 75Å cable-like extensions between the acrosome and the oolemma or possibly, penetrating the oolemma.
None of the seven fingerprint residues adjoining the SLLP1 groove is involved in filament organization, but part of the SLLP1 fingerprint regions (Glu46, Ala47, Gly49) are close to the top of the filament and may be partially blocked by stacking filament formation (Fig. 8B). Moreover, SLLP1 and SAS1B were observed to bind to each other with a 1:1 ratio (Sachdev et al., 2012) which predicts a preferred model involving SLLP1 monomers. Thus, it is more likely that the SLLP1 filament, if observed in vivo, would play a condensation role within the acrosome matrix to put SLLP1 molecules in a paracrystalline state rather than mediating the SLLP1/SAS1B interaction. In any case, finding SLLP1 filaments in crystallo opens the search for 75Å intra-acrosomal filaments both before and after the acrosome reaction.
Evolution of SLLP1 as a unique member of the c-type lysozyme family
The human genome encodes nine protein-coding genes that belong to the c-lysozyme family (Irwin et al., 2011), seven of which are predominantly expressed in testis, namely, SPACA3 (Mandal et al., 2003), LYZL1/LYZL2, LYZL4, LYZL6 (Zhang et al., 2005), and SPACA5/SPACA5B (unpublished data). Four of these seven testis-expressed genes, the sub-group of SLLP1, SLLP2 (SPACA5/SPACA5B), and LYZL4 (SLLP6), show a lack of at least one critical catalytic residue E35 or D52 required for bacteriolytic activity. Due to duplication of SLLP2 (SPACA5/SPACA5B) and LYZL1/LYZL2 in the human genome, five orthologues of the seven testis-expressed lysozyme-like genes found in human are also observed in other mammalian species including mouse, rabbit, cow, and elephant. The large number of the testis-expressed lysozyme-like genes as well as the recent duplications of the two protein coding genes SPACA5/SPACA5B and LYZL1/LYZL2 in humans suggest recent evolutionary pressure on this family and imply important biological functions for the SLLP sub-group as a whole.
Despite a very low overall sequence similarity of the c-type, g-type, and i-type lysozymes, they all share a strikingly similar fold (Callewaert & Michiels, 2010). The nine protein-coding genes that belong to the c-lysozyme family in the human genome includes one copy of lysozyme-C and eight other distinctive protein-coding genes, while the mouse genome encodes three copies of lysozyme-C and six other protein-coding genes classified as lysozyme-like that belong to the c-lysozyme family including α-lactalbumin, SLLP1, SLLP2, LYZL1, LYZL4, and LYZL6 (Irwin et al., 2011). With the exception of the LYZL1/LYZL2 (97% identity) duplication in primates and SPACA5/SPACA5B (100% identity) duplication in human, pairwise sequence identities (40%-47%) were observed between SLLP1, SLLP2, LYZL1, LYZL4, and LYZL6. Cross-species analysis revealed that lysozyme-like proteins are absent in amphibians, birds and reptiles but are present in primitive mammals such as platypus, leading to the conclusion that the lysozyme-like group likely does not exist in non-mammalian species and their role evolved with mammals. The sperm lysozyme-like protein SLLP1 has been localized to the acrosome (Mandal et al., 2003) and reported to be involved in fertilization (Herrero et al., 2005) as a ligand for oolemmal binding (Sachdev et al., 2012) suggesting that the SLLP subgroup of lysozyme-like proteins likely co-evolved with adaptations to internal fertilization in mammals and leading to the hypothesis that bacteriolytic lysozymes evolved first and the duplication of the multi-copied sperm lysozyme-like proteins evolved in early mammals as these proteins began to play a critical role in acrosome biogenesis and fertilization.
SLLP1 is a unique member of the diverse lysozyme family. Besides α-lactalbumin, SLLP1 is the only c-lysozyme like protein that lacks both of lysozyme’s critical catalytic residues, while SLLP2 and LYZL4 lacks only D52 but not E35. LYZ, LYZL1/LYZL2, and LYZL6 have both catalytic residues intact. SLLP1 exhibits a similar overall fold to c-lysozyme and α-lactalbumin (PDB code: 1ALC) (Acharya et al., 1989) structures in the PDB, providing an intriguing case where similar structural fold is employed to carry out dissimilar protein functions. Even though α-lactalbumin does not possess bacteriolytic activity, it influences lactose metabolism by binding to β-1,4-galactosyltransferase to lock it in a conformation that allows β-1,4-galactosyltransferase to catalyze glucose transfer from UDP-glucose to NAG (Qasba et al., 2008), thus exhibiting a distinct function differing from bacteriolytic c-lysozymes. SLLP1 is similar to HEWL in lacking bacteriolytic activity and displaying a unique function in molecular recognition as an intra-acrosomal ligand for an egg membrane receptor SAS1B (Sachdev et al., 2012). The different functions of these members of the c-lysozyme family suggest that both SLLP1 and α-lactalbumin have evolved different specificity towards partner proteins and imply that they evolved in parallel from a common ancestor congener to HEWL (Irwin et al., 2011).
Another core structural difference among SLLP1, HEWL and α-lactalbumin was evident from the analysis of the calcium-binding site in α-lactalbumin and analysis of its equivalent position in c-type lysozyme and the lysozyme-like proteins. The region near the N-termini of the linker helix Leu87-Val98 contains a potential calcium binding site in the lysozyme superfamily of proteins. The calcium binding site was evident in the α-lactalbumin crystal structure (PDB code: 1ALC) (Acharya et al., 1989), where three invariant aspartic acids (Asp82, Asp87, Asp88) coordinate the metal. However, this site was not observed in HEWL nor in any of the c-lysozyme-like proteins in human (seven proteins) and mouse (five proteins) including hSLLP1/mSLLP1 and hSLLP2/mSLLP2, indicating that none of these twelve proteins expressed in the testes possesses all three aspartic acids in the calcium coordinating domain and likely do not function as calcium binding proteins. Calcium-binding lysozymes with all three invariant aspartic acids conserved were indeed found in milk and introduction of the missing aspartic acids by mutagenesis in human c-lysozyme has created a fully-functional calcium binding site (Kuroki et al., 1989). These observations support the concept that the testis-expressed lysozyme-like proteins in mammals were evolved from non-calcium-binding lysozymes dissimilar to α-lactalbumin.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants GM53163, GM62414, NIH Fogarty International Center grant D43TW/HD00654, U01HD60491 from the Contraceptive Development Branch, and a Grand Challenges Grant from the Bill and Melinda Gates Foundation. The results shown in this report are derived from work performed at Argonne National Laboratory, at the Structural Biology Center of the Advanced Photon Source. Argonne is operated by University of Chicago Argonne, LLC, for the U.S.Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. We thank Maksymilian Chruszcz, Ivan G. Shabalin, David R. Cooper and Matthew D. Zimmerman for valuable discussion. We thank Daisuke Kihara and Juan Esquivel for help during the use of LZerD program for protein-protein docking. We thank CFG for carrying out the glycoarray binding assay. We thank Ruoya Ho and Jing Hou for help in the DLS experiment. We thank Hutton C. Chapman for the preparation of some figures. Atomic coordinates and experimental structure factors have been deposited to the Protein Data Bank (PDB) with accession code 4YF2.
Footnotes
Disclosures
None declared.
Author contributions
H.Z. and I.A.S. performed the crystallography and carbohydrate binding studies. A.M. and S.P. performed the oocyte binding studies. A.M. generated the mSLLP1 wild type and mutant constructs and performed the in gel bacteriolytic assay. H.Z. analyzed and interpreted the data. H.Z. and M.D.C. drafted the manuscript. A.M, J.C.H., and I.A.S. revised the initial draft and improved the intellectual content substantially. J.C.H. and W.M. provided overall supervision and critical review of the final manuscript. All authors approved the final submitted versions of the manuscript.
References
- Acharya KR, Stuart DI, Walker NP, Lewis M, Phillips DC. Refined structure of baboon alpha-lactalbumin at 1.7 A resolution. Comparison with C-type lysozyme. J Mol Biol. 1989;208:99–127. doi: 10.1016/0022-2836(89)90091-0. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Koonin EV. Iterated profile searches with PSI-BLAST--a tool for discovery in protein databases. Trends Biochem Sci. 1998;23:444–447. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanackovic D, Blum I, Cao Y, Wenzel S, Bartels K, Faltz C, Hossfeld DK, Hegewisch-Becker S, Bokemeyer C, Leuwer R. Expression of cancer-testis antigens as possible targets for antigen-specific immunotherapy in head and neck squamous cell carcinoma. Cancer Biol Ther. 2006;5:1218–1225. doi: 10.4161/cbt.5.9.3174. [DOI] [PubMed] [Google Scholar]
- Audy P, Grenier J, Asselin A. Lysozyme activity in animal extracts after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Comp Biochem Physiol B. 1989;92:523–527. doi: 10.1016/0305-0491(89)90126-0. [DOI] [PubMed] [Google Scholar]
- Avella MA, Xiong B, Dean J. The molecular basis of gamete recognition in mice and humans. Mol Hum Reprod. 2013;19:279–289. doi: 10.1093/molehr/gat004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkart AD, Xiong B, Baibakov B, Jimenez-Movilla M, Dean J. Ovastacin, a cortical granule protease, cleaves ZP2 in the zona pellucida to prevent polyspermy. J Cell Biol. 2012;197:37–44. doi: 10.1083/jcb.201112094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callewaert L, Michiels CW. Lysozymes in the animal kingdom. J Biosci. 2010;35:127–160. doi: 10.1007/s12038-010-0015-5. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzel E, Wessling J, Floehr J, Schafer C, Ensslen S, Denecke B, et Jahnen-Dechent W, et al. Fetuin-B, a liver-derived plasma protein is essential for fertilization. Dev Cell. 2013;25:106–112. doi: 10.1016/j.devcel.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue JP, Patel H, Anderson WF, Hawiger J. Three-dimensional structure of the platelet integrin recognition segment of the fibrinogen gamma chain obtained by carrier protein-driven crystallization. Proc Natl Acad Sci U S A. 1994;91:12178–12182. doi: 10.1073/pnas.91.25.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drickamer K, Taylor ME. Glycan arrays for functional glycomics. Genome Biol. 2002;3:REVIEWS1034. doi: 10.1186/gb-2002-3-12-reviews1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feolo M, Helmberg W, Sherry S, Maglott DR. NCBI genetic resources supporting immunogenetic research. Rev Immunogenet. 2000;2:461–467. [PubMed] [Google Scholar]
- Fokine A, Miroshnikov KA, Shneider MM, Mesyanzhinov VV, Rossmann MG. Structure of the bacteriophage phi KZ lytic transglycosylase gp144. J Biol Chem. 2008;283:7242–7250. doi: 10.1074/jbc.M709398200. [DOI] [PubMed] [Google Scholar]
- Frayne J, Taylor A, Cameron G, Hadfield AT. Structure of insoluble rat sperm glyceraldehyde-3-phosphate dehydrogenase (GAPDH) via heterotetramer formation with Escherichia coli GAPDH reveals target for contraceptive design. J Biol Chem. 2009;284:22703–22712. doi: 10.1074/jbc.M109.004648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frixione E. Recurring views on the structure and function of the cytoskeleton: a 300-year epic. Cell Motil Cytoskeleton. 2000;46:73–94. doi: 10.1002/1097-0169(200006)46:2<73::AID-CM1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Fukamizo T. Chitinolytic enzymes: catalysis, substrate binding, and their application. Curr Protein Pept Sci. 2000;1:105–124. doi: 10.2174/1389203003381450. [DOI] [PubMed] [Google Scholar]
- Goto T, Abe Y, Kakuta Y, Takeshita K, Imoto T, Ueda T. Crystal structure of Tapes japonica Lysozyme with substrate analogue: structural basis of the catalytic mechanism and manifestation of its chitinase activity accompanied by quaternary structural change. J Biol Chem. 2007;282:27459–27467. doi: 10.1074/jbc.M704555200. [DOI] [PubMed] [Google Scholar]
- Grant RP, Buttery SM, Ekman GC, Roberts TM, Stewart M. Structure of MFP2 and its function in enhancing MSP polymerization in Ascaris sperm amoeboid motility. J Mol Biol. 2005;347:583–595. doi: 10.1016/j.jmb.2005.01.054. [DOI] [PubMed] [Google Scholar]
- Han L, Monne M, Okumura H, Schwend T, Cherry AL, Flot D, Matsuda T, Jovine L. Insights into egg coat assembly and egg-sperm interaction from the X-ray structure of full-length ZP3. Cell. 2010;143:404–415. doi: 10.1016/j.cell.2010.09.041. [DOI] [PubMed] [Google Scholar]
- Herrero MB, Mandal A, Digilio LC, Coonrod SA, Maier B, Herr JC. Mouse SLLP1, a sperm lysozyme-like protein involved in sperm-egg binding and fertilization. Dev Biol. 2005;284:126–142. doi: 10.1016/j.ydbio.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin DM, Biegel JM, Stewart CB. Evolution of the mammalian lysozyme gene family. BMC Evol Biol. 2011;11:166. doi: 10.1186/1471-2148-11-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha KN, Shumilin IA, Digilio LC, Chertihin O, Zheng H, Schmitz G, Visconti PE, Flickinger CJ, Minor W, Herr JC. Biochemical and structural characterization of apolipoprotein A–I binding protein, a novel phosphoprotein with a potential role in sperm capacitation. Endocrinology. 2008;149:2108–2120. doi: 10.1210/en.2007-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissinger CR, Gehlhaar DK, Smith BA, Bouzida D. Molecular replacement by evolutionary search. Acta Crystallogr D. 2001;57:1474–1479. doi: 10.1107/s0907444901012458. [DOI] [PubMed] [Google Scholar]
- Kresge N, Vacquier VD, Stout CD. The crystal structure of a fusagenic sperm protein reveals extreme surface properties. Biochemistry. 2001;40:5407–5413. doi: 10.1021/bi002779v. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Kuroki R, Taniyama Y, Seko C, Nakamura H, Kikuchi M, Ikehara M. Design and creation of a Ca2+ binding site in human lysozyme to enhance structural stability. Proc Natl Acad Sci U S A. 1989;86:6903–6907. doi: 10.1073/pnas.86.18.6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Kihara D. Protein docking prediction using predicted protein-protein interface. BMC Bioinformatics. 2012;13:7. doi: 10.1186/1471-2105-13-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal A, Klotz KL, Shetty J, Jayes FL, Wolkowicz MJ, Bolling LC, Coonrod SA, Black MB, Diekman AB, Haystead TA, Flickinger CJ, Herr JC. SLLP1, a unique, intra-acrosomal, non-bacteriolytic, c lysozyme-like protein of human spermatozoa. Biol Reprod. 2003;68:1525–1537. doi: 10.1095/biolreprod.102.010108. [DOI] [PubMed] [Google Scholar]
- Mayrose I, Graur D, Ben-Tal N, Pupko T. Comparison of site-specific rate-inference methods for protein sequences: empirical Bayesian methods are superior. Mol Biol Evol. 2004;21:1781–1791. doi: 10.1093/molbev/msh194. [DOI] [PubMed] [Google Scholar]
- Miao L, Vanderlinde O, Liu J, Grant RP, Wouterse A, Shimabukuro K, Philipse A, Stewart M, Roberts TM. The role of filament-packing dynamics in powering amoeboid cell motility. Proc Natl Acad Sci U S A. 2008;105:5390–5395. doi: 10.1073/pnas.0708416105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution - from diffraction images to an initial model in minutes. Acta Crystallogr D. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Naaby-Hansen S, Mandal A, Wolkowicz MJ, Sen B, Westbrook VA, Shetty J, Coonrod SA, Klotz KL, Kim YH, Bush LA, Flickinger CJ, Herr JC. CABYR, a novel calcium-binding tyrosine phosphorylation-regulated fibrous sheath protein involved in capacitation. Dev Biol. 2002;242:236–254. doi: 10.1006/dbio.2001.0527. [DOI] [PubMed] [Google Scholar]
- Obita T, Ueda T, Imoto T. Solution structure and activity of mouse lysozyme M. Cell Mol Life Sci. 2003;60:176–184. doi: 10.1007/s000180300012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Anonymous Macromolecular Crystallography. San Diego, CA: Academic Press Inc.; 1997. Processing of X-ray diffraction data collected in oscillation mode; pp. 307–326. Part A. 525 B Street, Suite 1900 92101-4495. [DOI] [PubMed] [Google Scholar]
- Pike AC, Brew K, Acharya KR. Crystal structures of guinea-pig, goat and bovine alpha-lactalbumin highlight the enhanced conformational flexibility of regions that are significant for its action in lactose synthase. Structure. 1996;4:691–703. doi: 10.1016/s0969-2126(96)00075-5. [DOI] [PubMed] [Google Scholar]
- Pires ES, Hlavin C, Macnamara E, Ishola-Gbenla K, Doerwaldt C, Chamberlain C, Klotz K, Herr AK, Khole A, Chertihin O, Curnow E, Feldman SH, Mandal A, Shetty J, Flickinger C, Herr JC. SAS1B protein [ovastacin] shows temporal and spatial restriction to oocytes in several eutherian orders and initiates translation at the primary to secondary follicle transition. Dev Dyn. 2013;242:1405–1426. doi: 10.1002/dvdy.24040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primakoff P, Myles DG. Penetration, adhesion, and fusion in mammalian sperm-egg interaction. Science. 2002;296:2183–2185. doi: 10.1126/science.1072029. [DOI] [PubMed] [Google Scholar]
- Qasba PK, Ramakrishnan B, Boeggeman E. Structure and function of beta −1,4-galactosyltransferase. Curr Drug Targets. 2008;9:292–309. doi: 10.2174/138945008783954943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Sanchez LM, Alvarez J, Lopez-Otin C. Identification and characterization of human and mouse ovastacin: a novel metalloproteinase similar to hatching enzymes from arthropods, birds, amphibians, and fish. J Biol Chem. 2004;279:26627–26634. doi: 10.1074/jbc.M401588200. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan B, Shah PS, Qasba PK. alpha-Lactalbumin (LA) stimulates milk beta-1,4-galactosyltransferase I (beta 4Gal-T1) to transfer glucose from UDP-glucose to N-acetylglucosamine. Crystal structure of beta 4Gal-T1 × LA complex with UDP-Glc. J Biol Chem. 2001;276:37665–37671. doi: 10.1074/jbc.M102458200. [DOI] [PubMed] [Google Scholar]
- Romero A, Romao MJ, Varela PF, Kolln I, Dias JM, Carvalho AL, Sanz L, Topfer-Petersen E, Calvete JJ. The crystal structures of two spermadhesins reveal the CUB domain fold. Nat Struct Biol. 1997;4:783–788. doi: 10.1038/nsb1097-783. [DOI] [PubMed] [Google Scholar]
- Rosenbaum G, Alkire RW, Evans G, Rotella FJ, Lazarski K, Zhang RG, Ginell SL, Duke N, Naday I, Lazarz J, Molitsky MJ, Keefe L, Gonczy J, Rock L, Sanishvili R, Walsh MA, Westbrook E, Joachimiak A. The Structural Biology Center 19ID undulator beamline: facility specifications and protein crystallographic results. J Synchrotron Radiat. 2006;13:30–45. doi: 10.1107/S0909049505036721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdev M, Mandal A, Mulders S, Digilio LC, Panneerdoss S, Suryavathi V, Pires E, Klotz KL, Hermens L, Herrero MB, Flickinger CJ, van Duin M, Herr JC. Oocyte specific oolemmal SAS1B involved in sperm binding through intra-acrosomal SLLP1 during fertilization. Dev Biol. 2012;363:40–51. doi: 10.1016/j.ydbio.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Schrödinger L. The PyMOL Molecular Graphics System, Version~1.3r1. 2010 [Google Scholar]
- Stocker W, Karmilin K, Hildebrand A, Westphal H, Yiallouros I, Weiskirchen R, Dietzel E, Floehr J, Jahnen-Dechent W. Mammalian gamete fusion depends on the inhibition of ovastacin by fetuin-B. Biol Chem. 2014;395:1195–1199. doi: 10.1515/hsz-2014-0189. [DOI] [PubMed] [Google Scholar]
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, et al. et Mammalian Gene Collection Program Team. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranter R, Read JA, Jones R, Brady RL. Effector sites in the three-dimensional structure of mammalian sperm beta-acrosin. Structure. 2000;8:1179–1188. doi: 10.1016/s0969-2126(00)00523-2. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- Vjugina U, Evans JP. New insights into the molecular basis of mammalian sperm-egg membrane interactions. Front Biosci. 2008;13:462–476. doi: 10.2741/2693. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang Y, Mandal A, Zhang J, Giles FJ, Herr JC, Lim SH. The spermatozoa protein, SLLP1, is a novel cancer-testis antigen in hematologic malignancies. Clin Cancer Res. 2004;10:6544–6550. doi: 10.1158/1078-0432.CCR-04-0911. [DOI] [PubMed] [Google Scholar]
- Weaver LH, Grutter MG, Matthews BW. The refined structures of goose lysozyme and its complex with a bound trisaccharide show that the "goose-type" lysozymes lack a catalytic aspartate residue. J Mol Biol. 1995;245:54–68. doi: 10.1016/s0022-2836(95)80038-7. [DOI] [PubMed] [Google Scholar]
- Xu M, Arulandu A, Struck DK, Swanson S, Sacchettini JC, Young R. Disulfide isomerization after membrane release of its SAR domain activates P1 lysozyme. Science. 2005;307:113–117. doi: 10.1126/science.1105143. [DOI] [PubMed] [Google Scholar]
- Zhang K, Gao R, Zhang H, Cai X, Shen C, Wu C, Zhao S, Yu L. Molecular cloning and characterization of three novel lysozyme-like genes, predominantly expressed in the male reproductive system of humans, belonging to the c-type lysozyme/alpha-lactalbumin family. Biol Reprod. 2005;73:1064–1071. doi: 10.1095/biolreprod.105.041889. [DOI] [PubMed] [Google Scholar]
- Zheng H, Chruszcz M, Lasota P, Lebioda L, Minor W. Data mining of metal ion environments present in protein structures. J Inorg Biochem. 2008;102:1765–1776. doi: 10.1016/j.jinorgbio.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.