Abstract
It has been proposed that the copy number of mitochondria DNA (mtDNA) per cell reflects gene–environment interactions between unknown hereditary factors and exposures affecting levels of oxidative stress. However, whether copy number of mtDNA could be a risk predictor of oxidative stress-related human cancers, such as breast cancer, remains to be determined. To explore the role of mtDNA copy number in breast cancer etiology, we analyzed mtDNA copy number in whole blood from 103 patients with breast cancer and 103 matched control subjects and examined in relation to endogenous antioxidants. Case patients with breast cancer had a statistically significantly higher mtDNA copy number than control subjects (median: 1.29 vs. 0.80, P < 0.01). High mtDNA copy number (above the median in controls) was associated with a statistically significantly increased risk of breast cancer, compared with low copy number (Odds ratio (OR) = 4.67, 95% CI: 2.45–8.92), with a statistically significant dose–response relationship in trend analysis (P < 0.01). Moreover, mtDNA copy number was significantly inversely associated with several important endogenous oxidants and antioxidants in blood in either the cases (total glutathione, CuZn-SOD activity and myeloperoxidase (MPO)) or the controls (catalase (CAT) activity). These results suggest the mtDNA copy number could be associated with risk of breast cancer, perhaps through an oxidative stress mechanism.
Keywords: Mitochondrial DNA, Copy number, Oxidative stress, Breast cancer
1. Introduction
Mitochondria are semiautonomous organelles within cells that play an important role in cellular energy metabolism, generation of reactive oxygen species (ROS), and apoptosis (Crimi and Rigolio, 2008). Compared with nuclear DNA, mitochondria DNA (mtDNA) lacks protective histones and has diminished DNA repair capacity, and therefore is particularly susceptible to ROS and other types of genotoxic damage. Although the copy number of mtDNA per cell is maintained within a constant range according to the energy need of the cell to sustain normal physiological functions, it varies significantly among the population from 1000 to 10,000 per cell (Veltri et al., 1990). mtDNA also significantly varies by cell type, however, in general, there is good correlation of the amount of mtDNA in various cell types. Therefore, mtDNA copy number in surrogate tissues (such as, whole blood, etc.) might be a good indicator of mtDNA copy number in target tissues.
It is likely that the variations in the copy number of mitochondria reflect the net results of gene–environmental interactions between unknown hereditary factors and the levels of oxidative stress (an imbalance between ROS production and the antioxidant capacity), caused by a variety of endogenous and exogenous factors, such as, hormones, age, dietary and environmental oxidants/ antioxidants, and reaction to oxidative damage, all of which are thought to be risk factors for various types of cancer development (Lee et al., 1998; Renis et al., 1989; Verma et al., 2007). However, so far, only two studies have been carried out to investigate the role of mtDNA copy number in whole blood in human cancers, one in renal cell carcinoma (RCC) (Xing et al., 2008) and the other in non-Hodgkin lymphoma (NHL) (Lan et al., 2008). In RCC, Xing et al. found that low mtDNA copy number was associated with increased risk of RCC compared with high mtDNA copy number (OR = 1.53, 95% CI = 1.07–2.19). On the contrary, Lan et al. found that increased mtDNA copy number was associated with increased risk of NHL (OR = 1.3, 95% CI = 1.1–1.6). The contradictory results from these two studies suggest that the role of mtDNA copy number in human cancer is complicated and might be cancer site specific. Nonetheless, additional research in other cancer sites is warranted.
The copy number of mtDNA might be extremely relevant to breast cancer because oxidative stress has been suggested to play a significant role in breast cancer etiology (Gönenç et al., 2006; Sener et al., 2007; Nielsen and Grønbaek, 2006; Park and Kang, 2006; Zhang et al., 1999; Kushi et al., 1996). Although aberration of mtDNA copy number is observed in breast tumor tissues compared to adjacent normal counterparts (Yu et al., 2007; Ye et al. 2008; Wang et al., 2007; Putignani et al., 2008; Tseng et al., 2006), so far, whether copy number of mtDNA in nonneoplastic tissues, such as whole blood (leukocytes), is a possible risk factor for breast cancer is unknown. We thereby investigated mtDNA copy number measured in whole blood DNA as a possible biomarker for risk for breast cancer, as this biomarker is likely to reflect both exogenous and endogenous processes relevant for oxidative stress and consequently breast carcinogenesis.
2. Methods
2.1. Study population
De-identified biospecimens and questionnaire data used in this study were made available through the Roswell Park Cancer Institute’s (RPCI) Data Bank and BioRepository (DBBR) Ambrosone et al., 2006. The DBBR is a Cancer Center Shared Resource, and is a biorepository of blood samples collected, processed and stored in a rigorous, standardized manner, linked with clinical and epidemiological data. Patients are enrolled through site-specific clinics prior to surgery and/or chemotherapy, and controls are individuals who are free from cancer who are visitors or family members of patients. Relationships between patients and controls are carefully annotated, so that we avoid overmatching patients to their own family or friends. Patients and controls are consented to provide a non-fasting blood sample and to complete a questionnaire that collects data on family history of cancer, medical history, smoking history, menstrual and reproductive history, and lifestyle habits including diet, use of dietary supplements, smoking, physical activity, alcohol intake, as well as demographic data and height and weight from young adulthood to present. Blood samples are drawn in phlebotomy and transferred to the DBBR laboratory through the pneumatic tube system. In the laboratory, specimens are processed and aliquoted into 0.5 ml straws that are labeled with barcoded ID number and frozen. All samples are stored in liquid nitrogen and are available for use by RPCI and other researchers with Institutional Review Board (IRB)- approved protocols. Genomic DNA was extracted from whole blood for all the samples by use of Gentra Puregene Blood kit (Qiagen, Valencia, CA). In this study, a total of 103 women with breast cancer and 103 cancer-free women were included in the analysis.
2.2. Determination of mtDNA copy number by real-time quantitative PCR
This method is detailed in a recent publication by Xing et al. (2008). The method was shown to have high interassay reliability. In brief, two pairs of primers were used in the two steps of relative quantification for mtDNA content. One primer pair was used for the amplification of the MT-ND1 gene in mtDNA. Another primer pair was used for the amplification of the single-copy nuclear gene human globulin (HGB). In the first step, the ratio of mtDNA copy number to HGB copy number, which is also referred as mtDNA index, was determined for each sample from standard curves. This ratiois proportional to the mtDNA copy number in each cell and, for each sample, was normalized to a calibrator DNA (DNA sample from healthy control) in order to standardize between different runs. All samples were assayed in duplicate on a 96-well plate with an Applied Biosystems StepOne Plus System. The PCRs for ND-1 and HGB were always performed on separate 96-well plates with the same samples in the same well positions to avoid possible position effects. A standard curve of a diluted reference DNA, one negative control, and one calibrator DNA were included in each run. For each standard curve, one reference DNA sample was serially diluted 1:2 to produce a seven-point standard curve between 0.3125 and 20 ng of DNA. The R2 for each standard curve was 0.99 or greater. Standard deviations for the cycle of threshold (Ct) value were accepted at 0.25. Otherwise, the test was repeated.
2.3. Biochemical analyses of levels and activities of oxidants and antioxidants in blood
In this study, we measured levels of CuZn-Superoxide dismutase (SOD), catalase (CAT) activity, cellular glutathione peroxidase activity, glutathione reductase activity, and total glutathione in red blood cells, and levels of myeloperoxidase (MPO) in plasma in both cases and controls. Below is the brief summary for each assay.
2.3.1. Red blood cell CuZn-SOD
SOD levels were measured using the SOD-525 spectrophotometeric assay kit (Oxis International, Foster City, CA). The SOD-mediated increase in the rate of autoxidation of 5,6,6a,11b-tetrahydro- 3,9,10-trihydroxybenzo[c]fluorene (R1) in aqueous alkaline solution yields a chromophore with maximum absorbance at 525 nm. SOD activity is indicated by the ratio of auto-oxidation rates in the presence (VS) and absence (Vc) of SOD. Obtained SOD values were multiplied by the dilution factor and expressed as U per mg Hemoglobin (U/mg Hb).
2.3.2. Red blood cell CAT
CAT activity was measured using the Catalase-520 kit (Oxis International, Foster City, CA). The sample containing catalase was incubated in the presence of a known concentration of H2O2. After incubation for exactly 1 min, the reaction was quenched with sodium azide. The amount of H2O2 remaining in the reaction mixture was then determined by the oxidative coupling reaction of 4- aminophenzone and 3,5-dichloro-2-hydroxybenezensulfonic acid in the presence of H2O2 and catalyzed by horseradish peroxidase. The resulting quinoneimine dye was measured at 520 nm on SpectraMax UV–VIS microplate reader (Molecular Devices Corp., Union City, CA). Catalase activity was determined using standard curve data using a second order polynomial regression. The obtained value was multiplied by the appropriate dilution factor. Catalase activity was normalized to hemoglobin content in the sample and expressed as U per mg Hemoglobin (U/mg Hb).
2.3.3. Red blood cell TGSH
Total glutathione level in RBCs lysates was measured using the GSH-420 kit (Oxis International, Foster City, CA). The sample was buffered and the reducing agent tris (2-carboxyethyl) phosphate was added to reduce any oxidized glutathione. The chromogen 4- chloro-1-methyl-7-trifluoromethylquinolinium methylsulfate was added forming thioethers with all thiols present in the sample. Upon addition of base to raise the pH greater than 13, a β-elimination specific to the GSH-thioether resulted in the chromophoric thione measured at 420 nm which is directly proportional to the GSH concentration. Using linear regression, total GSH was determined using standard curve data, normalized to hemoglobin concentration, and expressed as nmol/mg Hb.
2.3.4. Red blood cell GPx
Cellular glutathione peroxidase activity was measured using the GPx-340 kit (Oxis International, Foster City, CA). Tert-Butyl Hydroperoxide was added as a substrate. Oxidized glutathione produced by the action of GPx present in the sample was reduced by glutathione reductase in the presence of NADPH. The decrease in NADPH was recorded at 340 nm using a Beckman DU-800 spectrophotometer (Beckman Coulter, Inc. Fullerton, CA), and the molar absorptivity of NADPH, 6.22 × 10−3 L mol−1 cm−1 was used to calculate enzyme activity. GPx activity was normalized to hemoglobin content in the sample and expressed as mU per mg Hemoglobin (mU/mg Hb).
2.3.5. Red blood cell GR
Glutathione reductase activity was measured using the GR-340 kit (Oxis International, Foster City, CA). Oxidized glutathione, in the presence of NADPH, was reduced by glutathione reductase present in the sample. The enzyme activity was calculated as described above for GPx. The GR activity was normalized to hemoglobin content in the sample and expressed as mU per mg hemoglobin (mU/ mg Hb).
2.3.6. Plasma MPO
Plasma levels of MPO were measured using the MPO-ELISA kit (BioCheck, Inc., Foster City, CA). The assay utilized a unique monoclonal antibody directed against a distinct antigenic determinant on the MPO molecule. This mouse monoclonal anti-MPO antibody was used for solid phase immobilization (on the micro-titter wells). Another mouse monoclonal anti-MPO antibody conjugated to horseradish peroxidase was in the enzyme conjugate solution. The test samples were allowed to react sequentially with these two antibodies, resulting in MPO molecules being sandwiched between the solid phase and enzyme-linked antibodies. After two separate 90- min incubation steps at room temperature with shaking, the wells were rinsed with wash buffer to remove unbound labeled antibodies. Tetramethylbenzidine (TMB) reagent was added and incubated for 20 min with shaking, resulting in the development of color. The color development was stopped with the addition of stop solution, changing the color to yellow. The concentration of MPO was directly proportional to the color intensity of the test samples. Absorbance was measured spectrophotometrically at 405 nm. MPO level was determined using the standard curve data using a four-parameter regression model and expressed as ng/ml.
For each assay, the reagents used were analytical grade, prepared each day, and stored in the refrigerator. Reagents were equilibrated at room temperature for 0.5 h before use. Levels and activities of antioxidants were measured in duplicates or triplicates. The reproducibility calculated as the coefficient of variation was <5% for all assays. No storage-related changes in enzyme activities were observed up to 6 months at −80 °C.
3. Statistical analysis
Statistical analyses were performed using SPSS statistical package (Version 16, SPSS Inc., Chicago, IL). Medians and frequencies of selected characteristics were compared between cases and controls using the Mann Whitney U test for continuous variables, the Fisher’s Exact Test for two level categorical variables, and the Pearson chi-square for all other categorical variables. To examine differences between cases and controls for the median point of mitochondrial DNA index associated with selected categorical characteristics, the Mann Whitney U test was used. To evaluate these median points for selected characteristics within cases and within controls, the Mann Whitney U test was used for two level categorical variables, and the Kruskal–Wallis test was used for variables with more than two levels. Correlations of host characteristics, including age and antioxidant enzyme levels, with case–control status were measured by Kendall’s Tau-b for continuous characteristics, by the phi coefficient for binary characteristics, and by Cramer’s V for characteristics with more than two levels. Analyses were stratified by menopausal status. Odd ratios (OR) and 95% confidence intervals (CI) were estimated with unconditional logistic regression for the main effect of mitochondrial DNA index on breast cancer risk. Potential confounders were tested at the P = 0.10 level and only age significantly affected risk and was included in the model. The mitochondrial DNA index variable was examined in several ways, including as a continuous variable, as a categorical variable based on tertile distributions in controls, and as a categorical variable divided by the median value. Cut-off points for all constructed categorical variables were based on the distribution within the control population. The dose response was tested for the tertile distribution of mitochondrial DNA index by inserting the median value of each tertile and then treating the variable as a continuous variable in the logistic regression model.
4. Results
The characteristics of the study population are summarized in Table 1. The case patients and control subjects were well-matched on ethnicity (P = 1.00), menopausal status (P = 1.00), and age (P = 0.68). There were no statistically significant differences between the case patients and the controls subjects in terms of daily alcohol intakes (P = 0.65), smoking history (P = 0.78), family history of breast cancer (P = 0.57), current fruit intake (P = 0.56), current vegetable intake (P = 0.54), current exercise (P = 0.74), and BMI (P = 0.32).
Table 1.
Distribution of selected characteristics of cases and controls.
| Characteristics | Subjects
|
P-value | |
|---|---|---|---|
| Cases n = 103 | Controls n = 103 | ||
| Median | Median | ||
| Ethnicity | |||
| White | 93 (90.3) | 93 (90.3) | 1.00a |
| Black | 10 (9.7) | 10 (9.7) | |
| Menopausal status | |||
| Premenopausal | 31 (30.1) | 32 (31.1) | |
| Postmenopausal | 72 (69.9) | 71 (68.9) | 1.00a |
| Daily alcohol intake | |||
| No drink per day | 28 (27.2) | 25 (24.3) | |
| 0.5–1 drink per day | 62 (60.2) | 73 (70.9) | |
| >1 drink per day | 13 (12.6) | 5 (4.9) | 0.65b |
| Ever smoked 100 cigarettes | |||
| No | 52 (50.5) | 51 (49.5) | |
| Yes | 50 (48.5) | 49 (47.6) | |
| Don’t’ Know | 0 (0) | 2 (1.9) | 0.78b |
| Family history of breast cancer | |||
| No | 84 (81.6) | 88 (85.4) | |
| Yes | 19 (18.4) | 15 (14.6) | 0.57a |
| Current vegetable intake | |||
| <Once per week | 6 (5.9) | 5 (4.9) | |
| 1–6 per week | 54 (52.4) | 55 (53.4) | |
| >1 per day | 41(39.8) | 42 (40.8) | 0.95b |
| Current fruit intake | |||
| <Once per week | 13 (12.6) | 11 (10.7) | |
| 1–6 per week | 44 (42.7) | 44 (42.8) | |
| >1 per day | 44 (42.7) | 47 (45.7) | 0.88b |
| Current exercise (20 min) | |||
| Never | 33 (32) | 37 (35.9) | |
| <Once per week | 30 (29.1) | 22 (21.4) | |
| 1–2 per week | 22 (21.4) | 34 (33) | |
| 3–4 per week | 16 (15.5) | 9 (8.7) | 0.74b |
| Age (years) | 58 | 56 | 0.68c |
| BMI | 27.79 | 26.43 | 0.32c |
Fisher Exact Analysis was used to examine differences.
Chi square was used to examine differences.
Mann Whitney U was used to examine difference.
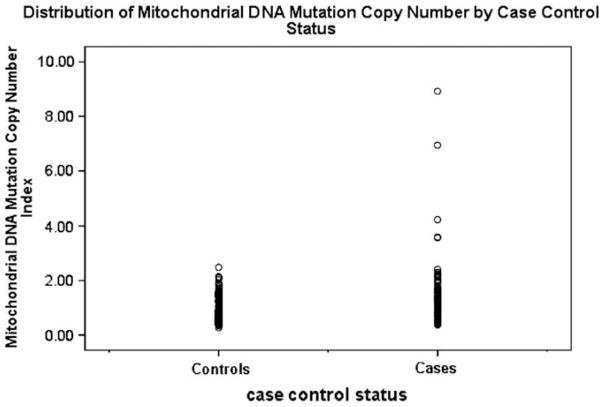
Overall, the relative median mtDNA copy number was statistically significantly higher in case patients with breast cancer (1.29 relative copies) than control subjects (0.80 relative copies) (P < 0.01) (Table 2). The distribution of mtDNA copy number was shown in Fig. 1. The significant difference between breast cancer cases and controls was only observed in Whites (1.31 vs. 0.78, P < 0.01), but not in Blacks (1.17 vs. 1.13, P = 0.88). The significant difference between breast cancer cases and controls was not affected by menopausal status. There was a trend of decreasing mtDNA copy number from premenopausal to postmenopausal in both cases and controls, although neither trend was statistically significant. For daily alcohol intake, the significant case–control difference was observed in women who consumed 0.5–1 drinks per day (1.32 vs. 0.77, P < 0.01), but not among those who consumed more than 1 drink per day. There was a marginal case–control difference among women who do not consume alcohol (1.28 vs. 0.91, P = 0.08). The significant case–control difference was not affected by smoking status, although associations were stronger in women who were neversmokers (1.36 vs. 0.80, P < 0.01) than in women who were ever smokers (1.15 vs. 0.79, P = 0.06). In cases, there was a borderline decrease of mtDNA copy number among women who were never smokers compared to smokers (1.36 vs. 1.15, P = 0.09). The case–control difference was not affected by family history of cancer. In terms of fruit and vegetable intake, the difference between cases and controls were significant in women who consumed vegetables or fruits 1–6 per week or more than 1 per day, but not among women who consumed vegetables or fruits less than once per week. There were significantly higher mtDNA copy number values among cases than among controls in women who exercised, regardless of their exercise frequency (less than once per week, 1–6 per week, or more than once per day).
Table 2.
Comparison of median mtDNA copy number in breast cancer patients and controls.
| Characteristics | Subjects
|
Pa | |
|---|---|---|---|
| Cases (n = 103) | Controls (n = 103) | ||
| Median mtDNA index | Median mtDNA index | ||
| Overall | 1.29 | 0.80 | <0.01 |
| By ethnicity | |||
| White | 1.31 | 0.78 | <0.01 |
| Black | 1.17 | 1.13 | 0.88 |
| Pb | 0.52 | 0.22 | |
| By menopausal status | |||
| Premenopausal | 1.39 | 0.96 | 0.01 |
| Postmenopausal | 1.23 | 0.72 | <0.01 |
| Pb | 0.14 | 0.2 | |
| By daily alcohol intake | |||
| No drinks per day | 1.28 | 0.91 | 0.08 |
| 0.5–1 drinks per day | 1.32 | 0.77 | <0.01 |
| >1 drinks per day | 0.78 | 0.84 | 0.88 |
| Pb | 0.45 | 0.19 | |
| By ever smoked 100 cigarettes | |||
| No | 1.36 | 0.8 | <0.01 |
| Yes | 1.15 | 0.79 | 0.06 |
| Pb | 0.09 | 0.24 | |
| By family history of breast cancer | |||
| No | 1.29 | 0.8 | <0.01 |
| Yes | 1.31 | 0.98 | 0.04 |
| Pb | 0.9 | 0.85 | |
| By current vegetable intake | |||
| <Once per week | 1.03 | 1.35 | 0.36 |
| 1–6 per week | 1.25 | 0.77 | <0.01 |
| >1 per day | 1.37 | 0.81 | <0.01 |
| Pb | 0.41 | 0.13 | |
| By current fruit intake | |||
| <Once per week | 1.29 | 0.8 | 0.2 |
| 1–6 per week | 1.13 | 0.91 | 0.01 |
| >1 per day | 1.39 | 0.82 | <0.01 |
| Pb | 0.12 | 0.8 | |
| By current exercise (20 min) | |||
| Never | 1.25 | 0.89 | 0.11 |
| <Once per week | 1.32 | 0.75 | <0.01 |
| 1–2 per week | 1.29 | 0.78 | 0.03 |
| 3–4 per week | 1.2 | 0.66 | 0.02 |
| Pb | 0.9 | 0.35 | |
Mann Whitney U was used to examine differences within two level categories. Kruskal Wallis was used to examine differences within three and four level categories.
The P-value comparing median mtDNA copy number between cases and controls.
The P-value comparing median mtDNA copy number between groups defined
Fig. 1.
Distribution of mtDNA copy number between cases and controls.
As shown in Table 3, the risk of developing breast cancer increased significantly with increasing mtDNA copy number. As a continuous variable, increasing one relative copy number of mtDNA was associated with 3.54-fold increased risk of breast cancer (95% CI: 1.96–6.46) after adjustment for age, which was the only significant covariate in the logistic regression analysis. After dichotomizing at the 50th percentile value (or median) of mtDNA copy number among control subjects, women with high mtDNA copy number were at significantly increased risk of breast cancer (adjusted OR = 4.67, 95% CI: 2.45–8.92). In further tertile analysis using 33% and 66% values of mtDNA copy number among control subjects as cut-off points, we found that women in the 2nd and 3rd tertiles were at an increased risk of breast cancer (adjusted ORs for the 2nd and 3rd categories = 3.29, 95% CI: 1.40–7.74, and 6.40, 95% CI: 2.74–14.94, respectively), when compared with women with the lowest category of mtDNA copy number. A statistically significant dose–response trend was observed (P < 0.01).
Table 3.
Risk of breast cancer as estimated by mtDNA index.
| mtDNA index (relative copy number) | Number of cases (%) | Number of controls (%) | OR (95% CI) a |
|---|---|---|---|
| Continuous variable | 103 (100) | 103 (100) | 3.54 (1.94–6.46) |
| By tertiles | |||
| ≤0.64 | 10 (9.7) | 33 (32) | 1.00 |
| 0.65–1.18 | 34 (33) | 36 (35) | 3.29 (1.40–7.74) |
| ≥1.19 | 59 (57.3) | 34 (33) | 6.40 (2.74–14.94) P for trend < 0.01 |
| Median | |||
| <0.80 | 19 (18.4) | 51 (49.5) | 1.00 (reference) |
| ≥0.80 | 84 (81.6) | 52 (50.5) | 4.67 (2.45–8.92) |
Odds ratio was adjusted by age only.
Because it is hypothesized that levels of mtDNA copy number might be affected by endogenous and exogenous levels of oxidants/ antioxidants, we investigated relationships between mtDNA copy number and levels and activities of six different oxidants and antioxidants. CuZn-SOD, CAT activity, cellular glutathione peroxidase activity, glutathione reductase activity, and total glutathione were measured in red blood cells, and levels of MPO in plasma (Table 4). In breast cancer cases, we found mtDNA copy number was significantly inversely associated with levels of total glutathione (Tau-b = −0.16, P = 0.02), CuZn-SOD (Tau-b = −0.2, P < 0.01), and MPO (Tau-b = −0.16, P = 0.02). In healthy controls, a significantly inverse association was observed between mtDNA copy number and CAT activities in red blood cells (Tau-b = −0.14, P = 0.03). When we stratified by menopausal status, in cases, the inverse associations between mtDNA copy number and total glutathione, CuZn-SOD, and MPO were still observed in both postmenopausal and premenopausal cases. However, only the association between mtDNA copy number and levels of CuZn-SOD in postmenopausal cases remained statistically significant (Tau-b = −0.22, P < 0.01). In controls, significant associations were only observed for CAT activity among postmenopausal women (Tau-b = −0.19, P = 0.02), and CuZn-SOD among premenopausal women (Taub = −0.26, P = 0.04). We also evaluated associations between mtDNA copy number and two oxidative stress related demographic variables: age and BMI. We observed significant inverse associations between mtDNA copy number and age overall (Taub = −0.18, P = 0.01) and among postmenopausal cases (Taub = −0.19, P = 0.02) and controls (Tau-b = −0.16, P = 0.05) when stratifying by menopausal status. Associations among controls were marginally significant (Tau-b = −0.13, P = 0.06) (Table 4). There were no associations between mtDNA copy number and BMI among cases or controls.
Table 4.
Correlation analyses between selected antioxidants/oxidants and others with mtDNA index stratified by menopausal status.
| Characteristics | Premenopausal
|
Postmenopausal
|
Overall
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases (n = 31) | P-value | Controls (n = 32) | P-value | Cases (n = 72) | P-value | Controls (n = 71) | P-value | Cases (n = 103) | P-value | Controls (n = 103) | P-value | |
| Total glutathione levels (mmol/mg Hgb) | −0.18 | 0.16 | 0.03 | 0.82 | −0.15 | 0.07 | 0.02 | 0.78 | −0.16 | 0.02 | 0.04 | 0.52 |
| Catalase activity (U/mg Hgb) | 0.12 | 0.36 | −0.14 | 0.27 | 0.01 | 0.9 | −0.19 | 0.02 | 0.04 | 0.58 | −0.14 | 0.03 |
| Glutathione reductase activity (mU/mg Hgb) | −0.14 | 0.26 | −0.11 | 0.38 | −0.02 | 0.85 | 0.11 | 0.16 | −0.06 | 0.38 | 0.05 | 0.44 |
| Glutathione peroxidase activity (mU/mg Hgb) | 0.03 | 0.84 | −0.04 | 0.72 | 0.01 | 0.89 | 0.04 | 0.64 | 0.01 | 0.99 | 0.01 | 0.9 |
| CuZn_Superoxide dismutase activity (U/mg Hgb) | −0.14 | 0.28 | −0.26 | 0.04 | −0.22 | <0.01 | 0.03 | 0.7 | −0.2 | <0.01 | 0.04 | 0.54 |
| Myeloperoxidase levels (ng/ml) | −0.2 | 0.11 | −0.02 | 0.85 | −0.12 | 0.13 | 0.1 | 0.21 | −0.16 | 0.02 | 0.07 | 0.31 |
| BMI | 0.04 | 0.73 | 0.08 | 0.54 | −0.07 | 0.42 | 0.03 | 0.76 | −0.05 | 0.45 | 0.05 | 0.46 |
| Age (years) | −0.06 | 0.63 | −0.02 | 0.85 | −0.19 | 0.02 | −0.16 | 0.05 | −0.18 | 0.01 | −0.13 | 0.06 |
Kendall’s Tau-b correlation coefficients (2-tailed) was used to assess the correlations.
5. Discussion
To the best of our knowledge, this is the first study of the relationship between mtDNA copy number measured in peripheral whole blood DNA and risk of breast cancer. In our study, we found that high mtDNA copy number was associated with a statistically significantly increased risk of breast cancer. In addition, we observed that mtDNA content was significantly associated with age and several important endogenous oxidants and antioxidants in cases (including total glutathione, CuZn-SOD activity and MPO) and in controls (CAT activity), suggesting a role of oxidative stress in determination of blood mtDNA copy number.
The relationship between mtDNA copy number in whole blood DNA and cancer risk has been examined in only two studies (Xing et al., 2008; Lan et al., 2008). Our results are consistent with findings in non-Hodgkin’s lymophoma (NHL) Lan et al., 2008, but contradictory to those in renal cell carcinoma (Xing et al., 2008). The inconsistency between mtDNA copy number and cancer risk may reflect findings of differences in mtDNA levels in different types of tumors (Yamada et al., 2006; Wang et al., 2005, 2006). For example, Yamada et al. reported that mtDNA copy number was statistically significantly decreased in hepatocellular carcinoma as compared with the corresponding noncancerous liver tissues (Yamada et al., 2006). However, Wang et al. observed increased mtDNA copy number in pure endometrial adenocarcinoma cells from 65 endometrial cancer patients compared to the normal endometrial glandular epithelial cells from 41 healthy controls (P = 0.001) (Wang et al., 2005). In ovarian tumors, Wang et al. showed that mtDNA copy number in ovarian tumor cells was significantly higher than that in normal ovary (Wang et al., 2006). Therefore, it is most likely that change of mtDNA copy number is not simply a function of enhanced cellular proliferation in neoplastic cells, but also has some degree of specificity for particular cancer type.
Considered the close probable role of oxidative stress in breast carcinogenesis, our finding that higher mtDNA content was associated with more than a threefold increased risk of breast cancer is not surprising at all. There is evidence that elevated mtDNA content is associated with some forms of oxidative stress (Shen et al., 2008; Gadaleta et al., 1992; Lee et al., 2000; Liu et al., 2003) in experimental models. It has been shown that cells under mild oxidative stress may increase mitochondrial production through a pathway that bypasses cell-cycle control. In cells treated with cell-cycle-arresting drugs or H2O2, even though overall cell division was under arrest, mitochondria continued to proliferate as if the cells were going to divide (Lee et al., 2000). During the process of oxidative phosphorylation involving ROS generated by endogenous and exogenous processes, accumulation of mutated mtDNA may occur (Fliss et al., 2000). In addition, certain mutated mtDNAs may obtain a replicative advantage that can generate increased superoxide and nitric oxide levels and lead to aberrant mitochondrial biogenesis, which has been associated with deficient or defective apoptosis (Carew et al., 2004). Therefore, in our study, the case–control difference in mtDNA copy number might reflect the possible case–control difference of oxidative stress.
This observation was further strengthened by our observations of associations between mtDNA copy number and the levels of antioxidants and oxidants in blood. Alteration of the copy number of mtDNA in tumor tissues has been considered as one of the factors involved in the pathogenesis of oxidative stress-related human cancers. Thus, the levels of antioxidants and prooxidants may play a role in the mechanism of adjusting mtDNA copy number in affected tissue cells. Human leukocyte is a good target cell for studying the relationship between the alteration of mtDNA copy number and the change of antioxidants and oxidants status, not only due to easy sampling but also because of the characteristic high demand on aerobic metabolism during the immune response (Tsai et al., 2001). For endogenous antioxidants, we observed statistically significant inverse associations between mtDNA copy number and RBC levels of total glutathione (P = 0.02) and CuZn- SOD activity (P < 0.01) in breast cancer cases, and RBC levels of CAT activity (P = 0.03) in healthy controls. Glutathione, CuZn- SOD, and catalase play important roles in the antioxidant system that is required for the defense against free radicals and detoxification of toxic compounds (Ambrosone, 2000; Halliwell and Gutteridge, 1999). The function of the CuZn-SOD protein is to dismutate superoxide to generate hydrogen peroxide. Then, catalase is required to convert hydrogen peroxide to water and molecular oxygen. Studies have suggested that the levels of key antioxidants in the blood, including total glutathione, CuZn-SOD, and catalase, are implicated in the diagnosis and prognosis of breast cancer (Kumaraguruparan et al., 2002; Seven et al., 1998; Polat et al., 2002). Decrease in antioxidants will lead to increased levels of oxidative stress. Consequently, the elevated oxidative stress may initiate the compensatory response of increasing mtDNA production in order to protect the mitochondrial genome from oxidative damage. Therefore, our observation of the inverse associations between mtDNA copy number and several antioxidants in blood may suggest that an increase of mtDNA copy number in cases may serve as an index of the increased cellular levels of oxidative stress. It also indicates that the cases might have higher levels of oxidative stress than the controls.
The inverse association between mtDNA copy number and plasma levels of MPO among women with breast cancer is intriguing. These associations may be due to the direct damage to mtDNA by MPO-dependent –OH production, and consequently the decrease of mtDNA copy number. MPO-generated reactive chlorinating species have been implicated in playing a role in carcinogenesis through both activation of procarcinogens to genotoxic intermediates and the potentiation of xenobiotic carcinogenicity (Shen et al., 2000), and we have observed increased risk of breast cancer among women with genotypes encoding higher levels of MPO. Studies have shown that MPO-dependent –OH production could cause significant damage in mtDNA (Shen et al., 2000; Roy et al., 2007). Compared to genomic DNA, RNA, and protein, mtDNA demonstrated the greatest levels of 8-OHG following exposure of cells to the MPO–H2O2–Cl– system. A likely explanation for the enhanced susceptibility of mtDNA to MPO-dependent DNA damage is that mitochondria serve as a primary source of intracellular – O2 production. This would facilitate –OH formation in close proximity to mtDNA. In addition, mtDNA does not bear a protective coat of highly basic histone proteins; thus, protection from reactive oxidants is far less than that observed in the nucleus. Finally, DNA repair mechanisms in the mitochondria are less efficient than those in the nucleus. Because the cases had slightly higher levels of MPO than the controls (median: 34.7 vs. 31.88, P = 0.08) in this study population, the mtDNA damage might be more severe in the cases than the controls. That might be the reason we only observed the inverse associations among the cases, but not in the controls.
Cellular oxidative stress is thought to play a role in the aging process and may affect mtDNA replication in the human cell. In this current study, we observed inverse associations between mtDNA copy number and age in both cases and controls. These associations were only observed among postmenopausal women. Our observation is consistent with several previous reports in neurons, muscle cells, and leukocytes (Chen et al., 2002; Hayakawa et al., 1991; Welle et al., 2003; Pollack and Leeuwenburgh, 2001; Drouet et al., 1999). The relationship between age and mtDNA copy number in leukocytes is thought to fit in the quadratic regression model. Before an individual reaches middle age, the mtDNA copy number in the leukocyte appears to correlate positively with age, and progressively shifts to negative correlation thereafter (Lee et al., 2000). Age from 48 to 60 seems to be a peak response of mtDNA copy number to aging. Because the median age of the cases is 58 and median age of the controls is 56 in this study, the mtDNA copy number for most of our study subjects has likely already reached or passed the peak, especially for postmenopausal women, and could account for the inverse associations. The decrease of mtDNA copy number at old age may be attributed to the enhanced oxidative stress in the advanced aging process.
In summary, our study provides the first evidence that mtDNA copy number is statistically significantly higher in breast cancer compared to the healthy controls. Results also indicate the role of endogenous antioxidants/oxidants in the determination of mtDNA copy number. Because the study is relatively small and also the nature of the case–control study design, replication in a large study is necessary, preferably in prospective cohort studies. Nevertheless, our study indicates that mtDNA copy number might be a potentially useful tool in studying the etiology of breast cancer.
References
- Ambrosone CB. Oxidants and antioxidants in breast cancer. Antioxid Redox Signal. 2000;2:903–917. doi: 10.1089/ars.2000.2.4-903. [DOI] [PubMed] [Google Scholar]
- Ambrosone CB, Nesline MK, Davis W. Establishing a cancer center data bank and biorepository for multidisciplinary research. Cancer Epidemiol Biomarkers Prev. 2006;15:1575–1577. doi: 10.1158/1055-9965.EPI-06-0628. [DOI] [PubMed] [Google Scholar]
- Carew JS, Nawrocki ST, Xu RH, Dunner K, McConkey DJ, Wierda WG, Keating MJ, Huang P. Increased mitochondrial biogenesis in primary leukemia cells: the role of endogenous nitric oxide and impact on sensitivity to fludarabine. Leukemia. 2004;18:1934–1940. doi: 10.1038/sj.leu.2403545. [DOI] [PubMed] [Google Scholar]
- Chen D, Cao G, Hastings T, Feng Y, Pei W, O’Horo C, Chen J. Age-dependent decline of DNA repair activity for oxidative lesions in rat brain mitochondria. J Neurochem. 2002;81:1273–1284. doi: 10.1046/j.1471-4159.2002.00916.x. [DOI] [PubMed] [Google Scholar]
- Crimi M, Rigolio R. The mitochondrial genome, a growing interest inside an organelle. Int J Nanomed. 2008;3:51–57. doi: 10.2147/ijn.s2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouet M, Lauthier F, Charmes JP, Sauvage P, Ratinaud MH. Age-associated changes in mitochondrial parameters on peripheral human lymphocytes. Exp Gerontol. 1999;34:843–852. doi: 10.1016/s0531-5565(99)00058-3. [DOI] [PubMed] [Google Scholar]
- Fliss MS, Usadel H, Caballero OL, Wu L, Buta MR, Eleff SM, Jen J, Sidransky D. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287:2017–2019. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- Gadaleta MN, Rainaldi G, Lezza AM, Milella F, Fracasso F, Cantatore P. Mitochondrial DNA copy number and mitochondrial DNA deletion in adult and senescent rats. Mutat Res. 1992;275:181–193. doi: 10.1016/0921-8734(92)90022-h. [DOI] [PubMed] [Google Scholar]
- Gönenç A, Erten D, Aslan S, Akinci M, Simŝek B, Torun M. Lipid peroxidation and antioxidant status in blood and tissue of malignant breast tumor and benign breast disease. Cell Biol Int. 2006;30:376–380. doi: 10.1016/j.cellbi.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge J. Free Radicals in Biology and Medicine. 3. Springer; Berlin: 1999. [Google Scholar]
- Hayakawa M, Torii K, Sugiyama S, Tanaka M, Ozawa T. Age-associated accumulation of 8-hydroxydeoxyguanosine in mitochondrial DNA of human diaphragm. Biochem Biophys Res Commun. 1991;179:1023–1029. doi: 10.1016/0006-291x(91)91921-x. [DOI] [PubMed] [Google Scholar]
- Kumaraguruparan R, Subapriya R, Kabalimoorthy J, Nagini S. Antioxidant profile in the circulation of patients with fibroadenoma and adenocarcinoma of the breast. Clin Biochem. 2002;35:275–279. doi: 10.1016/s0009-9120(02)00310-7. [DOI] [PubMed] [Google Scholar]
- Kushi LH, Fee RM, Sellers TA, Zheng W, Folsom AR. Intake of vitamins A, C, and E and postmenopausal breast cancer: the Iowa Women’s Health Study. Am J Epidemiol. 1996;144:165–174. doi: 10.1093/oxfordjournals.aje.a008904. [DOI] [PubMed] [Google Scholar]
- Lan Q, Lim U, Liu C, Weinstein SJ, Chanock S, Bonner MR, Virtamo J, Albanes D, Rothman N. A prospective study of mitochondrial DNA copy number and risk of non-Hodgkin lymphoma. Blood. 2008;112:4247–4249. doi: 10.1182/blood-2008-05-157974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Lu CY, Fahn HJ, Wei YH. Aging- and smoking-associated alteration in the relative content of mitochondrial DNA in human lung. FEBS Lett. 1998;441:292–296. doi: 10.1016/s0014-5793(98)01564-6. [DOI] [PubMed] [Google Scholar]
- Lee HC, Yin PH, Lu CY, Chi CW, Wei YH. Increase of mitochondria and mitochondrial DNA in response to oxidative stress in human cells. Biochem J. 2000;348(Pt. 2):425–432. [PMC free article] [PubMed] [Google Scholar]
- Liu CS, Tsai CS, Kuo CL, Chen HW, Lii CK, Ma YS, Wei YH. Oxidative stress-related alteration of the copy number of mitochondrial DNA in human leukocytes. Free Radical Res. 2003;37:1307–1317. doi: 10.1080/10715760310001621342. [DOI] [PubMed] [Google Scholar]
- Nielsen NR, Grønbaek M. Stress and breast cancer: a systematic update on the current knowledge. Nat Clin Pract Oncol. 2006;3:612–620. doi: 10.1038/ncponc0652. [DOI] [PubMed] [Google Scholar]
- Park NJ, Kang DH. Breast cancer risk and immune responses in healthy women. Oncol Nurs Forum. 2006;33:1151–1159. doi: 10.1188/06.ONF.1151-1159. [DOI] [PubMed] [Google Scholar]
- Polat MF, Taysi S, Gul M, Cikman O, Yilmaz I, Bakan E, Erdogan F. Oxidant/antioxidant status in blood of patients with malignant breast tumour and benign breast disease. Cell Biochem Funct. 2002;20:327–331. doi: 10.1002/cbf.980. [DOI] [PubMed] [Google Scholar]
- Pollack M, Leeuwenburgh C. Apoptosis and aging: role of the mitochondria. J Gerontol A Biol Sci Med Sci. 2001;56:B475–482. doi: 10.1093/gerona/56.11.b475. [DOI] [PubMed] [Google Scholar]
- Putignani L, Raffa S, Pescosolido R, Aimati L, Signore F, Torrisi MR, Grammatico P. Alteration of expression levels of the oxidative phosphorylation system (OXPHOS) in breast cancer cell mitochondria. Breast Cancer Res Treat. 2008;110:439–452. doi: 10.1007/s10549-007-9738-x. [DOI] [PubMed] [Google Scholar]
- Renis M, Cantatore P, Loguericio PP, Fracasso F, Gadaleta MN. Content of mitochondrial DNA and of three mitochondrial RNAs in developing and adult rat cerebellum. J Neurochem. 1989;52:750–754. doi: 10.1111/j.1471-4159.1989.tb02518.x. [DOI] [PubMed] [Google Scholar]
- Roy D, Cai Q, Felty Q, Narayan S. Estrogen-induced generation of reactive oxygen and nitrogen species, gene damage, and estrogen-dependent cancers. J Toxicol Environ Health B Crit Rev. 2007;10:235–257. doi: 10.1080/15287390600974924. [DOI] [PubMed] [Google Scholar]
- Sener DE, Gönenç A, Akinci M, Torun M. Lipid peroxidation and total antioxidant status in patients with breast cancer. Cell Biochem Funct. 2007;25:377–382. doi: 10.1002/cbf.1308. [DOI] [PubMed] [Google Scholar]
- Seven A, Erbil Y, Seven R, Inci F, Gülyaŝar T, Barutçu B, Candan G. Breast cancer and benign breast disease patients evaluated in relation to oxidative stress. Cancer Biochem Biophys. 1998;16:333–345. [PubMed] [Google Scholar]
- Shen Z, Wu W, Hazen SL. Activated leukocytes oxidatively damage DNA, RNA, and the nucleotide pool through halide-dependent formation of hydroxyl radical. Biochemistry. 2000;39:5474–5482. doi: 10.1021/bi992809y. [DOI] [PubMed] [Google Scholar]
- Shen M, Zhang L, Bonner MR, Liu CS, Li G, Vermeulen R, Dosemeci M, Yin S, Lan Q. Association between mitochondrial DNA copy number and occupational benzene exposure. Environ Mol Mutagen. 2008;49:453–457. doi: 10.1002/em.20402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K, Hsu TG, Lu FJ, Hsu CF, Liu TY, Kong CW. Age-related change in the mitochondrial depolarization induced by oxidative injury in human peripheral blood leukocytes. Free Radical Res. 2001;35:395–403. doi: 10.1080/10715760100300911. [DOI] [PubMed] [Google Scholar]
- Tseng LM, Yin PH, Chi CW, Chi CW, Hsu CY, Wu CW, Lee LM, Wei YH, Lee HC. Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosome Cancer. 2006;45:629–638. doi: 10.1002/gcc.20326. [DOI] [PubMed] [Google Scholar]
- Veltri KL, Espiritu M, Singh G. Distinct genomic copy number in mitochondria of different mammalian organs. J Cell Physiol. 1990;143:160–164. doi: 10.1002/jcp.1041430122. [DOI] [PubMed] [Google Scholar]
- Verma M, Naviaus RK, Tanaka M, Kumar D, Franceschi C, Singh KK. Meeting report: mitochondrial DNA and cancer epidemiology. Cancer Res. 2007;67:437–439. doi: 10.1158/0008-5472.CAN-06-4119. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu VW, Xue WC, Tsang PC, Cheung AN, Ngan HY. The increase of mitochondrial DNA content in endometrial adenocarcinoma cells: a quantitative study using laser-captured microdissected tissues. Gynecol Oncol. 2005;98:104–110. doi: 10.1016/j.ygyno.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu VW, Xue WC, Cheung AN, Ngan HY. Association of decreased mitochondrial DNA content with ovarian cancer progression. Brit J Cancer. 2006;95:1087–1091. doi: 10.1038/sj.bjc.6603377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Wang HW, Yao YG, Kong QP, Zhang YP. Somatic mutations of mitochondrial genome in early stage breast cancer. Int J Cancer. 2007;121:1253– 1256. doi: 10.1002/ijc.22822. [DOI] [PubMed] [Google Scholar]
- Welle S, Bhatt K, Shah B, Needler N, Delehanty JM, Thornton CA. Reduced amount of mitochondrial DNA in aged human muscle. J Appl Physiol. 2003;94:1479–1484. doi: 10.1152/japplphysiol.01061.2002. [DOI] [PubMed] [Google Scholar]
- Xing J, Chen M, Wood CG, Lin J, Spitz MR, Ma J, Amos CI, Shields PG, Benowitz NL, Gu J, de Andrade M, Swan GE, et al. Mitochondrial DNA content: its genetic heritability and association with renal cell carcinoma. J Nat Cancer Inst. 2008;100:1104–1112. doi: 10.1093/jnci/djn213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Nomoto S, Fujii T, Kaneko T, Takeda S, Inoue S, Kanazumi N, Nakao A. Correlation between copy number of mitochondrial DNA and clinico-pathologic parameters of hepatocellular carcinoma. Eur J Surg Oncol. 2006;32:303–307. doi: 10.1016/j.ejso.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Ye C, Shu XO, Wen W, Pierce L, Courtney R, Gao YT, Zheng W, Cai Q. Quantitative analysis of mitochondrial DNA 4977-bp deletion in sporadic breast cancer and benign breast diseases. Breast Cancer Res Treat. 2008;108:427–434. doi: 10.1007/s10549-007-9613-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Zhou Y, Shi Y, Ning L, Yang Y, Wei X, Zhang N, Hao X, Niu R. Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients. IUBMB Life. 2007;59:450–457. doi: 10.1080/15216540701509955. [DOI] [PubMed] [Google Scholar]
- Zhang S, Hunter DJ, Forman MR, Rosner BA, Speizer FE, Colditz GA, Manson JE, Hankinson SE, Willett WC. Dietary carotenoids and vitamins A, C, and E and risk of breast cancer. J Nat Cancer Inst. 1999;91:547–556. doi: 10.1093/jnci/91.6.547. [DOI] [PubMed] [Google Scholar]