Abstract
Purpose
Recent studies indicate that mitochondrial proteins may contribute to the pathogenesis of primary open-angle glaucoma (POAG). In this study, we examined the association between POAG and common variations in gene-encoding mitochondrial proteins.
Methods
We examined genetic data from 3430 POAG cases and 3108 controls derived from the combination of the GLAUGEN and NEIGHBOR studies. We constructed biological-system coherent mitochondrial nuclear-encoded protein gene-sets by intersecting the MitoCarta database with the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. We examined the mitochondrial gene-sets for association with POAG and with normal-tension glaucoma (NTG) and high-tension glaucoma (HTG) subsets using Pathway Analysis by Randomization Incorporating Structure.
Results
We identified 22 KEGG pathways with significant mitochondrial protein-encoding gene enrichment, belonging to six general biological classes. Among the pathway classes, mitochondrial lipid metabolism was associated with POAG overall (P = 0.013) and with NTG (P = 0.0006), and mitochondrial carbohydrate metabolism was associated with NTG (P = 0.030). Examining the individual KEGG pathway mitochondrial gene-sets, fatty acid elongation and synthesis and degradation of ketone bodies, both lipid metabolism pathways, were significantly associated with POAG (P = 0.005 and P = 0.002, respectively) and NTG (P = 0.0004 and P < 0.0001, respectively). Butanoate metabolism, a carbohydrate metabolism pathway, was significantly associated with POAG (P = 0.004), NTG (P = 0.001), and HTG (P = 0.010).
Conclusions
We present an effective approach for assessing the contributions of mitochondrial genetic variation to open-angle glaucoma. Our findings support a role for mitochondria in POAG pathogenesis and specifically point to lipid and carbohydrate metabolism pathways as being important.
Keywords: glaucoma, genetics, mitochondria
Although raised intraocular pressure (IOP) is a key risk factor for primary open-angle glaucoma (POAG), only a fraction of individuals with elevated IOP develop POAG, and approximately one-third of Caucasian (European ancestry) POAG patients have normal IOP levels (normal-tension glaucoma [NTG]) suggesting that other factors can influence susceptibility to optic nerve degeneration. There is growing evidence for mitochondrial dysfunction in optic nerve susceptibility to glaucoma.1–4 The characteristic visual field loss seen in glaucoma locates the site of retinal ganglion cell (RGC) injury to the optic nerve head. RGC axons coursing through the retina and into the prelaminar cribrosa optic nerve are not myelinated, providing the necessary transparency to incident light on its route to the retinal photoreceptors. This unmyelinated axonal segment represents the longest node of Ranvier in the body and to sustain action potentials is endowed with an ample supply of mitochondria necessary to generate sufficient adenosine triphosphate (ATP) to sustain action potential propagation in the absence of myelin.5 Because of this high energy requirement, the optic nerve head is a region that is particularly susceptible to mitochondrial dysfunction. For example, RGC loss or dysfunction is a feature of diseases caused by mitochondrial DNA mutations, including Leber's hereditary optic neuropathy and is also a common manifestation of many other mitochondrial disease syndromes.5 Mitochondrial dysfunction has been increasingly shown to play a role in other age-related neurodegenerative conditions such as Parkinson's disease and Alzheimer's disease.6
Genetic studies have suggested that nuclear genes encoding mitochondrial proteins may contribute to POAG risk.7 Recently, a large genome-wide association study identified significant association of POAG with single nucleotide polymorphisms (SNPs) in the TXNRD2 genomic region, a nuclear-encoded mitochondrial protein that functions to maintain redox homeostasis.4 In this study, we examined the association between genetic variation in a comprehensive set of nuclear-encoded mitochondrial proteins and POAG in a large case-control dataset. We conducted gene-set analyses of mitochondria-enriched biological pathways, examining the association with POAG as well as NTG and high-tension glaucoma (HTG) subgroups.
Methods
Participants and Definitions
We examined genetic data from 3430 POAG cases and 3108 controls derived from the combination of the Glaucoma Genes and Environment (GLAUGEN) and NEI Glaucoma Human Genetics collaBORation (NEIGHBOR) studies.8,9 A meta-analysis of the Genome-Wide Association Studies (GWAS) for POAG from these two studies has been previously published,10 and the results of this meta-analysis have been used for the analyses conducted in this study. Detailed methods for participant recruitment and the GWAS have been described.8–10 In brief, GLAUGEN consists of participants from two cohort studies, the Health Professionals Follow-up Study and the Nurses' Health Study, and one clinic-based study from the Massachusetts Eye and Ear Infirmary; NEIGHBOR consists of participants from 12 clinic-based studies in the United States. The research followed the Declaration of Helsinki and was approved by the institutional review boards of the Brigham and Women's Hospital; Duke University; Harvard T.H. Chan School of Public Health; Johns Hopkins University; the Marshfield Clinic; Massachusetts Eye and Ear Infirmary; Stanford University; University of California, San Diego; University of Miami; University of Michigan; University of Pittsburgh; and University of West Virginia. All participants provided written informed consent and all participants were Caucasians with European ancestry. Primary open-angle glaucoma cases were required to have visual field defects consistent with nerve fiber layer pathology, open angles, and no other significant findings on slit-lamp examination. Intraocular pressure was not part of the case definition, which allowed subgroup analyses of normal tension POAG (NTG, highest known screening IOP < 22 mm Hg) and high-tension POAG (HTG, history of IOP ≥ 22 mm Hg) in those participants with IOP data available. Visual field loss was required to be reproduced in the same region on a subsequent test, or if these data were not available, then the case definition required signs suggestive of glaucomatous optic neuropathy, namely a vertical cup-to-disc ratio > 0.7. Control participants were required to have a vertical cup-to-disc ratio < 0.6 and IOP < 22 mm Hg as evidenced at an eye examination during the 2 years prior to study enrollment.
Genotyping and Association Analysis
The Illumina Human 660WQuadv1C BeadChip array (Illumina, San Diego, CA, USA) was used to genotype all participants. GLAUGEN study samples were processed at the Broad Institute (Cambridge, MA, USA), and NEIGHBOR samples were processed at the Center for Inherited Disease Research (Baltimore, MD, USA). Details regarding data cleaning and quality control have been published previously.10 The association analyses of genotypes with POAG were carried out with logistic regression assuming an additive model using PLINK v1.07 (http://pngu.mgh.harvard.edu/~purcell/plink/, in the public domain).11 Analyses were conducted separately for GLAUGEN and NEIGHBOR (genomic inflation factors were 1.009 and 1.034, respectively) and then meta-analyzed, weighting by inverse variance using METAL.12 GLAUGEN analyses were adjusted for age, sex, study site, DNA source, DNA extraction method, and three principal components; NEIGHBOR analyses were adjusted for age, sex, study site, and two principal components.
Defining the Mitochondriome and Mitochondria-Enriched Biological Pathways
The mitochondrial proteome consists of more than 1000 proteins, only 13 of which are encoded by mitochondrial DNA; the vast majority of proteins that are important for mitochondrial structure and function are encoded by nuclear DNA.13 For the purpose of the current study, we confined our analysis to nuclear genes encoding the mitochondrial proteome because the platform used for genotyping does not include a sufficient number of mitochondrial DNA variants for analysis using pathway software programs, including the program Pathway Analysis by Randomization Incorporating Structure (PARIS; http://csg.sph.umich.edu/abecasis/metal/index.html, in the public domain) used for this study.14 We identified 1010 nuclear-encoded mitochondria genes using the Human MitoCarta database (www.broadinstitute.org/pubs/MitoCarta, in the public domian)15 and refer to these genes as the mitochondriome (Supplementary Fig. S1). In addition to the production of ATP by oxidative phosphorylation, mitochondria are essential for a diverse range of biological processes, and different genes in the mitochondriome may be responsible for contrasting biological pathways. We therefore identified subsets of the mitochondriome that were important for different biological pathways by intersecting the Human MitoCarta database with the Kyoto Encyclopedia of Genes and Genomes (KEGG) biological pathway database (version 58.1).16 We considered KEGG pathways to have a significant mitochondrial component if the gene enrichment P value of the MitoCarta genes in the overall pathway was significant using the hypergeometric test with Bonferroni correction (209 pathways yielded a significance threshold of 2.4 × 10−4).
Mitochondria-Enriched Biological Pathway Analyses
For each mitochondria-enriched biological pathway we identified, we defined a gene-set that consisted of only the MitoCarta genes within the pathway, aiming to represent the mitochondrial component of the biological process. We tested for association between these gene-sets and POAG overall, NTG and HTG using PARIS (version 1.1.1).14 An advantage of a pathway or gene-set approach is that variants of smaller effect sizes that individually did not meet strict multiple comparison criteria for genome-wide association may in aggregate show significant association with disease as a result of the overall effect of the cumulative variation of a pathway or set of genes.
The approach used by PARIS has been described in detail previously14 and has been used successfully for examining the association between the estrogen metabolism pathway and POAG17 as well as for examining the association of all KEGG pathways with POAG.18 Briefly, PARIS divides the specified gene-set into linkage disequilibrium (LD) blocks, and randomly selects similarly sized LD blocks from the rest of the genome for comparison. The number of LD blocks with at least one SNP significant at P < 0.05 is compared between the gene-set in question and the randomly selected set of LD blocks. This process is repeated a user-defined number of times (we completed 10,000 permutations per test), and a permuted P value is calculated based on the number of times the gene-set in question had more significant LD blocks than the randomly selected comparison.
PARIS also calculates gene P values by repeating the randomization and permutation process using just the LD blocks within each gene. Single nucleotide polymorphisms from the GWAS were considered to reside in a pathway gene if the SNP fell within the ENSEMBL genomic interval ±50 kb to either side of the gene. If the overlap included another gene in the pathway, the overlapping SNP(s) were counted once. Examining gene P values helps identify which parts of a larger pathway are driving an association. However, caution is required when interpreting gene P values, especially if only a small number of LD blocks are represented by the gene.14 Consider an extreme example of a gene with just one LD block, and that LD block has a SNP P < 0.05. PARIS will always produce a significant gene P value for such genes. We therefore only examined gene P values for genes represented by at least five LD blocks from our genotyping data.
We set a threshold for a single-allele P value of <0.05 as nominally significant for use within the PARIS analysis and used a permutation test of 10,000 permutations to determine the significance of the pathway analyses. A KEGG pathway was considered to be significant at the P < 0.0001 level if none of the 10,000 randomized pathways had more significant SNP signals than the actual pathway.
Results
We examined genetic data from 3,430 POAG cases (mean age 65.2 years, 54% women) and 3,108 controls (mean age 67.5 years, 56% women). For these cases, IOP data were available from 2354 participants (67%); 717 were classified as NTG and 1637 as HTG.
We identified 22 KEGG pathways with significant mitochondrial gene enrichment, belonging to six pathway classes: amino acid metabolism, carbohydrate metabolism, energy metabolism, lipid metabolism, metabolism of cofactors and vitamins, and nucleotide metabolism (Table 1). Pathway-specific mitochondrial gene-sets were created using these data; 6 gene-sets relating to the pathway classes and 22 gene-sets relating to the individual pathways within the classes (Supplementary Table S1). We excluded genes not present in the Human MitoCarta database from the gene sets.
Table 1.
KEGG Pathways With Significant Enrichment of MitoCarta Genes
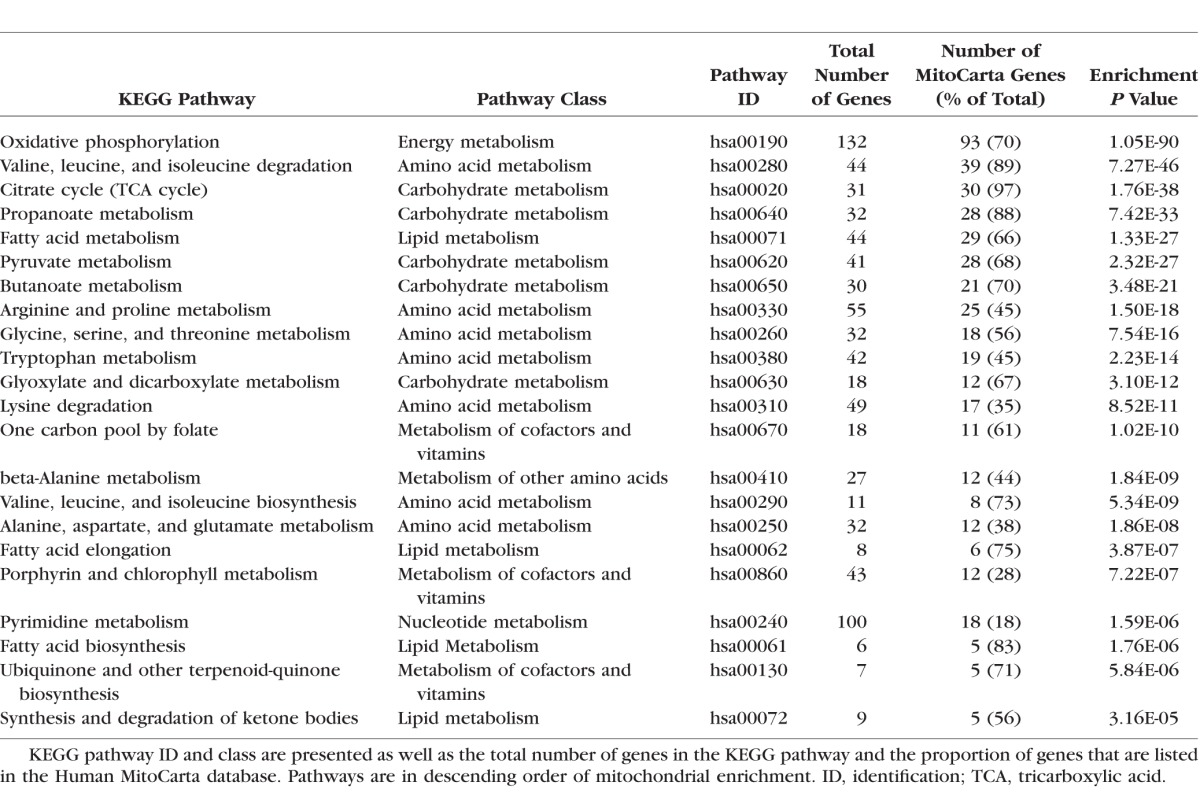
Table 2 presents the results of an examination of the associations between the six main pathway class gene-sets and POAG, NTG, and HTG using PARIS. The mitochondrial gene-set involved in lipid metabolism was nominally associated with POAG overall (P = 0.013); this association appeared to be driven by a significant association with NTG (P = 0.0006), with no significant association seen with HTG (P = 0.15). A nominal association was also observed between the carbohydrate metabolism mitochondrial gene-set and NTG and the nucleotide metabolism gene-set (that includes TXNRD2) with POAG overall (both P < 0.05).
Table 2.
Associations Between the Six Main Pathway Class Mitochondrial Gene-Sets and Primary Open-Angle Glaucoma (POAG, 3430 Cases), Normal Tension Glaucoma (NTG, 717 Cases), and High Tension Glaucoma (HTG, 1637 Cases) Versus Controls (3108) in the Combined GLAUGEN–NEIGHBOR Dataset
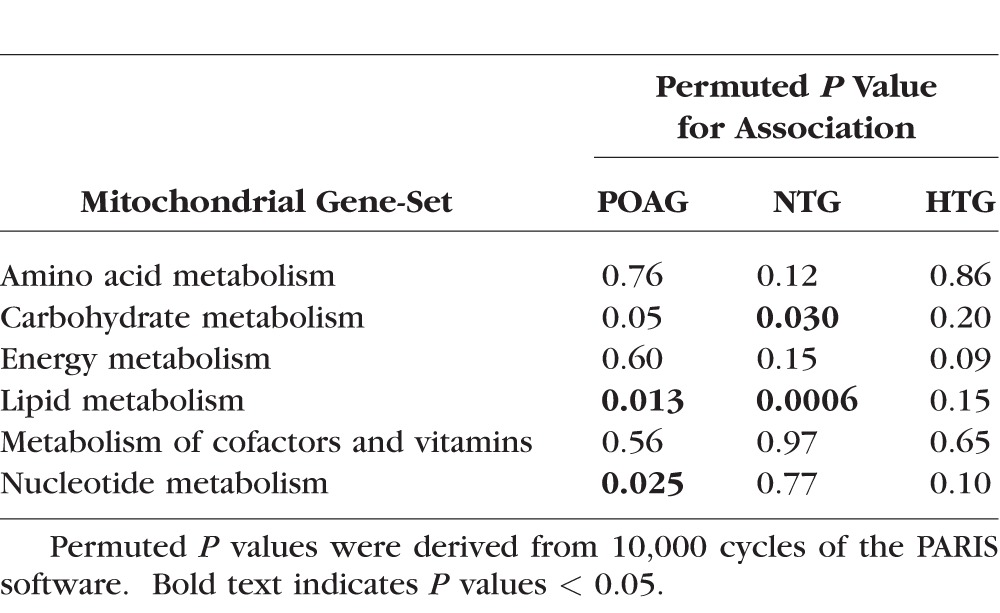
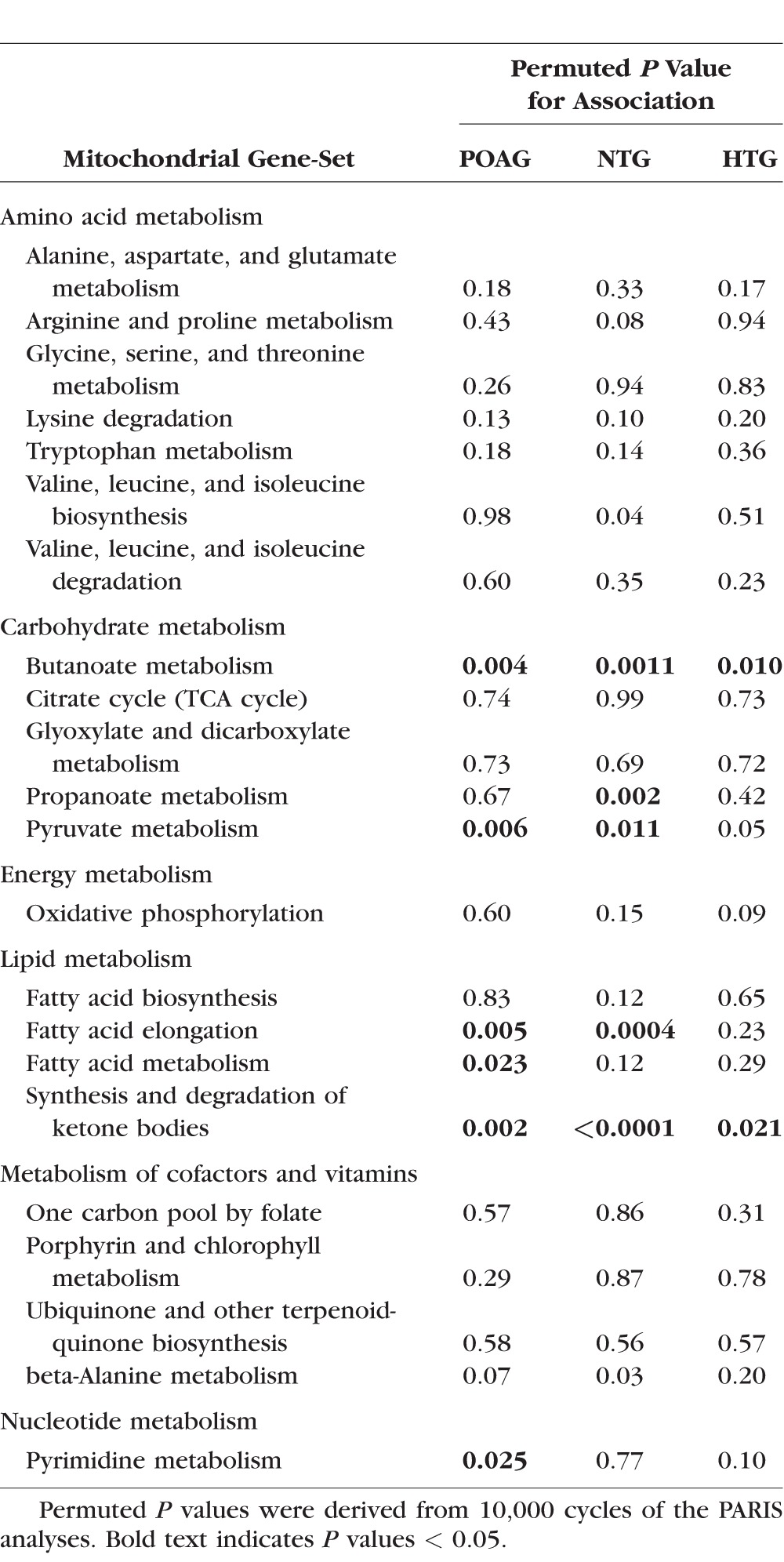
Table 3 presents the associations for the 22 individual KEGG pathway-defined mitochondrial gene-sets and POAG, NTG, and HTG. As expected, some lipid metabolism class gene-sets were significantly associated with POAG, and particularly with NTG. Specifically, the mitochondrial components of fatty acid elongation and synthesis and degradation of ketone bodies were significantly associated with POAG overall (P = 0.005 and P = 0.002), and significantly associated with NTG (P = 0.0004 and P < 0.0001). The mitochondrial component of fatty acid metabolism was nominally associated with POAG overall (P = 0.023). There was also evidence of association between the carbohydrate metabolism class gene-sets and NTG, especially the butanoate metabolism gene-set, which was significantly associated with NTG (P = 0.001) in addition to an interesting association between NTG and propanoate metabolism (P = 0.002) and pyruvate metabolism (P = 0.01).
Table 3.
Associations Between KEGG Pathway-Defined Mitochondrial Gene-Sets and Primary Open-Angle Glaucoma (POAG, 3430 Cases), Normal Tension Glaucoma (NTG, 717 Cases), and High Tension Glaucoma (HTG, 1637 Cases) Versus Controls (3108) in the Combined GLAUGEN–NEIGHBOR Dataset
We examined the component gene P values for the lipid metabolism and carbohydrate metabolism gene-sets (Supplementary Tables S2, S3). Genes driving both lipid and carbohydrate metabolism associations with POAG and NTG were BDH1 (POAG and NTG P < 0.0001), EHHADH (POAG P = 0.044, NTG P = 0.0009), and ECHS1 (POAG P = 0.004, NTG P = 0.002). In addition, ACAA2 (POAG P = 0.024, NTG P = 0.0008) from the lipid metabolism gene-set and ME3 (POAG P = 0.0003, NTG P = 0.30), ACYP2 (POAG P = 0.001, NTG P = 0.43), LDHD (POAG P = 1.00, NTG P = 0.004), and LDHB (POAG P = 1.00, NTG P = 0.003) from the carbohydrate metabolism gene-set were significantly associated with POAG and/or NTG. The gene P values for TXNRD2 were significant for an association with POAG overall (P < 0.001).
Discussion
In this large case-control study, we demonstrated significant associations between POAG and groups of nuclear genes important for specific aspects of mitochondrial function, namely, lipid metabolism (fatty acid elongation and synthesis and degradation of ketone bodies) and carbohydrate metabolism (butanoate metabolism). These associations were particularly apparent for the NTG subgroup.
Mitochondria serve many diverse biological roles, including generation of ATP, cellular homeostasis, signaling, neuronal excitability, and apoptosis.19 Challenges exist in examining the role of mitochondria in disease. It seems likely that for any particular disease, only specific mitochondrial functions may impact disease pathogenesis. For example, oxidative phosphorylation was notably not associated with POAG in our study despite being the pathway with the greatest nuclear-encoded mitochondrial gene enrichment. In our study, we present an effective approach to examining the role of mitochondria in disease by using variation in the mitochondriome as a proxy for mitochondrial function. After dividing the mitochondriome into biological system coherent gene-sets and examining each gene-set for collective association with disease, we demonstrated a significant association between POAG and mitochondrial gene-sets that are involved in lipid and carbohydrate metabolism. Our results were especially significant in the NTG subgroup, suggesting that the observed association is more likely to influence optic nerve susceptibility rather than elevated IOP.
The mechanisms by which mitochondrial impairment in these pathways may contribute to glaucoma are not clear, although our results suggest that they may be related to fatty acid metabolism and elongation, synthesis and degradation of ketone bodies, and butanoate and pyruvate metabolism. The association of the whole butanoate KEGG pathway (including nonmitochondrial genes) with POAG in our study population has been previously reported.18 Genes in the butanoate metabolism pathway contribute to the formation of 4-aminobutanoate (GABA). GABA plays an important role in the visual response, and the inhibition of GABA transaminase by the antiepileptic medication vigabatrin can result in visual field defects.20 Our gene-set analysis results are in agreement with a study that examined 65 candidate POAG genes using the Ingenuity knowledge database (www.ingenuity.com, in the public domain); the investigators reported that the genes take part in only four common functional molecular networks corresponding to lipid metabolism, developmental function, and inflammatory processes.21 Also consistent with our findings is a GWAS of NTG in Japanese patients that found an interesting association with ELOVL5, a gene involved in long-chain polyunsaturated fatty acid synthesis.22 Abnormal lipid23,24 and carbohydrate25 metabolism has been demonstrated in glaucomatous eyes, as observed in the trabecular meshwork and aqueous humor. These data are consistent with our pathway analysis data, although our results are stronger with NTG rather than POAG overall or HTG. Our study also identified nominal association with the pyrimidine pathway that includes TXNRD2, coding for a mitochondrial protein involved in managing mitochondrial oxidative stress. A KEGG pathway specifically including mitochondrial redox homeostasis proteins does not exist and therefore could not be evaluated in this study.
Using PARIS to evaluate individual constituent genes of a pathway helps identify any genes associated with the outcome, which may be the main drivers of the entire pathway association. We found strong signals of association with POAG for BDH1, EHHADH, ACAA2, and ME3. These genes have not been previously reported to be associated with POAG in other studies, and further work seeking replication of the associations in other populations would be of interest.
Our results could have important implications for future study of the role of mitochondria in the pathogenesis of glaucoma. Patients with an excess of risk alleles influencing mitochondrial proteins could have a unique natural history when compared with POAG overall and may have a different response to treatment. Resveratrol, caloric restriction, and physical activity have been suggested as interventions to improve mitochondrial function, and evidence exists for potential benefit in animal models of retinal ischaemia and acute intraocular pressure injury.26–28 One approach to identifying POAG patients with a mitochondrial component to their disease would be a genetic analysis of their mitochondriome. A genetic risk score (GRS) based on these gene-set analyses could be used in future studies to investigate the phenotypic features of glaucoma patients with an excess of mitochondrial risk variants. The GRS could also be used to examine interactions with other genetic or environmental factors. For example, no effect of dietary antioxidants was found on the development of POAG in a subset of the current study29; it would be of interest to know if an effect is evident when only examining participants with a high mitochondrial GRS score, especially considering the potential contributions of TXNRD2.
A significant strength of our study is the large sample size and number of POAG cases (both HTG and NTG), which provided the necessary power for our analyses. Our approach of testing mitochondrial genes in biological system groups collectively is novel and appears to be a useful method for probing specific hypotheses. There are some limitations to our approach. The completeness and accuracy of the gene-sets we tested were limited by the completeness of the MitoCarta and KEGG databases and current knowledge. We did not consider variation in mitochondrial DNA because of limited information for mitochondrial DNA variants in our genotype data. However, mitochondria DNA encode only 13 proteins and therefore represents a minor fraction of the overall mitochondrial proteome. Nevertheless, further study of mitochondrial DNA variants in glaucoma is warranted using other datasets. It is also not currently possible to replicate our findings in another population; our strongest findings were in NTG patients and there are no other similarly sized Caucasian NTG case-control datasets with genetic data available. Further work is required to determine the precise role of the pathways and genes we described in POAG.
In summary, we present evidence supporting a role for mitochondria in POAG pathogenesis. Specifically, genes encoding mitochondrial proteins that are involved in lipid and carbohydrate metabolism were found to be associated with POAG, and in particular NTG. Future studies stratifying POAG patients according to genetic variation in mitochondrial genes may help identify a pathologically distinct POAG subset related to mitochondrial dysfunction.
Supplementary Material
Acknowledgments
The MitoCarta KEGG pathway datasets were kindly provided by Sarah E. Calvo (Broad Institute, Harvard, and MIT). LRP and JLW are supported by the Harvard Glaucoma Center of Excellence and Margolis fund (Boston, MA, USA). DLB, LRP, JER, RNW, and JLW are also supported by Research to Prevent Blindness, Inc. (New York, NY, USA). The Arthur Ashley Foundation also supports LRP. The Glaucoma Research Foundation (San Francisco, CA, USA), Bright Focus Foundation (formerly called American Health Assistance Foundation; Clarksburg, MD, USA), the Glaucoma Foundation (New York, NY, USA), and National Eye Institute Grant R01EY023242 support YL. MB is supported by National Institutes of Health Grants UL1TR000427 and 1U01HG006389. National Institutes of Health/National Eye Institute Grant R01EY022305 supports the NEIGHBORHOOD consortium. The following grants from the National Human Genome Research Institute (Bethesda, MD, USA) supported GLAUGEN: HG004728 (LRP), HG004424 (Broad Institute to support genotyping), HG004446 (C. Laurie, University of Washington, to support genotype data cleaning and analysis). The National Eye Institute supported the formation of the Nurses' Health Study and Health Professional Follow-up Study subset of GLAUGEN (EY015473; LRP). Genotyping services for the NEIGHBOR study were provided by the Center for Inherited Disease Research and were supported by the National Eye Institute through Grant HG005259-01 (JLW). In addition, the Center for Inherited Disease Research is funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, Contract HHSN268200782096C. The National Eye Institute (Bethesda, MD, USA) through American Recovery and Reinvestment Act (ARRA) Grants EY015872 (JLW) and EY019126 (MAH) supported the collection and processing of samples for the NEIGHBOR dataset. APK received support from the Berkeley Fellowship and the Wellcome Trust (094791/Z/10/Z).
Disclosure: A.P. Khawaja, None; J.N. Cooke Bailey, None; J.H. Kang, None; R.R. Allingham, None; M.A. Hauser, None; M. Brilliant, None; D.L. Budenz, None; W.G. Christen, None; J. Fingert, None; D. Gaasterland, None; T. Gaasterland, None; P. Kraft, None; R.K. Lee, None; P.R. Lichter, None; Y. Liu, None; F. Medeiros, None; S.E. Moroi, None; J.E. Richards, None; T. Realini, None; R. Ritch, None; J.S. Schuman, None; W.K. Scott, None; K. Singh, None; A.J. Sit, None; D. Vollrath, None; G. Wollstein, None; D.J. Zack, None; K. Zhang, None; M. Pericak-Vance, None; R.N. Weinreb, None; J.L. Haines, None; L.R. Pasquale, None; J.L. Wiggs None
References
- 1. Lee S,, Van Bergen NJ,, Kong GY,, et al. Mitochondrial dysfunction in glaucoma and emerging bioenergetic therapies. Exp Eye Res. 2011; 93: 204–212. [DOI] [PubMed] [Google Scholar]
- 2. Chrysostomou V,, Rezania F,, Trounce IA,, Crowston JG. Oxidative stress and mitochondrial dysfunction in glaucoma. Curr Opin Pharmacol. 2013; 13: 12–15. [DOI] [PubMed] [Google Scholar]
- 3. Osborne NN,, del Olmo-Aguado S. Maintenance of retinal ganglion cell mitochondrial functions as a neuroprotective strategy in glaucoma. Curr Opin Pharmacol. 2013; 13: 16–22. [DOI] [PubMed] [Google Scholar]
- 4. Bailey JNC,, Loomis SJ,, Kang JH,, et al. Genome-wide association analysis identifies TXNRD2 ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nat Genet. 2016; 48: 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carelli V,, Ross-Cisneros FN,, Sadun AA. Mitochondrial dysfunction as a cause of optic neuropathies. Prog Retin Eye Res. 2004; 23: 53–89. [DOI] [PubMed] [Google Scholar]
- 6. Lin MT,, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006; 443: 787–795. [DOI] [PubMed] [Google Scholar]
- 7. Lascaratos G,, Garway-Heath DF,, Willoughby CE,, Chau K-Y,, Schapira AH. Mitochondrial dysfunction in glaucoma: understanding genetic influences. Mitochondrion. 2012; 12: 202–212. [DOI] [PubMed] [Google Scholar]
- 8. Cornelis MC,, Agrawal A,, Cole JW,, et al. The Gene Environment Association Studies consortium (GENEVA): maximizing the knowledge obtained from GWAS by collaboration across studies of multiple conditions. Genet Epidemiol. 2010; 34: 364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wiggs JL,, Hauser MA,, Abdrabou W,, et al. The NEIGHBOR consortium primary open-angle glaucoma genome-wide association study: rationale, study design, and clinical variables. J Glaucoma. 2013; 22: 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wiggs JL,, Yaspan BL,, Hauser MA,, et al. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet. 2012; 8: e1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Purcell S,, Neale B,, Todd-Brown K,, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Willer CJ,, Li Y,, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010; 26: 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Calvo SE,, Mootha VK. The mitochondrial proteome and human disease. Annu Rev Genomics Hum Genet. 2010; 11: 25–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yaspan BL,, Bush WS,, Torstenson ES,, et al. Genetic analysis of biological pathway data through genomic randomization. Hum Genet. 2011; 129: 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pagliarini DJ,, Calvo SE,, Chang B,, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008; 134: 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kanehisa M,, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000; 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pasquale LR,, Loomis SJ,, Weinreb RN,, et al. Estrogen pathway polymorphisms in relation to primary open angle glaucoma: an analysis accounting for gender from the United States. Mol Vis. 2013; 19: 1471–1481. [PMC free article] [PubMed] [Google Scholar]
- 18. Bailey JNC,, Yaspan BL,, Pasquale LR,, et al. Hypothesis-independent pathway analysis implicates GABA and Acetyl-CoA metabolism in primary open-angle glaucoma and normal-pressure glaucoma. Hum Genet. 2014; 133: 1319–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alberts B,, Johnson A,, Lewis J,, Raff M,, Roberts K,, Walter P. Molecular Biology of the Cell. 5th ed. New York, NY: Garland Science; 2008. [Google Scholar]
- 20. Clayton LM,, Stern WM,, Newman WD,, Sander JW,, Acheson J,, Sisodiya SM. Evolution of visual field loss over ten years in individuals taking vigabatrin. Epilepsy Res. 2013; 105: 262–271. [DOI] [PubMed] [Google Scholar]
- 21. Janssen SF,, Gorgels TGMF,, Ramdas WD,, et al. The vast complexity of primary open angle glaucoma: disease genes, risks, molecular mechanisms and pathobiology. Prog Retin Eye Res. 2013; 37: 31–67. [DOI] [PubMed] [Google Scholar]
- 22. Meguro A,, Inoko H,, Ota M,, Mizuki N,, Bahram S. Genome-wide association study of normal tension glaucoma: common variants in SRBD1 and ELOVL5 contribute to disease susceptibility. Ophthalmology 2010; 117: 1331–1338.e5. [DOI] [PubMed] [Google Scholar]
- 23. Babizhayev MA,, Bunin AYA. Lipid peroxidation in open-angle glaucoma. Acta Ophthalmol. 1989; 67: 371–377. [DOI] [PubMed] [Google Scholar]
- 24. Aljohani AJ,, Munguba GC,, Guerra Y,, Lee RK,, Bhattacharya SK. Sphingolipids and ceramides in human aqueous humor. Mol Vis. 2013; 19: 1966–1984. [PMC free article] [PubMed] [Google Scholar]
- 25. Junk AK,, Goel M,, Mundorf T,, Rockwood EJ,, Bhattacharya SK. Decreased carbohydrate metabolism enzyme activities in the glaucomatous trabecular meshwork. Mol Vis. 2010; 16: 1286–1291. [PMC free article] [PubMed] [Google Scholar]
- 26. Kong YX,, van Bergen N,, Bui BV,, et al. Impact of aging and diet restriction on retinal function during and after acute intraocular pressure injury. Neurobiol Aging. 2012; 33: 1126.e15–e25. [DOI] [PubMed] [Google Scholar]
- 27. Chrysostomou V,, Kezic JM,, Trounce IA,, Crowston JG. Forced exercise protects the aged optic nerve against intraocular pressure injury. Neurobiol Aging. 2014; 35: 1722–1725. [DOI] [PubMed] [Google Scholar]
- 28. Liu X-Q,, Wu B-J,, Pan WHT,, et al. Resveratrol mitigates rat retinal ischemic injury: the roles of matrix metalloproteinase-9, inducible nitric oxide, and heme oxygenase-1. J Ocul Pharmacol Ther. 2013; 29: 33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kang JH,, Pasquale LR,, Willett W,, et al. Antioxidant intake and primary open-angle glaucoma: a prospective study. Am J Epidemiol. 2003; 158: 337–346. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


