Abstract
Background
This study aimed to identify the potential key long non-coding RNAs (lncRNAs) and target genes associated with pneumonia using lncRNA sequencing (lncRNA-seq).
Material/Methods
A total of 9 peripheral blood samples from patients with mild pneumonia (n=3) and severe pneumonia (n=3), as well as volunteers without pneumonia (n=3), were received for lncRNA-seq. Based on the sequencing data, differentially expressed lncRNAs (DE-lncRNAs) were identified by the limma package. After the functional enrichment analysis, target genes of DE-lncRNAs were predicted, and the regulatory network was constructed.
Results
In total, 99 DE-lncRNAs (14 upregulated and 85 downregulated ones) were identified in the mild pneumonia group and 85 (72 upregulated and 13 downregulated ones) in the severe pneumonia group, compared with the control group. Among these DE-lncRNAs, 9 lncRNAs were upregulated in both the mild and severe pneumonia groups. A set of 868 genes were predicted to be targeted by these 9 DE-lncRNAs. In the network, RP11-248E9.5 and RP11-456D7.1 targeted the majority of genes. RP11-248E9.5 regulated several genes together with CTD-2300H10.2, such as QRFP and EPS8. Both upregulated RP11-456D7.1 and RP11-96C23.9 regulated several genes, such as PDK2. RP11-456D7.1 also positively regulated CCL21.
Conclusions
These novel lncRNAs and their target genes may be closely associated with the progression of pneumonia.
MeSH Keywords: Gene Regulatory Networks; Genes, vif; Pneumonia, Aspiration; RNA, Long Noncoding
Background
Pneumonia is defined as inflammation and consolidation of lung tissue due to an infectious agent [1]. It is the leading global cause of death, especially in children and elderly people [2,3]. The typical symptoms of pneumonia are fever, chills, pleuritic chest pain, and cough productive of purulent sputum [4]. A common type of pneumonia, community-acquired pneumonia (CAP), is responsible for high rates of morbidity and mortality worldwide, with an annual incidence of 1.5 to 1.7 per 1000 individuals among adults in Europe [5]. Severe pneumonia is defined as admission to the intensive care unit (ICU), and it results in an extremely high rate of mortality [6]. Therefore, it is very urgent to find more biomarkers associated with pneumonia, thus contributing to the clinical therapy of this disease.
Currently, several molecular mechanisms underlying pneumonia have been found. For instance, the genotype -174 GG of interleukin-6 (IL-6) is associated with lower severity and mortality in patients with pneumococcal CAP [7]. Four risk single-nucleotide polymorphisms (SNPs) located in chromosomes 1 and 17 have been found to be significantly correlated with the susceptibility to development of severe pneumonia in A/H1N1 infection [8]. Previous reports have indicated that severe pneumonia is associated with methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes and the staphylococcal cassette chromosome mec (SCCmec) type IV [9,10]. The activity of metalloproteinase-9 (MMP-9) in peripheral blood circulation in patients with CAP caused by Mycoplasma pneumoniae is increased in the acute phase of illness compared to the control group [11]. Furthermore, a recent study has reported that high expression of IL-10 and interferon-induced protein (IP)-10 in human immunodeficiency virus (HIV)-infected infants is associated with more severe hypoxic pneumonia [12]. However, currently, no study has reported the association of long non-coding RNAs (lncRNAs) with pneumonia.
LncRNAs have been previously found to be widely transcribed in the genome. Multiple evidence links dysregulations and mutations of lncRNAs to diverse human diseases [13], such as lung diseases (e.g., lung cancer [14,15] and pulmonary fibrosis [16]). Therefore, we suggest a hypothesis that lncRNAs are also correlated with the progression of pneumonia. Thus, in this study, we used a new sequencing technique, lncRNA sequencing (lncRNA-seq), to analyze the lncRNA expression profiling in peripheral blood from patients with mild and severe pneumonia and to identify the potential critical lncRNAs that are associated with the progression of pneumonia. These findings may provide some new information for understanding the molecular functions of lncRNAs in pneumonia and extend the knowledge of the molecular mechanisms underlying pneumonia.
Material and Methods
Clinical samples
This study was approved by the Medical Ethics Committee of the Chinese People’s Liberation Army General Hospital, Beijing, China. A total of 18 patients with pneumonia who received therapy in our hospital from June 2013 to December 2013 were included in this study, including 9 patients with mild pneumonia (MP group) and 9 patients with severe pneumonia (SP group). Another 9 volunteers without pneumonia were enrolled as normal controls (C group) in this study. Here, patients with severe pneumonia must meet at least one of the following criteria: (1) altered mental status; (2) respiratory rate ≥30/min; (3) diastolic blood pressure <60 mm Hg, PaO2/FiO2 <300, and mechanical ventilation; (4) systolic blood pressure ≤90 mm Hg; (5) septic shock; (6) bilateral or multilobar pneumonia by chest radiograph, or lesion enlargement within 48 h after admission ≥50%; (7) oliguria: urine volume <20 mL/h or <80 mL/4 h, or acute renal failure requiring dialysis treatment [17].
Peripheral blood was sampled from each patient and volunteer. Informed consent was signed before sampling.
RNA extraction
First, plasma was separated from each of the 9 sequencing samples. Total RNA was extracted and purified from the plasma samples using miRNeasy Serum/Plasma Kit (Qiagen, Germany). Subsequently, ribosome RNA (rRNA) was removed from the total RNA using Epicentre Ribo-Zero™ rRNA Removal Kit (Epicentre, Madison, Wisconsin, USA), and the remaining RNA was collected and purified. To obtain sufficient quantities of high-quality RNA for sequencing, three RNA samples of equal quantity were randomly pooled into one sample for sequencing. Thus, three samples were generated for each group: WLL1–3 for the SP group, WLL4–6 for the MP group, and WLL7–9 for the C group. The 9 RNA samples were interrupted into short fragments by fragmentation buffer (Agilent Technologies, California, USA). Afterwards, the RNA fragments were reverse transcribed into cDNAs. The concentration of cDNAs in the library was quantified into 1 ng/μL with a Qubit 2.0 fluorometer, and then cDNAs were detected using the Agilent Bioanalyzer 2100 (Agilent Technologies, California, USA). According to the data size and effective cDNA concentration, libraries were pooled. Clusters of the cDNA libraries were generated on an Illumina cBot. Finally, the cDNA libraries were sequenced on an Illumina HiSeq™ 4000 with the model of 2×150 bp. The raw sequencing data have been uploaded to the public database NCBI (the National Center for Biotechnology Information) under the BioProject Accession PRJNA324335.
Data filtering
Raw reads were cleaned by removing the empty reads, adapter sequences, reads with Q-value <10 in the both terminals, reads containing fewer than 80% of bases with Q-value >20, reads with length <50 nt, and reads with unknown sequences ‘N’. In addition, the reads of rRNA were removed. The above quality control was conducted using FASTX-Toolkit (available at http://hannonlab.cshl.edu/fastx_toolkit/).
Statistics and alignment of reads
Both Q20 and length of raw and clean reads were summarized to ensure the validity and reliability of the sequencing data. Furthermore, clean reads were aligned to the human genome (hg19) using TopHat 2.1.1 (available at http://ccb.jhu.edu/software/tophat/index.shtml).
Differential expression analysis of lncRNAs
Based on the annotation information of genes and lncRNAs in the GENCODE database (available at http://www.gencodegenes.org/), FPKM (fragments per kilobase of exon per million fragments mapped) of mRNAs and lncRNAs, as well as the read number of lncRNAs mapped, was calculated using the StringTie tool (available at http://ccb.jhu.edu/software/stringtie/).
Differentially expressed lncRNAs (DE-lncRNAs) in the comparison groups of SP versus C, MP versus C, and SP versus MP were identified using the limma package (available at http://www.bioconductor.org/packages/release/bioc/html/limma.html). Only the lncRNAs with the criteria of |log2FC (fold change)| >1 and p value <0.05 were identified as DE-lncRNAs.
Prediction of DE-lncRNA target genes
The Pearson correlation coefficient (PCC) was calculated to evaluate the coexpression relationships between DE-lncRNAs and mRNAs. The coexpression pairs with PCC >0.8 and p value <0.05 were selected for the construction of the regulatory network, which was visualized by Cytoscape 3.3.0 (available at http://www.cytoscape.org/).
Functional analysis of DE-lncRNA target genes
GO (Gene Ontology) functional and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analyses were performed for the target genes of DE-lncRNAs using clusterProfiler 3.0.1 in R (available at http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html). GO enrichment analysis contains three categories, including molecular function (MF), biological process (BP), and cellular component (CC). Only the GO and pathway terms with a p value <0.05 were considered significant.
Results
Data summary of quality control and sequence alignment
In total, 346 G of raw data were generated from the 9 samples. Q20 of reads in both terminals of all samples was at least 99.97% and 96.38%, respectively. The clean rate (clean reads/raw reads) of the both terminals was more than 95% and 75% (Table 1). The results indicated a high quality of the sequencing data.
Table 1.
Summary of the sequencing data after quality control.
| Sample | Raw reads | Raw base | Q20 | Clean reads | Clean base | Clean rate |
|---|---|---|---|---|---|---|
| WLL1 | 52831743 | 7928532000 | 99.99% | 50735720 | 7609666306 | 0.960326446 |
| WLL1 | 58572433 | 7928532000 | 97.10% | 44421351 | 6662121716 | 0.758400304 |
| WLL2 | 103035876 | 15455381400 | 99.98% | 98747743 | 14810817868 | 0.958382137 |
| WLL2 | 103035876 | 15455381400 | 97.50% | 87943761 | 13189434520 | 0.85352563 |
| WLL3 | 47936143 | 7190421450 | 99.97% | 45842678 | 6875831673 | 0.956328047 |
| WLL3 | 47936143 | 7190421450 | 96.67% | 39203922 | 5879662949 | 0.817836387 |
| WLL4 | 55117713 | 8267656950 | 99.97% | 52810671 | 7920931057 | 0.958143365 |
| WLL4 | 55117713 | 8267656950 | 96.86% | 45772099 | 6864690206 | 0.830442638 |
| WLL5 | 42414859 | 6362228850 | 99.97% | 40625314 | 6093303572 | 0.957808536 |
| WLL5 | 42414859 | 6362228850 | 96.49% | 34754331 | 5212336104 | 0.819390464 |
| WLL6 | 52831743 | 7924761450 | 99.97% | 50675720 | 7600736984 | 0.959190765 |
| WLL6 | 52831743 | 7924761450 | 96.38% | 43274947 | 6490230720 | 0.819108826 |
| WLL7 | 54142181 | 8121327150 | 99.99% | 51811388 | 7771187924 | 0.956950515 |
| WLL7 | 54142181 | 8121327150 | 97.28% | 44077521 | 6610710884 | 0.814106861 |
| WLL8 | 58572433 | 8785864950 | 99.99% | 56266066 | 8439365160 | 0.960623678 |
| WLL8 | 58572433 | 8785864950 | 97.30% | 48447586 | 7266131771 | 0.827139723 |
| WLL9 | 42047930 | 6307189500 | 99.99% | 40469826 | 6070080949 | 0.962468925 |
| WLL9 | 42047930 | 6307189500 | 97.23% | 34410808 | 5160914737 | 0.818371035 |
WLL1–3 represent the samples in the severe pneumonia; WLL4–6 represent the samples in the mild pneumonia; WLL7–9 represent the control samples. Clean rate – Clean reads/raw reads.
Furthermore, map rate of reads in most samples was about 70%; read coverage in most samples was more than 80%; and depth of sequencing was more than 3.4, 4–5.5 for most samples (Table 2).
Table 2.
Data summary of the sequence alignment.
| Sample | Mapped-reads | Unique-mapped reads | Left mapped reads | Right mapped reads | Map rate | Unique map rate | Coverage | Depth |
|---|---|---|---|---|---|---|---|---|
| WLL1 | 66658130 | 65814534 | 37288693 | 29369437 | 0.700506324 | 0.691641024 | 0.8124469 | 4.656258953 |
| WLL2 | 106477220 | 105906423 | 61493861 | 44983359 | 0.570337791 | 0.567280357 | 0.8897298 | 7.498857573 |
| WLL3 | 60484089 | 59601427 | 34143118 | 26340971 | 0.711187619 | 0.700809051 | 0.7997421 | 4.401589422 |
| WLL4 | 66873642 | 65760610 | 37192807 | 29680835 | 0.678350203 | 0.667059873 | 0.8028072 | 5.141637117 |
| WLL5 | 55225190 | 54350513 | 30940910 | 24284280 | 0.732627356 | 0.721023733 | 0.7059120 | 4.090939095 |
| WLL6 | 67240507 | 66192609 | 37900851 | 29339656 | 0.715700156 | 0.704546451 | 0.8202879 | 5.359257339 |
| WLL7 | 62030490 | 61253756 | 35429803 | 26600687 | 0.646899528 | 0.638799175 | 0.7911858 | 4.366865004 |
| WLL8 | 75880654 | 74793902 | 42041868 | 33838786 | 0.724649103 | 0.714270781 | 0.8280350 | 5.464377530 |
| WLL9 | 52286836 | 51342443 | 29460848 | 22825988 | 0.698269141 | 0.685657162 | 0.7566747 | 3.453845744 |
WLL1–3 represent the samples in the severe pneumonia; WLL4–6 represent the samples in the mild pneumonia; WLL7–9 represent the control samples.
Identification of DE-lncRNAs
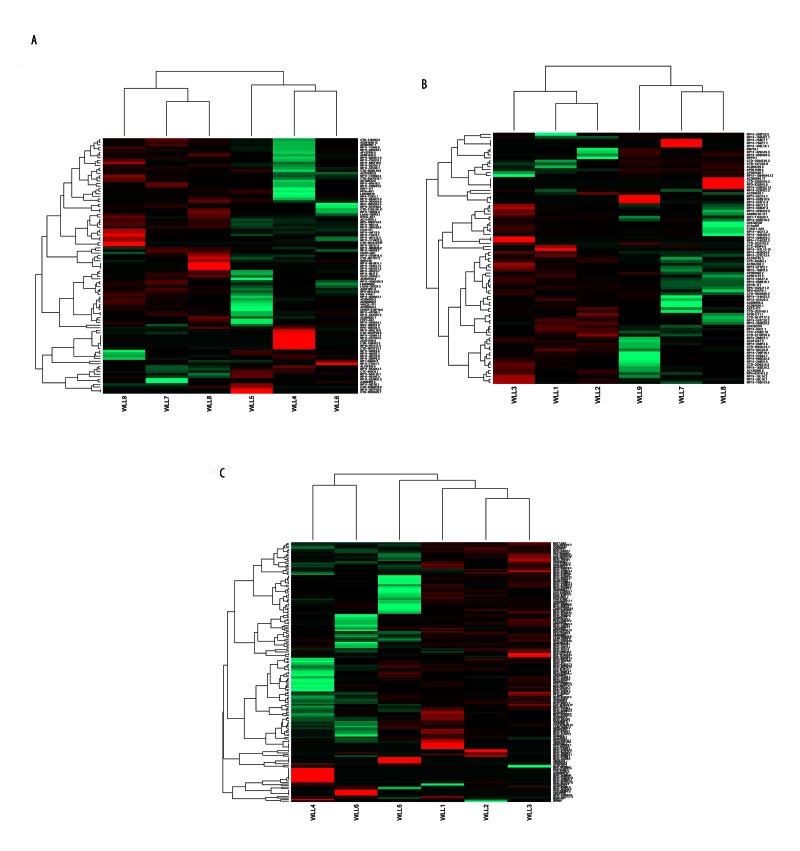
Among the 9 samples, there were 34,764 mRNAs and 5496 lncRNAs with FPKM >0 in at least one sample. Based on the criteria of differential expression analysis, 99 DE-lncRNAs (14 upregulated and 85 downregulated ones) were identified in the MP group and 85 (72 upregulated and 13 downregulated ones) in the SP group, compared with the C group. Nine lncRNAs were upregulated in both the MP and SP groups, compared with the C group. Furthermore, there were 159 upregulated and 8 downregulated lncRNAs in the SP group, compared with the MP group. These DE-lncRNAs were able to distinguish the two group samples (Figure 1A–1C).
Figure 1.
The heat maps of the differentially expressed long non-coding RNAs (DE-lncRNAs). (A) The heat map of DE-lncRNAs between the mild pneumonia and control groups. (B) The heat map of DE-lncRNAs between the severe pneumonia and control groups. (C) The heat map of DE-lncRNAs between the mild and severe pneumonia groups. Each row represents a lncRNA, and each column represents a sample. Green indicates downregulated and red indicates upregulated. WLL1–3 represent the severe pneumonia samples; WLL4–6 represent the mild pneumonia samples; and WLL7–9 represent the control samples.
Target genes of the DE-lncRNAs
To further reveal the potential regulatory relationships between DE-lncRNAs and downstream genes, target genes of the 175 DE-lncRNAs in the MP and SP groups were predicted with the PCC method. In total, 4908 genes were targeted by those DE-lncRNAs. The regulatory network consisted of 175 DE-lncRNAs, 4908 genes, and 17,385 regulatory relationships (Supplementary Figure 1).
Enrichment analyses of the target genes
To further investigate the potential biological functions of the identified DE-lncRNAs, GO and KEGG pathway enrichment analyses were carried out for the targets of these DE-lncRNAs. For the target genes of DE-lncRNAs in the MP group, the targets of the upregulated lncRNAs were mainly associated with GO functions, such as response to stimulus and regulation of cellular process; meanwhile, the targets of the downregulated lncRNAs were significantly correlated with multiple biological functions, such as protein binding and catalytic activity (Table 3).
Table 3.
The enriched Gene Ontology and pathway terms of differentially expressed lncRNAs in the mild pneumonia compared with the controls.
| Category of lncRNAs | Category of functional terms | ID | Term | Gene count | FDR |
|---|---|---|---|---|---|
| Upregulated | GO-BP | GO: 0008150 | Biological_process | 469 | 3.46E-22 |
| GO-BP | GO: 0009987 | Cellular process | 434 | 8.85E-12 | |
| GO-BP | GO: 0050896 | Response to stimulus | 261 | 0.0004501 | |
| GO-BP | GO: 0050794 | Regulation of cellular process | 303 | 0.003139471 | |
| GO-BP | GO: 0044699 | Single-organism process | 378 | 0.004578225 | |
| GO-CC | GO: 0005575 | Cellular_component | 510 | 2.68E-09 | |
| GO-MF | GO: 0003674 | Molecular_function | 481 | 1.52E-22 | |
| GO-MF | GO: 0005488 | Binding | 399 | 0.002334936 | |
| GO-MF | GO: 0060089 | Molecular transducer activity | 74 | 0.016441823 | |
| Downregulated | GO-BP | GO: 0008150 | Biological_process | 1852 | 2.03E-100 |
| GO-BP | GO: 0009987 | Cellular process | 1685 | 8.04E-41 | |
| GO-BP | GO: 0044699 | Single-organism process | 1517 | 2.59E-23 | |
| GO-BP | GO: 0044763 | Single-organism cellular process | 1386 | 2.07E-18 | |
| GO-BP | GO: 0008152 | Metabolic process | 1337 | 4.59E-18 | |
| GO-CC | GO: 0005575 | Cellular_component | 1965 | 2.83E-43 | |
| GO-CC | GO: 0005623 | Cell | 1758 | 1.73E-08 | |
| GO-CC | GO: 0044464 | Cell part | 1753 | 2.42E-08 | |
| GO-CC | GO: 0005622 | Intracellular | 1546 | 9.50E-07 | |
| GO-CC | GO: 0044424 | Intracellular part | 1504 | 1.05E-05 | |
| GO-MF | GO: 0003674 | Molecular_function | 1849 | 1.12E-96 | |
| GO-MF | GO: 0005488 | Binding | 1554 | 3.18E-22 | |
| GO-MF | GO: 0005515 | Protein binding | 1174 | 6.27E-10 | |
| GO-MF | GO: 0003824 | Catalytic activity | 675 | 6.37E-10 | |
| GO-MF | GO: 0016740 | Transferase activity | 292 | 9.63E-07 | |
| KEGG | hsa03010 | Ribosome | 32 | 0.009558616 |
LncRNA – long non-coding RNA; GO – Gene Ontology; MF – molecular function; CC – cellular component; BP – biological process; FDR – false discovery rate.
Additionally, the target genes of both upregulated and downregulated lncRNAs in the SP group were significantly related to GO functions, such as protein binding and catalytic activity. The targets of the downregulated lncRNAs were also implicated in the pathways of ribosome and apoptosis (Table 4).
Table 4.
The enriched Gene Ontology and pathway terms of differentially expressed lncRNAs in the severe pneumonia compared with the controls
| Category of lncRNAs | Category of functional terms | ID | Term | Gene count | FDR |
|---|---|---|---|---|---|
| Upregulated | GO-BP | GO: 0008150 | Biological_process | 1688 | 8.86E-91 |
| GO-BP | GO: 0009987 | Cellular process | 1543 | 4.84E-40 | |
| GO-BP | GO: 0044699 | Single-organism process | 1376 | 3.97E-19 | |
| GO-BP | GO: 0008152 | Metabolic process | 1218 | 5.83E-16 | |
| GO-BP | GO: 0071704 | Organic substance metabolic process | 1101 | 2.21E-14 | |
| GO-CC | GO: 0005575 | Cellular component | 1818 | 1.08E-39 | |
| GO-CC | GO: 0005623 | Cell | 1632 | 6.64E-09 | |
| GO-CC | GO: 0044464 | Cell part | 1628 | 6.82E-09 | |
| GO-CC | GO: 0044424 | Intracellular part | 1401 | 1.78E-06 | |
| GO-CC | GO: 0005622 | Intracellular | 1432 | 1.78E-06 | |
| GO-MF | GO: 0003674 | Molecular_function | 1698 | 3.69E-88 | |
| GO-MF | GO: 0005488 | Binding | 1421 | 1.71E-18 | |
| GO-MF | GO: 0003824 | Catalytic activity | 621 | 5.29E-09 | |
| GO-MF | GO: 0005515 | Protein binding | 1062 | 9.78E-07 | |
| GO-MF | GO: 0016740 | Transferase activity | 258 | 0.000259127 | |
| Downregulated | GO-BP | GO: 0008150 | Biological_process | 588 | 1.18E-28 |
| GO-BP | GO: 0009987 | Cellular process | 538 | 2.41E-12 | |
| GO-BP | GO: 0044699 | Single-organism process | 495 | 7.99E-10 | |
| GO-BP | GO: 0044763 | Single-organism cellular process | 456 | 2.16E-08 | |
| GO-BP | GO: 0044237 | Cellular metabolic process | 393 | 5.52E-08 | |
| GO-CC | GO: 0005575 | Cellular_component | 622 | 9.48E-12 | |
| GO-CC | GO: 0005622 | Intracellular | 527 | 1.20E-09 | |
| GO-CC | GO: 0044424 | Intracellular part | 512 | 2.29E-08 | |
| GO-CC | GO: 0005623 | Cell | 578 | 2.29E-08 | |
| GO-CC | GO: 0005737 | Cytoplasm | 421 | 2.29E-08 | |
| GO-MF | GO: 0003674 | Molecular_function | 587 | 4.87E-28 | |
| GO-MF | GO: 0005488 | Binding | 501 | 3.95E-08 | |
| GO-MF | GO: 0003735 | Structural constituent of ribosome | 22 | 1.40E-06 | |
| GO-MF | GO: 0005515 | Protein binding | 391 | 1.01E-05 | |
| GO-MF | GO: 0003824 | Catalytic activity | 232 | 1.05E-05 | |
| KEGG | hsa03010 | Ribosome | 21 | 5.76E-05 | |
| KEGG | hsa04210 | Apoptosis | 16 | 0.031885823 |
LncRNA – long non-coding RNA; GO – Gene Ontology; MF – molecular function; CC – cellular component; BP – biological process FDR – false discovery rate.
Analysis of the common 9 upregulated lncRNAs in the mild and severe pneumonia groups
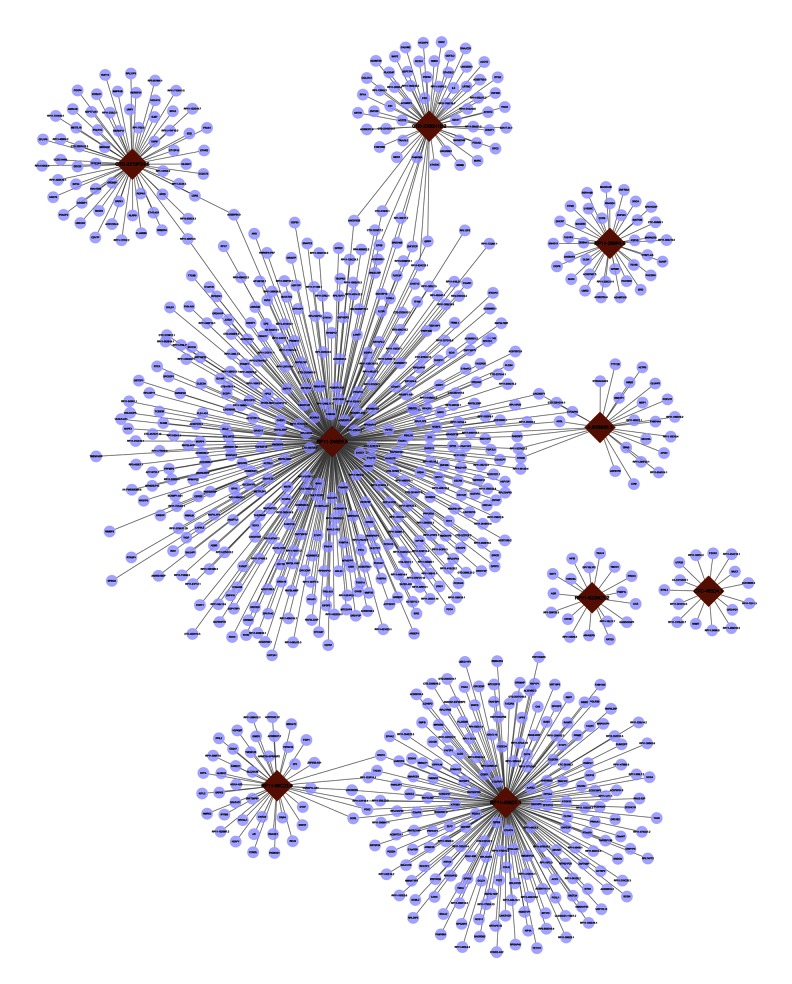
The 9 lncRNAs that were upregulated in both the MP and SP groups were analyzed in detail. A total of 868 genes were predicted to be targeted by these 9 DE-lncRNAs (Figure 2). Among them, RP11-248E9.5 targeted the most genes, such as QRFP and EPS8; these two genes were also targeted by CTD-2300H10.2. RP11-248E9.5 also targeted a set of genes encoding zinc finger proteins (ZFPs), such as ZNF717, ZNF460, ZNF687, and ZNF37CP. Furthermore, RP11-456D7.1 regulated a series of genes, such PDK2, which were also targeted by RP11-96C23.9. RP11-456D7.1 also regulated genes like CCL21.
Figure 2.
The regulatory network of the 9 long non-coding RNAs (lncRNAs) that are differentially expressed in both mild and severe pneumonia. Dark red nodes represent the lncRNAs, and purple nodes represent the target genes. Lines represent the regulatory relationships between lncRNAs and target genes.
In addition, according to the GO and pathway enrichment analyses, the target genes of RP11-248E9.5 (e.g., GPR75 and QRFP) were significantly enriched in the GO functions like G-protein coupled receptor signaling pathway; the target genes of RP11-456D7.1 were mainly enriched in the molecular function. Furthermore, the target genes of CTD-2300H10.2 (e.g., RC3H1, IHH, and IL4) were significantly enriched in the GO functions like negative regulation of alpha-beta T cell differentiation (Table 5).
Table 5.
The enriched Gene Ontology and pathway terms of lncRNAs that are differentially expressed in the both mild and severe pneumonia.
| LncRNA | Category | ID | Term | FDR | Gene count | Target genes |
|---|---|---|---|---|---|---|
| AJ006995.3 | BP | GO: 1901137 | Carbohydrate derivative biosynthetic process | 0.004964 | 5 | NME6, SEC23A, ADSL, POFUT1, ST6GALNAC5 |
| GO: 0006486 | Protein glycosylation | 0.016382 | 3 | SEC23A, POFUT1, ST6GALNAC5 | ||
| GO: 0043413 | Macromolecule glycosylation | 0.016382 | 3 | SEC23A, POFUT1, ST6GALNAC5 | ||
| GO: 0070085 | Glycosylation | 0.016382 | 3 | SEC23A, POFUT1, ST6GALNAC5 | ||
| GO: 1901135 | Carbohydrate derivative metabolic process | 0.019142 | 6 | NME6, SEC23A, ADSL, POFUT1, ARHGEF28, ST6GALNAC5 | ||
| CTD-2210P24.6 | MF | GO: 1901363 | Heterocyclic compound binding | 0.017896 | 16 | FOXP4, POU2F3, MAGI3, DDX25, HOXA13, NLRP9, MTA3, KIAA1586, BMPR1B, SRPK1, UBE2G2, UBP1, METTL16, CGGBP1, USP6, POLR1C |
| GO: 0097159 | Organic cyclic compound binding | 0.017896 | 16 | FOXP4, POU2F3, MAGI3, DDX25, HOXA13, NLRP9, MTA3, KIAA1586, BMPR1B, SRPK1, UBE2G2, UBP1, METTL16, CGGBP1, USP6, POLR1C | ||
| GO: 0005488 | Binding | 0.017896 | 24 | FOXP4, ADRA1B, NLGN4Y, POU2F3, MAGI3, DDX25, HOXA13, NLRP9, KRT8, RUFY2, MTA3, KIAA1586, BMPR1B, SRPK1, UBE2G2, UBP1, METTL16, FRAS1, CGGBP1, LGR5, EED, USP6, INTS4, POLR1C | ||
| GO: 0003700 | Sequence-specific DNA binding transcription factor activity | 0.019057 | 6 | FOXP4, POU2F3, HOXA13, MTA3, UBP1, CGGBP1 | ||
| GO: 0001071 | Nucleic acid binding transcription factor activity | 0.019057 | 6 | FOXP4, POU2F3, HOXA13, MTA3, UBP1, CGGBP1 | ||
| CTD-2300H10.2 | BP | GO: 0046639 | Negative regulation of alpha-beta T cell differentiation | 7.61E-05 | 3 | RC3H1, IHH, IL4 |
| GO: 0046636 | Negative regulation of alpha-beta T cell activation | 0.000187 | 3 | RC3H1, IHH, IL4 | ||
| GO: 0045581 | Negative regulation of T cell differentiation | 0.000269 | 3 | RC3H1, IHH, IL4 | ||
| GO: 0045620 | Negative regulation of lymphocyte differentiation | 0.000617 | 3 | RC3H1, IHH, IL4 | ||
| GO: 0046637 | Regulation of alpha-beta T cell differentiation | 0.00103 | 3 | RC3H1, IHH, IL4 | ||
| CC | GO: 0032587 | Ruffle membrane | 0.015463 | 3 | EPS8, PLA2G4F, PDE9A | |
| GO: 0031256 | Leading edge membrane | 0.033811 | 3 | EPS8, PLA2G4F, PDE9A | ||
| GO: 0001726 | Ruffle | 0.039361 | 3 | EPS8, PLA2G4F, PDE9A | ||
| MF | GO: 0052689 | Carboxylic ester hydrolase activity | 0.000439 | 4 | ACOT2, ESD, ACOT9, PLA2G4F | |
| GO: 0016790 | Thiolester hydrolase activity | 0.001389 | 3 | ACOT2, ESD, ACOT9 | ||
| GO: 0016788 | Hydrolase activity, acting on ester bonds | 0.027305 | 5 | ACOT2, ESD, ACOT9, PLA2G4F PDE9A | ||
| RP11-96C23.9 | MF | GO: 0016773 | Phosphotransferase activity, alcohol group as acceptor | 0.020183 | 6 | SRPK3, NMRK2, PDK2, CNTLN, RELN, PIP5K1B |
| GO: 0016301 | Kinase activity | 0.020183 | 6 | SRPK3, NMRK2, PDK2, CNTLN, RELN, PIP5K1B | ||
| GO: 0016772 | Transferase activity, transferring phosphorus-containing groups | 0.028775 | 6 | SRPK3, NMRK2, PDK2, CNTLN, RELN, PIP5K1B | ||
| GO: 0016740 | Transferase activity | 0.028775 | 9 | MBOAT2, PPIL2, SRPK3, NMRK2, PDK2, CNTLN, RELN, VCPKMT, PIP5K1B | ||
| RP11-248E9.5 | BP | GO: 0008150 | Biological process | 0.002259 | 101 | ZNF717, ARL4C, ZNF460, GPR75, UGT2A1, CHEK1, ERI2, CPE, DHRS13, ZNF418, TTC9B, IL23R, ADRA2B, SNRNP48, ADSL, DPH2, TIGIT, CERS3, EPS8, FANCF, OR4D11, LHFPL5, MYO16, CDY1B, PRPF40B, LRIG1, STEAP2, MSTN, OR2V1, ANG, CLEC9A, TMPRSS12, KCNRG, GPR22, FRYL, SCAI, GPX5, VN1R2, OR2AG2, TRIML1, IFNA14, ACTBL2, QRFP, OR6B2, OR4K17, OR13C6P, OR51A4, VN1R17P, CEP83, TCEB3B, SCARA3, LARP7, ASB1, PLCB4, SPIN2A, PCDH18, PPP2R3A, CHST12, TNFRSF19, FAM46A, KIF16B, EXOC1, PCDHA7, PRDM11, TAS2R38, RIMKLB, ZNF687, BAK1, HRH4, BCL2, RYR2, BLOC1S5, ARHGEF28, SH3GL3, SMYD3, RFX7, SNAPC3, SOX3, TFAM, THY1, KRTAP4–8, ZNF140, CEP97, OR4A16, OR12D3, FCRL4, C19orf12, HPS3, DUSP11, SORBS2, PPFIBP2, RTCA, NRP2, TGIF2LX, ANGPTL1, HTR3B, MMP20, RIN1, PCDHA9, TECPR2, JOSD1 |
| GO: 0007186 | G-protein coupled receptor signaling pathway | 0.002613 | 20 | GPR75, CPE, ADRA2B, OR4D11, OR2V1, GPR22, VN1R2, OR2AG2, QRFP, OR6B2, OR4K17, OR13C6P, OR51A4, VN1R17P, TAS2R38, HRH4, RYR2, OR4A16, OR12D3, HTR3B | ||
| GO: 0009593 | Detection of chemical stimulus | 0.017347 | 11 | UGT2A1, OR4D11, OR2V1, OR2AG2, OR6B2, OR4K17, OR51A4, TAS2R38, RYR2, OR4A16, OR12D3 | ||
| GO: 0007606 | Sensory perception of chemical stimulus | 0.017347 | 11 | UGT2A1, OR4D11, OR2V1, OR2AG2, OR6B2, OR4K17, OR13C6P, OR51A4, TAS2R38, OR4A16, OR12D3 | ||
| GO: 0007608 | Sensory perception of smell | 0.021759 | 10 | UGT2A1, OR4D11, OR2V1, OR2AG2, OR6B2, OR4K17, OR13C6P, OR51A4, OR4A16, OR12D3 | ||
| RP11-248E9.5 | MF | GO: 0003674 | molecular_function | 2.96E-05 | 99 | ZNF717, ARL4C, ZNF460, GPR75, UGT2A1, CHEK1, CHGB, ERI2, C1orf131, SLX4IP, CPE, DHRS13, ZNF418, TTC9B, IL23R, ADRA2B, SNRNP48, ADSL, TIGIT, CERS3, EPS8, FANCF, OR4D11, MYO16, CDY1B, STEAP2, MSTN, OR2V1, ANG, CLEC9A, TMPRSS12, KCNRG, GPR22, SCAI, GPX5, VN1R2, OR2AG2, TRIML1, IFNA14, ACTBL2, QRFP, OR6B2, OR4K17, OR13C6P, OR51A4, VN1R17P, CEP83, TCEB3B, SCARA3, LARP7, ASB1, PLCB4, SPIN2A, PCDH18, PPP2R3A, CHST12, TNFRSF19, FAM46A, KIF16B, EXOC1, PCDHA7, PRDM11, TAS2R38, RIMKLB, ZNF687, BAK1, HRH4, BCL2, RYR2, BLOC1S5, ANKEF1, ARHGEF28, CEP170P1, SH3GL3, SMYD3, RFX7, SNAPC3, SOX3, TFAM, THY1, ZNF140, CEP97, OR4A16, OR12D3, FCRL4, EFCAB7, DUSP11, SORBS2, PPFIBP2, RTCA, NRP2, TGIF2LX, ANGPTL1, HTR3B, MMP20, RIN1, PCDHA9, TECPR2, JOSD1 |
| GO: 0004930 | G-protein coupled receptor activity | 0.000121 | 17 | GPR75, ADRA2B, OR4D11, OR2V1, GPR22, VN1R2, OR2AG2, OR6B2, OR4K17, OR13C6P, OR51A4, VN1R17P, TAS2R38, HRH4, OR4A16, OR12D3, HTR3B | ||
| GO: 0004888 | transmembrane signaling receptor activity | 0.000206 | 20 | GPR75, IL23R, ADRA2B, OR4D11, OR2V1, GPR22, VN1R2, OR2AG2, OR6B2, OR4K17, OR13C6P, OR51A4, VN1R17P, TNFRSF19, TAS2R38, HRH4, OR4A16, OR12D3, NRP2, HTR3B | ||
| GO: 0038023 | signaling receptor activity | 0.000484 | 20 | GPR75, IL23R, ADRA2B, OR4D11, OR2V1, GPR22, VN1R2, OR2AG2, OR6B2, OR4K17, OR13C6P, OR51A4, VN1R17P, TNFRSF19, TAS2R38, HRH4, OR4A16, OR12D3, NRP2, HTR3B | ||
| GO: 0004872 | receptor activity | 0.001039 | 21 | GPR75, IL23R, ADRA2B, OR4D11, OR2V1, GPR22, VN1R2, OR2AG2, OR6B2, OR4K17, OR13C6P, OR51A4, VN1R17P, SCARA3, TNFRSF19, TAS2R38, HRH4, OR4A16, OR12D3, NRP2, HTR3B | ||
| KEGG | hsa04740 | Olfactory transduction | 0.001791994 | 7 | OR12D3, OR2AG2, OR4A16, OR4D11, OR4K17, OR51A4, OR6B2 | |
| RP11-456D7.1 | MF | GO: 0003674 | molecular_function | 0.007716221 | 61 | TANK, USP17L12, DSCR4, SCML2, SCGN, KHDRBS3, ZBTB6, OSBPL7, C1QTNF3, RBP7, CLCN4, GJD3, OR2T34, OR2T4, COX5B, CCBE1, UPP2, EPHA2, TXLNA, FOXL1, IQSEC2, TNFRSF13B, KIF4A, ATXN10, TRAF3IP1, GMFB, ANXA4, ZACN, LDHC, ATP2B3, OCRL, TAS2R8, PDK2, ACP5, PPP1R3D, PRIM2, PRKAA1, POLR3B, WWC3, PCDHGA7, TMX4, AARS2, NTN4, OPN1LW, RGS13, CCL21, PGA3, ZCCHC18, NMNAT1, SMARCD1, TACR1, CA6, OR51B2, COLQ, PPFIA4, ENDOU, BUB3, ZMYM3, GPR52, FEZ2, IKBKE |
LncRNA – long non-coding RNA; GO – Gene Ontology; MF – molecular function; CC – cellular component; BP – biological process; FDR – false discovery rate.
Discussion
In the present study, 99 DE-lncRNAs (14 upregulated and 85 downregulated ones) were identified in the MP group and 85 (72 upregulated and 13 downregulated ones) in the SP group, compared with the C group. Among these DE-lncRNAs, 9 lncRNAs were upregulated in the both the MP and SP groups, compared with the C group. According to the coexpression analysis between DE-lncRNAs and mRNAs, 868 genes were predicted to be targeted by the 9 lncRNAs. RP11-248E9.5 and RP11-456D7.1 targeted the majority of genes.
In the regulatory network, RP11-248E9.5 regulated several genes together with CTD-2300H10.2, such as QRFP and EPS8. QRFP encodes pyroglutamylated RFamide peptide, which is proteolytically processed to generate multiple protein products [18]. In this study, QRFP was predicted to be relevant to the G-protein coupled receptor signaling pathway. A previous study has found that G-protein coupled receptor kinase-5 (GRK5) deficiency improves pulmonary infection and inflammation in Escherichia coli-induced pneumonia [19]. Furthermore, G-protein coupled receptors have been suggested to be associated with inflammation [20–22]. Although there is no evidence to show the role of QRFP in pneumonia, we speculate that QRFP may participate in the progression of pneumonia via the G-protein coupled receptor signaling pathway. EPS8 encodes epidermal growth factor receptor (EGFR) pathway substrate 8 and functions as part of the EGFR pathway [23]. In mycoplasmal pneumonia, the EFGR pathway takes part in the IL-8 production by bronchial epithelial cells stimulated with Mp-Ag [24]. Therefore, EPS8 may be involved in the progression of pneumonia via the EFGR pathway. In addition to QRFP and EPS8, RP11-248E9.5 also targeted a series of ZFP coding genes, such as ZNF717, ZNF460, ZNF687, and ZNF37CP. ZNF37CP was also targeted by CTD-2300H10.2. Multiple studies have reported the associations of ZFPs with immunity [25–27], which is involved in pneumonia. In addition, in the network, CTD-2300H10.2 also targeted IL4, which is highly expressed in idiopathic interstitial pneumonias [28]. Currently, the associations of RP11-248E9.5 and CTD-2300H10.2 with pneumonia have not been previously reported, indicating they may be new potential molecules in pneumonia.
Furthermore, in the regulatory network, both upregulated RP11-456D7.1 and RP11-96C23.9 regulated several genes, such as PDK2, which encodes a member of the pyruvate dehydrogenase kinase family and is able to downregulate the activity of the mitochondrial pyruvate dehydrogenase complex. Inhibition of a homologue of PDK2, PDK4, can prevent multiorgan failure in severe influenza accompanied with pneumonia [29]. Moreover, pyruvate dehydrogenase E1 β subunit can act as fibronectin-binding protein in Mycoplasma pneumoniae, helping M. pneumoniae to locate in the host cells [30]. These evidences indicate that PDK2 may be related to the occurrence and development of pneumonia. In this study, RP11-456D7.1 also positively regulated CCL21, a high-affinity functional ligand for chemokine receptor 7 (CCR7) that is expressed on T and B lymphocytes and plays a key role in the inflammatory response [31,32]. CCL21 was detected at a significantly higher concentration in the bronchoalveolar lavage fluid of patients with eosinophilic pneumonia than in that of controls [33,34], which is similar to the results of this study. Taken together, although the roles of RP11-456D7.1 and RP11-96C23.9 have not been previously proved in pneumonia, we speculate that they may participate in the progression of pneumonia, likely via regulating their downstream genes PDK2 or CCL21.
In addition, according to the results of the enrichment analysis, functions of DE-lncRNAs in the SP group were similar to those in the MP group. However, 167 lncRNAs were identified to be differentially expressed between the SP and MP groups, indicating that lncRNA expression profiling between mild and severe pneumonia is different. In our future study, we will continue to focus on these DE-lncRNAs.
Despite the aforementioned results, this study has several limitations. In this study, the number of samples analyzed was small. Furthermore, the predicted results need to be validated by experimental data.
Conclusions
Based on the lncRNA-seq and bioinformatics analysis method, compared with the control, a set of DE-lncRNAs in patients with mild and severe pneumonia was identified. Nine lncRNAs were differentially expressed in both mild and severe pneumonia, such as RP11-248E9.5, CTD-2300H10.2, RP11-456D7.1, and RP11-96C23.9. All of them were predicted to target a set of downstream genes. At present, these lncRNAs have not been demonstrated to be associated with pneumonia by other studies; thus, they are novel lncRNAs that might be related to pneumonia. These results provided new information for further experimental studies.
Supplementary materials
The regulatory network of the differentially expressed long non-coding RNAs (DE-lncRNAs) and target genes. Light green nodes represent the downregulated lncRNAs in mild pneumonia; light red nodes represent the upregulated lncRNAs in mild pneumonia; green nodes represent the downregulated lncRNAs in severe pneumonia; red nodes represent the upregulated lncRNAs in severe pneumonia; dark red nodes represent the upregulated lncRNAs in both mild and severe pneumonia; purple nodes represent the target genes. Lines represent the regulatory relationships between lncRNAs and target genes.
Footnotes
Potential conflicts of interest
No benefits in any form have been received or will be received from a commercial party related directly or indirectly to subject of this article.
Source of support: This work was supported by grants from Welfare Industry Research Program of Ministry of Health (No. 201302017, 201502019), the National Natural Science Fund (No. 81272060, 81371561), the Hai Nan Natural Science Fund (20158315), the youth training program of the PLA (No. 13QNP171), Beijing Scientific and Technologic Supernova Supportive Project (Z15111000030000/XXJH2015B100), PLA General Hospital Science and Technology Innovation Nursery Fund Project (16KMM56), and PLA Logistic Major Science and Technology Project (14CXZ005, AWS15J004, BWS14J041)
References
- 1.Shahcheraghi F, Moezi H, Feizabadi MM. Distribution of TEM and SHV beta-lactamase genes among Klebsiella pneumoniae strains isolated from patients in Tehran. Med Sci Monit. 2007;13(11):BR247–50. [PubMed] [Google Scholar]
- 2.Thomas CP, Ryan M, Chapman JD, et al. Incidence and cost of pneumonia in Medicare beneficiaries. Chest. 2012;142:973–81. doi: 10.1378/chest.11-1160. [DOI] [PubMed] [Google Scholar]
- 3.Walker CL, Rudan I, Liu L, et al. Global burden of childhood pneumonia and diarrhoea. Lancet. 2013;381:1405–16. doi: 10.1016/S0140-6736(13)60222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lutfiyya MN, Henley E, Chang LF, Reyburn SW. Diagnosis and treatment of community-acquired pneumonia. Am Fam Physician. 2006;73:442–50. [PubMed] [Google Scholar]
- 5.Cillóniz C, Torres A, Niederman M, et al. Community-acquired pneumonia related to intracellular pathogens. Intensive Care Med. 2016;42(9):1374–86. doi: 10.1007/s00134-016-4394-4. [DOI] [PubMed] [Google Scholar]
- 6.Tagami T, Matsui H, Horiguchi H, et al. Low-dose corticosteroid use and mortality in severe community-acquired pneumonia patients. Eur Respir J. 2015;45:463–72. doi: 10.1183/09031936.00081514. [DOI] [PubMed] [Google Scholar]
- 7.Martín-Loeches I, Solé-Violán J, De Castro FR, et al. Variants at the promoter of the interleukin-6 gene are associated with severity and outcome of pneumococcal community-acquired pneumonia. Intensive Care Med. 2012;38:256–62. doi: 10.1007/s00134-011-2406-y. [DOI] [PubMed] [Google Scholar]
- 8.Zuniga J, Buendia-Roldan I, Zhao Y, et al. Genetic variants associated with severe pneumonia in A/H1N1 influenza infection. Eur Respir J. 2012;39:604–10. doi: 10.1183/09031936.00020611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillet Y, Vanhems P, Lina G, et al. Factors predicting mortality in necrotizing community-acquired pneumonia caused by Staphylococcus aureus containing Panton-Valentine leukocidin. Clin Infect Dis. 2007;45:315–21. doi: 10.1086/519263. [DOI] [PubMed] [Google Scholar]
- 10.Kilic A, Li H, Stratton CW, Tang Y-W. Antimicrobial susceptibility patterns and staphylococcal cassette chromosome mec types of, as well as Panton-Valentine leukocidin occurrence among, methicillin-resistant Staphylococcus aureus isolates from children and adults in middle Tennessee. J Clin Microbiol. 2006;44:4436–40. doi: 10.1128/JCM.01546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puljiz I, Markotić A, Cvetko KL, et al. Mycoplasma pneumoniae in adult community-acquired pneumonia increases matrix metalloproteinase-9 serum level and induces its gene expression in peripheral blood mononuclear cells. Med Sci Monit. 2012;18:500–5. doi: 10.12659/MSM.883270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green R, Terclanche A, Becker P, et al. Cytokine profile and clinical correlates in HIV-exposed infants with severe (hypoxic) pneumonia. S Afr Resp J. 2016;22:3–6. [Google Scholar]
- 13.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–61. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Liu X-h, Liu Z-l, Sun M, et al. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Hu L, Pan Q, et al. Detection and analysis of Wnt pathway related lncRNAs expression profile in lung adenocarcinoma. Pathol Oncol Res. 2016;22:609–15. doi: 10.1007/s12253-016-0046-9. [DOI] [PubMed] [Google Scholar]
- 16.Song X, Cao G, Jing L, et al. Analysing the relationship between lncRNA and protein-coding gene and the role of lncRNA as ceRNA in pulmonary fibrosis. J Cell Mol Med. 2014;18:991–1003. doi: 10.1111/jcmm.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Association RdbotCm. Diagnosis and treatment guideline of community-acquired pneumonia. Chin J Tuberc Respir Dis. 2006;29:651–55. [Google Scholar]
- 18.Fukusumi S, Fujii R, Hinuma S. Recent advances in mammalian RFamide peptides: the discovery and functional analyses of PrRP, RFRPs and QRFP. Peptides. 2006l;27:1073–86. doi: 10.1016/j.peptides.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 19.Packiriswamy N, Steury M, Parameswaran N. Critical involvement of GPCR kinase-5 in E coli induced pneumonia (INC7P 427) J Immunol. 2014;192:186.128. [Google Scholar]
- 20.Ohta A, Sitkovsky M. Role of G-protein-coupled adenosine receptors in downregulation of inflammation and protection from tissue damage. Nature. 2001;414:916–20. doi: 10.1038/414916a. [DOI] [PubMed] [Google Scholar]
- 21.Zabel BA, Agace WW, Campbell JJ, et al. Human G protein-coupled receptor GPR-9-6/CC chemokine receptor 9 is selectively expressed on intestinal homing T lymphocytes, mucosal lymphocytes, and thymocytes and is required for thymus-expressed chemokine–mediated chemotaxis. J Exp Med. 1999;190:1241–56. doi: 10.1084/jem.190.9.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dusaban SS, Purcell NH, Rockenstein E, et al. Phospholipase Cɛ links G protein-coupled receptor activation to inflammatory astrocytic responses. Proc Natl Acad Sci USA. 2013;110:3609–14. doi: 10.1073/pnas.1217355110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C, Liang Z, Huang W, et al. Eps8 regulates cellular proliferation and migration of breast cancer. Int J Oncol. 2015;46:205–14. doi: 10.3892/ijo.2014.2710. [DOI] [PubMed] [Google Scholar]
- 24.Arae K, Hirata M, Kurata S, et al. Mycoplasma pneumoniae induces interleukin-8 production via the epidermal growth factor receptor pathway. Microbiol Immunol. 2011;55:748–50. doi: 10.1111/j.1348-0421.2011.00375.x. [DOI] [PubMed] [Google Scholar]
- 25.Sun G, Liu X, Mercado P, et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–81. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 26.Cousins RJ, Lanningham-Foster L. Regulation of cysteine-rich intestinal protein, a zinc finger protein, by mediators of the immune response. J Infect Dis. 2000;182:S81–84. doi: 10.1086/315917. [DOI] [PubMed] [Google Scholar]
- 27.Evans PC, Huib O, Hamon M, et al. Zinc-finger protein A20, a regulator of inflammation and cell survival, has de-ubiquitinating activity. Biochem J. 2004;378:727–34. doi: 10.1042/BJ20031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jakubzick C, Choi E, Kunkel S, et al. Augmented pulmonary IL-4 and IL-13 receptor subunit expression in idiopathic interstitial pneumonia. J Clin Pathol. 2004;57:477–86. doi: 10.1136/jcp.2003.012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamane K, Indalao IL, Chida J, et al. Diisopropylamine dichloroacetate, a novel pyruvate dehydrogenase kinase 4 inhibitor, as a potential therapeutic agent for metabolic disorders and multiorgan failure in severe influenza. PLoS One. 2014;9:e98032. doi: 10.1371/journal.pone.0098032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kannan T, Blaylock MW, Dallo SF. Elongation factor Tu and E1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol Microbiol. 2001;44:1041–51. doi: 10.1046/j.1365-2958.2002.03207.x. [DOI] [PubMed] [Google Scholar]
- 31.Smigiel KS, Richards E, Srivastava S, et al. CCR7 provides localized access to IL-2 and defines homeostatically distinct regulatory T cell subsets. J Exp Med. 2014;211:121–36. doi: 10.1084/jem.20131142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hellmann J, Sansbury BE, Holden CR, et al. CCR7 maintains nonresolving lymph node and adipose inflammation in obesity. Diabetes. 2016;65:2268–81. doi: 10.2337/db15-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nureki S, Miyazaki E, Ishi T, et al. Elevated concentrations of CCR7 ligands in patients with eosinophilic pneumonia. Allergy. 2013;68:1387–95. doi: 10.1111/all.12243. [DOI] [PubMed] [Google Scholar]
- 34.Nureki S, Ishii T, Ando M, et al. Elevated concentrations Of Cc chemokine receptor 7 ligands in patients with eosinophilic pneumonia. Am J Respir Crit Care Med. 2012;185:A4156. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The regulatory network of the differentially expressed long non-coding RNAs (DE-lncRNAs) and target genes. Light green nodes represent the downregulated lncRNAs in mild pneumonia; light red nodes represent the upregulated lncRNAs in mild pneumonia; green nodes represent the downregulated lncRNAs in severe pneumonia; red nodes represent the upregulated lncRNAs in severe pneumonia; dark red nodes represent the upregulated lncRNAs in both mild and severe pneumonia; purple nodes represent the target genes. Lines represent the regulatory relationships between lncRNAs and target genes.