SUMMARY
Bacterial small RNAs (sRNAs) have been implicated in various aspects of post-transcriptional gene regulation. Here we demonstrate that sRNAs also act at the level of transcription termination. We use the rpoS gene, which encodes a general stress sigma factor σS, as a model system, and show that sRNAs DsrA, ArcZ, and RprA bind the rpoS 5′UTR to suppress premature Rho-dependent transcription termination, both in vitro and in vivo. sRNA-mediated antitermination markedly stimulates transcription of rpoS during the transition to the stationary phase of growth, thereby facilitating a rapid adjustment of bacteria to global metabolic changes. Next generation RNA sequencing and bioinformatic analysis indicate that Rho functions as a global “attenuator” of transcription, acting at the 5′UTR of hundreds of bacterial genes, and that its suppression by sRNAs is a widespread mode of bacterial gene regulation.
Graphical Abstract
Bacterial small RNAs balance the Rho-dependent termination pathway to prevent premature transcription termination, extending the role of these RNA regulators beyond post-transcriptional control.
INTRODUCTION
Regulatory RNAs actively control all aspects of gene expression in bacteria. Cis-acting riboregulators known as riboswitches are located in the 5′UTRs of many genes. They directly sense small metabolites, ions, temperature, or pH and adopt alternative conformations to control downstream events, such as transcription termination, translation initiation, RNA processing and stability (Mellin and Cossart, 2015; Serganov and Nudler, 2013). Another major class of bacterial riboregulators is comprised of relatively short RNAs, termed small RNAs (sRNAs). sRNAs act in trans by base-pairing directly with target mRNAs (Vogel et al., 2003; Wagner and Romby, 2015). They are the most abundant class of post-transcriptional regulators and are involved in numerous metabolic and stress response processes in the cell (Gogol et al., 2011; Gottesman and Storz, 2011; Mika and Hengge, 2014; Storz et al., 2011; Waters and Storz, 2009). RNA chaperone Hfq acts as a key mediator of sRNA-dependent gene expression by facilitating sRNA base pairing with its target mRNA (De Lay et al., 2013; Vogel and Luisi, 2011).
The 5′UTR of the E. coli rpoS gene is one of the most extensively studied targets of sRNAs. This gene encodes the σS subunit of RNA polymerase (RNAP), which is the general stress sigma factor that controls gene expression in response to nutrient limitation and other adverse conditions. Expression of rpoS is positively regulated by three sRNAs, DsrA, RprA and ArcZ (Battesti et al., 2011). During exponential growth, these sRNAs are poorly expressed, and translation of rpoS is inhibited by a stem-loop structure in the 5′UTR of its mRNA, which blocks access of the ribosome to the ribosome-binding site (RBS) (Majdalani et al., 1998). Upon transition to stationary phase or under stress, accumulated DsrA, RprA and ArcZ relieve translational inhibition by base-pairing to the rpoS leader sequence, rendering the RBS available to the ribosome (Majdalani et al., 1998, 2002; Mandin and Gottesman, 2010).
Rho is a multi-functional termination factor that together with its cofactor NusG is globally involved in transcription regulation (Nudler and Gottesman, 2002; Boudvillain et al., 2013; Grylak-Mielnicka et al., 2016). It is essential for generation of mRNA 3′ ends (Roberts, 1969), silencing horizontally transferred DNA elements (Cardinale et al., 2008), protecting genomic DNA from double-stranded breaks caused by transcription-replication collisions (Dutta et al., 2011), suppressing ubiquitous antisense transcription (Peters et al., 2012), and maintaining phage lysogeny (Menouni et al., 2013). Rho requires ~80 nt of untranslated, relatively unstructured, and preferably C-rich nascent RNA sequence to bind (Hart and Roberts, 1991; Richardson and Richardson, 1996). RNA binding stimulates Rho’s ATPase activity, enabling it to translocate in 5′→3′ direction to eventually dissociate the transcription elongation complex (Boudvillain et al., 2013; Epshtein et al., 2010). Given the pervasive, genome-wide action of Rho, and abundant 5′ untranslated leader regions (5′UTRs), it is surprising that only a few cases are known where Rho is active within 5′UTRs. Two notable examples are the riboswitches, ribB in E. coli and mgtA in Salmonella enterica, which control Rho-dependent termination in response, respectively, to cellular levels of FMN and Mn2+ (Hollands et al., 2012; Proshkin et al., 2014).
Here we establish that 5′UTRs are the common target of Rho-mediated termination in E. coli, and that many genes rely on sRNAs to control this process. sRNAs anneal within the leader sequences to inhibit premature Rho termination and activate the corresponding genes in response to changing growth conditions.
RESULTS
Rho-dependent Termination within the 5′UTR Occurs in Many E. coli Genes
5′UTRs longer than 80 nucleotides (“long 5′UTRs”) are present in 1203, over 27%, of the 4451 annotated E. coli genes (Salgado et al., 2013). We hypothesized that such leader sequences, likely to be devoid of ribosomes, are the natural substrates for Rho-dependent termination, and performed total RNA deep-sequencing (RNA-seq) to determine the changes in E. coli gene expression in response to brief exposure of the Rho-specific inhibitor bicyclomycin (BCM) (Zwiefka et al., 1993) during the exponential growth phase. Strikingly, BCM treatment resulted in markedly enhanced transcription in the first 300 nt from the transcription start site (TSS) of 635 genes that possess long 5′UTRs (Figure 1A), signifying strong Rho activity within their corresponding leader sequences. The majority of ORFs corresponding to 5′ UTRs identified as BCM-responsive in Figure 1A also become upregulated upon addition of BCM (Figure S1A). The complete list of these genes is provided in Table S1.
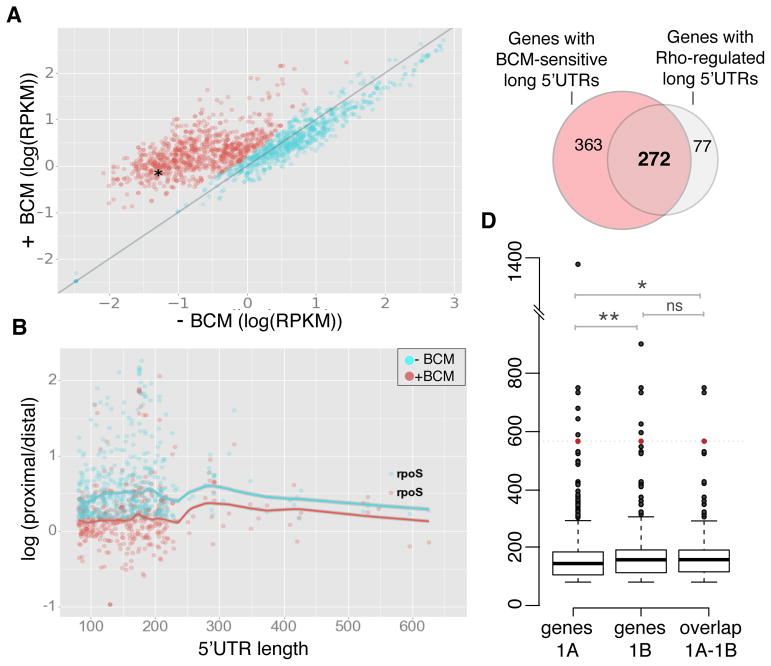
Figure 1. Long E. coli 5′UTRs are the Target by Rho.
(A) Scatter plot of transcription levels of first 300 nt in 1203 5′UTRs longer than 80 nt as Log(RPKM) (logarithm of the number of reads per kilobase of transcript per million mapped reads) in the exponentially growing cells with (+) vs without (−) BCM. The fraction of genes with BCM-upregulated long 5′UTRs (635 genes, marked in red) was identified using a Gaussian mixture model clustering (see Methods and Resources). rpoS 5′UTR is marked by an asterisk.
(B) Scatter plot of log(proximal/distal) (proximal and distal to the transcription start site) for long UTRs with the ratio (proximal/distal) >1.5 in the presence or absence of BCM. Solid lines represent lowess (local weighted regression) curve fitted to −BCM and +BCM samples with 95% CI (confidence intervals) indicated by shading. See Figure S1C for details.
(C) Venn diagram shows the overlap between the BCM-sensitive (BCM-upregulated) genes with long 5′UTRs (see 1A) and those with Rho-mediated termination within the 5′ UTRs (see 1B).
(D) Box and whisker plots showing the distribution of 5′UTR sizes for BCM-responsive genes with long leader sequences (labeled as “genes 1A”), 5′UTRs subject to Rho termination (labeled as “genes 1B”) and the set of overlapping genes between these two sets (labeled as “overlap 1A–1B”, see also C). Median for the 5′UTR sizes of all BCM-responsive genes with long leader sequences is 145 nt (genes 1A). The median size of identified 5′UTRs subject to Rho termination is 158 nt (genes 1B) and the overlap between the two sets is 158.5 nt (overlap 1A–1B). The significance was tested using non–parametric Wilcoxon-Mann-Whitney test; ** P < 0.01, * P < 0.05, ns – non-significant. rpoS (5′UTR length is 567 nt) is present in all three sets and is shown in red.
To identify 5′UTRs subject to Rho termination, and to discriminate between the readthrough from an upstream gene and BCM-mediated antitermination within a cognate 5′UTR, we analyzed the change in the ratio of reads between the two close regions, proximal and distal to TSS, for all the BCM-responsive genes from Table S1 (Figure 1B). We chose an 80 nt window immediately after TSS as the proximal window, as it provides the sequence sufficiently long for Rho to load, but yet too short for termination to occur, and another 80 nt window 200 nt downstream of TSS as the distal window (see schematics in Figure S1C). This distance (360 nt total) provides enough room for Rho to terminate after loading. The ratio of read counts between these windows (proximal/distal) and its decrease upon BCM treatment represent a measure of Rho termination within UTRs. For our analysis, we chose long (> 80 nt) UTRs with a proximal/distal ration > 1.5, signifying at least 50% termination. By implementing this strategy, we identified 349 genes that showed strong response to BCM treatment directly in their UTRs (Figure 1B and Table S1). Representative screenshots for some candidates from this list are shown in Figure S1C and D. Notably, the observed effect of BCM on the proximal/distal ratio is absent in genes with short 5′UTRs (<80 nt) (Figure S1B). In total, 272 unique genes were both upregulated and showed termination release within their 5′UTRs in response to BCM treatment (Figure 1C). We estimate the median size of such 5′UTRs to be 158,5 nt (Figure 1D). Taken together, these result show that Rho acts predominantly within long UTRs and the effect of BCM within those genes cannot be explained by readthrough from upstream genes.
Among the long 5′UTR genes responsive to BCM and directly regulated by Rho within their leader sequence was rpoS (Figure 1B and D, Table S1). Since rpoS is known to be expressed at a low level during exponential growth (Mika and Hengge, 2014; Tanaka et al., 1993), its activation by BCM suggested a Rho-dependent mechanism of repression that is relieved during transition to stationary phase or in response to stress.
Rho Terminates Transcription within the rpoS 5′UTR in vitro and in vivo
Quantitative RT-PCR within the first 240 nt of the rpoS ORF supported the RNA-seq data, demonstrating that exposure to BCM during exponential growth results in 1.7 fold increase in transcriptional output (Figure S2A–B). To test whether Rho indeed terminates transcription within the rpoS 5′UTR or early ORF, we performed a single round in vitro transcription assay. We amplified the transcription template, including the entire 5′UTR containing the natural rpoS promoter followed by 140 nt of ORF, directly from the E. coli genome (Figure 2A). In the absence of Rho, transcription of this template resulted in a full-length run-off product (Figure 2B, lane 1). Rho, with or without its cofactor NusG, caused robust termination (Figure 2B and Figure S2C), which was abolished by BCM, confirming the specificity of the assay (Figure 2B, lane 3). The most abundant Rho-dependent termination products were 450–540 nt long (Figure 2B, lanes 4–7), indicating that Rho loads within a ~+350 to +480 segment of the rpoS 5′UTR, beginning at the +1 transcription start site (TSS).
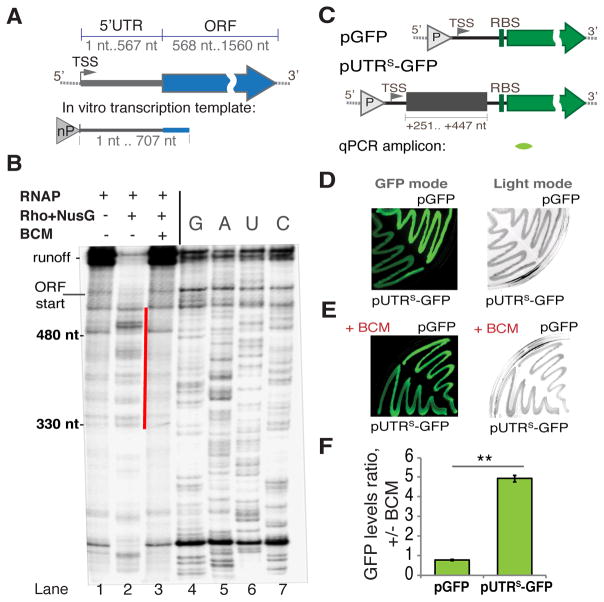
Figure 2. Rho-dependent Termination within the rpoS 5′UTR.
(A) A diagram of E. coli rpoS with its 5′UTR and ORF indicated. A genome-derived template for in vitro transcription (B) that includes the entire rpoS 5′UTR and the first 140 nt of ORF is depicted below, “nP” indicates the location of the native promoter driving rpoS transcription. TSS - transcription start site.
(B) A representative single round transcription (6% TBE-UREA gel) on the rpoS template (see A). Pre-formed elongation complexes were chased without (lane 1) or with Rho and NusG (without BCM - lane 2; with BCM - lane 3). Gel densitometry indicates that the efficiency of termination without BCM is ~87% (see also Figure S3). The most prominent termination products are marked with the red line. To determine the location of termination sites RNA sequencing was performed using 3′ dNTPs (lanes 4–7). The beginning of ORF and runoff are indicated.
(C) Reporter constructs. Upper panel: pGFP construct with the GFP reporter only. Lower panel: the transcriptional fusion pUTRS-GFP used to test the effect of the rpoS 5′UTR (fragment +251–447 nt) on Rho termination (see also Figure S2A). “P” indicates the location of a constitutive promoter. TSS – the transcription start site; RBS – the ribosome-binding site. The location of qRT-PCR amplicon is indicated below (green).
(D and E) Representative results from the GFP plate assay. DH5a cells transformed with pGFP or pUTRS-GFP grew on LB agar plates without (D) or with 8 μg/ml BCM (E) and the fluorescence intensity was measured (GFP mode, left panel). The same plates were also captured under visible light (Light mode, right panel).
(F) qRT-PCR data for exponentially growing cells transformed with pGFP or pUTRS-GFP. Ratio of gfp RNA levels +/− BCM is plotted. Values are means ±SD, n = 3; ** P < 0.01 (Student’s t-test, equal variance).
To monitor Rho-mediated termination at the rpoS 5′UTR in vivo, we designed a plasmid-based reporter containing the +251 – +447 segment of the rpoS 5′UTR, which was shown to be targeted by Rho in vitro, fused to GFP under a constitutive promoter (pUTRS-GFP; see also Figure S4). The same plasmid absent the rpoS 5′UTR region (pGFP) was used as a control (Figure 2C). Plated E. coli cells transformed with pUTRS-GFP displayed much weaker fluorescence compared to cells with pGFP (Figure 2D, left panel) or its derivatives with various upstream segments of rpoS 5′UTR (Figure S3A–C). All these strains had equal growth rates (Figure 2D and Figure S3B–C). Fluorescence of pUTRS-GFP cells increased to the control level if plates were supplemented with BCM (Figure 2E). qRT-PCR-based quantification of gfp shows that pUTRS-GFP cells respond to BCM ~5 times stronger than the pGFP control (Figure 2F). Notably, three representative long 5′UTRs from the non-BCM-responsive genes (absent in Table S1) display no significant difference in fluorescence from the UTR-less pGFP control, confirming the lack of Rho termination in vivo (Figure S3D and E). They also failed to support Rho termination in vitro (Figure S3F). Together these results validate the specificity of our assays and demonstrate that the rpoS 5′UTR contains a strong Rho-dependent termination signal that is active both in vivo and in vitro.
sRNAs Control Rho-dependent Termination at the rpoS 5′UTR
We noticed that Rho termination sites that we mapped within the rpoS 5′UTR overlap with and/or reside in close proximity to the known binding sites for sRNAs (DsrA, ArcZ, RprA), which positively regulate rpoS translation (see Introduction). To test whether these sRNAs influence Rho activity at the rpoS leader sequence, we performed a GFP assay similar to that described above in three E. coli strains: wild-type-like PM1409 (a parent strain carrying a chromosomal rpoS–lacZ translational fusion), a dsrA rprA arcZ triple deletion mutant PM1417 (derived from PM1409) and an hfq deletion mutant PM1419 (also derived from PM1409) (Mandin and Gottesman, 2010). These strains were transformed with the pUTRsRNA-GFP transcriptional fusion plasmid, containing the rpoS leader segment (+251 to +557 nt), which includes the well-characterized sRNAs-binding site (Figure 3A; see also Figure S4). Note that translation in this case should not be affected by sRNAs because the pUTRsRNA-GFP fusion lacks the RBS inhibitory hairpin (Figure S4C) (Battesti et al., 2011; Soper and Woodson, 2008).
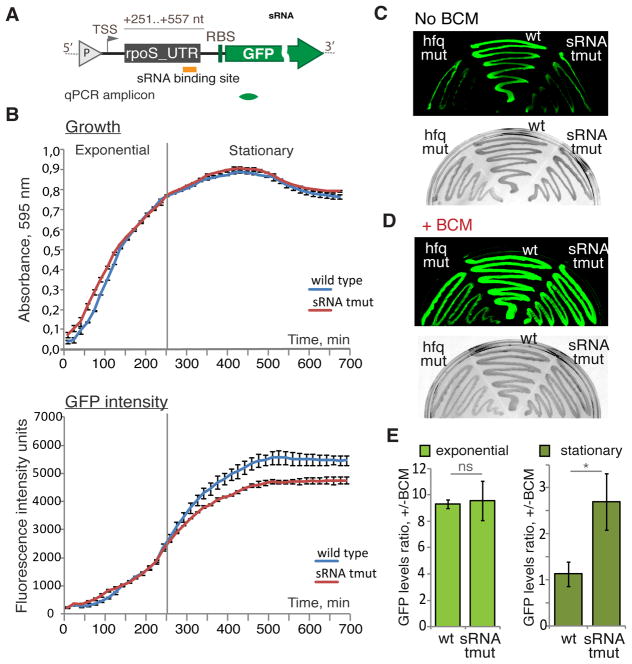
Figure 3. sRNAs Control Rho-dependent Termination within the rpoS 5′UTR Reporter.
(A) A diagram of the pUTRsRNA-GFP construct (see also Figure S2B) that includes the rpoS 5′UTR fragment (+251–557 nt) with the position of the sRNAs-binding site indicated (orange bar). “P” (grey triangle) indicates the location of the constitutive promoter. TSS - transcription start site; RBS – ribosome-binding site. The location of the qRT-PCR amplicon is indicated below (green).
(B) sRNA effect on transcription as detected by real-time fluorescence. E. coli strains PM1409 (wild type, blue) and PM1417 (dsrA rprA arcZ triple deletion mutant – tmut, red) transformed with pUTRsRNA-GFP (see also Figure S2B) grew in LB for more than 11 hours while cell density (upper panel) and fluorescence intensities (lower panel) were simultaneously monitored. The grey vertical line indicates the boundary between exponential and stationary phases. For each time point the values represent means ±SD, n = 3.
(C and D) Representative results from the GFP plate assay. PM1409 (wt), PM1417 (tmut), and the hfq deletion mutant (PM1419) transformed with pUTRsRNA-GFP grew on LB agar plates without (C) or with 8 μg/ml BCM (D) and the fluorescence intensity was measured (GFP mode – upper panel). The same plates were also captured under visible light (light mode – lower panel).
(E) qRT-PCR data for exponentially (left panel) and stationary (right panel) growing cell transformed with pUTRsRNA-GFP. Ratio of gfp RNA levels +/− BCM is plotted. Values are means ±SD, n = 3; * P < 0.05; ns - not significant (Student’s t-test, equal variance).
WT and sRNAs triple deletion cells transformed with the GFP reporter plasmid grew in rich liquid media at the same rate during the 12-hour experiment (Figure 3B, upper panel). Indeed, lack of rpoS does not cause a significant growth defect (Dong and Schellhorn, 2009; Dong et al., 2008). However, the intensity of the GFP signal progressively diverged between the WT and triple sRNAs deletion strains after the first 4 hours of growth (Figure 3B, lower panel). After 8 hours, the fluorescence in WT was about 20% higher than that in the sRNAs-deficient cells, consistent with the sRNAs stimulating transcription of the reporter upon the transition to stationary phase, which correlates with the induction of DsrA, RprA and ArcZ (Argaman et al., 2001). Considering that Rho is normally active within the rpoS 5′UTR sequence, these results suggest that inhibition of Rho termination was linked to the increased production of sRNAs, resulting in elevated GFP expression.
Likewise, after 18h of growth on plates of solid media the fluorescence of wild-type cells was much greater than that of sRNAs-deficient cells or Hfq-deficient cells (Figure 3C). Supplementing the plates with BCM substantially increased the fluorescence of the sRNAs-deficient strains (Figure 3D). In the exponential phase, qRT-PCR-based quantification shows an approximately 10-fold increase in the gfp expression in response to BCM in both the WT and sRNAs-deficient pUTRsRNA-GFP cells (Figure 3E, left panel). However, the response to BCM between the two cultures differs greatly in the stationary phase: the levels of gfp in WT pUTRsRNA-GFP cells changed only slightly, whereas sRNAs-deficient pUTRsRNA-GFP cells displayed almost a 3-fold increase in gfp expression (Figure 3E, right panel). As the ratio between sRNAs and their target is important (Levine et al., 2007), we confirmed that sRNAs could still induce endogenous rpoS upon transition from exponential to stationary phase in WT cells transformed with pUTRsRNA-GFP (Figure S4E). No difference in fluorescence was detected between the strains carrying the control plasmids lacking either the entire rpoS 5′UTR (Figure S4F) or its sRNAs binding sites (Figure S4G).
Together these results imply that sRNAs binding within the rpoS leader inhibits Rho termination.
To further support this conclusion, we quantified the effect of sRNAs and BCM using the chromosomal rpoS 5′UTR-lacZ fusion by qRT-PCR (Figure 4A and B). Reporter transcript levels were measured in the exponential phase with or without the plasmids expressing DsrA, RprA, or ArcZ (Soper et al., 2010)(Figure 4C and D). Induction of each individual sRNA led to the substantial accumulation of lacZ mRNA (Figure 4A). A similar increase of lacZ mRNA was observed in response to BCM (Figure 4A). Notably, after a brief pretreatment with BCM virtually no further induction of rpoS leader could be detected in response to sRNAs (Figure 4B). Since BCM per se does not induce any sRNAs accumulation in the exponential phase (Figure 4C), we conclude that inhibition of Rho by DsrA, RprA, or ArcZ within the rpoS 5′UTR accounts for the stimulating effect of these sRNAs on rpoS 5′UTR-lacZ transcription.
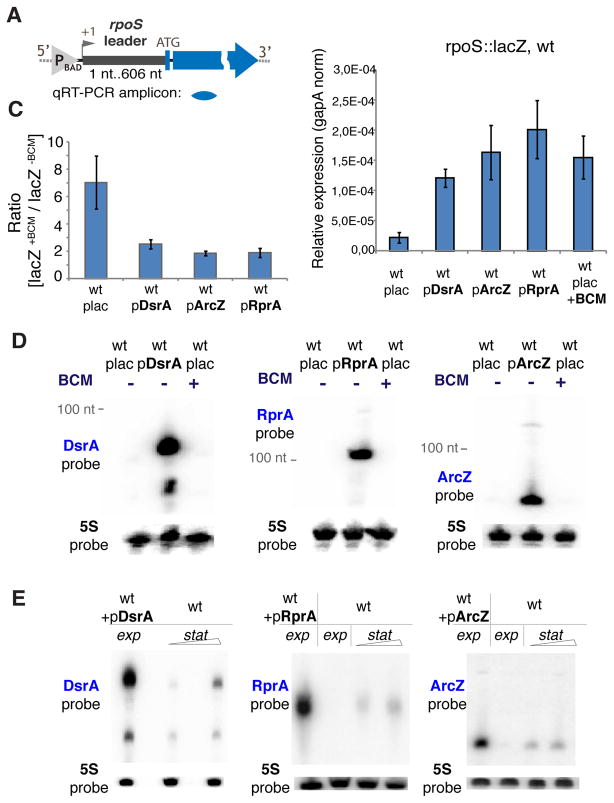
Figure 4. sRNAs Overexpression Stimulates rpoS Leader-driven Transcription by Inhibiting Rho.
(A) Upper panel: the rpoS::lacZ chromosomal reporter fusion. “P” indicates the inducible pBAD promoter. Position +1 indicates the location of transcription start site; ATG – translation start codon. The location of qRT-PCR amplicon is indicated below. Lower panel: transcript levels of lacZ measured in wt strain carrying the rpoS::lacZ chromosomal fusion during mid-exponential phase. Where indicated the expression of plasmid-borne sRNAs was induced (plac is an empty vector pBRplac), or BCM was added for 15 min. qRT-PCR values were normalization to that of a housekeeping gapA gene. Values represent means ±SD from three independent experiments.
(B) The effect of BCM on lacZ transcript levels. Where indicated DsrA, ArcZ and RprA were induced in the mid exponential phase. Afterwards, the corresponding strains were incubated with or without BCM followed by total RNA extraction. Ratios of lacZ transcript levels +/− BCM for each individual strain were calculated using qRT-PCR as in (A).
(C) Nothern blot analysis: levels of DsrA, RprA and ArcZ in the exponentially grown WT (PM1409) without sRNA overexpression (transformed with the parent empty pBRplac plasmid), after sRNA overexpression (wt+pDsrA/wt+pRprA/wt+pArcZ), and after treatment with BCM (wt+BCM). 5S probe – loading control.
(D) Nothern blot analysis: levels of overexpressed DsrA, RprA and ArcZ in the exponentially grown WT (PM1409 transformed with indicated pBRplac-derived sRNA construct) and endogenous sRNA accumulation in WT (PM1409 with empty pBRplac plasmid) grown to medium/late stationary phase. 5S probe – loading control.
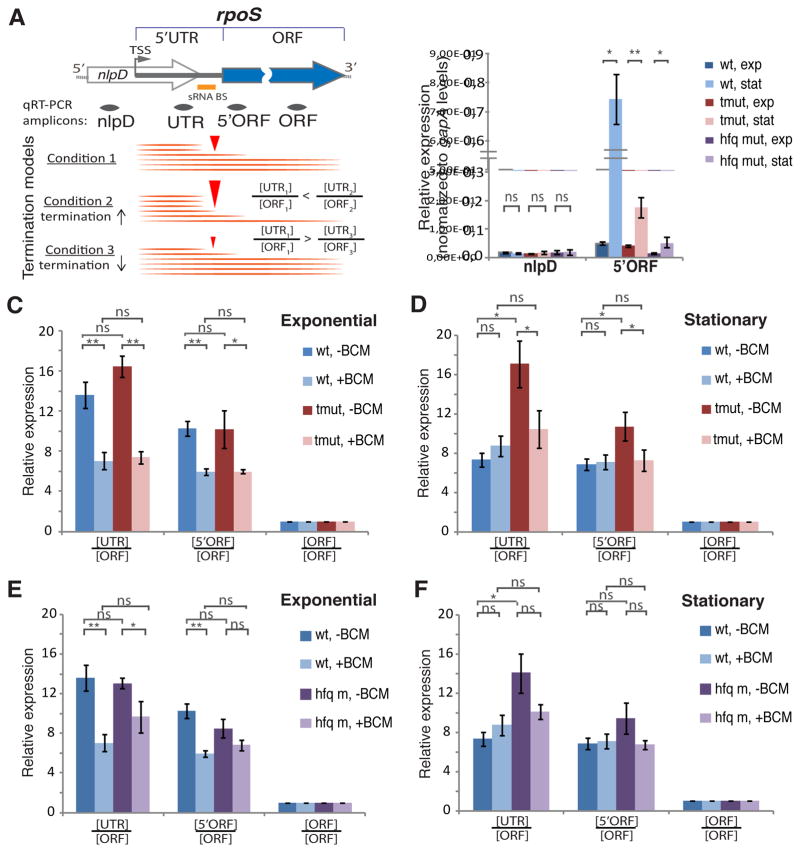
To estimate the efficiency of Rho-dependent termination and anti-termination effected by sRNAs in their natural environment, we used qRT-PCR to assay different regions of the native rpoS transcript in PM1409, PM1417 and PM1419 cells under various growth conditions. The selected amplicons covering parts of the rpoS 5′UTR and ORF are schematically shown in Figure 5A. Upon the transition to stationary phase, wild-type cells displayed a substantial induction of rpoS mRNA, whereas such induction was greatly compromised in cells deficient in sRNAs or Hfq (Figure 5B, see also Figure S5B). As the rpoS leader resides within the 3′-proximal part of the nlpD gene, we used the 5′ part of nlpD as negative control. This amplicon did not show any change in any of the three strains upon their transition to stationary phase (Figure 5B), validating the specificity of rpoS 5′UTR transcription regulation.
Figure 5. sRNAs Act on Chromosomal rpoS 5′UTR.
(A) A diagram of rpoS with its ORF and 5′UTR located within the nlpD gene. The locations of qRT-PCR amplicons are shown below. The orange bar marks the position of the DsrA/RprA/ArcZ binding site (sRNA BS). TSS - transcription start site. The lower panel shows the approach for an unbiased estimation of Rho-dependent termination within the rpoS leader that relies on changing [UTR]/[ORF] ratios (internal normalization within rpoS transcript). An increase in the [UTR]/[ORF] ratio corresponds to the increased termination efficiency, whereas a decrease in the [UTR]/[ORF] ratio corresponds to the decreased termination efficiency.
(B) Levels of nlpD and 5′-end-proximal rpoS ORF measured for the wt, triple sRNA deletion mutant (tmut), and hfq deletion mutant (hfq m) in exponential and stationary phases of growth. qRT-PCR values are normalized to that of the housekeeping gapA gene. Values represent means ±SD, n ≥ 3; ** P < 0.01; * P< 0.05; ns - not significant (Student’s t-test, equal variance).
(C and D) Transcript levels of different rpoS regions measured for the wt and triple sRNA deletion mutant (tmut) in exponential (C) and stationary (D) phase before and after BCM treatment (15 min, 50 μg/ml). qRT-PCR values are normalized to that of the rpoS ORF amplicon so that [ORF]/[ORF] equals to 1 for each strain/condition. Values represent means ±SD, n ≥ 3; *** P < 0.001; ** P < 0.01; * P< 0.05; ns - not significant (Student’s t-test, equal variance).
(E and F) Transcript levels of different rpoS regions measured for wt and hfq deletion mutant in exponential (E) and stationary (F) phase before and after BCM treatment (15 min, 50 μg/ml). qRT-PCR data with internal rpoS transcript normalization to ORF amplicon levels are shown. qRT-PCR values are normalized to that of the rpoS ORF amplicon. [ORF]/[ORF] equals to 1 for each strain/condition. Values represent means ±SD, n ≥ 3; *** P < 0.001; ** P < 0.01; * P< 0.05; ns - not significant (Student’s t-test, equal variance).
We then selected three rpoS regions to assess transcription termination and sRNA-mediated antitermination quantitatively: a segment upstream of the sRNA-binding site (“UTR” amplicon), the 5′-proximal part of the rpoS ORF (“5′ORF” amplicon), and the ORF segment ~ 500 nt downstream of the translation start (“ORF” amplicon). Internal normalization to the amount of “ORF” for every strain allowed us to directly compare the termination efficiency within rpoS among different strains and conditions, irrespective of strain-specific variations in rpoS amplicons, i.e. absolute levels of transcripts and their stabilities (Figure 5A; Figure S5A and B). In the exponential growth phase, a high ratio of [UTR]/[ORF] and [5′ORF]/[ORF] can be explained by efficient Rho-dependent termination for all three strains (Figure 5C, E). Indeed, addition of BCM during the exponential growth resulted in a substantial decrease of the [UTR]/[ORF] and [5′ORF]/[ORF] ratios for all three strains (Figure 5C, E). Notably, during exponential growth, the ratios were almost identical between the strains, either with or without BCM. However, the effects of the mutants become clearer when cells enter stationary phase. The WT strain has substantially lower [UTR]/[ORF] and [5′ORF]/[ORF] ratios, compared to exponential growth (compare dark blue bars in Figure 5C to 5D and 5F to 5E), consistent with less termination within the leader. Accordingly, BCM had essentially no effect on the ratios (compare light blue to dark blue bars in Figure 5D, F). In the sRNA deficient and hfq deficient cells, however, both the ratios in stationary phase and the effect of BCM were comparable to those in exponential phase (compare red bars in Figure 5C and 5D and purple bars in Figure 5E and 5F), consistent with Rho-dependent termination still occurring in these strains. Taken together, these results demonstrate that sRNAs suppress Rho-dependent termination within the rpoS 5′UTR upon the transition to stationary phase.
To roughly estimate the relative impact of sRNAs on transcription termination (Rho) and translation initiation of rpoS we utilized a strain carrying a C125T substitution that disrupts the RBS inhibitory hairpin of rpoS (Battesti et al., 2011; Brown and Elliott, 1997) (Figure S4C). C125T renders the rpoS translation independent of Hfq and thus, presumably also independent of the sRNAs. Therefore, the changes in rpoS (C125T) transcript levels should be primarily due to sRNA-mediated transcriptional regulation, whereas in WT cells such changes should involve both transcriptional and translational control (RNA stabilization due to ongoing translation). Treatment with BCM increased the levels of rpoS in exponential phase even more in the C125T strain than in WT, demonstrating that opening up the hairpin and thus allowing translation of RpoS is not sufficient to bypass the requirement for sRNA-mediated antitermination (Figure S5C–E).
Reconstitution of sRNA-mediated Antitermination
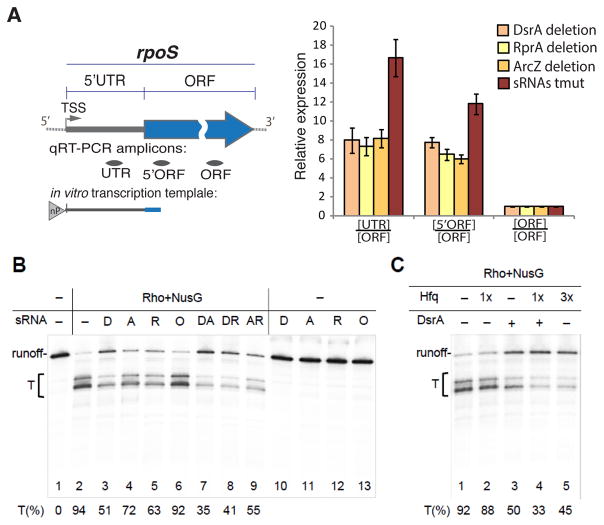
To determine whether sRNAs directly interfere with Rho-dependent termination and to study the mechanism of this process, we used an in vitro reconstituted system. The rpoS promoter template, containing the entire rpoS leader sequence, including the sRNAs-binding site, was used in the single round transcriptional assay (Figure 6). Transcription in the presence of Rho and NusG resulted in robust (94%) termination (Figure 6B, lane 2), which was suppressed by sRNAs. DsrA exhibited the strongest inhibitory effect, followed by RprA and ArcZ (Figure 6B, lanes 3–5). Addition of suboptimal amounts of two different sRNAs simultaneously resulted in the additive antitermination effect (Figure 6B, lanes 7–9). OxyS sRNA, which was previously shown to negatively regulate rpoS translation most likely via titration of Hfq (Moon and Gottesman, 2011; Zhang et al., 1998), displayed no antitermination activity at the same concentration (Figure 6B, lane 6). In the absence of Rho, neither of the sRNAs affected transcription elongation or stability of the full-length transcript (Figure 6B, lanes 10–13). The observed antitermination by sRNAs was specific to the rpoS leader sequence, because Rho-dependent termination on another template lacking any sRNA-binding sites was unaffected by sRNAs (Figure S6). This control rules out a possibility of non-specific inhibition of Rho by sRNAs. Thus, each of the three sRNAs (DsrA, ArcZ, and RprA) bind directly to rpoS 5′UTR to suppress Rho termination in vitro.
Figure 6. Reconstitution of sRNA-mediated Antitermination.
(A) A diagram of the E. coli rpoS gene with its 5′UTR and ORF indicated. The genome-derived template for in vitro transcription (B) includes the entire rpoS 5′UTR and first 140 nt of rpoS ORF is shown below. “nP” indicates the natural RNAP driving rpoS transcription. TSS - transcription start site. The locations of qRT-PCR amplicons are depicted below as grey bars.
(B) sRNAs inhibit Rho termination in vitro. Radiogram shows a representative single round transcription assay (see Experimental Procedures). Pre-formed elongation complexes were chased in the absence (lanes 1, 10–13) or presence of Rho and NusG. A bracket indicates the transcription termination zone (T). The efficiency of Rho-dependent termination was estimated in the absence (lanes 2) or in the presence of 1 M of different sRNAs (lanes 3–6), including DsrA (D), RprA (R), ArcZ (A), and OxyS (O). Pair combinations of sRNAs (each of 1 M) were used as indicated (lanes 7–9). The percentage of termination (T%) was calculated for each reaction as a ratio between the amount of radioactivity in the bands corresponding to the termination products (T) and the total radioactivity signal of the termination and readthrough bands. Values represent means from 3 independent experiments.
(C) Hfq assists DsrA-mediated antitermination. A single round transcription assay was performed as in (B), except for 25 nM (1x) or 75 nM (3x) Hfq were added during the chase reaction.
(D) Transcript levels of different rpoS regions measured for individual sRNA deletion strains and the triple sRNAs deletion (tmut) in stationary phase. qRT-PCR values are normalized to that of the rpoS ORF amplicon. Values represent means ±SD, n ≥ 3; *** P < 0.001; ** P < 0.01; * P< 0.05; ns - not significant (Student’s t-test, equal variance).
It has been shown that Hfq facilitates sRNAs binding within the rpoS leader in vitro and in vivo (Peng et al., 2014). Consistently, Hfq stimulates DsrA-mediated antitermination (Figure 6C, lanes 3 and 4). However, since Hfq itself inhibits Rho-mediated termination at the concentrations >50 nM in our reconstituted system (Figure 6C, lane 5), perhaps due to direct binding to Rho (Rabhi et al., 2011), it prevented us from examining its full potential with respect to sRNA-mediated antitermination in vitro. As it is not clear how much free Hfq is present in vivo, and how many of those molecules could bind Rho directly (to inhibit it), the in vitro experiment can only serve as an indication that at about a 1:1 molar ratio (with Rho hexamer) Hfq does not effectively inhibit Rho termination within the rpoS 5′UTR, unless sRNAs were present.
To examine whether the collective antitermination effect of sRNAs observed in vitro (Figure 6B, lanes 7–9) also occurs in vivo, we compared the efficiency of antitermination in the rpoS leader between the triple sRNA deletion strain and individual knockouts of dsrA (PM1411), rprA (PM1412) and arcZ (PM1413) by qRT-PCR. In the stationary phase, UTR/ORF and 5′ORF/ORF ratios for the triple sRNA deletion mutant significantly exceeded those of any single sRNA deletion mutant (Figure 6D), supporting the cumulative effect of sRNAs on Rho antitermination observed in vitro.
sRNA-mediated Antitermination is a General Phenomenon
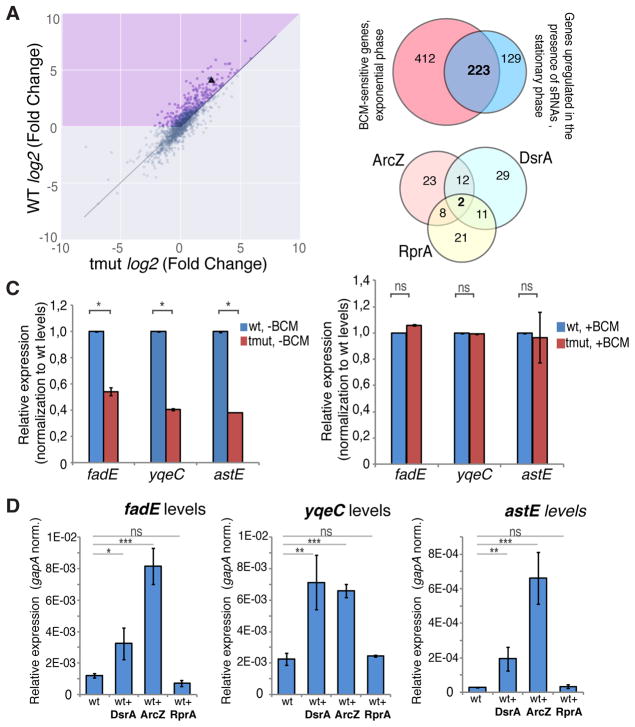
To examine whether sRNA-mediated antitermination occurs at genes other than rpoS, we compared RNA-seq expression profiles of WT and triple sRNAs deletion strains upon the transition to stationary phase. Following the rpoS example, we focused on a set of genes expressed more abundantly (q<0.05) in stationary phase in WT cells, but not in sRNAs-deficient cells. We identified 352 such genes with long 5′UTR, including rpoS (Figure 7A, blue sector; Table S2). Intriguingly, 223 of them (Figure 7B, upper panel) overlap with the long leader-containing genes that are BCM-sensitive in the exponential growth phase (Figure 1A). Furthermore, computational analysis using the RNApredator tool (Eggenhofer et al., 2011) suggests that 106 of these genes have a binding site(s) for at least one sRNAs (DsrA, RprA, or ArcZ) within their leader sequence (Table S2). The distribution of predicted sRNA-binding sites is shown in Figure 7B, lower panel. To validate these results, we further analyzed three representative genes from the candidate list (Table S2, marked in red), fadE, yqeC, and astE, carrying the binding sites for DsrA and ArcZ (Figure S7A). RT-qPCR analysis confirmed the RNA-seq results, indicating that all three genes were upregulated considerably less in sRNAs-deficient cells than in WT cells during the transition to stationary phase. BCM treatment of the sRNA-deficient cells caused a strong induction of all three genes whose mRNA levels rose to those of wild-type cells (Figure 7C; see also Figure S7C). Importantly, the overexpression of DsrA and ArcZ, but not RprA, in the exponential phase resulted in upregulation of fadE, yqeC and astE (Figure 7D). At the same time, the rpoS gene was upregulated by all three sRNAs (Figure S7B). These results validate our sRNA targets predictions and support sRNA-mediated antitermination within these three representative genes. It also indicates that sRNAs-mediated induction of fadE, yqeC and astE is likely to be independent of rpoS. Furthermore, overexpression of sRNAs in hfq-deficient cells did not lead to a comparable upregulation of fadE, yqeC and astE (Figure S7B), indicating the significance of Hfq for sRNA-mediated transcription antitermination. Taking together, these data show that Rho-mediated termination and sRNA-mediated antitermination at the 5′UTRs are common in E. coli.
Figure 7. sRNA-mediated Antitermination is a Global Phenomenon.
(A) Scatter plot of gene expression changes upon transition to the stationary phase (log2 Fold Change) in the wild type (WT) vs triple sRNAs deletion mutant. Differential gene expression analysis was performed using the DESeq2 software package for stationary vs exponential phase. Only the genes with long (>80 nt) 5′UTRs are shown. rpoS is marked by a triangle. The purple shaded area indicates genes with a greater upregulation during stationary phase in wt cells than in the sRNAs mutant strain.
(B) Upper panel: Venn diagram shows the overlap between the long leader genes upregulated in the exponential phase due to BCM treatment and those, which upregulation in the stationary phase depends on DsrA, RprA and ArcZ. Lower panel: Venn diagram shows the predicted sRNA target distribution in the 223 genes, which regulation depends on both Rho and mentioned sRNAs.
(C) Transcript levels of fadE, yqeC and astE in the wt and sRNAs triple deletion mutant (tmut) measured in stationary phase in the absence (left) or presence of BCM (right). qRT-PCR data with normalization against corresponding wt strain gene levels. Values represent means ±SD, n = 3; * P < 0.05; ns, not significant (Student’s t-test, equal variance).
(D) Transcript levels of fadE, yqeC and astE in the exponentially grown wt cells with or without overexpression of DsrA, ArcZ and RprA. qRT-PCR values are normalized to that of a housekeeping gapA gene. Values represent means ±SD, n = 3; ** P < 0.01, *** < 0.001, ns - not significant (Student’s t-test, equal variance).
DISCUSSION
Bacterial mRNAs frequently contain regulatory signals that govern their own transcription. For example, riboswitches, the 5′UTR-based cis-regulating elements, control transcription by alternatively forming terminator/antiterminator RNA hairpins in response to diverse metabolic and environmental cues (Nudler and Mironov, 2004). The general termination factor Rho was recently shown to act as an effector in riboswitch-mediated gene control (Hollands et al., 2012; Proshkin et al., 2014), a currently rare example of transcriptional regulation by Rho activity within a bacterial leader sequence.
Remarkably, more than a quarter of E. coli genes contain an annotated long 5′UTR (>80 nt), ribosome-free segment of the nascent transcript upstream of the translation start site that harbors a potential loading site for Rho. Here we provide evidence that such long 5′UTRs do indeed serve as common Rho-mediated regulatory signals. Our deep-sequencing data reveal that more than 50% of genes (635 out of 1203) with long 5′UTRs are subjected to premature Rho-dependent termination during exponential growth in rich media. In 272 of these genes Rho terminates transcription within their 5′UTRs with more than 50% efficiency (Figure 1B). This default repressive state can be at least partially relieved in response to changes in nutrient and metabolic conditions. For example, according to our RNA-seq data, Rho is active within the leader sequence of several well-characterized E. coli riboswitches (Table S1), including those of thiM and lysC, which respond, respectively, to intracellular levels of TPP and lysine. Thus, in addition to controlling translation initiation (Caron et al., 2012; Rentmeister et al., 2007; Serganov et al., 2006), these riboswitches are also likely to control Rho-dependent transcription termination, a mode of regulation similar to that described for E. coli ribB (Hollands et al., 2012).
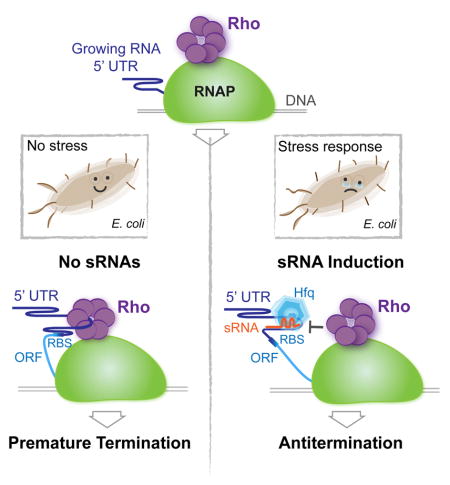
Importantly, our deep-sequencing analysis reveals a novel and surprisingly widespread mechanism to control Rho-dependent termination - sRNA-mediated antitermination. Traditionally, sRNAs have been implicated in the modulation of translation initiation and/or mRNA stability by base-pairing with regulatory motifs of mRNAs (Gogol et al., 2011; Mika and Hengge, 2014; Storz et al., 2011; Vogel and Luisi, 2011; Wagner and Romby, 2015). Trans-encoded ChiX from Salmonella has become the first example of a sRNA that regulates Rho-dependent termination by pairing within the 5′UTR: it activated Rho indirectly by inhibiting translation initiation, thereby uncoupling transcription from translation within the chiP operon (Bossi et al., 2012). Using the rpoS gene as a model, we present evidence that sRNAs, with the help of Hfq, directly control transcription by interfering with Rho loading and/or translocation along the nascent 5′UTRs in a genome-wide manner. sRNA-mediated antitermination contributes significantly to the induction of σS in response to stress, and it appears to work independently of previously described mechanisms involving sRNA-mediated regulation of σS that relied on inhibition of translation initiation and mRNA degradation (Battesti et al., 2011). The antitermination mechanism may also explain other known instances of sRNA-mediated gene activation, as, for example, in the case of RprA activation of RicI in Salmonella (Papenfort et al., 2015), that lacked obvious features of post-translation control. Indeed, our high-throughput data analysis and its experimental validation show that sRNA-mediated inhibition of Rho activity occurs within the leader sequences of many E. coli genes. It is likely that Hfq-dependent sRNAs other than DsrA, RprA, and ArcZ could also act in a similar fashion with respect to Rho.
Positioning of the termination sites within rpoS suggests that formation of the sRNA-5′UTR complex inhibits Rho by either interfering with its RNA loading or its translocation along RNA, serving as a “roadblock”. Indeed, while Rho can readily unwind RNA:DNA heteroduplexes, it is inefficient in displacing RNA from the RNA:RNA duplexes (Brennan et al., 1987; Steinmetz et al., 1990). By stabilizing the 5′UTR-sRNA complex, Hfq should further complicate Rho loading/translocation. Interestingly, the formation of an sRNA-5UTR complex could, in principle, stimulate Rho-mediated termination instead of inhibiting it. Indeed, instead of obscuring a loading site for Rho, sRNAs could unmask it directly via a 5′UTR structural rearrangement, or indirectly, by inhibiting translation and causing downstream polarity (Bossi et al., 2012). Such a pro-termination mode of action may be adopted by a large class of sRNAs known to inhibit their multiple targets, e.g. the highly conserved GcvB sRNA (Sharma et al., 2011). Whereas the pro-termination sRNAs may represent an interesting subject for future studies, the present results establish sRNAs as a new class of finely regulated anti-termination elements that function in trans throughout the bacterial genome in response to particular environmental changes.
METHODS AND RESOURCES
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Evgeny Nudler (evgeny.nudler@nyumc.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial Strains, Plasmids and Growth Conditions
All E. coli strains used in this study are derivatives of the wild-type MG1655 strain and are listed in Key Resource Table. Strains JC1085 and JC1088 were constructed by homologous recombination of wild type or C125T mutant rpoS leader sequences, which were synthesized using IDT geneblock service, into JC1078, a strain derived from NM541 that has a pBAD-ccdB-kan cassette inserted in the native rpoS leader, selecting recombinants resistant to arabinose induction of the CcdB toxin (Battesti et al., 2015).
Cloning steps were performed using DH5α E. coli strain [F-φ0lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rk−, mk+) phoA supE44 λ-thi-1 gyrA96 relA1]. The main plasmids used in this study are listed in the Key Resource Table.
For cloning, reporter assays and maintaining strains: cultures were grown in LB or LB-agar media and, when required, supplemented with ampicillin (100 μg/ml), kanamycin (30 μg/ml), tetracycline (10 μg/ml), chloramphenicol (10 μg/ml) and bicyclomycin (BCM) (8 μg/ml).
For measurements of rpoS-lacZ fusion transcript levels with or without DsrA, RprA, or ArcZ overexpression: strains transformed with empty vector pBRplac or pBRplac-derived plasmids overexpressing DsrA, RprA, or ArcZ, were grown at 37°C in LB supplemented with Amp (100 μg/ml), L-arabinose (0.2%) and IPTG (1 mM, induction for 20–25 min) to OD600 = 0.5–0.6 with constant aeration.
For measurements of endogenous transcript levels and preparation of deep-sequencing libraries: cells from the diluted overnight culture (starting OD600 ≈ 0.05) were grown in LB media (minimal volume – 50 ml) to exponential (OD600 = 0.5–0.6) or stationary phase (OD600 = 2.0–2.4) with constant aeration (shaking at 200 rpm) at 37°C.
For treatment with BCM, cultures were first grown to early exponential phase (OD600 ≈ 0.5), and then BCM was added to a final concentration of 50 μg/mL for 15 minutes before harvesting. For comparison of BCM response in different strains in stationary phase, cultures were first grown to OD600 ≈ 1.6–2.0, and then BCM was added to a final concentration of 50 μg/mL for 15 minutes before harvesting. We selected BCM conditions based on our previous studies (Cardinale et al., 2008; Dutta et al., 2011). 50 μg/ml was chosen to maximize the BCM-dependent changes at the transcriptome level, but at the same time not to significantly slow bacterial growth during the experiment. For extended treatments (e.g. overnight growth on agar plates) we used 8–10 μg/ml BCM – a maximum concentration that still supports bacterial growth.
METHOD DETAILS
Protein Reporter Assays
Plate GFP assay: to measure GFP fluorescence, E. coli strains transformed with GFP fusions were plated by streaking on LB-agar plates supplemented with ampicillin (100 μg/ml) and incubated for 16–20 hours at 37°C. Plates were photographed at GFP mode with excitation wavelength 460 nm (Fusion Fx7 Imager, PEQlab, Germany). To control cell density, plates were imaged at visible light mode with excitation wavelength 510 nm (Fusion Fx7 Imager, PEQlab, Germany).
GFP assay in liquid culture: corresponding strains harboring plasmids were grown in 5 ml LB media (supplemented with relevant antibiotic) to early stationary phase (OD600 = 1.0–1.2), then diluted to OD600 ≈ 0.05, and 150 μl was transferred to designated wells of a 96-well black plate (flat clear bottom) with a clear lid (Greiner, LOT 07290155). The plate was incubated at 37°C for 12–18 hours with constant orbital shaking (amplitude – 1 mm) in a Tecan Infinite F500 microplate reader. The plate was read from above and below every 15 min, with detection at 595 nm (Absorbance, 595 nm) using a fixed signal gain of 30% with excitation at 485 nm and detection at 535 nm (Fluorescence intensity). Measurements were taken at least in triplicates for each biological replicate.
RNA Isolation
Total RNA was isolated using the hot phenol method. Cells grown to the desired OD600 were harvested and mixed with 8 volumes of Stop Solution (95 % ethanol, 5 % phenol), followed by centrifugation at 3,000 × g at 4°C for 5 min. The collected bacterial pellets were frozen in liquid nitrogen, followed by resuspension in a lysis buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.5 mg/mL lysozyme) with 0.1 % SDS. The resulting lysate was incubated at 64°C for 2 min, followed by the addition of NaOAc (pH 5.2) to a final concentration of 0.1 M. Then an equal volume of phenol (water-saturated, pH ca 4.0, AppliChem GmbH) was added, gently mixed and incubated at 64°C for 6 min with inverting 6 – 8 times. After cooling on ice, cells were centrifuged at 16,100 × g (4°C, 10 min). The aqueous phase was mixed with the equal volume of phenol/chloroform/isoamyl alcohol (25:24:1, pH ca 4.0, AppliChem GmbH) and centrifuged for 5 min at 16,100 × g (4°C) in the Phase Lock Gel Heavy tube (5Prime). Then the aqueous phase was mixed with chloroform/isoamyl alcohol (24:1) followed by a 5 min centrifugation at 16,100 × g (4°C). RNA was ethanol-precipitated from the aqueous phase (3 volumes ethanol, 1/10 volume of 3 M sodium acetate, 1/100 volume 0.5 M EDTA). To remove traces of DNA, total RNA was treated with Recombinant DNase I (Roche) according to the manufacturer’s protocol. RNA integrity was monitored by electrophoresis in an agarose gel.
Library Preparation for Next-Generation Sequencing
Strand-specific libraries for NGC were prepared as described in (Lybecker et al., 2014) with some modifications. Briefly, total RNA from each strain/condition was depleted of rRNA (Ribo-Zero™ RNA removal kit for Gram-negative bacteria, Epicenter), and 500 ng of the rRNA-free transcripts were fragmented (Ambion® RNA Fragmentation Reagents) at 70 °C for 5 min. To remove 5′ tri- and monophosphates, fragmented RNA was first treated with Tobacco Acid Pyrophosphatase (TAP, Epicenter Biotechnologies), followed by Calf Intestinal Phosphatase (CIP, NEB) according to the manufacturer’s protocols. Then, to remove 2′–3′ cyclic-phosphates, RNA was treated with T4 polynucleotide kinase (T4 PNK, New England Biolabs) absent ATP at 37 °C for 4 h (reaction conditions - 100 mM Tris-HCl pH 6.5, 100 mM MgAc, 5 mM beta-Mercaptoethanol). 20 μM of 5′ end phosphorylated 3′ multiplex RNA adaptor (Illumina, 5′-GAUCGGAAGAGCACACGUCU [idT]-3′) was ligated to the 3′ ends of processed RNAs using T4 RNA ligase I (New England Biolabs). To allow further ligation of the 5′ RNA adaptor, RNAs were 5′-phosphorylated with T4 PNK (New England Biolabs) according to the manufacturer’s instructions. Then, RNA was purified and size-selected (150–300 nt) by electrophoresis in a denaturing 8% polyacrylamide-TBE gel (8% UREA). RNA was eluted from gel in polyacrylamide elution buffer (10 mM Tris-HCl pH 7.4, 2 mM EDTA, 0.3 M NaOAc, 0.1% SDS) with vigorous shaking for 2 hours at 37 °C. The resulting supernatant was filtered (Whatman® Cellulose Filter, Sigma Aldrich) and ethanol precipitated with 0.5 mkl of GlycoBlue Coprecipitant (ThermoFisher Scientific). Processed RNAs were ligated to Illumina small RNA 5′ adaptor (5′-GUUCAGAGUUCUACAGUCCG ACGAUC-3′), followed by one more step of size-selection (200–350 nt) and gel-purification as described above. The resulting RNA libraries were reverse-transcribed using random oligo 9-mers (Sigma) and SuperScript™ II Reverse Transcriptase (ThermoFisher Scientific), treated with RNase H (Promega) and then amplified by 15–18 cycles of PCR with Q5® High-Fidelity DNA Polymerase (New England Biolabs). Libraries were indexed with ScriptSeq Index PCR primers (Epicentre) to allow further multiplexing. Products from the major steps of library preparation, as well as the final libraries, were monitored with an Agilent 2100 Bioanalyzer.
Data Analysis
All custom scripts used for data analysis and visualization as well as instructions to setup the computational environment are available at: https://github.com/eco32i/rpoS (Langmead and Salzberg, 2012), see Key Resource Table. Raw reads were mapped to MG1655 reference (NC000913.3) using bowtie2 version 2.1.0. Annotations for 5′UTR regions were obtained at: http://regulondb.ccg.unam.mx/menu/download/datasets/files/UTR_5_3_sequence.txt, sRNA binding site prediction was performed using RNA predator version 1.55 (http://nylon.tbi.univie.ac.at/cgi-bin/RNApredator/target_search.cgi) and the resulting files were converted to the standard .bed format for further manipulations. General genomic interval operations were performed using bedtools version 2.17.0 (Quinlan and Hall, 2010). Counting of the reads mapped to the annotated 5′UTR regions and ORFs was performed using htseq-count version 0.6.1p1 (Anders et al., 2015) and the differential expression analysis was carried out in DESeq2 (Love et al., 2014, using R version 3.2.0 Platform: x86_64-pc-linux-gnu (64-bit)). BCM-responsive 5′UTRs were identified using a Gaussian mixture model (GMM) clustering implementation from scikit-learn version 0.16.1 (Pedregosa et al., 2011). GMM is a probabilistic model that assumes the data are drawn from a mixture of a (finite) number of Gaussian distributions with unknown parameters. It then performs the expectation minimization algorithm to perform the fitting.
RT-qPCR
To measure the levels of transcripts in RT-qPCR reactions, 1 μg total DNA-free RNA was reverse transcribed using random oligo 9-mers (Sigma) and SuperScript™ II Reverse Transcriptase (ThermoFisher Scientific)/ProtoScript® II Reverse Transcriptase (New England Biolabs) as suggested by the manufacturers. Depending on the target of interest, cDNA was amplified with the corresponding primers (see List of Oligonucleotides Used in This Study). For each primer pair the efficiency of real-time PCR amplification was estimated using the standard curve method in a one color detection system as described in (Pfaffl, 2004). qPCR was performed using Eppendorf Mastercycler® RealPlex2 and 5x HOT FIREPol® EvaGreen® qPCR Mix Plus (no ROX) from Medibena according to the manufacturer’s protocols. RT-qPCR primers used in the study are listed in Table S4 (“Supplemental Information”).
Northern Blot Analysis
To perform Northern blot analysis, 8 μg of total RNA was separated by gel electrophoresis using denaturing 8–10 % polyacrylamide-TBE-Urea (8M) gels in 1X TBE. RNA was loaded onto the gel after 15 min denaturation denaturing at 70°C in 2X RNA load dye (Thermo Scientific) followed by cooling on ice. Gel-separated RNA was transferred to HybondXL membranes (Ambion) by wet electro-blotting at 12 V for 1 hour in 0.5X TBE. The membranes were cross-linked by UV (150 mJ/cm2) and probed with 5′-end gamma32P-labeled DNA oligonucleotide probes (see Key Resource Table) in ULTRAhyb®-Oligo Hybridization Buffer (Ambion) according to the manufacturer’s instructions. The DNA oligonucleotide labeling reaction was performed with T4 PNK (New England Biolabs) according to the manufacturer’s instructions. After the incubation, membrane was washed two times for 35 minutes with 2xSSC/0.1%SDS solution at 42°C.
In vitro Transcription
To study transcription termination in vitro we first prepared the initial elongation complex stalled at position +20 (EC16). EC20 was immobilized on beads via biotin-tagged RNAP. Transcription in solid phase allows the reaction components to be washed out, which is useful in determining the released (terminated) RNA products.
In vitro transcription templates used in the study are listed in Table S3. To test Rho dependent termination at native rpoS promoter template (Figure 2B), 60 pmol of biotin tagged RNAP were mixed with 20μL NeutrAvidinin beads (Pierce) in TB50 [20mM Tris*HCl pH=8.0; 10mM MgCl2; 100mM NaCl; 0.003% Igepal-60] and the sample was shaken for 5 minutes at 22°C. The sample was washed 3 times in 1mL of TB50 and 20pmol rpoS DNA template with native promoter (see Table S3) were added. The sample was incubated for 5 minutes at 37°C. CTP, UTP and GTP were added to a final concentration of 1mM and incubation was continued at 37C for another 5 minutes. The sample was washed 2 times in 1mL TB1000 [40mM Tris*HCl pH=8.0; 10mM MgCl2; 1000mM NaCl; 0.003% Igepal-60] and 2 times in 1mL of TB100 [40mM Tris*HCl pH=8.0; 10mM MgCl2; 100mM NaCl; 0.003% Igepal-60] (EC9). 2μL of CTP-a-P32, 3μL of ATP-a-P32, 5μM GTP were added to the beads and incubation continued for 5 minutes at 22°C. The sample was washed 4 times in 1mL of TB100. The sample was mixed with 1.5mg/ml heparin and incubated for 5 minutes and washed as above (EC16). 70μL of TB100 plus 20 units of RNase inhibitor (RNasin, Promega) were added to the sample and 10μL aliquots were taken. Appropriate samples were mixed with 1μM Rho and 1μM NusG and/or with bicyclomycin. Samples were incubated for 5 minutes at 22°C and chased for 10 minutes at 37°C in 1mM UTP, 50μM ATP, 50μM GTP and 20μM CTP. Samples were quenched in 10μL of SB [1XTBE, 8M Urea, 20mM EDTA, 0.025% xylen cyanol, 0.025% bromophenol blue]. Four aliquots of the initial elongation complex (EC16) were also used for the sequencing reaction. The aliquots were chased for 10 minutes at room temperature in 25 μM of sequencing mixtures (3′dNTPs-NTPs - ratio 1:5 for 3′dATP and 3′dCTP; 1:10 for 3′dGTP and 1:3 for 3′dUTP) and quenched as above. All aliquots were phenol-chloroform extracted: 0.17mL of water plus 0.17 ml phenol-chloroform mixture (1:1) were added to each sample, mixed, centrifuged at maximum speed for 0.5 minutes and the supernatant was transferred to a fresh tube with 0.17 ml of chloroform. Samples were centrifuged briefly and each supernatant was mixed with 20 μl of 3M Na-Acetate pH=5.0 and 20 μg of glycogen each in a fresh tube. DNA was precipitated by addition of 0.75mL of ethanol, incubation for 10 minutes at −80°C, and centrifugation for 5 minutes at max speed, and the supernatant was discarded. Pellets were washed by 0.15 ml of 70% ethanol, spun down 4 minutes at top speed and supernatant was discarded. Samples were dissolved in 7μL of 50% SB/water mixture and were heated for 5 minutes at 100°C in a dry bath, centrifuged, and loaded on a 6% (20x40cm) (19:1) polyacrylamide gel containing 7M Urea and TBE pre-electrophoresed for 45 minutes. The loaded gel was electrophoresed for 4 hours at 50W. The gel was transferred at whatman paper, dried for 30 minutes at 80°C and exposed overnight to a phosphor-screen.
To test the effect of the sRNA on Rho-dependent termination (Figure 6B and C) the initial EC was formed with 75 nM of the linear DNA template (either rpoS-UTR or T7A1-rut) and 100 nM RNAP holoenzyme in 100 μl of TB (40 mM Tris-HCl pH=8.0; 20 mM MgCl2; 50 mM NaCl; 0.003% Igepal-60; 5 mM B-Me) with 80 units of RNasin (Promega). Transcription was initiated with 10 μM UUC primer and 25 μM of GTP and UTP for 5 min at 37°C. 2 μl of ATP-P-32 was added and further incubated for 5 min at 22°C. Rho (1μM) and NusG (1μM) were added with or without the indicated amounts of sRNAs and/or Hfq, and further incubated for 5 min at 22°C. The reactions were chased with 1 mM ATP and 100 μM other NTPs at 37°C for 10 min. Reactions were terminated by adding 2X STOP buffer and were heated for 5 min at 95°C and resolved on 6% urea-PAGE for 120 min at 50W. The gel was transferred to Whatman paper, dried for 30 minutes at 80°C, and exposed overnight to a phosphor-screen. Quantification was performed using Image-Quant software from GE.
QUANTIFICATION AND STATISTICAL ANALYSIS
General genomic interval operations were performed using bedtools version 2.17.0 (PMC2832824). Counting of the reads mapped to the annotated 5′UTR regions and ORFs was performed using htseq-count version 0.6.1p1 (PMC4287950) and the differential expression analysis was carried out in DESeq2 setting significance threshold at padj=0.05 (PMC4302049, using R version 3.2.0 Platform: x86_64-pc-linux-gnu (64-bit)).
RT-qPCR quantification was performed as described in Pfaffl, 2004. For each primer pair the efficiency of real-time PCR amplification was estimated using the standard curve method in a one-color detection system (Pfaffl, 2004).
Statistical parameters for each experiment are reported in the corresponding Figure Legends. The reported data were obtained from at least 3 biological replicates. The significance was tested using Student’s t-test or non-parametric Wilcoxon-Mann-Whitney test. In figures, asterisks indicate statistical significance between the compared datasets (specified by lines) as determined with mentioned tests where * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001; ns – non-significant.
Statistical analysis was performed using R Statistical Software and Simple Interactive Statistical Analysis Tools (http://www.quantitativeskills.com/sisa/index.htm).
DATA AND SOFTWARE AVAILABILITY
Raw RNA-Seq reads were deposited in SRA database as unaligned .bam files (SRP078327). Custom scripts, analysis notebooks and instructions to set up the computational environment are available at https://github.com/eco32i/rpoS.git (see Key Resource Table).
Supplementary Material
(A) Scatter plot of transcription levels of ORFs (in log(RPKMs) corresponding to long 5′UTRs identified in Figure 1A as BCM-responsive (red) or not (teal). The plot shows that the expression of the majority of ORFs corresponding to BCM-responsive long 5′ UTRs also become upregulated upon addition of BCM. There is, however, a relatively small number of genes in which the upregulation of 5′UTR expression does not result in the concomitant upregulation of ORF expression. Also, a number of ORFs that correspond to BCM-insensitive 5′UTRs in Figure 1A become upregulated upon the addition of BCM, possibly due to intrinsically poor translation and the polarity effect.
(B) Scatter plot of log(proximal/distal) for short UTRs in the presence or absence of BCM. Solid lines represent lowess curve fitted to −BCM and +BCM samples with 95% CI indicated by shading.
(C) Exemplary coverage plots for lysC 5′UTR and part of its ORF before (left panel) and after (right panel) treatment with BCM. Red shaded areas within the 5′UTR highlight set proximal and distal regions used for estimation of Rho effect within a leader sequence (the proximal/distal ratio) to build the dataset of Figure 1B.
(D) Exemplary coverage plots for thiM and moaA 5′UTRs and part of the ORFs before and after treatment with BCM. moaA is an example of a gene not upregulated upon BCM treatment, however showing a clear release of Rho-dependent termination within its 5′UTR.
(A) Schematics of E. coli rpoS with its 5′UTR and ORF. The location of the qRT-PCR amplicon within the rpoS ORF is indicated. TSS - transcription start site. The genome-derived template for in vitro transcription is shown below. It includes the whole rpoS 5′UTR and the first 140 nt of ORF (same as in Figure 2).
(B) Transcript levels of rpoS ORF measured for the wt in the exponential phase before and after BCM treatment (15 min, 50 μg/ml). qRT-PCR values were normalized to that of a housekeeping gapA gene. Values represent means ±SD, n = 3; ** P < 0.01 (Student’s t-test, equal variance).
(C) A representative single round in vitro transcription on the rpoS template. Pre-formed elongation complexes were chased with NTPs in the absence (lane 1) or presence of Rho (lane 2) or Rho+NusG (lane 3). A bracket indicates the transcription termination zone [T]. The percentage of termination (T%) was calculated for each reaction as a ratio between the amount of radioactivity in the bands corresponding to the termination products [T] and the total radioactivity signal of the termination and readthrough bands. Values represent means from 3 independent experiments.
(A) A schematic of the transcriptional reporter fusion pUTR+X+Y-GFP used to test the effect of different fragments of the rpoS 5′UTR on Rho termination. +X+Y coordinates indicate the beginning and the end of the tested leader fragment, counting from the TSS. “P” indicates the location of a constitutive promoter. TSS – the transcription start site; RBS – the ribosome-binding site.
(B and C) Representative results from the GFP plate assay. WT cells (DH5a) transformed with pUTR+X+Y-GFP plasmid grew on LB agar plates and the fluorescence intensity was measured (GFP mode, upper panels). The same plates were also captured under visible light (Light mode, lower panels). pUTR+251+447-GFP corresponds to pUTRS-GFP. Other pUTR+X+Y-GFP constructs contain fragments with indicated coordinates (counting from TSS) from the first 250 nts of rpoS 5′UTR.
(D) A schematic of the transcriptional reporters pGFP (left) and pgeneUTR-GFP (right). “P” shows the location of a constitutive promoter, TSS - transcription start site, RBS - ribosomal binding site. pgeneUTR-GFP represents the transcriptional fusion for testing various UTRs for the presence of Rho-dependent termination.
(E) Representative results from the GFP plate assay. WT cells (DH5a) transformed with pGFP or pgeneUTR-GFP plasmids grew on LB agar plates and the fluorescence intensity was monitored (GFP mode, left panels). The same plates were also captured under visible light (Light mode, right panels).
(F) A representative single round transcription (6% TBE-UREA gel) on the genome-derived templates for the genes tested in (B). Transcription templates for the marked genes contain natural promoter, the whole 5′UTR, and the proximal part of the ORF. Pre-formed elongation complexes were chased without or with Rho. The percentage of termination T(%) was calculated for each reaction as a ratio between the amount of radioactivity in the bands corresponding to the termination products and the total radioactivity signal of the termination and readthrough bands. A bracket indicates the readthrough bands (run-off). A template with known Rho-termination sites (Epshtein et al., 2010) was used as a positive control (run-off size 623 nts).
(A) A diagram of the pUTRS-GFP reporter used to test the effect of the rpoS 5′UTR (+251–447 nt segment) on Rho termination. “P” stands for the constitutive promoter; TSS - transcription start site; RBS – ribosome-binding site. Lower panel: the secondary structures of the full-length rpoS leader (light grey) (Peng et al., 2014). The +251–447 segment is highlighted in blue. This segment does not include the sRNAs-binding site (marked with orange line).
(B) A diagram of the pUTRsRNA-GFP construct used to test the role of the rpoS 5′UTR (+251–557 nt segment) on Rho termination. “P” (grey triangle) indicates the constitutive promoter; TSS - transcription start site; RBS – ribosome-binding site. Lower panel: the secondary structure of the full-length rpoS leader (shown in light grey (Peng et al., 2014)). The +251–557 segment is highlighted in blue and includes the sRNAs-binding site (orange line).
(C) A fragment of the rpoS 5′UTR-ORF with the inhibitory hairpin (shown in bold red lines), which sequesters the natural ribosome binding site (marked with green bar). The mutation C125T (red dot; position +471 counting from the TSS of rpoS) destabilizes the hairpin and renders rpoS translation independent of sRNAs and Hfq (Battesti et al., 2015; Brown and Elliott, 1997).
(D) Hfq-deficient strain demonstrates slower growth in comparison to WT in LB medium.
(E) The effect of E. coli sRNAs on endogenous rpoS transcript levels. Upper panel: schematics of E. coli rpoS with its 5′UTR and ORF. The location of the qRT-PCR amplicon within the rpoS ORF is indicated. TSS - transcription start site. Lower panel: transcript levels of endogenous rpoS ORF measured for the WT and sRNA tmut transformed with the pUTRsRNA-GFP construct in the exponential and stationary phases; these strains are the same as those used to monitor GFP as a function of sRNAs and Hfq (Figure 3). qRT-PCR values were normalized to that of a housekeeping gapA gene. Values represent means ±SD, n = 3; ** P < 0.01 (Student’s t-test, equal variance).
(F) – (G) The absence of sRNAs-binding sites within the rpoS 5′UTR renders its transcription insensitive to DsrA, RprA, ArcZ.
(F) Upper panel: a schematic of the transcriptional reporters pGFP. “P” shows the location of a constitutive promoter, TSS - transcription start site, RBS - ribosomal binding site. Middle/lower panels: representative results from the GFP plate assay. The wild type, triple sRNA deletion (tmut) and hfq deletion mutants transformed with pGFP construct grew on LB agar plates and the fluorescence intensity was measured (GFP mode). The same plates were also captured under visible light to compare bacterial growth (Light mode).
(G) Upper panel: a schematic of the transcriptional reporters pUTRS-GFP. “P” shows the location of a constitutive promoter, TSS - transcription start site, RBS - ribosomal binding site. pUTRS-GFP lacks the sRNAs binding site. Middle/lower panels: representative results from the GFP plate assay. The wild type, triple sRNA deletion (tmut) and hfq deletion mutants transformed with pUTRS-GFP construct grew on LB agar plates and the fluorescence intensity was measured (GFP mode). The same plates were also captured under visible light to compare bacterial growth (Light mode).
(A) – (B) The effect of sRNAs and Hfq on E. coli rpoS transcription.
(A) A diagram of the E. coli rpoS gene with its 5′UTR and ORF. The locations of qRT-PCR amplicons are depicted below. Orange bar marks the position of the DsrA/RprA/ArcZ binding site. TSS - transcription start site.
(B) Transcript levels of different rpoS regions measured for PM1409 (wt), PM1417 (tmut) and PM1419 (hfq mut) strains in the exponential (exp) and stationary (stat) phase. qRT-PCR values are normalized to that of the housekeeping gapA gene. Values represent means ±SD, n ≥ 3.
As expected, the effect of the hfq deletion is consistent with the loss of the sRNAs. If in fact Hfq inhibition of Rho via direct interaction (Rabhi et al., 2011) also contributes, this would be consistent with the effects in the same direction. However, the magnitude of the effects on Rho termination between the sRNAs triple mutant and the hfq(−) mutant are comparable (see also Figure 4). Therefore, Hfq is likely to inhibit Rho termination within the 5′UTR of rpoS (and other selected genes) predominantly via sRNAs. In addition to stabilizing sRNA-5′UTR interactions, Hfq likely also contributes to Rho antitermination by increasing the total levels of sRNAs (Møller et al., 2002; Schu et al., 2015).
(C) – (E) The effect of the “C125T” mutation and BCM on relative rpoS induction upon transition to the stationary phase.
(C) A diagram of rpoS WT (left panel) and the chromosomal rpoS mutant C125T (right panel) with the 5′UTR and ORF parts marked. C125T opens the inhibitory hairpin of rpoS 5′UTR, rendering rpoS translation independent of sRNAs (Brown and Elliott, 1997). The location of qRT-PCR amplicon for rpoS levels measurement is shown below. The orange bar marks the position of the DsrA/RprA/ArcZ binding site (sRNA BS). TSS - transcription start site.
(D) qRT-PCR data for the rpoS levels in exponential and stationary phase in the absence or presence of BCM (normalized to the housekeeping gapA levels). Values represent means (labeled on the chart) ±SD, n = 3.
(E) qRT-PCR data for the rpoS levels change upon transition from the exponential to stationary phase in the absence or presence of BCM. Values represent means ±SD, n = 3; *** P < 0.001; ns - not significant (Student’s t-test).
Radiogram shows a representative single round transcription assay (see Experimental Procedures). Pre-formed elongation complexes were chased in the absence (lanes 1, 7–10) or presence of Rho and NusG. A bracket indicates the transcription termination zone (T). The efficiency of Rho-dependent termination was estimated in the absence (lanes 2) or in the presence of 1 μM of individual sRNAs (lanes 3–6), including DsrA (D), RprA (R), ArcZ (A), and OxyS (O). The percentage of termination (T%) was calculated for each reaction as a ratio between the amount of radioactivity in the bands corresponding to the termination products (T) and the total radioactivity signal of the termination and readthrough bands. Values are from 3 independent experiments.
(A) sRNAs control Rho-dependent termination of various E. coli genes with long 5′UTRs. Transcript levels of BCM-sensitive long leader genes, fadE, yqeC and astE in PM1409 (wt) and PM1417 (tmut) strains measured in the stationary phase without or with BCM. qRT-PCR values were normalization to that of gapA. Values represent means ±SD, n = 3; * P < 0.05; ns - not significant (Student’s t-test, equal variance).
(B) Hfq effect on sRNAs-mediated control of Rho termination of various E. coli genes with long 5′UTRs. Transcript levels of fadE, yqeC, astE and rpoS in the exponentially grown wt and hfq mutant cells with or without overexpression of DsrA, ArcZ and RprA. qRT-PCR values are normalized to that of a housekeeping gapA gene.
The accumulation of DsrA, RprA, and ArcZ in hfq(−) cells and in cells containing the hfq mutant (Q8), which binds sRNAs poorly, was partially compromised (Schu et al., 2015). The function of these three overexpressed sRNAs in an hfq null mutant has been examined (Soper et al., 2010): DsrA has a reduced but measurable ability to stimulate rpoS synthesis, the others have none. Therefore, the effect the hfq mutant on the expression of rpoS and the genes studied here is likely to be a combination of weaker sRNAs-5′UTR interaction and lower levels of sRNAs.
(C) IntaRNA (Freiburg RNA Tools) predictions of sRNAs interactions with fadE and yqeC targets (related to Figure 6). Left panel: potential interaction of ArcZ and fadE RNA within ORF (upper) or 5′UTR (lower). Right panel: potential interaction of yqeC 5′UTR transcript with ArcZ (upper) or DsrA (lower).
Table S1. E. coli genes with long 5′UTRs responsive to BCM in exponential phase (related to Figure 1).
Table S2. Genes upregulated in stationary phase in WT, but not in sRNAs-deficient cells (dsrA, arcZ, rprA triple deletion mutant) (related to Figure 7).
Table S3. Templates for in vitro transcription (related to the Methods and Resources section: In vitro Transcription).
Table S4. List of RT-qPCR primers (related to the Methods and Resources section: RT-qPCR).
Table S5. 5′UTR fragments of BCM-insensitive genes tested in GFP reporter assay (related to the Methods and Resources section: Protein Reporter Assays and Figure S3).
HIGHLIGHTS.
Rho termination factor acts within 5′UTRs of many bacterial genes
Rho functions as a global attenuator of gene expression
Small RNAs interfere with Rho-mediated termination by base-paring within 5′UTRs
sRNA-mediated antitermination is a widespread mode of bacterial gene regulation
Acknowledgments
We thank the Vienna BioCenter Core Facilities (VBCF) for NGS. This work was supported by the NIH grant R01 GM107329 and by the Howard Hughes Medical Institute (E.N.) and by the Austrian Science Fund FWF Grants I538-B12, F4301 and F4308 (R.S.). S. G. and J. C. were supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
AUTHOR CONTRIBUTION
E.N. conceptualized the study. N.S. and E.N. designed the experiments. N.S., I.S., B.B., V.E., and J.C. conducted the experimental work, discussed the results and commented on the manuscript; I.S. performed the bioinformatics analysis; E.N., R.S., and S.G. supervised the research and edited the manuscript; N.S. and E.N. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EGH, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A, Majdalani N, Gottesman S. Stress sigma factor RpoS degradation and translation are sensitive to the state of central metabolism. Proc Natl Acad Sci U S A. 2015;112:5159–5164. doi: 10.1073/pnas.1504639112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L, Schwartz A, Guillemardet B, Boudvillain M, Figueroa-Bossi N. A role for Rho-dependent polarity in gene regulation by a noncoding small RNA. Genes Dev. 2012;26:1864–1873. doi: 10.1101/gad.195412.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudvillain M, Figueroa-Bossi N, Bossi L. Terminator still moving forward: expanding roles for Rho factor. Curr Opin Microbiol. 2013;16:118–124. doi: 10.1016/j.mib.2012.12.003. [DOI] [PubMed] [Google Scholar]
- Brennan CA, Dombroski AJ, Platt T. Transcription termination factor rho is an RNA-DNA helicase. Cell. 1987;48:945–952. doi: 10.1016/0092-8674(87)90703-3. [DOI] [PubMed] [Google Scholar]
- Brown L, Elliott T. Mutations that increase expression of the rpoS gene and decrease its dependence on hfq function in Salmonella typhimurium. J Bacteriol. 1997;179:656–662. doi: 10.1128/jb.179.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale CJ, Washburn RS, Tadigotla VR, Brown LM, Gottesman ME, Nudler E. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320:935–938. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron MP, Bastet L, Lussier A, Simoneau-Roy M, Massé E, Lafontaine DA. Dual-acting riboswitch control of translation initiation and mRNA decay. Proc Natl Acad Sci U S A. 2012;109:E3444–E3453. doi: 10.1073/pnas.1214024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong T, Schellhorn HE. Control of RpoS in global gene expression of Escherichia coli in minimal media. Mol Genet Genomics. 2009;281:19–33. doi: 10.1007/s00438-008-0389-3. [DOI] [PubMed] [Google Scholar]
- Dong T, Kirchhof MG, Schellhorn HE. RpoS regulation of gene expression during exponential growth of Escherichia coli K12. Mol Genet Genomics. 2008;279:267–277. doi: 10.1007/s00438-007-0311-4. [DOI] [PubMed] [Google Scholar]
- Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell. 2011;146:533–543. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenhofer F, Tafer H, Stadler PF, Hofacker IL. RNApredator: fast accessibility-based prediction of sRNA targets. Nucleic Acids Res. 2011;39:W149–W154. doi: 10.1093/nar/gkr467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers CH, Caimano MJ, Clawson ML, Miller WG, Samuels DS, Radolf JD. Identification of loci critical for replication and compatibility of a Borrelia burgdorferi cp32 plasmid and use of a cp32-based shuttle vector for the expression of fluorescent reporters in the lyme disease spirochaete. Mol Microbiol. 2002;43:281–295. doi: 10.1046/j.1365-2958.2002.02758.x. [DOI] [PubMed] [Google Scholar]
- Epshtein V, Dutta D, Wade J, Nudler E. An allosteric mechanism of Rho-dependent transcription termination. Nature. 2010;463:245–249. doi: 10.1038/nature08669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogol EB, Rhodius VA, Papenfort K, Vogel J, Gross CA. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc Natl Acad Sci U S A. 2011;108:12875–12880. doi: 10.1073/pnas.1109379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grylak-Mielnicka A, Bidnenko V, Bardowski J, Bidnenko E. Transcription termination factor Rho: a hub linking diverse physiological processes in bacteria. Microbiology. 2016;162:433–447. doi: 10.1099/mic.0.000244. [DOI] [PubMed] [Google Scholar]
- Hart CM, Roberts JW. Rho-dependent transcription termination. Characterization of the requirement for cytidine in the nascent transcript. J Biol Chem. 1991;266:24140–24148. [PubMed] [Google Scholar]
- Hollands K, Proshkin S, Sklyarova S, Epshtein V, Mironov A, Nudler E, Groisman EA. Riboswitch control of Rho-dependent transcription termination. Proc Natl Acad Sci U S A. 2012;109:5376–5381. doi: 10.1073/pnas.1112211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N, Schu DJ, Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem. 2013;288:7996–8003. doi: 10.1074/jbc.R112.441386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine E, Zhang Z, Kuhlman T, Hwa T. Quantitative Characteristics of Gene Regulation by Small RNA. PLOS Biol. 2007;5:281–297. doi: 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci U S A. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol Microbiol. 2002;46:813–826. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010;29:3094–3107. doi: 10.1038/emboj.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellin JR, Cossart P. Unexpected versatility in bacterial riboswitches. Trends Genet. 2015;31:150–156. doi: 10.1016/j.tig.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Menouni R, Champ S, Espinosa L, Boudvillain M, Ansaldi M. Transcription termination controls prophage maintenance in Escherichia coli genomes. Proc Natl Acad Sci U S A. 2013;110:14414–14419. doi: 10.1073/pnas.1303400110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika F, Hengge R. Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA Biol. 2014;11:494–507. doi: 10.4161/rna.28867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WG, Bates AH, Horn ST, Brandl MT, Wachtel MR, Mandrell RE. Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl Environ Microbiol. 2000;66:5426–5436. doi: 10.1128/aem.66.12.5426-5436.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K, Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol Microbiol. 2011;82:1545–1562. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E, Gottesman ME. Transcription termination and anti-termination in E. coli. Genes to Cells. 2002;7:755–768. doi: 10.1046/j.1365-2443.2002.00563.x. [DOI] [PubMed] [Google Scholar]
- Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Papenfort K, Espinosa E, Casadesús J, Vogel J. Small RNA-based feedforward loop with AND-gate logic regulates extrachromosomal DNA transfer in Salmonella. Proc Natl Acad Sci U S A. 2015;112:E4772–E4781. doi: 10.1073/pnas.1507825112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. Quantification strategies in real-time PCR. International University Line (IUL); La Jolla, CA, USA: 2004. [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. Scikit-learn: Machine learning in Python. JMLR. 2011;12:2825–2830. [Google Scholar]
- Peng Y, Soper TJ, Woodson SA. Positional effects of AAN motifs in rpoS regulation by sRNAs and Hfq. J Mol Biol. 2014;426:275–285. doi: 10.1016/j.jmb.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Mooney RA, Grass JA, Jessen ED, Tran F, Landick R. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 2012;26:2621–2633. doi: 10.1101/gad.196741.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proshkin S, Mironov A, Nudler E. Riboswitches in regulation of Rho-dependent transcription termination. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbagrm.2014.04.002. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabhi M, Espéli O, Schwartz A, Cayrol B, Rahmouni AR, Arluison V, Boudvillain M. The Sm-like RNA chaperone Hfq mediates transcription antitermination at Rho-dependent terminators. EMBO J. 2011;30:2805–2816. doi: 10.1038/emboj.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentmeister A, Mayer G, Kuhn N, Famulok M. Conformational changes in the expression domain of the Escherichia coli thiM riboswitch. Nucleic Acids Res. 2007;35:3713–3722. doi: 10.1093/nar/gkm300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson LV, Richardson JP. Rho-dependent termination of transcription is governed primarily by the upstream Rho utilization (rut) sequences of a terminator. J Biol Chem. 1996;271:21597–21603. doi: 10.1074/jbc.271.35.21597. [DOI] [PubMed] [Google Scholar]
- Roberts JW. Termination factor for RNA synthesis. Nature. 1969;224:1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- Salgado H, Peralta-Gil M, Gama-Castro S, Santos-Zavaleta A, Muñiz-Rascado L, García-Sotelo JS, Weiss V, Solano-Lira H, Martínez-Flores I, Medina-Rivera A, et al. RegulonDB v8.0: omics data sets, evolutionary conservation, regulatory phrases, cross-validated gold standards and more. Nucleic Acids Res. 2013;41:D203–D213. doi: 10.1093/nar/gks1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf HJ, Hinton JCD, Vogel J. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol Microbiol. 2011;81:1144–1165. doi: 10.1111/j.1365-2958.2011.07751.x. [DOI] [PubMed] [Google Scholar]
- Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14:1907–1917. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci U S A. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Brennan CA, Platt T. A short intervening structure can block Rho factor helicase action at a distance. J Biol Chem. 1990;265:13408–13413. [PubMed] [Google Scholar]
- Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Takayanagi Y, Fujita N, Ishihama A, Takahashi H. Heterogeneity of the principal sigma factor in Escherichia coli: the rpoS gene product, sigma 38, is a second principal sigma factor of RNA polymerase in stationary-phase Escherichia coli. Proc Natl Acad Sci U S A. 1993;90:8303. doi: 10.1073/pnas.90.17.8303a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jäger JG, Hüttenhofer A, Wagner EGH. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EGH, Romby P. Small RNAs in Bacteria and Archaea: Who They Are, What They Do, and How They Do It. Adv Genet. 2015;90:133–208. doi: 10.1016/bs.adgen.2015.05.001. [DOI] [PubMed] [Google Scholar]
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiefka A, Kohn H, Widger WR. Transcription termination factor rho: the site of bicyclomycin inhibition in Escherichia coli. Biochemistry. 1993;32:3564–3570. doi: 10.1021/bi00065a007. [DOI] [PubMed] [Google Scholar]
SUPPLEMENTAL REFERENCES
- Battesti A, Majdalani N, Gottesman S. Stress sigma factor RpoS degradation and translation are sensitive to the state of central metabolism. Proc Natl Acad Sci U S A. 2015;112:5159–5164. doi: 10.1073/pnas.1504639112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L, Elliott T. Mutations that increase expression of the rpoS gene and decrease its dependence on hfq function in Salmonella typhimurium. J Bacteriol. 1997;179:656–662. doi: 10.1128/jb.179.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epshtein V, Dutta D, Wade J, Nudler E. An allosteric mechanism of Rho-dependent transcription termination. Nature. 2010;463:245–249. doi: 10.1038/nature08669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller T, Franch T, Højrup P, Keene DR, Bächinger HP, Brennan RG, Valentin-Hansen P. Hfq: A Bacterial Sm-like Protein that Mediates RNA-RNA Interaction. Mol Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- Peng Y, Soper TJ, Woodson SA. Positional effects of AAN motifs in rpoS regulation by sRNAs and Hfq. J Mol Biol. 2014;426:275–285. doi: 10.1016/j.jmb.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabhi M, Espéli O, Schwartz A, Cayrol B, Rahmouni AR, Arluison V, Boudvillain M. The Sm-like RNA chaperone Hfq mediates transcription antitermination at Rho-dependent terminators. EMBO J. 2011;30:2805–2816. doi: 10.1038/emboj.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schu DJ, Zhang A, Gottesman S, Storz G. Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J. 2015;34:2557–2573. doi: 10.15252/embj.201591569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci U S A. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Scatter plot of transcription levels of ORFs (in log(RPKMs) corresponding to long 5′UTRs identified in Figure 1A as BCM-responsive (red) or not (teal). The plot shows that the expression of the majority of ORFs corresponding to BCM-responsive long 5′ UTRs also become upregulated upon addition of BCM. There is, however, a relatively small number of genes in which the upregulation of 5′UTR expression does not result in the concomitant upregulation of ORF expression. Also, a number of ORFs that correspond to BCM-insensitive 5′UTRs in Figure 1A become upregulated upon the addition of BCM, possibly due to intrinsically poor translation and the polarity effect.
(B) Scatter plot of log(proximal/distal) for short UTRs in the presence or absence of BCM. Solid lines represent lowess curve fitted to −BCM and +BCM samples with 95% CI indicated by shading.
(C) Exemplary coverage plots for lysC 5′UTR and part of its ORF before (left panel) and after (right panel) treatment with BCM. Red shaded areas within the 5′UTR highlight set proximal and distal regions used for estimation of Rho effect within a leader sequence (the proximal/distal ratio) to build the dataset of Figure 1B.
(D) Exemplary coverage plots for thiM and moaA 5′UTRs and part of the ORFs before and after treatment with BCM. moaA is an example of a gene not upregulated upon BCM treatment, however showing a clear release of Rho-dependent termination within its 5′UTR.
(A) Schematics of E. coli rpoS with its 5′UTR and ORF. The location of the qRT-PCR amplicon within the rpoS ORF is indicated. TSS - transcription start site. The genome-derived template for in vitro transcription is shown below. It includes the whole rpoS 5′UTR and the first 140 nt of ORF (same as in Figure 2).
(B) Transcript levels of rpoS ORF measured for the wt in the exponential phase before and after BCM treatment (15 min, 50 μg/ml). qRT-PCR values were normalized to that of a housekeeping gapA gene. Values represent means ±SD, n = 3; ** P < 0.01 (Student’s t-test, equal variance).
(C) A representative single round in vitro transcription on the rpoS template. Pre-formed elongation complexes were chased with NTPs in the absence (lane 1) or presence of Rho (lane 2) or Rho+NusG (lane 3). A bracket indicates the transcription termination zone [T]. The percentage of termination (T%) was calculated for each reaction as a ratio between the amount of radioactivity in the bands corresponding to the termination products [T] and the total radioactivity signal of the termination and readthrough bands. Values represent means from 3 independent experiments.
(A) A schematic of the transcriptional reporter fusion pUTR+X+Y-GFP used to test the effect of different fragments of the rpoS 5′UTR on Rho termination. +X+Y coordinates indicate the beginning and the end of the tested leader fragment, counting from the TSS. “P” indicates the location of a constitutive promoter. TSS – the transcription start site; RBS – the ribosome-binding site.
(B and C) Representative results from the GFP plate assay. WT cells (DH5a) transformed with pUTR+X+Y-GFP plasmid grew on LB agar plates and the fluorescence intensity was measured (GFP mode, upper panels). The same plates were also captured under visible light (Light mode, lower panels). pUTR+251+447-GFP corresponds to pUTRS-GFP. Other pUTR+X+Y-GFP constructs contain fragments with indicated coordinates (counting from TSS) from the first 250 nts of rpoS 5′UTR.
(D) A schematic of the transcriptional reporters pGFP (left) and pgeneUTR-GFP (right). “P” shows the location of a constitutive promoter, TSS - transcription start site, RBS - ribosomal binding site. pgeneUTR-GFP represents the transcriptional fusion for testing various UTRs for the presence of Rho-dependent termination.
(E) Representative results from the GFP plate assay. WT cells (DH5a) transformed with pGFP or pgeneUTR-GFP plasmids grew on LB agar plates and the fluorescence intensity was monitored (GFP mode, left panels). The same plates were also captured under visible light (Light mode, right panels).
(F) A representative single round transcription (6% TBE-UREA gel) on the genome-derived templates for the genes tested in (B). Transcription templates for the marked genes contain natural promoter, the whole 5′UTR, and the proximal part of the ORF. Pre-formed elongation complexes were chased without or with Rho. The percentage of termination T(%) was calculated for each reaction as a ratio between the amount of radioactivity in the bands corresponding to the termination products and the total radioactivity signal of the termination and readthrough bands. A bracket indicates the readthrough bands (run-off). A template with known Rho-termination sites (Epshtein et al., 2010) was used as a positive control (run-off size 623 nts).
(A) A diagram of the pUTRS-GFP reporter used to test the effect of the rpoS 5′UTR (+251–447 nt segment) on Rho termination. “P” stands for the constitutive promoter; TSS - transcription start site; RBS – ribosome-binding site. Lower panel: the secondary structures of the full-length rpoS leader (light grey) (Peng et al., 2014). The +251–447 segment is highlighted in blue. This segment does not include the sRNAs-binding site (marked with orange line).
(B) A diagram of the pUTRsRNA-GFP construct used to test the role of the rpoS 5′UTR (+251–557 nt segment) on Rho termination. “P” (grey triangle) indicates the constitutive promoter; TSS - transcription start site; RBS – ribosome-binding site. Lower panel: the secondary structure of the full-length rpoS leader (shown in light grey (Peng et al., 2014)). The +251–557 segment is highlighted in blue and includes the sRNAs-binding site (orange line).
(C) A fragment of the rpoS 5′UTR-ORF with the inhibitory hairpin (shown in bold red lines), which sequesters the natural ribosome binding site (marked with green bar). The mutation C125T (red dot; position +471 counting from the TSS of rpoS) destabilizes the hairpin and renders rpoS translation independent of sRNAs and Hfq (Battesti et al., 2015; Brown and Elliott, 1997).
(D) Hfq-deficient strain demonstrates slower growth in comparison to WT in LB medium.
(E) The effect of E. coli sRNAs on endogenous rpoS transcript levels. Upper panel: schematics of E. coli rpoS with its 5′UTR and ORF. The location of the qRT-PCR amplicon within the rpoS ORF is indicated. TSS - transcription start site. Lower panel: transcript levels of endogenous rpoS ORF measured for the WT and sRNA tmut transformed with the pUTRsRNA-GFP construct in the exponential and stationary phases; these strains are the same as those used to monitor GFP as a function of sRNAs and Hfq (Figure 3). qRT-PCR values were normalized to that of a housekeeping gapA gene. Values represent means ±SD, n = 3; ** P < 0.01 (Student’s t-test, equal variance).
(F) – (G) The absence of sRNAs-binding sites within the rpoS 5′UTR renders its transcription insensitive to DsrA, RprA, ArcZ.
(F) Upper panel: a schematic of the transcriptional reporters pGFP. “P” shows the location of a constitutive promoter, TSS - transcription start site, RBS - ribosomal binding site. Middle/lower panels: representative results from the GFP plate assay. The wild type, triple sRNA deletion (tmut) and hfq deletion mutants transformed with pGFP construct grew on LB agar plates and the fluorescence intensity was measured (GFP mode). The same plates were also captured under visible light to compare bacterial growth (Light mode).
(G) Upper panel: a schematic of the transcriptional reporters pUTRS-GFP. “P” shows the location of a constitutive promoter, TSS - transcription start site, RBS - ribosomal binding site. pUTRS-GFP lacks the sRNAs binding site. Middle/lower panels: representative results from the GFP plate assay. The wild type, triple sRNA deletion (tmut) and hfq deletion mutants transformed with pUTRS-GFP construct grew on LB agar plates and the fluorescence intensity was measured (GFP mode). The same plates were also captured under visible light to compare bacterial growth (Light mode).
(A) – (B) The effect of sRNAs and Hfq on E. coli rpoS transcription.
(A) A diagram of the E. coli rpoS gene with its 5′UTR and ORF. The locations of qRT-PCR amplicons are depicted below. Orange bar marks the position of the DsrA/RprA/ArcZ binding site. TSS - transcription start site.
(B) Transcript levels of different rpoS regions measured for PM1409 (wt), PM1417 (tmut) and PM1419 (hfq mut) strains in the exponential (exp) and stationary (stat) phase. qRT-PCR values are normalized to that of the housekeeping gapA gene. Values represent means ±SD, n ≥ 3.
As expected, the effect of the hfq deletion is consistent with the loss of the sRNAs. If in fact Hfq inhibition of Rho via direct interaction (Rabhi et al., 2011) also contributes, this would be consistent with the effects in the same direction. However, the magnitude of the effects on Rho termination between the sRNAs triple mutant and the hfq(−) mutant are comparable (see also Figure 4). Therefore, Hfq is likely to inhibit Rho termination within the 5′UTR of rpoS (and other selected genes) predominantly via sRNAs. In addition to stabilizing sRNA-5′UTR interactions, Hfq likely also contributes to Rho antitermination by increasing the total levels of sRNAs (Møller et al., 2002; Schu et al., 2015).
(C) – (E) The effect of the “C125T” mutation and BCM on relative rpoS induction upon transition to the stationary phase.
(C) A diagram of rpoS WT (left panel) and the chromosomal rpoS mutant C125T (right panel) with the 5′UTR and ORF parts marked. C125T opens the inhibitory hairpin of rpoS 5′UTR, rendering rpoS translation independent of sRNAs (Brown and Elliott, 1997). The location of qRT-PCR amplicon for rpoS levels measurement is shown below. The orange bar marks the position of the DsrA/RprA/ArcZ binding site (sRNA BS). TSS - transcription start site.
(D) qRT-PCR data for the rpoS levels in exponential and stationary phase in the absence or presence of BCM (normalized to the housekeeping gapA levels). Values represent means (labeled on the chart) ±SD, n = 3.
(E) qRT-PCR data for the rpoS levels change upon transition from the exponential to stationary phase in the absence or presence of BCM. Values represent means ±SD, n = 3; *** P < 0.001; ns - not significant (Student’s t-test).
Radiogram shows a representative single round transcription assay (see Experimental Procedures). Pre-formed elongation complexes were chased in the absence (lanes 1, 7–10) or presence of Rho and NusG. A bracket indicates the transcription termination zone (T). The efficiency of Rho-dependent termination was estimated in the absence (lanes 2) or in the presence of 1 μM of individual sRNAs (lanes 3–6), including DsrA (D), RprA (R), ArcZ (A), and OxyS (O). The percentage of termination (T%) was calculated for each reaction as a ratio between the amount of radioactivity in the bands corresponding to the termination products (T) and the total radioactivity signal of the termination and readthrough bands. Values are from 3 independent experiments.
(A) sRNAs control Rho-dependent termination of various E. coli genes with long 5′UTRs. Transcript levels of BCM-sensitive long leader genes, fadE, yqeC and astE in PM1409 (wt) and PM1417 (tmut) strains measured in the stationary phase without or with BCM. qRT-PCR values were normalization to that of gapA. Values represent means ±SD, n = 3; * P < 0.05; ns - not significant (Student’s t-test, equal variance).
(B) Hfq effect on sRNAs-mediated control of Rho termination of various E. coli genes with long 5′UTRs. Transcript levels of fadE, yqeC, astE and rpoS in the exponentially grown wt and hfq mutant cells with or without overexpression of DsrA, ArcZ and RprA. qRT-PCR values are normalized to that of a housekeeping gapA gene.
The accumulation of DsrA, RprA, and ArcZ in hfq(−) cells and in cells containing the hfq mutant (Q8), which binds sRNAs poorly, was partially compromised (Schu et al., 2015). The function of these three overexpressed sRNAs in an hfq null mutant has been examined (Soper et al., 2010): DsrA has a reduced but measurable ability to stimulate rpoS synthesis, the others have none. Therefore, the effect the hfq mutant on the expression of rpoS and the genes studied here is likely to be a combination of weaker sRNAs-5′UTR interaction and lower levels of sRNAs.
(C) IntaRNA (Freiburg RNA Tools) predictions of sRNAs interactions with fadE and yqeC targets (related to Figure 6). Left panel: potential interaction of ArcZ and fadE RNA within ORF (upper) or 5′UTR (lower). Right panel: potential interaction of yqeC 5′UTR transcript with ArcZ (upper) or DsrA (lower).
Table S1. E. coli genes with long 5′UTRs responsive to BCM in exponential phase (related to Figure 1).
Table S2. Genes upregulated in stationary phase in WT, but not in sRNAs-deficient cells (dsrA, arcZ, rprA triple deletion mutant) (related to Figure 7).
Table S3. Templates for in vitro transcription (related to the Methods and Resources section: In vitro Transcription).
Table S4. List of RT-qPCR primers (related to the Methods and Resources section: RT-qPCR).
Table S5. 5′UTR fragments of BCM-insensitive genes tested in GFP reporter assay (related to the Methods and Resources section: Protein Reporter Assays and Figure S3).