Abstract
Enterocytozoon bieneusi is a common opportunistic pathogen causing diarrhea and enteric disease in a variety of animal hosts. Although it has been reported in many animals, there is no published information available on the occurrence of E. bieneusi in red-bellied tree squirrels. To understand the occurrence, genetic diversity, and zoonotic potential of E. bieneusi in red-bellied tree squirrels, 144 fecal specimens from Sichuan province, China, were examined by PCR amplification and sequencing of the internal transcribed spacer (ITS) region of the ribosomal RNA (rRNA) gene of E. bieneusi. The overall infection rate of E. bieneusi 16.7% (24/144) was observed in red-bellied tree squirrels. Altogether five genotypes of E. bieneusi were identified: three known genotypes D (n = 18), EbpC (n = 3), SC02 (n = 1) and two novel genotypes CE01, CE02 (one each). Multilocus sequence typing (MLST) analysis employing three microsatellite (MS1, MS3, MS7) and one minisatellite (MS4) revealed 16, 14, 7 and 14 positive specimens were successfully sequenced, and identified eight, three, three and two genotypes at four loci, respectively. In phylogenetic analysis, the three known genotypes D, EbpC, and SC02 were clustered into group 1 with zoonotic potential, and the two novel genotypes CE01 and CE02 were clustered into group 6. The present study firstly reported the occurrence of E. bieneusi in red-bellied tree squirrels in China, and the E. bieneusi genotypes D and EbpC were found in humans previously. These results indicate that red-bellied tree squirrels may play a potential role in the transmission of E. bieneusi to humans.
Introduction
Microsporidia, obligate intracellular eukaryotic pathogens, are composed of approximately 1300 species in 160 genera [1, 2]. Currently, at least 14 microsporidia species in eight genera have been detected in humans [3]. Enterocytozoon bieneusi is the most prevalent microsporidian species and accounts for more than 90% of the cases of human microsporidiosis [4, 5]. Generally, infective spores of E. bieneusi are excreted through feces of infected animals into the environment, and are capable of infecting susceptible humans, especially children, via consumption of contaminated food and water [3]. In humans, clinical symptoms caused by E. bieneusi in immunocompetent individuals are self-limiting diarrhea and malabsorption. Most seriously, E. bieneusi cause a life-threatening diarrhea in immune-compromised patients, particularly in AIDS patients and organ transplant recipients [3, 6]. Apart from humans, E. bieneusi has been observed in many vertebrates species, including mammals, reptiles, and birds [7–10].
Sequence analysis of the internal transcribed spacer (ITS) region of the ribosomal RNA (rRNA) gene is the standard method for genotyping E. bieneusi due to a high degree of gene tic polymorphism within E. bieneusi isolates in humans and animals [4, 9]. Thus far, molecular epidemiological surveys of E. bieneusi in different parts of the world demonstrate over 240 genotypes in animals and humans [11–16]. All the published genotypes of E. bieneusi have been divided into eight different groups via the phylogenetic analysis [17]. Group 1, considered as the human pathogenic group, contains almost all the E. bieneusi genotypes from humans and some genotypes from animals [18]. In contrast, the remaining clusters that form the groups 2 to 8 are mostly found in specific hosts and wastewater [5, 19]. However, the use of single ITS maker may be inadequate in identifying genotypes of E. bieneusi due to the uncertainty about whether meiotic recombination occurs in E. bieneusi lifecycle [15]. Recently, a multilocus sequencing typing (MLST) analysis employing three microsatellites (MS1, MS3, MS7) and one minisatellite (MS4) makers has been developed to better know the route of transmission, genetic diversity and host specificity of E. bieneusi [15, 20, 21].
In China, E. bieneusi has been confirmed in humans, animals, and water samples [13, 16, 17, 22–26], but only limited reports about rodents are available, and their role as reservoirs of infection for humans and other animals are still unknown. The red-bellied tree squirrels, as commercial and companion animals, are known to be closely associated with humans. In recent years, they have gained more popularity among various groups of people, especially children. Nevertheless, there has been no research conducted on the infection rates and genetic characterization of E. bieneusi in red-bellied tree squirrels. To the best of our knowledge, the current study is the first to explore the occurrence and genetic diversity of E. bieneusi in red-bellied tree squirrels, and to evaluate the zoonotic potential in transmission of human microsporidiosis.
Materials and Methods
Ethics statement
This study was conducted in accordance with the Guide for the Care and Use of Laboratory Animals of the Ministry of Health, China. The research protocol was reviewed and approved by the Research Ethics Committee of the Sichuan Agricultural University, Sichuan, China. Permission was obtained from the animal owners or managers before collecting the fecal specimens. During the collection of fecal specimens, the animals were not subjected to any kind of injury.
Collection of specimens
From March 2014 to September 2015, a total of 144 fecal specimens were collected from pet owners, pet shops, and breeding facility in Ya'an, Nanchong, and Chengdu, of Sichuan province, southwestern China (Table 1) (S1 Table). We used sterile disposable latex gloves during the collection of each fresh specimen. The samples were placed into individual 30 ml plastic containers, and then were transported to the laboratory with ice packs within 24 h of collection. At the same time, data on the source, age, gender, and physical condition of each animal were recorded. The red-bellied tree squirrels were grouped according to their age as follows: <3 months (n = 55), 3 to 12 months (n = 67), and >12 months (n = 22) (Table 2). None of these experimental animals presented with any diarrheic or gastrointestinal conditions.
Table 1. Occurrence and genotypes of E. bieneusi in red-bellied tree squirrels from different cities and sources of southwest China.
| City | Source | No. of animals | No. of positive (%) | Genotypes (n) |
|---|---|---|---|---|
| Ya'an | Pet shop1 | 35 | 7(20.0) | D(3), EbpC(2), CE01(1), SC02(1) |
| Nanchong | Pet shop2 | 23 | 4(17.4) | D(4) |
| Chengdu | Pet shop3 | 12 | 2(16.7) | D(1), EbpC(1) |
| Owner | 25 | 3(12.0) | D(3) | |
| Breeding facility | 49 | 8(16.3) | D(7), CE02(1) | |
| Total | 144 | 24(16.7) | D(18), EbpC(3), SC02(1) CE01(1), CE02(1), |
Table 2. Occurrence and genotypes of E. bieneusi in red-bellied tree squirrels by age and gender.
| Group | No. of animals | No. of positive (%) | Genotypes(n) |
|---|---|---|---|
| Age(month) | |||
| <3 | 55 | 11(20.0) | D(8), EbpC(2), SC02(1) |
| 3–12 | 67 | 9(13.4) | D(7), CE01(1), CE02(1) |
| >12 | 22 | 4(18.2) | D(3), EbpC(1) |
| Gender | |||
| Male | 61 | 10(16.4) | D(8), CE01(1), SC02(1) |
| Female | 83 | 14(16.9) | D(10), EbpC(3), CE02(1) |
DNA extraction
Each fecal specimen was sieved, and the filtrates were concentrated and washed three times with distilled water by centrifugation for 10 min at 1500 g. Genomic DNA was extracted from approximately 200 mg of each processed fecal specimen using the E.Z.N.A.® Stool DNA Kit (D4015–02; OMEGA Biotek Inc., Norcross, GA, USA) according to the manufacturer’s instructions. The extracted DNA was stored at -20°C until PCR analysis.
PCR amplification
All the DNA preparations were examined for the presence of E. bieneusi by nested PCR amplification of a fragment of 389 bp in size from the rRNA gene of E. bieneusi, and positive specimens were further determined by MLST analyses using the MS1, MS3, MS4, and MS7 loci. The primers and cycling parameters employed for these reactions were as previously described (Table 3) [20, 27]. TaKaRa Taq DNA Polymerase (TaKaRa Bio Inc., Tokyo, Japan) was used for the PCR amplifications. A negative control with no DNA was set up for all the PCR tests. Secondary PCR products were subjected to electrophoresis in a 1.5% agarose gel and visualized by staining the gel with ethidium bromide.
Table 3. Gene locus, primer sequences, annealing temperatures and fragment length for the identification of E. bieuensi used in this study.
| Gene locus | Primer sequence (5'-3') | Annealing temperature (°C) | Fragment length (bp) | References |
|---|---|---|---|---|
| ITS | F1:GATGGTCATAGGGATGAAGAGCTT | 55 | 410 | [27] |
| R1:AATACAGGATCACTTGGATCCGT | ||||
| F2:AGGGATGAAGAGCTTCGGCTCTG | 55 | 392 | ||
| R2:AATATCCCTAATACAGGATCACT | ||||
| MS1 | F1: CAAGTTGCAAGTTCAGTGTTTGAA | 58 | 843 | [20] |
| R1: GATGAATATGCATCCATTGATGTT | ||||
| F2:TTGTAAATCGACCAAATGTGCTAT | 58 | 676 | ||
| R2:GGACATAAACCACTAATTAATGTAAC | ||||
| MS3 | F1:CAAGCACTGTGGTTACTGTT | 55 | 702 | [20] |
| R1:AAGTTA GGGCATTTAATAAAATTA | ||||
| F2:GTTCAAGTAATTGATACCAGTCT | 55 | 537 | ||
| R2:CTCATTGAATCTAAATGTGTATAA | ||||
| MS4 | F1:GCATATCGTCTCATAGGAACA | 55 | 965 | [20] |
| R1:GTTCATGGTTATTAATTCCAGAA | ||||
| F2:CGA AGTGTACTACATGTCTCT | 55 | 885 | ||
| R2: GGACTTTAATAAGTTACCTATAGT | ||||
| MS7 | F1:GTTGATCGTCCAGATGGAATT | 55 | 684 | [20] |
| R1:GACTATCAGTATTACTGATTATAT | ||||
| F2:CAATAGTAAAGGAAGATGGTCA | 55 | 471 | ||
| R2:CGTCGCTTTGTTTCATAATCTT |
Nucleotide sequencing and analysis
The secondary PCR products of the expected size were directly sequenced at Life Technologies (Guangzhou, China) using an ABI Big Dye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Carlsbad, CA, USA). The accuracy of the sequences was confirmed by bidirectional sequencing, and a new PCR secondary product was re-sequenced, if necessary.
The sequences generated in this study were respectively aligned with known reference sequences downloaded from the National Center for Biotechnology Information (NCBI) GenBank database by using BLAST (http://www.ncbi.nlm.nih.gov) to determine their genotype identity. The genotypes, identified as identical to the known genotypes, were assigned the already published names. Meanwhile, the genotypes with single nucleotide substitutions, deletions or insertions compared to the known genotypes were considered novel genotypes, and then named according to the established nomenclature system [9].
Phylogenetic relationship of E. bieneusi
To assess the genetic relationship of ITS genotypes of E. bieneusi obtained in the present study and those published in the previous studies, a phylogenetic analysis was performed by constructing a neighboring-joining tree using the software Mega 6 (http://www.megasoftware.net/), based on the evolutionary distances calculated by a Kimura 2-parameter model. The reliability of these trees was assessed using bootstrap analysis with 1000 replicates.
Statistical analysis
The χ2 test was used to compare the E. bieneusi infection rates between the sex, age and different sampling areas, and differences were considered significant when p < 0.05.
Nucleotide sequence accession numbers
Representative nucleotide sequences have been deposited into GenBank database with the following accession numbers: KU847350 to KU847351 for the rRNA gene ITS sequences of two novel genotypes obtained in the present study (CE01, CE02), and KX259505 to KX259519 for the microsatellite loci (MS1, MS3, MS7) and minisatellite (MS4).
Results
Occurrence of E. bieneusi in red-bellied tree squirrels
Among the 144 fecal samples, 24 were positive for E. bieneusi (16.7%) by PCR amplification of the ITS gene. The infection rates of E. bieneusi in different sources ranged from 12.0% (3/25) in owners to 20.0% (7/35) in pet shop 1 (Table 1), but the difference were not found to be significant (P>0.05). Infection rates of E. bieneusi in red-bellied tree squirrels of different ages and genders have been presented in Table 2; the highest infection rate was observed in <3 months (20.0%, 11/55), followed by 18.2% (4/22) in >12 months, and 13.4% (9/67) in 3–12 months of age (with non-significant differences, P>0.05). Male and female red-bellied tree squirrels showed an infection rate of 16.4% (10/61) and 16.9% (14/83), respectively; however, the difference was not significant (P>0.05).
Genotype distribution and genetic characterizations of E. bieneusi in red-bellied tree squirrels
DNA sequencing and subsequent analysis of the ITS-PCR products from the 24 E. bieneusi-positive specimens revealed the existence of three known E. bieneusi genotypes (D, EbpC, SC02), and two novel genotypes, which were named as CE01 and CE02 (Table 1). Genotype D was the most prevalent (75.0%, 18/24), and was observed in samples from all the three cities, followed by EbpC, which was detected in three specimens from Chengdu and Ya'an cities (12.5%, 3/24); and the genotypes, SC02, CE01, and CE02 were found in one specimen each collected from Ya'an and Chengdu (4.7%, 1/24) (Table 1).
With regard to the novel genotypes, CE01 displayed five single nucleotide polymorphisms (SNPs) within the 243 bp of the ITS gene sequence of E. bieneusi (transversions: T/G, G/T; transitions: A/G, C/T, G/A), when compared to the genotype horse 2 (KU194600), with 99% homology; CE02 had one SNP (transition: C/T) in comparison with genotype horse 2, with 99% homology.
Phylogenetic relationship of E. bieneusi
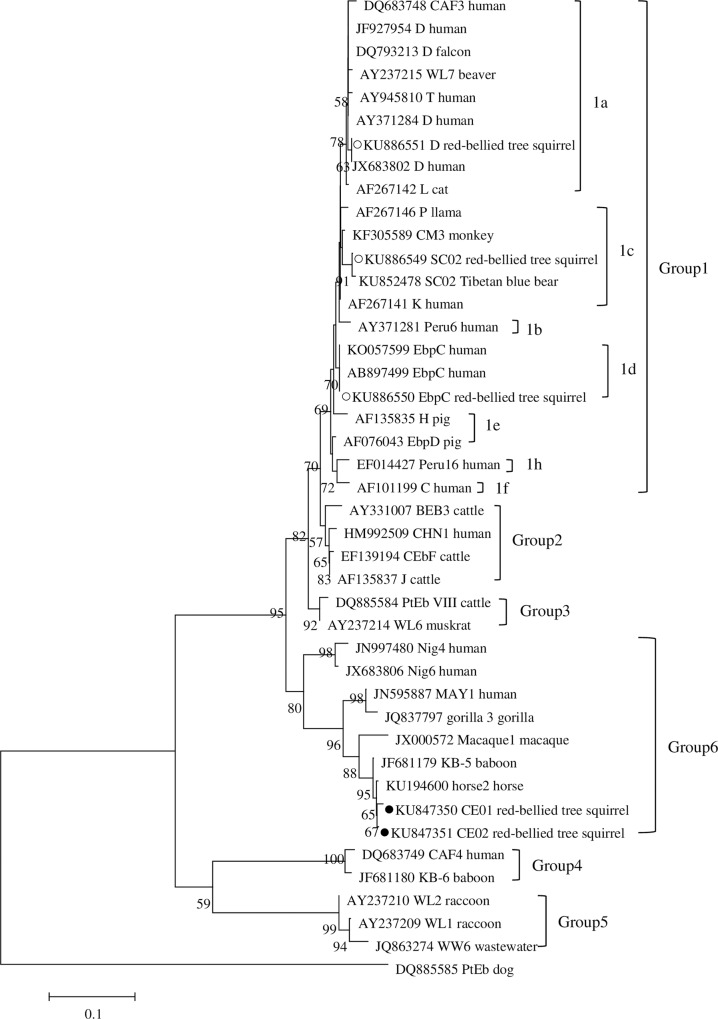
A phylogenetic analysis using neighbor-joining method based on the ITS gene sequences of E. bieneusi showed that all positive samples found in the present study belonged to two groups. Genotypes D, EbpC, and SC02 were clustered into group 1 and were further classed into subgroup 1a, 1d, and 1c, respectively (Fig 1). The two novel genotypes, CE01 and CE02, were clustered into group 6 (Fig 1).
Fig 1. Phylogenetic relationship of Enterocytozoon bieneusi groups, the relationship between E. bieneusi genotypes identified in this study and other known genotypes deposited in the GenBank was inferred by a neighbor-joining analysis of ITS sequences based on genetic distance by the Kimura-2-parameter model.
The numbers on the branches represent percent bootstrapping values from 1,000 replicates, with more than 50% shown in tree. Each sequence is identified by its accession number, genotype designation, and host origin. The group terminology for the clusters is based on the work of Zhao et al [28]. Genotypes with black circles and open circles are novel and known genotypes identified in this study, respectively.
Multilocus sequence typing of E. bieneusi
ITS-positive specimens were further characterized using one minisatellite (MS4) locus and three microsatellites (MS1, MS3 and MS7) loci. A total of 16, 14, 7 and 14 fecal samples were successfully amplified at the MS1, MS3, MS4, and MS7 loci, respectively, and then sequencing analysis revealed 8, 3, 3 and 2 genotypes at the MS1, MS3, MS4, and MS7 loci, respectively. Only 6 samples were simultaneously amplified and sequenced at four loci, sequencing analysis formed five distinct MLGs, namely MLG1-5 (Table 4). All MLGs (MLG1-5) were observed in genotypes D (Table 4).
Table 4. Multilocus characterization of E. bieneusi isolates from red-bellied tree squirrels in Sichuan, southwestern China.
| ITS genotype | Multilocus genotypes | MLGs | No. of MLGs | ||||
|---|---|---|---|---|---|---|---|
| MS1 | MS3 | MS4 | MS7 | GenBank accession Nos. | |||
| D | Type3 | Type1 | Type2 | Type2 | KX259510, KX259513, KX259516, KX259519 | MLG1 | 1 |
| D | Type3 | Type1 | Type2 | Type1 | KX259510, KX259513, KX259516, KX259518 | MLG2 | 1 |
| D | Type1 | Type1 | Type2 | Type1 | KX259508, KX259513, KX259516, KX259518 | MLG3 | 2 |
| D | Type8* | Type2* | Type2 | Type1 | KX259509, KX259514, KX259516, KX259518 | MLG4 | 1 |
| D | Type7* | Type2* | Type3 | Type1 | KX259511, KX259514, KX259517, KX259518 | MLG5 | 1 |
*Novel genotypes
Discussion
E. bieneusi is an emerging zoonotic pathogen and has been reported in humans as well as many animals, such as cattle, pigs, dogs, cats, horses, goats, birds, giant pandas, red pandas, deer, snakes, and golden takins [6, 16, 18, 23, 26, 28–30]. To our knowledge, the present study is the first to reveal the presence of E. bieneusi in red-bellied tree squirrels in China, with an infection rate of 16.7% (24/144). Although the rate of infection of E. bieneusi in red-bellied tree squirrels was frequent in <3 months (20.0%), no age-associated differences were observed in this study. This finding, which was in accordance with a previously published report on chinchillas in China [31], may be attributed to the fact that young animals have incomplete immune system and are prone to the intensive breeding environments. Despite considerable research on this pathogen, only a few genetic studies have documented the occurrence of E. bieneusi in rodents (Table 5); for instance, the highest infection rate of E. bieneusi was observed in wild small rodents (38.9%) in Poland [32], followed by 26.8% in wild rodents from New York [11], 15.3% in beavers in Maryland [27], 10.7% in wild mice from Czech Republic [33], 3.6% in chinchillas in China [31], and 1.0% in wild mice from Slovakia [34]. The observed infection rate was lower than that reported in Poland and New York City, and it was higher than that estimated in Czech Republic, China, and Slovakia.
Table 5. Distribution of E. bieneusi genotypes in red-bellied tree squirrels from different countries.
| Country | Host | No. positive/no. examined (%) | Genotypes | Reference |
|---|---|---|---|---|
| Poland | Wild rodents | 121/311 (38.9%) | D, gorilla 1, WR1-WR10 | [31] |
| United States | Wild rodents | 38/142 (26.8%) | Peru11, Type IV, WL4, WW6, PtEbV, WL20, WL21, WL22, WL23, WL25 | [11] |
| United States | Beavers | 13/85 (15.3%) | WL7, WL8, WL9, WL12, WL13, WL15 | [27] |
| Czech Republic | Wild mice | 31/289 (10.7%) | D, EpbA, PigEBITS5, C, H, CZ3, Peru 8, S6 | [32] |
| China | Chinchillas | 5/140 (3.6%) | D, BEB6 | [33] |
| Slovakia | Wild mice | 3/280 (1.0%) | Peru16 | [34] |
Analysis of the ITS region of the ribosomal RNA revealed a total of five distinct genotypes out of the 24 E. bieneusi isolates, which comprised of three known genotypes (D, EbpC, and SC02) and two novel genotypes (CE01 and CE02). The genotype D showed the highest percentage of E. bieneusi-positive specimens in the present study, accounting for 75.0% (18/24), followed by genotype EbpC (12.5%; 3/24) (Table 1). In China, genotype D has an extensive host range and has been examined in humans [35], non-human primate, pigs, dogs, foxes, and cats [13, 36–39], as well as in waste water [22]. Besides, the genotype EbpC has been previously observed in many animals, including humans, cattle, pigs, sheep, dogs, non-human primates, deer, beavers, otters, muskrats, raccoons, and foxes, even giant panda [23, 24, 40]. Interestingly, the two genotypes, D and EbpC, which were examined for the first time in red-bellied tree squirrels in the current study, presented an expanded host range. These results indicated that red-bellied tree squirrels may play a potential role in the transmission of E. bieneusi to humans.
Genetic relationship of two novel genotypes of E. bieneusi to the known ones was observed in a phylogenetical analysis. The two novel genotypes (CE01 and CE02) were clustered into group 6. Genotypes WW7, WW8 were first detected in urban wastewater belonging to group 6 [22] and then recent studies have revealed that certain genotypes from other animals were also clustered into group 6, including gorilla 3 in gorillas, KB-5, and Macaque1 in non-human primates, Horse 2 in horses [33, 41, 42]. Meanwhile, other members of this group, such as genotypes Nig4, Nig6, MAY1, have also been reported in humans [10, 35, 43, 44]. Therefore, it is not completely known whether the two novel genotypes belonging to group 6 have an ability to cause microsporidiosis in humans, and the potential of zoonotic transmission need to be confirmed by future extensive genotyping research in large samples of human microsporidiosis.
In order to better understand route of transmission, genetic diversity and host specificity of E. bieneusi, the MLST tool for subtyping E. bieneusi was developed [20]. In the present study, sequencing analysis indicated 8, 3, 3 and 2 genotypes at the MS1, MS3, MS4, and MS7 loci, respectively, and identified three, one, one novel genotypes in loci MS1, MS3, MS4, respectively. A total of five distinct MLGs (MLG1-5) were observed in genotypes D. These results showed the genetic diversity of E. bieneusi in red-bellied tree squirrels.
In conclusion, this is the first report on the occurrence of three known human-pathogenic E. bieneusi genotypes (D, EbpC, SC02) and two novel genotypes (CE01, CE02) in red-bellied tree squirrels in Sichuan province, China. Genetic diversity was observed by MLST tool, and five MLGs were found in red-bellied tree squirrels. The fact that genotypes D and EbpC have been previously reported in humans, suggest that red-bellied tree squirrels can serve as potential reservoir hosts for the zoonotic transmission of human microsporidiosis. Due to the high frequency of human contact with pet animals in China, proper advice should be given to the susceptible human populations in order to reduce the zoonotic transmission of this neglected disease.
Supporting Information
(XLSX)
Acknowledgments
The study was financially supported by the Chengdu giant panda breeding research foundation (CPF2014-14) and the National Natural Science Foundation of China (No. 31370407).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The study was financially supported by the Chengdu giant panda breeding research foundation project (CPF research 2014-14) and the National Natural Science Foundation of China 3137040 Huailiang Xu.
References
- 1.Patrick K. Five questions about microsporidia. PloS Pathog. 2009; 5(9): e1000489 doi: 10.1371/journal. ppat.1000489 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Didier ES, Weiss LM. Microsporidiosis: current status. Cur Opin Infect Dis. 2006; 19(5): 485–92. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Didier ES, Weiss LM. Microsporidiosis: not just in AIDS patients. Cur Opin Infect Dise. 2011; 24(5):490–5. 10.1097/QCO.0b013e32834aa152 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matos O, Lobo ML, Xiao L. Epidemiology of Enterocytozoon bieneusi Infection in Humans. J Parasitol Res. 2012; 2012(4): 981424 10.1155/2012/981424 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thellier M, Breton J. Enterocytozoon bieneusi in human and animals, focus on laboratory identification and molecular epidemiology. Parasite. 2008; 15(3): 349–58. . [DOI] [PubMed] [Google Scholar]
- 6.Galván-Díaz AL, Magnet A, Fenoy S, Henriques-Gil N, Haro M, Gordo FP, et al. Microsporidia detection and genotyping study of human pathogenic E. bieneusi in animals from Spain. PloS One. 2014; 9(3): e92289 10.1371/journal.pone.0092289 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akiyoshi DE, Morrison HG, Shi L, Xiaochuan F, Quanshun Z, Nicolas C, et al. Genomic survey of the non-cultivatable opportunistic human pathogen, Enterocytozoon bieneusi. PloS Pathog. 2009; 5(1): 1000261 10.1371/journal.ppat.1000261 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexander M, Rainer W, Peter D. Zoonotic potential of the microsporidia. Clin Microbiol Rev. 2005; 18(3): 423–45. 10.1128/CMR.18.3.423-445.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santín M, Fayer R. Enterocytozoon bieneusi Genotype Nomenclature Based on the Internal Transcribed Spacer Sequence: A Consensus. J Eukaryot Microbiol. 2009; 56(1): 34–8. 10.1111/j.1550-7408.2008.00380.x . [DOI] [PubMed] [Google Scholar]
- 10.Sokolova OI, Demyanov AV, Bowers LC, Didier ES, Yakovlev AV, Skarlato SO, et al. Emerging microsporidian infections in Russian HIV-infected patients. J Clin Microbiol. 2011; 49(6): 2102–8. 10.1128/JCM.02624-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo Y, Alderisio KA, Yang W, Cama V, Feng Y, Xiao L. Host specificity and source of Enterocytozoon bieneusi genotypes in a drinking source watershed. Appl Environ Microbiol. 2014; 80(1): 218–25. 10.1128/AEM.02997-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haro M, Izquierdo F, Henriques-Gil N, Andres I, Alonso F, Fenoy S, et al. First detection and genotyping of human-associated microsporidia in pigeons from urban parks. Appl Environ Microbiol. 2005; 71(6): 3153–7. 10.1128/AEM.71.6.3153-57 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Li Y, Li W, Yang J, Song M, Diao R, et al. Genotypes of Enterocytozoon bieneusi in livestock in China: high prevalence and zoonotic potential. PloS One. 2014; 9(5): e97623 10.1371/journal.pone.0097623 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Hao Y, Wang L, Xiang H, Zhou Z. Genome-wide identification and comprehensive analyses of the kinomes in four pathogenic microsporidia species. PloS One. 2014; 9(12): e115890 10.1371/journal.pone.0115890 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Widmer G, Akiyoshi DE. Host-specific segregation of ribosomal nucleotide sequence diversity in the microsporidian Enterocytozoon bieneusi. Infect Genet Evol 2010; 10(1): 122–8. 10.1016/j.meegid.2009.11.009 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao W, Zhang W, Yang D, Zhang L, Wang R, Liu A. Prevalence of Enterocytozoon bieneusi and genetic diversity of ITS genotypes in sheep and goats in China. Infect Genet Evol 2015; 32: 265–70. 10.1016/j.meegid.2015.03.026 . [DOI] [PubMed] [Google Scholar]
- 17.Karim MR, Wang R, Dong H, Zhang L, Li J, Zhang S, et al. Genetic polymorphism and zoonotic potential of Enterocytozoon bieneusi from nonhuman primates in China. Appl Environ Microbiol. 2014; 80(6): 1893–8. 10.1128/AEM.03845-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santin M, Fayer R. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res Vet Sci]. 2011; 90(3): 363–71. 10.1016/j.rvsc.2010.07.014 . [DOI] [PubMed] [Google Scholar]
- 19.Li W, Cama V, Feng Y, Gilman RH, Bern C, Zhang X, et al. Population genetic analysis of Enterocytozoon bieneusi in humans. Int J Parasitol. 2012; 42(3): 287–93. 10.1016/j.ijpara.2012.01.003 . [DOI] [PubMed] [Google Scholar]
- 20.Feng Y, Li N, Dearen T, Lobo ML, Matos O, Cama V, et al. Development of a multilocus sequence typing tool for high-resolution genotyping of Enterocytozoon bieneusi. Appl Environ Microbiol. 2011; 77(14): 4822–8. 10.1128/AEM.02803-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang XT, Wang RJ, Ren GJ, Yu ZQ, Zhang LX, Zhang SY, et al. Multilocus genotyping of Giardia duodenalis and Enterocytozoon bieneusi in dairy and native beef (Qinchuan) calves in Shaanxi province, northwestern China. Parasitol Res. 2016; 115(3): 1–7. 10.1007/s00436-016-4908-6 . [DOI] [PubMed] [Google Scholar]
- 22.Li N, Xiao L, Wang L, Zhao S, Zhao X, Duan L, et al. Molecular surveillance of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi by genotyping and subtyping parasites in wastewater. PLoS Negl Trop Dis. 2012; 6(9): e1809 10.1371/journal.pntd.0001809 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian GR, Zhao GH, Du SZ, Hu XF, Wang HB, Zhang LX, et al. First report of Enterocytozoon bieneusi from giant pandas (Ailuropoda melanoleuca) and red pandas (Ailurus fulgens) in China. Infect Genet Evol 2015; 34: 32–5. 10.1016/j.meegid.2015.06.015 . [DOI] [PubMed] [Google Scholar]
- 24.Zhang Z, Huang J, Karim MR, Zhao J, Dong H, Ai W, et al. Zoonotic Enterocytozoon bieneusi genotypes in Pere David's deer (Elaphurus davidianus) in Henan, China. Experiment Parasitol. 2015; 155: 46–8. 10.1016/j.exppara.2015.05.008 . [DOI] [PubMed] [Google Scholar]
- 25.Zhao W, Zhang W, Yang Z, Liu A, Zhang L, Yang F, et al. Genotyping of Enterocytozoon bieneusi in Farmed Blue Foxes (Alopex lagopus) and Raccoon Dogs (Nyctereutes procyonoides) in China. PloS One. 2015; 10(11): e0142611 10.1371/journal.pone.0142611 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao GH, Du SZ, Wang HB, Hu XF, Deng MJ, Yu SK, et al. First report of zoonotic Cryptosporidium spp., Giardia intestinalis and Enterocytozoon bieneusi in golden takins (Budorcas taxicolor bedfordi). Infect Genet Evol. 2015; 34: 394–401. 10.1016/j.meegid.2015.07.016 . [DOI] [PubMed] [Google Scholar]
- 27.Sulaiman IM, Ronald F, Lal AA, Trout JM, Schaefer FW, Lihua X. Molecular characterization of microsporidia indicates that wild mammals Harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Appl Environ Microbiol. 2003; 69(8): 4495–501. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rinder H, Thomschke A, Dengjel B, Gothe R, Löscher T, Scher M, et al. Close genotypic relationship between Enterocytozoon bieneusi from humans and pigs and first detection in cattle. J Parasitol. 2000;86(1):185–8. 10.1645/0022-3395(2000)086[0185:CGRBEB]2.0.CO;2 . [DOI] [PubMed] [Google Scholar]
- 29.Santin M, Fayer R. Enterocytozoon bieneusi, giardia, and Cryptosporidium infecting white-tailed deer. J Eukaryot Microbiol. 2015; 62(1): 34–43. 10.1111/jeu.12155 . [DOI] [PubMed] [Google Scholar]
- 30.Liu W, Nie C, Zhang L, Wang R, Liu A, Zhao W, et al. First detection and genotyping of Enterocytozoon bieneusi in reindeers (Rangifer tarandus): a zoonotic potential of ITS genotypes. Parasit Vectors. 2015; 8: 526 10.1186/s13071-015-1155-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi M, Luo N, Wang H, Yu F, Wang R, Huang J, et al. Zoonotic Cryptosporidium spp. and Enterocytozoon bieneusi in pet chinchillas (Chinchilla lanigera) in China. Parasitol Int. 2015; 64(5): 339–41. 10.1016/j.parint.2015.05.007 . [DOI] [PubMed] [Google Scholar]
- 32.Perec-Matysiak A, Bunkowska-Gawlik K, Kvac M, Sak B, Hildebrand J, Lesnianska K. Diversity of Enterocytozoon bieneusi genotypes among small rodents in southwestern Poland. Vet Parasitol. 2015; 214(3–4): 242–6. 10.1016/j.vetpar.2015.10.018 . [DOI] [PubMed] [Google Scholar]
- 33.Sak B, Kvac M, Kvetonova D, Albrecht T, Pialek J. The first report on natural Enterocytozoon bieneusi and Encephalitozoon spp. infections in wild East-European House Mice (Mus musculus musculus) and West-European House Mice (M. m. domesticus) in a hybrid zone across the Czech Republic-Germany border. Vet Parasitol. 2011; 178(3–4): 246–50. 10.1016/j.vetpar.2010.12.044 . [DOI] [PubMed] [Google Scholar]
- 34.Danisova O, Valencakova A, Stanko M, Luptakova L, Hasajova A. First report of Enterocytozoon bieneusi and Encephalitozoon intestinalis infection of wild mice in Slovakia. Ann Agric Environ Med. 2015; 22(2): 251–2. 10.5604/12321966.1152075 . [DOI] [PubMed] [Google Scholar]
- 35.Wang L, Zhang H, Zhao X, Zhang L, Zhang G, Guo M, et al. Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J Clin Microbiol. 2013; 51(2): 557–63. 10.1128/JCM.02758-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du SZ, Zhao GH, Shao JF, Fang YQ, Tian GR, Zhang LX, et al. Cryptosporidium spp., Giardia intestinalis, and Enterocytozoon bieneusi in Captive Non-Human Primates in Qinling Mountains. Korean J Parasitol. 2015; 53(4): 395–402. 10.3347/kjp.2015.53.4.395 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karim MR, Wang R, He X, Zhang L, Li J, Rume FI, et al. Multilocus sequence typing of Enterocytozoon bieneusi in nonhuman primates in China. Vet Parasitol. 2014; 200(1–2): 13–23. 10.1016/j.vetpar.2013.12.004 . [DOI] [PubMed] [Google Scholar]
- 38.Wei L, Ruinan D, Jinping Y, Lihua X, Yixin L, Yijing L, et al. High diversity of human-pathogenic Enterocytozoon bieneusi genotypes in swine in northeast China. Parasitol Res. 2014; 113(3): 1147–53. 10.1007/s00436-014-3752-9 [DOI] [PubMed] [Google Scholar]
- 39.Ma J, Pei L, Zhao X, Xu H, Wu W, Wang Y, et al. Occurrence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi in dairy cattle, beef cattle and water buffaloes in China. Vet Parasitol. 2014; 207(3–4): 220–7. 10.1016/j.vetpar.2014.10.011 . [DOI] [PubMed] [Google Scholar]
- 40.Jiang Y, Tao W, Wan Q, Li Q, Yang Y, Lin Y, et al. Zoonotic and Potentially Host-Adapted Enterocytozoon bieneusi Genotypes in Sheep and Cattle in Northeast China and an Increasing Concern about the Zoonotic Importance of Previously Considered Ruminant-Adapted Genotypes. Appl Environ Microbiol. 2015; 81(15): 5278 10.1128/aem.01928-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sak B, PetržElková KJ, KvěToňOvá D, MynářOvá A, Pomajbíková K, Modrý D, et al. Diversity of Microsporidia, Cryptosporidium and Giardia in Mountain Gorillas (Gorilla beringei beringei) in Volcanoes National Park, Rwanda. PloS One. 2014; 9(11): e109751 10.1371/journal.pone.0109751 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laatamna AE, Wagnerová P, Sak B, Květoňová D, Xiao L, Rost M, et al. Microsporidia and Cryptosporidium in horses and donkeys in Algeria: Detection of a novel Cryptosporidium hominis subtype family (Ik) in a horse. Vet Parasitol. 2015; 208(3–4): 135–42. 10.1016/j.vetpar.2015.01.007 . [DOI] [PubMed] [Google Scholar]
- 43.Akinbo FO, Okaka CE, Richard O, Haileeyesus A, Lihua X. Unusual Enterocytozoon bieneusi genotypes and Cryptosporidium hominis subtypes in HIV-infected patients on highly active antiretroviral therapy. Ame J Trop Med Hyg. 2013; 89(1): 157–61. 10.4269/ajtmh.12-0635 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Wang Z, Su Y, Liang X, Sun X, Peng S, et al. Identification and genotyping of Enterocytozoon bieneusi in China. J Clin Microbiol. 2011; 49(5): 2006–8. 10.1128/JCM.00372-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.