Abstract
Adverse effect of alcohol on neural function has been well documented. Especially, the teratogenic effect of alcohol on neurodevelopment during embryogenesis has been demonstrated in various models, which could be a pathologic basis for fetal alcohol spectrum disorders (FASDs). While the developmental defects from alcohol abuse during gestation have been described, the specific mechanisms by which alcohol mediates these injuries have yet to be determined. Recent studies have shown that alcohol has significant effect on molecular and cellular regulatory mechanisms in embryonic stem cell (ESC) differentiation including genes involved in neural development. To test our hypothesis that alcohol induces molecular alterations during neural differentiation we have derived neural precursor cells from pluripotent human ESCs in the presence or absence of ethanol treatment. Genome-wide transcriptomic profiling identified molecular alterations induced by ethanol exposure during neural differentiation of hESCs into neural rosettes and neural precursor cell populations. The Database for Annotation, Visualization and Integrated Discovery (DAVID) functional analysis on significantly altered genes showed potential ethanol’s effect on JAK-STAT signaling pathway, neuroactive ligand-receptor interaction, Toll-like receptor (TLR) signaling pathway, cytokine-cytokine receptor interaction and regulation of autophagy. We have further quantitatively verified ethanol-induced alterations of selected candidate genes. Among verified genes we further examined the expression of P2RX3, which is associated with nociception, a peripheral pain response. We found ethanol significantly reduced the level of P2RX3 in undifferentiated hESCs, but induced the level of P2RX3 mRNA and protein in hESC-derived NPCs. Our result suggests ethanol-induced dysregulation of P2RX3 along with alterations in molecules involved in neural activity such as neuroactive ligand-receptor interaction may be a molecular event associated with alcohol-related peripheral neuropathy of an enhanced nociceptive response.
Introduction
Alcohol consumption is recognized as the leading preventable cause of birth defects and mental retardation. High levels of alcohol consumption during pregnancy can result in fetal alcohol spectrum disorders (FASDs), which is characterized by prenatal and postnatal growth restriction, craniofacial dysmorphology and structural abnormalities of the central nervous system [1]. Depending on conditions and manifestations, these damages are referred as fetal alcohol syndrome (FAS), alcohol-related birth defects (ARBDs), and alcohol-related neurodevelopmental disorder (ARND). While the developmental defects from alcohol abuse during gestation have been described, it is still unanswered about what are the specific mechanisms by which alcohol mediates these injuries [2, 3]. This is important question to address to identify affected children at an early age and intervene to prevent or mitigate the damage.
The effect of alcohol on development has been widely studied in many different animal species [4]. Adverse effect of alcohol on brain function has been well documented. Especially, the teratogenic effect of alcohol on neurodevelopment during embryogenesis has been demonstrated in animal models, which could be a pathologic basis for FASDs [1, 3]. It has been demonstrated that alcohol exposure during preimplantation period has significant effect on embryo development [5]. Reports have demonstrated genetic, cellular, and biochemical association of alcohol with teratogenesis [6–9]. The wide range of physiological and morphological defects associated with in utero alcohol exposure suggest that the etiology of FASDs involve a high degree of cellular and molecular heterogeneity. Gastrulation period is considered to be the most sensitive to teratogenic insult, suggesting that differentiating cells might be especially vulnerable to the teratogenic effects of alcohol [7].
Currently, it is not clearly established what causes FASDs. Recently, epigenetic regulations have emerged as potential mechanisms associated with alcohol teratogenesis. Epigenetic imprinting or genome-wide epigenetic reprogramming has been proposed as a mechanism responsible for alcohol-induced teratogenesis in preimplantation embryos [2, 3]. Interestingly, even paternal or maternal alcohol consumption prior to conception has been shown to result in a wide range of birth defects and fetal abnormalities. It is likely that alcohol-induced epigenetic changes in the gametes or within germ line are responsible for pre-conceptional effects of alcohol [10]. Considering the importance of epigenetic factors in development, especially in central nervous system development and dysfunction, it is quite reasonable to link epigenetic mechanisms as potential regulatory events involved in alcohol teratogenesis [2, 11–13].
Embryonic stem cells (ESCs) are pluripotent cells that can be derived into all lineages of cells in the organism [14]. Due to this biological competency of ESCs, beneficial utility of ESCs for regenerative medicine has been suggested in many applications [15]. In addition, ESC has been proven to be a useful tool to study mechanisms associated with the pathogenesis of genetic disorders, especially disease-associated molecular alterations at the early stage of fetal development [16]. ESCs provide us with an opportunity to establish an experimental model to study the functional effects of genetic alterations on normal embryo development and further to test tools to intervene deleterious effects of genetic alterations on the later stage of life. Stem cell models are beneficial to developmental studies especially where in vivo molecular/cellular study models are not available.
Stem cells are especially vulnerable to ethanol (EtOH) toxicity through decreases in pluripotency, survival capacity, and/or altered differentiation [7]. Studies have shown that alcohol has significant effect on molecular and cellular regulatory mechanisms in ESC differentiation [17]. More interestingly, it has been demonstrated that alcohol induces alteration in genes involved in neural development in ESCs [18, 19]. It is known that gastrulation periods of ESCs including neuronal differentiation process require epigenetic controls, especially DNA methylation [2]. Our recent studies have described the molecular signatures of EtOH’s effects on stem cell potency and differentiation in human embryonic stem cells (hESCs) [20]. Studies also showed that EtOH exposure reduces neuronal stem cell numbers in developing and adult brains [21, 22].
As a surrogate model for adult neural stem cells, pluripotent hESCs can be differentiated into neural progeny and used to elucidate mechanisms underlying early human neurogenesis. An established in vitro neural stem cell model will be useful for the evaluation of developmental regulatory mechanisms and also the significance of their alterations in pathological conditions such as developmental disorders or exposure to neuroteratogens [19]. We have derived neural stem cells from human embryonic stem cells as a model to study the effect of teratogen in neural development. We have successfully derived neural precursor cells exhibiting key molecular features of neural stem cells, which will be useful for experimental application [23]. Our established model has further been used to assess the molecular effect of alcohol treatment on neural stem cell population. It was found that alcohol exposure of neural precursor cells resulted in significant alterations of molecular signatures that may have functional role in neural stem cell maintenance and development and may be further involved in deleterious cellular effect of alcohol exposure.
Materials and Methods
Human embryonic stem cell culture and derivation of neural stem cells
Human embryonic stem cells (H1 and H9 lines) obtained through license agreement with WiCell Research Institute (Madison, WI) were cultured on mouse embryonic fibroblast (MEF) feeder layer and were transferred to mTeSR1 serum free human embryonic stem cell (hESC) culture system (STEMCELL Technologies Inc., Vancouver, Canada). Neural differentiation of hESCs was performed by using STEMdiff Neural System (STEMCELL Technologies Inc., Vancouver, Canada) according to the manufacturer’s instruction as described in our previous publications [23, 24]. After 7 day differentiation, morphological assessment and scoring of neural rosettes were done to ensure 50% or more of the area of each aggregate was filled with neural rosettes (as shown in Fig 1). On day 7, neural rosettes were selected away from contaminating flat cells and collected. The rosettes were resuspended in pre-warmed NIM and cultured on 6-well plates precoated with poly-L-Ornithine and laminin (PLO/L) with daily full medium changes using pre-warmed STEMdiff NIM (without or with 20 mM ethanol) for 5 days with alternating treatment for a day and withdrawal for a day. Cells were EtOH concentration was chosen for its physiological relevance in that 20 mM is equivalent to DUI level and 50 mM falls within levels measured in alcoholics [25].
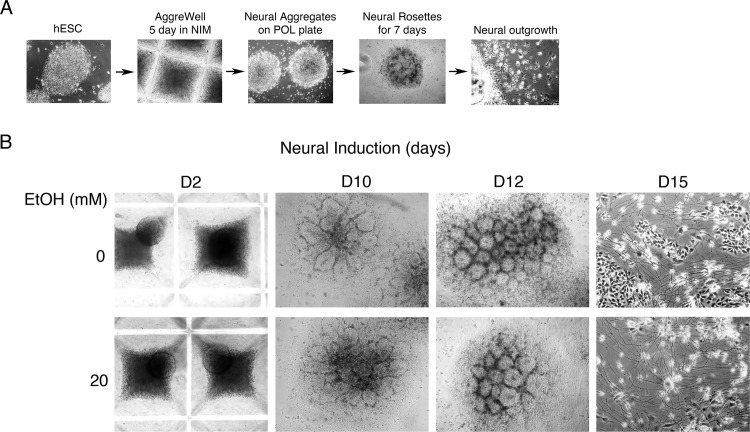
Fig 1. Neural differentiation of human embryonic stem cells in vitro.
A. Neural differentiation of hESC in vitro. Human embryonic stem cells were subjected to embryoid body (EB) formation using AggreWell for 5 days in neural induction medium (NIM). Neural aggregates were seeded on plates coated with poly-L-ornithine/laminin (PLO/L) and cultured with NIM for additional 7 days to develop neural rosette structure. After 7 days, the neural rosettes were dislodged and then replated for the expansion of neural precursor cells for 3–5 days. B. Human embryonic stem cells were subjected to embryoid body formation using AggreWell for 5 days in neural induction medium. Neural aggregates were seeded on poly-L-ornithine/laminin coated plates and cultured with NIM for 7 days to develop neural rosette structure. Ethanol treatment was initiated a day after plating the neural aggregates onto PLO/L plates. For ethanol treatment cells were fed with fresh medium every day by alternating a treatment with 20 mM ethanol for 1 day and a withdrawal for 1 day. Treatment was continued till the end of neural expansion. After 7 days, the neural rosettes were dislodged and then re-plated for the expansion of neural precursor cells for 5 days.
Immunofluorescence analysis
To ensure proper neural differentiation of hESCs, the same experimental procedure was applied to a set of cells plated on the coverslips. The level of neural markers (Nestin, Sox2, Musashi and beta-3 tubulin) was assessed by immunofluorescence microscopy. For IF analysis cells were seeded on a coverslip coated with Matrigel in a 6 well plate. The cells were fixed with 4% paraformaldehyde/PBS for 30 min at room temperature and washed with PBS. IF analysis for neural stem cell markers was done by using Human Neural Stem Cell Characterization Kit (EMD Millipore, Billerica, MA). Samples were incubated with blocking solution (5% normal donkey serum, 0.3% Triton X-100 in 1X PBS) for 30 min at room temperature. After washing three times (5 min each) with 1X PBS, samples were incubated with primary antibody [mouse anti-Nestin (1:500), rabbit anti-Sox2 (1:1000), mouse anti-βIII tubulin (1:1000), or rabbit anti-Musashi1 (1:500)] diluted in 1X PBS (or blocking solution) and incubated overnight at 4°C. As a negative control, we have included the same isotype rabbit IgG or mouse IgG at equivalent concentrations. After washing three times for 5 min each with 1X PBS, samples were incubated with appropriate secondary antibody [donkey anti-rabbit IgG-FITC (1:500) or donkey anti-mouse IgG-rhodamine (1:500)] for 2 hrs at RT. After washing, the slips were mounted on the slide with mounting medium containing DAPI (Vector Labs, Inc., Burlingame, CA) and fluorescence imaging was performed with Olympus IX81 imaging microscope.
RNA isolation and microarray analysis
RNA isolation and microarray analysis was done as described [20, 24]. Briefly, the neural aggregates at D10 for the formation of rosettes and at D15 (5 days after re-plating the rosette clusters for the expansion of neural precursor cells) were used for total RNA isolation by using RNeasy mini kit (Qiagen, Valencia, CA). RNA purity and concentration was determined by NanoDrop, ND-1000 spectrophotometer (Thermo Scientific, Indianapolis, IN) and microfluidics-based platform 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Samples in biological duplicate were hybridized to Affymetrix Human Genome Plus 2.0 (Cat. No. 900469). Standard quality control metrics recommended by Affymetrix including image quality, signal distribution and pair wise scatter plots were used for all arrays. Mas5.CHP files were generated for each array by MAS 5.0 (Affymetrix, Santa Clara, CA) and combined to a final RESULTS.MAS5.TXT file [20, 24].
Quantitative RT-PCR analysis
For validation of gene expression by quantitative RT-PCR analysis was performed as describe previously [20]. Total RNA was first subjected to DNase digestion with Turbo DNA-free kit (Life Technologies, Grand Island, NY). 2 μg of total RNA treated with DNase was used to synthesize cDNA by using iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) in 40 μl reaction mixture. The resulting cDNA was diluted (mixed with H2O by 1:4) and 1.5 μl of diluted cDNA was used per well (in 10 μl reaction volume) in a 384 well plate using LightCycler 480 SYBR Green I master mix (Roche Diagnostics, Indianapolis, IN).
Results
Derivation of neural precursor cells from human embryonic stem cells
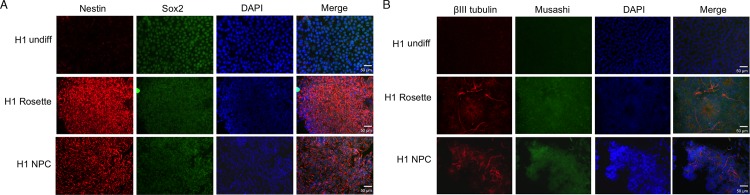
To establish a model system to examine the molecular effects of EtOH on neural differentiation and maintenance of neural stem cells, we used a commercial culture system developed for neural differentiation of human embryonic stem cells (STEMCELL Technologies Inc., Vancouver, Canada) as we have previously reported [23, 24]. Human embryonic stem cells (hESCs) exponentially growing on mouse embryonic fibroblast (MEF) feeder were first adapted to the feeder-free culture system using mTeSR1 medium (STEMCELL Technologies Inc., Vancouver, Canada) and subjected to embryoid body formation in neural differentiation medium as described in Materials and Methods. Fig 1A shows the overall scheme for the derivation of neural precursor cells from hESCs. Neural differentiation was performed in the presence or absence of 20 mM EtOH (Fig 1B). Cells were collected for RNA purification at day 12 (rosette structure) and day 15 (neural precursor cells). Morphological observation clearly showed the formation or rosette structures and neural differentiation (Fig 1B). However, EtOH’s effect on the process could not be quantitatively assessed by microscopic observation. To examine if hESCs were properly specified into neural cells the expression of neural lineage markers was assessed by immunofluorescence microscopy (Fig 2). Compared to undifferentiated hESCs, rosette and NPC showed induced expression of neural maker Nestin, neuron-specific class III beta-tubulin (TBB3 or Tuj1) and Musashi (Fig 2). Expression of stemness factor Sox2 was maintained in undifferentiated, rosette and NPC as expected (Fig 2A). Even though immunofluorescence microscopy analysis was not proper to quantitatively compare the expression level of markers, it seems that EtOH may have inhibitory effect on the expression of Nestin and Musashi, notably in rosette structure (Fig 2A).
Fig 2. Immunofluorescence analysis to confirm neural differentiation.
Induced expression of neural markers in hESC-derived neural stem cells. Neural differentiation of hESCs was confirmed by examining the expression of neural markers. A. Nestin and SOX7 and B. beta-tubulin and Musashi expressions were noticeably increased in rosettes and NPCs. SOX2 expression remained high in pluripotent hESCs and also hESC-derived neural lineage cells. Cells were counterstained with DAPI for nuclei (scale bar, 50 μm).
To further quantitatively characterize the neural specification of hESCs into rosette and NPCs, we have performed quantitative RT-PCR analysis on neural markers (S1 Fig). We found significant induction of Nestin, Musashi and Tuj1 mRNA in neural rosettes and NPCs (S1A Fig). Our analysis showed that 20 mM EtOH treatment resulted in significant reduction of Nestin and Musashi mRNA (S1B Fig). But this reduction seems marginal and may not cause dramatic decrease in the steady state level of mRNA’s considering highly induced levels of these mRNA’s after neural differentiation, which may have marginal effect on the level of corresponding protein as shown in Fig 2. We have previously demonstrated that EtOH treatment resulted in significant reduction of pluripotent markers such as Oct4 and Sox2 in undifferentiated hESCs [20]. It is known that Sox2 plays a role in the neural specification and proliferation of NPCs while Oct4 and Nanog is involved in mesendoderm differentiation [26]. We found that neural differentiation resulted in dramatic downregulation of Oct4 mRNA, but the level of Sox2 was comparably maintained and was not greatly affected by EtOH treatment in rosettes and NPCs (S1C Fig).
Profiling of gene signatures affected by ethanol treatment during differentiation of hESCs to neural precursors
We have used our hESC neural differentiation model to profile gene signatures significantly altered by alcohol exposure, specifically during directed differentiation into neural precursors in vitro. H1 and H9 hESCs were independently directed to neuronal differentiation in the presence or absence of 20 mM ethanol in biological duplicates. Mixed population of cells pooled from each replicate was harvested at day 12 for neural rosette cells and at day 15 for neural precursor cells (NPC). Gene expression microarray analysis was done using samples from H1 hESC line. Total RNA was prepared and subjected to gene expression microarray as described in Materials and Methods by using Affymetrix Human U133A 2.0 arrays (data accession number GSE56906). We have analyzed the processed data by Weighted Gene Correlation Network Analysis (WGCNA) and also by simple fold change comparisons against control samples.
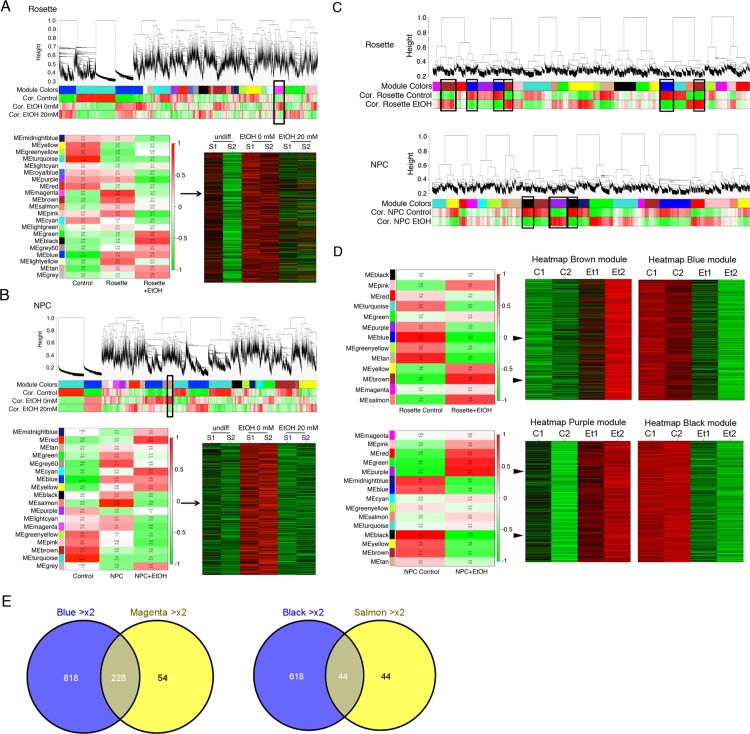
We have initially performed WGCNA on datasets from both rosette and NPC samples altogether and found that the effects of differentiation (hESCs to neural rosette and further to NPC) were too overwhelming to see the effect of EtOH treatment on gene signatures (data not shown). Correlation of gene expression to EtOH treatment was not strong enough over the effect of differentiation (data not shown). Therefore, we decided to analyze the dataset for the effect of EtOH on rosette cells separately from on NPC. We performed WGCNA on undifferentiated hESCs (undiff), differentiated without EtOH treatment (EtOH 0 mM) and differentiated with EtOH treatment (EtOH 20 mM). We examined correlations to differentiation and EtOH treatment in neural rosette (Fig 3A) or NPC (Fig 3B) from the biological duplicate samples (S1 and S2). We identified modules that are altered by differentiation or altered by EtOH treatment during differentiation. As an example, we have shown in Fig 3 the most significantly associated module with EtOH treatment in rosettes and NPCs. Fig 3A shows the magenta module of genes that were upregulated in rosettes, but downregulated by EtOH treatment. Fig 3B shows the salmon module of genes that were upregulated in NPCs, but downregulated by EtOH treatment. This analysis allowed us to identify gene signatures that were differentially regulated during differentiation into neural rosette or NPC and affected by EtOH treatment.
Fig 3. Gene expression analysis and WGCNA.
WGCNA analysis on EtOH-induced transcriptomic alterations in neural rosettes and NPC populations derived from hESCs compared to undifferentiated parental hESCs. A. Result from rosettes identified the magenta module of genes that were upregulated in rosettes, but downregulated by EtOH treatment. B. Analysis for NPCs showed the salmon module of genes that were upregulated in NPCs, but downregulated by EtOH treatment. Heatmaps are shown for modules with most significant association with EtOH treatment identified from the module trait map of the module eigengene (ME) from two biological duplicates (S1 and S2) to each treatment. Rosettes showed less extent of association than NPCs. C. In addition, we have performed WGCNA analysis on gene expression microarray data to directly assess EtOH’s effect on transcriptomic alterations in neural rosettes and NPCs with and without EtOH treatment. D. Heatmaps are shown for modules with most significant association upon EtOH treatment (biological duplicate of Et1 and Et2) compared to without EtOH (biological duplicate of C1 and C2). The Brown module (upregulated- red) and the blue module (downregulated- green) with EtOH treatment in neural rosettes and the purple module (upregulated- red) and the black module (downregulated- green) upon EtOH exposure in NPCs. E. Common genes that demonstrated consistent alterations were identified. For example, genes in the blue module (downregulated >2 fold in EtOH-treated rosettes compared to untreated rosettes) were compared to the magenta module (>2 fold upregulated during differentiation into rosettes and then downregulated >2 fold upon EtOH treatment). Similar analysis was done with the black and salmon modules to identify genes truly downregulated in NPC with EtOH treatment.
It was noted that correlation to EtOH treatment was not strong and only a very limited number of modules showed EtOH-dependent association. We reasoned that this was due to the strong effect of neural differentiation on the gene regulation in our model. Next we examined correlative gene expression simply associated with EtOH treatment in differentiated cells. We used datasets from differentiated rosette cells or NPCs with or without EtOH treatment (Fig 3C and 3D). WGCNA on rosette cells (control vs. EtOH treated) showed that the brown module was most significantly correlated to genes upregulated and the blue module was most significantly correlated to genes downregulated upon EtOH treatment (Fig 3C and 3D). A heatmap for each representative module is shown. Similar analysis was done with dataset from NPC cells. The purple module was most significantly correlated to genes upregulated upon EtOH treatment and the black was most significantly correlated to genes downregulated upon EtOH treatment (Fig 3C and 3D). To examine concordance between these two different analysis settings (Fig 3A/3B vs. Fig 3C/3D), we examined genes that were coherently altered by EtOH treatment (Fig 3E). The magenta module (Fig 3A) and the blue module (Fig 3D) represent genes downregulated in rosette cells upon EtOH exposure. Likewise, the salmon module (Fig 3B) and the black module (Fig 3D) represent genes downregulated in NPCs upon treatment with EtOH. The result showed list of genes that were similarly affected by EtOH treatment in two analysis settings.
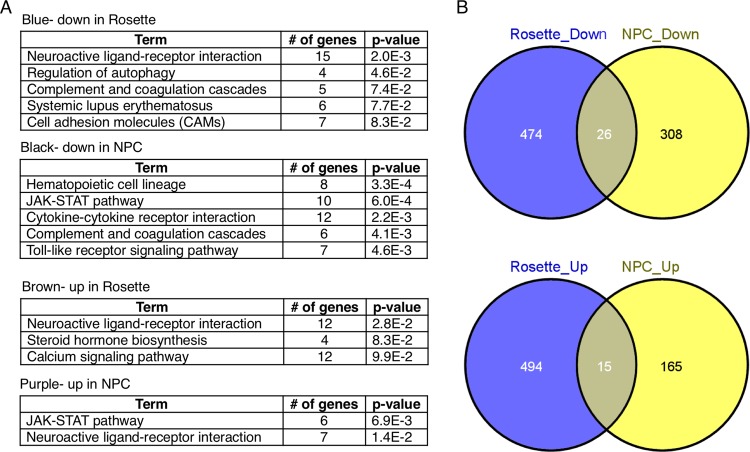
To examine potential effects of EtOH on biological process, we have performed DAVID (The Database for Annotation, Visualization and Integrated Discovery) on genes from the blue (downregulated in neural rosette), the black (downregulated in NPC), the brown (upregulated in neural rosette) and the purple (upregulated in NPC). EtOH treatment in neural rosettes showed alterations of genes potentially involved in neuroactive ligand-receptor interaction, cell adhesion molecules and calcium signaling pathway (Fig 4A). The black module of downregulated genes in NPC upon EtOH treatment was associated with several important signaling pathways, such as JAK-STAT, cytokine-cytokine receptor interaction and Toll-like receptor (TLR) signaling (Fig 4A). On the other hand, the purple module of upregulated genes showed association with neuroactive ligand-receptor interaction (Fig 4A). To examine if there is any concordance in molecular networks in NPCs and neural rosettes affected by EtOH treatment, we have identified common genes in rosettes and NPCs that showed greater than 3-fold changes upon exposure to EtOH by using Venny 2.0.2 (Computational Genomics Service) (Fig 4B). We identified core genes that are similarly regulated by EtOH treatment in rosette cells and NPCs by combining the genes in the blue and the black module for commonly downregulated genes (>3-fold) and by combining the genes in the brown and the purple module for commonly upregulated genes (>3-fold). List of genes are shown in Table 1.
Fig 4. DAVID function analysis of identified genes.
A. DAVID functional analysis was performed to examine potential functional association of identified genes. Genes in the four major modules (black and purple modules from NPC model and blue and brown modules from neural rosette model) were analyzed. It was found that genes dysregulated upon EtOH treatment in both NPCs and neural rosettes are significantly associated with neuroactive ligand-receptor interaction, JAK-STAT pathway and cytokine-cytokine receptor interaction and TLR signaling pathway. B. Genes that are commonly affected by EtOH (greater than 3-fold) in neural rosette and NPC were identified by Venny plot. List of genes are shown in Table 1.
Table 1. List of genes that are commonly dysregulated in NPC and neural rosette after EtOH treatment.
| Down >3 NPC and Rosette | Up >3 NPC and Rosette |
|---|---|
| C11orf21 | TSHR |
| C19orf23 | ALS2CR11 |
| CD36 | DDO |
| CD84 | DOC2B |
| CLCN1 | DOCK4 |
| CNGB3 | FKSG83 |
| COL4A3 | KCNE3 |
| FLJ32255 | KLF14 |
| GPR84 | LIN9 |
| HNRNPD | MAP1LC3B2 |
| IFNA2 | OPRM1 |
| ITPRIPL2 | RHAG |
| LOC100506517 | SCARA5 |
| LOC100507226 | TNP2 |
| LOC253039 | UPB1 |
| LOC283079 | |
| LOC283674 | |
| MACC1 | |
| NBLA00301 | |
| NPAS3 | |
| OFCC1 | |
| PCBD2 | |
| TLR3 | |
| TNS1 | |
| TRD@ | |
| ZAN |
Validation of candidate genes differentially regulated by EtOH treatment in NPCs
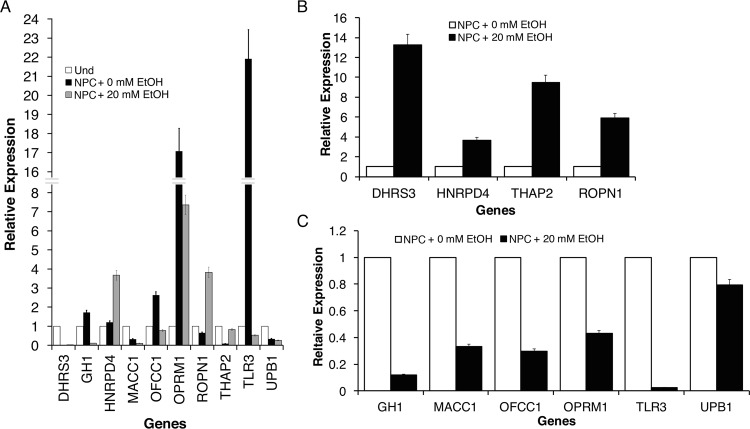
Candidate genes potentially regulated by EtOH treatment during neural differentiation were validated by qRT-PCR analysis (Fig 5). Selected genes were first validated in undifferentiated hESC, NPC with 0 mM EtOH and NPC with 20 mM EtOH. As shown in Fig 5A, we observed significant alterations associated with neural differentiation (neural diff + 0 mM EtOH) compared to undifferentiated control. To better examine the effect of EtOH on gene regulation, NPCs were treated with 20 mM EtOH and relative expression levels of candidate genes were compared to their respective controls (0 mM EtOH) (Fig 5B and 5C). Genes DHRS3, HNRPD4, THAP2 and ROPN1 demonstrated alcohol induced upregulated expression (Fig 5B). By contrast, downregulated expression was seen in the EtOH treatment of GH1, MACC1, OFCC1, OPRM1, TLR3 and UPB1 (Fig 5C).
Fig 5. Identification of candidates and validation.
qRT-PCR validation of candidate genes that are potentially regulated by EtOH treatment during neural differentiation of hESCs. Some of validation analysis results are shown. From WGCNA we have identified genes that are significantly affected by EtOH treatment in neural stem cells. A. Top candidate genes identified showed the effect of EtOH on gene levels in validation analysis by comparing undifferentiated hESCs, untreated NPCs and EtOH treated NPCs. Some of validation result by comparing the level of genes in NPCs with and without EtOH treatment showed B. upregulation and C. downregulation upon EtOH treatment. We have identified DHRS3, MACC1, THAP2, TLR3 and DRD4 are most significantly and consistently validated genes among candidates. Bars are mean ± SEM from triplicates; significant (p<0.05) difference from control (one-way ANOVA).
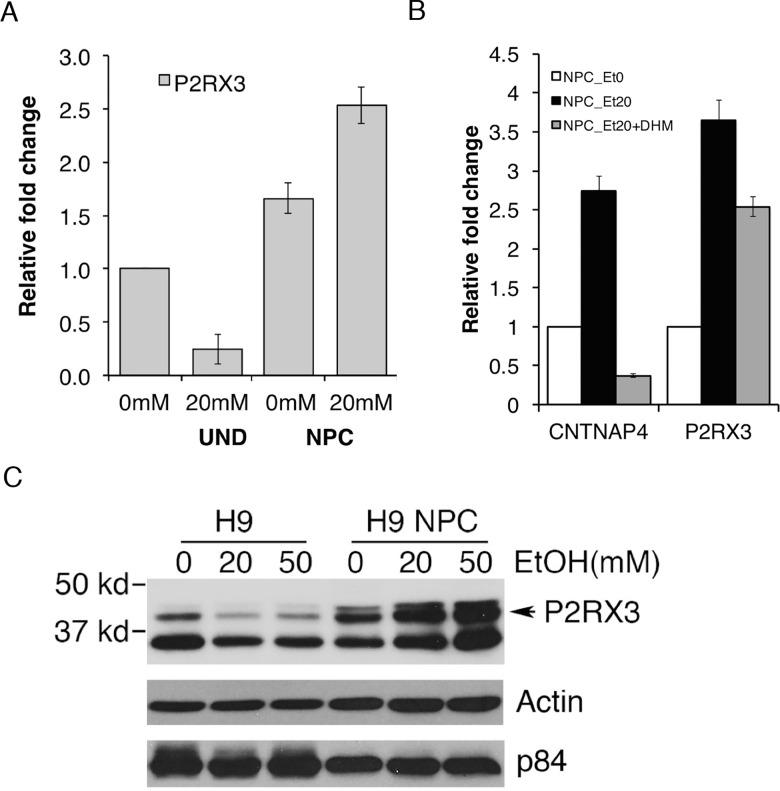
The effect of EtOH on the expression of one of the candidate genes, P2RX3, was validated by both qRT-PCR analysis and western analysis. The P2RX3 gene encodes a purinergic receptor which functions as a ligand-gated ion channel and mediates peripheral pain responses [27]. EtOH has been shown to epigenetically alter P2RX3, which may have downstream effects on the regulation of ion transport and may lead to reductions in hESC pluripotency [20]. We suggest it may also have a functional significance in alcohol-induced neuropathy. qRT-PCR analysis was performed to compare the relative fold changes of P2RX3 expression in undifferentiated H1 hESC and hESC-derived NPC treated with EtOH. P2RX3 in undifferentiated hESC was downregulated and conversely upregulated in NPC with increased EtOH concentration. (Fig 6A). We included 50 mM EtOH to examine if there is a dose dependency or any potential variations in response to different doses of EtOH. CNTNAP4 and P2RX3 genes are among upregulated genes that are upregulated by EtOH treatment. To examine if the alteration is due to EtOH’s effect we have concomitantly treated NPCs with EtOH and dihydromyricetin (DHM), a flavonoid component of herbal medicines that counters acute EtOH intoxication [28]. As shown in Fig 6B, NPCs treated with 20 mM EtOH showed induced level of CNTNAP4 and P2RX3 mRNA compared to untreated NPC. The induction was reduced by co-treatment of 1 μM DHM. Furthermore, undifferentiated H9 hESCs and H9 NPC cell lines were treated with 0, 20, and 50 mM EtOH for 48 hrs. From Western analysis, we have confirmed alcohol-induced downregulation of P2RX3 in undifferentiated hESCs and upregulation in hESC-derived NPCs compared to beta-actin or p84 expression (Fig 6C). This result demonstrates P2RX3 gene as one of molecular targets that are potentially deregulated by EtOH during neural differentiation process.
Fig 6. Ethanol induced-alteration of P2RX3 in undifferentiated hESCs and NPCs.
A. Comparison of fold changes of P2RX3 expression in undifferentiated H1 hESCs and hESC-derived NPCs treated with EtOH. P2RX3 in undifferentiated hESCs was downregulated and conversely upregulated in NPCs with increased EtOH concentration. Bars are mean ± SEM from triplicates; significant (p<0.05) difference from control (one-way ANOVA). B. The level of CNTNAP4 and P2RX3 in H1 NPC was induced by treatment with 20 mM EtOH. Co-treatment with 1 μM dihydromyricetin (DHM), a flavonoid component of herbal medicines that counters acute EtOH intoxication, significantly interfered the effect of EtOH, demonstrating the specificity of EtOH’s effect on the gene expression. Bars are mean ± SEM from triplicates; significant (p<0.05) difference from control (one-way ANOVA). C. Relative protein levels of P2RX3 in undifferentiatied H9 hESCs and H9 NPC cell lines treated with 0, 20, and 50 mM EtOH for 48 hrs. From Western analysis, we have confirmed alcohol-induced downregulation of P2RX3 in undifferentiated hESCs and upregulation in NPCs.
Discussion
Various factors, signaling molecules, and other medias have been applied to differentiate ESCs into various neural precursor cells (NPCs). Some of them include: stromal-derived inducing cells, retinoic acid, bone morphogenetic protein, fibroblast growth factor receptors, and neurobasal medium [29]. ESCs can differentiate into the columnar neuroectodermal cells known as neural rosettes and can mimic in vivo neuroectodermal development in terms of timing and morphology [30]. Neural rosette cells can proliferate by self-renewal over a certain period of time and contain neural progenitor cells, which can further differentiate into more restricted progenitors leading to neuronal and glial cell lines[31, 32]. NPCs express high levels of neuroepithelial markers such as nestin, Pax6, Sox1, and maintain expression of Sox2 [33]. Differentiation potential of rosette cells diminishes with proliferation, occurring in vivo during neural development. Currently, there are many studies that focus on differentiation of ESCs for specific regions of neural cells as the nervous system consists of different regions.
Neural differentiation of ESC and neurogenesis pathway integrates complex epigenetic processes that require strict spatial and temporal cues. Major epigenetic mechanisms including DNA methylation, covalent histone posttranscriptional modifications, chromatin organization, and non-coding regulatory RNA play critical role in pluripotency maintenance and cell fate determination[34]. For instance, decreased DNA methylation activates neuronal genes such as Sox2 during ESC differentiation. More importantly, high levels of DNMT1 have been found to be crucial in DNA methylation during neural development. Furthermore, histone acetyl transferases and histone deacetylases are also known to be key players in regulation of ESC differentiation. Yet, the exact role of histone acetylation in embryonic development in terms of NSCs and brain development has not been clearly elucidated [35].
In relation to this study, epigenetic regulation of the neural transcriptome and the alcohol interference has recently studied and discussed widely. Resendiz et al. have elaborated on the alterations in neural stem cell development and on how alcohol leads to neurodevelopment deficits through neuroepigenetics [36]. As alcohol inhibits the differentiation of NSCs, cellular growth, migration, and cell viability are all negatively affected [37]. In specific, NSCs treated with EtOH exhibit cell cycle delays, reduced NSC proliferation and increased DNA fragmentation. In addition, EtOH are found to block the intrinsic hypermethylation of the cell cycle genes such as Adra1a, Tnf, Pik3r1, and Sh3bp2 during differentiation [38]. Moreover, the pluripotency genes Oct4, Sox2, and Nanog have demonstrated an EtOH-specific delay of down-regulation in NSC models [36].
Additionally, gene expression analysis of hESCs that differentiate into neural cells has facilitated in further defining the molecular mechanisms of neural development. Nestin and βIII-tubulin (TUBB3), which are cytoskeleton proteins, are marker proteins of neural stem cells and neurons respectively [39]. Nestin, expressed by neural progenitor cells, is specifically involved in the cytoskeleton remodeling and central/peripheral nervous system development. It has been found that nestin deficiency leads to embryonic lethality through the developmental defect of neural tubes with low NSCs. On the other hand, βIII-tubulin is an identifier for neurons that is also involved in neural development. In fact, mutations in the tubulin-encoding gene TUBB3 are found to yield a set of disease symptoms via microtubule dynamics in neurons [40]. Interestingly, it has been reported that there is a contrast in expression patterns of the two genes in the NPCs derived from the hESC H9 cell line: Nestin showed high expression while βIII-tubulin showed low expression, as well as random distribution[39]. It has also been noted that nestin is highly expressed during early stages of neural development but becomes downregulated in mature neurons whereas βIII-tubulin is expressed robustly in the later stages of the neurogenesis in both the cell soma and the neurite-like structures. Besides nestin and tubulin, there are other classic pluripotency markers such as Oct4, Sox2, ALP, and Nanog, as well as Sox1 and Pax6, which are upregulated during neural induction.
Our study was aimed to assess molecular effect of alcohol on the process of neural differentiation. Potential teratogenic effects of alcohol on fetal development have been documented. Especially studies have demonstrated deleterious effect of ethanol exposure on neuronal development in animal models and on the maintenance and differentiation of neuronal precursor cells derived from stem cells. Molecular and cellular effects of alcohol on neural stem cells and neural progenitors have been demonstrated [17, 20, 37, 38]. Several studies have shown that alcohol has a significant molecular effect on neural physiology. However, it has not been studied if alcohol exposure in utero has any molecular effect on the onset of the lineage specification of hESCs into neural precursors. Our recent study using hESCs demonstrated significant ethanol-induced molecular alterations in hESCs, especially through epigenetic mechanism [20].
In this study, we have generated neural stem cells from pluripotent hESCs and examined global transcriptomic signature changes affected by ethanol treatment (Gene Expression Omnibus under GSE56906), which will provide scientific insight on potential molecular effects of fetal alcohol exposure on neural differentiation of early embryo development [24]. In particular, we have identified and verified several candidate genes that alcohol could interfere in neural stem cell regulation. Major molecular pathways in NPCs affected by EtOH have been demonstrated as associated with alcohol exposure, such as JAK-STAT signaling pathway [41–43], neuroactive ligand-receptor interaction [20, 44, 45]and Toll-like receptor (TLR) signaling pathway [46, 47]. Among verified genes, we have further examined EtOH’s effect on CNTNAP4 and P2RX3. CNTNAP4 is Contactin Associated Protein-Like 4 and belongs to the neurexin family, members of which function in the vertebrate nervous system as cell adhesion molecules and receptors. It is a presynaptic protein involved in both dopaminergic synaptic transmission and GABAergic system, thereby participating in the structural maturation of inhibitory interneuron synapses. Involved in the dopaminergic synaptic transmission by attenuating dopamine release through a presynaptic mechanism. Studies have demonstrated a potential association of CNTNAP4 autism [48–50] or alcohol dependence. P2RX3, Purinergic Receptor P2X, Ligand Gated Ion Channel 3, belongs to the family of purinoceptors for ATP. It functions as a ligand-gated ion channel and may transduce ATP-evoked nociceptor activation [51, 52]. It needs to be demonstrated how these genes are dysregulated in NPCs upon EtOH exposure and what are the significance of their dysregulation in mediating EtOH’s effect on NPC development. Further functional assessment using our model will help us delineate molecular mechanisms and understand cellular consequence of alcohol’s effect on neural stem cell regulation.
Supporting Information
A. The level of neural makers (Nestin, Tuj1 and Musashi1) was assessed by qRT-PCR in rosette and NPCs and compared to undifferentiated hESCs. B. The effect of 20 mM EtOH treatment on neural markers (Nestin, Tuj1 and Musashi1) in neural rosette and NPCs was determined by qRT-PCR assay. C. The level of pluripotent markers, Oct4 and Sox2, was assessed in undifferentiated hESC (Und) and NPCs without (0 mM) or with (20 mM) EtOH treatment. Bars are mean ± SEM from triplicates; the asterisk denotes significant (p<0.05) difference from control (one-way ANOVA).
(TIFF)
Acknowledgments
We acknowledge the UCLA Broad Stem Cell Institute for providing us with human embryonic stem cell cultures. This work was supported by NIH awards from NIAAA (R01AA021301, R21AA024210), CIRM Basic Biology Award II (RB02-1562), and UCLA School of Dentistry Faculty Seed Grant Award to Y.K.
Data Availability
Data are available from the Gene Expression Omnibus (GEO) database under accession number GSE56906.
Funding Statement
This study was supported by NIH awards from NIAAA (R01AA021301, R21AA024210) (https://www.niaaa.nih.gov), CIRM Basic Biology Award II (RB02-1562) (https://www.cirm.ca.gov) and UCLA School of Dentistry Faculty Seed Grant Award to YK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hoyme HE, May PA, Kalberg WO, Kodituwakku P, Gossage JP, Trujillo PM, et al. A practical clinical approach to diagnosis of fetal alcohol spectrum disorders: clarification of the 1996 institute of medicine criteria. Pediatrics. 2005;115(1):39–47. Epub 2005/01/05. doi: 115/1/39 [pii] 10.1542/peds.2004-0259 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haycock PC. Fetal alcohol spectrum disorders: the epigenetic perspective. Biology of reproduction. 2009;81(4):607–17. Epub 2009/05/22. doi: biolreprod.108.074690 [pii] 10.1095/biolreprod.108.074690 . [DOI] [PubMed] [Google Scholar]
- 3.Haycock PC, Ramsay M. Exposure of mouse embryos to ethanol during preimplantation development: effect on DNA methylation in the h19 imprinting control region. Biology of reproduction. 2009;81(4):618–27. Epub 2009/03/13. doi: biolreprod.108.074682 [pii] 10.1095/biolreprod.108.074682 . [DOI] [PubMed] [Google Scholar]
- 4.Cudd TA. Animal model systems for the study of alcohol teratology. Exp Biol Med (Maywood). 2005;230(6):389–93. Epub 2005/06/16. doi: 230/6/389 [pii]. . [DOI] [PubMed] [Google Scholar]
- 5.Padmanabhan R, Hameed MS. Effects of acute doses of ethanol administered at pre-implantation stages on fetal development in the mouse. Drug and alcohol dependence. 1988;22(1–2):91–100. Epub 1988/10/01. doi: 0376-8716(88)90042-7 [pii]. . [DOI] [PubMed] [Google Scholar]
- 6.Resnicoff M, Rubini M, Baserga R, Rubin R. Ethanol inhibits insulin-like growth factor-1-mediated signalling and proliferation of C6 rat glioblastoma cells. Laboratory investigation; a journal of technical methods and pathology. 1994;71(5):657–62. Epub 1994/11/01. . [PubMed] [Google Scholar]
- 7.Armant DR, Saunders DE. Exposure of embryonic cells to alcohol: contrasting effects during preimplantation and postimplantation development. Seminars in perinatology. 1996;20(2):127–39. Epub 1996/04/01. . [DOI] [PubMed] [Google Scholar]
- 8.Goodlett CR, Horn KH. Mechanisms of alcohol-induced damage to the developing nervous system. Alcohol research & health: the journal of the National Institute on Alcohol Abuse and Alcoholism. 2001;25(3):175–84. Epub 2002/01/29. . [PMC free article] [PubMed] [Google Scholar]
- 9.Wozniak DF, Hartman RE, Boyle MP, Vogt SK, Brooks AR, Tenkova T, et al. Apoptotic neurodegeneration induced by ethanol in neonatal mice is associated with profound learning/memory deficits in juveniles followed by progressive functional recovery in adults. Neurobiology of disease. 2004;17(3):403–14. Epub 2004/12/02. doi: S0969-9961(04)00191-3 [pii] 10.1016/j.nbd.2004.08.006 . [DOI] [PubMed] [Google Scholar]
- 10.Abel E. Paternal contribution to fetal alcohol syndrome. Addiction biology. 2004;9(2):127–33; discussion 35–6. Epub 2004/06/30. 10.1080/13556210410001716980 NFQYKAH2ML7W08GW [pii]. . [DOI] [PubMed] [Google Scholar]
- 11.Garro AJ, McBeth DL, Lima V, Lieber CS. Ethanol consumption inhibits fetal DNA methylation in mice: implications for the fetal alcohol syndrome. Alcoholism, clinical and experimental research. 1991;15(3):395–8. Epub 1991/06/01. . [DOI] [PubMed] [Google Scholar]
- 12.Bielawski DM, Zaher FM, Svinarich DM, Abel EL. Paternal alcohol exposure affects sperm cytosine methyltransferase messenger RNA levels. Alcoholism, clinical and experimental research. 2002;26(3):347–51. Epub 2002/03/30. . [PubMed] [Google Scholar]
- 13.Kaminen-Ahola N, Ahola A, Maga M, Mallitt K-A, Fahey P, Cox TC, et al. Maternal ethanol consumption alters the epigenotype and the phenotype of offspring in a mouse model. PLoS genetics. 2010;6(1):e1000811 Epub 2010/01/20. 10.1371/journal.pgen.1000811 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–7. . [DOI] [PubMed] [Google Scholar]
- 15.Mountford JC. Human embryonic stem cells: origins, characteristics and potential for regenerative therapy. Transfus Med. 2008;18(1):1–12. 10.1111/j.1365-3148.2007.00807.x . [DOI] [PubMed] [Google Scholar]
- 16.Verlinsky Y, Strelchenko N, Kukharenko V, Rechitsky S, Verlinsky O, Galat V, et al. Human embryonic stem cell lines with genetic disorders. Reprod Biomed Online. 2005;10(1):105–10. . [DOI] [PubMed] [Google Scholar]
- 17.Arzumanyan A, Anni H, Rubin R, Rubin E. Effects of ethanol on mouse embryonic stem cells. Alcoholism, clinical and experimental research. 2009;33(12):2172–9. Epub 2009/09/22. doi: ACER1057 [pii] 10.1111/j.1530-0277.2009.01057.x . [DOI] [PubMed] [Google Scholar]
- 18.Krishnamoorthy M, Gerwe BA, Scharer CD, Heimburg-Molinaro J, Gregory F, Nash RJ, et al. Low ethanol concentration alters CHRNA5 RNA levels during early human development. Reproductive toxicology (Elmsford, NY). 2010;30(3):489–92. Epub 2010/05/05. doi: S0890-6238(10)00089-4 [pii] 10.1016/j.reprotox.2010.04.011 . [DOI] [PubMed] [Google Scholar]
- 19.Talens-Visconti R, Sanchez-Vera I, Kostic J, Perez-Arago MA, Erceg S, Stojkovic M, et al. Neural differentiation from human embryonic stem cells as a tool to study early brain development and the neuroteratogenic effects of ethanol. Stem Cells Dev. 2010. Epub 2010/05/25. 10.1089/scd.2010.0037 . [DOI] [PubMed] [Google Scholar]
- 20.Khalid O, Kim JJ, Kim HS, Hoang M, Tu TG, Elie O, et al. Gene expression signatures affected by alcohol-induced DNA methylomic deregulation in human embryonic stem cells. Stem Cell Res. 2014;12(3):791–806. 10.1016/j.scr.2014.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrera DG, Yague AG, Johnsen-Soriano S, Bosch-Morell F, Collado-Morente L, Muriach M, et al. Selective impairment of hippocampal neurogenesis by chronic alcoholism: protective effects of an antioxidant. Proc Natl Acad Sci U S A. 2003;100(13):7919–24. 10.1073/pnas.1230907100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nixon K, Crews FT. Binge ethanol exposure decreases neurogenesis in adult rat hippocampus. J Neurochem. 2002;83(5):1087–93. . [DOI] [PubMed] [Google Scholar]
- 23.Roubal I, Park SJ, Kim Y. Derivation of Neural Precursor Cells from Human Embryonic Stem Cells for DNA Methylomic Analysis. Methods Mol Biol. 2014. Epub 2014/12/19. 10.1007/7651_2014_152 . [DOI] [PubMed] [Google Scholar]
- 24.Kim JJ, Duan L, Tu TG, Elie O, Kim Y, Mathiyakom N, et al. Molecular effect of ethanol during neural differentiation of human embryonic stem cells in vitro. Genom Data. 2014;2:139–43. 10.1016/j.gdata.2014.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones AW, Sternebring B. Kinetics of ethanol and methanol in alcoholics during detoxification. Alcohol Alcohol. 1992;27(6):641–7. Epub 1992/11/01. . [PubMed] [Google Scholar]
- 26.Thomson M, Liu SJ, Zou LN, Smith Z, Meissner A, Ramanathan S. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145(6):875–89. 10.1016/j.cell.2011.05.017 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fabbretti E, Nistri A. Regulation of P2X3 receptor structure and function. CNS Neurol Disord Drug Targets. 2012;11(6):687–98. . [DOI] [PubMed] [Google Scholar]
- 28.Shen Y, Lindemeyer AK, Gonzalez C, Shao XM, Spigelman I, Olsen RW, et al. Dihydromyricetin as a novel anti-alcohol intoxication medication. J Neurosci. 2012;32(1):390–401. 10.1523/JNEUROSCI.4639-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dhara SK, Stice SL. Neural differentiation of human embryonic stem cells. J Cell Biochem. 2008;105(3):633–40. 10.1002/jcb.21891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li XJ, Zhang SC. In vitro differentiation of neural precursors from human embryonic stem cells. Methods Mol Biol. 2006;331:169–77. 10.1385/1-59745-046-4:168 . [DOI] [PubMed] [Google Scholar]
- 31.Elkabetz Y, Panagiotakos G, Al Shamy G, Socci ND, Tabar V, Studer L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22(2):152–65. 10.1101/gad.1616208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19(12):1129–33. 10.1038/nbt1201-1129 . [DOI] [PubMed] [Google Scholar]
- 33.Noisa P, Ramasamy TS, Lamont FR, Yu JS, Sheldon MJ, Russell A, et al. Identification and characterisation of the early differentiating cells in neural differentiation of human embryonic stem cells. PLoS One. 2012;7(5):e37129 10.1371/journal.pone.0037129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toraño EG, Fernandez, A. F., Urdinguio, R. G., & Fraga, M. F. Role of Epigenetics in Neural Differentiation: Implications for Health and Disease. 2014.
- 35.Lilja AM, Luo Y, Yu QS, Rojdner J, Li Y, Marini AM, et al. Neurotrophic and neuroprotective actions of (-)- and (+)-phenserine, candidate drugs for Alzheimer's disease. PLoS One. 2013;8(1):e54887 10.1371/journal.pone.0054887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Resendiz M, Mason S, Lo CL, Zhou FC. Epigenetic regulation of the neural transcriptome and alcohol interference during development. Front Genet. 2014;5:285 10.3389/fgene.2014.00285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell JC, Stipcevic T, Flores RE, Perry C, Kippin TE. Alcohol exposure inhibits adult neural stem cell proliferation. Exp Brain Res. 2014;232(9):2775–84. 10.1007/s00221-014-3958-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou FC, Balaraman Y, Teng M, Liu Y, Singh RP, Nephew KP. Alcohol alters DNA methylation patterns and inhibits neural stem cell differentiation. Alcohol Clin Exp Res. 2011;35(4):735–46. 10.1111/j.1530-0277.2010.01391.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu C, Zhong Y, Apostolou A, Fang S. Neural differentiation of human embryonic stem cells as an in vitro tool for the study of the expression patterns of the neuronal cytoskeleton during neurogenesis. Biochem Biophys Res Commun. 2013;439(1):154–9. 10.1016/j.bbrc.2013.07.130 . [DOI] [PubMed] [Google Scholar]
- 40.Tischfield MA, Baris HN, Wu C, Rudolph G, Van Maldergem L, He W, et al. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell. 2010;140(1):74–87. 10.1016/j.cell.2009.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeVito WJ, Stone S. Ethanol inhibits prolactin-induced activation of the JAK/STAT pathway in cultured astrocytes. J Cell Biochem. 1999;74(2):278–91. Epub 1999/07/15. [pii]. . [DOI] [PubMed] [Google Scholar]
- 42.Ward RJ, Lallemand F, de Witte P. Biochemical and neurotransmitter changes implicated in alcohol-induced brain damage in chronic or 'binge drinking' alcohol abuse. Alcohol Alcohol. 2009;44(2):128–35. Epub 2009/01/22. 10.1093/alcalc/agn100 agn100 [pii]. . [DOI] [PubMed] [Google Scholar]
- 43.Nicolas CS, Amici M, Bortolotto ZA, Doherty A, Csaba Z, Fafouri A, et al. The role of JAK-STAT signaling within the CNS. JAKSTAT. 2013;2(1):e22925 Epub 2013/09/24. 10.4161/jkst.22925 2012JAKS0028R [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biernacka JM, Geske J, Jenkins GD, Colby C, Rider DN, Karpyak VM, et al. Genome-wide gene-set analysis for identification of pathways associated with alcohol dependence. Int J Neuropsychopharmacol. 2013;16(2):271–8. Epub 2012/06/22. 10.1017/S1461145712000375 S1461145712000375 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang R, Miao Q, Wang C, Zhao R, Li W, Haile CN, et al. Genome-wide DNA methylation analysis in alcohol dependence. Addict Biol. 2013;18(2):392–403. Epub 2013/02/08. 10.1111/adb.12037 . [DOI] [PubMed] [Google Scholar]
- 46.Tang SC, Arumugam TV, Xu X, Cheng A, Mughal MR, Jo DG, et al. Pivotal role for neuronal Toll-like receptors in ischemic brain injury and functional deficits. Proc Natl Acad Sci U S A. 2007;104(34):13798–803. Epub 2007/08/19. 0702553104 [pii] 10.1073/pnas.0702553104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin L, Crews FT. Chronic ethanol increases systemic TLR3 agonist-induced neuroinflammation and neurodegeneration. J Neuroinflammation. 2012;9:130 Epub 2012/06/20. 10.1186/1742-2094-9-130 1742-2094-9-130 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Roak BJ, State MW. Autism genetics: strategies, challenges, and opportunities. Autism Res. 2008;1(1):4–17. Epub 2008/02/01. 10.1002/aur.3 . [DOI] [PubMed] [Google Scholar]
- 49.O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485(7397):246–50. Epub 2012/04/13. 10.1038/nature10989 nature10989 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karayannis T, Au E, Patel JC, Kruglikov I, Markx S, Delorme R, et al. Cntnap4 differentially contributes to GABAergic and dopaminergic synaptic transmission. Nature. 2014;511(7508):236–40. Epub 2014/05/30. nature13248 [pii] 10.1038/nature13248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.North RA. Molecular physiology of P2X receptors. Physiol Rev. 2002;82(4):1013–67. Epub 2002/09/25. 10.1152/physrev.00015.2002 . [DOI] [PubMed] [Google Scholar]
- 52.Lewis C, Neidhart S, Holy C, North RA, Buell G, Surprenant A. Coexpression of P2X2 and P2X3 receptor subunits can account for ATP-gated currents in sensory neurons. Nature. 1995;377(6548):432–5. Epub 1995/10/05. 10.1038/377432a0 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. The level of neural makers (Nestin, Tuj1 and Musashi1) was assessed by qRT-PCR in rosette and NPCs and compared to undifferentiated hESCs. B. The effect of 20 mM EtOH treatment on neural markers (Nestin, Tuj1 and Musashi1) in neural rosette and NPCs was determined by qRT-PCR assay. C. The level of pluripotent markers, Oct4 and Sox2, was assessed in undifferentiated hESC (Und) and NPCs without (0 mM) or with (20 mM) EtOH treatment. Bars are mean ± SEM from triplicates; the asterisk denotes significant (p<0.05) difference from control (one-way ANOVA).
(TIFF)
Data Availability Statement
Data are available from the Gene Expression Omnibus (GEO) database under accession number GSE56906.