Abstract
The Virus Pathogen Resource (ViPR; www.viprbrc.org) and Influenza Research Database (IRD; www.fludb.org) have developed a metadata-driven Comparative Analysis Tool for Sequences (meta-CATS), which performs statistical comparative analyses of nucleotide and amino acid sequence data to identify correlations between sequence variations and virus attributes (metadata). Meta-CATS guides users through: selecting a set of nucleotide or protein sequences; dividing them into multiple groups based on any associated metadata attribute (e.g. isolation location, host species); performing a statistical test at each aligned position; and identifying all residues that significantly differ between the groups. As proofs of concept, we have used meta-CATS to identify sequence biomarkers associated with dengue viruses isolated from different hemispheres, and to identify variations in the NS1 protein that are unique to each of the 4 dengue serotypes. Meta-CATS is made freely available to virology researchers to identify genotype-phenotype correlations for development of improved vaccines, diagnostics, and therapeutics.
Keywords: Dengue, DENV, bioinformatics, virology, comparative genomics, statistical comparison, virus, database
Introduction
The symbiotic relationship between traditional wet-bench research and bioinformatics is being fueled by a large amount of available sequence data and the need to analyze and interpret such a wealth of information. To aid in the analysis of this data deluge, it is imperative that standardized annotations describing the characteristics of the isolated strains (metadata) are recorded, reported and associated with the corresponding sequence record. For example, interrogations into the emergence and seasonality of new infectious disease outbreaks can be conducted only if metadata describing the time and place of isolation is available (Bloom-Feshbach et al., 2013; Patel et al., 2013); identification of sequence substitutions that correlate with host specificity requires pathogen host species information (Shi and Hu, 2008); and investigations of sequence biomarkers that contribute to pathogen virulence and drug resistance can only be performed if the host’s health status and medical history are available (Ansari et al., 2013; Kaminski et al., 2013; Schvoerer et al., 2013). Access to additional metadata will lead to increased data exploration and hypothesis generation that can then be subjected to ‘wet lab’ experiments to validate the observations, biochemically identify the mechanism of actions and augment the existing knowledge base of the systems being studied.
To anticipate the magnitude of data produced by modern experimentation, the National Institute of Allergy and Infectious Diseases (NIAID) currently supports five Bioinformatics Resource Centers (BRC) for Infectious Diseases, each of which is charged with storing and integrating various types of data about human pathogens and disease vectors from multiple resources, and providing them to the research communities in intuitive ways. The Virus Pathogen Database and Analysis Resource (ViPR, www.viprbrc.org) contains information for viruses categorized as either select agents or as public health threats belonging to the Arenaviridae, Bunyaviridae, Caliciviridae, Coronaviridae, Flaviviridae, Filoviridae, Hepeviridae, Herpesviridae, Paramyxoviridae, Picornaviridae, Poxviridae, Reoviridae, Rhabdoviridae, and Togaviridae families (Pickett et al., 2012). Similarly, the Influenza Research Database (IRD, www.fludb.org) contains information relating specifically to influenza viruses (Squires et al., 2012). The data, together with the integrated analytical and visualization tools are made freely available to the scientific community to support both hypothesis generation and data mining to enable the research and development of diagnostics, prophylactics, vaccines and therapeutics against the supported viruses.
Dengue virus (DENV), a member of the Flaviviridae family, has emerged as a significant public health threat. DENV is transmitted among humans by the Aedes aegypti mosquito vector (Vasilakis and Weaver, 2008), and is responsible for approximately 50–100 million human infections every year (Weaver and Vasilakis, 2009). DENV is now endemic in more than 100 tropical and subtropical countries, putting more than 1/3 of the world population at risk of infection with disease incidence and severity increasing over the past few decades. There are currently 4 antigenically distinct serotypes of DENV, with each serotype further subdivided into multiple genotypes. Although multiple serotypes and genotypes can co-circulate in the same geographic area, introduction of a new serotype or genotype into an immunologically ‘naïve’ area can result in a new disease outbreak, accompanied by the establishment of a new genetic lineage with limited sequence diversity, termed a ‘founder effect’ (Allicock et al., 2012; Lee et al., 2010). In contrast to viruses like influenza that are capable of direct human-to-human transmission, the introduction of new DENV lineages in geographically dispersed regions would be expected to be relatively rare due to the vector transmission requirement.
In order to quickly and reliably identify viral sequence variations that are associated with differences in virus or host attributes, we developed the metadata-driven Comparative Analysis Tool for Sequences (meta-CATS), which segregates genomic or protein sequences into groups based on associated metadata, performs a multiple sequence alignment of all sequences (if they are not already aligned), and then calculates a chi-squared statistic for each aligned position to identify amino acids or nucleotides that significantly differ between the defined sequence groups. Here we report the application of the meta-CATS pipeline to identify statistically significant sequence biomarkers of distinct DENV geographic lineages and serotypes.
Methods
Meta-CATS method
The underlying statistical algorithm for the meta-CATS method was written in the R programming language (R Development Core Team, 2010) and consists of: 1) identifying and segregating the sequence records of interest based on the associated metadata annotations through the search interface available in ViPR or IRD; 2) performing a multiple sequence alignment (if necessary) using the Multiple Sequence Comparison by Log-Expectation (MUSCLE) and UCLUST algorithms (Edgar, 2004, 2010); 3) performing the chi-square test of independence and Pearson’s chi-square test in tandem to calculate a p-value; 4) displaying the results for the positions identified as statistically significant in a webpage; 5) permitting additional validation and orthogonal analyses using the suite of additional analysis tools integrated into the ViPR and IRD systems.
To facilitate ease-of-use for all researchers employing this statistical workflow, we designed the meta-CATS analytical pipeline to be fully automated following the segregation of the sequence records. The tabular report that is generated from a meta-CATS analysis allows fast browsing of the results according to aligned position, chi-square value, degrees of freedom, and residue diversity between groups. When protein sequences are being analyzed, links to the characterized biological function(s) that overlap with the significant residue(s) are also displayed in the results table in taxa where sequence features have been defined (Noronha et al., 2012).
Sequence data
The sequence data, together with the associated metadata, used throughout this study were obtained from the NIAID Virus Pathogen Database and Analysis Resource system (ViPR, www.viprbrc.org). The ViPR system parses GenBank records to capture sequence data and extract semi-structured metadata available in the structured comments fields. These metadata are then translated and parsed into structured database tables in the ViPR database and associated with the extracted sequence data. Searches involving host species, country of origin, virus type, year of isolation, sample source, clinical diagnosis and other criteria can then be used to retrieve and group relevant sequences for subsequent analysis with the meta-CATS analytical tool.
DENV sequence sets examined in the current study include: 1) 364 total polyprotein amino acid sequences comprised of 267 strains from the Western hemisphere and 97 strains from the Eastern hemisphere isolated between 2001 and 2010 (Supplementary Table 1), and 2) all ViPR-annotated NS1 amino acid sequences available as of January 2012. Upon further examination, groups of sequences that were 100% identical across the region being analyzed in the latter dataset were identified. All but one representative strain from each of these groups were removed from the original dataset to reduce statistical bias due to phylogenetic relatedness and over-sampling. After this process, 700 strains remained for analysis (259 DENV1, 238 DENV 2, 163 DENV 3 and 40 DENV 4) (Supplementary Table 3). In addition to the automated meta-CATS pipeline, manual edits and exploration of the multiple sequence alignment as well as verification of the reported sequence variations were performed using the ViPR implementation of Jalview (Waterhouse et al., 2009). The Immune Epitope Database (IEDB) was used to determine all regions that correspond to a curated immune epitope record (Kim et al., 2012).
Results
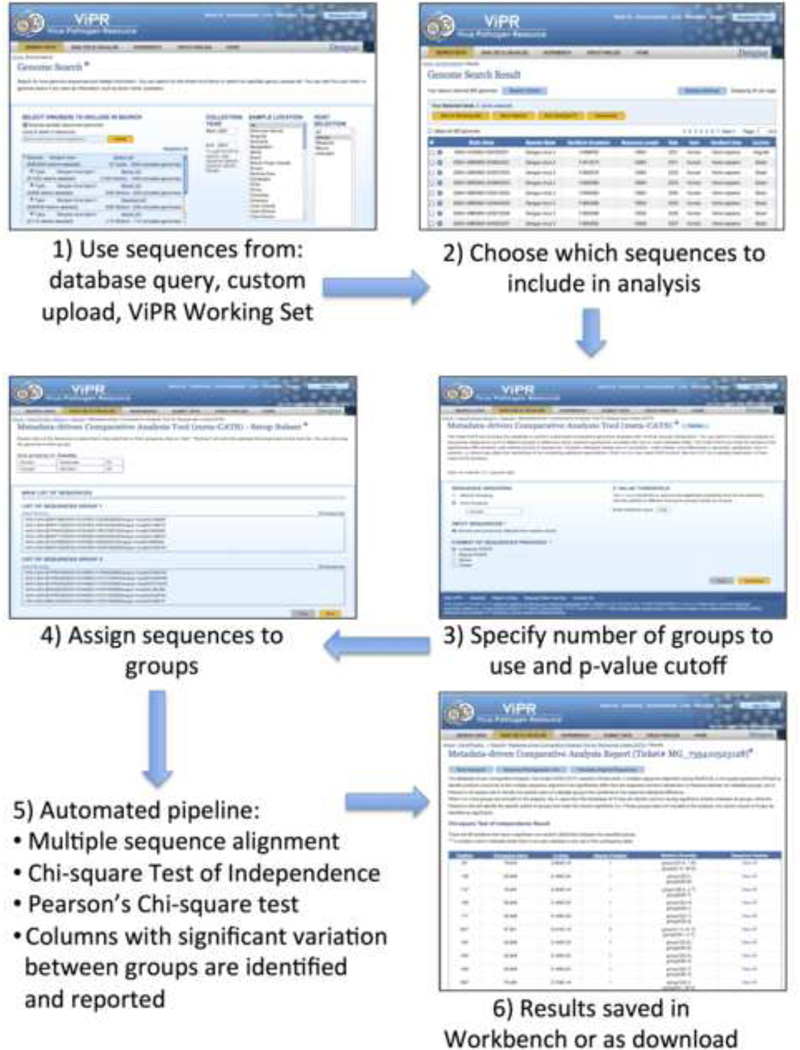
We set out to develop a universally applicable, freely available, automated tool that could be used to identify residues that have significant variation between groups of nucleotide or amino acid sequences. Meta-CATS fulfills this goal through an easy-to-navigate web-based user interface in both ViPR and IRD. The process begins by taking sequence input either from: 1) a database query, 2) existing working sets of sequences, 3) an uploaded file containing custom sequences, or a combination of these options. After selecting the desired sequences the user either divides them into 2 to 5 groups manually or instructs the system to automatically divide them based on the desired metadata criterion, and specifies a maximum probability cutoff for statistical significance (default = 0.05). If the input sequences are unaligned, a multiple sequence alignment is performed prior to calculating the statistical values. A chi-square test of independence is then performed on each non-conserved position of the alignment to identify those that differ in a statistically significant manner. For this step, the null hypothesis being tested is that the group assignment and residue are independent (i.e. knowing the nucleotide or amino acid residue at any given position will not help to predict the group to which the sequence is assigned). If the input sequences are at the nucleotide level then all 4 nucleotide possibilities are included in the calculations since, in theory, any nucleotide could be present at any given position. If gaps are present at a particular position in the alignment, then it is counted as a “5th” base in the calculations. If the input sequences are at the amino acid level, then only the residues that are observed across all sequences at that position are taken into account to determine the correct degrees of freedom for the analysis—due to the additional biological constraints that exist at the protein level. At each position where the null hypothesis is rejected (i.e. there exists statistical evidence of the dependence of the group assignment on the nucleotide or amino acid residue at that position) the data are subsequently subjected to a pairwise chi-square calculation to determine which pairs of user-defined groups contribute to the statistically significant result. In this second step, the null hypothesis being tested is that no relationship exists between group assignment and the nucleotide or amino acid residue composition at each position. Logically, the second chi-square calculation is helpful only if sequences are assigned to three or more groups since positions found to have significant variation between sequences assigned to only two groups will be identical with the results from the chi-square test of independence. In the case where three or more groups are included in the analysis, a significant result from the pairwise Pearson’s chi-square test suggests that the group assignments and sequence composition are dependent. The results from the various statistical tests are then summarized, together with the sequence variation observed for each group at that position, in a tabular form within a webpage and are made available for download in comma-separated value (csv) format (Figure 1).
Figure 1. Screenshots of the meta-CATS tool.
This diagram shows how to navigate the workflow for a meta-CATS analysis in ViPR (or IRD). The user begins by 1) searching for sequences by their associated metadata, selecting sequences to include in the analysis, specifying the number of groups and a probability cutoff value, assigning sequences to groups, and viewing the results.
Once meta-CATS development was completed, we validated the automated pipeline by using a dataset comprised of Hepatitis C virus sequences that had been subjected to a similar statistical analysis after being divided into two groups based on phylogenetic topology (Pickett et al., 2010). Meta-CATS identified the same nucleotide positions as those that were published from the original analysis.
Genotypic Differences in DENV-3 Genomes Isolated from Different Hemispheres
Phylogenetic analysis of DENV sequences derived from strains isolated from around the world has provided evidence that a combination of founder effect and geographic barriers have contributed to the establishment of distinct DENV lineages (Araujo et al., 2009; Schmidt et al., 2011). We applied the meta-CATS pipeline to a dataset of DENV type 3 (DENV-3) sequences to determine if sequence-based biomarkers could be identified that could distinguish these geographic lineages. All available DENV-3 polyprotein sequences in the ViPR database were assigned to either the Eastern hemisphere group or Western hemisphere group based on the geographical location of virus isolation metadata associated with the respective sequence records, and the sequence and group assignments submitted to the meta-CATS statistical tool.
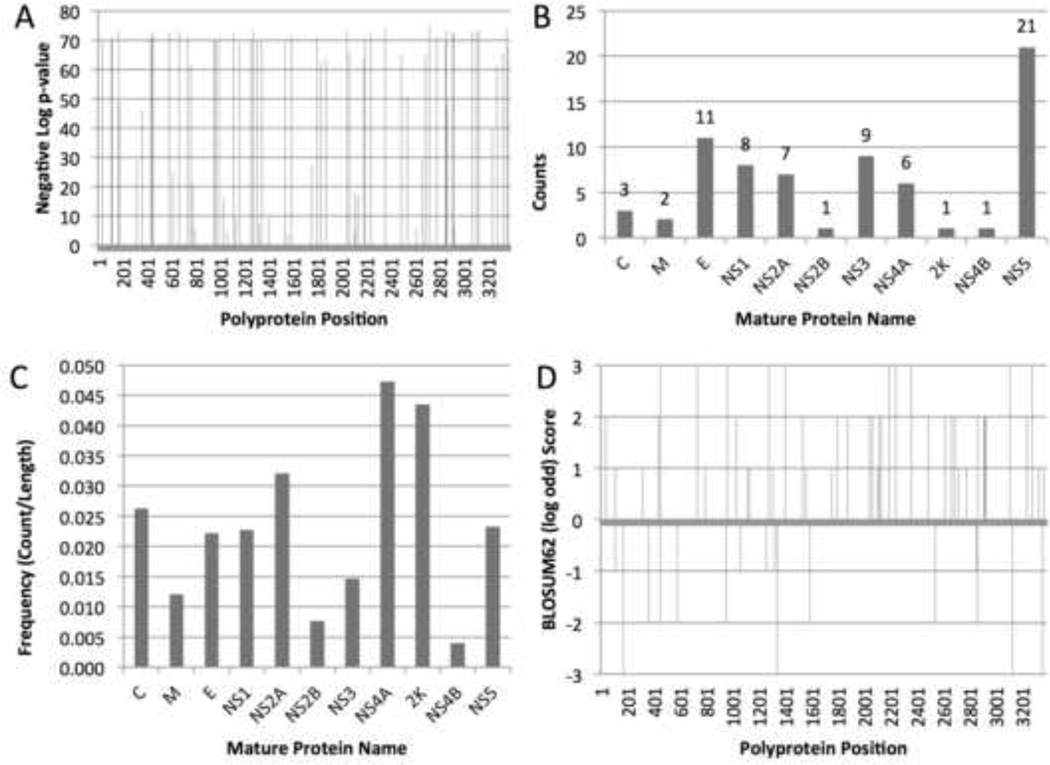
The chi-squared analysis performed by meta-CATS identified 70 amino acid positions within the polyprotein which displayed statistically significant variation between isolates from both the Eastern and Western hemispheres (Supplementary Table 1, Figure 2A). Bonferroni-adjusted probabilities from the chi-squared analysis for these positions ranged from 3.32 × 10−75 to 4.20 × 10−4 reflecting the tremendous variation in the composition of residues found in sequences isolated from both hemispheres (Bonferroni, 1936). When these amino acid positions were mapped back to the respective polyprotein cleavage products, at least one significant amino acid position was identified within each of the mature proteins, with the highest number of counts located in the NS5, E, and NS3 proteins (Figure 2B). When normalized for length, the NS2B and NS4B proteins had the lowest number of observed sequence differences between viruses isolated from the two hemispheres. This suggests that functional constraints may limit the available amino acid substitutions in these two proteins (Figure 2C). A majority of statistically significant positions (43 of 70) were also found to have positive log-odds scores from the BLOSUM62 matrix (Figure 2D), indicating that the necessary physicochemical properties of each position are preserved. This result would be expected for lineage variants that are maintained over time (Henikoff and Henikoff, 1992).
Figure 2. Statistical results for significant positions.
(A) Shows all amino acid positions that significantly differed (after Bonferroni adjustment) between the polyprotein of strains isolated from the Eastern and Western Hemispheres. (B) The number of significant amino acid positions (counts) identified by the meta-CATS tool segregated by mature proteins. (C) The frequency of significant amino acid positions identified by meta-CATS, normalized by length of the respective mature proteins. (D) The BLOSUM62 matrix scores for comparing all substitutions observed in the polyprotein sequences of strains from both hemispheres. All polyprotein positions refer to positions in the DENV-3/US/BID-V1473/2002 strain.
While inspecting the group-specific amino acid composition in each aligned position, many were found to have incomplete residue segregation—particularly within the sequences isolated from the Eastern hemisphere. The multiple sequence alignment was examined to determine whether the same sequences were consistently responsible for the segregation anomaly across multiple columns. After examination, we identified two Eastern hemisphere isolates that possessed amino acid sequences characteristic of isolates from the Western hemisphere at the vast majority of the significantly different positions. The strains responsible for this effect are: GZ1D3 isolated from Guangzhou, China in August 2009 (GU363549), and SGEHI(D3)0235Y07 isolated from Singapore in May 2007 (GU370053). When a phylogenetic tree was constructed, these isolates from the Eastern hemisphere were found in the same clade as the Western hemisphere isolates, although they form a distinct sub-clade that branches off earlier than the rest of the Western hemisphere sequences (Figure 3, Supplementary Figure 1). This implies that the two sequences are closely related to the most recent common ancestor (MRCA) for extant sequences in the two hemispheres.
Such a pattern, in which “outlier” sequences lie between two well-defined clades in a phylogenetic tree, has been attributed to genomic recombination in some cases (Pickett and Lefkowitz, 2009; Wang et al., 2010). However, the fact that the relevant amino acid variations were distributed across the entire polyprotein suggests that it is unlikely that recombination would be the cause of the topological discordance for a Western Hemisphere genotype being isolated in the Eastern Hemisphere. Indeed, an in silico recombination analysis performed using the Recombination Detection Program 3 (RDP3) (Martin et al., 2010) provided no evidence for a recombination event (data not shown).
The most likely explanation is that these “outlier” sequences are due to one or more international transmission events that occurred from the transport of either an infected host or vector between hemispheres. Given that the GZ1D3 sequence was taken from a patient living in China who had no history of recent international travel, at least two possible explanations exist: i) a separate lineage, which is an ancestor of “Western” strains, is co-circulating with extant “Eastern” viruses in Asia, or ii) a virus from the Western hemisphere was transferred to the Eastern hemisphere more recently and subsequently infected the patient of origin.
Further, the amount of DENV-3 sequence data currently available from this region of the world is insufficient to determine: i) whether these sequences are direct descendants of either the virus that was originally transferred or a more direct descendant of the MRCA, ii) whether these sequences were isolated at a point in time close to the primary transmission event, or iii) what the ancestral virus sequence might have been. Additional sequences will assist in determining which of these possibilities is most probable.
NS1 Amino Acid Residue Variations Correlating with DENV Serotypes
In a subsequent study, we used the meta-CATS pipeline to identify NS1 amino acid positions that significantly vary between all four DENV serotypes. The intent of this analysis was to identify serotype-specific NS1 peptides that could then be used to produce serotype-specific antibodies and reagents for use in a rapid clinical test. While determining the serotype responsible for a given infection will not directly affect patient treatment, subsequent infection by a different serotype can increase the risk of clinical severity, thus making it important to capture this information. In addition, multiple serotypes may be co-circulating in a given area, but only one of the serotypes will be the cause of an outbreak. Knowing the serotype in this case will support epidemiology studies and aid in tracking. The NS1 protein was selected as the target for this analysis because of its presence in the host serum, making rapid detection by early immunological tests possible. Such tests would be complementary to current and more expensive nucleic acid methodologies that require isolation of nucleic acid, enzymatic expansion of viral genome using either PCR or a similar amplification method. The detection of genome copies in a clinical sample is a technically difficult process due to the instability of RNA genomes in whole blood or serum and storage of the sample using a cold chain until later analysis. In addition, sophisticated laboratory equipment and expertise in data interpretation are also currently required to distinguish the specific virus serotype in the infected individual.
Detection of NS1 using either immunological methods or aptamers that are both specific and sensitive to each serotype in a point-of-care setting could not only improve early detection of dengue fever but also contribute to better control measures of dengue epidemics. To perform this NS1 comparative analysis using the ViPR meta-CATS implementation, we first assigned 700 non-redundant NS1 protein sequences (259 serotype 1, 238 serotype 2, 163 serotype 3 and 40 serotype 4) to four separate groups, based on their serotype classification. Analysis of the results revealed 165 amino acid residues that were dependent on group assignment; however, only 19 of these residues had significant variation existing between all possible pairwise combinations of the 4 serotypes when the Pearson chi-squared test was applied (Table 1).
Table 1.
Nineteen amino acid positions identified as significantly varying between NS1 sequences from all 4 DENV serotypes.
| NS1 Position12 |
Polyprotein Position |
P-value | Type 1 Consensus Residue (%) |
Type 2 Consensus Residue (%) |
Type 3 Consensus Residue (%) |
Type 4 Consensus Residue (%) |
|---|---|---|---|---|---|---|
| 47 | 823 | <1.0E-340 | G (100) | Q (99) | A (100) | L (98) |
| 83 | 859 | <1.0E-340 | D (97) | E (100) | N (99) | G (100) |
| 84 | 860 | <1.0E-340 | M (97) | V (97) | I (99) | H (100) |
| 98 | 874 | <1.0E-340 | A (91) | Q (99) | E (99) | T (77) |
| 103 | 879 | <1.0E-340 | M (97) | S (98) | T (98( | A (95) |
| 124 | 900 | <1.0E-340 | I (100) | L (98) | V (99) | F (100) |
| 128 | 904 | <1.0E-340 | V (66) | S (77) | T (89) | A (83) |
| 139 | 915 | 7.07E-193 | N (70) | E (99) | N (97) | D (100) |
| 146 | 922 | <1.0E-340 | D (81) | T (94) | A (90) | E (100) |
| 147 | 923 | <1.0E-340 | Q (100) | N (100) | S (97) | R(100) |
| 152 | 928 | <1.0E-340 | I (100) | S (100) | V (100) | F (50) |
| 174 | 950 | <1.0E-340 | S (100) | K (87) | V (83) | G (100) |
| 191 | 967 | <1.0E-340 | S (99) | N (95) | E (100) | Q (100) |
| 205 | 981 | <1.0E-340 | E (100) | A (94) | Q (100) | S (100) |
| 208 | 984 | <1.0E-340 | E (100) | D (100) | G (100) | Q (100) |
| 224 | 1000 | <1.0E-340 | I (79) | H (96) | T (99) | L (100) |
| 246 | 1022 | 1.62E-280 | I (95) | N (92) | S (99) | S (75) |
| 279 | 1055 | <1.0E-340 | L (99) | F (91) | Y (100) | E (100) |
| 324 | 1100 | 4.90E-324 | R (53) | R (99) | M (98) | L (100) |
NS1 amino acid position 1 corresponds to polyprotein amino acid position 776 of the DENV-2 S1 vaccine strain.
Significant positions that overlap with experimentally-determined immune epitopes from the IEDB database are highlighted in bold.
To determine if any of these positions were located in any known immune epitopes, we compared the list of 19 significant positions against information about experimentally determined immune epitopes curated by the IEDB and supported in ViPR. We found a total of 3 positions that were located in known immune epitopes characterized using various serotypes. Two of these significant positions, located at NS1 positions 124 and 128, were found within 2 separate curated B-cell epitope regions (IEDB ID 2246 and 13267) (Falconar, 2007). The remaining significant position is located within a MHC-binding epitope at NS1 position 191 (IEDB ID 29978) (Lund et al., 2011). Experimentation is currently underway to determine if peptides containing these residues can be used to develop serotype-specific reagents for serology-based diagnostics.
Discussion
In this work, we report the development of an online publicly available analysis pipeline called the metadata-driven Comparative Analysis Tool for Sequences (meta-CATS), hosted by the ViPR and IRD Bioinformatics Resource Centers, for use by the virus research community. Through the use of this tool, we have identified multiple positions located throughout the DENV-3 viral polyprotein that differ between viruses isolated in the Eastern and Western hemispheres. We also identified two sequences most likely representing at least one case of trans-continental transmission of dengue. We identified multiple positions in the DENV NS1 mature protein that significantly differ between the four virus serotypes, for potential use in the future development of a serotype-specific diagnostic tool or product.
Incorporating this web-based meta-CATS tool into a comparative genomics analytical workflow allows researchers to perform complex bioinformatics genotype-phenotype correlation analyses regardless of their training or background, and without the need to develop their own customized code. A few examples of metadata that could be used to segregate sequence groups include: clinical severity of disease, measures of antiviral drug resistance, host source, tissue tropism, geospatial point of isolation, time of isolation, virus protein binding affinity, antibody avidity, association with an epidemic, phylogenetic clade assignment, other taxonomic assignments (subtype, serotype, genotype, genogroup), etc. We anticipate the virology research community will use tools such as meta-CATS with increased frequency as the amount of sequence data and metadata continues to increase.
Meta-CATS is a data exploration and hypothesis generation tool. However, as is the case with all statistical analyses, the results are highly dependent on the quality of the data that are used as input. For this reason, we align the sequences used as input (if necessary) with the MUSCLE alignment algorithm as the default since it provides a balance between speed and accuracy with viral sequences. However, if a user pre-aligns the input sequences with a different alignment algorithm, then the chi-square statistical analysis will proceed directly without the alignment step. Additionally, it is extremely difficult for current statistical methods to account for all possible causes of sequence variations such as: co-variation, codon bias, taxonomic relationships, sampling bias, etc. while simultaneously ensuring that the underlying assumptions for the statistical test(s) are met. Consequently, users should not only educate themselves about any anomalies in the data to avoid statistical bias that could give rise to false-positive results, but they should also subject the results from any statistical analysis, including meta-CATS, to more in-depth orthogonal analyses and ‘wet-lab’ experimentation for validation.
The biological meaning of the statistical output from the meta-CATS pipeline will be dependent on at least two criteria: 1) the biological relevance of the defined groups, and 2) the sequence variation present between those groups. If insufficient sequence variation is present due either to sampling bias or viral biological constraints, the number of residues identified as divergent between the groups will be minimal or nonexistent. An increase in the number of sequences, and the relevant metadata, available in GenBank will improve the predictive ability of bioinformatics tools such as meta-CATS.
In this study, group assignments used in the current study to assess the variation-between-hemispheres were based strictly on geography. The observed scattering of DENV-3 positions that were identified as significantly different between hemispheres throughout the viral polyprotein is expected. This result could reflect a founder effect in which distinct viruses established independent lineages in each hemisphere, with many of their lineage-specific sequence differences maintained over time. However, our work also identified two sequences arising from at least one occurrence of trans-continental disease transmission between the Western and Eastern hemispheres, perhaps reflecting the ease and rapidity of long-distance travel that is currently possible. Since only two such sequences have been isolated to date, it is likely that this event happened in the not-too-distant past, when transportation of the infected host could have occurred, while viral titers were sufficiently to continue the virus transmission cycle after arriving at the new location. As additional sequence data becomes available for this group of ‘transplanted’ viruses, the evolution of the virus in both hemispheres and its ability to compete with the original endemic lineage can be better understood.
The second scientific use case described in this work, which addresses divergence in the NS1 protein between serotypes, demonstrates the application of meta-CATS to a problem with direct clinical relevance. Combining meta-CATS with other publicly available data, including 3D protein structures and immune epitope information, has helped to further refine the results and identify regions potentially capable of producing serotype-specific NS1 antibodies for use in diagnostic assays. Thus as existing computational and bioinformatics analyses move to incorporate meta-CATS into their workflows, the amount of effort required for downstream validation of the predicted high confidence “hits” will be reduced.
The amalgam between biology, statistics, and computer science to perform bioinformatics analyses contributes to an ever-growing knowledge base for scientific research. The meta-CATS tool can assist researchers in performing metadata-driven comparative genomics statistical analyses on any virus sequences within the multiple viral taxa supported by ViPR and IRD. Although we chose to explore the more obvious genetic differences existing between DENV geographic clades and serologic types as a proof-of-concept, we expect that meta-CATS can also be applied to identify more subtle genotype-phenotype correlations. Indeed, one of the main advantages of meta-CATS is that it provides a finer degree of granularity (individual residues within a sequence) than that derived from traditional phylogenetic analysis alone (average across an entire sequence). Thus the results obtained from meta-CATS can be used to generate hypotheses and optimize candidate identification prior to applying such knowledge to the development of novel diagnostics, prophylactics, and therapeutics.
Supplementary Material
Supplementary Figure 1 Evolutionary divergence between strains isolated from different hemispheres. Maximum likelihood phylogenetic tree reconstructions demonstrating the underlying sequence relationship between hemispheres. The RAxML algorithm, using the General Time Reversible model with other parameters as default, was used to construct the tree. (A) Strains isolated from the Western Hemisphere, Eastern Hemisphere, as well as the descendants of the inter-hemisphere transfer event are outlined by the blue bracket, cyan bracket, and red line respectively. (B) Strain names for representative sequences from the Western Hemisphere, Eastern Hemisphere, as well as the descendants of the inter-hemisphere transfer event are labeled in blue, cyan, and red respectively. A Dengue virus serotype 2 sequence (16681) is included at the bottom of each tree as an outgroup. Branch lengths for the trees are proportional to distance, or number of amino acid changes, observed between sequences. Each scale bar represents the number of observed substitutions per amino acid site.
Supplementary Table 1: DENV-3 strains used for the hemisphere analysis.
Supplementary Table 2: List of 70 positions that significantly differ between hemispheres.
Supplementary Table 3: List of non-redundant DENV strains used in the NS1 analysis.
Supplementary Table 4: List of positions, scores, and Pearson’s chi-square statistical information for positions that significantly differ between NS1 sequences from all four DENV serotypes.
Highlights.
We have developed an analysis tool for identifying significant sequence differences.
70 amino acids differ between eastern and western hemisphere dengue viruses.
19 amino acids in the NS1 protein differ between the four dengue virus serotypes.
Two of the NS1 differences overlap known B cell epitopes.
Meta-CATS tool is freely available at www.viprbrc.org.
Acknowledgments
We thank the primary data providers for the sequence data that was used throughout this study, notably the Genome Resources in Dengue (GRID) consortium. We appreciate discussions with Fuchun Zhang regarding the patient history behind the GZ1D3 strain. We acknowledge Dr. Alison Yao for expert oversight of the BRC program. ViPR is wholly supported with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272200900041C. The funding source had no involvement in the design, analysis, or interpretation of this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
BE Pickett, Email: bpickett@jcvi.org.
M Liu, Email: mengyasmu@gmail.com.
EL Sadat, Email: eva.sadat@yahoo.com.
RB Squires, Email: richard.squires@nih.gov.
JM Noronha, Email: jyothi.noronha@gmail.com.
S He, Email: fangxue.he@ngc.com.
W Jen, Email: wei.jen@ngc.com.
S Zaremba, Email: Sam.Zaremba@ngc.com.
Z Gu, Email: zhiping.gu@ngc.com.
L Zhou, Email: Liwei.Zhou@ngc.com.
CN Larsen, Email: clarsen@vecna.com.
I Bosch, Email: ibosch@mit.edu.
L Gehrke, Email: lgehrke@mit.edu.
M McGee, Email: mmcgee@smu.edu.
EB Klem, Email: Ed.Klem@ngc.com.
RH Scheuermann, Email: rscheuermann@jcvi.org.
References
- Allicock OM, Lemey P, Tatem AJ, Pybus OG, Bennett SN, Mueller BA, Suchard MA, Foster JE, Rambaut A, Carrington CV. Phylogeography and population dynamics of dengue viruses in the Americas. Mol Biol Evol. 2012;29:1533–1543. doi: 10.1093/molbev/msr320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari IU, Allen T, Berical A, Stock PG, Barin B, Striker R. Phenotypic analysis of NS5A variant from liver transplant patient with increased cyclosporine susceptibility. Virology. 2013;436:268–273. doi: 10.1016/j.virol.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo JM, Nogueira RM, Schatzmayr HG, Zanotto PM, Bello G. Phylogeography and evolutionary history of dengue virus type 3. Infect Genet Evol. 2009;9:716–725. doi: 10.1016/j.meegid.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Bloom-Feshbach K, Alonso WJ, Charu V, Tamerius J, Simonsen L, Miller MA, Viboud C. Latitudinal variations in seasonal activity of influenza and respiratory syncytial virus (RSV): a global comparative review. PloS one. 2013;8:e54445. doi: 10.1371/journal.pone.0054445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonferroni CE. Teoria statistica delle classi e calcolo delle probabilit à. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze. 1936;8:3–62. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Falconar AK. Antibody responses are generated to immunodominant ELK/KLE-type motifs on the nonstructural-1 glycoprotein during live dengue virus infections in mice and humans: implications for diagnosis, pathogenesis, and vaccine design. Clinical and vaccine immunology : CVI. 2007;14:493–504. doi: 10.1128/CVI.00371-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski MM, Ohnemus A, Staeheli P, Rubbenstroth D. Pandemic 2009 H1N1 influenza A virus carrying a Q136K mutation in the neuraminidase gene is resistant to zanamivir but exhibits reduced fitness in the guinea pig transmission model. Journal of virology. 2013;87:1912–1915. doi: 10.1128/JVI.02507-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Ponomarenko J, Zhu Z, Tamang D, Wang P, Greenbaum J, Lundegaard C, Sette A, Lund O, Bourne PE, Nielsen M, Peters B. Immune epitope database analysis resource. Nucleic Acids Res. 2012;40:W525–W530. doi: 10.1093/nar/gks438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Lai YL, Lo S, Barkham T, Aw P, Ooi PL, Tai JC, Hibberd M, Johansson P, Khoo SP, Ng LC. Dengue virus surveillance for early warning, Singapore. Emerging infectious diseases. 2010;16:847–849. doi: 10.3201/eid1605.091006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund O, Nascimento EJ, Maciel M, Jr, Nielsen M, Larsen MV, Lundegaard C, Harndahl M, Lamberth K, Buus S, Salmon J, August TJ, Marques ET., Jr Human leukocyte antigen (HLA) class I restricted epitope discovery in yellow fewer and dengue viruses: importance of HLA binding strength. PloS one. 2011;6:e26494. doi: 10.1371/journal.pone.0026494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noronha JM, Liu M, Squires RB, Pickett BE, Hale BG, Air GM, Galloway SE, Takimoto T, Schmolke M, Hunt V, Klem E, Garcia-Sastre A, McGee M, Scheuermann RH. Influenza virus sequence feature variant type analysis: evidence of a role for NS1 in influenza virus host range restriction. Journal of virology. 2012;86:5857–5866. doi: 10.1128/JVI.06901-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MM, Pitzer VE, Alonso WJ, Vera D, Lopman B, Tate J, Viboud C, Parashar UD. Global seasonality of rotavirus disease. The Pediatric infectious disease journal. 2013;32:e134–e147. doi: 10.1097/INF.0b013e31827d3b68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett BE, Lefkowitz EJ. Recombination in West Nile Virus: minimal contribution to genomic diversity. Virol J. 2009;6:165. doi: 10.1186/1743-422X-6-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett BE, Sadat EL, Zhang Y, Noronha JM, Squires RB, Hunt V, Liu M, Kumar S, Zaremba S, Gu Z, Zhou L, Larson CN, Dietrich J, Klem EB, Scheuermann RH. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40:D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett BE, Striker R, Lefkowitz EJ. Evidence for separation of HCV subtype 1a into two distinct clades. J Viral Hepat. 2010 doi: 10.1111/j.1365-2893.2010.01342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. 1.35. Vienna, Austria: R Foundation for Statistical Computing; 2010. [Google Scholar]
- Schmidt DJ, Pickett BE, Camacho D, Comach G, Xhaja K, Lennon NJ, Rizzolo K, de Bosch N, Becerra A, Nogueira ML, Mondini A, da Silva EV, Vasconcelos PF, Munoz-Jordan JL, Santiago GA, Ocazionez R, Gehrke L, Lefkowitz EJ, Birren BW, Henn MR, Bosch I. A phylogenetic analysis using full-length viral genomes of South American dengue serotype 3 in consecutive Venezuelan outbreaks reveals a novel NS5 mutation. Infect Genet Evol. 2011;11:2011–2019. doi: 10.1016/j.meegid.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvoerer E, Moenne-Loccoz R, Murray JM, Velay A, Turek M, Fofana I, Fafi-Kremer S, Erba AC, Habersetzer F, Doffoel M, Gut JP, Donlin MJ, Tavis JE, Zeisel MB, Stoll-Keller F, Baumert TF. Hepatitis C virus envelope glycoprotein signatures are associated with treatment failure and modulation of viral entry and neutralization. The Journal of infectious diseases. 2013;207:1306–1315. doi: 10.1093/infdis/jit032. [DOI] [PubMed] [Google Scholar]
- Shi Z, Hu Z. A review of studies on animal reservoirs of the SARS coronavirus. Virus research. 2008;133:74–87. doi: 10.1016/j.virusres.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires RB, Noronha J, Hunt V, Garcia-Sastre A, Macken C, Baumgarth N, Suarez D, Pickett BE, Zhang Y, Larsen CN, Ramsey A, Zhou L, Zaremba S, Kumar S, Deitrich J, Klem E, Scheuermann RH. Influenza Research Database: an integrated bioinformatics resource for influenza research and surveillance. Influenza and other respiratory viruses. 2012 doi: 10.1111/j.1750-2659.2011.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N, Weaver SC. The history and evolution of human dengue emergence. Advances in virus research. 2008;72:1–76. doi: 10.1016/S0065-3527(08)00401-6. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhang W, Ni B, Shen H, Song Y, Wang X, Shao S, Hua X, Cui L. Recombination analysis reveals a double recombination event in hepatitis E virus. Virol J. 2010;7:129. doi: 10.1186/1743-422X-7-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Vasilakis N. Molecular evolution of dengue viruses: contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease. Infect Genet Evol. 2009;9:523–540. doi: 10.1016/j.meegid.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Evolutionary divergence between strains isolated from different hemispheres. Maximum likelihood phylogenetic tree reconstructions demonstrating the underlying sequence relationship between hemispheres. The RAxML algorithm, using the General Time Reversible model with other parameters as default, was used to construct the tree. (A) Strains isolated from the Western Hemisphere, Eastern Hemisphere, as well as the descendants of the inter-hemisphere transfer event are outlined by the blue bracket, cyan bracket, and red line respectively. (B) Strain names for representative sequences from the Western Hemisphere, Eastern Hemisphere, as well as the descendants of the inter-hemisphere transfer event are labeled in blue, cyan, and red respectively. A Dengue virus serotype 2 sequence (16681) is included at the bottom of each tree as an outgroup. Branch lengths for the trees are proportional to distance, or number of amino acid changes, observed between sequences. Each scale bar represents the number of observed substitutions per amino acid site.
Supplementary Table 1: DENV-3 strains used for the hemisphere analysis.
Supplementary Table 2: List of 70 positions that significantly differ between hemispheres.
Supplementary Table 3: List of non-redundant DENV strains used in the NS1 analysis.
Supplementary Table 4: List of positions, scores, and Pearson’s chi-square statistical information for positions that significantly differ between NS1 sequences from all four DENV serotypes.