Abstract
The longitudinal relaxation properties of NMR active nuclei carry useful information about the site-specific chemical environments and about the mobility of molecular fragments. Molecular mobility is in turn a key parameter reporting both on stable properties like size, as well as on dynamic ones such as transient interactions and irreversible aggregation. In order to fully investigate the latter, a fast sampling of the relaxation parameters of transiently formed molecular species may be needed. Nevertheless, the acquisition of longitudinal relaxation data is typically slow, being limited by the requirement that the time for which the nucleus relaxes be varied incrementally until a complete build-up curve is generated. Recently a number of single-shot inversion recovery methods have been developed capable of alleviating this need; still, these may be challenged by either spectral resolution restrictions or when coping with very fast relaxing nuclei. Here we present a new experiment to measure the T1s of multiple nuclear spins that experience fast longitudinal relaxation, while retaining full high-resolution chemical shift information. Good agreement is observed between T1s measured with conventional means and T1s measured using the new technique. The method is applied to the real time investigation of the reaction between D-xylose and sodium borate, which is in turn elucidated with the aid of ancillary ultrafast and conventional 2D TOCSY measurements.
Keywords: Longitudinal relaxation, T1, Ultrafast 2D NMR, Real-time NMR
Introduction
Nuclear Magnetic Resonance (NMR) relaxation parameters are useful in a variety of contexts, and in particular as probes of molecular dynamics occurring on a wide range of timescales –from ps to ms. These dynamics can in turn reveal unique information about the conformation and functioning of molecules, including vistas into how motions can enable enzymatic or biological function.1, 2 Moreover, fast motions are, in themselves, important in the activity of a variety of biomolecules.3, 4 Longitudinal relaxation time (T1) measurements are an important probe of these faster timescale motions.5, 6 This method is complementary to Pulsed Field Gradient Spin Echo (PGSE) techniques, enabling the study of overall molecular sizes and local dynamics by the distinct T1s displayed by shift-resolved sites.7–9 Moreover, when applied to monitor unidirectional dynamics by rapid mixing of liquid samples, T1 measurements are free from turbulent motion effects that are often encountered and may jeopardize real-time NMR studies based on PGSE.
T1 relaxation measurements are usually accomplished using the inversion recovery (IR) method. In this experiment, spins are inverted by a π-pulse, are allowed to relax for a period ΔIR, and are then brought into the transverse plane for detection by a π/2-pulse. Importantly, this experiment requires incrementally varying ΔIR, which incurs a time cost. Traditional inversion recovery methods are therefore unsuitable for studying in real-time the changes occurring in a molecule’s structure or flexibility as it interacts with its environment. These changes in flexibility are important in the thermodynamics of molecular interactions, reporting on the essential entropic contribution of changes in free energies.10–18 This was realized early on in the progress of NMR,19 and is routinely exploited in instances which, like MRI, have no need to include a high-resolution chemical shift dimension.20–22 A number of recent studies have been aimed at developing fast inversion recovery experiments, which can also describe site specific chemical shifts. Bhattacharya et al have developed a single scan inversion recovery experiment,23 but which is ill suited to measuring short T1s. Loening et al have also developed alternatives to measure T1 relaxation in a single shot,24 but which once again are challenged by short relaxation times or by spectral resolution restrictions.
To address the need for a single-shot longitudinal relaxation measurement capable of handling short relaxation times without suffering from resolution restrictions, we propose a new pulse sequence where T1 relaxation is encoded spatially and then the sample is imaged using a detection scheme akin to echo planar spectroscopic imaging (EPSI).25 The new inversion recovery sequence, which falls within the domain of ultrafast methods, is applied to study the D-xylose and borate reaction, a process that is relevant to the design of affinity ligands for the separation of carbohydrates and to the construction of saccharide sensors.26–29 A comparison is made between the new ultrafast inversion recovery (UFIR) method and the multiple acquisitions inversion recovery (MA-IR) experiment of Loening et al;24 it is shown that each sequence functions optimally in its own niche –ultrafast sequences when T1s are short and MA-IR when T1s are longer– thus providing an optimal complement for each other. Additional insight into the interaction between D-xylose and borate was garnered by monitoring in real time their reaction by ultrafast TOCSY,30 demonstrating how single-shot 2D methods probing molecular flexibility and structure in real time, can function synergistically in dynamic real-time investigations.
Materials and Methods
Pulse-sequencing Considerations
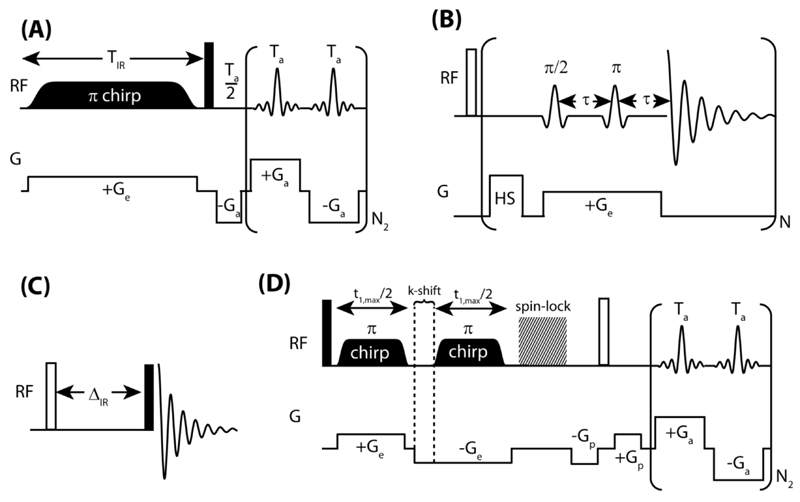
The UFIR approach utilized here to measure multiple sites’ longitudinal relaxation recoveries in a single scan, begins with the simultaneous application of a z-gradient and of a frequency-swept adiabatic inversion pulse, as shown in Figure 1A. The z-gradient Ge will create a position-dependent addition to the resonance frequencies of the spins equal to γGez. The action of the linearly swept π-pulse covering all spins over a span L can be approximated as an instantaneous inversion of the spins at a particular z, occurring whenever their spatial + spectral resonance frequencies match the offset of the chirped RF. Therefore, as the frequency-swept pulse is applied, it will invert the spins’ magnetizations starting at one end of the sample (e.g., at ≈+L/2) and continue to do so along the length of the sample until reaching z ≈ -L/2. As soon as spins are inverted, however, they begin to recover back to their equilibrium Mo magnetizations. This in effect can map every site’s T1 inversion recovery profile along the length of the sample; given that the swept RF pulse imposes an a priori known relation between position and inversion time, one can then recover the individual IR profiles by reading out the ensuing spatial profiles in a shift resolved fashion. Such readout can be carried out in a single shot using an EPSI acquisition,25 of the kind shown in brackets at the end of the sequence in Figure 1A. This begins with a π/2 pulse putting all the sites’ magnetizations in the xy-plane, and a pre-winding gradient that prepares the signal for detection. During the course of the ensuing oscillating train of acquisition gradients Ga, spins with a chemical shift Ω2 and at a position z will precess in the transverse plane, giving rise to a signal of the form:
| (1) |
where is a wavenumber describing the acquisition gradient’s action following the π/2 excitation pulse. The overall sample’s signal will equal the sum of the contributions from all sites over the entire length of the sample:
| (2) |
where is the profile of a particular site’s magnetization for t2=0, depending on its rate of longitudinal relaxation and possessing its shortest (in principle null) recovery time at zo ≈ L/2. It is apparent then that the signal in k-t2 space is the Fourier conjugate of the shift-resolved distribution of the various sites’ magnetizations along z. The desired IR maps should therefore become available by 2D Fourier transform of the data, following their suitable rearrangement in the 2D k-t2 space. Proper line shapes, however, would only become then available if the k(t2) = 0 signal is fully and symmetrically sampled. Ensuring this is the purpose of the preliminary gradient that shifts the imaging echoes to the middle of each acquisition window (a bipolar gradient with a refocusing pulse in its center may be used instead of this preliminary gradient, to remove first order phase distortions in situations where these may affect spectral quality/phasing). Suitable data processing according to the recipes for EPSI, involving data rearrangement, weighting and 2D FT, leads to the shift-resolved IR curves being sought; an important factor in recovering faithful T1 recovery estimates includes mapping the NMR coil’s sensitivity profile, as described elsewhere.31
Figure 1.
Pulse sequences employed in this study; filled rectangles indicate π/2 pulses and empty ones indicate π pulses. (A) The ultrafast inversion recovery experiment. A linear frequency swept (chirped) inversion pulse is applied with a gradient. Spins at one end of the sample –e.g., at its bottom–that are on resonance at the start of the frequency sweep are inverted first and experience longitudinal relaxation for a longer period, than spins that are on resonance later and are therefore inverted later. After T1 relaxation is spatially encoded with a 1s WURST-40 π chirp pulse, the precession frequencies of the spins are monitored using an echo planar spin imaging (EPSI) type procedure. (B) The multiple acquisitions inversion recovery (MA-IR) technique:24 a hard π pulse inverts all the sample magnetization, and spins from single slices throughout the sample are rendered observable by the combined effect of a gradient and frequency selective π/2 and π pulses. (C) Conventional inversion-recovery experiment. (D) Ultrafast 2D TOCSY experiment; a DIPSI-2 sequence was used for spin-lock during the mixing period [30].
Materials
All spectral results described in this work were obtained on a Varian 600 MHz VNMRS spectrometer equipped with a multi-resonance inverse gradient probehead. The experiments were carried out at 24°C. Although a triple axis gradient set was available only the longitudinal z-axis, endowed with the strongest field gradients and associated to the longest spatial dimensions, was used to spatially encode the 2D T1/shift NMR correlations. For the ultrafast inversion recovery sequence, a gradient field strength of 0.5 G/cm was used to spatially encode longitudinal relaxation and ±10 G/cm gradients were used for the EPSI-type acquisition with typical switching times of 10 µs. A 1s WURST-40 adiabatic inversion pulse with a bandwidth of 4.785 kHz was used to invert sample magnetization. In order to minimize artifacts a ~0.1 kHz presaturation pulse was applied on the HDO resonance prior to execution of the sequence; a sinc frequency selective pulse followed by a purge gradient (17 G/cm) was also applied after the spatial encoding to minimize artifacts arising from HDO that relaxed back to equilibrium during the spatial encoding of longitudinal relaxation.
These ultrafast IR (UFIR) experiments were complemented by MA-IR acquisitions (Fig. 1B). Here, Gaussian pulses with a 1% truncation level were used for slice-selective excitation and refocusing. The frequency offsets for the selective pulses were varied from 51.1 kHz to -51.1 kHz in increments of ~11.4 kHz and they were applied synchronously with 16 G/cm magnetic field gradient. To ensure complete inversion of the sample magnetization, a short (2 ms) frequency-swept adiabatic inversion pulse was applied. Ten acquisition periods were used to sample the IR curves, with each acquisition period being 1.5 s long, totaling 15 s. A recycle delay of 30 s was employed to ensure that the magnetization of all the slices fully recovered to equilibrium (the longest T1 was ~ 6 s). Traditional IR experiments (Fig. 1C) were also applied to the investigated systems.
The usefulness of the UFIR and MA-IR methods to monitor real-time transformations, was explored by analyzing the nature of the xylose-borate reaction. Further insight regarding the connectivity of the various species intervening in this mixture was obtained using conventional and ultrafast (Fig. 1D) 2D TOCSY NMR. These spectra are represented in magnitude mode.
A variety of Matlab 8.0 software programs (The Mathworks) were written for processing the data. All chemicals and solvents were obtained from Aldrich and used as purchased.
Results and Discussion
The ultrafast inversion recovery sequence
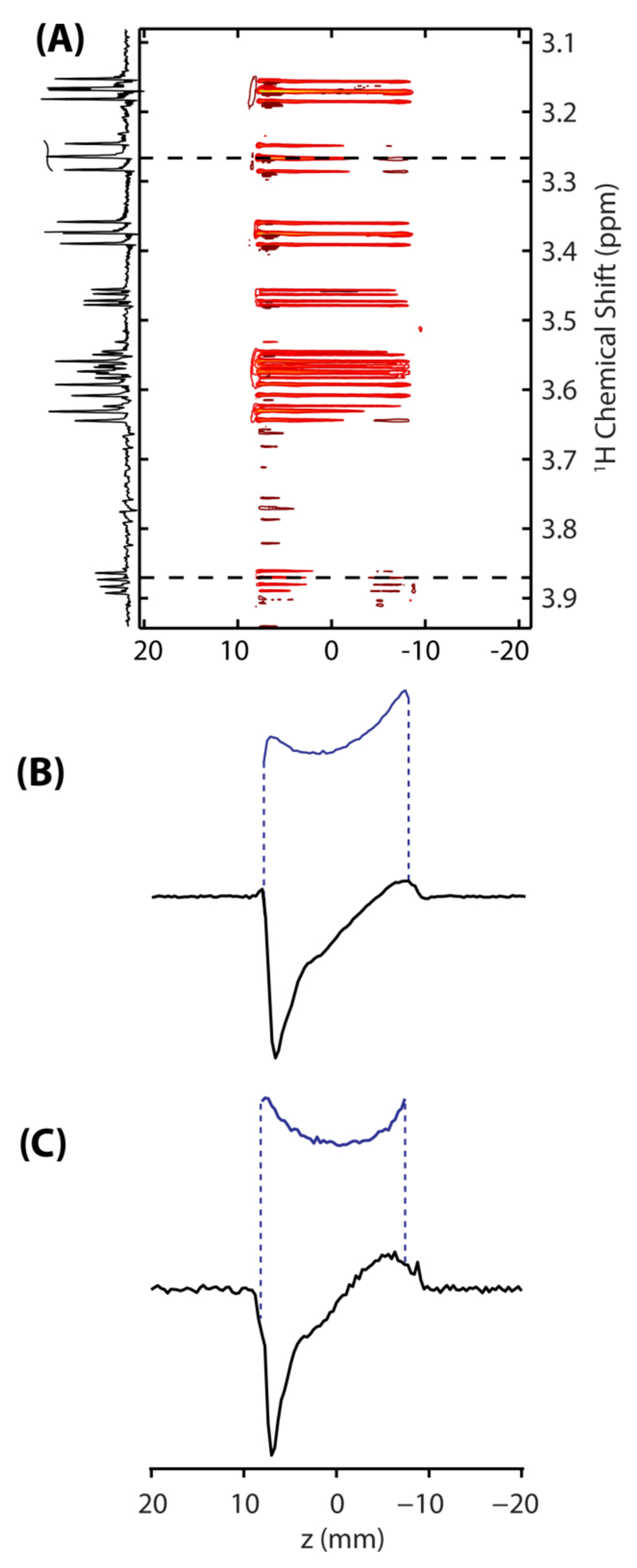
To test the performance of the UFIR method just outlined, the inversion-recovery behavior of 100 mM D-xylose dissolved in D2O was monitored. Figure 2A illustrates representative results of these tests, as a 2D plot displaying the recovery contours observed for each chemical site as a function of position corresponding to a ΔIR of 100 ms is shown. Two slices show the spatial variation in transverse magnetization induced by longitudinal relaxation at chemical shifts of ~3.3 and ~3.9 ppm (Figs. 2B and 2C respectively). Inversion recovery curves can be reconstructed from these spatial profiles, by normalization against a priori known coil sensitivity profiles (blue curves in Figs. 2B and 2C). The peaks shown in Figure 2 arise from the axial and equatorial β5 proton sites of D-xylose (β5,a and β5,e, see Figure 4A), which exhibit faster relaxation times due to their proximity to each other. Note that the individual sites and their splitting patterns are resolved in the 2D plot after EPSI processing. This illustrates some of the advantages of the UFIR technique, including its ability to provide single-shot longitudinal relaxation measurements with high time resolution and without sacrificing spectral resolution. This feature would not be available if the MA-IR experiment would be used to analyze peaks like these ones, for which T1 and T2 values are comparable. On the other hand, the range of relaxation times that can be measured by UFIR may be limited by the length of time for which a suitable encoding gradient can be applied, or compromised if dealing with sites dealing with very different relaxation times.
Figure 2.
(A) Ultrafast inversion recovery (UFIR) spectrum acquired on 100 mM D-xylose dissolved in D2O; illustrating contours of the spins’ recovery as a function of position as well as the 1D spectrum emerging of the data at ∆IR ≈ 100 ms. These data arise from the sequence shown in Figure 1A, after processing its signal in an EPSI-like fashion. Shown in (B) and (C) are slices taken at ~3.3 ppm (B) and ~3.9 ppm (C), illustrating the variation of magnetization along the length of the sample set up by an adiabatic inversion pulse applied synchronously with a magnetic field gradient. Shown in blue are profiles arising from the same sites after full recovery; these are needed as reference for an accurate reconstruction of the inversion-recovery curves.
Figure 4.
Overview of the xylose-borate reaction. (A) Series of 1D 1H NMR spectra of the D-xylose solution/xylose-borate reaction mixture as a function of time (shown on the right side of each spectrum) after sodium borate injection. The protons of uncomplexed D-xylose are assigned as labeled on the α (green) and β (blue) anomeric forms shown at the top; “e” refers to an equatorial proton and “a” refers to an axial proton (assignments taken from Ref. 46). Primed numbers correspond to xylose peaks in the 1:1 xylose-borate complex (assigned via TOCSY and inversion-recovery spectra) and unprimed peaks correspond to uncomplexed xylose. Note that in the text, the anomeric form of the resonance is referenced with a Greek letter and the number is indicated with a subscript (e.g. α1 indicates the anomeric peak of the α anomer of xylose). Green dashed lines indicate new peaks associated with the 1:1 borate:xylose complex and the red dashed line indicates a new peak associated with the 1:2 borate:xylose complex. (B) The integral intensity of the HDO peak as a function of time after the injection of sodium borate. The inset shows the mechanism of borate-diol interaction.41.
At the edges of the sample images shown in Figures 2B and C, at ~ ±9 mm, anomalous decreases in signal intensities are observed. These effects –which are often seen in gradient shimming experiments– reflect variations in the main magnetic field homogeneity throughout the sample, non-linearities in the applied field gradient as well as RF inhomogeneities throughout the length of the excitation/detection coil. Of all these the inhomogeneous detection factor is probably most important, as slight deviations in the RF pulse delivered throughout the adiabatic sweep or in Bo’s ideal profile, are not expected to affect significantly a signal’s strength. In order to account for all these variations, the points along an UFIR curve need to be divided by a reference image (shown in blue in Figs. 2B and 2C). Note that such normalizations were also required for processing the MA-IR and the single-scan DOSY data.24, 31
A comparison of the relaxation time measurements thus emerging from single-scan and from conventional IR methods is given in Figure 3. There is a close correspondence between the T1s measured by single-scan and conventional IR techniques for all the sites except the β5,e site, the results not differing by more than 10%. Note that in the UFIR experiment the encoding gradient was applied for a second, limiting the range of T1s observable using this single-scan technique to ≤ 1s. The relatively larger disagreement between conventional IR and MA-IR for the β5,e site (16%) may be reflecting cross-correlations between the dipolar and chemical shift mediated T1 relaxation mechanisms, leading to relaxation behavior which is not strictly monoexponential. Then, as for the MA-IR experiment larger ΔIR values are sampled on the IR curve, slowly relaxing components exert a larger influence on the T1 fit value.
Figure 3.
T1 values derived from fits to the IR curves obtained from conventional IR (blue diamonds), MA-IR (red squares), and UFIR (green triangles) plotted against chemical shift. The markers are labeled with resonance assignments (given in Fig. 4). Note that only T1s ≤ 1 can be reliably observed using UFIR because the encoding gradient was applied for no longer than one second.
Monitoring xylose-borate reaction kinetics with the ultrafast IR and 2D TOCSY techniques
In order to explore the ability of these single-shot IR methods to monitor real-time chemical changes, they were applied to study the xylose-borate reaction. This process has been investigated by a number of means,32–34 including NMR. Boronic acids bind with high affinity to compounds containing diol moieties through reversible ester formation.35 This tight binding enables the use of boronic acids and their derivatives in the construction of sensors for saccharides,26, 27, 36, 37 as affinity ligands for the separation of carbohydrates and glycoproteins,28, 29, 38 and as antibody mimics for cell-surface polysaccharides.39, 40 Shown in Figure 4A is a time series of 1H NMR spectra collected upon adding 100 µL of 50 mM sodium borate to a 600 µL solution of 100 mM D-xylose under basic (pD > 10) conditions. Both of these solutions were prepared in D2O, and the reaction was triggered by injecting the sodium borate into the D-xylose with a custom-made apparatus. D-Xylose has two interconverting pyranose (6 membered ring) and furanose (5 membered ring) forms; the pyranose forms are more stable than the furanose one due to their reduced intra-ring strains, and peaks in the spectrum of pure D-xylose (Fig. 4A, top trace) are therefore assigned to these pyranose forms. As the reaction proceeds water is evolved, as is evidenced by the increasing intensity of the HDO peak (Fig. 4B) and as would be predicted on the basis of the borate anion’s interaction with polyols.41 To interpret the remaining spectral changes it is worth reminding that: (i) it is known that the borate anion only interacts with the α anomers of xylose;34 (ii) there is some evidence that it interacts with the furanose form of D-xylose;32, 42 and (iii) it was recently shown that under certain conditions, some simple sugars will associate with the borate anion while in their pyranose form.43 Because of the proximity of the α5’ protons to each other in the borate-xylose complex, they exhibit short T1s and may be easily identified with inversion recovery methods. If the borate-xylose complex is exclusively associated with D-xylose’s furanose forms, two resonances for the diastereotopic α5 protons might be expected at ~3.65 and ~3.80 ppm; this is observed for example in the 1H NMR spectrum of similar D-xylofuranose compounds such as 1,2-cyclohexylidene-3-O-tosyl-α-D-xylofuranose.44 On the other hand, if the borate anion complexes with the pyranose form of D-xylose, one would expect to observe peaks corresponding to the α5 axial and equatorial protons, which, in the case of 1,4-anhydro-2,3-di-O-benzyl-α-D-xylopyranose, appear at 3.39 and 4.06 ppm (separated by more than 0.6 ppm).45 Given the fact that two equal intensity peaks with short T1 values appear initially at 3.74 and 3.84 ppm, separated by less than 0.2 ppm (Fig. 4A), the borate-xylose complexes formed are most likely associated with the furanose form of D-xylose. This result is in agreement with the one reported by Chapelle and Verchere.32
The borate anion associates with a maximum of two diol moieties.41 Early in the reaction a set of new 1H NMR peaks appear simultaneously, as indicated by green dashed lines in Fig. 4A. These are probably associated with a 1:1 borate-xylose complex: it is unlikely that, early on in the reaction, the borate ion will complex with two D-xylose molecules simultaneously. The results of Pepi et al suggest that the borate anion associates with D-xylose at the O1 and O2 oxygens and that the tridentate boronate ester structure, where the borate anion associates with the O1, O3, and O5 oxygens, is not formed.43 Further along in the time course of the reaction a new peak appears (shown by a red dashed line) indicating the presence of a new species and strongly suggesting the formation of a 1:2 borate-xylose complex.32 To get further insight into this possibility, TOCSY experiments on the equilibrated reaction mixture were recorded. This conventional 2D spectrum suggests the presence of at least two species, since at a mixing time of 40 ms, resonances appear to be separated into two groups with no common cross-peaks evident (Fig. 5A). This, however, could be misleading, as some of the peaks in the various borate-xylose complexes formed at the conclusion of the reaction may overlap. One possible way to alleviate this problem is to follow the cross-peaks arising in the 2D trace in real time, using the ultrafast TOCSY sequence presented in Fig. 1D. Representative ultrafast TOCSY spectra are shown 0.7, 9.2, and 46.9 minutes after the injection of borate, in Fig. 5B. These traces allow us to distinguish peaks that are uniquely identified with the 1:1 borate-xylose complex, from those associated with either the 1:2 complex or from both the 1:1 and 1:2 complexes (identified with green and red dashed lines in Fig. 4B, respectively). These data also show that the “α1’” peak shares a cross peak with the 1:2 peak as well as with the 1:1 borate-xylose complex, and is not associated with a single species in the reaction mixture. Nevertheless, when the co-added peak volumes of all the peaks associated with the 1:1 borate-xylose complex are compared with the co-added peak volumes of all the peaks uniquely assigned to the 1:2 borate-xylose complex, it is clear that there is a 1:1 borate-xylose species forming first, and that further along in the course of the reaction a 1:2 borate-xylose species forms; these data are summarized by the green and red data points shown in the time progression of Fig. 5C.
Figure 5.
(A) Conventional TOCSY experiment acquired on the equilibrated reaction product. Peaks uniquely assigned to the 1:2 borate-xylose complex are shown against a red background and peaks associated with the 1:1 (and possibly 1:2) borate-xylose complex(es) are shown against a green background. A 40 ms DIPSI mixing sequence was used and 1024 points were acquired in the t1 dimension with a sw1 of 2500 Hz. (B) Representative ultrafast TOCSY spectra acquired 0.7, 9.2, and 46.9 minutes after the injection of borate. These data were collected using the sequence in Fig. 1D with chirped pulses spanning 79 kHz; Ge = 10 G/cm; t1max/2 = 17.5 ms; Ga = 14.76 G/cm; Ta = 542 µs; N2 = 180. A bipolar purge gradient (Gp) of 25 G/cm was applied for 1.1 ms prior to acquisition and, in order to ensure all peaks fell within the f1 spectral window, an extra gradient (k-shift in Fig. 1D) was applied at –Ge for 3.15 ms. Data were sampled every 2 µs and each data set was subject to a suitable shearing transform and zero filling along both dimensions before t2 Fourier transformation. A 40ms long DIPSI-2 mixing pattern was used. (C) The reaction products are shown to the left. Shown to the right (in green) is the sum of peak volumes associated with the 1:1 and 1:2 borate-xylose peaks (shown is a green highlight in panels A and B), and (in red) the sum of peak volumes uniquely associated with the 1:2 borate-xylose peaks.
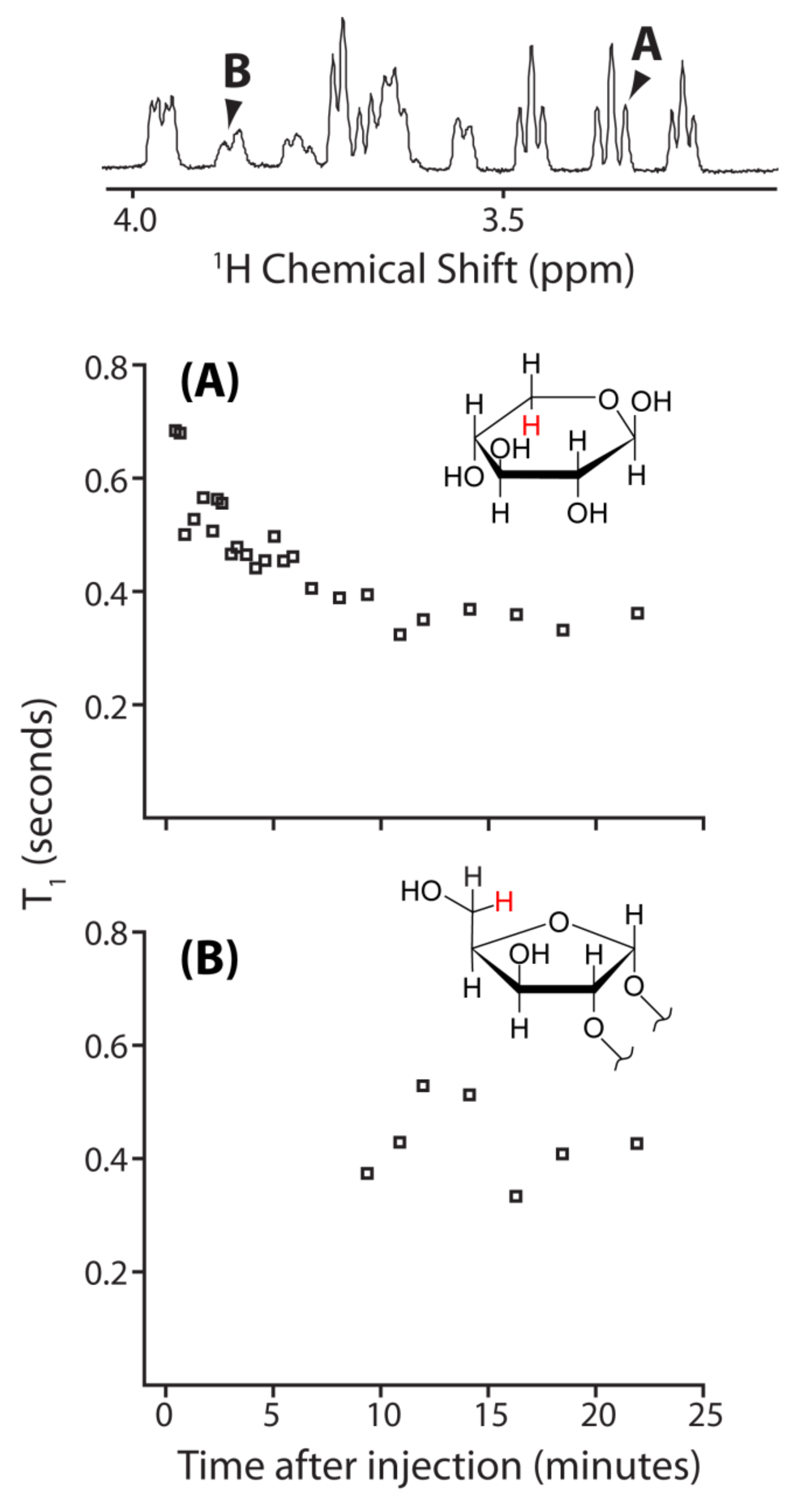
The use of UFIR to track the reaction of D-xylose with the borate anion could also be helpful to establish the temporal precedence with which new peaks appear in the xylose-borate mixture. Figure 6A shows the variation of the D-xylose monomer β5,a proton’s longitudinal relaxation as a function of time, as monitored by UFIR. Changes in the magnetic susceptibility of the sample occur upon mixing the reactants. These susceptibility changes create magnetic field inhomogeneity, which manifests as a variation in the resonance frequency of the spins throughout the length of the sample and a “warping” of the sample images. However, this effect was easily corrected for by using a reference (unwarped) sample image.47 Note the correspondence between the decrease in this proton’s T1 and the formation of the borate complex; an increase in molecular weight, plus the higher concentration of solvent protons as the reaction proceeds, probably lead to this decrease in the T1 for the 1H nuclei associated with the D-xylose molecules and the borate-xylose complexes. This finding also highlights the sensitivity of UFIR to changes in the nuclear environment, such as changes in molecular dynamics or in the location of nearby protons that serve as relaxation sinks. The T1 variation of the peak at 3.84 ppm (second furthest upfield among the new peaks) as a function of the time after the reaction is initiated, is shown in Figure 6B. This peak is associated with the formation of the 1:1 borate-xylose complex. Because an adequate signal-to-noise ratio to measure the T1 of the new peak is only available later in the course of the reaction, when the increased density of the 1H nucleus already exerts an effect on the T1s of all the resonances, a significant change in this peak’s T1 is not observed.
Figure 6.
The variation of T1 as a function of time after the injection of borate, monitored using UFIR. The peaks for which T1s are provided are identified in the 1H spectrum given above the plots. (A) Variation of T1 as a function of time for the axial β5 hydrogen of uncomplexed D-xylose (depicted in red in the inset). (B) Variation of T1 as a function of time for the 1:1 xylose-borate complex peak at 3.84 ppm (depicted in red in the inset).
In order to investigate the trend of a decrease in T1 associated with the reaction’s progression, the xylose-borate reaction was monitored with MA-IR. Figure 7A shows a series of T1s obtained using MA-IR for the α1 proton of uncomplexed D-xylose, which may be compared with the T1 obtained using MA-IR on uncomplexed D-xylose prior to the reaction’s initialization (indicated with a dashed horizontal line). In order to measure these protons’ T1s, a longer recycle delay (~5 X 6 s = 30 s) had to be utilized and furthermore, the acquisition of 1D 1H spectra for 10 slices, each with an acquisition time of 1.5 s, took 15 s. A decrease in T1 relative to the α1 proton’s T1 prior to reaction initialization is clearly evident, however T1 values remain relatively constant after the reaction initialization. Although the fact that the reaction is proceeding while the T1 of these protons is being measured (a process which lasts 15 s) complicates interpretation of these T1 values, it is nevertheless clear that after the T1 values stabilize, they remain shorter than the T1 value observed prior to reaction initialization. Figure 7B shows a series of T1s obtained using MA-IR for the β1 proton of uncomplexed D-xylose, which is again shorter than the T1 obtained for the β1 proton prior to reaction initialization (horizontal dashed line). In Fig. 7C, T1 values associated α1’ resonance of the borate-xylose complex are plotted. Notably, the T1s observed for this peak are ~0.5 s shorter than those observed for the α1 resonance, which illustrates the feasibility of distinguishing different molecular species, such as uncomplexed D-xylose and the borate-xylose complex, on the basis of T1. Even for challenging, broad resonances, such as α2’, MA-IR can be used to extract T1s in real time, as shown in Fig 7D.
Figure 7.
Variation of T1 as a function of time after the injection of borate, monitored using MA-IR. The peaks for which T1s are provided are identified in the 1H spectrum given above the plots. (A) The variation of T1 as a function of time for the α1 hydrogen of uncomplexed D-xylose, (B) for the β1 hydrogen of uncomplexed D-xylose, (C) for the α1’ hydrogen of the borate-xylose complex, and (D) for the α2’ hydrogen of the borate-xylose complex. The associated protons are depicted in insets in red.
Conclusion
Information regarding the longitudinal relaxation of NMR active nuclei can yield valuable insights into the flexibility of molecules. Of particular importance would be new methods to monitor the changes as complexation and aggregation occur in real time, when molecules undergo reactions or biophysical functions. Here, a new ultrafast method to monitor the flexibility changes occurring in molecules in real-time is developed and its use is demonstrated on the reaction of D-xylose with borate. Results obtained with other ultrafast 2D spectroscopic methods complement well the insight obtained from inversion recovery methods. For those spins exhibiting a range from long to short longitudinal relaxation values, UFIR may be supplemented with the MA-IR technique. Using the combined methods, the changes in relaxation parameters associated with a uni-directional reaction can be investigated for an array of timescales; further efforts are under way to investigate similar phenomena in pathological and non-pathological biological processes.
Acknowledgements
P.E.S.S. is grateful to the Fulbright Foundation for a postdoctoral fellowship. Financial support from Agilent Technologies (Research Gift #2305), ERC Advanced Grant #246754, EU’S BioNMR Grant #261863, and the generosity of the Perlman Family Foundation, are also acknowledged.
References
- 1.Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M, Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450(7171):913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 2.Palmer AG, Williams J, McDermott A. Nuclear Magnetic Resonance Studies of Biopolymer Dynamics. J Phys Chem. 1996;100(31):13293–13310. [Google Scholar]
- 3.Guallar V, Jacobson M, McDermott A, Friesner RA. Computational Modeling of the Catalytic Reaction in Triosephosphate Isomerase. J Mol Biol. 2004;337(1):227–239. doi: 10.1016/j.jmb.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 4.Williams JC, McDermott AE. Dynamics of the Flexible Loop of Triose-Phosphate Isomerase: The Loop Motion Is Not Ligand Gated. Biochemistry. 1995;34(26):8309–8319. doi: 10.1021/bi00026a012. [DOI] [PubMed] [Google Scholar]
- 5.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28(23):8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 6.Bertini I, Luchinat C, Niikura Y, Presenti C. Model-free analysis of a thermophilic Fe7S8 protein compared with a mesophilic Fe4S4 protein. Proteins. 2000;41(1):75–85. doi: 10.1002/1097-0134(20001001)41:1<75::aid-prot100>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 7.García a de la Torre J, Huertas ML, Carrasco B. HYDRONMR: Prediction of NMR Relaxation of Globular Proteins from Atomic-Level Structures and Hydrodynamic Calculations. J Magn Reson. 2000;147(1):138–146. doi: 10.1006/jmre.2000.2170. [DOI] [PubMed] [Google Scholar]
- 8.Ghose R, Fushman D, Cowburn D. Determination of the Rotational Diffusion Tensor of Macromolecules in Solution from NMR Relaxation Data with a Combination of Exact and Approximate Methods – Application to the Determination of Interdomain Orientation in Multidomain Proteins. J Magn Reson. 2001;149(2):204–217. doi: 10.1006/jmre.2001.2295. [DOI] [PubMed] [Google Scholar]
- 9.Soong R, Brender JR, Macdonald PM, Ramamoorthy A. Association of Highly Compact Type II Diabetes Related Islet Amyloid Polypeptide Intermediate Species at Physiological Temperature Revealed by Diffusion NMR Spectroscopy. J Am Chem Soc. 2009;131(20):7079–7085. doi: 10.1021/ja900285z. [DOI] [PubMed] [Google Scholar]
- 10.Ishima R, Torchia DA. Protein dynamics from NMR. Nat Struct Mol Biol. 2000;7(9):740–743. doi: 10.1038/78963. [DOI] [PubMed] [Google Scholar]
- 11.Cavanagh J, Akke M. May the driving force be with you – whatever it is. Nat Struct Mol Biol. 2000;7(1):11–13. doi: 10.1038/71202. [DOI] [PubMed] [Google Scholar]
- 12.Forman-Kay JD. The 'dynamics' in the thermodynamics of binding. Nat Struct Mol Biol. 1999;6(12):1086–1087. doi: 10.1038/70008. [DOI] [PubMed] [Google Scholar]
- 13.Akke M, Bruschweiler R, Palmer AG. NMR order parameters and free-energy – an analytical approach and its application to cooperative Ca2+ binding by calbindin-D9k. J Am Chem Soc. 1993;115(21):9832–9833. [Google Scholar]
- 14.Li Z, Raychaudhuri S, Wand AJ. Insights into the local residual entropy of proteins provided by NMR relaxation. Protein Sci. 1996;5(12):2647–2650. doi: 10.1002/pro.5560051228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang D, Kay LE. Contributions to Conformational Entropy Arising from Bond Vector Fluctuations Measured from NMR-Derived Order Parameters: Application to Protein Folding. J Mol Biol. 1996;263(2):369–382. doi: 10.1006/jmbi.1996.0581. [DOI] [PubMed] [Google Scholar]
- 16.Zidek L, Novotny MV, Stone MJ. Increased protein backbone conformational entropy upon hydrophobic ligand binding. Nat Struct Mol Biol. 1999;6(12):1118–1121. doi: 10.1038/70057. [DOI] [PubMed] [Google Scholar]
- 17.Lee AL, Kinnear SA, Wand AJ. Redistribution and loss of side chain entropy upon formation of a calmodulin-peptide complex. Nat Struct Mol Biol. 2000;7(1):72–77. doi: 10.1038/71280. [DOI] [PubMed] [Google Scholar]
- 18.Bracken C, Carr PA, Cavanagh J, Palmer AG. Temperature dependence of intramolecular dynamics of the basic leucine zipper of GCN4: implications for the entropy of association with DNA. J Mol Biol. 1999;285(5):2133–2146. doi: 10.1006/jmbi.1998.2429. [DOI] [PubMed] [Google Scholar]
- 19.Look DC, Locker DR. Time Saving in Measurement of NMR and EPR Relaxation Times. Rev Sci Instrum. 1970;41(2):250–251. [Google Scholar]
- 20.Kay I, Henkelman RM. Practical Implementation and Optimization of One-shot T1 imaging. Magnet Reson Med. 1991;22(2):414–424. doi: 10.1002/mrm.1910220249. [DOI] [PubMed] [Google Scholar]
- 21.Gowland P, Mansfield P. Accurate measurement of T1 in vivo in less than 3 seconds using echo-planar imaging. Magnet Reson Med. 1993;30(3):351–354. doi: 10.1002/mrm.1910300312. [DOI] [PubMed] [Google Scholar]
- 22.McKenzie CA, Pratoa FS, Thornhill RE, Drosta DJ. T1 Fast Acquisition Relaxation Mapping (T1-FARM): Optimized Data Acquisition. Magn Reson Imaging. 2000;18(2):129–138. doi: 10.1016/s0730-725x(99)00133-2. [DOI] [PubMed] [Google Scholar]
- 23.Bhattacharyya R, Kumar A. A fast method for the measurement of long spin-lattice relaxation times by single scan inversion recovery experiment. Chem Phys Lett. 2004;383(1-2):99–103. [Google Scholar]
- 24.Loening NM, Thrippleton MJ, Keeler J, Griffin RG. Single-scan longitudinal relaxation measurements in high-resolution NMR spectroscopy. J Magn Reson. 2003;164(2):321–328. doi: 10.1016/s1090-7807(03)00186-1. [DOI] [PubMed] [Google Scholar]
- 25.Mansfield P. Spatial mapping of the chemical shift in NMR. Magnet Reson Med. 1984;1(3):370–386. doi: 10.1002/mrm.1910010308. [DOI] [PubMed] [Google Scholar]
- 26.James TD, Sandanayake KRAS, Iguchi R, Shinkai S. Novel Saccharide-Photoinduced Electron Transfer Sensors Based on the Interaction of Boronic Acid and Amine. J Am Chem Soc. 1995;117(35):8982–8987. [Google Scholar]
- 27.James TD, Sandanayake KRAS, Shinkai S. Saccharide Sensing with Molecular Receptors Based on Boronic Acid. Angew Chem Int Ed. 1996;35(17):1910–1922. [Google Scholar]
- 28.Liu X-C, Hubbard JL, Scouten WH. Synthesis and structural investigation of two potential boronate affinity chromatography ligands catechol [2-(diisopropylamino)carbonyl]phenylboronate and catechol [2-(diethylamino)carbonyl, 4-methyl]phenylboronate. J Organomet Chem. 1995;493(1-2):91–94. [Google Scholar]
- 29.Adamek V, Liu XC, Zhang YA, Adamkova K, Scouten W. New aliphatic boronate ligands for affinity chromatography. J Chromatogr. 1992;625(2):91–99. [Google Scholar]
- 30.Shrot Y, Shapira B, Frydman L. Ultrafast 2D NMR spectroscopy using a continuous spatial encoding of the spin interactions. J Magn Reson. 2004;171(1):163–170. doi: 10.1016/j.jmr.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 31.Shrot Y, Frydman L. Single-scan 2D DOSY NMR spectroscopy. J Magn Reson. 2008;195(2):226–231. doi: 10.1016/j.jmr.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 32.Chapelle S, Verchere J-F. A 11B and 13C NMR determination of the structures of borate complexes of pentoses and related sugars. Tetrahedron. 1988;44(14):4469–4482. [Google Scholar]
- 33.Verchere JF, Hlaibi M. Stability constants of borate complexes of oligosaccharides. Polyhedron. 1987;6(6):1415–1420. [Google Scholar]
- 34.Ciobanu L, Jayawickrama DA, Zhang X, Webb AG, Sweedler JV. Measuring Reaction Kinetics by Using Multiple Microcoil NMR Spectroscopy. Angew Chem Int Ed. 2003;115(38):4817–4820. doi: 10.1002/anie.200351901. [DOI] [PubMed] [Google Scholar]
- 35.Springsteen G, Wang B. A detailed examination of boronic acid – diol complexation. Tetrahedron. 2002;58(26):5291–5300. [Google Scholar]
- 36.Yang W, He H, Drueckhammer DG. Computer-Guided Design in Molecular Recognition: Design and Synthesis of a Glucopyranose Receptor. Angew Chem Int Ed. 2001;40(9):1714–1718. [PubMed] [Google Scholar]
- 37.Arimori S, Bosch LI, Ward CJ, James TD. Fluorescent internal charge transfer (ICT) saccharide sensor. Tetrahedron Lett. 2001;42(27):4553–4555. [Google Scholar]
- 38.Westmark PR, Valencia LS, Smith BD. Influence of eluent anions in boronate affinity chromatography. J Chromatogr A. 1994;664(1):123–128. [Google Scholar]
- 39.Sugasaki A, Sugiyasu K, Ikeda M, Takeuchi M, Shinkai S. First Successful Molecular Design of an Artificial Lewis Oligosaccharide Binding System Utilizing Positive Homotropic Allosterism. J Am Chem Soc. 2001;123(42):10239–10244. doi: 10.1021/ja010806e. [DOI] [PubMed] [Google Scholar]
- 40.Yang W, Gao S, Gao X, Karnati VVR, Ni W, Wang B, Hooks WB, Carson J, Weston B. Diboronic acids as fluorescent probes for cells expressing sialyl lewis X. Bioorg Med Chem Lett. 2002;12(16):2175–2177. doi: 10.1016/s0960-894x(02)00339-6. [DOI] [PubMed] [Google Scholar]
- 41.Sinton SW. Complexation chemistry of sodium borate with poly(vinyl alcohol) and small diols: a 11B NMR study. Macromolecules. 1987;20(10):2430–2441. [Google Scholar]
- 42.Eggert H, Frederiksen J, Morin C, Norrild JC. A New Glucose-Selective Fluorescent Bisboronic Acid. First Report of Strong a-Furanose Complexation in Aqueous Solution at Physiological pH. J Org Chem. 1999;64(11):3846–3852. [Google Scholar]
- 43.Pepi F, Garzoli S, Tata A, Giacomello P. Low-energy collisionally activated dissociation of pentose-borate complexes. Int J Mass Spectrom. 2010;289(2-3):76–83. [Google Scholar]
- 44.Hadžić PA, Vukojević NS. Selective desulfonylation in D-xylofuranose 3, 5-disulfonates. J Serb Chem Soc. 2001;66(5):289–295. [Google Scholar]
- 45.Uryu T, Yamanouchi J, Hayashi S, Tamaki H, Matsuzaki K. Selective ring-opening polymerization of 1,4-anhydro-2,3-di-O-benzyl-a-D-xylopyranose and synthesis of stereoregular (1 → 5)-a-D-xylofuranan. Macromolecules. 1983;16(2):320–326. [Google Scholar]
- 46.Benesi AJ, Falzone CJ, Banerjee S, Farber GK. NMR assignments for the aldopentoses. Carbohyd Res. 1994;258(0):27–33. [Google Scholar]
- 47.Irarrazabal P, Meyer CH, Nishimura DG, Macovski A. Inhomogeneity correction using an estimated linear field map. Magnet Reson Med. 1996;35(2):278–282. doi: 10.1002/mrm.1910350221. [DOI] [PubMed] [Google Scholar]