Abstract
Mitochondrial dysfunction is at the core of many diseases, ranging from inherited metabolic diseases to common conditions that are associated with ageing. While associations between ageing and mitochondrial function have been identified using mammalian models, much of the mechanistic insight has emerged from C. elegans. Mitochondrial respiration is recognized as an indicator of mitochondrial health. Seahorse XF96 respirometers are the state-of-the-art platform to assess respiration in cells, and we adapted the technique for applications involving C. elegans. Here, we provide a detailed protocol to optimise and measure respiration in C. elegans with the XF96 respirometer, including the interpretation of parameters and results. The protocol takes ~2 days to complete, excluding time spent culturing C. elegans, and includes (i) the preparation of C. elegans samples, (ii) selection and loading of compounds to be injected, (iii) preparing and executing a run with the XF96 respirometer, and (iv) post-experimental data-analysis, including normalization. In addition, we compare our XF96 application with other existing techniques, including the 8-well Seahorse XFp. The main benefits of the XF96 include the limited number of worms required and high-throughput capacity due to 96-well format.
Keywords: C. elegans, oxygen consumption, mitochondria, screens, high throughput biology
Introduction
Mitochondria have long been considered the crucial organelles responsible for the production of ATP1. To perform their key roles in energy production, mitochondria use an intricate system that encompasses the breakdown of substrates – such as glucose, fatty acids, and amino acids – coupled to the generation of ATP via oxidative phosphorylation2,3 (Figure 1a, b). The last few years it has become increasingly apparent that mitochondria are not only involved in the production of cellular energy. The ‘powerhouses’ of the cell are also intimately involved in a variety of processes and signalling pathways including, but not limited to, apoptosis and calcium homeostasis (as reviewed in ref 4–6). As a consequence, it does not come as a surprise that mitochondrial dysfunction is associated with a broad spectrum of diseases, including inherited metabolic diseases as well as common conditions such as ageing, neurodegenerative diseases, cancer and diabetes7–14.
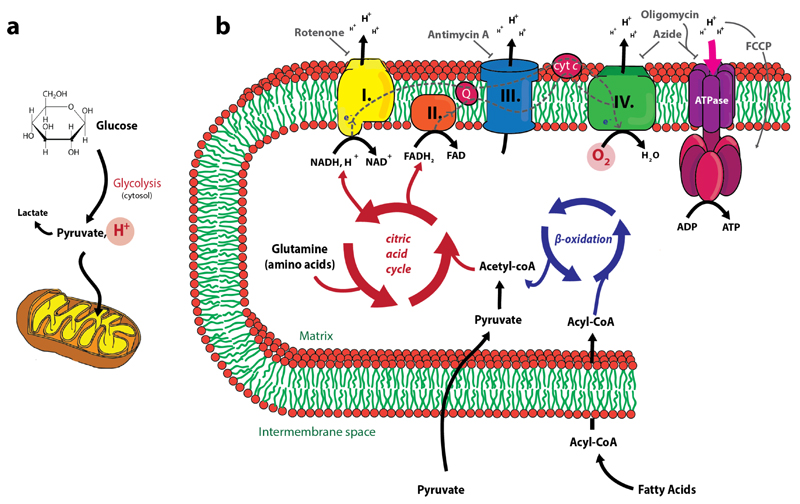
Figure 1. A schematic overview of glycolysis and oxidative phosphorylation.
(a) The mitochondrion with its characteristic structure and the translocation of pyruvate, which is the result of glycolysis. During glycolysis, H+ is generated, which causes extracellular acidification (ECAR) (b) Energy related metabolic pathways that take place in the mitochondrial matrix. Oxidative phosphorylation requires the consumption of O2 (OCR). Compounds known to inhibit the different complexes are illustrated in grey (rotenone, antimycin A, oligomycin and azide). FCCP is an uncoupler reagent and transfers H+ back to the mitochondrial matrix without generating ATP.
Numerous in vitro, in situ and in vivo methodologies have been developed to assess various aspects of mitochondrial function1,15–16. Assays include the estimation of mitochondrial membrane potential17, determining the production and/or concentration of ATP with bioluminescence18 and calcium retention19. While these assays measure one specific aspect of mitochondrial function, they do not cover the full extent of mitochondrial health. The oxygen consumption rate of mitochondria provides an additional layer of complexity that is dependent on many sequential reactions in mitochondria. As such, the measurement of mitochondrial respiration at the level of the electron transport chain (Figure 1b) has emerged as a proxy for mitochondrial health20–22. Indeed, alterations in the rate of oxygen consumption are described as an informative indicator of mitochondrial dysfunction23. It was the introduction of the Seahorse Extracellular Flux Analysers (hereafter called XF respirometers) that allowed measurement of O2 consumption in multiple parallel wells without the need to lyse cells or to isolate mitochondria1,24.
Although initially developed for cultured or isolated cells, XF respirometers can also be used for whole organisms such as Caenorhabditis elegans, when taking some important adjustments into account25–28. C. elegans is often used as a model organism to study mitochondria in the context of longevity29. It is the ease of genetic modifications (e.g. RNAi, CRISPR) that renders the exploration of mitochondria-related pathways in worms readily accessible30–32, especially considering that many of the genes involved in human metabolism are conserved in C. elegans33–36. Consequently, we can use C. elegans as a simple model to study the complex interplay between genes and mitochondrial function, which is usually measured at the level of gene expression (e.g. heat-shock protein induction) or phenotypic characterization of live animals (e.g. locomotion or pharyngeal pumping rates). Nevertheless, the progress of the mitochondrial field in C. elegans was long delayed by the limited options to dedicatedly measure metabolism, in particular mitochondrial activity. Hence, we set out to develop a platform using Seahorse respirometry, which was successfully applied in several projects15,26–27. Here, we describe in detail how Seahorse XF96 respirometers can be used to study mitochondrial biology in C. elegans. Moreover, we also provide detailed information about how parameters can by optimised for individual worm strains.
Theoretical background of XF96 respirometers
The XF96 respirometers provide researchers with the possibility to do real-time measurements of the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR). The OCR is predominantly the result of mitochondrial respiration through oxidative phosphorylation (Figure 1b), while ECAR is predominantly the result of glycolysis (Figure 1a). The system uses a 96-well plate combined with a sensor cartridge containing an equal number of individual sensor probes. At the tip of each of the sensor probes, there are two separate polymer-embedded fluorophores that are sensitive to either O2 or H+. The sensitivity of theses probes is based on the quenching chemistry of the fluorophores in response to O2 and H+ in the assay medium. The concentration of O2 and H+ is therefore directly linked to the intensity of the fluorescence signal relative to a standard solution. Moreover, any changes in the signal can be inferred to be proportional to changes in O2 or H+ concentration, and therefore rates of change in O2 or H+ concentration can be obtained by following the fluorescent signal over time.
During a measurement cycle, the sensor cartridge is lowered into the wells creating a transient microchamber of a defined volume (Figure 2a). Fiber optic bundles are inserted into the probes, and light is pumped into the sensor to excite the embedded fluorophores. The resulting emitted light is transmitted back through the fiber optic bundle and measured by the detector. The fluorescent emission is measured for a specified period (measuring time; Figure 2b). The slope of the linear decline (for O2 concentration over time) or incline (for H+ concentration over time) provides the basis for calculating the OCR (in pmol/min) and ECAR (mpH/min), respectively (Figure 2b). The exact algorithm that is used to calculate the OCR in XF respirometers is described in ref (37).
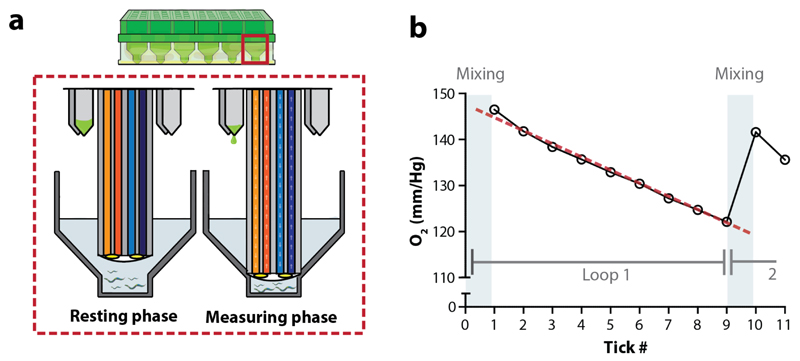
Figure 2. The measurement units of XF respirometers.
(a) A close-up of the probes of the sensor cartridge. During the measuring phase probes of the sensor cartridge are lowered, thereby creating a transient microchamber (from resting phase to measuring phase). Light is transmitted through the fiber optic bundles to excite the embedded fluorophores, that are sensitive to O2 and H+, at the tip of the probe. Emitted light then passes back up the optic bundle to the detector. Compounds can be injected during the experimental assay via ports at the base of the probe. (b) During a measuring cycle oxygen is consumed and the concentration of the analyte drops. A representative measurement is shown. The mixing phase represents the stage where the probe is repeatedly moving between the resting position and measuring position, allowing for mixing of the medium. During the measuring phase, O2 (mm/Hg) concentration is estimated 9 times per loop, when measuring for 2 minutes. These 9 estimations of the oxygen concentration (or ticks: as directly derived from the ‘level data’ in the Excel file) together are used to calculate one OCR per loop. Here, the linear decline of O2 (mm/Hg) concentration (red dotted line) provides the basis for the OCR estimation.
After several minutes of continuous measurement, the sensor cartridge is raised and subsequently allowed to move up and down for a specified time interval, causing the larger volume of medium above to mix with the medium in the transient microchamber, thereby restoring the O2 and H+ concentration (mixing time; Figure 2b). In order to get multiple, steady estimations of the OCR and ECAR, the measuring and mixing cycle can be repeated (looped). In addition, the presence of an integrated drug delivery system in the sensor cartridge plate allows sequential addition of up to four compounds/solutions per well at user-defined time points. Selection of the compounds to be added, and the sequence in which they are added, makes it possible to differentiate between several aspects of cellular respiration, such as determining non-mitochondrial respiration by blocking mitochondrial respiration with a compound (e.g. rotenone or antimycin A)38–41 (Figure 1b).
Comparison with other in vivo respirometry methods
There are several methods available to measure respiration of living samples, which can be globally divided into two groups: O2-dependent quenching of porphyrin-based phosphors (Seahorse Bioscience XF respirometer and Luxcel MitoXpress) and amperometric O2 sensors (Clark electrodes, including the widely adopted Oroboros system)1,42. Historically, the amperometric approach has been the main method used to assess mitochondrial respiration in C. elegans. For the amperometric approach, nematodes are delivered into a single respiratory chamber, which is separated from two half-cells by O2-permeable material. In this way, only O2 can diffuse from the assay medium through the membrane. When a small voltage is applied to the half-cells, O2 is reduced by electrons at the cathode yielding hydrogen peroxide. Subsequently, H2O2 oxidizes the Ag (silver) of the Ag/AgCl anode, which results in an electrical current that is proportional to the O2 pressure – and thus concentration – in the experimental respiratory chamber.
Apart from the detection modality, differences of the XF respirometric method appear at the level of number of worms per assay, replicates, multiple, or real-time measurements and the ability to inject compounds during an experiment (Table 1). The Clark electrode approach requires thousands (~2000-5000) of worms in a single chamber to obtain an estimation of the oxygen consumption rate43. Performing multiple measurements, biological replicates and comparing conditions provide the biggest challenges within the Clark electrode method as the traditional set-up only allows the measurements of one sample at a time. In contrast, a XF96 respirometer requires ~10-20 worms per well to acquire a reproducible oxygen consumption rate, measurements can be easily and quickly (in the order of minutes) repeated in an automated way and since XF respirometers can analyse whole plates at the same time, about 96 conditions/replicates can be tested at once. An additional difference is the presence of drug-injection ports that can be programmed to inject compounds in all 96 wells at time points that are specified a priori during an XF respirometer experiment. Clark electrode systems also allow injection of compounds, and even offer flexibility with respect to the timing, dosing and number of additions as compounds are injected manually during the course of the assay. However, precise timing of manual additions between replicate experiments may be challenging.
Table 1. Differences between in vivo respirometric methods.
| Clark-electrode | Seahorse XF96 Flux analyser | Luxcel MitoXpress | |
|---|---|---|---|
| Type of approach | Amperometric | Porphyrin-based phosphors | Porphyrin-based phosphors |
| Required # of worms | ~2000-5000 | ~2-25 | ~2-25a |
| Time required for measurement of OCR | 2-15 minutes | 2-3 minutes | >90 minutes |
| Multiple measurements/loops | No | Yes, automated | No |
| Maximal conditions per run | 1 | 96 wells per plate (92 without background correction wells) | 96 or 384 (92 or 380 without background correction wells) |
| Injection of compounds during run | Yes, manualb | Yes, automated | Noc |
Provides an estimation of the number of worms per well, since we did not test this method ourselves and specific numbers are not mentioned in literature yet. Numbers are derived from those in the XF96 respirometers.
Compounds can be injected manually, not in an automated fashion.
The influence of compounds can be assessed in this method, but they should be injected prior to the start of the assay.
More similar to the Seahorse XF respirometer method is the Luxcel MitoXpress O2 consumption assay, which relies on O2-dependent quenching of porphyrin-based phosphor. The MitoXpress kits provide a way of performing real-time analysis of cellular respiration, via an oxygen-quenching fluorophore system. Worms are placed into the wells of a 96- or 384-well plate, the kit reagents are added, and measurements are made in a fluorometric plate reader. Multiple conditions and replicates can be tested side-by-side in the wells of a single plate, but repeated measurements over time are more challenging as there is typically no automatized mixing system integrated in the plates or plate-readers to restore basal O2 levels. In addition, single estimation of the OCR takes >90 minutes, while careful estimations of the OCR in the XF respirometer approach takes only 2-5 minutes of measuring time. Finally, the use of compounds to assess multiple aspects of mitochondrial function related to oxygen consumption is limited since compounds need to be injected manually immediately prior to the start of the experiment.
Advantages and limitations of XF respirometry
Based on the comparisons with other respirometric methods, the XF respirometer approach offers important benefits to measure mitochondrial respiration in C. elegans, especially when considering the ease to perform repeated measurements, comparing conditions, replicates and investigating several aspects of mitochondrial function. This allows a higher throughput for screening-based applications; e.g. genome-wide screens to find genetic interactors affecting mitochondrial bioenergetics and compound screens for mitochondrial toxicity. Indeed, XF respirometry was already successfully applied to study the effects of compounds or gene knockdowns on mitochondrial function in C. elegans15,26–27.
In vivo assessment of mitochondrial function in C. elegans has, however, also some clear limitations. First of all, oxygen consumption of an intact organism (e.g. basal respiration) does not strictly reflect mitochondrial respiration. For instance, cells possess a variety of oxygenases that also consume oxygen and thereby contribute to the oxygen consumption rate44. Hence, residual respiration should be estimated in the presence of effective electron transport inhibitors in order to distinguish non-mitochondrial respiration from mitochondrial respiration (Figure 3a). In cells, it is well-established to use four compounds (making use of the drug injection ports; Figure 3b) to distinguish between and quantify several aspects of cellular respiration including, but not limited to: (1) respiration linked to ATP production (using oligomycin); (2) maximal respiration (using carbonyl cyanide-4(ptrifluormethoxy)phenylhydrazone: FCCP); and (3) mitochondrial and non-mitochondrial respiration (using antimycin A and rotenone) (Table 2; Figure 1b)38–41. Nevertheless, C. elegans has traditionally been considered as a poor candidate for drug-related assays due to the relatively inefficient uptake of compounds caused by impermeability of the cuticle to non-water soluble compounds45–47. Consequently, it does not come as a surprise that, except for FCCP, the compounds typically used in cultured cells are inefficient in acutely inhibiting specific aspects of mitochondrial function in C. elegans (Supplementary Figure 1)28 . However, there are alternative compounds, such as the complex IV/V inhibitor sodium azide, which can be used to help decipher aspects of mitochondrial respiration in C. elegans28,48 (Figure 1b).
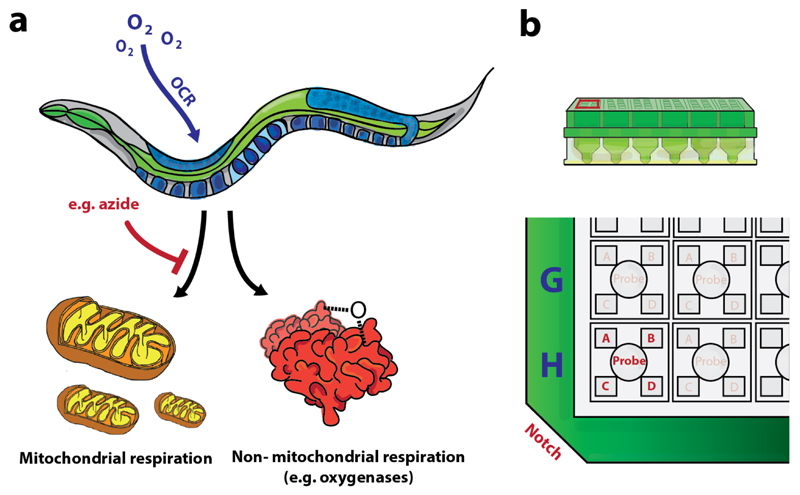
Figure 3. Distinguishing non-mitochondrial respiration from mitochondrial respiration.
(a) The oxygen consumption in cells and organisms is not only due to mitochondrial respiration. Therefore, one should add a compound (e.g. sodium azide) to block mitochondrial respiration, in order to distinguish mitochondrial oxygen consumption from other oxygen consuming processes. (b) Lay-out of the drug injection ports per well; indexed A-D when considering the notch at the bottom left.
Table 2. Compounds used to measure key parameters of mitochondrial function.
| Compound | Function | Description |
|---|---|---|
| Oligomycina | ATP synthase inhibitor | Inhibits ATP synthesis by blocking the proton channel of the F0 portion ATP synthase (complex V) |
| FCCPa | ETC accelerator | An ionophore that disrupts the mitochondrial membrane potential and thus ATP synthesis while still allowing proton pumping, electron transport and oxygen consumption |
| Antimycin Aa and Rotenone | Mito inhibitors | Rotenone is a complex I inhibitor and antimycin a complex III inhibitor. Together inhibits total mitochondrial respiration |
| Sodium azideb | Complex IV and V inhibitor | Inhibits complex IV (cytochrome c oxidase) and ATP-synthase (complex V) |
Are commonly used in cells (Mito Stress kit, Bioscience).
Is not used in cell assays, but is used to inhibit mitochondrial respiration in C. elegans and is a good alternative to antimycin A and rotenone (which are inefficient in C. elegans).
One can argue that isolating mitochondria of C. elegans would represent a more sophisticated way to assess mitochondrial function. Isolated mitochondria make exploration of the molecular (super)complexes involved in oxidative phosphorylation possible49. Changes in respiratory chain complexes and anaplerotic enzymes can be identified using specific combinations of substrates and inhibitors to the isolated mitochondria50, allowing careful dissection of the respiratory chain. Nevertheless, the clear advantages of isolated mitochondria are also accompanied by several disadvantages. First of all, existing methods often require large amounts of sample due to a poor yield of mitochondrial content, and do not prevent biased selection towards specific mitochondrial populations51–54. Second, the isolation process may decrease mitochondrial membrane integrity and does disrupt both the mitochondrial environment and the cellular/organismal context, which affects mitochondrial morphology, respiration and ROS production53–54. In addition, mitochondrial isolation procedures are technically challenging in C. elegans compared to tissue samples or even cells, precluding the use of such a strategy for routine biochemical analysis55. As such, it is clear that the XF respirometers provide the user with a platform to do relatively quick high-throughput screens in an organismal context. In this way, interactions of the mitochondria with the rest of the cell are maintained and so is the physiological relevance.
Finally, next to the aforementioned considerations of using XF respirometers for assessing mitochondrial function in C. elegans, some critical technical ‘limitations’ should be mentioned. Since XF respirometers were originally developed for cells, the temperature control/heater of the machine is designed to work at 37°C. Considering that nematodes are typically cultured at 20-25°C, and their physiology is dependent on the environmental temperature, the lack of tight temperature control could affect the outcome of the experiments. Depending on the environmental setting of the XF respirometers, turning the heater off will result in temperatures of around 20-25°C, which will increase during the run with about 2-4°C. We have found that temperatures in the range of 20-25°C will not cause any significant changes in the respiration rate of wild type C. elegans (Figure 4). Nevertheless, it is strongly advised to only compare strains within one plate to account for masked effects of temperature on respiration. Another aspect to consider is that ECAR cannot be assessed in worms when using the protocol we describe here, since worms are assayed in buffered medium (M9, see below) that precludes changes in H+ concentration. A method for measuring ECAR in C. elegans with XFe24 respirometer has been described recently, which uses unbuffered water as assay medium28. Nonetheless, even when it is possible to estimate the ECAR, it is unclear whether this represents a valid marker for glycolytic rate. Further validation of these parameters will hopefully shed light on this option and expand the toolkit for measuring metabolism in C. elegans.
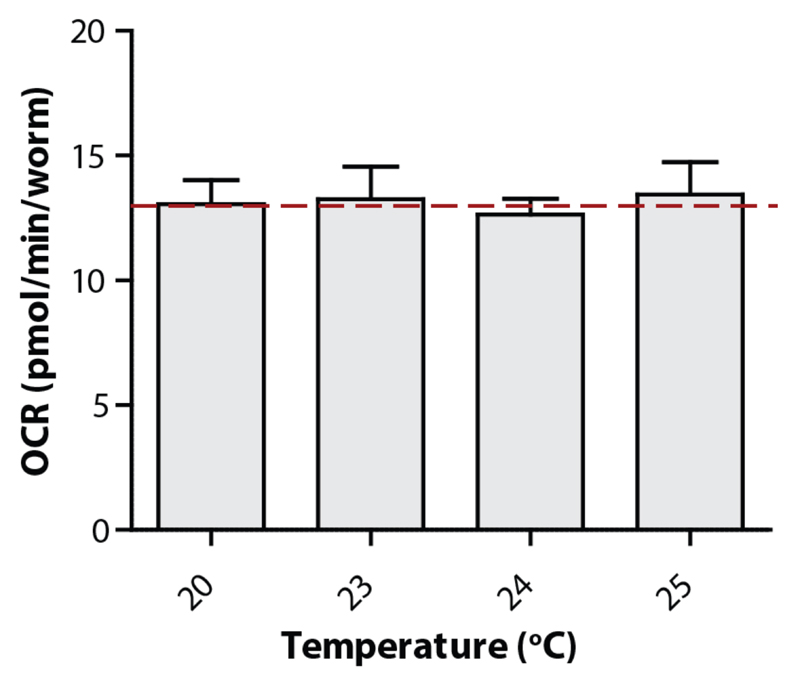
Figure 4. Temperature does not influence the OCR of C. elegans .
Up to 25°C, the basal respiration of nematodes (N2) is not influenced by the temperature. Bars are mean ± SEM. One-way ANOVA (ns), n = 7-9 wells.
Alternative applications
The XF96 respirometer method provides a flexible platform for performing genome-wide screens or compound-screens in relation to mitochondrial respiration. As the experimental set-up only requires organisms that are viable in liquid culture and have a size that fits the transient microchamber (XF96: 3 μl, XF24: 7 μl), mitochondrial respiration can also be estimated in species other than C. elegans. Therefore, Seahorse XF respirometers might open new avenues for other well-established or upcoming species that may be used in metabolic or aging studies, such as the flatworm Macrostomum lignano56, Danio Rerio embryos57–58 and even Nothobranchius furzeri embryos59. While XF respirometers are, to our knowledge, not used in Macrostomum lignano or Nothobranchius furzeri research yet, it has been shown that respiratory kinetics in embryos from Danio Rerio can be investigated with XF respirometers58.
Experimental design
Overview of the procedure
Performing a XF respirometer experiment with C. elegans is technically a simple procedure that consists of six main steps: (a) pre-experimental procedures; (b) Preparing the XF respirometer; (c) loading compounds; (d) collecting and loading worm samples; (e) the actual XF experiment; and (f) post-experimental data analysis. An overview of the experimental procedure is given in Figure 5. Optimisation of steps a-d is required in order to get reliable estimations of the OCR and to reduce variation within runs. Some of the preparatory steps are very similar to those used in cells50. However, many of the exact parameters and steps cannot be translated one-on-one to C. elegans, and it is therefore highly recommended to take note of our proposed optimisation steps before continuing to the Procedure section.
Figure 5. A schematic overview of a XF respirometer experiment.
Overview of experimental procedures involved in preparing and performing a XF assay with C. elegans. The procedure consists of (1) synchronizing worms, (2) ageing worms, (3) preparing compounds to be injected, (4) hydrating the probes of the cartridge (step 1-4 in PROCEDURE), (5) preparing the XF respirometer (step 5-14 in PROCEDURE), (6) loading the compounds in the injection ports (step 15-20 in PROCEDURE), (7) collecting, washing and loading worm samples (step 21-35 in PROCEDURE), and (8) executing the actual respirometer run and performing data analysis (step 36-47 in PROCEDURE). See main text for a more detailed explanation of all steps involved.
(a). Pre-experimental procedures
The pre-experimental procedures should be performed one day before the actual XF experiment, but will in general take more than one day of preparations. Comparing OCR of different worm strains requires strict age synchrony since respiration changes significantly during development and ageing (Figure 6). Hence, it is important that careful phenotyping is performed prior to the respiration experiments in order to anticipate potential differences in rate of development and worm size. When assessed, worm strains can be age synchronized (hypochlorite treatment, or egg laying) several days before the experiment60. To avoid offspring or eggs from hatching one can use plates containing 5-Fluoro-2'-deoxyuridine (FUdR) (starting from young adults)61. Here, it is important to realize that FUdR can alter the biology of the worms, as evidenced by reported changes to lifespan, worm size and metabolite levels62–63. Other techniques to maintain synchronized worm cultures are described in ref (64). In addition to preparing the worm samples, the cartridge needs to be prepared in advance in order to hydrate the sensor probes. The sensor cartridges should be incubated with Seahorse Bioscience XF96 calibrant solution pH 7.4 at 37°C without CO2 (to keep the calibration temperature constant and optimal for XF respirometers). It is recommended to hydrate the probes overnight, but a minimum hydration time of 4h is required, and if necessary the hydration can be prolonged to 72h, in which case we advise to seal the plate with parafilm and store at 4°C. One day prior to the assay, one can also prepare stock solutions of the compounds to be injected during the seahorse experiment, such as FCCP (10mM in DMSO: 1000x the intended working concentration) and sodium azide (400mM in dH2O: 10x working concentration). It is important to realise that compounds are ten times diluted when they are injected in the wells (due to the volume already present). Therefore, stock solutions need to have a concentration that is 10-fold higher than the intended working concentration. Moreover, for compounds that are dissolved in DMSO it is highly suggested to make a stock that is 1000x concentrated, to ensure that the DMSO concentration is ≤1% (dilute 100x to get the stock required for injection)65.
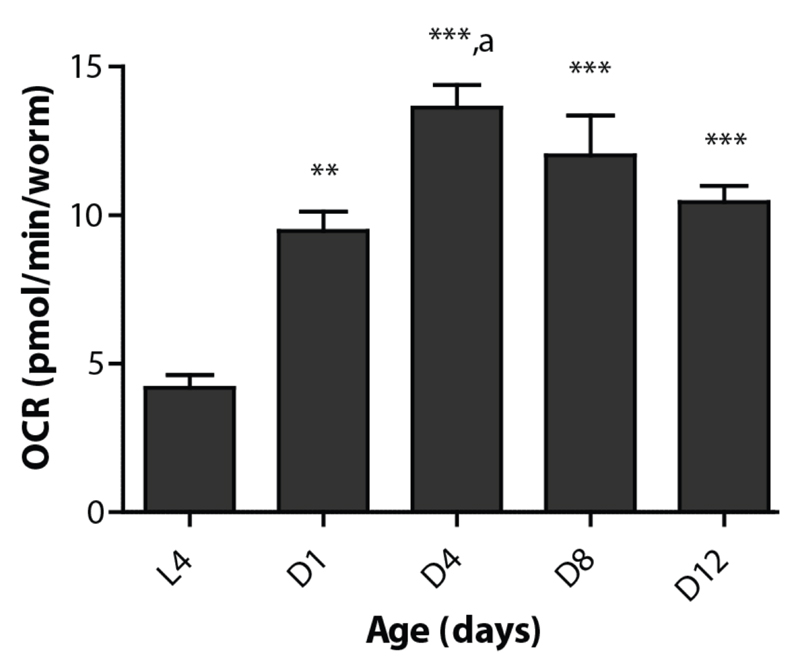
Figure 6. Respiration rates change during development and with age in C. elegans .
Basal respiration changes during development (from L4 to adult stage) and ageing in N2 worms. All adult stages show significantly higher respiration than the larval stage L4. Also, respiration is significantly higher at day 4 of adulthood (D4) compared to day 1 (D1) (p < 0.05) as indicated with ‘a’. Bars are mean ± SEM. One-way ANOVA (p < 0.001) with post-hoc Tukey, n = 6-8 wells. ** = p < 0.01, *** = p < 0.001.
(b). Preparing the XF respirometer
In anticipation of the respiration measurements, the XF96 software should be prepared and programmed. Although the exact steps are described in the PROCEDURE, there are some important aspects that require clarification. Since XF respirometers were initially developed for cell-related research, the machine has an automatic heater and temperature control system to maintain a temperature of 37°C. For Seahorse experiments with C. elegans it is essential that the temperature control system and heater are turned off in advance allowing the machine to adapt to room temperature (~20-25 °C). Be aware that every time you restart the respirator, the temperature control system and the heater have to be turned off.
The ‘assay wizard’ embedded in the Seahorse software provides a user-friendly interface to generate an experimental template with all the commands to control the XF96 respirometer during the experiment. Many of the tabs and fields are to the user’s discretion to fill, including for instance user info, medium info, and plate layout. While these items can be helpful in tracing the experimental conditions, they do not affect the XF run. Three tabs are, however, important to adjust: ‘General’ where you fill in the date and name of your experiment; ‘Background correction’, where you select the wells that function as correction wells (these contain buffer only); and ‘Protocol’, where you specify the order and duration of mixing, measuring and waiting time as well as the injection of compounds. The ‘Protocol’ tab provides you with several commands that can be executed by the machine. Although most of them are self-explanatory, we provide here a brief overview:
Calibrate: This is always the first command of every XF assay; it calibrates the sensors for the planned experiment. Takes about 15-20 minutes.
Equilibrate: Is always the second step after calibration. It consists of 3 loops of 2 minutes of mixing and waiting respectively. Equilibration is performed to stabilize the temperature of the plate.
Mix: Mixes the volume of the wells by gentle up/down motion of the sensor sleeves. Mixing is necessary to restore analyte (O2 and H+) distribution and concentration in the wells and to mix injected compounds.
Wait: Instructs the machine to wait for an x number of minutes.
Measure: Instructs the machine to do the actual measurements to determine the OCR and ECAR.
Inject: Instructs the machine to inject compounds that are loaded in the ports into the wells. You can choose here also which port should be injected (A, B, C or D).
Loop: Is used to repeat a subset of commands for x number of times.
The duration of the commands mix, wait and measure should be carefully optimised to ensure a stable OCR over time, see Optimisation of parameters. Also, multiple loops should be performed to get not one, but multiple estimations of the OCR. When the experimental template is ready, the actual run can be directly started, however, to avoid unnecessary heating, it is important to only start the run whenever the samples are almost ready.
(c). Loading compounds
The benefit of working with XF96 respirometers is the possibility to use the integrated drug delivery system that allows sequential injection of up to four compounds. With the use of specific mitochondrial toxins a more complete profile of mitochondrial function can be generated. When a XF experimental template is generated, it is possible to include the command ‘Injection of port X’, where X refers to a port indexed with a specific letter: A-D. The drug ports are organized in the following way around each probe: A = top left, B = top right, C = lower left, D = lower right (Figure 3b). To facilitate the loading procedure, the sensor cartridge is accompanied by two loading guides, i.e. lids with holes that allow easy access to pipet only in the port of choice. When the sensor cartridge is loaded with the compound(s), the plate can be placed back in an incubator at 37°C without CO2. It is important to realize that the injection is initiated through a pneumatic mechanism. As a consequence, when using a specific injection port, this port should be filled for all 96 wells, even if those wells are not used for the oxygen consumption measurement.
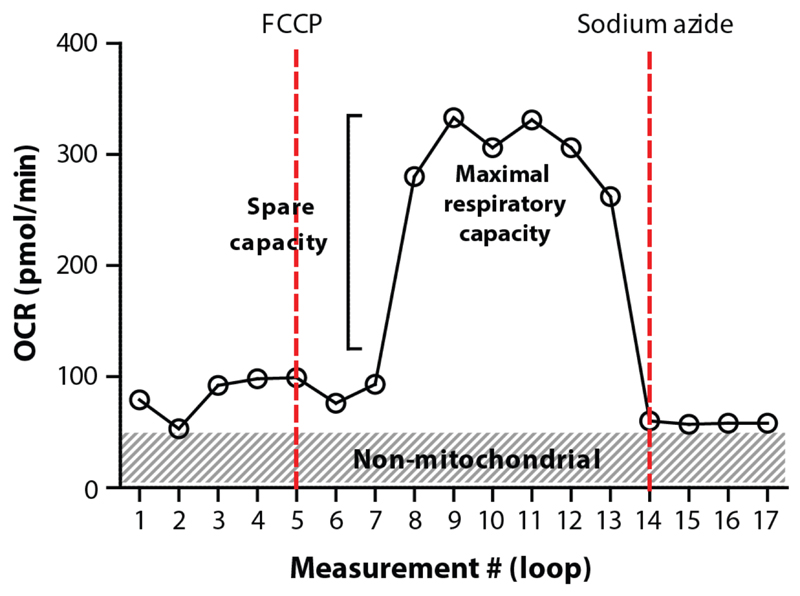
There are several compounds that can be injected to assess different aspects of respiration. A drug that we have extensively tested and that is often used in combination with XF96 respirometers is FCCP. FCCP is a proton ionophore, indicating that it is able to transport hydrogen ions across membranes. FCCP is therefore an uncoupling agent, since it disrupts ATP synthesis by transporting hydrogen ions across the inner mitochondrial membrane instead of the proton channel of the ATP synthase (complex V)66. Consequently, administration of FCCP results in the collapse of the mitochondrial membrane potential, thereby inducing a rapid increase in the consumption of energy and oxygen without the generation of ATP. FCCP injection can be used to calculate the maximal and spare respiratory capacity of worms. The latter is defined as the quantitative difference between the initial basal OCR and the maximal uncontrolled OCR (induced by FCCP) (Figure 7). Another drug that we have tested is sodium azide. Sodium azide blocks both cytochrome c oxidase (complex IV) and the ATP synthase (complex V), thereby shutting down the whole electron transport chain48. Therefore, we use sodium azide to estimate non-mitochondrial respiration, i.e. respiration caused by other cellular processes than oxidative phosphorylation (Figure 3a, 7).
Figure 7. Typical example of OCR profiles in C. elegans respirometry.
Starting with the basal respiration, the spare capacity and maximal (mitochondrial) respiratory capacity (plateau phase) can be estimated by injecting FCCP. Non-mitochondrial respiration can be distinguished from mitochondrial respiration by injecting sodium azide.
(d). Collecting and loading worm samples
When collecting the nematodes for the XF respirometric assay, several aspects should be taken into account. First of all, before removing worms from their plates, it is important to make note of any abnormalities, e.g. infections by fungi and the amount of food present (starvation). These observations may be critical in the interpretation of the data. Furthermore, collected worms should be washed multiple times to get rid of E. coli. However, we have shown that the contribution of the E. coli to respiration is marginal and therefore will not affect the respiration data extensively (Supplementary Figure 2). By estimating the number of worms per volume unit, one can estimate the volume required for a specific number of worms per well. This number needs to be optimised per strain and age, see optimisation of parameters.
Worms are loaded on a freshly opened cell culture microplate of Seahorse Bioscience (the utility plate), keeping the total volume per well at 200 μl (= μl of worms + μl M9). Use a XF96 plate layout (Supplementary Figure 3) to keep track of which samples were placed in which wells. Visual inspection of the wells should show approximately the same number of worms in each experimental well and no worms in the background correction wells (typically the four corner wells: A1, A12, H1 and H12). The time between collecting the worms from their plate and loading them into a XF96 utility plate should be as short as possible, but not shorter than 30 minutes to ensure that the worms have cleared their gut and the marginal oxygen consumption due to E. coli is further reduced. The OCR in the worms has a slight tendency to drop over time, therefore a much longer waiting time, e.g. 2h, should be avoided as much as possible.
(e). Starting the XF experiment
When the XF respirometer is prepared in advance, one can start the assay directly. We suggest starting the XF experimental assay when worms are collected, but not loaded yet (see Procedure), to avoid unnecessary heating of the machine before running the actual experiment. Calibration, the first command of an experimental template, takes around 20 minutes during which the worms can be transferred to the utility plate. Just before starting calibration, we advise to briefly place the sensor cartridge with calibrant solution at 37°C, so that the calibration will take place at approximately 37°C, which is the optimal temperature for the instrument’s determination of actual oxygen concentrations. However, since the protocol is set to room temperature (i.e. the heater is turned off), the interior of the wells and machine will actually be around 20-25°C during the measurement. Once the worms are loaded into the wells and the calibration has completed, the experimental part of the XF assay can start. The results of the XF assay can be followed in real-time, but it should be mentioned that the OCR is represented per well and not per worm or μg protein. In addition to that, the XF software can be unstable during the assay causing it to crash, which is why we discourage using the software during the run.
(f). Post-experimental data-analysis
In order to interpret the data and compare strains or treatments, the OCR per well should be converted in OCR per worm (or OCR per μg protein). Therefore, the number of worms per wells needs to be counted. This can either be done directly, or one can capture images of each well using a microscope and count the number of worms per well at a later date Use the XF96 plate layout sheet (Supplementary Figure 3) to fill in the number of worms per well. Some mutant or RNAi-treated worms are smaller than age-matched controls. In this case, correcting for protein levels can provide additional information, although mitochondrial deficiency through RNAi leads to a reduced OCR with both normalisation protocols67. Additionally, differences in mitochondrial activity can be picked up by normalisation to mitochondrial DNA content68.
While inspecting the wells and/or images, make also note of any anomalies observed in the wells, such as dust or dirt, bacteria, embryos or larvae, as these may be important when interpreting the respiration data in the next step. An Excel spreadsheet is generated by the program after the XF assay is done. There will be different tabs in this spreadsheet, starting with ‘Data viewer’. Although most of the information within the tabs is self-explanatory, a quick overview:
Data viewer: when the Seahorse XF96 software is installed on the same computer that you use for analysis, there will be a Seahorse plugin in the ‘data viewer’ tab (Windows only). Here, the data representation is identical to that on the XF respirometer computer. Although it gives a quick summary of the run, it is not corrected for worm number or protein concentration.
Assay configuration: the XF assay template is summarized here. The protocol, groups, background wells and all other parameters that were filled in are collected in this overview tab.
Levels: This is one of the most important tabs, as it provides an overview of the oxygen concentration, pH and the temperature per well and per time point.
Rate data: the tab with the actual measurements of OCR and ECAR per well and time point. For C. elegans only the OCR column is of importance. Wells 1-12 are at the first row of the plate (A1-A12), wells 13-24 are on the second row (B1-B12) and so on. Please note that as well 1, 12, 85, 96 are empty, these values should all be at zero.
Rate date (Plate view): also showing the actual measurements of OCR and ECAR, but this time in a plate view in which respiration rates match the appropriate wells.
Time events: a written overview of all the steps in the Seahorse protocol
Calibration: an overview of the calibration parameters
The tab with ‘rate data’ is needed to do the actual calculations. The OCR values per well and time point can be divided by the number of worms in that specific well. Please note that the OCR and ECAR values are typically not stable until the second or third measurement cycle (or loop), thus it is strongly recommended that the first basal measurement is excluded from analysis in order to establish a consistent baseline. Take the average of all the basal measurements per well for a good estimation of the OCR.
Overview of parameter optimisation
There might be differences between C. elegans strains with respect to size and respiration rates. Therefore, it is essential to first optimize some important parameters for the strains of interest. These parameters include, but are not limited to: the timing of commands in the XF respirometer experimental template (i.e. measuring, mixing and waiting time), the number of worms per well, the stability of the OCR over time and the concentrations of compounds to be injected. By taking into account the parameters and optimisation steps, one can adjust the protocol to each individual worm strain and also adapt the protocol to other organisms. The step-by-step PROCEDURE can easily be used to do optimisation runs when parameters in specific steps are adjusted as described in the following sections.
Table 3 shows the suggested start conditions, including optimal drug concentrations and timing, the XF template and the number of worms when working with the N2 strain. This set of parameters also works for worms treated with RNAi, the mev-1(kn1) mutant, and several other mutants.
Table 3. Optimized parameters for N2.
| Parameter/protocol | Optimized values |
|---|---|
| XF template | (1) Calibrate, (2) Equilibrate, (3) Loop 5 times, (3a) Mix 2 minutes, (3b) Wait 0.5 minutes, (3c) Measure 2 minutes (4) End loop. Optional: (5) Inject port A/FCCP, (6) Loop 9 times, (6a) Mix 2 minutes, (6b) Wait 0.5 minutes, (6c) Measure 2 minutes, (7) End loop, (8) Inject port B/Sodium azide, (9) Loop 4 times, (9a) Mix 2 minutes, (9b) Wait 0.5 minutes, (9c) Measure 2 minutes, (10) End loop, (11) End programme. |
| Maximal loops | 12 – 20a |
| Number of worms/well | Minimum of 2 worms. Maximum for L4: up to 50, D1: up to 25 , D3: up to 30, D5: up to 30, D8: up to 30. |
| Drug concentrations | FCCP: final concentration 10 μM (injection concentration: 100 μM), Sodium azide: final concentration 40 mM (injection concentration 400 mM). |
When the loop consisting of 2 minutes measuring, 2 minutes mixing and 0.5 minutes of waiting is used, the OCR remains stable up till 20 loops, although there might be some fluctuations. Up till 12 loops the OCR is consistently stable.
When only interested in basal respiration, one can increase 5 loops to e.g. 10. However, this is not necessary, 5 loops are sufficient to get a reliable estimation of the OCR.
Optimisation of assay timing
Optimizing the timing of the XF assay commands mixing, measuring and waiting requires interpretation of two aspects of the OCR: the course of the changing oxygen concentration and the stability of OCR over time. Based on our experience, we advise to start with the following protocol: (1) Calibrate, (2) Equilibrate, (3) Loop X times, (3a) Mix for 2 minutes, (3b) Measure for 3 minutes, (4) Loop end, (5) Programme end. Set X in loops at 20, pipette 10-25 worms per well (see optimisation number of worms), start the run (PROCEDURE) and look for the factors mentioned above (course of the changing oxygen concentration and the stability of OCR over time).
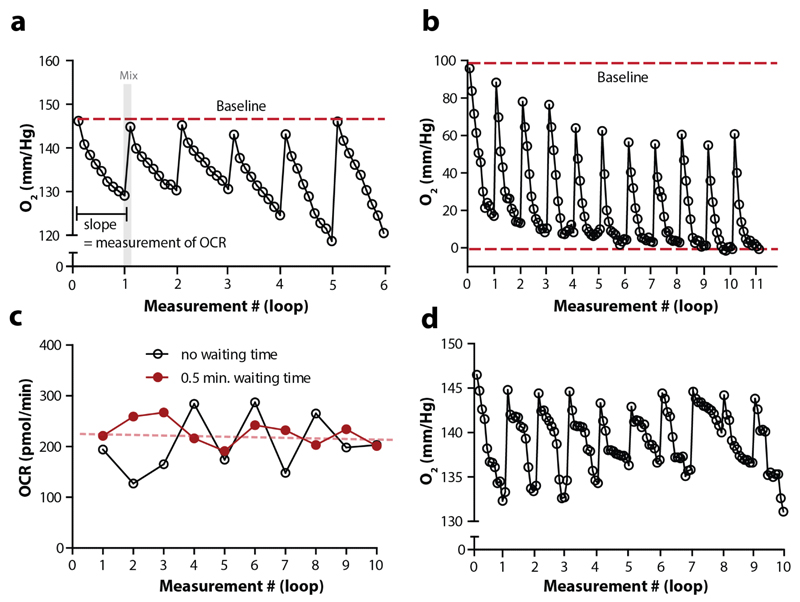
When investigating the course of changing oxygen concentration, it is essential to determine whether the parameters within a loop (measuring, mixing and, when applicable, waiting time) are optimal. During the measurement step, oxygen is consumed in the transient microchamber, and the oxygen concentration drops. During mixing, the oxygen concentration should go back to baseline level (Figure 8a). When O2 concentration does not reach the baseline after each mixing cycle this often indicates that the mixing time is too short. Alternatively, if oxygen concentration drops to zero during the measurement phase (Figure 8b), this may cause an underestimation of the OCR and the measuring time or the number of worms per well should be decreased. The raw data concerning oxygen concentration per time unit can be found in the ‘levels’ tab of the generated Excel result sheet. Add a filter to the column ‘well’ in order to follow the changes in oxygen concentration over time per specific well.
Figure 8. Detailed tracing of oxygen levels is required to identify experimental issues.
(a) The oxygen concentration at the start of each loop (measure cycle) should be the same, i.e. it should touch the imaginary baseline (red dotted line). The O2 concentration is estimated 9 times per loop, when measuring for 2 minutes. These 9 measurements (ticks) together are used to calculate the OCR per loop. After each measure cycle, mixing takes place (as indicated with the grey area) to restore the initial O2 concentration. (b) The graph shows data from a single well, in which the oxygen concentration does not return back to baseline after a loop due to an excess of worms. Moreover, in the last 3 loops the oxygen concentration drop below 0, which causes an underestimation of the OCR. Loop: 2 min. mixing - 3 min. of measuring. (c) The OCR over time when a protocol with or without waiting time is used (both 2 minutes mixing and measuring). Clearly, introducing a waiting step decreases the fluctuation over time. Moreover, when applying linear regression to the OCR data with a waiting step (red dotted line), the slope does not significantly differ from 0, indicating that the OCR is stable over time (single well data). (d) Data from a single well that shows clear fluctuation in the steepness of the slopes per loop. Loop: 2 min. mixing - 3 min. of measuring. The variation in these slopes is likely caused by worms not always being in the transient microchamber.
The stability of OCR over time can be studied by looking at the OCR estimations over the duration of the experiment. Since there are no compounds injected during the optimisation runs, basal respiration should stay stable over time (at least up to 60 minutes, Figure 8c, red dotted line). Small fluctuations in OCR typically occur across the different time-points, usually without any overall upward or downward trend, but more drastic variation indicates that something is not optimal (Figure 8c, black line). Outlier time points may be attributed to worms that occasionally stick to the probe during mixing, or remain outside the transient microchamber during measuring (Figure 8d). To reduce this possibility, one can introduce the command ‘waiting time’ between the mixing and measuring time, allowing worms to sediment after mixing and ensuring that all worms are in the transient microchamber during the measurements. In our hands, a ‘waiting time’ of 30s is optimal for N2 worms.
Optimisation of worm number
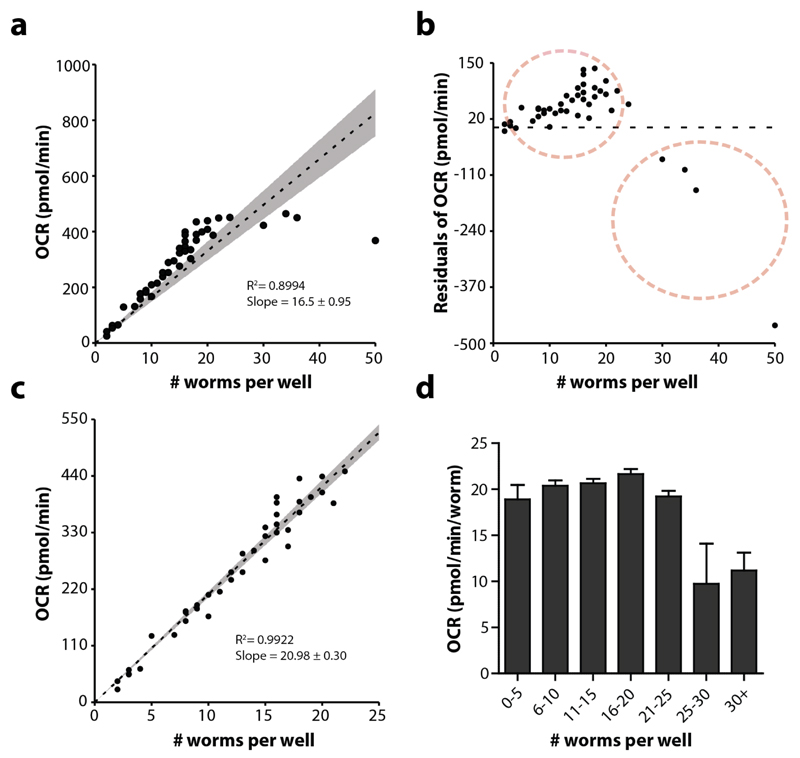
When the values for the commands measuring, mixing and waiting are optimized, the influence of the number of worms per well can be studied. Using too many worms will exhaust the oxygen pool, thereby contributing to an underestimation of the OCR. In order to establish the appropriate number of worms per well, load a 96-well plate with different numbers of worms, e.g. ~5-10-15-20-25-30 worms per well. Start an XF assay with the optimised XF template (measuring, mixing and waiting time) and loop ~10 times. Calculate the OCR per worm for each well (average over the last 9 loops) and plot this against the number of worms. For N2, adult day 1, a clear regression with a good fit can be generated (Figure 9a), although two non-stochastic subpopulations can be distinguished (Figure 9b). With segmented regression, one can distinguish a clear cut-off around 23 worms per well, implicating that the OCR is reliable up to 23 worms, at least for N2 worms at day 1 of adulthood (Figure 9c,d). The optimal number does change with age, e.g. for L4s up to 50 worms can be used (Supplementary Figure 4).
Figure 9. The OCR correlates with the number of worms.
(a) Linear regression of the OCR as dependent variable of the number of worms per well, n = 48 wells. Although the R2 is relatively high, the fit is not optimal as shown in the residual plot (b). There is no stochastic distribution, but a clear biased distribution towards two populations (red dotted circles). (c) When excluding the negative residual population, the fit of the regression line is much better. (d) More than 25 worms per well (adult day 1) renders OCR measurement inaccurate. In a and c, the grey area shows the confidence interval (95%) and the dotted line shows the linear regression. Bars are mean ± SEM.
Optimisation of compound concentrations
Titration assays of compounds of interest should be performed prior to XF assays, to ensure the optimal working concentration. When using new compounds, one can often find commonly used concentrations of the drug of interest in cells, worms or other organisms. Use this concentration as starting point and generate a dose-response curve (half-logarithmic scale concentrations) around this value, keeping in mind that worms often require higher doses. Programme the machine to loop 5 times (with the just optimised numbers for mixing, waiting and measuring and worm number), inject the compound and loop another 15 times to see its effect on respiration. It is highly suggested to pick the specific drug concentration that maximizes the effect at the lowest concentration possible. By looping 15 times the kinetics of the compound effect can be determined. As previously mentioned, C. elegans has a cuticle that shows clear impermeability towards non-water soluble compounds. Consequently, if a compound is injected and no clear respiratoric response is observed, it does not exclude involvement of the targeted cellular process in C. elegans respiration, it may just be a matter of uptake efficiency. Here, using bus mutants, which have increased cuticle permeability69, may help distinguishing between these two possibilities. In addition, actual uptake-efficiency could be determined with HPLC or liquid chromatography–tandem MS (LC-MS/MS).
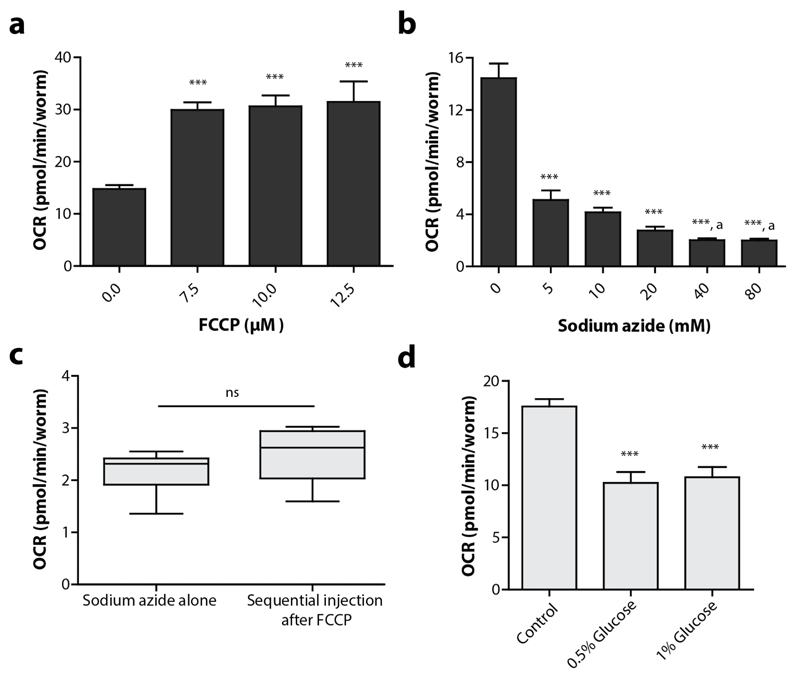
We have already shown that the optimal concentration of FCCP and sodium azide in N2 worms are 10 μM and 40 mM respectively (Figure 10a, b). Other compounds, like rotenone and antimycin A, that are typically used for XF experiments with cultured cells appear to be highly inefficient in our experimental setup (Supplementary Figure 1). Possible interference of sequential treatments should be carefully assessed when new compound combinations are used. For instance, it was reported that sodium azide is less effective when injected in sequence with FCCP28, although we could not reproduce this effect (Figure 10c). Finally, we have tested the effect of glucose pre-treatments on the OCR of C. elegans. In multiple model organisms, increasing the glucose content causes a shift from oxidative phosphorylation towards glycolysis, resulting in a decreased OCR43,70. With the XF96 respirometer we could also detect a decreased oxygen consumption rate when worms were fed with 1 or 2% glucose starting from the L1 stage until day 4 (Figure 10d). Clearly, pre-feeding with compounds can affect cellular respiration and therefore provide researchers with a long-term intervention method of which effects can be analysed with XF respirometers.
Figure 10. The effects of specific compounds on the OCR in C. elegans .
(a) Titration of FCCP and its effect on the oxygen consumption rate. 7.5-12.5 μM FCCP causes a significant increase in the OCR. One-way ANOVA (p < 0.001) with post-hoc Tukey, n = 8-16 wells. (b). Titration of sodium azide and its effect on the oxygen consumption rate. 5-80 mM sodium azide causes a significant decrease in the OCR. One-way ANOVA (p < 0.001) with post-hoc Tukey. ‘a’ indicates that 40-80 mM causes a significant decrease in OCR compared to 5 mM sodium azide treatment, n = 14-16 wells. (c) There is no significant difference in the effect of sodium azide when the compound is solely injected or in sequence with FCCP; Student t-test, n = 15-16 wells. (d) Feeding worms from L1 on with different concentrations of glucose (in the agar medium), causes a significant decrease in basal respiration at adult day 4. Bars are mean ± SEM. One-way ANOVA (p < 0.001) with post-hoc Tukey, n = 8 wells. *** = p < 0.001.
Controls and replicates
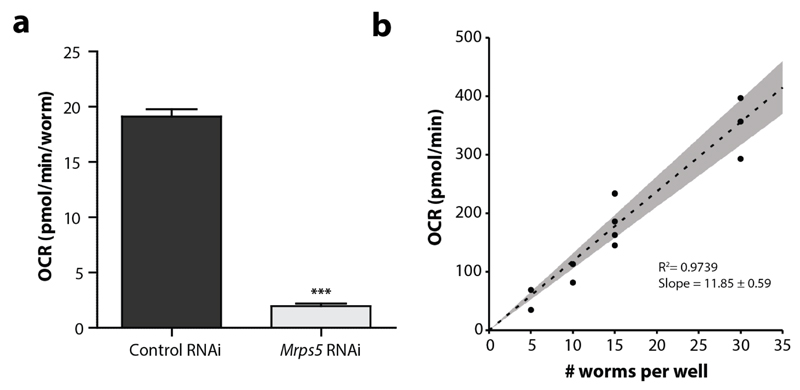
Experimental control groups are helpful for determining whether the experiment worked and to assess variability from assay-to-assay. Typically, including an established mutant/RNAi condition with decreased respiration helps to determine whether the experimental set-up is sensitive enough. Moreover, such a control may facilitate the relative comparison between different experiments, although we discourage directly comparing conditions between different plates or experiments. Established controls are gas-1(fc21)/CW152, mev-1(kn1)/TK22 and mrps-5 RNAi. Around 6-8 replicates per group (per individual experiment) are sufficient to estimate OCR with relatively small well-to-well intra-assay variability. It is strongly recommended that experimental set-ups are repeated at least three times, so one does not have to rely on one experiment with 6-8 technical replicates when drawing conclusions.
Materials
Equipment
CRITICAL For equipment and protocols related to C. elegans maintenance we kindly refer to ref. 71.
-
-
Bioscience Seahorse XF96 Flux Analyser (or XFe96)
-
-
Heratherm Incubator (Thermo Scientific: 50125882) or similar
-
-
XFe96 extracellular flux assay kits (Seahorse Bioscience: Part #102416-100)
-
-
XF96 cell culture microplates (Seahorse Bioscience: Part #101085-004)
-
-
Centrifuge (e.g. Thermo scientific: SL40R)
-
-
15 ml tubes (e.g. Cellstar: 188271)
-
-
1.5 ml tubes (e.g. Greiner bio-one: 616201)
-
-
Serological pipettes 10-25 ml (e.g. Sarstedt)
-
-
Multichannel pipette 20-200 μl (e.g. Eppendorf)
-
-
Pipette boy
-
-
Pipettors 20, 200 μl
-
-
Freezer -20°C
-
-
Vortex
-
-
Dissection microscope
-
-
Optional: parafilm
Reagents
-
-
Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) (e.g. Abcam: ab120081)
CAUTION FCCP can be acutely toxic even at low doses. Personal protective equipment should be worn at all times while handling this reagent; wear gloves and protective clothing.
-
-
Calibration solvent, pH 7.4 (Seahorse Bioscience Part # 100840-000)
-
-
Sodium azide (e.g. Sigma-Aldrich: S2002)
CAUTION The ETC inhibitor sodium azide can be acutely toxic even at low doses. Personal protective equipment should be worn at all times while handling this reagent; wear gloves and protective clothing. Sodium azide requires extra caution as it changes rapidly into a toxic gas when mixed with water or acids.
-
-
Dimethyl sulfoxide (DMSO) (e.g. Thomas Scientific: C987Y85)
-
-
KH2PO4 (e.g. Merck: 1048731000)
-
-
Na2HPO4 (e.g. Acros Organics: 424380010)
-
-
NaCl (e.g. Merck: 1064041000)
-
-
KCl (e.g. Sigma: P9333-500)
-
-
MgSO4 (e.g. Fisher Chemicals: M1000/60)
-
-
dH2O
-
-
Triton-X100 (e.g. Thermo Fisher: 215680010)
-
-
C. elegans N2 (Caenorhabditis Genetics Center: C. elegans wild isolate)
-
-
C. elegans mev-1(kn1) (Caenorhabditis Genetics Center: TK22)
Reagent Set-Up
M9 buffer (1X)
-
-
3 g KH2PO4, 6 g Na2HPO4, 5 g NaCl, H2O to 1 L. Sterilize by autoclaving and add 1 ml of 1 M MgSO4 after cooling. Store at room temperature for several months.
FCCP stock (10mM and 100 μM)
-
-
2.54 mg FCCP in 1 ml DMSO, make 25 μl aliquots and store at -20°C, up to 2-4 months. Dilute 25 μl of the FCCP stock (10 mM) in 2475 μl M9 buffer up to 24 hours before use (= 100 μM). ~2.5 ml of 100 μM FCCP solution is required per assay (25 μl of the stock in DMSO).
Sodium azide stock (400 mM)
-
-
26 mg sodium azide per 1 ml dH2O. Store at room temperature, up to 24 hours before use. 2.5 ml of 400 mM of sodium azide solution is required per assay.
1x PBS + 0.05% Triton-X100 (v/v)
-
-
To make PBS: add 8g NaCl, 0.2 g KCl, 1.44 g Na2HPO4 and 0.24 g KH2PO4 (pH 7.4) in dH2O to 1 L. Store PBS at room temperature or 4°C, up to 6 months. Add 0.5 ml Triton-X100 to 1 L PBS (0.05%) and store at 4°C, up to 1 month.
Procedure
CRITICAL STEP: C. elegans age-synchronisation should take place in advance depending on the age of interest. Before starting the experimental procedures on day 2 of the protocol make sure that the worms are at the appropriate age.
Step 1-4: Hydrating probes TIMING: 12-24 hours (Day 1)
-
1.
Open an unused flux package (green lid). In the package you will find a utility plate with a green sensor cartridge on top, a lid, and two drug loading guides.
-
2.
Remove the green sensor cartridge, but place it upside down so that the probe surface will not be scratched.
-
3.
Using a multichannel pipet, add 200 μl of the Seahorse Bioscience XF96 Calibrant pH 7.4 solution in each well of the utility plate.
-
4.
Place the sensor cartridge back on the utility plate so that the probes dip into the wells. Place the lid on top and incubate the cartridge overnight at 37°C without CO2.
CRITICAL STEP One should be careful not to disturb the sensor probes’ surface.
CRITICAL STEP Cartridges cannot be reused. When performing multiple runs make sure to hydrate multiple cartridges.
PAUSE POINT Hydration can take as little as 4h and as long as 72h (when using >24h, seal the plate with parafilm and store at 4°C. Re-warm to 37°C before use).
Step 5-8: Preparing the XF96 respirometer: Temperature control and background correction TIMING: 5 minutes (Day 2)
-
5.
Launch the XF software and when prompted select the “standard” software settings. Either create a new account or proceed by clicking on ‘Seahorse guest’.
CRITICAL STEP Always use the standard software package and not the software associated with one of the so-called “stress kits”.
-
6.
Turn the heater and temperature control off. Click on the instrument icon in the right lower corner and subsequently select administration > temperature > heater and temperature control. The buttons should be grey when they are turned off.
CRITICAL STEP This is a critical step for working with C. elegans as use of the heater is designed for cell culture at 37°C. The temperature of the machine can be followed on its display directly.
? | TROUBLESHOOTING
-
7.
In the same screen (administration) click on background correction. Make sure that background correction is on and the correction wells are set to A1, A12, H1 and H12.
-
8.
Leave the instrumental set-up mode by clicking on ‘end instrument set-up mode’.
Step 9 - 14: Preparing the XF96 respirometer: Creating a XF-assay TIMING: 10-20 minutes (Day 2)
-
9.
Click on the assay wizard icon (next to the instrument icon). In the tab general, fill in at least the assay name and results file name, for instance using the following format: ‘YearMonthDate_Strains/conditions’ e.g. ’20160515_N2_L4’
-
10.
The tabs ‘Cells’, ‘Media’ and ‘Compounds’ can be ignored. However, make sure to write down the compounds to be injected, their concentrations and the port they will be loaded in.
-
11.
Proceed to the ‘background correction’ tab and double-check that the correction is ‘on’ and the corner wells are selected (A1, A12, H1 and H12).
-
12.
OPTIONAL: Wells can be labelled in the tab ‘Groups and labels’. Select the wells with technical replicates and give them a name and colour. During the run you can see the average OCR of the groups involved. When labelling is not done prior to the assay, it can still be done afterwards.
CRITICAL STEP The OCR values that can be followed in real time are not corrected for the number of worms and are therefore not directly comparable.
-
13.
Go to the tab ‘Protocol’. By clicking on the commands, you can add instructions to the current protocol. For commands “measure”, “mix” and “wait” you also have to fill in the duration. Always start with (1) Calibrate and (2) Equilibrate. For N2 worms, use the parameters shown in Table 3. For advice on designing assay protocols for different strains, see EXPERIMENTAL DESIGN.
CRITICAL STEP When not injecting, or injecting multiple compounds, make sure to exclude or include these commands in your protocol, respectively.
-
14.
When the appropriate protocol is generated, go to the last tab ‘End’ and click the button ‘End Assay Wizard’. A screen with a green start button will appear under the “Run” tab.
CRITICAL STEP Do not click the start button yet. To avoid unnecessary heating of the machine, you should postpone the start of the machine until <20 min before the worm assay plate will be ready.
Step 15 - 20: Loading compounds to be injected (optional) TIMING: 30 minutes (Day 2)
-
15.
Take the hydrated utility plate from step 4 with the sensor cartridge from the incubator.
-
16.
If desired, thaw one 25 μl aliquot of FCCP (10 mM) and dilute it 100-fold in M9 buffer (2475 μl) as described in REAGENT SETUP (Table 4).
-
17.
Slowly pipette 22 μl diluted FCCP (100 μM) into each ‘port A’ on the cartridge (Table 4), including those belonging to the background correction wells. During pipetting, insert the pipette tips into the holes and dispense the content. Optional: the loading guide can be used to ensure pipetting in the right port.
CRITICAL STEP A multichannel pipette can be used for this purpose. Make sure to avoid that the liquid adheres to the sides of the port and avoid creating bubbles since compounds are injected into the XF96 in a pneumatic fashion. Also, be aware that the ports have a hole at the bottom for injection.
CRITICAL STEP Since the compounds are injected using a pneumatic mechanism it is essential to add the same volume of liquid in all injection ports, i.e. when injecting FCCP into port A, all of these ports should contain the compound or a vehicle.
CRITICAL STEP Do not use automated pipettes to load the compounds, as this will increase leakage through the bottom of the ports.
-
18.
Load 24 μl sodium azide (400 mM) in the same fashion as described for FCCP into each ‘port B’ of the green sensor cartridge.
CRITICAL STEP Since FCCP is injected first, the volume in the well is not 200 μl anymore, but 220 μl. Therefore, the injection volume of sodium azide is higher to ensure a correct final concentration.
-
19.
Visually inspect the injection ports A and B for even loading; the liquid should be down at the bottom of the port.
-
20.
Place the lid on top of the cartridge. Carefully transfer the sensor cartridge (still on top of the utility plate) back into the non-CO2 incubator at 37°C, and keep it there until calibration is started.
CRITICAL STEP Keep the plate at least 10 minutes at 37°C to ensure that the compounds are warmed-up. In this way, calibration will take place at around 37°C, which is the optimal calibration temperature for XF respirometers (the heater is still off).
Table 4. Stock, working and final concentrations of FCCP and sodium azide.
| Compound | (1) Final well concentration | (2) Concentration drug injection port | (3) Stock concentration | (4) Stock volume required per plate (96 wells)a |
|---|---|---|---|---|
| FCCP | 10 μM | 100 μM (1:100 of stock in M9)a | 10 mM in DMSOb | 25 μl |
| Sodium azide | 40 mM | 400 mMa | 400 mM in dH2O | 2.5 ml |
The volume required per assay depends on the volume that is pipetted per drug injection port. We normally use 22 μl per port for FCCP and 24 μl per port for sodium azide when injected sequentially. Since there are 96 wells/ports, ~2.5 ml of the drug (with a concentration equal to (2)) is required per full plate.
The stock concentration of compounds in DMSO should be higher to ensure final DMSO concentration being <1% DMSO.
Step 21 – 40: Preparing samples and loading them, starting the XF assay TIMING: 2 – 3 hours (Day 2)
-
21.
Make note of the developmental stages seen on the worm agar plates
CRITICAL STEP Strict age-synchrony is required for reliable estimations of the OCR. Make sure that worms are synchronized (by hypochlorite treatment, or egg laying and/or FUdR) and that they are the age-of-interest on the day of the XF assay.
-
22.
Using sufficient (2-4 ml per 9 cm plate) M9 buffer, rinse the worms of the plate with gentle agitation and collect the worms of each sample in a 15 ml tube.
-
23.
Spin the worms down in a centrifuge (20°C, ~500 x g for 2 minutes).
-
24.
Remove the supernatant and add fresh M9 buffer. Repeat step 23-24 two more times.
-
25.
Because there may be eggs or even small larvae present in your samples when you are testing aged worms (even when using FUdR), add M9 to the tube, mix gently, and allow the worms to settle for a few minutes by gravity. When the worms have sunken into a loose pellet, aspirate the supernatant that usually contain smaller animals and eggs.
-
26.
Repeat step 25
-
27.
Resuspend the worms in 1.5-3 ml M9, depending on the density and the size of the worm pellet.
-
28.
Mix the worm suspension carefully and pipet two 3 μl drops on an empty petri dish and count the number of worms per drop using a microscope.
CRITICAL STEP Wash the pipette tip with PBS + 0.05 % (v/v) Triton X-100 before you pipette the worms, or use low-retention tips to avoid worms sticking to the tip. Otherwise, one may use wide-bore tips or tips with the ends cut off using a razor blade.
-
29.
Calculate the volume of sample that should be loaded to acquire 10-20 worms per well. Subtract this volume from 200 μl (the maximal volume per well); this is the volume of M9 that will be added per well.
CRITICAL STEP The volume of M9 that should be loaded into the wells needs to be calculated for every sample separately. Make a clear pipetting scheme and fill in the scheme shown in Supplementary Figure 3. Also, include control strains (e.g. mev-1 mutant, gas-1 mutant, mrps-5 RNAi) to ensure correct interpretation of the results later on.
-
30.
Before loading the worms click ‘Start’ to start the actual XF assay. Make sure to fill in the ‘Save directory’ and ‘Save name’ before you click ‘Start’ again.
-
31.
Follow the prompts on the screen for sensor cartridge calibration. When the loading door opens, take the sensor cartridge from the non-CO2 incubator (step 20) and place it on the tray.
CRITICAL STEP The notch of the plate should point to the front of the machine.
CRITICAL STEP Make sure to remove the lid before inserting the plate into the machine.
? | TROUBLESHOOTING
-
32.
Open a fresh cell culture microplate (utility plate; blue lid) and load the appropriate volume of M9 per well, followed by the calculated volume of the samples.
CRITICAL STEP Wash the pipette tip with PBS + 0.05 % (v/v) Triton X-100 before pipetting the worms, or use low-retention tips to avoid worms sticking to the tip. Otherwise, one may use wide-bore tips or tips with the ends cut off using a razor blade.
-
33.
Pipette 200 μl M9 in the background correction wells: A1, A12, H1 and H12.
-
34.
By visual inspection with a dissection microscope, inspect all wells and make sure that the wells A1, A12, H1 and H12 are empty and that the experimental wells contain ~10-25 worms (see Table 3 for appropriate worm numbers per well) worms.
-
35.
Wait at least 2 minutes from when the worms are pipetted until the plate is loaded, to allow the worms to settle by gravity to the bottom of the wells.
-
36.
By now, the calibration should be finished. Note: when calibration is finished before samples are ready, the instrument stays closed and waits until samples are ready to load. Open the loading door by clicking “OK” on the screen, this will release the utility plate containing calibration buffer from the XF respirometer, while the cartridge stays inside the machine. Replace the calibrant plate with the worm plate.
CRITICAL STEP The green sensor cartridge is not ejected, but stays in the machine. Only the utility plates are swopped. Do not forget to remove the lid of the worm plate.
-
37.
Close the door by clicking on ‘continue’, the next step of the protocol will now be executed: equilibrate.
-
38.
Once the run is completed, follow the prompts to remove the cell plate and the sensor cartridge from the XF96 respirometer.
-
39.
Keep the worm plate and use a dissection microscope to count the number of worms per well. Make note of the numbers on the scheme shown in Supplementary Figure 3. Optional: capture images of each well using a microscope and count the number of worms per well at a later date.
CRITICAL STEP When sodium azide is injected during the XF assay, worms are paralyzed which makes counting easier. Without injection of sodium azide it is suggested to cool down the plate before counting the worms, as this also immobilizes the worms, or alternately add sodium azide to the wells manually
PAUSE POINT The experimental part of the PROCEDURE is finished, the post-experimental analysis can be performed at any moment from here on.
? | TROUBLESHOOTING
-
40.
While inspecting the number of worms, also make note of any anomalies observed in the wells (e.g. larvae, embryos, bacteria) as this likely influenced the OCR measurements.
4. Step 41 – 47 : Post-experimental analysis TIMING: 1 - 2 hours (Day X)
-
41.
Open the Excel spreadsheet that is automatically exported to the directory selected by the user.
CRITICAL STEP When doing the analysis on a computer without XF software, copy the important values in the tab ‘rate data’ to another Excel file to avoid error messages.
? | TROUBLESHOOTING
-
42.
Copy the OCR values per well (with all separate measurement points) to a new Excel tab in such a way that the OCR values of a single well are collected below each other (in a column).
-
43.
Divide all OCR values for each well at each time point by the number of worms in that well. Optional: Determine protein content with a BCA assay in exactly 1000 worms to estimate the protein concentration per nematode. Subsequently divide the OCR per worm by this number.
CRITICAL STEP When nematode strains differ in size, the optional protein determination step is recommended, although we suggest reporting both OCR per worm and OCR per mg protein.
-
44.
Estimate the basal OCR for each well by averaging the OCR per worm of measurement point (loop) 2 – 5.
CRITICAL STEP The basal OCR values are typically not stable until the second measurement loop. Also the first measurement after injection is typically not stable. Exclude these measuring points when calculating average OCRs.
CRITICAL STEP The specific measurement points are following the settings of our proposed protocol, as shown in Table 3.
-
45.
To estimate the FCCP-induced OCR per worm, find a stable plateau phase of 3 points within measurements 7 – 14 and average these numbers.
? | TROUBLESHOOTING
-
46.
To calculate non-mitochondrial respiration per worm (sodium azide-induced), take the average of measurements 17-18.
? | TROUBLESHOOTING
-
47.
Using values obtained in steps 44-46, calculate mitochondrial respiration for each well by subtracting the average sodium azide-induced OCR from the average basal OCR. To calculate the maximal respiratory capacity, subtract the sodium azide-induced OCR from the FCCP-induced average OCR. To calculate spare respiratory capacity, subtract the average of the basal OCR from the FCCP-induced average OCR. Non-mitochondrial respiration is defined as the sodium-azide induced OCR.
? | TROUBLESHOOTING
Timing
Age-synchronization and ageing of worms: X days.
Step 1-4: Hydrating probes: 12-24 hours
Step 5-8: Preparing the XF96 respirometer: Temperature control and background correction: 5 minutes
Step 9 - 14: Preparing the XF96 respirometer: Creating a XF-assay : 10-20 minutes
Step 15 - 20: Loading compounds to be injected (optional): 30 minutes
Step 21 – 40: Preparing samples and loading them, starting the XF assay: 2 – 3 hours
Step 41 – 47: Post-experimental analysis: 1 – 2 hours
Troubleshooting
Please see Table 5 for troubleshooting advice.
Table 5. Troubleshooting.
| Step | Problem | Possible cause | Solution |
|---|---|---|---|
| 6 | The temperature is too high | The heater of the machine is still on | Make sure to turn both the heater and the temperature control off before starting the experiment. |
| 31 | The cartridge is not recognized/barcode cannot be scanned | Condensation within the machine sometimes interferes with reading the barcode | Restart the device; make sure to turn off the heater/temperature control again. |
| 39, 46 | Worms still move after injection of sodium azide | The concentration of the compound it too low | Increase the concentration sodium azide. Make a doseresponse curve for sodium azide in the strain of interest. |
| The drug was loaded in the wrong port. | Check whether the drug is still in one of the injection ports. Make sure to load the compound next time in the right port. | ||
| The drug is not properly loaded into the injection ports | Visual inspection could show whether the compounds are still in the injection ports. Make sure to pipette carefully, in one stream, and avoid inducing air bubbles. | ||
| The drug has gone bad | Make a fresh stock from recently purchased reagent in sterile deionized H2O | ||
| 41 | The temperature heavily increases during the experimental assay | The environmental temperature is too high | Place the XF respirometer in a temperature-controlled room or the change the lab temperature. |
| 41 | The basal OCR heavily fluctuates over time/ decreases over time. | Nematodes are outside the transient microchamber during the measuring time | Introduce a waiting step, or increase the duration of the waiting step. |
| Mixing time is insufficient | Increase the mixing time or decrease the measuring time | ||
| The time between collecting the worms and the end of the assay is too long | Increase the working speed or ask somebody to help you preparing the samples. Alternatively, reduce the length of the experimental protocol (number of loops) | ||
| 41 | Oxygen levels drop to 0 mm/Hg during a measure cycle | Mixing time is insufficient | Increase the mixing time |
| Too many worms in the well | Decrease the number of nematodes per well. | ||
| Measuring time is too long | Decrease the measuring time | ||
| 41 | The OCR is much lower in the second measure cycle | The first estimation of the OCR is in general not reliable | Exclude the first measure cycle from analysis |
| 45 | Worms do not respond to FCCP injection | The used FCCP concentration is too low | Increase the concentration FCCP. Make a dose-response curve for FCCP in the strain of interest. |
| The starting oxygen concentration – before the start of the measuring cycle - is too low. During the cycle, the oxygen concentration turns into zero and the OCR is underestimated. | Decrease the measuring time or decrease the number of worms per well. | ||
| The drug is loaded in the wrong injection port | Check whether the drug is still in one of the injection ports. Make sure to load the compound next time in the right port. | ||
| The drug is not properly loaded into the injection ports | Visual inspection could show whether the compounds are still in the injection ports. Make sure to pipette carefully, in one stream, and avoid inducing air bubbles. | ||
| The drug has gone bad | Make a fresh stock from recently purchased reagent in a freshly opened anhydrous vial of DMSO | ||
| 47 | The variation between technical replicates (within one run) is large | There are too many or too few worms in some of the wells | Plot the total OCR versus the number of worms and add a linear trend line. Create a residual plot and look for abnormalities. Exclude outliers caused by a high or low number of worms. |
Anticipated results
Interpretation and considerations
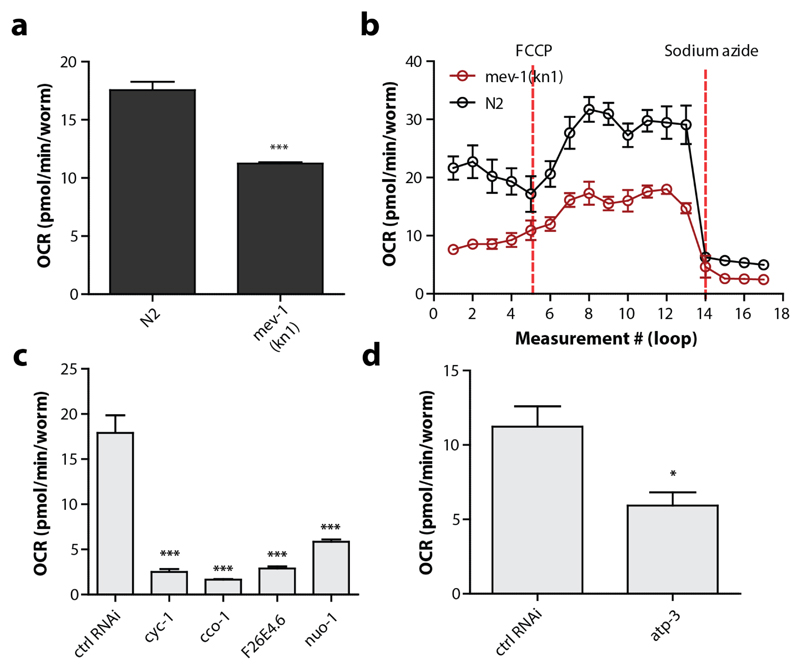
The goal of using the XF96 respirometers for C. elegans is to estimate the OCR, which mirrors cellular respiration of the worm. Clearly, including established mitochondrial mutants such as gas-1 or mev-1, or mrps-5 RNAi in the experimental set-up allows the user to interpret whether the assay is sensitive enough to detect differences in respiration. Basal OCR should therefore differ between N2 and a mitochondrial mutant (Figure 11a,b). Note that absolute OCR values can differ between assays due to changes in temperature and the use of different sensor cartridges. Therefore, mutants are ideally analysed side-by-side on the same plate and on the same day. The injection of FCCP typically increases the OCR 2 to 3-fold, but the effect is somewhat delayed when compared to experiments performed with cells, which is likely due to a difference in uptake efficiency. The OCR in N2 worms drops about 75-90% when sodium azide is added, which highlights that most of the respiration is caused by mitochondrial function (Figure 11b). The quality of the different OCR readings can be verified by looking at the raw data, e.g. the variation in different time points and the course of the oxygen concentration as described in detail in the Optimisation section. The variation between different time points should be small (<15%), except for initial time points after compound injection (particularly FCCP), and the oxygen concentration should restore itself every loop (Figure 8).
Figure 11. Genetic manipulation of the electron transport chain lowers the OCR in C. elegans .
(a) Basal respiration is significantly lower in the mev-1 strain compared to N2s. Student t-test (p < 0.001), n = 8 wells. (b). Mev-1 mutants have lower basal respiration, lower maximal respiration, lower spare capacity, but comparable non-mitochondrial respiration (n = 8 wells). (c) Basal respiration is significantly lower when proteins in the electron transport chain are knocked down. One-way ANOVA (p<0.001) with post-hoc Tukey, n = 6-8 wells. (d) A knockdown of atp-3 in complex V causes a significant decrease in basal respiration. Bars are mean ± SEM. Student t-test (p=0.032), n = 8 wells.
To further verify and test the ability of XF96 respirometers to measure oxygen consumption and especially differences in the oxygen consumption rates, we tested RNAi directed towards several proteins in the electron transport chain such as cyc-1 (cytochrome c1, complex III), cco-1 (cytochrome c oxidase, complex IV), nuo-1 (NADH:ubiquinone oxidoreductase, complex I), atp-3 (ATP synthase subunit δ, complex V) and F26E4.6 (cytochrome c oxidase subunit VIIc, complex IV) (Figure 1b, 11c,d). Clearly, similar to compounds that inhibit specific electron transport chain proteins (Figure 1b, Table 2), knocking down proteins associated with oxidative phosphorylation cause a striking decrease in basal respiration. These results underline the strength of XF respirometers in combination with C. elegans, as the effects of genetic interventions on cellular respiration can be assessed side-by-side in a quick and straightforward manner when following our developed protocol.
One important consideration that should be taken into account is the interpretation of mitochondrial respiration, spare respiratory capacity, maximal respiratory capacity and non-mitochondrial respiration. These terms were once coined to describe respiration effects in isolated mitochondria with exogenously controlled energy sources. It is likely that the more complex intact animal should be interpreted differently than isolated mitochondria in a defined medium. The question that directly arises is whether treatment with FCCP and sodium azide indeed accurately reflects these different aspects of cellular respiration. This argues caution against over-interpretation of the OCR values in terms of selective association with aspects such as maximal respiratory capacity and would argue for relying on basal respiration only. Clearly, basal respiration provides the user with the most readily interpretable number from a XF96 experiment.
Scaling down: from an XF96 to XFp respirometer
The possibility to assess respiration in 96 parallel wells makes the XF96 respirometer attractive for moderately large screens and comparing many conditions at the same time. However, when for example only two conditions should be compared, 8-12 wells may be sufficient. In this specific case it is not convenient to only fill a small fraction of the 96-wells, as it is not suggested to reuse plates. Here, the recently launched XFp respirometers provide an alternative. The XFp respirometer works with 8-well plates (including two wells for background correction), in which the separate wells have the same size as the wells of the XF96 plates. The user interface of the XFp respirometer is intuitive, the machine is easy to handle and due to the limited number of wells, the manual transfer time from worms into the 8-well plate and eventually starting the assay is relatively short. Using the XFp, operated in a cold room so that the operating temperature was stable at 20°C, we have confirmed that knockdown of mrps-5 reduces OCR in C. elegans (Figure 12a), similar to what we observed before in XF96 respirometer26. Furthermore, the optimal FCCP concentrations are similar to those used in XF96 respirometers and the number of worms correlates well with the OCR (Figure 12b). The main limitation of the XFp respirometer is, however, the number of assay wells. Sample-to-sample variation in the XFp respirometer is similar to that observed in XF96 respirometers, but the limited number of wells (6 experimental wells) can preclude sufficient technical replicates when analysing mobile samples such as worms. As a consequence, although massive differences in respiration are easily picked up using the XFp respirometer (Figure 12a), small changes in OCR might be missed. As such, the XF96 is more practical than the XFp respirometer for measuring worms, and these benefits outweigh the difference in investment that should be made when purchasing the XF96 respirometer and its consumables. As a final consideration, the same optimisation steps can be used to measure respiration with the 24-well respirometer (XF24), which has larger wells that require more worms per well74.
Figure 12. XFp respirometers provide an alternative to XF96 respirometers.
Data acquired in XFp respirometers. (a) Nematodes treated with mrps5 RNAi show significant lower respiration than worms treated with control RNAi. Student t-test (p <0.0001). This is comparable to what is previously described in XF96 respirometers26. Bars are mean ± SEM. (b) Linear regression of the OCR as dependent variable of the number of worms per well. Data from two separate runs are combined (n=10 wells). Grey area shows the confidence interval (95%) and the dotted line shows the linear regression.
Supplementary Material
Editor Summary.
Mitochondrial dysfunction is linked to a range of common disorders including inherited metabolic diseases, diabetes and cancer. Here, Koopman et al. describe a protocol to study mitochondrial function in C. elegans using the XF96 respirometer.
Acknowledgments
We would like to thank Tobias Ackermann for advice and Cornelis F. Calkhoven for allowing us to make use of the Seahorse XF96 machine. Work in the Nollen group is financially supported by an ERC starting grant (no. 281622). Work in the Houtkooper group is financially supported by an ERC Starting grant (no. 638290), and a VIDI grant from ZonMw (no. 91715305). The Seahorse XF96 at AMC was supported by a grant from NWO-Middelgroot (no. 91112009).
Footnotes
Contributions
M.K. performed most experiments, optimized the final experimental pipeline and wrote the manuscript together with R.H.H. and contributions from all authors. H.M. and R.K performed experiments on the XFp/XF96 respirometers. L.M., R.H.H., B.M.D., J.A. pioneered the use of XF respirometers for C. elegans and have developed the initial protocols. M.K., H.M, E.A.A.N, B.M.D. and R.H.H. have been extensively involved in discussions and interpretations of results.
Competing financial interests
The authors declare that they have no competing financial interests.
References
- 1.Perry CGR, Kane DA, Lanza IR, Neufer PD. Methods for assessing mitochondrial function in diabetes. Diabetis. 2013;62:1041–1053. doi: 10.2337/db12-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hatefi Y. The mitochondrial electron transport and oxidative phosphorylation system. Annu Rev Biochem. 1985;54:1015–1069. doi: 10.1146/annurev.bi.54.070185.005055. [DOI] [PubMed] [Google Scholar]
- 3.Brand MD, Nicholls DG. Assessing mitochondrial dysfunction in cells. Biochem J. 2011;435:297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldenthal MJ, Marín-García J. Mitochondrial signaling pathways: a receiver/integrator organelle. Mol Cell Biochem. 2004;262:1–16. doi: 10.1023/b:mcbi.0000038228.85494.3b. [DOI] [PubMed] [Google Scholar]
- 5.Galluzzi L, Kepp O, Kroemer G. Mitochondria: master regulators of danger signalling. Nat Rev Mol Cell Biol. 2012;13:780–788. doi: 10.1038/nrm3479. [DOI] [PubMed] [Google Scholar]
- 6.Rizzuto R, De Stefani D, Raffaello A, Mammucari C. Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol. 2012;13:566–578. doi: 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- 7.Fernyhough P, Roy Chowdhury SK, Schmidt RE. Mitochondrial stress and the pathogenesis of diabetic neuropathy. Expert Rev Endocrinol Metab. 2010;5:39–49. doi: 10.1586/eem.09.55. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreira IL, Resende R, Ferreiro E, Rego AC, Pereira CF. Multiple defects in energy metabolism in Alzheimer’s disease. Curr Drug Targets. 2010;11:1193–1206. doi: 10.2174/1389450111007011193. [DOI] [PubMed] [Google Scholar]
- 9.Jarrett SF, Lewin AS, Boulton ME. The importance of mitochondria in age-related and inherited eye disorders. Opthalmic Res. 2010;44:179–190. doi: 10.1159/000316480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawamata H, Manfredi G. Mitochondrial dysfunction and intracellular calcium dysregulation in ALS. Mech Ageing Dev. 2010;131:517–526. doi: 10.1016/j.mad.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren J, Pulakat L, Whaley-Connell A, Sowers JR. Mitochondrial biogenesis in the metabolic syndrome and cardiovascular disease. J Mol Med. 2010;88:993–1001. doi: 10.1007/s00109-010-0663-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenstock TR, Duarte AI, Rego AC. Mitochondrial-associated metabolic changes and neurodegeneration in Huntington’s disease – from clinical features to the benc. Curr Drug Targets. 2010;11:1218–1236. doi: 10.2174/1389450111007011218. [DOI] [PubMed] [Google Scholar]
- 13.Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- 14.Andreux PA, Houtkooper RH, Auwerx J. Pharmacological approaches to restore mitochondrial function. Nat Rev Drug Discov. 2013;12:465–483. doi: 10.1038/nrd4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andreux PA, et al. A method to identify and validate mitochondrial modulators using mammalian cells and the worm C. elegans. Sci Rep. 2014;4:05285. doi: 10.1038/srep05285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felkai S, et al. CLK-1 controls respiration, behaviour and aging in the nematode Caenorhabditis elegans. EMBO J. 1999;18:1783–1792. doi: 10.1093/emboj/18.7.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamo N, Muratsugu M, Hongoh R, Kobatake Y. Membrane potential of mitochondria measured with an electrode sensitive to tetraphenyl phosphonium and relationship between proton electrochemical potential and phosphorylation potential in steady state. J Membr Biol. 1979;49:105–121. doi: 10.1007/BF01868720. [DOI] [PubMed] [Google Scholar]
- 18.Drew B, Leeuwenburgh C. Method for measuring ATP production in isolated mitochondria: ATP production in brain and liver mitochondria of Fischer-344 rats with age and caloric restriction. Am J Physiol Regul Integr Comp Physiol. 2003;285:R1259–R1267. doi: 10.1152/ajpregu.00264.2003. [DOI] [PubMed] [Google Scholar]
- 19.Pan X, et al. The physiological role of mitochondrial calcium revealed by mice lacking the mitochondrial calcium uniporter. Nat Cell Biol. 2013;15:1464–1472. doi: 10.1038/ncb2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chance B, Williams GR. Respiratory enzymes in oxidative phosphorylation. I. Kinetics of oxygen utilization. J Biol Chem. 1955;217:383–393. [PubMed] [Google Scholar]
- 21.Wu M, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetics function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292:C125–C136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 22.Gerenscer AA, et al. Quantitative microplate-based respirometry with correction for oxygen diffusion. Anal Chem. 2009;81:6868–6878. doi: 10.1021/ac900881z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Will Y, Hynes J, Ogurtsov VI, Papkovsky DB. Analysis of mitochondrial function using phosphorescent oxygen-sensitive probes. Nat Protoc. 2006;1:2563–2572. doi: 10.1038/nprot.2006.351. [DOI] [PubMed] [Google Scholar]
- 24.Nadanaciva S, et al. Assessment of drug-induced mitochondrial dysfunction via altered cellular respiration and acidification measured in 96-well platform. J Bioenerg Biomembr. 2012;44:421–437. doi: 10.1007/s10863-012-9446-z. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto H, et al. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell. 2011;147:827–839. doi: 10.1016/j.cell.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Houtkooper RH, et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mouchiroud L, et al. The NAD(+)/sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luz AL, et al. Mitochondrial morphology and fundamental parameters of the mitochondrial respiratory chain are altered in Caenarhanditis elegans strains deficient in mitochondrial dynamics and homeostasis processes. PLoS One. 2015;10:e0130940. doi: 10.1371/journal.pone.0130940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SS, et al. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2002;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 30.Fire AZ. Gene silencing by double-stranded RNA. Cell Death Differ. 2007;14:1998–2012. doi: 10.1038/sj.cdd.4402253. [DOI] [PubMed] [Google Scholar]
- 31.Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science. 2000;287:2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- 32.Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wadsworth WG, Riddle DL. Developmental regulation of energy metabolism in Caenorhabditis elegans. Dev Biol. 1989;132:167–173. doi: 10.1016/0012-1606(89)90214-5. [DOI] [PubMed] [Google Scholar]
- 34.Li J, et al. Proteomics analysis of mitochondria from Caenorhabditis elegans. Proteomics. 2009;9:4539–4554. doi: 10.1002/pmic.200900101. [DOI] [PubMed] [Google Scholar]
- 35.Mullaney BC, Ashrafi K. C. elegans fat storage and metabolic regulation. Biochim Biophys Acta. 2009;1791:474–478. doi: 10.1016/j.bbalip.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dancy BM, Sedensky MM, Morgan PG. Mitochondrial bioenergetics and disease in Caenorhabditis elegans. Front Biosci (Landmark Ed) 2015;20:198–228. doi: 10.2741/4305. [DOI] [PubMed] [Google Scholar]
- 37.Gerencser AA, et al. Quantitative microplate-based respirometry with correction for oxygen diffusion. Anal Chem. 2009;81:6868–6878. doi: 10.1021/ac900881z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hill BG, Dranka BP, Zou L, Chatham JC, Darley-Usmar VM. Importance of the bioenergetics reserve capacity in response to cardiomyocyte stress induced by 4-hydroxynonenal. Biochem J. 2009;424:99–107. doi: 10.1042/BJ20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dranka BP, et al. Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radic Biol Med. 2011;51:1621–1635. doi: 10.1016/j.freeradbiomed.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sansbury BE, Jones SP, Riggs DW, Darley-Usmar VM, Hill BG. Bioenergetic function in cardiovascular cells: the importance of the reserve capacity and its biological regulation. Chem Biol Interact. 2011;191:288–295. doi: 10.1016/j.cbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Readnower RD, Brainard RE, Hill BG, Jones SP. Standardized bioenergetics profiling of adult mouse cardiomyocytes. Physiol Genomics. 2012;44:1208–1213. doi: 10.1152/physiolgenomics.00129.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson DF, Vinogradov S, Lo LW, Huang L. Oxygen dependent quenching of phosphorescence: a status report. Adv Exp Med Biol. 1996;388:101–107. doi: 10.1007/978-1-4613-0333-6_12. [DOI] [PubMed] [Google Scholar]
- 43.Schulz TJ, et al. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 44.Ehrismann D, et al. Studies on the activity of the hypoxia-inducible-factor hydroxylases using an oxygen consumption assay. Biochem J. 2007;401:227–234. doi: 10.1042/BJ20061151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rand JB, Johnson CD. Genetic pharmacology: interactions between drugs and gene products in Caenorhabditis elegans. Methods Cell Biol. 1995;48:187–204. doi: 10.1016/s0091-679x(08)61388-6. [DOI] [PubMed] [Google Scholar]
- 46.Burns AR, et al. A predictive model for drug bioaccumulation and bioactivity in Caenorhabditis elegans. Nat Chem Biol. 2010;6:549–557. doi: 10.1038/nchembio.380. [DOI] [PubMed] [Google Scholar]
- 47.Zheng S-Q, Ding A-J, Li G-P, Wu G-S, Luo H-R. Drug absorption efficiency in Caenorhabditis elegans delivered by different methods. PLoS One. 2013;8:e56877. doi: 10.1371/journal.pone.0056877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Massie MR, Lapoczka EM, Boggs KD, Stine KE, White GE. Exposure to the metabolic inhibitor sodium azide induces stress protein expression and thermotolerance in the nematode Caenorhabditis elegans. Cell Stress Chaperones. 2003;8:1–7. doi: 10.1379/1466-1268(2003)8<1:ettmis>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lanza IR, Nair KS. Functional assessment of isolated mitochondria in vivo. Methods Enzymol. 2009;457:349–372. doi: 10.1016/S0076-6879(09)05020-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salabei JK, Gibb AA, Hill BG. Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis. Nat Protoc. 2014;9:421–438. doi: 10.1038/nprot.2014.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piper HM, et al. Development of ischemia-induced damage in defined mitochondrial subpopulations. J Mol Cell Cardio. 1985;17:885–896. doi: 10.1016/s0022-2828(85)80102-4. [DOI] [PubMed] [Google Scholar]
- 52.Picard M, et al. Mitochondrial functional impairment with aging is exaggerated in isolated mitochondria compared to permeabilized myofibers. Aging Cell. 2010;9:1032–1046. doi: 10.1111/j.1474-9726.2010.00628.x. [DOI] [PubMed] [Google Scholar]
- 53.Picard M, et al. Mitochondrial structure and function are disrupted by standard isolation methods. PLoS One. 2011;6:e18317. doi: 10.1371/journal.pone.0018317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Picard M, Taivassalo T, Gouspillou G, Hepple RT. Mitochondria: isolation, structure and function. J Physiol. 2011;598:4413–4421. doi: 10.1113/jphysiol.2011.212712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kayser EB, Morgan PG, Hoppel CL, Sedensky MM. Mitochondrial expression and function of GAS-1 in Caenorhabditis elegans. J Biol Chem. 2001;276:20551–20558. doi: 10.1074/jbc.M011066200. [DOI] [PubMed] [Google Scholar]
- 56.Rivera-Ingraham GA, Bickmeyer U, Abele D. The physiological response of the marine platyhelminth Macrostomum ligano to different environmental oxygen concentrations. J Exp Biol. 2013;216:2741–2751. doi: 10.1242/jeb.081984. [DOI] [PubMed] [Google Scholar]
- 57.Steele SL, Prykhozhij SV, Berman JN. Zebrafish as a model system for mitochondrial biology and diseases. Transl Res. 2014;163:79–98. doi: 10.1016/j.trsl.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 58.Gibert Y, McGee SL, Ward AC. Metabolic profile analysis of zebrafish embryos. J Vis Exp. 2013;14:e4300. doi: 10.3791/4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hartman N, et al. Mitochondrial DNA copy number and function decrease with age in the short-lived fish Nothobranchius furzeri. Aging Cell. 2011;10:824–831. doi: 10.1111/j.1474-9726.2011.00723.x. [DOI] [PubMed] [Google Scholar]
- 60.Porta-de-la-Riva M, Fontrodona L, Villanueva A, Cerón J. Basic Caenorhabditis elegans methods: synchronization and observation. J Vis Exp. 2012;10:e4019. doi: 10.3791/4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mitchell DH, Stiles JW, Santelli J, Sanadi DR. Synchronous growth and aging of caenorhabditis elegans in the presence of fluorodeoxyuridine. J Gerontol. 1979;34:28–36. doi: 10.1093/geronj/34.1.28. [DOI] [PubMed] [Google Scholar]
- 62.Davies SK, Leroi AM, Bundy JG. Fluorodeoxyuridine affects the identification of metabolic responses to daf-2 status in Caenorhabditis elegans. Mech Ageing Dev. 2012;133:46–49. doi: 10.1016/j.mad.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 63.Rooney JP, et al. Effects of 5’-fluoro-2-deoxyuridine on mitochondrial biology in Caenorhabditis elegans. Exp Gerontol. 2014;56:69–76. doi: 10.1016/j.exger.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gruber J, Ng LF, Poovathingsal SK, Halliwell B. Deceptively simple but simply deceptive – Caenorhabditis elegans lifespan studies: consideration for aging and antioxidant effects. FEBS Lett. 2009;583:3377–3387. doi: 10.1016/j.febslet.2009.09.051. [DOI] [PubMed] [Google Scholar]
- 65.Boyd WA, et al. A high-throughput method for assessing chemical toxicity using a Caenorhabditis elegans reproduction assay. Toxicol Appl Pharmacol. 2010;245:153–159. doi: 10.1016/j.taap.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zubovych IO, Straud S, Roth MG. Mitochondrial dysfunction confers resistance to multiple drugs in Caenorhabditis elegans. Mol Biol Cell. 2010;21:956–969. doi: 10.1091/mbc.E09-08-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu Y, et al. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell. 2014;158:1415–1430. doi: 10.1016/j.cell.2014.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weimer S, et al. D-glucosamine supplementation extends life span of nematodes and ageing mice. Nat Commun. 2014;5:3563. doi: 10.1038/ncomms4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Partridge FA, Tearle AW, Gravato-Nobre MJ, Schafer WR, Hodgkin J. The C. elegans glycosyltransferase BUS-8 has two distinct and essential roles in epidermal morphogenesis. Dev Biol. 2008;317:549–559. doi: 10.1016/j.ydbio.2008.02.060. [DOI] [PubMed] [Google Scholar]
- 70.MacVicar TD, Lane JD. Impaired OMA1-dependent cleavage of OPA1 and reduced DRP1 fission activity combine to prevent mitophagy in cells that are dependent on oxidative phosphorylation. J Cell Sci. 2014;127:2313–2325. doi: 10.1242/jcs.144337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stiernagle T. Maintenance of C. elegans. In: The C. elegans Research Community, editor. WormBook. 2006. [DOI] [Google Scholar]
- 72.Fraser A, et al. Functional genomic analysis of C. elegans Chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 73.Kamath RS, et al. Systematic Functional Analysis of the C. elegans Genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 74.Dancy BM, et al. Glutathione S-transferase mediates an ageing response to mitochondrial dysfunction. Mech Ageing Dev. 2015;153:14–21. doi: 10.1016/j.mad.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.