Abstract
Introduction
The last two decades have witnessed revolutionary changes in clinical diagnostics, fueled by the Human Genome Project and advances in high throughput, Next Generation Sequencing (NGS). We review the current state of sequencing-based pediatric diagnostics, associated challenges, and future prospects.
Areas Covered
We present an overview of genetic disease in children, review the technical aspects of Next Generation Sequencing and the strategies to make molecular diagnoses for children with genetic disease. We discuss the challenges of genomic sequencing including incomplete current knowledge of variants, lack of data about certain genomic regions, mosaicism, and the presence of regions with high homology.
Expert Commentary
NGS has been a transformative technology and the gap between the research and clinical communities has never been so narrow. Therapeutic interventions are emerging based on genomic findings and the applications of NGS are progressing to prenatal genetics, epigenomics and transcriptomics.
Keywords: Genomic diagnostics, Next generation sequencing, Exome sequencing, Genome sequencing, Variant interpretation, Pediatric diagnostics
1. Introduction
1.1 Pediatric Genetic Disease
As our understanding of the human genome has grown in the Post Human Genome Project era (since 2001), so has our understanding of the contribution of genetics to human disease. Genetic disorders are due to changes in an individual’s DNA either as a result of the inheritance of a genetic variant from a parent, or the result of a new genomic alteration. Genetic disease can affect any organ system, and problems can be developmental or emerge later in childhood or adulthood, or can cause cancer occurring in any organ system. They can be caused by chromosome-level abnormalities (e.g. extra, missing, deleted, duplicated, translocated, inverted, chromosomes) or sequence-based changes (e.g. nucleotide deletions, duplications, substitutions), and follow several different patterns of inheritance. Some genetic disorders have simple patterns of inheritance following the laws originally described by Gregor Mendel and are termed Mendelian disorders, or they exhibit one of the several non-Mendelian forms of inheritance (e.g. mitochondrial or imprinting disorders) but in other cases it is not a single gene responsible for the patients phenotype but rather several genes (polygenic inheritance) or a genetic susceptibility coupled with an environmental factor (multifactorial). A number of studies have been conducted to estimate the impact of genetics in pediatric illness [1–5] and the percentage of patients whose disease etiology is genetic has been found to depend on the clinical setting (regional hospital or tertiary center with an active intensive care unit), ranging from a low of 5% of pediatric illness (in a General hospital, [3]) to 70% of children admitted to an acute neonatal intensive care unit [2]. While genetic and developmental disorders are individually rare, collectively they are the leading cause of infant mortality and childhood disability.
Genetic disorders may present at birth (or in the prenatal period) in an infant or fetus with congenital anomalies, in the neonatal period with disease affecting any organ system, or during childhood with recognition of an emerging phenotype such as developmental delays, seizures or growth disorders. These disorders often have a profound impact on the affected children, their families, health care systems and society with an estimated 50–80% of resources used to manage disease in pediatric in-patient hospitals allocated to patients with a recognized genetic condition [4; 6–8]. The ability to provide optimal clinical management is dependent on identification of the underlying genetic cause of disease and the strategy for selection of a genomic diagnostic test is highly dependent on the patient’s presenting clinical phenotype and the differential diagnosis. [9].
1.2 Historical perspective of sequencing based diagnostics
Genetic diagnostics began in 1959 when Down Syndrome was shown to be caused by an abnormal number of chromosomes. The role of genetic changes in cancer was identified soon after with the finding of the Philadelphia chromosome in chronic myelogeneous leukemia [10]. DNA-based diagnostics began almost 2 decades later, following the discoveries of tools for DNA analysis such as restriction enzymes [11], hybridization-based analysis [12] and the Southern blot [13]. The first DNA sequence-based diagnosis was reported in 1978, with the prenatal diagnosis of sickle cell disease, using restriction enzyme digestion and Southern blotting [14]. In 1977, the now classic technique of DNA sequencing using chain terminating nucleotides was published by Fred Sanger and colleagues [15]. Early barriers to effective sequencing for medical diagnostics included the difficulties inherent in obtaining the gene of interest (which often required cloning), and the relatively few human disease genes known in the mid 1980’s. This changed dramatically with the introduction of the polymerase chain reaction (PCR) in 1986 [17] and soon after that in 1991, the Human Genome Project was launched, resulting in an explosion of tools for human gene identification. Today, there are over 4600 known human disease genes (OMIM.org statistics), most of which were identified since 1991, resulting from a number of strategies derived from the Human Genome Project, including positional cloning, linkage analysis, and genome-wide association studies.
Genomic diagnostics today is based on a combination of tests that look for DNA abnormalities that cause human disease. Cytogenetics based testing (chromosome analysis, fluorescence in situ hybridization (FISH) and chromosomal microarray analysis (CMA)), searches for abnormalities of the amount of DNA, be it whole chromosome or smaller deletions or duplications, as well as structural variation such as translocations or inversions. High-resolution copy number analysis using chromosomal microarray platforms is now standard of care in the diagnostic work-up of children with congenital anomalies and neurologic findings, but does not find all mutation types as it is unable to identify sequence variants and sub exon-level copy number alterations. Full gene Sanger sequencing has been the gold standard for sequencing but it is expensive, labor intensive and time consuming. The recent application of next generation sequencing (NGS) based technology to diagnostics has revolutionized our ability to achieve diagnoses in children with a broad array of clinical presentations, many of whom would have previously represented unsolvable cases. With reasonable sequencing costs, and improved informatics approaches to analysis, exome and genome sequencing has become a reality in the clinic and will likely progress towards a nearly routine tool for the care of pediatric illness.
2. Next Generation Sequencing
In 2005, several papers were published that presented alternative strategies to Sanger sequencing, allowing much faster sequencing of the entire human genome; these technologies have collectively been termed Next Generation Sequencing (NGS). NGS was quickly recognized as being able to overcome the limitations of Sanger sequencing and has demonstrated rapid uptake since its introduction [19; 20]. The power of next generation sequencing (NGS) lies in its unprecedented cost–effective scalability, ultimately leading to democratization of large scale sequencing, (including whole genome sequencing) to individual investigators and diagnosticians beyond well-resourced institutions. NGS is sometimes referred to as Massively Parallel Sequencing, and involves physical separation and immobilization of millions or billions of DNA molecules, and simultaneous generation of sequence reads for each one of those molecules. As such, NGS is quantitative in nature: while each read represents one DNA fragment, read or allele fraction can be used to infer zygosity and/or level of mosaicism in samples known to have mixed population of cells such as tumor samples [21; 22]. The number of reads at any given nucleotide refers to “coverage” at that position and heterozygous and homozygous germline variants are theoretically represented by 50% and 100% of the aligned reads, respectively (Figure 1). Using NGS technology, a human whole genome sequence can be generated within a week, for a cost of a few thousand dollars on a relatively small machine that can be operated by one trained technologist. This is in contrast to Sanger sequencing whereby a similar task involved multiple machines, and several hundred million dollars over several years.
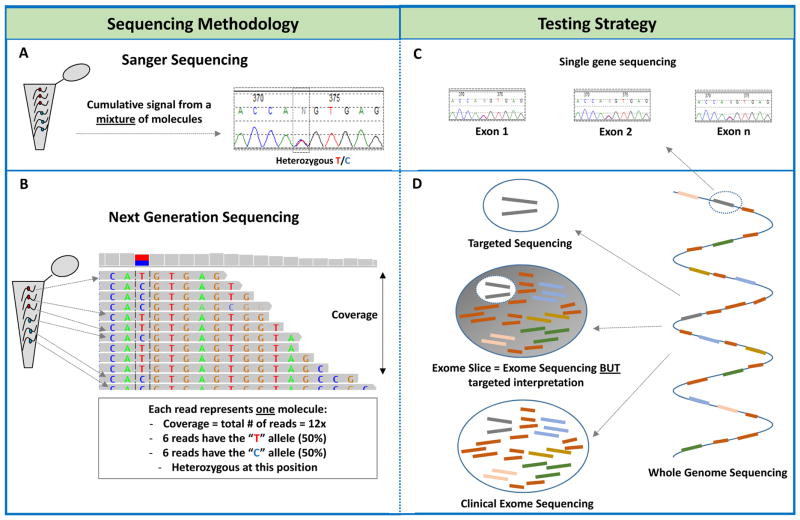
Figure 1. Sequencing technologies and associated testing strategies.
A) Schematic demonstrating how a specific genomic region is amplified from a mixture of DNA molecules and translated into a “Sanger trace”. For heterozygous variants (marked by a dashed box), 50% of the DNA molecules will have the reference allele (“T” in this example) while the other 50% carry the variant allele “C”. Overall, this heterozygous variant will appear as two overlapping peaks of relatively equal size and height on the Sanger trace. A homozygous allele (100% reference or variant allele) will be represented by one peak.
B) NGS is digital in nature and each DNA molecule is represented by one read or fragment. The number of reads at any nucleotide is referred to as the coverage at that position. The same “T” to “C” heterozygous variant (as with the Sanger example above) is represented by 12 reads (12x coverage) where 6 of them carry either a “T” or a “C”. This also appears on the top coverage bar graph. A homozygous allele will be covered by reads with the same nucleotide at that position.
C) Single gene testing is primarily done by Sanger sequencing where each exon in the gene of interest is amplified by PCR and then separately sequenced and analyzed. For large genes, NGS might offer a faster and cost-effective alternative to Sanger sequencing.
D) NGS allows more content to be simultaneously sequenced in one assay. On the right: The whole genome is represented by a single DNA strand whereby each colored thick bar represents a gene in the genome. The blue connectors represent intronic or noncoding regions. Top left: For targeted gene panels, only a few genes (only two in this example) are captured, sequenced and analyzed. Bottom left: in clinical exome sequencing, all gene-coding regions are captured and sequenced. Note, however, that certain captures have overrepresentation of probes towards clinically associated genes, so called the “medical exome”. Middle left: for an “exome slice” or “virtual panel”, the exome is sequenced but the interpretation is focused on a select of genes. This approach allows for faster reflexive interpretation of other content if the exome slice result was negative.
For detailed technical information about NGS and its applications, we refer the readers to excellent recently published reviews [21–27]. Here, we provide a brief overview of the technology before we discuss its clinical implementation, successes and challenges.
At its core, NGS includes four major steps: library preparation, sequencing, data processing, and interpretation. In the first step, a pool of fragmented input DNA or RNA (most commonly 200–400bp) is generated and ligated to oligonucleotide adaptors specific to each sequencing platform. Adaptors are needed for subsequent binding to the sequencer flow cell or beads, and also contain sequencing primer binding sites and can include molecular “barcodes” to allow sample pooling or multiplexing [26]. To limit sequencing to specific regions of interest in the genome (as would be the case for targeted gene panels and exome sequencing), target enrichment can be performed using several approaches such as multiplex PCR, molecular inversion probes, or solid- or liquid-phase hybridization capture [29]. For large targeted sequencing content, such as the exome, liquid-phase hybridization capture has been predominantly used for clinical applications [30]. PCR-based enrichment has also been used clinically especially when the desired gene content is limited, such as single large genes [31], small gene panels or targeted genotyping across many genes such as in cancer hotspot panels [32]. Once an adaptor-ligated library is prepared, it can now be “clonally” amplified, using adaptor specific primers, before proceeding to sequencing.
In the second step, millions or billions of clonally amplified fragments are physically separated and sequenced in parallel using one of several available chemistries including most commonly sequencing by synthesis (Illumina), sequencing by ligation (SOLiD), pyrosequencing (Roche 454) or semiconductor sequencing (ion Torrent) [31; 33]. These technologies vary by run time, data output, read lengths, and error rates.
The third step involves converting raw signal data (images, light intensity, or PH values) into actual base calls with acceptable quality followed by read alignment to a reference genome and subsequent variant calling and annotation. Several pipelines are currently available each with their own advantages and limitations. Given the amount of data and type of analysis (exome or genome versus targeted panels, inherited versus somatic, etc.), this step requires high computing power and sophisticated bioinformatics support [26].
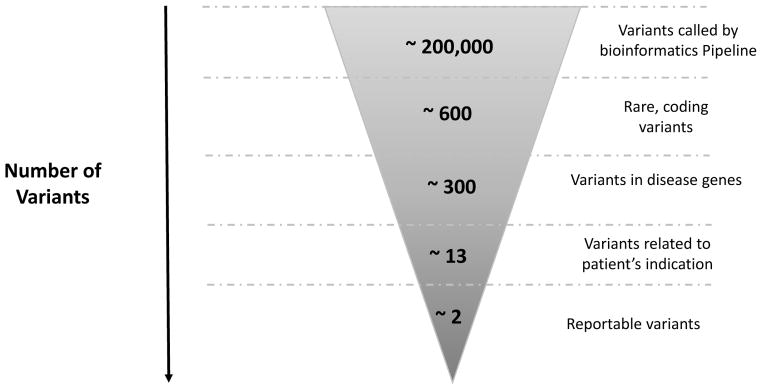
In the last step, annotated variants go through a filtration strategy including, most commonly, variant allele frequency cutoffs (see below) to remove variants unlikely to associate with the patient’s phenotype based on known disease prevalence, age of onset, penetrance, and mode of inheritance. This is a critical step in the process especially for whole exome and whole genome sequencing where hundreds of thousands of variants are generated. Other filters might be used to include variants of certain type (loss of function, for example) or those that have been previously reported in the literature or in disease databases. If parental samples are included, so-called trio analysis, filtration can be used to identify variants that fit a suspected inheritance model (de novo dominant, or compound heterozygous or homozygous recessive). The choice of filtration strategy might vary for each case and, therefore, requires skilled individuals with expertise in genetics and the technical aspects of NGS testing. An example of the number of variants obtained following sequencing and then after each filtration step is demonstrated in Figure 2.
Figure 2.
Number of variants at each step in the clinical exome bioinformatics filtration pipeline and interpretation process. As shown, ~200,000 variants are called by the bioinformatics pipeline following alignment of the captured sequence to a reference genome. Initial filtration is to select rare variants affecting exons ± 6bp. Variants that are high frequency in the general population (and therefore likely to be polymorphisms) are removed by filtering using population databases, primarily ExAC. Variants that are known to be disease causing are identified by filtering against databases with known mutations such as the Human Gene Mutation Database (HGMD). After this, on average, ~600 variants are retained, of which ~300 are in known disease genes. Subsequently, the remaining variants are manually correlated with the patient phenotype to identify genes that may be associated with the patient’s clinical features. An average of 13 variants remain, of which ~2 variants are likely to be clinically significant and therefore reportable (These are either pathogenic, likely pathogenic variants or variants of uncertain clinical significance). Incidental findings in the 56 ACMG gene list are reported in ~5% of cases. Data was derived from 182 clinical exome sequencing tests performed at the Division of Genomic Diagnostics, Children’s Hospital of Philadelphia (approximate averages are represented).
Despite the widespread application of NGS in several research areas including genomics, transcriptomics, metagenomics and epigenomics (26,27), clinically it has mostly been used for the diagnosis of inherited or acquired (e.g. cancer) genetic disease. Several considerations regarding the distinction between research and clinical NGS testing are beyond the scope of this review though it is important to mention that given its complexity and the significant impact it might have on patient’s management and care, a clinical NGS test has to be first adequately optimized and then carefully validated end-to-end, wet bench to bioinformatics pipeline, to determine key performance specifications such as reproducibility, accuracy, sensitivity and specificity [34]. In addition, as discussed below, clinically significant regions with low coverage – determined during test optimization and validation – are usually filled in using Sanger sequencing to ensure the highest possible clinical sensitivity.
3. Testing Strategies
Traditional Sanger-based diagnostic testing is based on analysis of a single gene at a time, starting with the gene most commonly associated with the patient’s presenting features and progressing to other possible genes if the first gene test was negative. This approach is often expensive, with long turnaround times and lack of scalability. The massively parallel nature of NGS has enabled the simultaneous analysis of many – or even all genes, eliminating the sequential candidate gene approach to testing and dramatically improving diagnostic yield and reducing the time to diagnosis for many patients. Currently, sequencing based diagnostic tests can be performed by 1) traditional Sanger-based sequencing of single genes; 2) Analysis of small groups of genes (up to 50 gene panels) that can be captured and analyzed simultaneously by NGS based on the clinical diagnosis (Noonan syndrome, microcephaly, ataxia, cancer predisposition genes, solid tumor causing genes etc.); 3) exome slice panels (> 50 genes where the entire exome is captured but a specific subset of genes are analyzed 4) exome sequencing, where the entire collection of exons is captured and analyzed and 5) whole genome sequencing, where the entire genome is sequenced (not just the coding regions), and structural variation may also be analyzed. Targeted panels (large or small) are most useful when there is strong clinical suspicion of the diagnosis, with a well-known list of candidate genes. If disease heterogeneity is high, larger exome slice panels may be quicker and more cost effective (hearing loss, epilepsy, mitochondrial disease)..
Optimal testing strategies are primarily dictated by the complexity of a patient’s clinical presentation and anticipated diagnostic yield of targeted diagnostic tests for that condition. For clinically recognizable disorders with limited locus heterogeneity or where other laboratory tests strongly suggest a single genetic etiology, such as familial adenomatous polyposis or medium-chain acyl-CoA dehydrogenase deficiency, Sanger-based single gene sequencing remains the method of choice. However, it is currently more cost effective to sequence large single genes by NGS, a trend that will extend into smaller genes as NGS costs continue to decline and we anticipate a continued shift towards NGS for most single gene testing. For disorders that have a relatively static, defined set of genes with a high diagnostic yield, such as Noonan syndrome for which an 8-gene panel has a diagnostic yield of approximately 80% [35], targeted gene panels are ideal. These panels are designed to yield full, deep coverage of the coding region of all targeted genes, offering very high sensitivity for sequence variants and improved detection of somatic mosaicism. Panels of genes are also being used to great effectiveness to sequence liquid or solid tumors to look for genetic changes associated with specific treatments or outcomes [36]. The clinical sensitivity of this approach is limited by the gene content in the panel and requires periodic updates as new gene-disease associations emerge, but still offers an advantage of cost and reduced turnaround time compared to methods detailed below.
The second diagnostic approach is the analysis of a restricted set of genes following exome sequencing, most appropriate for disorders with high locus heterogeneity and rapidly evolving genetic associations. These ‘virtual’ or ‘in silico’ panels offer the advantage of rapid content updates and reflex analysis of whole exome data for negative cases. While this strategy is attractive for the clinical laboratory workflow – a single laboratory assay that requires a single wet bench validation for several clinical tests –there are caveats to this approach. Most notably, sequencing coverage of the captured exome is not as high as for targeted panel based tests, and incomplete coverage of many clinically relevant genes within the exome panel can potentially result in a high burden of Sanger based fill in of sequencing gaps.
Clinical exome sequencing (CES) has become the standard sequencing based test for children with novel clinical presentations, suspected diagnoses for which there is no clinically available diagnostic test, those who have exhausted all other appropriate clinical genetic testing, or a clinical presentation with poorly understood genetic etiologies. However, some clinically defined disease groups with extreme locus heterogeneity, such as mitochondrial disorders or nonsyndromic intellectual disability, may also be best suited for CES instead of a gene panel approach. It is important to be aware that CES is likely to uncover findings unrelated to patient’s indication – so-called incidental findings including variants in medically actionable genes – autosomal recessive carrier status, and pharmacogenetic variants. Furthermore, given the expanded set of genes to be analyzed with this type of testing the number of variants of uncertain clinical significance (VUS) is expected to be much higher relative to targeted testing. In 250 clinical exomes, an indication-focused report included 0–2 deleterious variants and 4–9 VUSs, while an expanded report included an additional 1–3 deleterious variants and 17–41 VUSs in genes unrelated to patient’s phenotype. Furthermore, 17–25 truncating variants were identified in genes with no known disease association (39). In 2000 clinical exomes, the rate of medically actionable incidental findings was 3–4.6% (40).
While sequencing of the full genome (whole genome sequencing or GS) is clinically available, it is still more costly than CES, tools for simultaneous analysis of copy number variation from WGS are not yet mature and CGS has not yet become a primary diagnostic test. As the cost differential between CES and CGS diminishes and studies quantify the differential diagnostic yield of these approaches, a shift towards CGS in the future is anticipated.
4. Diagnostic Utility
Molecular diagnosis of rare inherited disorders can be challenging due to genetic heterogeneity, atypical presentations (variable expressivity and reduced penetrance), poorly delineated clinical features, and the presence of ultra-rare disorders in the differential diagnosis. Accordingly, around half of the individuals with a genetic disorder never receive an accurate diagnosis, while for those who do it is often significantly delayed [37]. The ability to simultaneously test all known genes associated with a particular disorder through gene panels results in significantly higher rates of molecular diagnoses than the traditional Sanger based approach [38]. Clinical diagnostic laboratories have reported a diagnostic yield of 25–40% for clinical exome sequencing in patients that have generally explored traditional avenues of diagnostic testing [39–43].
Diagnostic utility varies with the clinical indication for testing with some phenotypes exhibiting higher diagnostic yields (retinal disorders, specific neurological findings and multiple congenital anomalies, while autism spectrum disorder and disorders of the immune system may have lower yield [41; 43](Table 1).. There are numerous anecdotal reports of CES and CGS results that revealed a diagnosis and subsequent therapeutic intervention [44], however, larger-scale reports of the impact of CES on the care of patients with rare diseases are only beginning to emerge. Across a series of 105 families with positive CES results, Sawyer reported 6 that had dramatically altered medical management as the result of a positive finding [45]. A study of CGS in 35 acutely ill neonates reported 65% of molecular diagnoses were useful for acute clinical management with clinical outcome improving in 11% of these individuals [46]. In a study of 41 probands with intellectual disability and metabolic disorders, diagnoses were made in 68% by exome sequencing, with the possibility of medical intervention based on the underlying medical defect [47]. Other striking examples of how NGS diagnoses have impacted management include a report of a 15 month-old boy with intractable inflammatory bowel disease who was found to have a potentially causative missense variant in a gene not previously associated with this condition, the X-linked inhibitor of apoptosis (XIAP) gene by ES. This finding led to successful allogenic hematopoietic progenitor cell transplant, a recommended treatment for XIAP deficiency, and he recovered from the gastrointestinal disease [44]. A19 month old with a progressive neurodegenerative condition identified biallelic loss of function variants in SLC52A2, a cellular riboflavin transporter associated with Brown-Vialetto-Van Laere syndrome 2 (http://molecularcasestudies.cshlp.org/content/1/1/a000257.full). High dose riboflavin has previously been shown to normalize biochemical abnormalities and to stabilize or improve clinical outcomes in affected individuals [48]. Indeed, riboflavin treatment resulted in both short-term response and continued improvements of all disease manifestations of this usually fatal condition after 8 month follow up (http://molecularcasestudies.cshlp.org/content/1/1/a000265.full.pdf).
Table 1.
Pediatric Exome Sequencing Diagnostic Yield Based on Indication
| Indication | Diagnostic Yield |
|---|---|
| Inherited cancer53 | 10% |
| Somatic cancer53 | 47%* |
| Neurologic disorder40 | 27.2% |
| Specific neurologic disorder40 | 36.1% |
| Neurologic and organ-system disorder40 | 24.6% |
| Hearing disorder41 | 55.0% |
| Vision disorder41 | 47% |
| Skeletal muscle disorder41 | 40% |
| Skeletal disorder41 | 39% |
| Multiple congenital anomalies41 | 36% |
| Cardiovascular system41 | 28% |
Clinical utility of genomic testing is very high in cancer diagnostics, where identification of key molecular players in well-defined signaling pathways provides personalized prognostic and therapeutic avenues to cancer patients. The prime examples are a group of kinase inhibitors used in the treatment of lung, skin, gastrointestinal tumors, and leukemias [49–52]. Lung cancer patients with activating mutations in the epidermal growth factor (EGFR) show significant and durable responses to the tyrosine kinase inhibitor erlotinib [51]. Furthermore, continuous molecular monitoring in those patients enables identification of new tumor-specific drug resistance mutations and, therefore, altered management [52]. Imatinib (also known as Gleevec) have also shown significant impact on patients with chronic myeloid leukemia (CML) due to its ability to inhibit the Bcr-Abl tyrosine kinase, a fusion protein formed as result of the reciprocal translocation between chromosomes 9 and 22 – also known as the Philadelphia chromosome [10]. Identification of somatic and/or germline causative mutations in the RB1 gene can have significant impact on treatment, management, surveillance and recurrence risk of retinoblastoma, the most common ocular malignancy in childhood [53]. In addition, the identification of activating mutations in the ALK tyrosine kinase in children with neuroblastoma provided an avenue for potential targeted therapy in those patients [54]. More recently, using either exome sequencing [55] or targeted cancer panels [56], two pilot studies identified actionable mutations in 27–31% of children with several cancer diagnoses (n=100–150). Whole genome sequencing has also started to delineate the genetic landscapes across several pediatric cancer subtypes, and to identify rare structural alterations undetectable by targeted panels or whole exome sequencing [57] More interestingly, epigenetic analyses alongside whole genome sequencing of retinoblastoma showed that upregulation of the proto-oncogene SYK is required for tumor survivor, while its inhibition leads to retinoblastoma tumor cell death [58] Additional studies have started to uncover the complex interplay between genomic alterations and the cancer epigenomic landscape, opening new avenues for targeted therapy [59]. Integration of genomic, transcriptomic, and epigenomic data will certainly revolutionize the field of precision medicine, and unravel novel biological pathways with new insights into targeted therapy.
Widespread adoption of clinical CES has also revealed previously unappreciated phenomena, such as multiple molecular diagnoses in 3–4.6% of individuals with positive exome results [42; 43]. These individuals represent very complex clinical pictures that likely would never have been solved in the absence of CES/CGS evaluation. As cost benefit analyses emerge, it has been suggested that CES may be optimally employed as a first tier diagnostic test, where a recent study described a diagnostic rate of 57.5% across 80 clinical cases in a cohort of infants with a suspected genetic diagnosis [60].; an application that would likely reduce diagnosis time and facilitate optimal medical management for numerous individuals. Due to the success of universal newborn screening, necessity for a rapid diagnosis, and nonspecific early presentation of many life threatening neonatal disorders, it has been proposed that genomic testing in acutely ill neonates is a high yield application [61]. Indeed, methods to return results within 1–3 days have been developed [62] with a diagnostic yield of 47% [46].
Reflective of the rapidly evolving field of clinical genomics, CES has identified potential novel disease causing genes in 7.5–24% of cases, with one cohort reporting subsequent validation of a candidate gene in 8.3% of cases [39; 43; 60]. In this way, clinical genomic testing has utility as both a clinical diagnostic test and tool for genomic research.
The emerging application of NGS to fetal genomic sequencing promises to have a significant impact on newborn screening and early detection of pediatric-onset disorders. The ability to study placental cell-free nucleic acids circulating in maternal blood has revolutionized prenatal diagnosis by substituting a maternal blood draw for the invasive amniocentesis or chorionic villous sampling procedulres. Maternal serum can be screened for trisomies 13, 18, and 21 in the fetus. Although NGS is used, the data analysis is quite different than for standard sequencing, as the goal is analysis of chromosomal copy number, rather than sequence variation. Sequenced fragments are aligned back to the relevant chromosomes (e.g. 21), counted and then compared to the counts of fragments aligning to a reference chromosome. This is then used to infer chromosomal copy number states [63; 64]. Finally, fetal whole genome sequencing has been performed in research settings, though aided with highly sophisticated bioinformatics algorithms that are still not mature enough to be applied clinically [65; 66].
5. Technical challenges
The introduction of NGS to clinical genomics has been met with great successes; significantly improving the diagnostic yields in several clinical scenarios, most notably in pediatric constitutional and somatic disorders (Table 1), and is also steadily progressing to prenatal diagnostic applications [67; 68]. Despite all the success, NGS has limitations including both technical and interpretive challenges.
5.1 Variant Type
Although it can accurately detect single nucleotide variants (SNVs) and small insertions and deletions (indels) of up to 25 – 50bp depending on read length and the alignment and variant calling pipeline, NGS is still not as reliable in detecting small, exon-level copy number variants (CNVs). Such variants can significantly contribute to the overall test’s clinical sensitivity especially for genes associated with a loss of function disease mechanism. For instance, CNVs were shown to account for ~20% of all positive cases diagnosed using a targeted hearing loss gene panel [69]. Although coverage data from NGS have been used to infer copy number variation, such an approach has not yet reached the necessary performance metrics for a clinical-grade assay[70]. Copy number calling using whole genome sequencing is much more reliable due to homogeneous coverage (see below) and the likelihood of detecting CNV breakpoints in intergenic regions. However, WGS cost remains a major obstacle. Until more reliable and cost-effective NGS-based CNV tools become available, a secondary method will be needed to complement NGS panels with robust copy number detection especially for several diseases such as hearing loss and Charcot-Marie-Tooth Type 1 (CMT1) where, for example, PMP22 duplication represent 70–80% of such cases [71].
In addition to copy number variants, “traditional” targeted NGS strategies are blind to novel structural variants such as inversions, as those seen in hemophilia A for example [72], and balanced translocations such as BCR-ABL seen in chronic myeloid leukemia. Only in the rare scenario where the breakpoint of a structural event happens to be within a read or within an insert (where the insert size is larger than the read length) can such events be detected through identification of “split reads” or mapping of paired end reads to unexpected loci, respectively. Nonetheless, clever library preparations alongside bioinformatics solutions have been shown to reliably detect such aberrations, including, most recently, robust detection of gene fusions – without prior knowledge of fusion partners followed by sequencing [26; 68; 73; 74]. This is an area that is rapidly evolving and it is expected that several robust approaches will be at hand soon. Aside from the cost limitation, WGS might be a better strategy since the breakpoints of such events are likely to be intronic.
Finally, coding and non-coding repeat expansions, known to cause a large number of neurodegenerative and intellectual disorders, such as fragile X and dementias, cannot be reliably detected using NGS and, therefore, other PCR-based assays are currently needed to capture this type of variation [75]. With the ongoing sequencing and bioinformatics improvements – such as increased read length – however, this and other variant types described above are expected to be routinely detected using this technology.
5.2 Regulatory and extra-nuclear DNA
Unlike whole genome sequencing, most gene panels and the exome test target coding regions and ~20bp surrounding exon/intron boundaries. As such, deep intronic and noncoding regulatory regions – potentially affecting gene expression – will not be captured by this approach unless baits targeting those regions were included at the enrichment step. Currently, interpreting variants in such regions is difficult and, therefore, only variants with well-established clinical validity and/or utility in those regions should be considered for capture. In addition to noncoding regions, the mitochondrial genome (~16Kb) is normally not present in the exome capture unless it is separately isolated – most commonly by long range PCR (LR-PCR) – and then “spiked” into the exome. Although it can be sequenced through WGS, a separate pipeline specific for mitochondrial sequence alignment and variant calling should be employed.
5.3 Regions of homology
Highly homologous regions including pseudogenes pose a major challenge for short read NGS. This is mainly due to the inability to obtain unique alignments; homologous reads erroneously align to more than one region leading to false positive and/or false negative calls. Using long reads can be useful only if the read is longer than the homology or repeat region. Similarly, using paired end sequencing might be helpful since unique alignment of one read can help direct unique alignment of the second read. Nonetheless, a significant number of homologous regions are large enough (> 0.5 Kb) such that sequence read lengths currently obtained by NGS cannot reliably detect them. Molecular laboratories are thus required to develop ancillary assays using a different technology to reliably detect variants in regions of homology. There are several examples of such complementary assays in the literature including PMS2 [76], a known cause of Lynch syndrome, and STRC [77] associated with hearing loss. Both have pseudogenes with sequence identity > 98%, but contribute significantly to the test detection rate which, in the case of STRC, can be increased by ~10%; recently appreciated after designing a test that can resolve variants in STRC from its pseudogene [77].
Although LR-PCR was used in the cases of PMS2 and STRC, this approach is not scalable to the roughly over 100 pseudogenes with high homology to genes in the “medical” exome – the portion of the exome currently associated with clinical disorders. Innovative high throughput approaches are needed to tackle all homologous regions, the “PseudoGenome”, and to enhance diagnostic yield using NGS technology. Meanwhile, laboratories should investigate if genes on their targeted panels have any homology issues and/or if repeat expansions are a common cause of disease such as in neurodegenerative disorders.
Coverage gaps
Target enrichment usually includes a PCR step, often leading to unequal amplification across targeted regions, especially in “GC-rich” areas or in other complex, hard to amplify genomic structures. This is known to explain the coverage fluctuations or “gaps” seen in targeted gene panels and, more significantly, in the exome due to its larger gene content and overall lower coverage [34; 78]. Again, to attain the highest clinical sensitivity for panel based testing, such “gaps” should be filled in using a different technology. This is currently done by Sanger sequencing, which is somewhat effective for limited gene panels, but certainly not scalable to the exome. Whole genome sequencing, on the other hand, does not include a target enrichment step and thus generates a more uniform coverage across sequenced regions including the exome, a property that makes WGS more reliable for NGS-based CNV calling (see above).
5.4 Low-level mosaicism
Detecting variants present at very low levels – lower than what would be expected for heterozygous (50%) or homozygous (100%) variants – is essential for the detection of mosaic mutations causing disease including somatic variants in mixed populations of tumor and normal cells, and low level heteroplasmic mitochondrial variants. In both scenarios, relaxed allelic fraction ratios should be employed by the variant calling pipeline though careful validation is needed to establish the limit of detection, and to eliminate potential false positives that can be abundant at low allele fractions. For germline variants, most bioinformatics pipelines use cutoffs that assume either heterozygous (20–60%) or homozygous (>80%) variant calls. Interestingly, several new studies have challenged this assumption and presented examples where inherited disorders can be caused by tissue-specific, low level mosaic variants [79–81] that can be easily missed if DNA from blood was sequenced and/or if the variant was present at an allele fraction below the preset variant caller cutoff.
In summary, given all of the above limitations, it is likely that the reported exome detection rate (25–30%) is an underestimate of what would be expected if the NGS technology improves to reliably detect exon-level CNVs, structural variants, repeat expansions, and variants present at low level mosaicism or found with homology regions.
6. Interpretation challenges
By virtue of enabling simultaneous analysis of a large number of genes in one assay, next generation sequencing generates a large amount of data, necessitating the establishment of bioinformatics infrastructure in molecular genetics laboratories in order to extract meaningful information from such data. Even following bioinformatics filtration, there remain a large number of variants each requiring manual curation by a genomic analyst to determine their clinical significance in the context of the patient’s clinical findings. This is a time-consuming process in which information about each variant is gathered from commercial and publically available databases and the scientific literature, and synthesized to determine its clinical significance, ranging from benign to pathogenic [82]. Relevant variant evidence typically includes its allele frequency in the general population, evolutionary conservation, presence or absence in affected individuals and functional effect on the encoded protein. In the absence of functional or segregation data to support classification for each variant, a significant proportion are classified as variants of uncertain clinical significance (VUS). As such, variant interpretation is recognized to be the major bottleneck facing the widespread adoption of genomic sequencing, and is further exacerbated by the fact that most clinically significant variants are private, i.e. unique to each family [83].
As more data has accumulated since the implementation of NGS, it has become clear that key elements are needed in the genetics community to facilitate variant interpretation and to maximize the benefit of genomic sequencing. We emphasize that our discussion is centered on coding and splice site (10–20bp of the exon/intron boundaries) variants associated with monogenic, highly penetrant Mendelian disorders where rare variants conferring significant effects are expected. We will not address common variants with modest effects associated with complex genetic disorders, which – for the major part – remain immature, at least, from a molecular diagnostic perspective. We do not discuss the interpretation of regulatory and/or deep intronic variants that are extremely challenging to assess mainly due to our limited knowledge about their functional impact. Strictly speaking, we only focus on sequence variants identified in the 1–2% coding regions of the genome (so-called exome) claimed to house ~87% of the variants leading to genetic disorders.
6.1 Population databases
To assess the impact of variants in affected individuals, normal variation in the general population has to be catalogued to the best extent possible. The primary assumption is that a pathogenic variant is more likely to be absent or very rare – depending on several factors such as disease prevalence, mode of inheritance, penetrance and age of onset – in controls, defined as a group of individuals not selectively enriched for the phenotype at hand. On the other hand variants that are common in the general population are expected to be benign, and can thus be excluded from further analysis. Not long ago, clinical and research laboratories used in-house “control” datasets compromising a few hundred individuals sequenced for the genes of interest. Such control sets fail to capture all possible human variation within and across different ethnicities, and cannot be reliably used to estimate the true allele frequencies of the variants. However, two large scale, publicly accessible sequencing datasets have greatly improved our knowledge of allele frequencies. These are the 1000 Genomes project and the Exome Sequencing Project, which started accumulating data since 2010 [84; 85], although there are still limitations of overall numbers and/or ethnic representation. More recently, the ambitious Exome Aggregation Consortium (ExAC) data was made publicly accessible (http://exac.broadinstitute.org/). In this study, uniformly called exome sequencing data from ~60,000 individuals was obtained, with the individuals representing a wide spectrum of ethnic backgrounds.
Such large “population” variation datasets are now an integral component of the bioinformatics filtration strategy. Bioinformatics filters are set to exclude common, benign variants, ones with allele frequencies in the general population exceeding what would be expected given disease prevalence, mode of inheritance, penetrance, age of onset, besides other disease specific information. For example, hypertrophic cardiomyopathy (HCM) is a genetically heterogeneous, autosomal dominant disorder with an estimated prevalence of 1/500, but with reduced penetrance. Assuming one variant is the cause of all HCM cases, its allele frequency should not exceed 0.1% (1/1000 chromosomes) if the disease is 100% penetrant. However, assuming 50% penetrance, this allele frequency can be 0.2% and a safe filter of 0.3% can be employed to account for other variables such as disease age of onset. In addition to variant filtration, these control datasets have also been utilized to support clinical variant interpretation in many interesting ways including the quantification of genic and intragenic tolerance to variation, and better understanding the impact of loss of functions variants [86; 87] [88].
However, cautionary notes should be made with regard to variant interpretation. The presence or absence of a variant from the general population should be carefully interpreted. Although high allele frequency variants can be assumed to be benign in the context of Mendelian disorders, rare variants cannot be considered pathogenic. This is especially important in light of the finding that >50% of the variants identified in the ExAC cohort (>3.5 million variants) were seen only once [88], implying that saturation of variant detection is not yet reached with this sample size, and that sequencing more individuals is likely to reveal additional variation.
Finally, despite its comprehensiveness, the ExAC database still does not include sequence variation from certain groups such as the Ashkenazi Jewish and Middle Eastern populations, in which genetic disorders represent a significant public health concern. Thus, it is more challenging to interpret variants from patients in either group. Nonetheless, there is no doubt that sequencing datasets from these groups and others will soon be available.
6.2 Clinical variant databases
A significant proportion of variants survive the filtration step; most lack functional and/or segregation data to support their classification, and are thus classified as VUSs. Notably, despite the fact that most clinically significant variants and VUSs are extremely rare or even unique to most probands [83], information about these variants remain inaccessible, mostly residing in local databases within each diagnostic or research laboratory. Data sharing is essential. As more clinical laboratories move from targeted disease panels to whole exome sequencing, every laboratory will be faced with the challenge of interpreting variants in genes and diseases where they do not possess an expertise or internal proprietary knowledge base.
Recognizing the importance of variant sharing, the National Institute of Health (NIH) has recently funded the Clinical Genome Resource (ClinGen) Program with the goal of establishing and maintaining a publically accessible clinical grade genomic knowledge base – ClinVar – to promote variant data sharing between clinical laboratories, researchers, and clinicians. To date, contributing laboratories have submitted over 100,000 variants with information about each variant’s classification and the supporting evidence [83]. Despite its usefulness, ClinVar is still a work in progress. Significant discrepancies exist between submitters regarding variant classification; this is to be expected in the absence of universal guidelines, at least, until very recently [89].
6.3 Guidelines
To establish variant pathogenicity for a given disease, first the affected gene has to be associated with disease (gene-disease correlation), then the variant must affect protein function in a way that fits what is known about the disease mechanism for that gene (gain or loss of function, for example). Finally, the patient’s phenotype should, for the most part, fit the expectation at least for severe, early onset Mendelian disorders. Uncertainty at the variant, gene, and less so at the phenotype levels can lead to a VUS call. In the absence of the appropriate standards, wrong assertions about variant and gene causality have been made in the literature and current databases [90; 91]. This is a serious hindrance to effective data sharing. In fact, the second major goal of the ClinGen program is the establishment and dissemination of standards and guidelines for variant interpretation and gene-disease association. While several initiatives and recent guidelines have started to address the challenge, it will be a while before full harmonization is achieved amongst laboratories [92].
6.4 Clinical and research networks
Most genetic disorders are extremely rare, making it hard for researchers and clinicians to identify additional unrelated cases to establish causality of novel genes. It is equally challenging to perform functional studies on every identified novel variant or gene. A potentially useful solution is the establishment of internet networks allowing diagnostics laboratories, researchers, and clinicians to deposit phenotypic and/or gene information so that matches can be identified and shared amongst contributors. In fact, several such networks have recently been launched, and have already led to successful establishment of a plethora of gene-disease relationships [93–95].
We emphasize that current efforts, at least within the clinical genetics community, are mostly geared towards rare Mendelian disorders. Complex or multifactorial disorders such as most types of diabetes, obesity and blood pressure, where rare and common variants in multiple genes combined with environmental and lifestyle factors contribute to disease and its progression, are still not appropriate for clinical molecular diagnostics. Different statistical and variant filtration approaches, interpretation guidelines, and infrastructure – including rich longitudinal phenotype-genotype databases – are needed before sequencing-based approaches can be clinically applied to complex traits or diseases.
7. Ethical Challenges
Genomic diagnostics is fraught with ethical concerns for several reasons including 1) the potential for provision of predictive information regarding an individual’s future health status, 2) the fact that providing genetic information to one individual has implications for family members, 3) the fact that genetic diagnoses are often not absolute, but provide information regarding risks, which can be complex and difficult to interpret without expert assistance, 4) the possibility that a genome-wide test will reveal information about health conditions other than the reason for study, and these “incidental” or “secondary” findings can have grave implications regarding future health, 5) the possibility that there will be information obtained regarding family relationships (non-paternity or incest) which may be difficult for the family psychologically and 6) the potential for misuse of genetic information by insurance companies, employers or others (in spite of legislation designed to prevent this) [96]. Recognition of all of these issues has led to the practice of obtaining informed consent for genetic studies to insure that individuals undergoing testing understand all of these risks, however, since children usually cannot provide informed consent themselves, there are special considerations in the pediatric setting. For all of these reasons, it is imperative that clinicians delivering genetic information are superbly well educated, have access to professionals who can provide assistance with complex ethical decisions, and are willing to provide the necessary education and obtain informed consent from the families of their patients.
8. Expert commentary
The widespread adoption of genome-wide testing via next generation sequencing technologies is changing the practice of medicine [97]. The use of NGS in the clinical laboratory has facilitated a rapid proliferation of diagnostic tests for nearly every inherited and acquired genetic disorder imaginable. Even smaller diagnostic laboratories can now offer a multitude of NGS-based single gene and gene panel tests, with some offering clinical exome or genome sequencing. Clinical exome sequencing has changed the role of the clinical molecular genetics laboratory from primarily confirmation of suspected diagnoses to making new and unexpected diagnoses in some of the most challenging clinical cases. Genomic diagnostic tests such as clinical exome or genome sequencing are now simpler to order (as they don’t require specific diagnostic hypotheses), but may be more difficult to interpret, as variants of uncertain significance and incidental findings pose unique challenges. While therapeutic interventions resulting from new diagnoses in the inherited disorders are relatively few (although growing rapidly), the therapeutic implications in oncology are substantial and will also continue to grow as more genetically determined treatments are evaluated. These transformative technologies have made clinical diagnostic laboratories major players in the explosion in new gene discoveries and a vital member of the personalized medicine team. The importance of sequencing based diagnostics in the diagnosis, management and treatment of pediatric disease will increase as these technologies and their clinical applications mature and continue to accurately detect additional types of variants such as structural rearrangements and copy number changes [98].
9. Five year-view
Continually declining sequencing costs will undoubtedly drive further proliferation of sequencing based diagnostic tests and move the field towards the adoption of a single integrated genomic test – whole genome sequencing which will have the capabiiity to diagnose single nucleotide changes, as well as structural variation. Indeed, the $1000 genome may have already been achieved [99], most recently with the claim that this price includes interpretation and genetic counseling [100]. In time WGS will query all types of genetic variation. Increased read lengths, sequencing depth and enhanced bioinformatics approaches will enable WGS to sensitively detect exon and sub-exon level copy number variations, structural rearrangements and analyze repetitive elements in the human genome, such as those responsible for the repeat expansion disorders. Ideally, these data will be housed in electronic medical records (EMR) and can be periodically queried for new clinical indications, pharmacogenomic consultation and to inform lifestyle, healthcare and family planning decisions throughout an individual’s lifetime. However, one of the primary limitations of sequencing based diagnostics is our limited understanding of the human genome and consequent difficulty in interpreting the significance of variations therein. A first step towards addressing this limitation is the integration of genomic data with the EMR in searchable databases where genetic variations can be linked to detailed phenotypes and clinical histories, which has been initiated at several institutions including Geisinger Medical Center [22]. The immediate availability of familial clinical and genetic data will enable thorough investigation of variants in real time, improving the clinical interpretation and prognostic value of genomic testing results. Data sets like these combined with environmental and lifestyle information will also prove critical for the investigation of disorders with complex genetics and significantly reduced penetrance, enabling the assemblage of large case-control studies on the fly. Furthermore, these efforts will facilitate the identification of genotype specific therapeutics and truly move us towards the practice of personalized medicine.
Sequencing based diagnostics will also begin to explore additional levels of variation, such as the epigenome and transcriptome. Concurrent RNA sequencing will directly facilitate the interpretation of variants that may impact splicing or gene regulation, which likely play a larger than appreciated role in human disease [101], or that are exclusively expressed as a result of allelic bias [102]. Alternatively, this information may present a molecular signature (e.g. rearranged or overexpressed transcript) that is diagnostic or suggestive of a treatment modality in Mendelian disorders. Querying genome-wide DNA methylation and histone modifications – through bisulfite treatment and chromatin immunoprecipitation (ChIP), respectively, followed by NGS – will shed light on epigenetic changes affecting gene regulation and leading to or modifying human disease [25].
Moreover, as genome-editing tools continue to mature, their potential application in large scale, high throughput functional genomics – the study of the functional impact of genetic variants – will have a significant impact on clinical variant interpretation.
Whole exome and genome based newborn screening (NBS) has been discussed recently, with pilot studies suggesting this approach may have specific advantages over traditional NBS with potentially faster return of results and clarification of indeterminate results [103–106]. With sufficiently fast and cheap methods of sequencing and automated means of variant and genome interpretation, these approaches may become routine in time. However, significant advances need to be made in each component of genomic testing to make this a reality. As the entire fetal genome can be sequenced from maternal cell free DNA [65], one could imagine a prenatal fetal genome screen to identify those at high risk for complications soon after birth. Where appropriate, such an approach could facilitate therapeutic interventions in utero, such as is currently practiced for a very limited subset of disorders like congenital adrenal hyperplasia. Alternatively, a prenatally identified risk could allow for the timely treatment of infants with disorders that may manifest before traditional NBS results are available or in those where false negatives are known to occur.
Key Issues.
Next-Generation Sequencing (NGS) enables simultaneous analysis of a large number of genes in one diagnostic test. NGS based tests have been increasingly utilized to replace the time-consuming and expensive single gene reflexive tests that are based on Sanger sequencing.
NGS testing strategies include targeted gene panels, clinical exome sequencing, “in silico” or “virtual” panels, and genome sequencing.
Clinical exome sequencing offers high – up to 50% – diagnostic yields for several pediatric disorders, with a growing number of cases where an exome finding led to a successful treatment or management.
Technical challenges inherent to the NGS technology include coverage gaps and the inability to query certain variant types such as repeat expansions, copy number and structural variants, in addition to sequence variants in regions of high homology (pseudogenes).
Variant interpretation is the major challenge facing large-scale genomic sequencing. This is mainly due to lack of functional and segregation data to interpret identified variants thus leading to a large number of variants and genes of uncertain clinical significance. Data sharing alongside interpretation standardization are essential.
Several ethical issues, such as the identification of incidental or secondary findings, have to be thoroughly considered in the context of clinical exome and genome sequencing.
Due to its consistent coverage and inclusion of intronic regions, genome sequencing promises to query most types of variations if the appropriate bioinformatics tools are employed.
NGS applications are expanding to other clinical settings (besides the pediatric clinic) such as newborn screening and prenatal genetics. It is also being employed in studies of the transcriptome and the epigenome.
Acknowledgments
Funding
This manuscript has been supported by The Children’s Hospital of Philadelphia and The Perelman School of Medicine at The University of Pennsylvania Department of Pathology and Laboratory Medicine. NB Spinner has been supported by the following NIH/NHGRI Grant U01-HG006546 Applying genome sequencing in Pediatrics; NIH/NIDDK: 1 R01KD090045-01 Genetic susceptibility to biliary atresia; NIH NIDDK/2 U01 DK062481 Advancing our understanding of rare pediatric liver disease.
Footnotes
Declaration of Interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Contributor Information
Ahmad Abou Tayoun, Email: aboutayoua@email.chop.edu, Assistant Professor and Assistant Laboratory Director, Division of Genomic Diagnostics, Department of Pathology and Laboratory Medicine, The Children’s Hospital of Philadelphia and The Perelman School of Medicine at The University of Pennsylvania, 716D Abramson Research Center, 3501 Civic Center Blvd, Philadelphia, PA 19104, 215-590-3264; fax: 215-590-2156.
Bryan Krock, Email: krockb@email.chop.edu, Assistant Professor and Assistant Laboratory Director, Division of Genomic Diagnostics, Department of Pathology and Laboratory Medicine, The Children’s Hospital of Philadelphia and The Perelman School of Medicine at The University of Pennsylvania, 706B Abramson Research Center; 3501 Civic Center Blvd, Philadelphia, PA 19104; 267-426-7588; fax: 215-590-2156.
Nancy B. Spinner, Email: spinner@upenn.edu, Professor and Division Chief, Division of Genomic Diagnostics Department of Pathology and Laboratory Medicine, The Children’s Hospital of Philadelphia and The Perelman School of Medicine at The University of Pennsylvania, 716B Abramson Research Center; 3501 Civic Center Blvd, Philadelphia, PA 19104, 215-590-4177’; fax: 215-590-2156
References
Reference annotations
*Of interest
**Of considerable interest
- 1.Costa T, Scriver CR, Childs B. The effect of Mendelian disease on human health: a measurement. Am J Med Genet. 1985;21:231–242. doi: 10.1002/ajmg.1320210205. [DOI] [PubMed] [Google Scholar]
- 2.FitzPatrick DR, Skeoch CH, Tolmie JL. Genetic aspects of admissions to a paediatric intensive care unit. Arch Dis Child. 1991;66:639–641. doi: 10.1136/adc.66.5.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lialiaris T, Mantadakis E, Kareli D, Mpountoukas P, et al. Frequency of genetic diseases and health coverage of children requiring admission in a general pediatric clinic of northern Greece. Ital J Pediatr. 2010;36:9. doi: 10.1186/1824-7288-36-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCandless SE, Brunger JW, Cassidy SB. The burden of genetic disease on inpatient care in a children’s hospital. Am J Hum Genet. 2004;74:121–127. doi: 10.1086/381053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scriver CR, Neal JL, Saginur R, Clow A. The frequency of genetic disease and congenital malformation among patients in a pediatric hospital. Can Med Assoc J. 1973;108:1111–1115. [PMC free article] [PubMed] [Google Scholar]
- 6.Liu L, Oza S, Hogan D, Perin J, et al. Global, regional, and national causes of child mortality in 2000–13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet. 2015;385:430–440. doi: 10.1016/S0140-6736(14)61698-6. [DOI] [PubMed] [Google Scholar]
- 7.Shevell M, Ashwal S, Donley D, Flint J, et al. Practice parameter: evaluation of the child with global developmental delay: report of the Quality Standards Subcommittee of the American Academy of Neurology and The Practice Committee of the Child Neurology Society. Neurology. 2003;60:367–380. doi: 10.1212/01.wnl.0000031431.81555.16. [DOI] [PubMed] [Google Scholar]
- 8.Hall JG. The impact of birth defects and genetic diseases. Arch Pediatr Adolesc Med. 1997;151:1082–1083. doi: 10.1001/archpedi.1997.02170480012002. [DOI] [PubMed] [Google Scholar]
- 9.Coulter ME, Miller DT, Harris DJ, Hawley P, et al. Chromosomal microarray testing influences medical management. Genet Med. 2011;13:770–776. doi: 10.1097/GIM.0b013e31821dd54a. [DOI] [PubMed] [Google Scholar]
- 10.Nowell PC. The minute chromosome (Phl) in chronic granulocytic leukemia. Blut. 1962;8:65–66. doi: 10.1007/BF01630378. [DOI] [PubMed] [Google Scholar]
- 11.Dancey GF, Shapiro BM. Specific phospholipid requirement for activity of the purified respiratory chain NADH dehydrogenase of Escherichia coli. Biochim Biophys Acta. 1977;487:368–377. doi: 10.1016/0005-2760(77)90013-3. [DOI] [PubMed] [Google Scholar]
- 12.Kan YW, Golbus MS, Dozy AM. Prenatal diagnosis of alpha-thalassemia. Clinical application of molecular hybridization. N Engl J Med. 1976;295:1165–1167. doi: 10.1056/NEJM197611182952104. [DOI] [PubMed] [Google Scholar]
- 13.Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 14.Kan YW, Dozy AM. Antenatal diagnosis of sickle-cell anaemia by D.N.A. analysis of amniotic-fluid cells. Lancet. 1978;2:910–912. doi: 10.1016/s0140-6736(78)91629-x. [DOI] [PubMed] [Google Scholar]
- 15.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith LM, Sanders JZ, Kaiser RJ, Hughes P, et al. Fluorescence detection in automated DNA sequence analysis. Nature. 1986;321:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 17.Mullis K, Faloona F, Scharf S, Saiki R, et al. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):263–273. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- 18.Metzker ML. Emerging technologies in DNA sequencing. Genome Res. 2005;15:1767–1776. doi: 10.1101/gr.3770505. [DOI] [PubMed] [Google Scholar]
- 19.Shendure J, Porreca GJ, Reppas NB, Lin X, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 20.Margulies M, Egholm M, Altman WE, Attiya S, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 22.Voelkerding KV, Dames S, Durtschi JD. Next generation sequencing for clinical diagnostics-principles and application to targeted resequencing for hypertrophic cardiomyopathy: a paper from the 2009 William Beaumont Hospital Symposium on Molecular Pathology. J Mol Diagn. 2010;12:539–551. doi: 10.2353/jmoldx.2010.100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23*.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. This paper provided one of the first methods to accomplish highly parallel sequencing, albeit in a bacterial system. Polony refers to “Polymerase Colony” which describes the polymerase driven amplification of a complex library of sequencing templates. This method was a forerunner of the Life Technologies/ABI SOLiD sequencing system. [DOI] [PubMed] [Google Scholar]
- 24.Martin JA, Wang Z. Next-generation transcriptome assembly. Nat Rev Genet. 2011;12:671–682. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- 25.Ku CS, Naidoo N, Wu M, Soong R. Studying the epigenome using next generation sequencing. J Med Genet. 2011;48:721–730. doi: 10.1136/jmedgenet-2011-100242. [DOI] [PubMed] [Google Scholar]
- 26.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 27.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 28.Caruccio N. Preparation of next-generation sequencing libraries using Nextera technology: simultaneous DNA fragmentation and adaptor tagging by in vitro transposition. Methods Mol Biol. 2011;733:241–255. doi: 10.1007/978-1-61779-089-8_17. [DOI] [PubMed] [Google Scholar]
- 29.Mamanova L, Coffey AJ, Scott CE, Kozarewa I, et al. Target-enrichment strategies for next-generation sequencing. Nat Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- 30.Gnirke A, Melnikov A, Maguire J, Rogov P, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abou Tayoun AN, Tunkey CD, Pugh TJ, Ross T, et al. A comprehensive assay for CFTR mutational analysis using next-generation sequencing. Clin Chem. 2013;59:1481–1488. doi: 10.1373/clinchem.2013.206466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsongalis GJ, Peterson JD, de Abreu FB, Tunkey CD, et al. Routine use of the Ion Torrent AmpliSeq Cancer Hotspot Panel for identification of clinically actionable somatic mutations. Clin Chem Lab Med. 2014;52:707–714. doi: 10.1515/cclm-2013-0883. [DOI] [PubMed] [Google Scholar]
- 33.Voelkerding KV, Dames SA, Durtschi JD. Next-generation sequencing: from basic research to diagnostics. Clin Chem. 2009;55:641–658. doi: 10.1373/clinchem.2008.112789. [DOI] [PubMed] [Google Scholar]
- 34.Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, et al. ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013;15:733–747. doi: 10.1038/gim.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allanson JE, Roberts AE. Noonan Syndrome. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, et al., editors. GeneReviews(R) Seattle (WA): 1993. [Google Scholar]
- 36.Marrone M, Filipski KK, Gillanders EM, Schully SD, et al. Multi-marker Solid Tumor Panels Using Next-generation Sequencing to Direct Molecularly Targeted Therapies. PLoS Curr. 2014:6. doi: 10.1371/currents.eogt.aa5415d435fc886145bd7137a280a971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shashi V, McConkie-Rosell A, Rosell B, Schoch K, et al. The utility of the traditional medical genetics diagnostic evaluation in the context of next-generation sequencing for undiagnosed genetic disorders. Genet Med. 2014;16:176–182. doi: 10.1038/gim.2013.99. [DOI] [PubMed] [Google Scholar]
- 38.Neveling K, Feenstra I, Gilissen C, Hoefsloot LH, et al. A post-hoc comparison of the utility of sanger sequencing and exome sequencing for the diagnosis of heterogeneous diseases. Hum Mutat. 2013;34:1721–1726. doi: 10.1002/humu.22450. [DOI] [PubMed] [Google Scholar]
- 39.Farwell KD, Shahmirzadi L, El-Khechen D, Powis Z, et al. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet Med. 2015;17:578–586. doi: 10.1038/gim.2014.154. [DOI] [PubMed] [Google Scholar]
- 40.Lee H, Deignan JL, Dorrani N, Strom SP, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41**.Yang Y, Muzny DM, Reid JG, Bainbridge MN, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. This paper presented results of exome sequencing of the first 250 patients from a large clinical laboratory. A molecular diagnosis was reported in 25% of patients referred with a range of phenotypes, with 4/250 having two non-overlapping diagnoses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y, Muzny DM, Xia F, Niu Z, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Retterer K, Juusola J, Cho MT, Vitazka P, et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med. 2015 doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 44.Worthey EA, Mayer AN, Syverson GD, Helbling D, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13:255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 45.Sawyer SL, Hartley T, Dyment DA, Beaulieu CL, et al. Utility of whole-exome sequencing for those near the end of the diagnostic odyssey: time to address gaps in care. Clin Genet. 2016;89:275–284. doi: 10.1111/cge.12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willig LK, Petrikin JE, Smith LD, Saunders CJ, et al. Whole-genome sequencing for identification of Mendelian disorders in critically ill infants: a retrospective analysis of diagnostic and clinical findings. Lancet Respir Med. 2015;3:377–387. doi: 10.1016/S2213-2600(15)00139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarailo-Graovac M, Shyr C, Ross CJ, Horvath GA, et al. Exome Sequencing and the Management of Neurometabolic Disorders. N Engl J Med. 2016;374:2246–2255. doi: 10.1056/NEJMoa1515792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foley AR, Menezes MP, Pandraud A, Gonzalez MA, et al. Treatable childhood neuronopathy caused by mutations in riboflavin transporter RFVT2. Brain. 2014;137:44–56. doi: 10.1093/brain/awt315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goetz EM, Ghandi M, Treacy DJ, Wagle N, et al. ERK mutations confer resistance to mitogen-activated protein kinase pathway inhibitors. Cancer Res. 2014;74:7079–7089. doi: 10.1158/0008-5472.CAN-14-2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paez JG, Janne PA, Lee JC, Tracy S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 51.Pao W, Miller V, Zakowski M, Doherty J, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aerts I, Lumbroso-Le Rouic L, Gauthier-Villars M, Brisse H, et al. Retinoblastoma. Orphanet J Rare Dis. 2006;1:31. doi: 10.1186/1750-1172-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bresler SC, Weiser DA, Huwe PJ, Park JH, et al. ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer Cell. 2014;26:682–694. doi: 10.1016/j.ccell.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parsons DW, Roy A, Yang Y, Wang T, et al. Diagnostic Yield of Clinical Tumor and Germline Whole-Exome Sequencing for Children With Solid Tumors. JAMA Oncol. 2016 doi: 10.1001/jamaoncol.2015.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harris MH, DuBois SG, Glade Bender JL, Kim A, et al. Multicenter Feasibility Study of Tumor Molecular Profiling to Inform Therapeutic Decisions in Advanced Pediatric Solid Tumors: The Individualized Cancer Therapy (iCat) Study. JAMA Oncol. 2016 doi: 10.1001/jamaoncol.2015.5689. [DOI] [PubMed] [Google Scholar]
- 57.Downing JR, Wilson RK, Zhang J, Mardis ER, et al. The Pediatric Cancer Genome Project. Nat Genet. 2012;44:619–622. doi: 10.1038/ng.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J, Benavente CA, McEvoy J, Flores-Otero J, et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature. 2012;481:329–334. doi: 10.1038/nature10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu F, Wang L, Perna F, Nimer SD. Beyond transcription factors: how oncogenic signalling reshapes the epigenetic landscape. Nat Rev Cancer. 2016;16:359–372. doi: 10.1038/nrc.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stark Z, Tan TY, Chong B, Brett GR, et al. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet Med. 2016 doi: 10.1038/gim.2016.1. [DOI] [PubMed] [Google Scholar]
- 61**.Saunders CJ, Miller NA, Soden SE, Dinwiddie DL, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med. 2012;4:154ra135. doi: 10.1126/scitranslmed.3004041. Demonstration of the ability to utilize genome sequencing to make rapid diagnoses in the neonatal intensive care unit. Early diagnosis has the potential to allow early treatment, with potential to impact clinical course dramatically. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller NA, Farrow EG, Gibson M, Willig LK, et al. A 26-hour system of highly sensitive whole genome sequencing for emergency management of genetic diseases. Genome Med. 2015;7:100. doi: 10.1186/s13073-015-0221-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chiu RW, Chan KC, Gao Y, Lau VY, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci U S A. 2008;105:20458–20463. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, et al. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci U S A. 2008;105:16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kitzman JO, Snyder MW, Ventura M, Lewis AP, et al. Noninvasive whole-genome sequencing of a human fetus. Sci Transl Med. 2012;4:137ra176. doi: 10.1126/scitranslmed.3004323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lo YM, Chan KC, Sun H, Chen EZ, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2:61ra91. doi: 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- 67*.Bianchi DW, Parker RL, Wentworth J, Madankumar R, et al. DNA sequencing versus standard prenatal aneuploidy screening. N Engl J Med. 2014;370:799–808. doi: 10.1056/NEJMoa1311037. Results of a study comparing noninvasive prenatal testing for fetal aneuploidy (trisomy 13,18 and 21) using NGS of cell free fetal DNA (obtained in maternal serum) with the then standard approach for trisomy screening by serum biochemical assays with or without nuchal translucency measurements. The results demonstrated that the sequencing approach had higher positive predictive values and lower false positive rates for trisomies of chromosome 18 and 21. [DOI] [PubMed] [Google Scholar]
- 68.Talkowski ME, Ordulu Z, Pillalamarri V, Benson CB, et al. Clinical diagnosis by whole-genome sequencing of a prenatal sample. N Engl J Med. 2012;367:2226–2232. doi: 10.1056/NEJMoa1208594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shearer AE, Kolbe DL, Azaiez H, Sloan CM, et al. Copy number variants are a common cause of non-syndromic hearing loss. Genome Med. 2014;6:37. doi: 10.1186/gm554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pugh TJ, Amr SS, Bowser MJ, Gowrisankar S, et al. VisCap: inference and visualization of germ-line copy-number variants from targeted clinical sequencing data. Genet Med. 2015 doi: 10.1038/gim.2015.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bird TD. Charcot-Marie-Tooth Neuropathy Type 1. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, et al., editors. GeneReviews(R) Seattle (WA): 1993. [Google Scholar]
- 72.Konkle BA, Josephson NC, Nakaya Fletcher S. Hemophilia A. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, et al., editors. GeneReviews(R) Seattle (WA): 1993. [Google Scholar]
- 73.Zheng Z, Liebers M, Zhelyazkova B, Cao Y, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20:1479–1484. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 74.Talkowski ME, Ernst C, Heilbut A, Chiang C, et al. Next-generation sequencing strategies enable routine detection of balanced chromosome rearrangements for clinical diagnostics and genetic research. Am J Hum Genet. 2011;88:469–481. doi: 10.1016/j.ajhg.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Renton AE, Majounie E, Waite A, Simon-Sanchez J, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li J, Dai H, Feng Y, Tang J, et al. A Comprehensive Strategy for Accurate Mutation Detection of the Highly Homologous PMS2. J Mol Diagn. 2015;17:545–553. doi: 10.1016/j.jmoldx.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 77.Mandelker D, Amr SS, Pugh T, Gowrisankar S, et al. Comprehensive diagnostic testing for stereocilin: an approach for analyzing medically important genes with high homology. J Mol Diagn. 2014;16:639–647. doi: 10.1016/j.jmoldx.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 78.Rehm HL. Disease-targeted sequencing: a cornerstone in the clinic. Nat Rev Genet. 2013;14:295–300. doi: 10.1038/nrg3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu X, Yang X, Wu Q, Liu A, et al. Amplicon Resequencing Identified Parental Mosaicism for Approximately 10% of “de novo” SCN1A Mutations in Children with Dravet Syndrome. Hum Mutat. 2015;36:861–872. doi: 10.1002/humu.22819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Acuna-Hidalgo R, Bo T, Kwint MP, van de Vorst M, et al. Post-zygotic Point Mutations Are an Underrecognized Source of De Novo Genomic Variation. Am J Hum Genet. 2015;97:67–74. doi: 10.1016/j.ajhg.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamaguchi K, Komura M, Yamaguchi R, Imoto S, et al. Detection of APC mosaicism by next-generation sequencing in an FAP patient. J Hum Genet. 2015;60:227–231. doi: 10.1038/jhg.2015.14. [DOI] [PubMed] [Google Scholar]
- 82.Duzkale H, Shen J, McLaughlin H, Alfares A, et al. A systematic approach to assessing the clinical significance of genetic variants. Clin Genet. 2013;84:453–463. doi: 10.1111/cge.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83*.Rehm HL, Berg JS, Brooks LD, Bustamante CD, et al. ClinGen--the Clinical Genome Resource. N Engl J Med. 2015;372:2235–2242. doi: 10.1056/NEJMsr1406261. Presentation of the NIH’s Clinical Genome Resource (ClinGen) program, which was established in 2013 as the primary site for deposition and retrieval of genomic variant data and annotations to guide diagnostics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tennessen JA, Bigham AW, O’Connor TD, Fu W, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Genomes Project C. Abecasis GR, Altshuler D, Auton A, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gussow AB, Petrovski S, Wang Q, Allen AS, et al. The intolerance to functional genetic variation of protein domains predicts the localization of pathogenic mutations within genes. Genome Biol. 2016;17:9. doi: 10.1186/s13059-016-0869-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Petrovski S, Wang Q, Heinzen EL, Allen AS, et al. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lek M, Karczewski K, Minikel E, et al. Analysis of protein-coding genetic variation in 60,706 humans. bioRxiv. 2015 doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Richards S, Aziz N, Bale S, Bick D, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Abou Tayoun AN, Al Turki SH, Oza AM, Bowser MJ, et al. Improving hearing loss gene testing: a systematic review of gene evidence toward more efficient next-generation sequencing-based diagnostic testing and interpretation. Genet Med. 2015 doi: 10.1038/gim.2015.141. [DOI] [PubMed] [Google Scholar]
- 91.McLaughlin HM, Ceyhan-Birsoy O, Christensen KD, Kohane IS, et al. A systematic approach to the reporting of medically relevant findings from whole genome sequencing. BMC Med Genet. 2014;15:134. doi: 10.1186/s12881-014-0134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92*.MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. Authoritative proposal of guidelines for diagnosis of genomic variants as pathogenic, benign or somewhere in between. The guidelines addressed the factors that go into considering both genes and variants within genes as diagnostic for a patients clinical phenotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Philippakis AA, Azzariti DR, Beltran S, Brookes AJ, et al. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum Mutat. 2015;36:915–921. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sobreira N, Schiettecatte F, Boehm C, Valle D, et al. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum Mutat. 2015;36:425–431. doi: 10.1002/humu.22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Botkin JR, Belmont JW, Berg JS, Berkman BE, et al. Points to Consider: Ethical, Legal, and Psychosocial Implications of Genetic Testing in Children and Adolescents. Am J Hum Genet. 2015;97:6–21. doi: 10.1016/j.ajhg.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bowdin S, Gilbert A, Bedoukian E, Carew C, et al. Recommendations for the integration of genomics into clinical practice. Genet Med. 2016 doi: 10.1038/gim.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dong Z, Zhang J, Hu P, Chen H, et al. Low-pass whole-genome sequencing in clinical cytogenetics: a validated approach. Genet Med. 2016 doi: 10.1038/gim.2015.199. [DOI] [PubMed] [Google Scholar]
- 99.Illumina. HiSeq X Ten System. 2016. [Google Scholar]
- 100.Genetics, V. Veritas Genetics Launches $999 Whole Genome. 2016. [Google Scholar]
- 101.Maurano MT, Humbert R, Rynes E, Thurman RE, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Leung D, Jung I, Rajagopal N, Schmitt A, et al. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature. 2015;518:350–354. doi: 10.1038/nature14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bodian DL, Klein E, Iyer RK, Wong WS, et al. Utility of whole-genome sequencing for detection of newborn screening disorders in a population cohort of 1,696 neonates. Genet Med. 2016;18:221–230. doi: 10.1038/gim.2015.111. [DOI] [PubMed] [Google Scholar]
- 104.Kingsmore SF. Newborn testing and screening by whole-genome sequencing. Genet Med. 2016;18:214–216. doi: 10.1038/gim.2015.172. [DOI] [PubMed] [Google Scholar]
- 105.Tang H, Feuchtbaum L, Neogi P, Ho T, et al. Damaged goods?: an empirical cohort study of blood specimens collected 12 to 23 hours after birth in newborn screening in California. Genet Med. 2016;18:259–264. doi: 10.1038/gim.2015.154. [DOI] [PubMed] [Google Scholar]
- 106.Baker MW, Atkins AE, Cordovado SK, Hendrix M, et al. Improving newborn screening for cystic fibrosis using next-generation sequencing technology: a technical feasibility study. Genet Med. 2016;18:231–238. doi: 10.1038/gim.2014.209. [DOI] [PMC free article] [PubMed] [Google Scholar]