Abstract
Background: Neural stem cells (NSCs) are pluripotent and self-renewing cells which could differentiate into diverse types of neural cells, such as dopaminergic (DA) neurons, the loss of which is the typical characteristic of Parkinson’s disease (PD). This study aimed to examine the molecular mechanisms of BMP2-mediating NSCs differentiation into DA neurons. Methods: Different concentrations of BMP2 were used to induce the differentiation of NSCs into DA neurons, which were characterized by the number and the neurite lengths of tyrosine hydroxylase (TH)+ and dopamine transporter (DAT)+ neurons by immunocytochemistry. qRT-PCR and Western blot were performed to explore the expression of miR-145 and Nurr1. The methylation level of miR-145 promoter was examined by DNA methylation analyses. The regulation of miR-145 on Nurr1 was detected by Dual-Luciferase reporter assay. Results: The number of TH+ and DAT+ neurons were significantly increased in NSCs treated with 20 and 100 ng/ml of BMP2, as well as the neurite lengths of TH+ and DAT+ neurons. The reduced level of miR-145 and up-regulated Nurr1 were observed in NSCs induced by BMP2. The hypermethylation level of miR-145 promoter down-regulated the expression of miR-145 in NSCs pretreated with BMP2, which was regulated by DNMT3b. Luciferase reporter assay showed that Nurr1 was a direct target of miR-145. miR-145 overexpression restrained the differentiating effect of BMP2. Moreover, overexpression of Nurr1 abrogated this effect of miR-145 overexpression. Conclusion: Our results showed that BMP2 promoted the differentiation of NSCs into DA neurons in vitro and miR-145 and Nurr1 were involved in the neurotrophic effects of BMP2.
Keywords: Neural stem cells, dopaminergic neurons, miR-145, BMP2, Nurr1, differentiation
Introduction
Neural stem cells (NSCs) are pluripotent and self-renewing cells which could differentiate into diverse types of neural cells following asymmetrical cell division, such as dopaminergic (DA) neurons, astrocytes, and oligodendroglia [1]. The central nervous system (CNS) and spinal cord are the source of NSCs. Among them, mesencephalic NSCs are the precursor cells of DA neurons and show strong proliferative capacity and multipotentiality [2]. Moreover, they could be induced into DA neurons under certain conditions in vivo and in vitro [2]. Parkinson’s disease (PD), as a serious degenerative disorder of CNS, is characterized by Tthe progressive degeneration and necrosis of DA neurons. Recently, DA cell replacement therapy is considered as a promising treatment for PD, especially using NSCs as new donor cells for midbrain DA neurons [3]. Therefore, the study of NSCs cultured in vitro and the mechanism of induced differentiation is in the ascendant nowadays, which will facilitate the clinical application of NSCs-based therapies for PD.
Nurr1 is a transcription factor belonging to the orphan nuclear receptor family 4 (NR4A) [4]. The expression of Nurr1 is confined predominantly to the CNS in limbic areas and the ventral midbrain, including DA neurons [5]. Mounting evidence indicate that Nurr1 in the ventral midbrain starts to be expressed at embryonic day 10.5 and just before tyrosine hydroxylase (TH) which is the dopaminergic marker enzyme [6,7]. Nurr1 is expressed in DA neurons throughout life, suggesting its essential role for normal functions of DA neurons. Nurr1 could directly bind to a NGFI-B response element on the promoter of TH, resulting in the induction of TH transcription and activating the expression of TH [8]. In addition, Nurr1 could induce the transcription of dopamine transporter (DAT) [9]. In the absence of Nurr1, neural stem cells were found to fail to differentiate into a fully DA phenotype, indicating that Nurr1 affected the survival and differentiation of midbrain DA neurons [7]. However, the regulatory mechanism of Nurr1 in the differentiation of NSCs into DA neurons has not been established.
MicroRNAs (miRNAs) are a class of small non-coding RNAs (18-24 nucleotides) that negatively regulate the expression of several genes through interacting with the 3’-UTRs of target mRNAs, which either blocks translation or promotes the degradation of the mRNA target [10]. miRNAs are involved in the regulation of several cellular processes, such as cell proliferation, cycle, apoptosis, and development [11]. In the CNS, ectopic expression of specific miRNAs is associated with some physiological processes and neurological disorders, such as PD, stroke, Alzheimer’s disease and brain injury [12]. Among the identified miRNAs so far, miR-145 can direct vascular smooth muscle cell (VSMC) differentiation from multipotent stem cells [13]. Lee et al. reported that exogenous miR-145 could be delivered to neural cells by mesenchymal stem cells and induced neuronal differentiation of the neural progenitor cells (NPCs) [14]. In addition, miR-145 has been considered as a tumor suppressor and is downregulated in various types of cancers [15].
In this study, we induced the differentiation of NSCs into DA neurons with different concentration of bone morphogenetic protein (BMP) and detected the expression of miR-145 and Nurr1. We found that knockdown of miR-145 promoted the differentiation of NSCs into DA neurons, which was abrogated by the knockdown of Nurr1. Moreover, we explored the effect of miR-145 and Nurr1 on the NSCs differentiation induced by BMP.
Materials and methods
Cell cultures
The ventral midbrain from E14 SD rat was harvested and NSC cultures were generated as described previously [16]. The ventral midbrain were minced with scalpels and digested in 0.25% of trypsase at 37°C for 5 min. Dulbecco’s modified Eagle’s medium (DMEM) containing 10% of fetal bovine serum (FBS) (Invitrogen) was used to stop the digestion. After centrifugation, the supernatant was filtered with 150-200 mesh and seeded in density of 4×105 cells/mL in culture medium containing DMEM/F12, knockout serum replacement, B27, basic fibroblast growth factor (FGF, 20 ng/mL), and epidermal growth factor (EGF, 20 ng/mL) to form neurospheres. Neurospheres were passaged into small clumps and were then cultured in maintenance medium containing DMEM/F12, B27, basic FGF (20 ng/mL) and EGF (20 ng/mL). The neurospheres were passaged every 7-14 days by changing the medium every 3-4 days. Cells were cultured at 37°C in a humidified atmosphere containing 5% CO2. BMP2 (R&D) was used to induce the differentiation of NSC at the concentrations of 0, 0.1, 1, 10, 20, 100 ng/ml for two weeks.
HT22 neuronal cells were purchased from American Type Culture Collection (ATCC) and cultured in DMEM containing 10% fetal bovine serum (FBS) and penicillin/streptomycin (all media from Invitrogen, Carlsbad, CA, USA) at 37°C in a humidified atmosphere containing 5% CO2.
Quantitative real-time PCR (qRT-PCR)
Total RNA from cultured cells was extracted by using TRIzol Reagent (Life Technologies, USA) according to the instruction. For mRNA detection, total RNA was reverse-transcribed to yield cDNA by using the Transcriptor First Strand cDNA Synthesis Kit (Roche). The expression of Nurr1 and GAPDH were measured by real-time PCR using the SYBR Green PCR Master (Roche). For miRNA detection, reverse transcription was performed using the TaqMan microRNA Reverse Transcription Kit (Applied Biosystem). qRT-PCR was performed by using TaqMan MicroRNA® Assays (Applied Biosystems). U6 was used as the endogenous control to normalize miR-145 expression. Samples were performed by using Bio-Rad Chromo 4 System Real-Time PCR detector (Bio-Rad), and the 2-ΔΔCt method was used to calculate the fold change. All reactions were performed in duplicate.
Western blot analysis
Cells were lysed on ice in RIPA buffer with Protease Inhibitor (Roche) for 10 min. Protein Assay (Bio-Rad) was performed to measure the total protein concentrations. Protein was loaded in 10% SDS-polyacrylamide gel for electrophoresis, and then transferred to a PVDF membrane (Millipore, Bedford, MA, USA). After blocked with 5% non-fat milk for 2 h at room temperature, the membrane was incubated with primary antibody Nurr1 (1:1000, Abcam) and β-atcin (1:1000, Abcam, Cambridge, UK) for 3 h at room temperature. The HRP labeled goat-anti-rabbit secondary antibody (1:5000, Abcam, Cambridge, UK) was incubated for 1 h at room temperature.
DNA extraction and methylation analyses
Genomic DNA was extracted from the cells by using Puregene reagents (Qiagen) according to the manufacturer’s instructions. Bisulfite treatment was performed on 1 µg DNA using EpiTect Fast Bisulfite Conversion Kit (Qiagen), according to the manual. Amplicons and primers within the promoter area were designed using SequenomEpiDesigner (www.epidesigner.com). An amplicon starting 353 bp, and ending 90 bp upstream of miR-145 was used for the methylation analysis. PCR amplification of the amplicon was carried out with 50 ng of bisulfite-treated DNA in a total volume of 25 µl. PCR conditions were 1 cycle for 10 min at 95°C followed by 35 cycles for 20 s at 94°C, 30 s at 60°C and 1 min at 72°C, then later for 3 min at 72°C. The PCR products were analyzed by 2% agarose gel electrophoresis and purified using the QIAquick PCR purification kit (Qiagen). PCR products were subcloned into T-vector and transformed into Escherichia coli. The cloned PCR products were sequenced by Sangon Biotech (Shanghai, China). Analysis and representation of CpG dinucleotide methylation was carried out using BISMA (http://services.ibc.uni-stuttgart.de/BDPC/BISMA).
Cell transfection
MiRNA-145 mimics or inhibitor and its negative control were purchased from RiboBio (Guangzhou, China). SiRNA that target DNMT3b or Nurr1 and the negative control were purchased from Invitrogen (USA). A DNA fragment encoding Nurr1 was amplified by PCR and cloned into pcDNA-3.1 vector (Ambion), yielding the recombinant vector pcDNA-3.1-Nurr1. NSCs were cultured the day before transfection at a confluence of 50% and were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instruction.
Dual-Luciferase reporter assay
Nurr1-3’-UTR fragment containing predicted target site of miR-145 and Nurr1-3’-UTR fragment with mutated site were synthesized and purchased from Sangon Biotech (Shanghai, China). These fragments were cloned into the pmirGLO Vector, a dual-luciferase miRNA target expression reporter vector (Promega). HT22 cells were then co-transfected with 200 ng reporter vector and 50 nM miR-145 mimic or inhibitor by using Lipofectamine 2000. Cells were harvested 48 h post transfection and luciferase activities were analyzed by the Dual-Glo Luciferase Assay System (Promega, Madison, WI). Firefly luciferase was used to normalize the Renilla luciferase.
Immunocytochemistry
Primary antibodies included mouse anti-TH (1:200, Millipore) and rat anti-DAT (1:5000, Millipore). Alexa Fluor 568-conjugated goat anti-mouse IgG (1:500, Invitrogen) was the secondary antibody. The expression of TH and DAT was observed under a fluorescent confocal microscope (Olympus, Japan). We then calculated the percentage of cells double labeled with GFP/TH or GFP/DAT in the Hoechst labeled cells (total cells). The number of GFP, TH, DAT, and Hoechst-positive cells were counted, respectively, in 10 randomly selected microscopic visual fields per well, 6 wells per group, and the neurite lengths of TH+ and DAT+ neurons were analyzed using QWin automatic image analyzer (Leica, Germany).
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed using SimpleChIP Enzymatic Chromatin IP kit (Cell Signaling Technology, Beverly, MA) as per the manufacturers’ instructions. Briefly, cells were cross-linked with 1% formaldehyde at room temperature for 10 min and then 2.5 M glycine added to stop the reaction. After a series of pretreatments, lysates were incubated with 50 μl of Protein A agarose/salmon sperm DNA preloaded with 1 μg of DNMT3a, or DNMT1, or DNMT3b or IgG antibody overnight at 4°C. Protein A agarose/salmon sperm DNA was incubated at 65°C to reverse the covalent histone-DNA bonds. The released DNA was purified and then used for analysis by RT-PCR. The fold induction over input was calculated using 2-ΔΔCt.
Statistical analysis
The data in this study were presented as the mean ± standard deviation (SD). Statistical significance was assessed by ANOVA followed by the Bonferroni’s post hoc test. Each experiment was repeated at least three times. Data were analyzed using Origin 7.0 Software. Statistical significance was defined as P<0.01.
Results
BMP2 promotes the differentiation of NSCs into DA neurons
NSCs were treated with different concentration (0, 0.1, 1, 10, 20, 100 ng/ml) of BMP2 for two weeks. The number of TH+ and DAT+ neurons in the NSCs grew with increasing BMP2 concentration. The differentiation rate in NSCs pretreated with 20 or 100 ng/ml of BMP2 reached at 41.8% and 59.1% (Figure 1A). In addition, the average neurite lengths in neurons were significantly increased in NSCs pretreated with 1, 10, 20, 100 ng/ml of BMP2 (Figure 1A). These results suggested that BMP2 promoted the differentiation of NSCs into DA neurons. To explore the possible molecular mechanism involving this process, we examined the miR-145 level in these NSCs by qRT-PCR. As demonstrated in Figure 1B, the miR-145 level was downregulated with the increasing BMP2 concentration. The mRNA and protein level of Nurr1 significantly increased in a concentration-dependent manner of BMP2. These results showed the reduced level of miR-145 and up-regulated Nurr1 in NSCs induced by higher concentration of BMP2.
Figure 1.
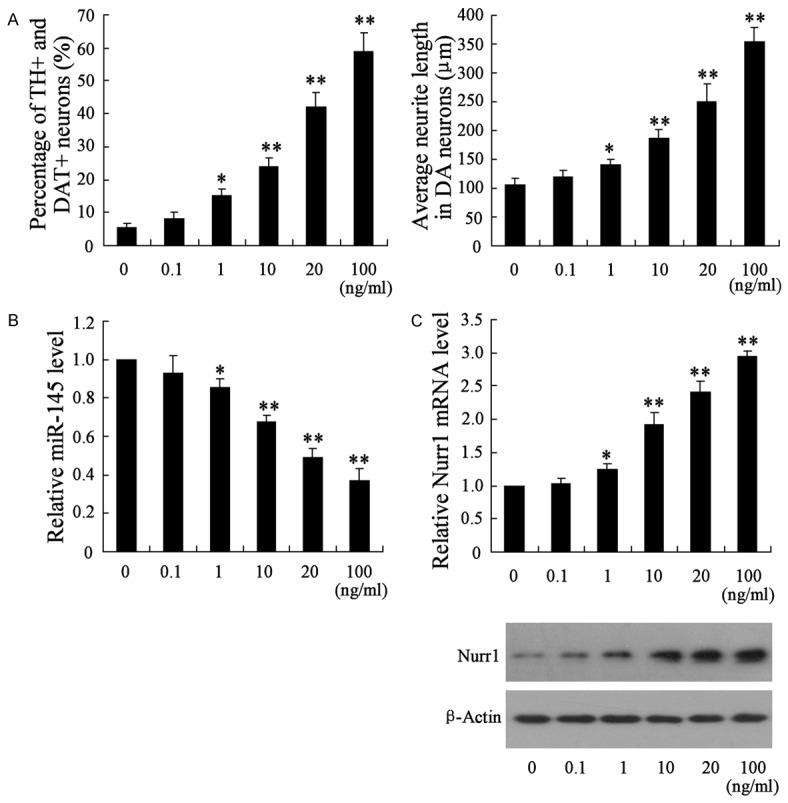
Expression of miR-145 was decreased in the differential NSCs induced by BMP2. To induce the differentiation of NSCs into DA neurons, BMP2 were used to treat NSCs with different concentration (0, 0.1, 1, 10, 20, 100 ng/ml) for two weeks. A. The differentiation rate and neurite lengths of TH+ and DAT+ neurons were detected by immunocytochemistry and flow cytometry; B. The miR-145 level in these differential NSCs; C. The mRNA and protein level of Nurr1 in these differential NSCs. *VS control NSCs, P<0.05; **VS control NSCs, P<0.01; n=6.
DNA methylation level of miR-145 in BMP2-inducing NSCs
To dissect the molecular mechanisms underlying the decreased abundance of miR-145 in NSCs induced by BMP2, we assessed the methylation status of CpG island in miR-145 promoter by direct bisulfite sequencing. As shown in Figure 2A, NSCs pretreated with 20 ng/ml of BMP2 for two weeks showed remarkably elevated methylation level of miR-145 CpG island comparing to that in NSCs with no treatment. As DNA methylation was typically regulated by methyltransferases (DNMTs), we detected the binding of miR-145 promoter with DNMT1, DNMT3a, or DNMT3b by ChIP. Consequently, there was little difference in the binding of miR-145 promoter with DNMT1 (or DNMT3a) between the NSCs pretreated with 0 and that pretreated with 20 ng/ml of BMP2 (Figure 2B). However, increasing binding of miR-145 promoter with DNMT3b was observed in NSCs pretreated with 20 ng/ml of BMP2. Altogether these findings proposed that CpG island methylation within the miR-145 promoter contributed to the down-regulation of miR-145 in NSCs pretreated with BMP2.
Figure 2.
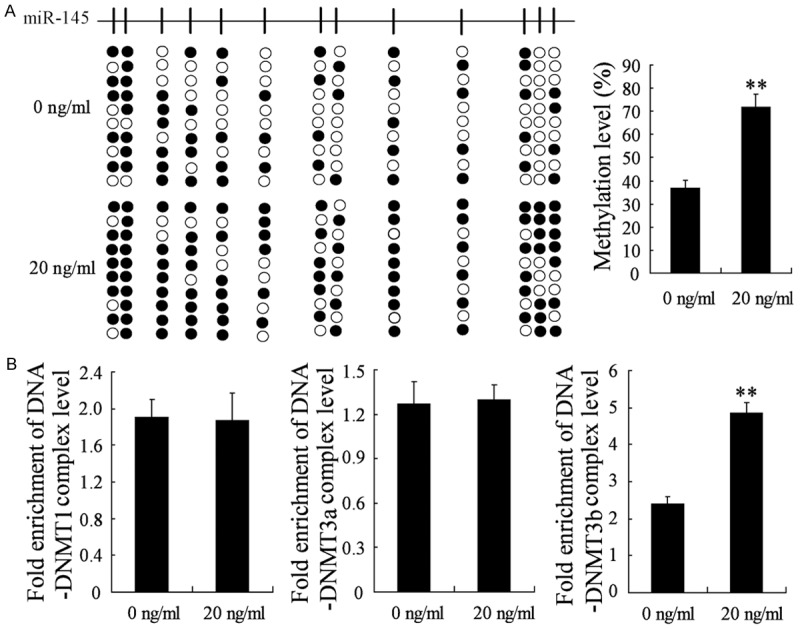
DNA methylation level of miR-145 promoter was increased in BMP2-inducing NSCs. NSCs were treated with 0 or 20 ng/ml of BMP2 for two weeks. A. Sequencing of bisulfite-treated DNA encoding miR-145 in cells and the methylation level of miR-145 promoter in the NSCs. B. The binding of miR-145 promoter with DNMT1, DNMT3a, or DNMT3b were examined by ChIP. Each line represents a single clone. Black boxes, methylated CpG sites; white boxes, unmethylated CpG sites; **VS control NSCs, P<0.01; n=6.
DNMT3b knockdown prevents BMP2-induced downregulation of miR-145 in NSCs
As DNMT3b was found to be involved in the down-regulation of miR-145 in NSCs pretreated with BMP2, we detected the impact of DNMT3b knockdown on the expression of miR-145. NSCs were divided into four groups: treated with 0 ng/ml of BMP2; treated with 20 ng/ml of BMP2; treated with 20 ng/ml of BMP2 and transfected with si-NC; treated with 20 ng/ml of BMP2 and transfected with si-DNMT3b. It has been found that knockdown of DNMT3b abrogated the down-regulation of miR-145 induced by BMP2 in NSCs (Figure 3).
Figure 3.
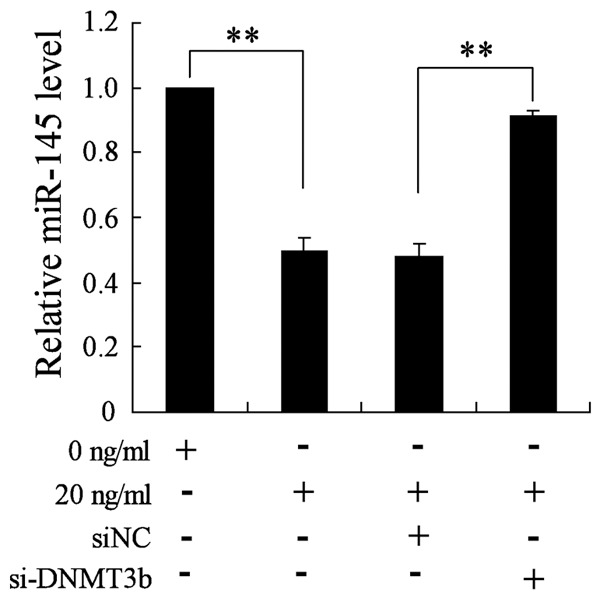
Abrogating effect of DNMT3b knockdown on the expression of miR-145 induced by BMP2. NSCs transfected with si-DNMT3b or si-NC were treated with 0 or 20 ng/ml of BMP2 for two weeks. qRT-PCR were performed to detect the level of miR-145. **VS control, P<0.01; n=6.
Knockdown of miR-145 promotes the differentiation of NSCs into DA neurons
To test the effect of miR-145 on the differentiation of NSCs into DA neurons, NSCs were transfected with miR-145 inhibitor or NC and then treated with differentiation liquid without BMP2. Immunocytochemistry assay revealed that the percentage of TH+ and DAT+ neurons has increased substantially in NSCs transfected with miR-145 inhibitor (Figure 4). Congruously, the average neurite lengths in neurons were also significantly increased in NSCs transfected with miR-145 inhibitor (Figure 4). These results suggested that knockdown of miR-145 promoted the differentiation of NSCs into DA neurons.
Figure 4.
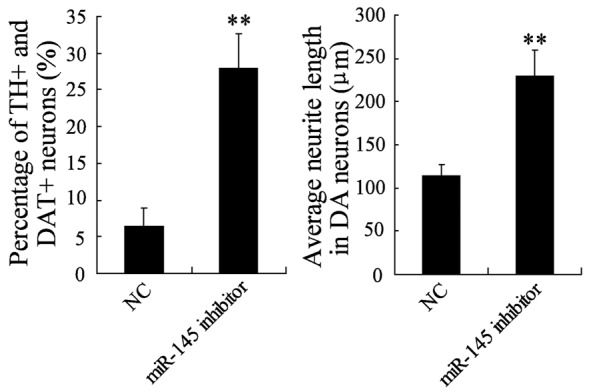
The effect of miR-145 inhibitor on the differentiation of NSCs into DA neurons. NSCs were transfected with miR-145 inhibitor or NC and then treated with differentiation liquid without BMP2. The differentiation rate and neurite lengths of TH+ and DAT+ neurons were detected by immunocytochemistry and flow cytometry. **VS control NSCs, P<0.01; n=6.
Nurr1 is a direct target of miR-145
To further dissect the contribution of miR-145 down-regulation to the differentiation of NSCs, we searched for additional, yet undiscovered, mRNA target of miR-145. Finally, we noted Nurr1, the annotated functions of which were well-known to be related to the differentiation of NSCs, among the putative mRNA targets based on the biological prediction software online. As shown in Figure 5A, the 3’-UTR of Nurr1 contained one consensus binding site of miR-145. Thus we constructed the dual-luciferase reporter vectors containing the Nurr1 3’-UTR-WT or Nurr1 3’-UTR-MU oligonucleotide pairs, which were corresponding to miR-145 targeting site or mutating site, respectively. HT22 cells were co-transfected with miR-145 mimics and the dual-luciferase reporter vectors. The dual-luciferase reporter assay showed that the luciferase activity of Nurr1 wild-type 3’-UTR was significantly reduced after miR-145 mimics transfection, whereas the luciferase activity of Nurr1 3’-UTR-MU was not affected (Figure 5B). Subsequently, the qRT-PCR experiment and Western blot showed that mRNA and protein level of Nurr1 both decreased after HT22 cells transfected with miR-145 mimics. Moreover, we also found that miR-145 inhibitor enhanced the luciferase activity of Nurr1 wild-type 3’-UTR and the expression of Nurr1 in HT22 cells (Figure 5C). Altogether, these findings identified Nurr1 as a novel target of miR-145.
Figure 5.
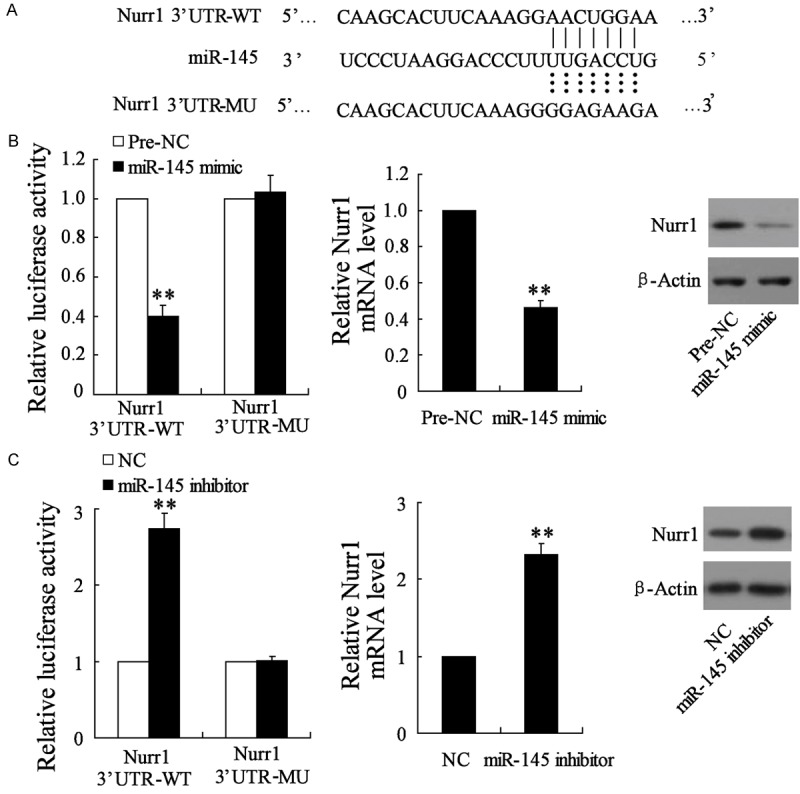
miR-145 impaired the expression of Nurr1 through targeting the 3’-UTR sequences of Nurr1. A. The putative miR-145 binding site within the human Nurr1 3’-UTR (WT) and the 3’-UTR mutated sequence (MU) were showed; B. Luciferase reporter assay were performed in HT22 cells with Nurr1 3’-UTR-WT or Nurr1 3’-UTR-MU vectors together with miR-145 mimics or Pre-NC. In addition, qRT-PCR assay and Western blot assay were performed to detect the expression of Nurr1; C. Luciferase reporter assay were performed in HT22 cells with Nurr1 3’-UTR-WT or Nurr1 3’-UTR-MU vectors together with miR-145 inhibitors or NC. In addition, qRT-PCR assay and Western blot assay were performed to detect the expression of Nurr1. **VS the control, P<0.01, n=6.
Nurr1 involves in the differentiation of NSCs induced by the knockdown of miR-145
To identify miR-145 and Nurr1 implicated in the differentiation of NSCs, NSCs were transfected with NC, or miR-145 inhibitor, or miR-145 inhibitor and si-NC, or miR-145 inhibitor and si-Nurr1. Then differentiation liquid without BMP2 was utilized to treat these NSCs. As shown in Figure 6, the percentage of TH+ and DAT+ neurons and the average neurite lengths in neurons have increased substantially in NSCs transfected with miR-145 inhibitor. However, siRNA-mediated knock down of Nurr1 markedly impaired this catalytic role of miR-145 inhibitor on differentiation of NSCs (Figure 6). These results suggest that Nurr1 involved in the differentiation of NSCs into DA neurons induced by the knockdown of miR-145.
Figure 6.
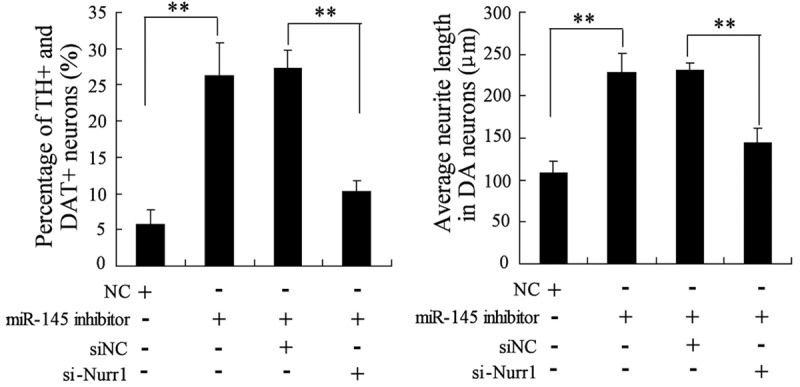
The effect of miR-145 inhibitor and si-Nurr1 on the differentiation of NSCs into DA neurons. NSCs were transfected with NC, or miR-145 inhibitor, or miR-145 inhibitor and si-NC, or miR-145 inhibitor and si-Nurr1. Then differentiation liquid without BMP2 was utilized to treat these NSCs. The differentiation rate and neurite lengths of TH+ and DAT+ neurons were detected by immunocytochemistry and flow cytometry. **VS control NSCs, P<0.01; n=6.
The impact of miR-145 overexpression and Nurr1 on the differentiation of NSCs induced by BMP2
To further elucidate the effect of miR-145 overexpression and Nurr1 overexpression on the differentiation of NSCs induced by BMP2, NSCs were treated with 0 ng/ml of BMP2, or 20 ng/ml of BMP2, or 20 ng/ml of BMP2+NC, or 20 ng/ml of BMP2+miR-145 mimics, or 20 ng/ml of BMP2+miR-145 mimics+pcDNA-3.1, 20 ng/ml of BMP2+miR-145 mimics+pcDNA-3.1-Nurr1. miR-145 overexpression significantly reduced the percentage of TH+ and DAT+ neurons and the average neurite lengths in neurons induced by BMP2, which indicated that miR-145 overexpression restrained the differentiating effect of BMP2 (Figure 7). Moreover, overexpression of Nurr1 abrogated this effect of miR-145 overexpression, as demonstrated in Figure 7.
Figure 7.
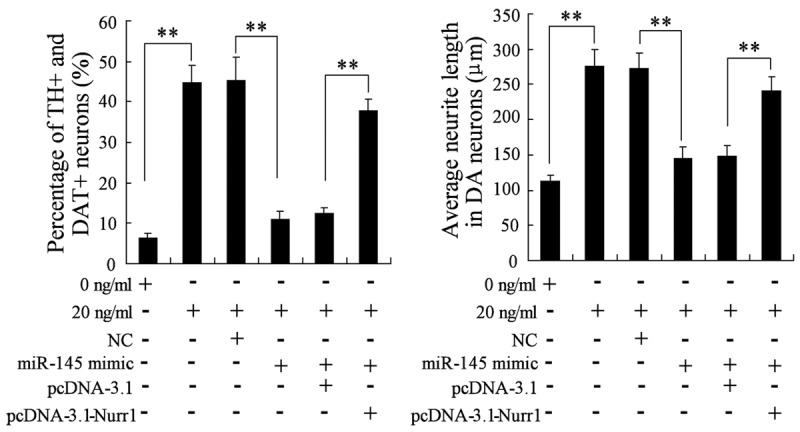
The impact of miR-145 overexpression and Nurr1 on the differentiation of NSCs induced by BMP2. NSCs were treated with 0 ng/ml of BMP2, or 20 ng/ml of BMP2, or 20 ng/ml of BMP2+NC, or 20 ng/ml of BMP2+miR-145 mimics, or 20 ng/ml of BMP2+miR-145 mimics+pcDNA-3.1, 20 ng/ml of BMP2+miR-145 mimics+pcDNA-3. 1-Nurr1. The differentiation rate and neurite lengths of TH+ and DAT+ neurons were detected by immunocytochemistry and flow cytometry. **VS control NSCs, P<0.01; n=6.
Discussion
PD has traditionally been regarded as a common neurodegenenerative disorder. As the gradual loss of nigostriatal DA neurons is the pathological hallmark of PD, it is a logical treatment for PD to replace the lost DA neurons and accompanied tissues. In previous clinical studies, fresh human fetal ventral mesencephalic (hfVM) tissue transplantation was developed and showed safety and efficacy in cell therapy trials for PD patients [17,18]. However, the elevated cell death rate of the transplanted cells and a lack of reproducibility depleted the application of this therapy [19]. Owing to the tremendous progress in understanding the stem cell biology, human NSCs (hNSCs) are considered to help to avoid these problems [20]. NSCs are self-renew cells and can differentiate into all neural lineage cells, which may replace lost DA neurons and reverse the degenerative process of PD [21]. Based on the experience with hfVM tissue engraftment and the properties of NSC, NSC transplantation is a promising and attractive treatment option for PD. A study reported by Redmond et al. showed that non-differentiated NSCs had a functional impact when implanted into PD primates [22]. Differentiated cells derived from fetal NSCs improved the motor deficits in a rat model of PD. Nowadays, the key challenges for NSCs transplantation might be the induction of the differentiation of NSCs into DA neurons and their functional survival in dopamine secretion.
Bone morphogenetic protein (BMP) is a member of the transforming growth factor (TGF)-β superfamily, which have emerged as critical regulators of NSC self-renewal and its maintenance in adulthood [23]. Among the member of these families, BMP2 was the most extensively studied member. Mounting evidence indicated that BMP2 promoted the survival and growth of midbrain dopaminergic neurons in vitro and in vivo [24,25]. In this study, we treated the NSCs with different concentration (0, 0.1, 1, 10, 20, 100 ng/ml) of BMP2 for two weeks. Immunocytochemistry showed that the differentiation rate of NSCs treated with 20 or 100 ng/ml of BMP2 reached at 41.8% and 59.1%. In addition, the average neurite lengths in neurons were significantly increased in NSCs treated with BMP2. These results suggested that BMP2 promoted the differentiation of NSCs into DA neurons in a concentration-dependent manner. It has been found that the function of BMP2 in the developing nervous system was various, which might depend upon the developmental stage of the progenitor cells. For example, BMP2 treatment has been found to induce neuronal differentiation of neural crest progenitor cells, but conversely has been reported to result in inhibition of neuronal differentiation in other populations of progenitor cells [26,27]. In this study, we found BMP2 promoted the differentiation of NSCs into DA neurons.
To date, several miRNAs have been identified that can regulate the neural cell lineage during differentiation by modulating the BMP signaling pathway, such as miR-17, miR-22 and miR-134 [28]. To further elucidate the molecular mechanisms that mediated the neurotrophic effects of BMP2 on NSCs differentiation, the miR-145 level was detected in the NSCs pre-treated with BMP2. miR-145 was downregulated with the increasing BMP2 concentration. In addition, hyper-methylation level of miR-145 promoter was observed and regulated by DNMT3b. Caruso et al. suggested that miR-145 was the downstream of BMP signaling in mice and pulmonary arterial hypertension (PAH) patients [29]. BMP signal was also found to activate the transcription of the miRNA-143/145 gene cluster through a consensus sequence [28]. In general, miR-145 acted as a tumor suppressor in several cancers [30-32]. miR-145 also regulated the VSMC differentiation via repressing Kruppel-like factor 4 (KLF4) [33]. In this study, we found that overexpression of miR-145 inhibited the differentiation of NSCs into DA neurons induced by BMPs.
Through the prediction of biological software, we found that the 3’-UTR of Nurr1 contained one consensus binding site of miR-145. Dual-luciferase reporter assay further showed that Nurr1 was a novel target of miR-145. Nurr1 has been found to play important roles in midbrain DA neuron development [7]. In familial PD, mutations in the human Nurr1 gene have been identified, suggesting its direct function in neurodegenerative disease [34]. Kim et al. indicated that Nurr1 repressed the expression of TH via SIRT1 in hNSCs [35]. Nurr1 could also induce midbrain DA neuron from expanded NSCs in synergy with Ngn2 [36]. In this study, we found that overexpression of Nurr1 abrogated the effect of miR-145 overexpression on NSCs differentiation induced by BMPs.
In conclusion, our study demonstrated the differentiation of NSCs into DA neurons induced by BMPs. In addition, miR-145 and Nurr1 were associated with the neurotrophic effects of BMP2 on NSCs differentiation. Furthermore, we found that BMP2 promoted the differentiation of NSCs into DA neurons in vitro via miR-145 upregulating the expression of Nurr1. These findings provide potential novel therapeutic targets for the cell replacement therapy of PD.
Acknowledgements
This work was supported by a grant from Projects of medical and health technology development program in Zhejiang province (NO. 2014kya198).
Disclosure of conflict of interest
None.
References
- 1.Temple S. The development of neural stem cells. Nature. 2001;414:112–117. doi: 10.1038/35102174. [DOI] [PubMed] [Google Scholar]
- 2.Morrison SJ. Neuronal potential and lineage determination by neural stem cells. Curr Opin Cell Biol. 2001;13:666–672. doi: 10.1016/s0955-0674(00)00269-6. [DOI] [PubMed] [Google Scholar]
- 3.Chou CH, Fan HC, Hueng DY. Potential of Neural Stem Cell-Based Therapy for Parkinson’s Disease. Parkinsons Dis. 2015;2015:571475. doi: 10.1155/2015/571475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodríguez-Traver E, Solís O, Díaz-Guerra E, Ortiz Ó, Vergaño-Vera E, Méndez-Gómez HR, García-Sanz P, Moratalla R, Vicario-Abejón C. Role of Nurr1 in the Generation and Differentiation of Dopaminergic Neurons from Stem Cells. Neurotox Res. 2016;30:14–31. doi: 10.1007/s12640-015-9586-0. [DOI] [PubMed] [Google Scholar]
- 5.Sakurada K, Ohshima-Sakurada M, Palmer TD, Gage FH. Nurrl, an orphan nuclear receptor, is a transcriptional activator of endogenous tyrosine hydroxylase in neural progenitor cells derived from the adult brain. Development. 1999;126:4017–4026. doi: 10.1242/dev.126.18.4017. [DOI] [PubMed] [Google Scholar]
- 6.Bäckman C, Perlmann T, Wallén Å, Hoffer BJ, Morales M. A selective group of dopaminergic neurons express Nurr1 in the adult mouse brain. Brain Res. 1999;851:125–132. doi: 10.1016/s0006-8993(99)02149-6. [DOI] [PubMed] [Google Scholar]
- 7.Saucedo-Cardenas O, Quintana-Hau JD, Le WD, Smidt MP, Cox JJ, De Mayo F, Burbach JP, Conneely OM. Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc Natl Acad Sci U S A. 1998;95:4013–4018. doi: 10.1073/pnas.95.7.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim KS, Kim CH, Hwang DY, Seo H, Chung S, Hong SJ, Lim JK, Anderson T, Isacson O. Orphan nuclear receptor Nurr1 directly transactivates the promoter activity of the tyrosine hydroxylase gene in a cell-specific manner. J Neurochem. 2003;85:622–634. doi: 10.1046/j.1471-4159.2003.01671.x. [DOI] [PubMed] [Google Scholar]
- 9.Sacchetti P, Mitchell TR, Granneman JG, Bannon MJ. Nurr1 enhances transcription of the human dopamine transporter gene through a novel mechanism. J Neurochem. 2001;76:1565–1572. doi: 10.1046/j.1471-4159.2001.00181.x. [DOI] [PubMed] [Google Scholar]
- 10.Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Kamps JA, Krenning G. Micromanaging cardiac regeneration: Targeted delivery of microRNAs for cardiac repair and regeneration. World J Cardiol. 2016;8:163–179. doi: 10.4330/wjc.v8.i2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee HK, Finniss S, Cazacu S, Xiang C, Brodie C. Mesenchymal stem cells deliver exogenous miRNAs to neural cells and induce their differentiation and glutamate transporter expression. Stem Cells Dev. 2014;23:2851–2861. doi: 10.1089/scd.2014.0146. [DOI] [PubMed] [Google Scholar]
- 15.Almeida MI, Calin GA. The miR-143/miR-145 cluster and the tumor microenvironment: unexpected roles. Genome Med. 2016;8:29. doi: 10.1186/s13073-016-0284-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azari H, Sharififar S, Rahman M, Ansari S, Reynolds BA. Establishing embryonic mouse neural stem cell culture using the neurosphere assay. J Vis Exp. 2011 doi: 10.3791/2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Politis M, Wu K, Loane C, Quinn NP, Brooks DJ, Rehncrona S, Bjorklund A, Lindvall O, Piccini P. Serotonergic neurons mediate dyskinesia side effects in Parkinson’s patients with neural transplants. Sci Transl Med. 2010;2:38ra46. doi: 10.1126/scitranslmed.3000976. [DOI] [PubMed] [Google Scholar]
- 18.Politis M, Oertel WH, Wu K, Quinn NP, Pogarell O, Brooks DJ, Bjorklund A, Lindvall O, Piccini P. Graft-induced dyskinesias in Parkinson’s disease: High striatal serotonin/dopamine transporter ratio. Mov Disord. 2011;26:1997–2003. doi: 10.1002/mds.23743. [DOI] [PubMed] [Google Scholar]
- 19.Ramos-Gómez M, Martínez-Serrano A. Tracking of iron-labeled human neural stem cells by magnetic resonance imaging in cell replacement therapy for Parkinson’s disease. Neural Regen Res. 2016;11:49. doi: 10.4103/1673-5374.169628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindvall O, Kokaia Z, Martinez-Serrano A. Stem cell therapy for human neurodegenerative disorders-how to make it work. Nat Med. 2004;(Suppl):S42–50. doi: 10.1038/nm1064. [DOI] [PubMed] [Google Scholar]
- 21.Suksuphew S, Noisa P. Neural stem cells could serve as a therapeutic material for age-related neurodegenerative diseases. World J Stem Cells. 2015;7:502. doi: 10.4252/wjsc.v7.i2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Redmond DE, Bjugstad KB, Teng YD, Ourednik V, Ourednik J, Wakeman DR, Parsons XH, Gonzalez R, Blanchard BC, Kim SU. Behavioral improvement in a primate Parkinson’s model is associated with multiple homeostatic effects of human neural stem cells. Proc Natl Acad Sci U S A. 2007;104:12175–12180. doi: 10.1073/pnas.0704091104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Espejo M, Cutillas B, Ventura F, Ambrosio S. Exposure of foetal mesencephalic cells to bone morphogenetic protein-2 enhances the survival of dopaminergic neurones in rat striatal grafts. Neurosci Lett. 1999;275:13–16. doi: 10.1016/s0304-3940(99)00708-9. [DOI] [PubMed] [Google Scholar]
- 25.Jordan J, Böttner M, Schluesener HJ, Unsicker K, Krieglstein K. Bone morphogenetic proteins: neurotrophic roles for midbrain dopaminergic neurons and implications of astroglial cells. Eur J Neurosci. 1997;9:1699–1710. doi: 10.1111/j.1460-9568.1997.tb01527.x. [DOI] [PubMed] [Google Scholar]
- 26.Li W, Cogswell CA, LoTurco JJ. Neuronal differentiation of precursors in the neocortical ventricular zone is triggered by BMP. J Neurosci. 1998;18:8853–8862. doi: 10.1523/JNEUROSCI.18-21-08853.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shou J, Rim PC, Calof AL. BMPs inhibit neurogenesis by a mechanism involving degradation of a transcription factor. Nat Neurosci. 1999;2:339–345. doi: 10.1038/7251. [DOI] [PubMed] [Google Scholar]
- 28.Hata A, Kang H. Functions of the bone morphogenetic protein signaling pathway through microRNAs (Review) Int J Mol Med. 2015;35:563–568. doi: 10.3892/ijmm.2015.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caruso P, Dempsie Y, Stevens HC, McDonald RA, Long L, Lu R, White K, Mair KM, McClure JD, Southwood M. A role for miR-145 in pulmonary arterial hypertension evidence from mouse models and patient samples. Circ Res. 2012;111:290–300. doi: 10.1161/CIRCRESAHA.112.267591. [DOI] [PubMed] [Google Scholar]
- 30.Xue G, Ren Z, Chen Y, Zhu J, Du Y, Pan D, Li X, Hu B. A feedback regulation between miR-145 and DNA methyltransferase 3b in prostate cancer cell and their responses to irradiation. Cancer Lett. 2015;361:121–127. doi: 10.1016/j.canlet.2015.02.046. [DOI] [PubMed] [Google Scholar]
- 31.Pekow J, Meckel K, Dougherty U, Butun F, Mustafi R, Lim J, Crofton C, Chen X, Joseph L, Bissonnette M. Tumor suppressors miR-143 and miR-145 and predicted target proteins API5, ERK5, K-RAS, and IRS-1 are differentially expressed in proximal and distal colon. Am J Physiol Gastrointest Liver Physiol. 2015;308:G179–G187. doi: 10.1152/ajpgi.00208.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee HK, Bier A, Cazacu S, Finniss S, Xiang C, Twito H, Poisson LM, Mikkelsen T, Slavin S, Jacoby E. MicroRNA-145 is downregulated in glial tumors and regulates glioma cell migration by targeting connective tissue growth factor. PLoS One. 2013;8:e54652. doi: 10.1371/journal.pone.0054652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis-Dusenbery BN, Chan MC, Reno KE, Weisman AS, Layne MD, Lagna G, Hata A. Down-regulation of Krüppel-like Factor-4 (KLF4) by microRNA-143/145 is critical for modulation of vascular smooth muscle cell phenotype by transforming growth factor-β and bone morphogenetic protein 4. J Biol Chem. 2011;286:28097–28110. doi: 10.1074/jbc.M111.236950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jankovic J, Chen S, Le W. The role of Nurr1 in the development of dopaminergic neurons and Parkinson’s disease. Prog Neurobiol. 2005;77:128–138. doi: 10.1016/j.pneurobio.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 35.Kim TE, Seo JS, Yang JW, Kim MW, Kausar R, Joe E, Kim BY, Lee MA. Nurr1 represses tyrosine hydroxylase expression via SIRT1 in human neural stem cells. PLoS One. 2013;8:e71469. doi: 10.1371/journal.pone.0071469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersson EK, Irvin DK, Ahlsiö J, Parmar M. Ngn2 and Nurr1 act in synergy to induce midbrain dopaminergic neurons from expanded neural stem and progenitor cells. Exp Cell Res. 2007;313:1172–1180. doi: 10.1016/j.yexcr.2006.12.014. [DOI] [PubMed] [Google Scholar]


