Abstract
Exosomes containing microRNA-155 act as molecule carriers during immune cell-cell communication and play an important role in the inflammatory response of H. pylori infection macrophages. Previous reports have found that miR-155 was over-expressed in H. pylori infection macrophages, but the significance of which is still unknown. In this study, we analyzed the impact of miR-155 loaded in exosomes derived from macrophages to the inflammatory response of H. pylori infection macrophages and possible mechanisms. We found that miR-155 promoted the expression of inflammatory cytokines including TNF-a, IL-6, IL-23, but also increased the expression of CD40, CD63, CD81, and MCH-I. Meanwhile, inflammatory signal pathways proteins, such as MyD88, NF-κB in H. pylori infection macrophages were down-regulated due to the over-expression of miR-155. Experiments in vitro or in vivo revealed that miR-155 promoted macrophages to inhibit or kill H. pylori by regulating the inflammatory response of cells to prevent the gastritis caused by H. pylori infection. These findings contribute to the understanding of miR-155 contained in exosomes in inflammatory responses of H. pylori infection macrophages.
Keywords: MicroRNA-155, helicobacter pylori, macrophage, inflammatory response
Introduction
Helicobacter pylori (H. pylori) infection causes a chronic gastric mucosal inflammation, leading to pepticulcer disease in 5-10% of the infection people [1]. In acute or chronic H. pylori infection, inflammation is thought to be a major determinant of both peptic ulceration and gastric malignancy [2], so immune cells including macrophages, dendritic cells (DC) and mucosa infiltrating lymphocytes take part in the innate and adaptative immune responses to the bacteria, but the regulatory mechanisms of H. pylori-induced inflammation are still not well understood. Host defense against H. pylori requires the induction of appropriate innate immune responses, various immune regulators, for instance microRNAs (miRNA), also take part in the immune responses. MicroRNAs attracted the considerable attention because of their implication in maintaining homeostasis in fundamental biological processes in non-pathological states, and their deregulation in pathological states [3]. Changes in miRNA expression in response to H. pylori infection have been reported in immune cells such as macrophages, dendritic cells [4,5].
Exosomes are lipid bilayer vesicles of 30 to 100 nm in size, which are released from various cell types including dendritic cells, mast cells, platelets, macrophages and so on. The composition of exosomes can vary depending on the cell type of origin but some common protein components have been defined [6]. Exosomes have been implicated in cellular immune responses, however, it also has become evident that exosomes contain substantial amounts of RNA such as mRNA, microRNA and tRNA and were involved in immune-independent regulatory mechanisms recently. Pioneering studies have established that exosomes are enriched in mRNA and could perform intercellular transfer of miRNAs, participating in miRNA-based signaling mechanisms [7]. Recent studies also indicated that exosomes have a selective subset of miRNA which can be functionally transferred as a consequence of fusion with recipient cells [8]. Together, all data suggest that the function of exosomes as carriers of genetic information and that this genetic material plays an important role in cells to cells communication.
Recently, miR-155 has been indicated to play a key role in the regulation of normal immunity or inflammation response [9,10]. miRNA-155 exhibits crucial roles during innate or adaptive immune responses [11,13]. Significantly, miR-155 is known as a prototype multifunctional miRNA and is recognized for its inducible expression in activated T cells, macrophages, and dendritic cells [14]. Recent reports have showed novel roles for miR-155 in Th1 or Th17 differentiation during microbial infections as well as augmented expression of miR-155 in T cells during pathogenic microbial challenge [15,16].
In our previous study, we reported the increased expression of miR-155 in macrophages cells infected with H. pylori, and over-expression of miR-155 negatively regulated the release of proinflammatory cytokines [17]. Here, we intend to investigate the functions of miR-155 contained in exosomes in the inflammatory response of H. pylori infected macrophages, and its mechanisms.
Materials and methods
Cultures of macrophages and H. pylori
Mouse RAW264.7 cell line was purchased from the Center for Type Culture Collection of Shanghai Academy of Sciences. Cells were cultured in wells or flasks at 37°C under 5% CO2, in RPMI 1640-GlutaMAX™ (HyClone Laboratories , USA) containing 10% (v/v) fetal bovine serum (HyClone Laboratories, USA), 100 U/ml penicillin, 0.1 mg/ml streptomycin and 0.25 μg/ml amphotericin B.
The wild-type H. pylori strain 26695 obtained from the Center for Type Culture Collection of Shanghai Academy of Sciences, was cultivated for 48 h at 37°C under microaerobic conditions (5% O2) on selective agar consisting of 21.5 g of Wilkins Chalgren agar, 50 ml of human blood, 10 μg/ml of vancomycin, 10 μg/ml of cefsulodin, 5 μg/ml of trimethoprim, and 10 μg/ml of amphotericin B. And resuspended in RPMI-1640 growth medium at an optical density of 0.6 at 600 nm, which corresponds to 3×107 CFU/ml. The bacteria were added to the macrophages at the indicated infection (MOI) 10 and the co-cultures were further incubated at 37°C in a 5% CO2 atmosphere for 24 h.
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA including miRNAs and mRNAs were extracted using Trizol reagent (Invitrogen, USA). cDNAs were synthesized from enriched total RNA that annealed with random primers and tem-loop reverse transcriptional (RT) primers and then reverse transcribed with M-MLV reverse transcriptase (TaKaRa, China). The synthesized cDNAs were stored at -70°C.
Real-time quantitative PCR was performed using the Roche lightcycler (LightCycler 2.0) System with primers designed by Primer Express 3.0 and synthesized in Invitrogen. miRNA levels were normalized by U6 snRNA, while mRNA levels were normalized by GAPDH. All reactions were performed in triplicate. The specificity of PCR was determined by sequencing of the PCR products. All primers were following miR-155: 5’-ACGCTCAGTTAATGCTAATCGTGATA-3’, U6: 5’-TTCCTCCGCAAGGATGACACGC-3’, which were synthetized by Invitrogen company (New York, US).
Exosomes purification
Macrophages culture supernatants were centrifuged at 3000 ×g for 15 min to remove cells and cell debris, and transferred to sterile tubes. The appropriate ExoQuick Exosome Precipitation Solution (System Biosciences, San Francisco, USA) was added, and tubes were mixed by inverting and then refrigerated for 30 min. The mixture was then sedimented at 1,500 ×g for 30 min, and the supernatant was removed by aspiration. Residual ExoQuick solution was removed by re-centrifugation at 1,500 ×g for 5 min followed by aspiration. Finally, exosomes pellets were resuspended in 1/10 original volume of nuclease-free water, 25 μl 9% sucrose containing protease inhibitors (GenScript Biotechnology, Nanjing, China) was added, and the preparation was stored at -4°C.
MicroRNA-155 loading in exosomes
To transfer miR-155 into exosomes successfully, we optimized loading conditions for exosomes derived from macrophages. Re-suspended exosomes were diluted in Gene Pulser® Electroporation buffer (Bio-Rad Laboratories, Berkeley, CA, USA) in 1:1 ratio. miRNA-155 at final amount of 200 pmol was added to 1 μg/μl of exosomes sample. The mixtures were transferred into cold 0.2 cm electroporation cuvettes and electroporated at various voltages (0.150-0.200 kV) at 120 μF. Finally, the results of different capacitance were assessed. The exosomes were treated with one unit of RNase H to eliminate free floating miRNA-155 outside the exosomes and re-isolated using Exoquick-TC™. The relative amount of encapsulated miRNA-155 was determined using RT-PCR Assays as described previously.
Analysis of inflammatory cytokines
Macrophages in six-well plates were treated with medium, exosomes (50 μg/ml), miR-155 and exosomes+miR-155 (50 μg/ml) for 24 h. The concentrations of TNF-a, IL-6, IL-23 in cell supernatants of stimulated macrophages cells were analyzed by enzyme-linked immunoabsorbent assay (ELISA), according to the manufacturer’s instructions (Boster Biotechnology Company, WuHan, China). Cytokines concentrations were calculated by standard curves.
Flow cytometry analysis
Macrophages treated with medium, exosomes, miR-155 and exosomes+miR-155 were cultured in six well plates for 24 h, then were harvested and washed twice with PBS containing 0.2% BSA. Then macrophages were stained with fluorescein isothiocyanate (FITC)- or phycoerythrin (PE)-labeled monoclonal antibodies (BD, Franklin Lakes, NJ, USA) to CD40, CD63, CD81, and MCH-I respectively, or the appropriate isotype controls. Macrophages were washed and fixed in 10% (v/v) formaldehyde-PBS, and cell sorting and analysis were performed on a flow cytometry (Beckman MOFLO XDP, USA). Median fluorescence intensities (MFI) and the percentages of positively expressing cells were determined after subtraction of the values for the isotype controls.
Western blotting analysis
Macrophages stimulated with medium, exosomes, miR-155 and exosomes+miR-155 were analyzed by western blotting. For western blotting, equal concentration of proteins from cell lysates, as quantitated by the Micro BCA Protein Assay (Merck Millipore, USA) were loaded on 10% SDS-PAGE gels, electrophoresed, and transferred onto polyvinylidene difluoride membrane (Milipore, Bedford, MA). The membranes were probed for MyD88 (R&D Systems, Minneapolis, MN, USA, 1:600 dilution), NF-κB (R&D Systems, Minneapolis, MN, USA, 1:500 dilution) and IRAK-1 (R&D Systems, Minneapolis, MN, USA, 1:800 dilution). The membranes were incubated at room temperature for 1 h with appropriate HRP-conjugated secondary antibodies and visualized with Plus-ECL (PerkinElmer, Shelton, CA) according to the manufacturer’s protocol. Immune-detected protein bands were quantified with Image J and statistically analyzed by ANOVA software.
The proliferation of H. pylori influenced by miR-155 loaded in exosomes
Macrophages were stimulated with medium, exosomes, miR-155 and miR-155 loaded in exosomes for 24 h, then were infection with H. pylori (1×107 CFU) for 24 h. H. pylori phagocytosed by macrophages were extracted to cultured on H. pylori culture medium for eight days. Meanwhile, H. pylori (1×105 CFU) were inoculated in H. pylori culture medium containing cell supernatant of macrophages treated with medium, exosomes, miR-155 and miR-155 loaded in exosomes, respectively for eight days. The titration of H. pylori was determined every two days to analyze their proliferative ability by bacteria plate count.
HE staining of mice gastric tissues
All mice were housed at the institutional animal facility under specific-pathogen-free conditions during the experiment. H. pylori infection was carried out in the biosafety level 2 laboratory. All animal procedures were approved by the Institutional Animal Care and Use Committee of JiangSu University. Briefly, mice (5 male/group) were administered with medium, exosomes, miR-155 and exosomes+miR-155, at a dose of 30 µg/mouse by tail vein injection. The mice were immunized three times every 2 weeks for 2 weeks between vaccinations. Then all experimental groups were infected with H. pylori 1×106 CFU/mouse two times every weeks for 2 weeks by intragastric administration.
Small sections of gastric tissues of normal mice and H. pylori infection mice administered with medium, exosomes, miR-155 or exosomes+miR-155, were fixed in 10% neutral buffered formalin (Fisher Scientific, Fair Lawn, NJ) at room temperature overnight, and then embedded in paraffin (Leica, Richmond, IL). Gastric tissue slices were analyzed by HE staining kit (Beyotime, JiangSu, China) according to the manufacturer’s instructions.
Statistical analysis
All data were expressed as mean ± SEM. Comparisons were performed with One-way ANOVA with SNK post hoc. P<0.05 was considered statistically significant. All analyses were performed using SPSS16.0 (SPSS).
Results
The profile of miRNAs in exosomes from H. pylori infection macrophages
In order to investigate the potential function of miRNAs in exosomes originated from macrophages infected by H. pylori, the expression profile of seven miRNAs known to be related to innate immunity in macrophage cells following a H. pylori infection was determined by qRT-PCR (Table 1). The expressions of three miRNAs, miR-155, miR-146a, let-7a increased obviously in exosomes from macrophages after H. pylori infection (*P<0.05). Results show that miRNAs especially miR-155 might play an important regulatory function in exosomes from macrophages against H. pylori infection.
Table 1.
MicroRNAs differentially regulated in exosomes from H. pylori 26695-activated macrophages cells
| Up-regulated miRNA | Fold change upon infection | Relative level in infected cells |
|---|---|---|
| hsa-miR-21 | 18.6547 | 0.0587 |
| hsa-miR-155 | 22.3215 | 13.2548 |
| hsa-miR-146a | 8.7451 | 15.6213 |
| hsa-miR-126 | 4.6251 | 0.8513 |
| hsa-miR-106 | 3.1582 | 0.4568 |
| hsa-miR-151 | 3.0215 | 0.3014 |
| has-let-7a | 2.1253 | 4.6534 |
The expression of miR-155 in exosomes and macrophages
MicroR-155 in exosomes derived from H. pylori infection macrophages or in H. pylori infection macrophages was determined by RT-PCR (Figure 1), findings show that miR-155 was increased remarkably in exosomes derived from H. pylori infection macrophages or in macrophages infected with H. pylori compared with H. pylori un-infection groups (*P<0.05).
Figure 1.
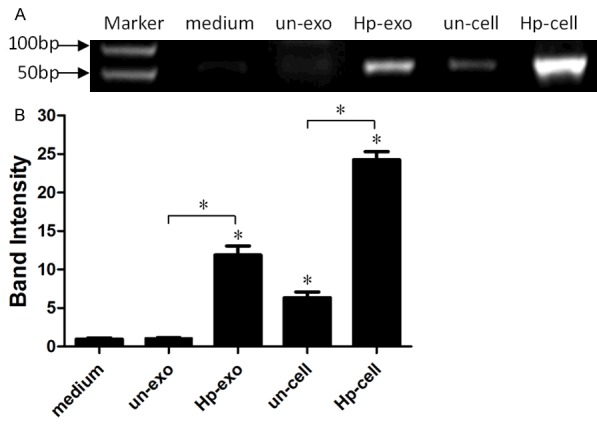
Analysis of the expression of miR-155 in exosomes or macrophages cells. A: The expression of miR-223 in exosomes (un-exo, Hp-exo) secreted from H. pylori un-infection or infection macrophages and in H. pylori un-infection or infection macrophages by a RT-PCR assay. B: The fold changes of miR-155 expression relative to exosomes from H. pylori un-infection macrophages was used for analysis. A significantly increased expression of miR-155 was detected in Hp-exo and Hp macrophages. (*P<0.05), compared with un-exo group and control (medium group, *P<0.05). Results represented the mean ± SE from three independent triplicated experiments.
The expression of miR-155 loaded in exosomes
Exosomes derived from H. pylori non-infection macrophages were loaded with miR-155 using the optimal conditions (like voltage and concen-187 trations), as determined in the previous section, then miR-155 in exosomes was analyzed by RT-PCR to assay the transfection efficiency (Figure 2), Experimental results indicated that miR-155 was transported into exosomes from H. pylori non-infection macrophages successfully.
Figure 2.
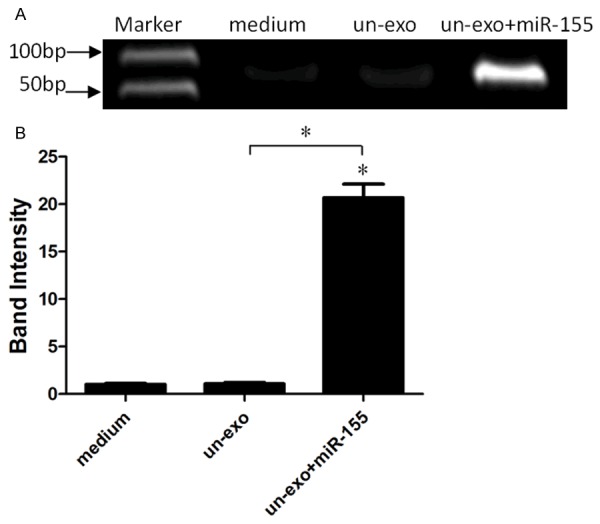
The expression of miR-155 in exosomes using the optimal conditions. A: H. pylori un-infection exosomes were transfected with miR-155 using the optimal conditions, then the expression of miR-155 was analyzed by RT-PCR. B: The fold changes of miR-155 expression relative to the miR-155 un-transfected exosomes was used for analysis. A significantly increased expression of miR-155 was detected in exosomes transfected with miR-155 (*P<0.05), compared with H. pylori un-infection exosomes group and control (medium group, *P<0.05). Results represented the mean ± SE from three independent triplicated experiments.
The expressions of TNF-a, IL-6, IL-23 regulated by miR-155
Exosomes contained antigens of H. pylori and miR-155 both could regulate the inflammatory response in H. pylori infection macrophages. In order to understand the functions of miR-155 loaded in exosomes, the expression of inflammatory cytokines TNF-a, IL-6, IL-23 were detected by ELISA method (Figure 3). As expected, the expression of TNF-a, IL-6, IL-23 were increased significantly in macrophages compared with macrophages transfected with miR-155 and control groups (*P<0.05), suggestting that exosomes could a good information carrier to transport miR-155 between macrophages to regulate pro-inflammatory mediators.
Figure 3.
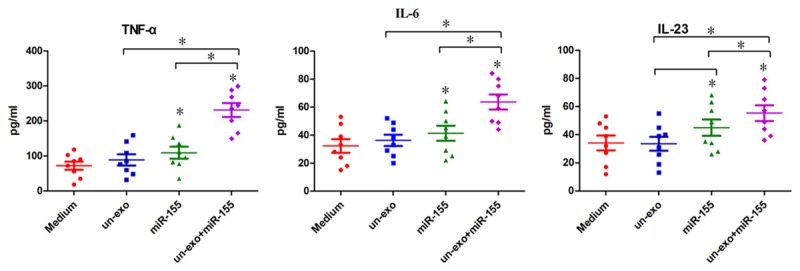
The influence of exosomes+miR-155 on cytokine expressions of TNF-a, IL-6, IL-23. Macrophages were stimulated with exosomes, miR-155, exosomes+miR-155 and medium respectively. The supernatants were harvested at 24 h post stimulation, and were determined by ELISA. Each symbol per condition represents the data obtained with cells from one well. Horizontal lines show the median values of 8 experiments. MicroR-155 loaded in exosomes enhanced the expression of TNF-a, IL-6, IL-23 in macrophages significantly compared with control groups, (mean ± SE, 6 independent experiments), and statistical analysis was by SPSS 16.0, with asterisks indicating the pairs of values compared for which significant differences were observed (*P≤0.05).
Expression of CD40, CD63, CD81, and MCH-I in macrophages (flow cytometry)
To study the common function of miR-155 and exosomes on molecular signaling proteins in cell surface, CD40, CD63, CD81, and MCH-I levels in H. pylori infection macrophages were quantified by flow cytometry (Figure 4). Data show that the expression of CD40, CD63, CD81, and MCH-1 levels were enhanced in macrophages stimulated with exosomes+miR-155 in contrast with control groups (*P<0.05).
Figure 4.
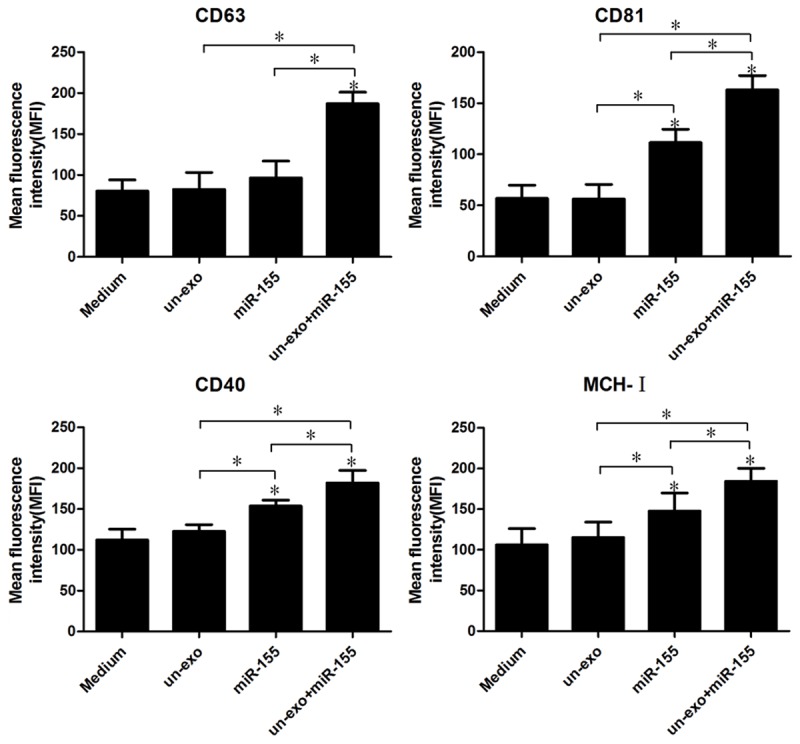
The expressions of CD40, CD63, CD81, and MCH-I were analyzed by flow cytometry in macrophages. Macrophages were treated with medium, exosomes, miR-155 and exosomes+miR-155, then 106 macrophage cells were analyzed on FACS and obtained fluorescence intensities of CD40, CD63, CD81, and MCH-I. Bar graphs show the mean fluorescence intensities (MFI) of CD40, CD63, CD81, and MCH-I (mean ± SE, 3 independent experiments), and statistical analysis was by SPSS 16.0, with asterisks indicating the pairs of values compared for which significant differences were observed (*P≤0.05).
Inflammation-related proteins influenced in macrophages by MicroiRNA-155 loaded in exosomes
To explore whether miR-155 loaded in exosomes is capable of regulating inflammation-related proteins such as MyD88, NF-κB and IRAK-1 in H. pylori infection macrophages. Total proteins of all experimental groups were harvested at 24 h and analyzed with western blotting (Figure 5). Interestingly, miR-155 loaded in exosomes showed an stronger ability to down-regulate the expression of MyD88, NF-κB proteins in H. pylori infection macrophages compared with control groups (*P<0.05); but IRAK-1 proteins were unchanged. Those displayed that miR-155 regulate the immune responses in H. pylori infection macrophages by MyD88, NF-κB Inflammatory response pathways.
Figure 5.
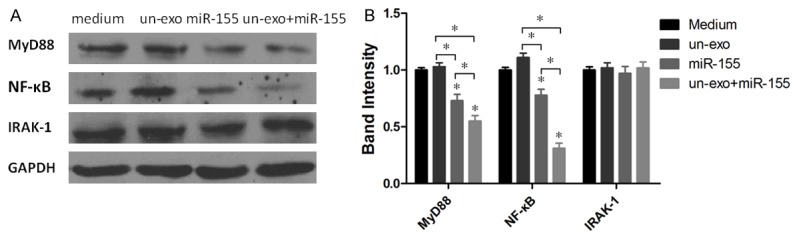
The expression of MyD88, NF-κB and IRAK-1 proteins was analyzed by western blotting. A: MyD88, NF-κB and IRAK-1 proteins in macrophages stimulated with medium, un-exosomes, miR-155 and un-exosomes+miR-155 respectively were detected by western blotting; B: The fold changes of MyD88, NF-κB and IRAK-1 proteins expression relative to the H. pylori un-infection exosomes group and control (medium group) were used for analysis. MyD88 and NF-κB proteins were significantly decreased in macrophages stimulated with miR-155 and un-exosomes+miR-155 (*P<0.05), compared with H. pylori un-infection exosomes group and control (medium group, *P<0.05). Results represented the mean ± SE from three independent triplicated experiments.
The proliferation analysis of H. pylori
The proliferation of H. pylori obtained from the phagocytosis in macrophages treated with miR-155 loaded in exosomes was assessed, and the proliferation of H. pylori incubated with the cell supernatant of macrophages treated with miR-155 loaded in exosomes was evaluated. Every two days, the CFUs of H. pylori were recorded by OD value measurement (Figure 6). Out of surprise, the proliferation of H. pylori acquired from macrophages stimulated with miR-155 loaded in exosomes was retarded observably compared with control groups, meaningly the cell supernatant of macrophages treated with miR-155 loaded in exosomes has certain inhibitory effect on the proliferation of H. pylori.
Figure 6.
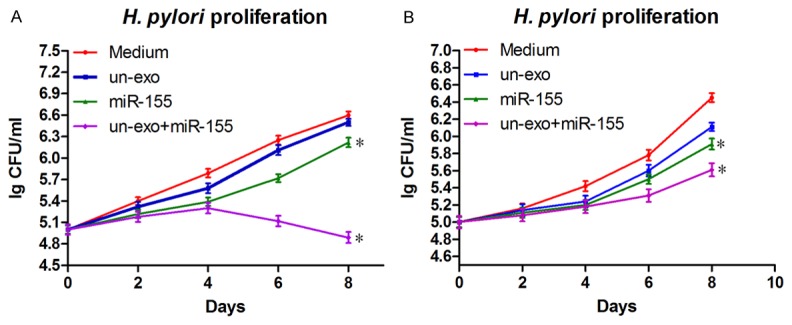
The effect of miR-155 loaded in exosomes on H. pylori proliferation. H. pylori obtained from the phagocytosis in macrophages treated with miR-155 loaded in exosomes (A), or H. pylori incubated with the cell supernatant of macrophages treated with miR-155 loaded in exosomes (B) were cultured on bacterial culture medium, and H. pylori proliferation was monitored every two days by OD600 nm value determination. Data showed mean ± SD of three independent experiments, and Student’s t test was used for statistical analysis (*P≤0.05).
Analysis of HE staining
Gastric tissue slices from normal mice and H. pylori infection mice immuned with medium, exosomes, miR-155 and exosomes+miR-155 were analyzed by HE staining (Figure 7). Experimental data presented that the number of inflammatory cells such as neutrophils, monocytes, et al. decreased in the gastric tissue of H. pylori infection mice immuned with miR-155 loaded in exosomes, compared with other groups that H. pylori infection mice treated with medium, exosomes or miR-155.
Figure 7.
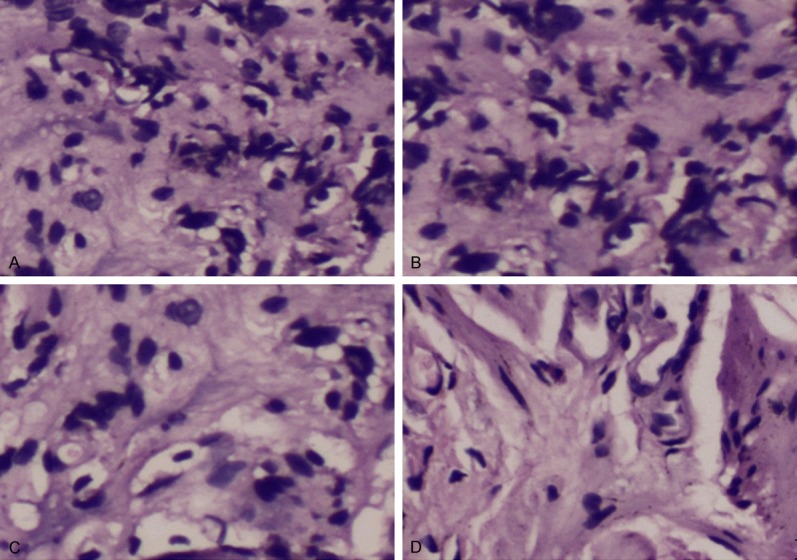
Typical HE staining of gastric tissue of H. pylori infection mice treated with different immune complexes (100×). A: H. pylori infection mice treated with medium; B: H. pylori infection mice immunized with exosomes; C: H. pylori infection mice treated with miR-155; D: H. pylori infection mice stimulated with miR-155 loaded in exosomes.
Discussion
Helicobacter pylori plays an important role in the development of Gastric ulcer [18,19], however, the relationships between miRNAs and the immune or inflammatory responses in H. pylori infection are still unknown.
Numerous studies reported that exosomes isolated from cells infected with various intracellular pathogens contain microbial components and could promote antigen presentation and macrophage activation [20,21]. Exosomes not only contain proteins, but also are rich in miRNAs, which could be functionally transferred from parent cells to recipient cells [22].
There are multiple studies describing various roles of exosomal miRNAs in cancer. For example, miRNAs transferred in exosomes from T lymphocytes to antigen-presenting cells at immune synapses were shown to be capable to regulate gene expression in recipient cells [23]. Similarly, the transfer of miRNAs between mouse dendritic cells via exosomes was found to be functional by repressing target mRNAs, and miRNAs or cytokines carried by dendritic cell-derived exosomes interact with and influence immune cells [24,25].
H. pylori infection can stimulate the expression of miR-155 in gastric epithelial cells and in gastric mucosal tissues. The up-expression of miR-155 in macrophages infected by H. pylori has been reported, indicating that miR-155 may function as a novel negative regulator in inflammation responses to H. pylori infection [26]. In this study, miR-155 in exosomes from H. pylori infection macrophages was up-expressed significantly, so miR-155 may regulated the inflammation of host cells by exosomes between different immune cells.
H. pylori activated human DCs and promoted a strong inflammatory response characterized by the early production of pro-inflammatory TNF-a or IL-6 cytokines via MyD88 in a TLR-dependent manner [27,28], followed by IL-10, IL-1β, and IL-23 secretion. DCs activation and cytokine production were accompanied by a strong miR-155 induction, which mainly controlled TNF-a production. Furthermore, the over-expression of miR-155 modulate the release of the proinflammatory cytokines IL-8 and GRO-a negatively [29]. Our results also show that miR-155 loaded in exosomes were deliveried into macrophages successfully and induced a strong inflammatory response that cytokines TNF-a, IL-6, IL-23 or cell signal transduction proteins CD40, CD63, CD81, and MCH-I all increased remarkablely, suggesting that miR-155 may function as novel negative regulator that help to fine-tune the inflammation response of H. pylori infection.
In gastric epithelial cell lines, H. pylori was shown to induce miR-155 expression in an TLR/MyD88-dependent NF-κB activation manner, but the over-expression of miRNAs could play an inhibitory effect to this signal transduction pathway [30,32]. Consistent with previous findings, we found that the over-expression miR-155 transfected into macrophages by loading in exosomes regulate the production of pro-inflammatory mediators by targeting MyD88, NF-κB proteins expression in the present study.
In the current study, we found out that macrophages transfected with miR-155 loaded in exosomes could damage H. pylori, which were inhibited on culture medium in vitro. Meanwhile, the cell culture supernatant of macrophages transfected with miR-155 loaded in exosomes also could inhibit the proliferation of H. pylori, indicating cytokines secreted from macrophages playing an important role in the inhibition. Interestingly, animal experiments in vivo show the inflammatory reaction of gastric epithelial tissue in H. pylori infection mice treated with miR-155 loaded in exosomes were be reduced significantly, revealed that miR-155 loaded in exosomes may have an potential application prospect in gastrointestinal diseases caused by the infection of H. pylori, but further research to investigate the underlying mechanisms is warranted.
Acknowledgements
This work was supported by Science and technology project of Suzhou in China (NO: SYSD2015022) and by Medical science and technology development fund of Jiangsu University in China (JLY20140047). We thank Xiang Ya Hospital and Xiangya Medical college for technical assistance.
Disclosure of conflict of interest
None.
References
- 1.Yin M, Hu Z, Tan D, Ajani JA, Wei Q. Molecular epidemiology of genetic susceptibility to gastric cancer: focus on single nucleotide polymorphisms in gastric carcinogenesis. Am J Transl Res. 2009;1:44–54. [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson K, Argent RH, Atherton JC. The inflammatory and immune response to Helicobacter pylori infection. Best Pract Res Clin Gastroenterol. 2007;21:237–259. doi: 10.1016/j.bpg.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Sonkoly E, Pivarcsi A. microRNAs in inflammation. Int Rev Immunol. 2009;28:535–561. doi: 10.3109/08830180903208303. [DOI] [PubMed] [Google Scholar]
- 4.Belair C, Baud J, Chabas S, Sharma CM, Vogel J, Staedel C, Darfeuille F. Helicobacter pylori interferes with an embryonic stem cell microRNA cluster to block cell cycle progression. Silence. 2011;2:7. doi: 10.1186/1758-907X-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsushima K, Isomoto H, Inoue N, Nakayama T, Hayashi T, Nakayama M, Nakao K, Hirayama T, Kohno S. MicroRNA signatures in Helicobacter pylori infected gastric mucosa. Int J Cancer. 2011;128:361–370. doi: 10.1002/ijc.25348. [DOI] [PubMed] [Google Scholar]
- 6.Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8:4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 7.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:1237–1248. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan ML, Karlsson JM, Baty CJ, Gibson GA, Erdos G, Wang Z, Milosevic J, Tkacheva OA, Divito SJ, Jordan R, Lyons-Weiler J, Watkins SC, Morelli AE. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756–766. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Worm J, Stenvang J, Petri A, Frederiksen KS, Obad S, Elmén J, Hedtjärn M, Straarup EM, Hansen JB, Kauppinen S. Silencing of microRNA-155 in mice during acute inflammatory response leads to derepression of c/ebp Beta and down-regulation of G-CSF. Nucleic Acids Res. 2009;37:5784–5792. doi: 10.1093/nar/gkp577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matta R, Wang X, Ge H, Ray W, Nelin LD, Liu Y. Triptolide induces anti-inflammatory cellular responses. Am J Transl Res. 2009;1:267–282. [PMC free article] [PubMed] [Google Scholar]
- 11.Ruggiero T, Trabucchi M, De Santa F, Zupo S, Harfe BD, McManus MT, Rosenfeld MG, Briata P, Gherzi R. LPS induces KH-type splicing regulatory protein dependent processing of microRNA-155 precursors in macrophages. FASEB J. 2009;23:2898–2908. doi: 10.1096/fj.09-131342. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 14.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, Smith KG, Rada C, Enright AJ, Toellner KM, Maclennan IC, Turner M. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wallet MA, Sen P, Tisch R. Immunoregulation of dendritic cells. Clin Med Res. 2005;3:166–175. doi: 10.3121/cmr.3.3.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murugaiyan G, Beynon V, Mittal A, Joller N, Weiner HL. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J Immunol. 2011;187:2213–2221. doi: 10.4049/jimmunol.1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao Y, Li G, Wu J, Zhang X, Wang J. Inflammatory response of macrophages cultured with Helicobacter pylori strains was regulated by miR-155. Int J Clin Exp Pathol. 2015;8:4545–4554. [PMC free article] [PubMed] [Google Scholar]
- 18.Oertli M, Engler DB, Kohler E, Koch M, Meyer TF, Müller A. MicroRNA-155 is essential for the T cell-mediated control of Helicobacter pylori infection and for the induction of chronic gastritis and colitis. J Immunol. 2011;187:3578–3586. doi: 10.4049/jimmunol.1101772. [DOI] [PubMed] [Google Scholar]
- 19.Yin M, Hu Z, Tan D, Ajani JA, Wei Q. Molecular epidemiology of genetic susceptibility to gastric cancer: focus on single nucleotide polymorphisms in gastric carcinogenesis. Am J Transl Res. 2009;1:44–54. [PMC free article] [PubMed] [Google Scholar]
- 20.BLASER MJ, BERG DE. Helicobacter pylori genetic diversity and risk of human disease. J Clin Invest. 2001;107:767–773. doi: 10.1172/JCI12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schorey JS, Bhatnagar S. Exosome function: from tumor immunology to pathogen biology. Traffic. 2008;9:871–881. doi: 10.1111/j.1600-0854.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Camussi G, Deregibus MC, Bruno S, Grange C, Fonsato V, Tetta C. Exosome/microvesicle-mediated epigenetic reprogramming of cells. Am J Cancer Res. 2011;1:98–110. [PMC free article] [PubMed] [Google Scholar]
- 23.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez MA, Bernad A, Sanchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nature Comm. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan MLG, Karlsson JM, Baty CJ, Gibson GA, Erdos G, Wang Z, Milosevic J, Tkacheva OA, Divito SJ, Jordan R, Lyons-Weiler J, Watkins SC, Morelli AE. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756–766. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pitt JM, Charrier M, Viaud S, Andre F, Besse B, Chaput N, Zitvogel L. Dendritic cell-derived exosomes as immunotherapies in the fight against cancer. J Immunol. 2012;193:1006–1011. doi: 10.4049/jimmunol.1400703. [DOI] [PubMed] [Google Scholar]
- 27.Xiao B, Liu Z, Li BS, Tang B, Li W, Guo G, Shi Y, Wang F, Wu Y, Tong WD, Guo H, Mao XH, Zou QM. Induction of microRNA-155 during Helicobacter pylori infection and its negative regulatory role in the inflammatory response. J Infect Dis. 2009;200:916–925. doi: 10.1086/605443. [DOI] [PubMed] [Google Scholar]
- 28.Janssens S, Beyaert R. A universal role for MyD88 in TLR/IL-1Rmediated signaling. Trends Biochem Sci. 2002;27:474–482. doi: 10.1016/s0968-0004(02)02145-x. [DOI] [PubMed] [Google Scholar]
- 29.Rad R, Ballhorn W, Voland P, Eisenächer K, Mages J, Rad L, Ferstl R, Lang R, Wagner H, Schmid RM, Bauer S, Prinz C, Kirschning CJ, Krug A. Extracellular and intracellular pattern recognition receptors cooperate in the recognition of Helicobacter pylori. Gastroenterology. 2009;136:2247–2257. doi: 10.1053/j.gastro.2009.02.066. [DOI] [PubMed] [Google Scholar]
- 30.Xiao B, Liu Z, Li BS, Tang B, Li W, Guo G, Shi Y, Wang F, Wu Y, Tong WD, Guo H, Mao XH, Zou QM. Induction of microRNA-155 during Helicobacter pylori Infection and Its Negative Regulatory Role in the Inflammatory Response. J Infect Dis. 2009;200:916–925. doi: 10.1086/605443. [DOI] [PubMed] [Google Scholar]
- 31.Mathias O, Daniela BE, Esther K, Manuel K, Thomas FM, Anne M. MicroRNA-155 Is Essential for the T Cell-Mediated Control of Helicobacter pylori Infection and for the Induction of Chronic Gastritis and Colitis. J Immunol. 2011;187:3578–3586. doi: 10.4049/jimmunol.1101772. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Wu J, Cheng Y, Jiang Y, Li G. Over-expression of microRNA-223 inhibited the proinflammatory responses in Helicobacter pylori-infection macrophages by down-regulating IRAK-1. Am J Transl Res. 2016 Epub ahead of print. [PMC free article] [PubMed] [Google Scholar]


