Abstract
To explore the effects of microRNA-218 (miR-218) on glioma cell lines and the related mechanism. U251 and U87 cells were transfected with negative control, miR-218 mimic or miR-218 inhibitor using lipofectamine 2000. The expressions of mRNA and proteins were detected with qRT-PCR and Western blotting. The cell proliferation, apoptosis, migration and invasion were studied using MTT, flow cytometry, Transwell assay and scratch-wound assay, respectively. The targeting effect of HMGB1 by miR-218 was measured with luciferase reporter assay. The results showed that miR-218 was significantly downregulated while HMGB1 was upregulated in both glioma cell lines. Transfection of miR-218 significantly reduced the cell viability and colony formation, increased cell apoptosis and arrested cell in G0/G1 phase. Transfection of miR-218 also decreased the invasion and migration of glioma cells. The expressions of HMGB1, RAGE, cyclin D1 and MMP-9 were downregulated while the expression of caspase-9 was upregulated by miR-218. Silencing HMGB1 increased the expression of RAGE, cyclin D1, MMP-9 but decreased the expression of caspase-9 in U251 and U87 cells. Co-transfection with pcHMGB1 and miR-218 significantly decreased the growth inhibition and increased the apoptosis of glioma cells while these effects were abolished in glioma cells co-transfected with HMGB1 siRNA and miR-218 inhibitor. In addition, co-transfection with pcHMGB1 and miR-218 inhibitor increased the invasiveness of U251 and U87 cells. These findings suggested that miR-218 may negatively regulate HMGB-mediated suppression of RAGE to regulate cell proliferation, apoptosis and invasion, and that intervention of miR-218-HMGB1-RAGE may be useful for developing potential clinical strategies.
Keywords: microRNA-218, glioma, proliferation, apoptosis, invasion, high mobility group box 1 (HMGB1)
Introduction
Glioma is the most common type of malignant tumors in brain and the median survival of patients with glioma depends largely on the histological grade of the tumor [1]. Despite of improvements of surgical skills, radiotherapy and chemotherapy, the treatment of patients with gliomas remains one of the greatest challenges in oncology [2]. The reason of high mortality of glioma is partially because of local invasion of tumor into normal brain tissues, which prevents complete surgical resection [3]. Thus, understanding the potential mechanism accounting for the invasion and blocking the invasive process of glioma may be an effective therapeutic strategy.
MicroRNAs (miRNAs) are a group of endogenous small noncoding RNAs (22-25 nucleotides in length) and generally regulate the mRNA expression at the post-transcriptional level by negatively regulating their target mRNAs to either degradation or translational repression. Increasing studies have proved that miRNAs are downregulated in various diseases, including cardiovascular diseases, neurological diseases and human cancers [4-6]. Among of these miRNAs, miRNA-218 (miR-218) is found to be greatly suppressed in several kinds of solid tumors while the downregulation of miR-218 can enhance invasion and proliferation of tumor cells by altering the expression of target genes [7-10]. However, if the miR-218 is involved in the activities of glioma is not clear.
As one member of the high mobility group box (HMGB) of non-histone chromosomal proteins, HMGB1 is indicated to be overexpressed in various tumors including human glioma [11] and is associated with pathological grade and poor prognosis [12], acting as oncogene [13,14]. In contrast, Zhang et al. [15] found that knockdown of HMGB1 can increase the apoptosis and suppress the proliferation and invasion of glioma cells, suggesting the involvement of HMGB1 in glioma. Recent studies using Targetscan software indicated that HMGB1 is a target gene of miR-218, and that upregulation of miR-218 can suppress the proliferation and invasion, promote the apoptosis of pancreatic cancer cells [16], and suppress the cell migration and invasion of non-small cell lung cancer [17]. These effects of HMGB1 may be achieved through the interaction with receptor for advanced glycation end-products (RAGE) which constitutes a signaling pathway with HMGB1 [6,18]. However, whether miR-218 could regulate the expression of target gene HMGB1 through RAGE in glioma and the underlying mechanisms are still unclear.
In the present study, we investigated the effects of miR-218 on the activities of glioma cells and related mechanisms, which would provide theoretic basis for target therapy strategies of glioma.
Materials and methods
Cell lines and transfection
Normal human astrocytes (NHA) were purchased from Sciencell Research Laboratories (Carlsbad, CA, USA) as control. Glioma U118, U251, U373, U87, SNB19 and LN229 cell lines were obtained from the Institute of Biochemistry and Cell Biology (Shanghai Institutes for Biological Sciences, Chinese Academy of Science, Shanghai, China). The cells at density of 1×105 were seeded in 6-well plates in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin, and incubated in 5% CO2 atmosphere at 37°C. The cells were separately treated with negative control (NC), miR-218 mimic, miR-218 inhibitor, HMGB1 and RAGE-targeted small interfering RNA (HMGB1 siRNA and RAGE siRNA) and siRNA negative control (GenePharma, Shanghai, China), respectively, through transfection with Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. After 24 h of transfection, the medium was removed and the cells were placed in the complete medium and maintained at 37°C in 5% CO2 incubator.
qRT-PCR
The total RNA of glioma cells was extracted with TRIzol (Invitrogen) and the total miRNAs were extracted using miRVana kits (Ambion, Austin, TX, USA), according to the manufacturer’s instructions. cDNA was generated with the High-Capacity cDNA Reverse Transcription kit (Roche Diagnostics GmbH, Mannheim, Germany). The expression level of mature miR-218 in the glioma cells was confirmed with TaqMan microRNA assay (Applied Biosystems, Foster City, CA, USA). The expression level of miR-218 and HMGB1 was examined with qRT-PCR with SYBR Green PCR Master Mix kit (Applied Biosystems, USA) in conjunction with ABI-Prism 7300 System. The following primers were used: HMGB1, forward 5’-GCTCCATAGAGACAGCGCCGGG-3’ and reverse 5’-CCTCAGCGAGGCACAGAGTCGC-3’; GAPDH, forward 5’-TCGGAGTCAACGGATTTGG-3’ and reverse 5’-CATGGGTGGAATC ATATTGGA-3’. The relative expression levels of mature miR-218 and HMGB1 mRNA were calculated with the 2-ΔΔCt method and normalized to U6 snRNA and GAPDH mRNA levels, respectively.
Western blot
The total cell lysates from different groups were obtained by lysing the cells in RIPA buffer and the protein concentration was determined using the BCA protein assay (Pierce Biotechnology). Forty µg protein from each sample was resolved by 10% SDS-PAGE gel and transferred to PVDF membranes (Millipore) which was incubated with primary antibodies and subsequently with secondary antibodies. The primary antibodies included polyclonal rabbit of HMGB1 antibody (Abgent, San Diego, CA; 1:1,000 dilution) and RAGE antibody (R&D Systems; 1:200), polyclonal mouse cyclin D1 antibody (Abcam, 1:500 dilution), polyclonal rabbit matrix metallopeptidase 9 (MMP9 antibody (Abcam, Cambridge, UK; 1:500 dilution), monoclonal mouse caspase-9 antibody (Life Technologies, Carlsbad, CA; 1:500 dilution) and polyclonal rabbit β-actin antibody (1:1000 dilution; Abcam, San Francisco, USA). The secondary antibodies were HRP-conjugated goat anti-rabbit/mouse IgG polyclonal antibodies (Zymed, San Diego, CA; 1:1000 dilution). Autoradiography was conducted using ECL chemiluminescent substrate. The signal intensity was determined with gel analysis software ImageJ (National Institutes of Health, Bethesda, MA, USA).
Luciferase reporter assay
The 3’-UTR sequence of HMGB1 which was predicted to interact with miR-218 or a mutated sequence with the predicted target sites was synthesized and inserted into the XbaI and FseI sites of pGL3 control vector (Promega Corporation, Madison, WI, USA). For reporter assay, U251 and U87 cells were plated in 24-well plates and transfected with pGL3-HMGB1-3’UTR or pGL3-HMGB1-3’UTR-mut and miR-218 mimic or NC vectors using FuGENE HD transfection reagent (Promega Corporation). Renilla luciferase vector pRL-SV50 (Promega Corporation) was co-transfected to normalize the differences in transfection efficiency. After 24 h of transfection, the cells were collected and assayed using the Dual-Luciferase Reporter Assay System (Promega Corporation) according to the manufacturer’s instructions. Transfection was repeated in three independent experiments.
MTT assay
The U251 and U87 cells were inoculated in 96-well plates with density of 5×103 cells/well in 200 µL DMEM and incubated for 24, 48, 72 and 96h after transfection and another 3 h after addition of MTT (10 mg/ml), according to the manufacturer’s protocol. The cell proliferation was documented every 24 h for 4 days using a MTT assay. The absorbance at wavelength of 570 nm (OD 570) was detected using the microplate reader (Bio-Rad).
Colony formation assay
After stable transfection, the glioma cells were seeded in 6-well plates at the density of 1×103/well and cultured for 2 weeks to allow colony formation. The culture medium was changed every 3 days. The colonies were then fixed in 100% methanol and stained with crystal violet solution. Subsequently, the number of macroscopically observable colonies was recorded.
Cell cycle and apoptosis assay
The effects of miR-218 on cell cycle and apoptosis of U251 and U87 glioma cells were examined with flow cytometry. In brief, U251 and U87 cells after transfection for 48 h were collected and rinsed twice with PBS, fixed in 70% ethanol and re-suspended in staining solution (50 lg/ml of propidium iodide, 1 mg/ml of RNase A, 0.1% Triton X-100 in PBS) at -20°C for 30 min and then stored at 4°C overnight. The multiplication cycle and apoptotic rate were measured with flow cytometry (BD Biosciences, Mansfield, MA, USA) and the data were analyzed using CellQuest software (BD biosciences, San Jose, CA, USA).
Invasion assay
Cell invasion was determined using Transwell assays. Transfected cells at density of 3×104 were transferred to the top of Matrigel-coated invasion chambers (BD Biosciences, San Jose, CA, USA) in serum-free DMEM. DMEM containing 10% FBS was added to the lower chamber. After 24 h of incubation and removal of non-invading cells with rinse, the invading cells were fixed with 95% ethanol and stained with 0.1% crystal violet. The number of invading cells was manually counted in 5 randomly chosen fields under a microscope and images were captured under ×100 magnification. Experiments were repeated three times.
Statistical analysis
All data were shown as mean ± SD. Statistical analysis was performed using SPSS 12.0 software (SPSS Inc., Chicago, IL). Inter-group differences were analyzed with Student’s t-test and multiple comparisons among several groups were conducted by analysis of variance. P<0.05 was set as significant level.
Results
MiR-218 was downregulated and HMGB1 was upregulated in human glioma cell lines
As shown in Figure 1A, qRT-PCR analysis indicated that miR-218 was downregulated in all glioma cell lines (U118, U251, U373, U87, SNB19 and LN229) when compared with that in the NHA cells (P<0.05); on the contrary, the expression levels of HMGB1 in 6 glioma cell lines were higher than that in the NHA cells (P<0.05) (Figure 1B). Moreover, western blotting results showed that the expression of HMGB1 protein also significantly increased in all glioma cell lines compared with that in the NHA cells (P<0.05; Figure 1C).
Figure 1.
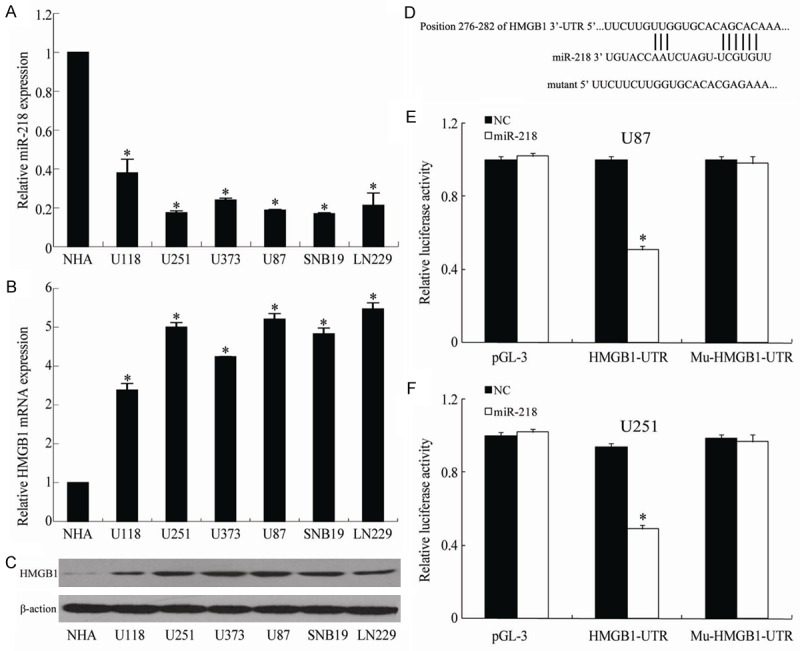
Showing the expression of miR-218 (A), the expressions of HMGB1 mRNA (B) and protein (C) in glioma cell lines, the sequence matching between HMGB1 and miR-218 (D), the luciferase activities in U85 cells (E) and U251 cells (F) under different treatments. *P<0.05 vs NHA.
HMGB1 was the direct target of miR-218 in glioma cells
Using the bioinformatic database TargetScan, we identified HMGB1 as a potential target of miR-218 and confirmed the direct interaction between miR-218 and the 3’-UTR of HMGB1 mRNA. Figure 1D showed that the seed sequence was located at nucleotides 276-282 of the 3’-UTR region of the HMGB1 gene. We constructed a luciferase plasmid containing the wild-type 3’-UTR of HMGB1 and the mutated 3’-UTR of HMGB1 to co-transfect the HEK293 cells with miR-218 mimic or NC. Compared to constructs containing mutated 3’-UTRs, the luciferase activity for the wild-type 3’-UTR of HMGB1 was significantly inhibited by co-transfection with miR-218 mimics (P<0.01) (Figure 1E and 1F).
Aberrant expression of miR-218 regulated proliferation and apoptosis of glioma cells
To confirm the effect of miR-218 on the proliferation of glioma cells, we transfected U251 and U87 cells with NC, miR-218 mimic and miR-218 inhibitor, respectively. MTT assay showed that the viability of U251 and U87 cells was significantly lower in the miR-218 mimic transfected group compared with NC group at 48, 72 and 96 hours after transfection (P<0.05). In contrast, the viability of glioma cells was significantly increased following transfection with miR-218 inhibitor when compared to miR-218 mimic (P<0.05; Figure 2A and 2B). Similarly, compared to the NC, the results of the colony formation assay suggested that miR-218 mimic significantly inhibited while miR-218 inhibitor enhanced colony formation in U251 and U87 ells (P<0.05; Figure 2C and 2D).
Figure 2.
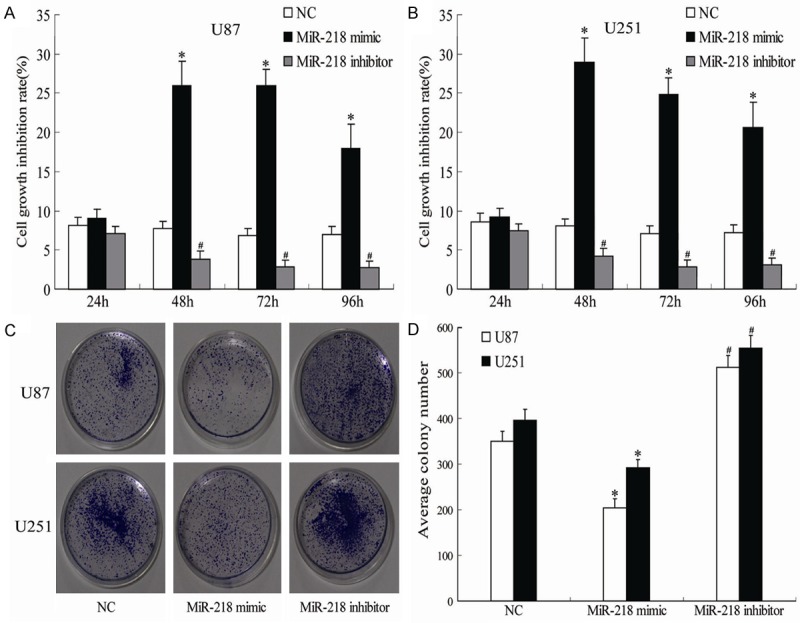
Showing the effects of different treatments on the growth inhibition (A, B) and colony formation (C, D) of U85 and U251 cells. *P<0.05 vs NC, #P<0.05 vs miR-218 mimic.
Flow cytometry analysis indicated that, compared to NC group, U251 and U87 ells transfected with the miR-218 mimic were arrested in G0/G1 phase while miR-218 inhibitor significantly decreased the percentage of cells in the G0/G1 phase (Figure 3A-D). In addition, compared with NC, the cell apoptosis rate in U251 and U87 cells was greatly increased by the miR-218 mimic and decreased by transfection with miR-218 inhibitor (P<0.05; Figure 3E and 3F).
Figure 3.
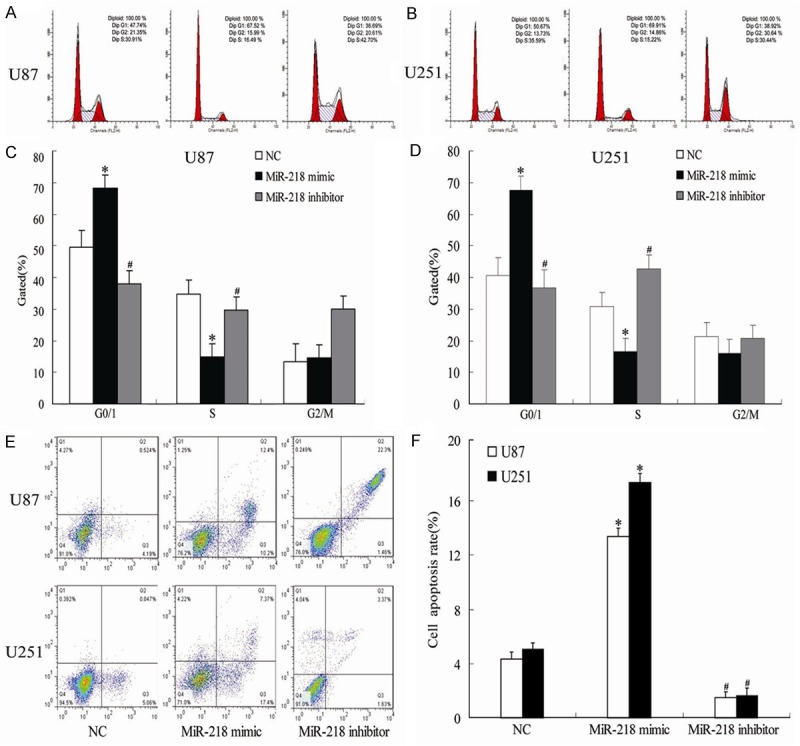
Showing the effects of different treatments on the cell proliferation (A-D) and apoptosis (E, F) of U85 and U251 cells. *P<0.05 vs NC, #P<0.05 vs miR-218 mimic.
Aberrant expression of miR-218 regulated invasion and migration of glioma cells
Invasion is not only one of the pathophysiological features of human glioma, but also one of the most important reasons for the poor prognosis of glioma. Transwell and Scratch assays was used to check the effects of miR-218 on the invasion and migration of glioma cells. Transwell assay showed that, after transfection for 24 h, the invasion of U251 and U87 cells transfected with miR-218 mimic was significantly suppressed compared with NC group; however, transfection with miR-218 inhibitor significantly enhanced the invasion of U251 and U87 cells (P<0.05; Figure 4A and 4B). Similarly, the results of migration showed that miR-218 mimic decreased the migration while miR-218 inhibitor increased the migration of glioma cells the scratch assay (Figure 4C and 4D). These results suggested that the aberrant expression of miR-218 may have ability to regulate glioma cells invasion in vitro.
Figure 4.
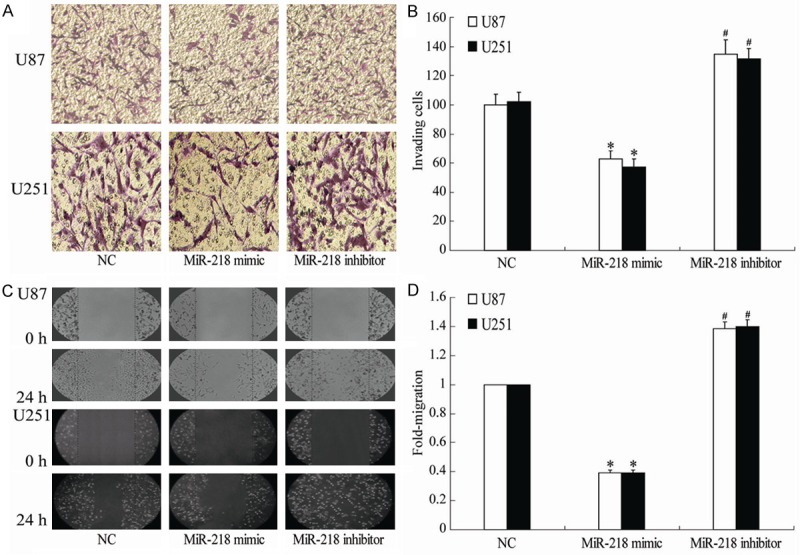
Showing the effects of different treatments on the cell invasion (A, B) and migration (C, D) of U85 and U251 cells. *P<0.05 vs NC, #P<0.05 vs miR-218 mimic.
MiR-218 regulated expression of HMGB1-RAGE axis0
Several studies have shown that HMGB1-RAGE interaction is involved in tumor growth, migration and metastases [19,20] and HMGB1 can regulate expression of some downstream genes related to proliferation, apoptosis and invasion of tumor, such as cyclin D1, caspase-9 and MMP-9. So we transiently transfected U251 and U87 cells NC, miR-218 mimic and miR-218 inhibitor, respectively. After 48 hours of transfection, total cellular proteins were extracted for detecting the expression of HMGB1-RAGE axis. The results showed that, compared to NC group, U251 and U87 cells transfected with miR-218 mimic demonstrated downregulation of HMGB1, RAGE, cyclin D1 and MMP-9; however, caspase-9 was significantly upregulated by miR-218 mimic. The expression of HMGB1, RAGE, cyclin D1, MMP-9 and caspase-9 in miR-218 inhibitor group was significantly different from miR-218 mimic (all P<0.05; Figure 5A and 5B).
Figure 5.
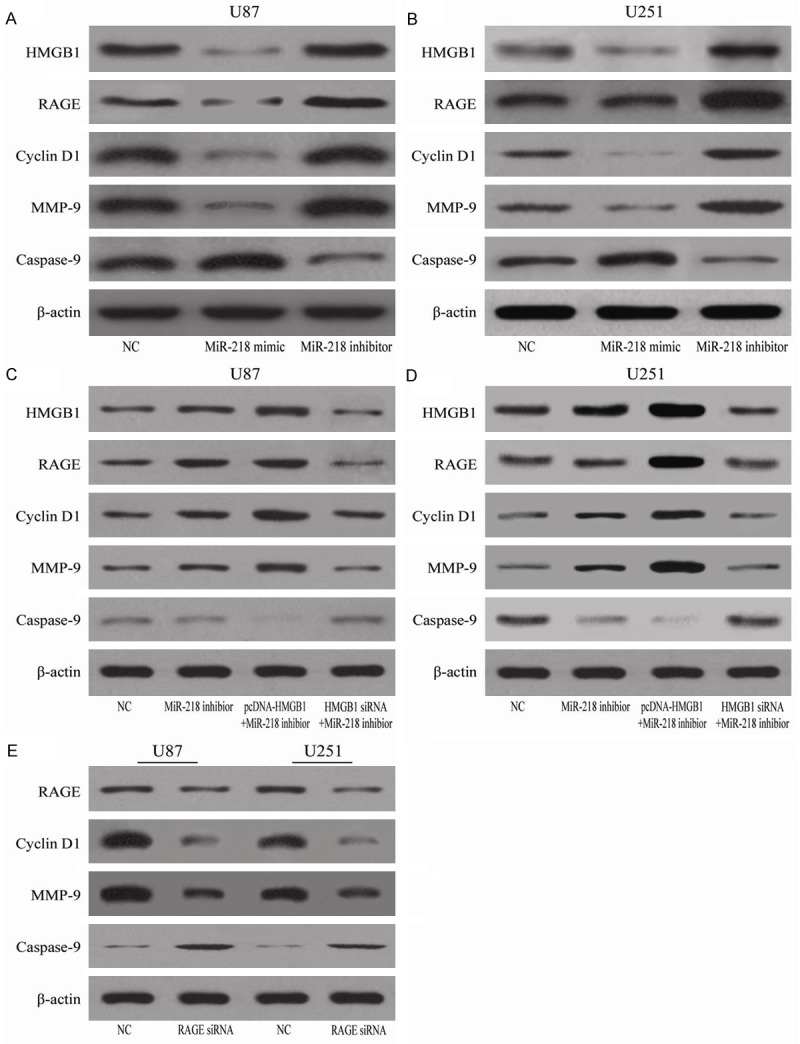
Showing the effects of different treatments on the expression of different proteins (A-E) in U85 and U251 cells.
Aberrant expression of HMGB1 modulated the effects of miR-218 on glioma cells
In order to determine whether aberrant expression of HMGB1 can influence the role of miR-218 in glioma cells, we constructed pcDNA-HMGB1 and HMGB1 siRNA co-transfected into glioma cells with the miR-218 inhibitor. The results showed that pcDNA-HMGB1 co-transfected with miR-218 inhibitor in glioma cells increased the expression of RAGE, cyclin D1, MMP-9 but decreased the expression of caspase-9 in U251 and U87 cells (Figure 5C and 5D). However, these changes were not significantly different in cells co-transfected with HMGB1 and siRNA-218 when compared to NC or miR-218 inhibitor (Figure 5C and 5D). In addition, glioma cells treated with RAGE siRNA showed decrease of the expression of cyclin D1 and MMP-9 but increase of caspase-9 in U251 and U87 cells (Figure 5E).
The flow cytometry showed that co-transfection with pcHMGB1 and miR-218 significantly decreased the growth inhibition and increased the apoptosis of U251 and U87 cells (P<0.05; Figure 6A-D). And these effects were abolished in U251 and U87 cells co-transfected with HMGB1 siRNA and miR-218 inhibitor (P<0.05; Figure 6A-D). In addition, Transwell assay showed that co-transfection with pcHMGB1 and miR-218 inhibitor increased the invasion of U251 and U87 cells (Figure 6E and 6F).
Figure 6.
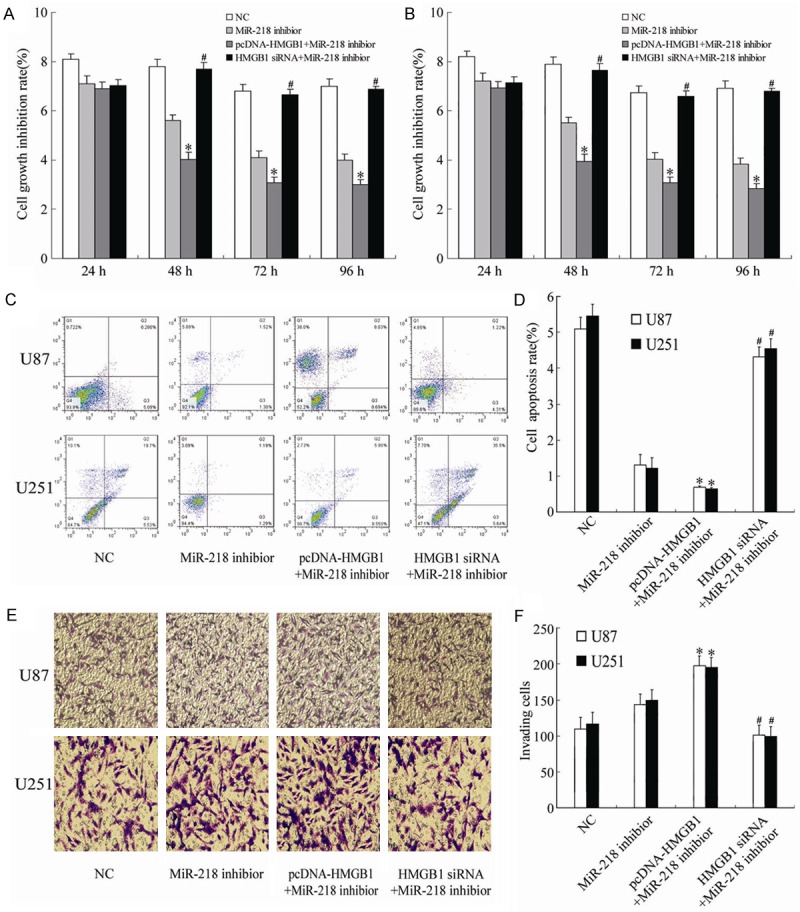
Showing the effects of different treatments on the cell growth inhibition (A, B), apoptosis (C, D) and invasion (E, F) of U85 and U251 cells. *P<0.05 vs NC, #P<0.05 vs pcDNA-HMGB1+miR-218 inhibitor.
Discussion
The heterogeneity, aggressive nature and angiogenic behaviors of glioma make it have the features of high morbidity, high recurrence rate, high mortality and low cure rate [1-3], which has severe threat to the life and health of patients. Current studies focus on the role of activation of oncogenes and loss of tumor suppressor genes in the development and progression of glioma [21,22]. Although there are some progress, the therapy of glioma is still an intractable problem; the median survival value of patients with glioma has not changed much over the past several years and is about only 14.6 months [23]. Therefore, it is urgent for us to investigate deeper mechanism involved in the development and progression of glioma.
As a member of the non-coding miRNAs, miR-218 is found to be involved in cancer pathogenesis such as cell differentiation, growth, and death and tumor development [7-10]. Recent studies indicated that miR-218 is down-regulated and has been regarded as a tumor suppressor in glioma and a potential valuable biomarker for glioma patients [24]. While miR-218 over-expression can inhibit the invasion, migration, proliferation and tumorigenicity, and induce apoptosis of glioma cells through its target genes including cyclin D1 [10,25-27]. Consistently, the present study indicate that the expressions of miR-218 in glioma cell lines is significantly decreased while over-expression of miR-218 decreases the viability, suppresses the proliferation and invasion, and promotes the apoptosis of glioma cells. However, downregulation of miR-218 in glioma cells produces the opposite results. Therefore, the study supports the proposal that miR-218 is a tumor suppressor of glioma cells.
HMGB1 gene, through combining with the minor groove of double-stranded DNA and prompting the local deformation of double-stranded DNA, can cause DNA bending and promote nuclear transcription of a variety of genes [28]. Once secreted outside of the cells, HMGB1 can function as extracellular signal molecule involved in the inflammation, the differentiation, proliferation and migration of cells, and tumor metastasis [29]. Previous studies suggested HMGB1 is involved in the pathology of glioma [11,15] as oncogene [13,14] and is associated with the pathological grades and poor prognosis of tumors [12]. Recent studies using Targetscan software indicated that HMGB1 is a target gene of miR-218 [16,17]. In the present study, we found that miRNA mimic significantly inhibits the expression of HMGB1 in glioma cells, confirming that HMGB1 is target gene of miR-218 in glioma cells.
The targeting effects of miR-218 on HMGB1 in glioma cells may be achieved through multiple pathways, such as RAGE [18]. Recurrent studies found that extracellular HMGB1 mainly functions through RAGE and related signaling pathways to play its biological effects [30]. HMGB1 modulates the RAGE-mediated multiple signaling pathways and activates a variety of downstream signaling molecules including MMP-9, caspase-9 and cyclin D1 to mediate inflammation, apoptosis and tumor behaviors [31,32]. The present study indicated that the expression of MMP-9, RAGE and cyclin D1 in glioma cells is decreased while caspase-9 is increased by miR-218 mimic; the miR-218 inhibitor displays opposite effects on the expression of these proteins. Furthermore, we found that HMGB1-siRNA blocks the change of these proteins, increases the apoptosis of glioma cells and decreases the invasion/migration of glioma cells. These results confirm the involvement of these molecules in the modulation of glioma cells by miR-218 and HMGB1. However, the detailed mechanism remains to be further studied.
In conclusion, the present study demonstrates that miR-218 plays an important role in regulating the proliferation, apoptosis and invasion of glioma cells and confirms that HMGB1 is the target gene of miR-218 in glioma cells. These results present a novel mechanism of miRNA-mediated direct suppression of HMGB1-RAGE pathway in glioma and blocking this pathway may be used as an important strategy for the treatment of glioma.
Disclosure of conflict of interest
None.
References
- 1.Paw I, Carpenter RC, Watabe K, Debinski W, Lo HW. Mechanisms regulating glioma invasion. Cancer Lett. 2015;362:1–7. doi: 10.1016/j.canlet.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi Q, Bao S, Song L, Wu Q, Bigner DD, Hjelmeland AB, Rich JN. Targeting SPARC expression decreases glioma cellular survival and invasion associated with reduced activities of FAK and ILK kinases. Oncogene. 2007;26:4084–4094. doi: 10.1038/sj.onc.1210181. [DOI] [PubMed] [Google Scholar]
- 3.Giese A, Bjerkvig R, Berens ME, Westphal M. Cost of migration: invasion of malignant gliomas and implications for treatment. J. Clin. Oncol. 2003;21:1624–1636. doi: 10.1200/JCO.2003.05.063. [DOI] [PubMed] [Google Scholar]
- 4.Zhong JF, He Y, Chen WJ. Circulating microRNA-19a as a Potential Novel Biomarker for Diagnosis of Acute Myocardial Infarction. Int J Mol Sci. 2014;15:20355–20364. doi: 10.3390/ijms151120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yelamanchili SV, Chaudhuri AD, Chen LN, Xiong H, Fox HS. MicroRNA-21 dysregulates the expression of MEF2C in neurons in monkey and human SIV/HIV neurological disease. Cell Death Dis. 2010;1:e77. doi: 10.1038/cddis.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen RC, Yi PP, Zhou RR, Xiao MF, Huang ZB, Tang DL, Huang Y, Fan XG. The role of HMGB1-RAGE axis in migration and invasion of hepatocellular carcinoma cell lines. Mol Cell Biochem. 2013;390:271–280. doi: 10.1007/s11010-014-1978-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J, Wang Y, Dong R, Huang X, Ding S, Qiu H. Circulating microRNA-218 was reduced in cervical cancer and correlated with tumor invasion. J Cancer Res Clin Oncol. 2012;138:671–674. doi: 10.1007/s00432-012-1147-9. [DOI] [PubMed] [Google Scholar]
- 8.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 9.Nishikawa R, Goto Y, Sakamoto S, Chiyomaru T, Enokida H, Kojima S, Kinoshita T, Yamamoto N, Nakagawa M, Naya Y, Ichikawa T, Seki N. Tumor-suppressive microRNA-218 inhibits cancer cell migration and invasion via targeting of LASP1 in prostate cancer. Cancer Sci. 2014;105:802–811. doi: 10.1111/cas.12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Yan W, Zhang W, Chen L, You G, Bao Z, Wang Y, Wang H, Kang C, Jiang T. MiR-218 reverses high invasiveness of glioblastoma cells by targeting the oncogenic transcription factor LEF1. Oncol Rep. 2012;28:1013–1021. doi: 10.3892/or.2012.1902. [DOI] [PubMed] [Google Scholar]
- 11.Gupta P, Ghosh S, Nagarajan A, Mehta VS, Sen E. β-defensin-3 negatively regulates TLR4-HMGB1 axis mediated HLA-G expression in IL-1β treated glioma cells. Cell Signal. 2013;25:682–689. doi: 10.1016/j.cellsig.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Wang XJ, Zhou SL, Fu XD, Zhang YY, Liang B, Shou JX, Wang JY, Ma J. Clinical and prognostic significance of high-mobility group box-1 in human gliomas. Exp Ther Med. 2014;9:513–518. doi: 10.3892/etm.2014.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao J, Ding Y, Huang J, Li Q, Liu Y, Ni W, Zhang Y, Zhu Y, Chen L, Chen B. The association of HMGB1 gene with the prognosis of HCC. PLoS One. 2014;9:e89097. doi: 10.1371/journal.pone.0089097. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Zhang J, Kou YB, Zhu JS, Chen WX, Li S. Knockdown of HMGB1 inhibits growth and invasion of gastric cancer cells through the NF-ĸB pathway in vitro and in vivo. Int J Oncol. 2014;44:1268–1276. doi: 10.3892/ijo.2014.2285. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Liu C, Hou R. Knockdown of HMGB1 improves apoptosis and suppresses proliferation and invasion of glioma cells. Chin J Cancer Res. 2014;26:658–668. doi: 10.3978/j.issn.1000-9604.2014.12.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z, Xu Y, Long J, Guo K, Ge C, Du R. MicroRNA-218 suppresses the proliferation, invasion and promotes apoptosis of pancreatic cancer cells by targeting HMGB1. Chin J Cancer Res. 2014;27:247–257. doi: 10.3978/j.issn.1000-9604.2015.04.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C, Ge S, Hu C, Yang N, Zhang J. MiRNA-218, a new regulator of HMGB1, suppresses cell migration and invasion in non-small cell lung cancer. Acta Biochim Biophys Sin. 2013;45:1055–1061. doi: 10.1093/abbs/gmt109. [DOI] [PubMed] [Google Scholar]
- 18.Bassi R, Giussani P, Anelli V, Colleoni T, Pedrazzi M, Patrone M, Viani P, Sparatore B, Melloni E, Riboni L. HMGB1 as an autocrine stimulus in human T98G glioblastoma cells: role in cell growth and migration. J Neurooncol. 2008;87:23–33. doi: 10.1007/s11060-007-9488-y. [DOI] [PubMed] [Google Scholar]
- 19.Taguchi A, Blood DC, del Toro G, Canet A, Lee DC, Qu W, Tanji N, Lu Y, Lalla E, Fu C, Hofmann MA, Kislinger T, Ingram M, Lu A, Tanaka H, Hori O, Ogawa S, Stern DM, Schmidt AM. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature. 2000;405:354–360. doi: 10.1038/35012626. [DOI] [PubMed] [Google Scholar]
- 20.Volp K, Brezniceanu ML, Bosser S, Brabletz T, Kirchner T, Göttel D, Joos S, Zörnig M. Increased expression of high mobility group box1: HMGB1; is associated with an elevated level of the antiapoptotic c-IAP2 protein in human colon carcinomas. Gut. 2006;55:234–242. doi: 10.1136/gut.2004.062729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee EK, Lee MJ, Abdelmohsen K, Kim W, Kim MM, Srikantan S, Martindale JL, Hutchison ER, Kim HH, Marasa BS, Selimyan R, Egan JM, Smith SR, Fried SK, Gorospe M. MiR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol. 2011;31:626–638. doi: 10.1128/MCB.00894-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li B, Qi XQ, Chen X, Huang X, Liu GY, Chen HR, Huang CG, Luo C, Lu YC. Expression of targeting protein for Xenopus kinesin-like protein 2 is associated with progression of human malignant astrocytoma. Brain Res. 2010;1352:200–207. doi: 10.1016/j.brainres.2010.06.060. [DOI] [PubMed] [Google Scholar]
- 23.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2014;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 24.Cheng MW, Wang LL, Hu GY. Expression of microRNA-218 and its clinicopathological and prognostic significance in human glioma cases. Asian Pac J Cancer Prev. 2015;16:1839–1843. doi: 10.7314/apjcp.2015.16.5.1839. [DOI] [PubMed] [Google Scholar]
- 25.Gu JJ, Gao GZ, Zhang SM. MiR-218 inhibits the migration and invasion of glioma U87 cells through the Slit2-Robo1 pathway. Oncol Lett. 2015;9:1561–1566. doi: 10.3892/ol.2015.2904. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Xia H, Yan Y, Hu M, Wang Y, Wang Y, Dai Y, Chen J, Di G, Chen X, Jiang X. MiR-218 sensitizes glioma cells to apoptosis and inhibits tumorigenicity by regulating ECOP-mediated suppression of NF-κb activity. Neuro Oncol. 2013;15:413–422. doi: 10.1093/neuonc/nos296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu JJ, Gao GZ, Zhang SM. miR-218 inhibits glioma U87 cells proliferation via CDK6/Cyclin D 1/p21Cip1/Waf1 pathway. Oncol Lett. 2015;9:2743–2749. [Google Scholar]
- 28.Muller S, Scaffidi P, Degryse B, Bonaldi T, Ronfani L, Agresti A, Beltrame M, Bianchi ME. New EMBO members’ review: the double life of HMGB1 chromatin protein: architectural factor and extracellular signal. EMBO J. 2001;20:4337–4340. doi: 10.1093/emboj/20.16.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoetzer OJ, Fersching DM, Salat C, Steinkohl O, Gabka CJ, Hamann U, Braun M, Feller AM, Heinemann V, Siegele B, Nagel D, Holdenrieder S. Circulating immunogenic cell death biomarkers HMGB1 and RAGE in breast cancer patients during neoadjuvant chemotherapy. Tumour Biol. 2013;34:81–90. doi: 10.1007/s13277-012-0513-1. [DOI] [PubMed] [Google Scholar]
- 30.Moser B, Janik S, Schiefer AI, Müllauer L, Bekos C, Scharrer A, Mildner M, Rényi-Vámos F, Klepetko W, Ankersmit HJ. Expression of rage and HMGB1 in thymic epithelial tumors, thymic hyperplasia and regular thymic morphology. PLoS One. 2014;9:e94118. doi: 10.1371/journal.pone.0094118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donato R. RAGE: a single receptor for several ligands and different cellular responses: the case of certain S100 proteins. Curr Mol Med. 2007;7:711–724. doi: 10.2174/156652407783220688. [DOI] [PubMed] [Google Scholar]
- 32.Meng Q, Zhao J, Liu H, Zhou G, Zhang W, Xu X, Zheng M. HMGB1 promotes cellular proliferation and invasion, suppresses cellular apoptosis in osteosarcoma. Tumour Biol. 2014;35:12265–12274. doi: 10.1007/s13277-014-2535-3. [DOI] [PubMed] [Google Scholar]


