Abstract
Gastric cancer (GC) causes nearly one million deaths worldwide each year. However, the molecular pathway of GC development remains unclear. Increasing evidences have shown that microRNAs (miRNAs) are highly associated with tumor development. However, relative little is known about the potential role of miRNAs in gastric cancer development. In the present study, we showed that miR-561 was down-regulated frequently in human GCs cell lines and tissues, and its expression was associated with tumor-node-metastasis (pTNM) stage. Enforced expression of miR-561 in GC cells inhibited cell proliferation and invasion in vitro. In contrast, knockdown of miR-561 had the opposite effect on cell proliferation and invasion. Moreover, c-Myc was identified as a potential miR-561 target. Further studies confirmed that miR-561 suppressed the expression of c-Myc by directly binding to its 3’-untranslated region. Restoration of c-Myc in miR-561-overexpressed GC cells reversed the suppressive effects of miR-561 and c-Myc was inversely correlated with miR-561 expression in GC tissues. These results demonstrate that miR-561 acts as a novel tumor suppressor in GC by targeting c-Myc gene and inhibiting GC cells proliferation and invasion. These findings contribute to current understanding of the functions of miR-561 in GC.
Keywords: Gastric cancer, microRNA-561, c-Myc, tumor suppressor
Introduction
Gastric cancer (GC) is the fourth most prevalent cancer worldwide and the second most frequent cause of cancer death, with an estimated 850,000 newly diagnosed cases and 650,000 deaths per year [1,2]. Despite recent advances in GC therapy, the 5-year overall survival of respectable GC in specialized centers in Europe or the United States wasonly approximately 35% [3,4]. The 5-year survival rate of metastatic or advanced GC wasnearly 5-20%, with median overall survival only less than 1 year [5]. However, the method for early detection and prediction of lymph node metastasis has not yet been well established. Therefore, it is necessary to identify novel biomarkers toaccurately predict GC development, prognosis, or response to treatment [5,6].
MicroRNAs (miRNAs) are a new class of endogenous small non-coding regulatory RNAs, which regulate gene expression at the posttranscriptional level [7-9]. MiRNAs play important roles in various biological processes such as cell proliferation, apoptosis, metabolism, and differentiation [2,10-12]. It is reported that miRNAs are aberrantly expressed in many cancers, affecting cellmigration, invasion, epithelial-to-mesenchymal transition, and metastasis in tumor [13-15]. Some miRNAs act as tumor suppressor genes or oncogenes in human malignancies [16-18]. However, the role of miRNAs in GC remains largely unknown.
Deregulation of miR-561 is a constant event in various cancers, suggesting that miR-561 may play an important role in tumor progression [19,20]. Yu et al. showed that miR-561 was down-regulated human colon cancer stem cells [21]. However, the expression and function of miR-561 in GC remain uninvestigated. In our study, miR-561 was downregulated in human GC. In addition, miR-561 was associated with lymph node metastasis in an independent GC cohort. Functional investigation indicated that miR-561 might act as a novel antimetastaticmiRNA in GC. Moreover, the c-Myc was identified as a direct and functional target of miR-561.
Materials and methods
Ethics statement
All patients agreed to participate in the study and gave written informed consent. Both this study and consent were approved by the ethical board of the The First Hospital of Chongqing University and complied with the Declaration of Helsinki.
Human samples
Human GCs and their corresponding non-tumorous gastric samples were collected duringsurgical resection from The First Hospital of Chongqing University from 2008 to 2009. Samples collected were immediately frozen in liquid nitrogen and subsequently stored in -80°C freezer. None of the patients had received radiotherapy or chemotherapy before surgery. The characteristics of the patients are described in Table S1.
Cell lines and cell culture
The following human cell lines were used: SGC-7901, HGC-27, MKN-45, MGC-803, GES and HEK293T. These cell lines were purchased from the Cell Resource Center of the Institute of Basic Medical Sciences at the Chinese Academy of Medical Sciences and the Peking Union Medical College (Beijing, China). Cells were propagated in Dulbecco’s modified Eagle’s medium (Gibco; Invitrogen; Life Technologies, Germany), which wassupplemented with 10% fetal bovine serum (GIBCO, NY, USA), streptomycin (100 μg/ml), and penicillin (100 U/ml).
Plasmids and cell transfection
MiR-561 mimic/inhibitor and controls were purchased from RiboBio (Guangzhou, China). HGC-27 cells were incubated in the six-well plates at 30% confluence one day before transfection. Transfection with miR-561 mimic/inhibitor or controls was performed using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Luciferase assay
The 3’UTRs of human c-Myc mRNAs were amplified by PCR and cloned into pRL-TK to generate the c-Myc reporter. Mutations in these mRNA sequences were prepared using the Quick Change SiteDirected Mutagenesis kit (Stratagene, CA, USA). The plasmid wascreated using the Plasmid Maxi Kit (Qiagen, CA, USA). Cells were first transfected in 12-well plates using appropriate plasmids. After transfection for 48 hours, cells were harvested and lysed for luciferase assay. Luciferase assays were performed by luciferase assay kit (Promega, Madison, WI). Renilla luciferase was chosenas the normalization.
RNA extraction and quantitative real-time PCR
Total RNA was isolated using TRIzol reagent (Invitrogen). To measure mRNA expression, quantitative real-time PCR (qRT-PCR) assays were performed using the PrimeScript RT Reagent Kit (TaKaRa) and SYBR Premix Ex Taq (TaKaRa). The expression of c-Myc was detectedby Real-Time PCR System real-time PCR System (Applied Biosystems, Carlsbad, USA) with SYBR Premix Ex Taq (Takara, Dalian, China). GAPDH was used as an internal control. The expression of mature miR-561 was detectedby TaqMan miRNA assays (Applied Biosystems). U6 small nuclear RNA was used as an internal control (Table S2).
Northern blot analysis
RNAs were electrophoresed on 15% acrylamide and 8 M urea denatures gels, and then transferred onto Hybond N membrane (Amersham Biosciences). The membranes were baked at 80°C for 2 hours. Subsequently, the membranes were hybridized with oligo-nucleotide probes of the miRNAs. The probes of the miRNA-561 were as follows: 5’-CAAAGUUUAAGAUCCUUGAAGU-3’. Probes were 5-end labeled using the polynucleotide kinase in the presence of [r-32P] ATP. Hybridization was done at 39°C in ULTRAhybTMOligo Hybridization Buffer (Ambion) for 16 hours. The membranes werewashed with SSC at 42°C for 3 times and subsequently rehybridized after stripping the oligo nucleotides used as probes in 1% SDS for 30 minutes at 65°C. The U6 RNA was used as an internal control.
Cell proliferation assay
Cells were plated in 96-well plates before transfection. Cells were cultured for 24 h in normal conditionsand then transfected with miR-561 mimic or antimiR-561 inhibitor along with paired negative controls. Cell proliferation was assessed using Cell Counting Kit 8 assay (Dojindo, Tokyo, Japan) according to manufacturer’s protocol.
Invasion assays
Cell invasion was assayed using a transwell chamber (Millipore, USA) with Matrigel (BD, Franklin Lakes, USA). Thetranswell chamber was coated with 30 μl Matrigel, placed into a 24-well plateand incubated for 40 minutes at 37°C. After 48 hours of transfection, cells (8 × 104 cells per well) were trypsinized and seeded in chambers, cultured inRPMI 1640 medium containing 2% serum and 600 μl of 10% FBS-1640 in the lower chamber for 24 h. Invasive cells were fixed with 100% methanol for 30 min and non-migrated cells were removed by cotton swabs. Cells on the bottom of the membrane were stained by 0.1% crystal violet (Sigma) for 20 min. Cell images were observed under a phase-contrast microscope (Olympus, Tokyo, Japan).
Western blot analysis
Western blot analysis was performed as follows. Proteins were separated by 10% SDS-PAGE and then transferred to nitrocellulose membrane (Bio-Rad). Membranes were blocked with 5% nonfat milk and incubated with the following primary antibodies: rabbit anti-c-Myc monoclonal antibody (1:500; Santa Cruz Biotechnology, USA), mouse anti-GAPDH mAb (1:10,000; Sigma). Enhanced Chemiluminescence (ECL) system (Pierce Biotechnology Inc, USA) was used to detect the bound secondary antibody.
Rescue assays of c-Myc gene expression
The full length of c-MyccDNAs (ORF and 3’UTR) was PCR-amplified and then cloned into the pcDNA 3.1 vector to generate the pcDNA-c-Myc constructs. The primers were used as follows: c-Myc-F (5’-TGCTAAGAAGATTGGTGCTGTA-3’) and c-Myc-R (5’-GCGAAGGGCTGAGACATTTAC). The plasmid was prepared using the Plasmid Maxi Kit (Qiagen, CA, USA). HGC-27 cells were first transfected with miR-561 or a scrambled dsRNAs (60 nM). Cells were cultured for 24 h and thenco-transfected with miR-561 (20 nM) and pcDNA-c-Myc (2.0 μg), miR-561 (20 nM) and pcDNA-empty (2.0 μg). At the indicated time points after hemin addition, cells were harvested and assayed as required.
Statistical analysis
Each experiment was repeated at least three times. Statistical analyses were performed using SPSS 15.0. Data are presented as mean ± standard deviation. Statistical analyses were performed with either an analysis of variance (ANOVA) or Student’s t-test. α = 0.05 (two-side) was set as the statistical significance level
Results
miR-561 was down-regulated in GC cell lines
As shown in Figure 1A, miR-561 was down-regulated in all the GC cell lines compared with GES. Northern blot also showed that the expression level of miR-561 was generally lower in the GC cell lines compared with GES (Figure 1B). Thus, we speculated that miR-561 might be a tumor suppressor in GC.
Figure 1.
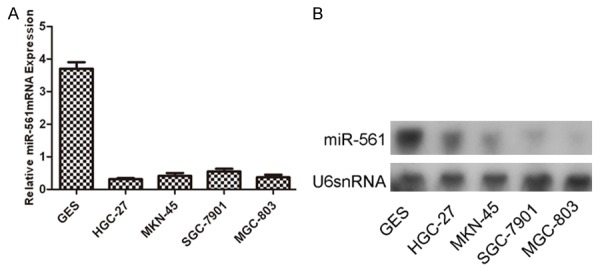
The expression of miR-561 is down-regulated in GC cell lines. A. Down-regulation of miR-561 expression in GC cell lines (SGC-7901, HGC-27, MKN-45, MGC-803) compared with the corresponding controls. Relative expression of miR-561 in four GC cell lines and one normal gastric epithelial-1 cells (GES) was determined by qRT-PCR. U6 snRNA was used as internal control. B. Northern blot analysis of miR-561 expression in four GC cell lines and one normal gastric epithelial-1 cells (GES). U6 was also detected as a loading control.
miR-561 expression was down-regulated in both GC tissues
In general, the expression of miR-561 in GC tissues was lower than in adjacent tissues (Figure 2A, p<0.001). miR-561 was down-regulated in 44 cases (44/50, 88%) compared with normal adjacent tissues (Figure 2B). In addition, the lower level expression of miR-561 was associated with pTNM stage (Figure 2C) and tumor metastasis (p≤0.001, metastasis vs. no metastasis) in GC patients (Figure 2D). These data suggested that alterations of miR-561 might be involved in GC progression and metastasis.
Figure 2.
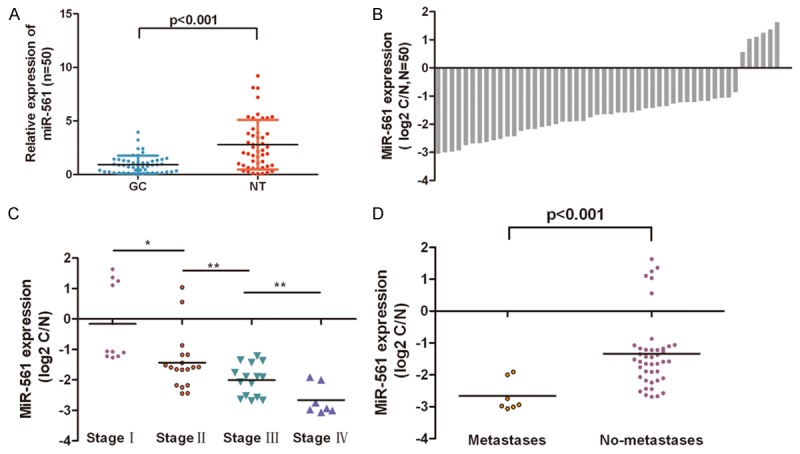
miR-561 expression was down-regulated in both GC tissues. A. Relative miR-561 expression levels in GC tissues and adjacent normal tissues (NT) were determined by qRT-PCR. U6 snRNA was used as internal control. B. miR-561 was detected in 50 pairs of GC tissues and its adjacent normal controls by quantitative RT-PCR. Data are presented as log2 of fold change of GC tissues relative to adjacent normal regions; C and D. The Statistical analysis of the association between miR-561 level and pTNM stage (I, II, III and IV) and pM stage (No metastasis and Metastasis); *p<0.05, and **p<0.01, ***p<0.001.
miR-561 inhibited GC cell proliferation and invasion
Cells were transfected with scrambled control oligo or miR-561 mimics, control and inhibitor, withhigh transfection efficiency (Figure 3A and 3C). CCK-8 proliferation assay showed that cell proliferation was inhibited in miR-561 mimics-transfected GC cells compared with scrambled oligo-transfected cells or untreated cells in the both HGC-27 cells and MGC-803 cells (Figure 3B). Conversely, miR-561 inhibitor increased cell proliferation of the HGC-27 cells and MGC-803 cells (Figure 3D). Intriguingly, invasion assay showed that overexpression of miR-561 inhibited the invasion of HGC-27 cells and MGC-803 cells compared with the control whereas miR-561 inhibitor promoted cell invasion (Figure 3E).
Figure 3.
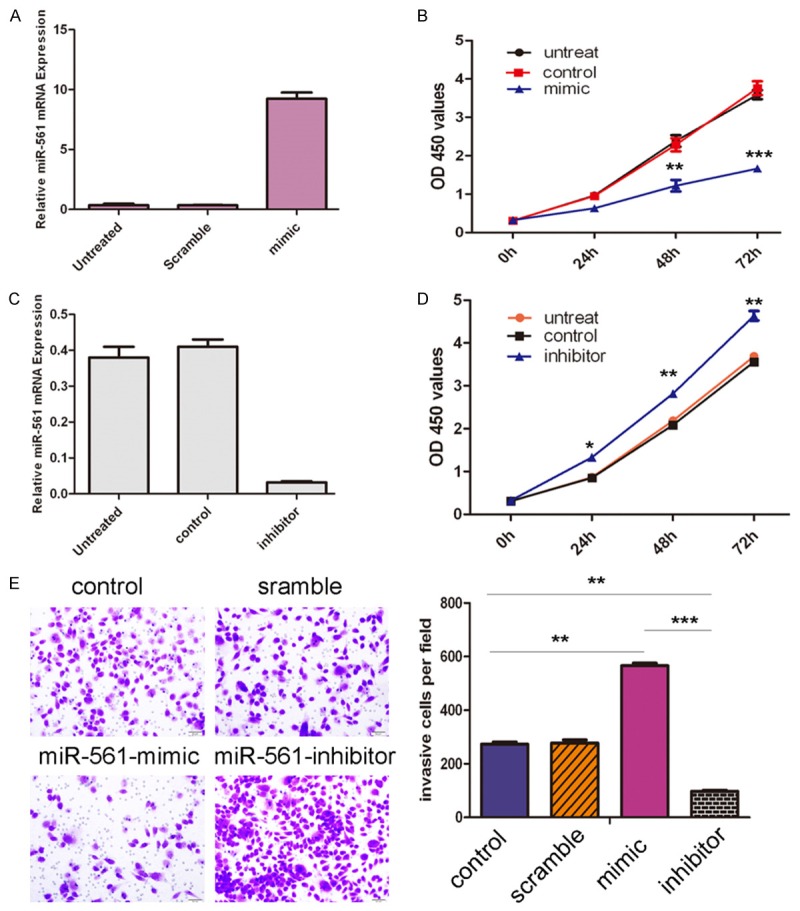
miR-561 inhibited GC cell proliferation and invasion. A. Expression levels of miR-561 were examined by real-time PCR after transfection of miR-561 mimics or scramble or no transfection. B. Growth of HGC-27 cells and MGC-803 cells were shown after transfection with miR-561 mimics or sramble or no transfection. The growth index as assessed at 0, 24, 48 and 72 h. C. Expression levels of miR-561 were examined by real-time PCR after transfection of miR-561 inhibitor or control or no transfection. D. Growth of HGC-27 cells and MGC-803 cells were shown after transfection with miR-561 inhibitor or control or no transfection. The growth index as assessed at 0, 24, 48 and 72 h. E. Transwell analysis of HGC-27 cells and MGC-803 cells after treatment with miR-561 mimics, inhibitors or scramble or control; the relative ratio of invasive cells per field is shown below, *p<0.05, **p<0.01, and ***p<0.001.
miR-561 downregulated c-Myc through interaction with its 3’UTR
Bioinformatics analysis using TargetScan showed that c-Myc was a potential target of miR-561 (Figure 4A). Real-time RT-PCR and Western blot analysis of c-Myc in HGC-7 cell lines showed that miR-561 mimic transfection inhibited c-Myc mRNA and protein expression (Figure 4C and 4D). The effect of miR-561 on the translation of c-Myc mRNA into protein was assessed by luciferase reporter assay (Figure 4B). Overexpression of miR-561 reduced luciferase activity of reporter gene with wild type, but not the mutant c-type, indicating that miR-561 directly targeted c-Myc 3’UTR.
Figure 4.
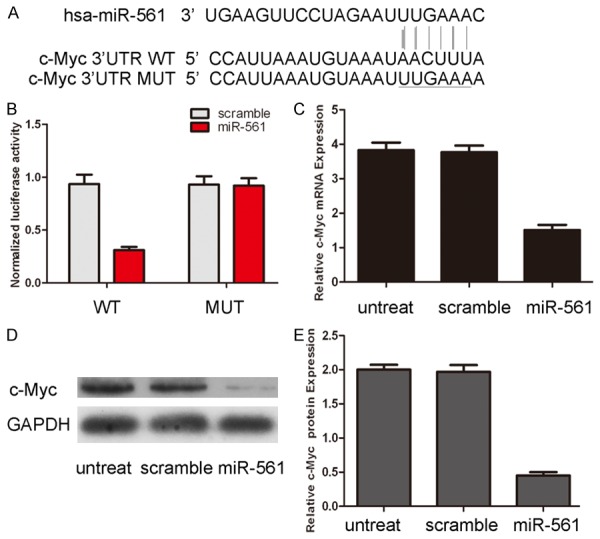
miR-561 downregulates c-Myc through interaction with its 3’UTR. A. The sequences of miR-561 binding sites within the human c-Myc 3’UTRs and schematic reporter constructs, in this panel, c-Myc-WT represent the reporter constructs containing the entire 3’UTR sequences of c-Myc. C-Myc-MUT represent the reporter constructs containing mutated nucleotides. B. The analysis of the relative luciferase activities of c-Myc-WT, c-Myc-MUT. The error bars are derived from triplicate experiments. C. qRT-PCR analysis of c-Myc mRNA expression in HGC-27 cells after treatment with miRNA mimics or scramble or no transfection. The expression of c-Myc was normalized to GAPDH. D. Western blot analysis of c-Myc expression in HGC-27 cells transfected with miR-561 mimics or scramble or no transfection. GAPDH was also detected as a loading control. E. The relative expression of c-Myc was shown.
Overexpression of c-Myc partially rescued miR-561-inhibited cell proliferation and invasion
C-Myc expression was restored by cotransfection of a construct expressing c-Myc and miR-561 in HGC-7 cells, as confirmed by Western blot (Figure 5A). This restoration of c-Myc increased the proliferation and invasive capabilities of GC cells. More importantly, restoration of c-Myc could reverse the proliferation and invasion inhibition imposed by miR-561 (Figure 5B and 5C). In summary, these data indicate that inhibition of c-Myc by miR-561 is responsible, at least in part, for the miR-561 inhibition of cell proliferation and invasion in human GC.
Figure 5.
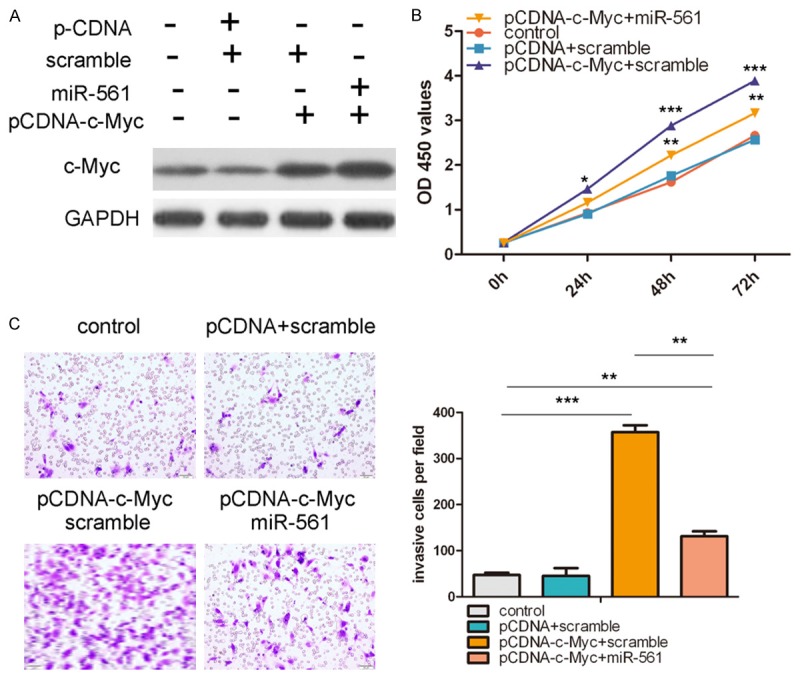
Overexpression of c-Myc partially rescued miR-561-inhibited cell proliferation and invasion. A. Western blot analysis of c-Myc in HGC-27 cells co-transfected with either miR-561 mimic or scramble and 2.0 μg pCDNA-c-Myc or pCDNA empty vector. GAPDH was also detected as a loading control. B. Cell growth curves in HGC-27 cells transfected with different combinations at 0, 24, 48 and 72 h. C. Transwell analysis of HGC-27 cells treated with different combinations. The relative ratio of invasive cells per field is shown right. *p<0.05, **p<0.01, ***p<0.001.
C-Myc was inversely expressed with miR-561 in GC
As shown in Figure 6A and 6B, the mRNA and protein expression of c-Myc was much higher in 4 GC cell lines than that of gastric epithelial-1 cells. We also found that the expression of c-Myc in GC tissues was higher than in the corresponding adjacent normal tissues (Figure 6C and 6D). The scatter plot and the Pearson correlation analysis further showed that miR-561 expression was negatively correlated with the level of c-Myc mRNA in the GC samples (Figure 6E). These data suggested that decreased miR-561 expression was related to increased c-Myc levels in most GC patients.
Figure 6.
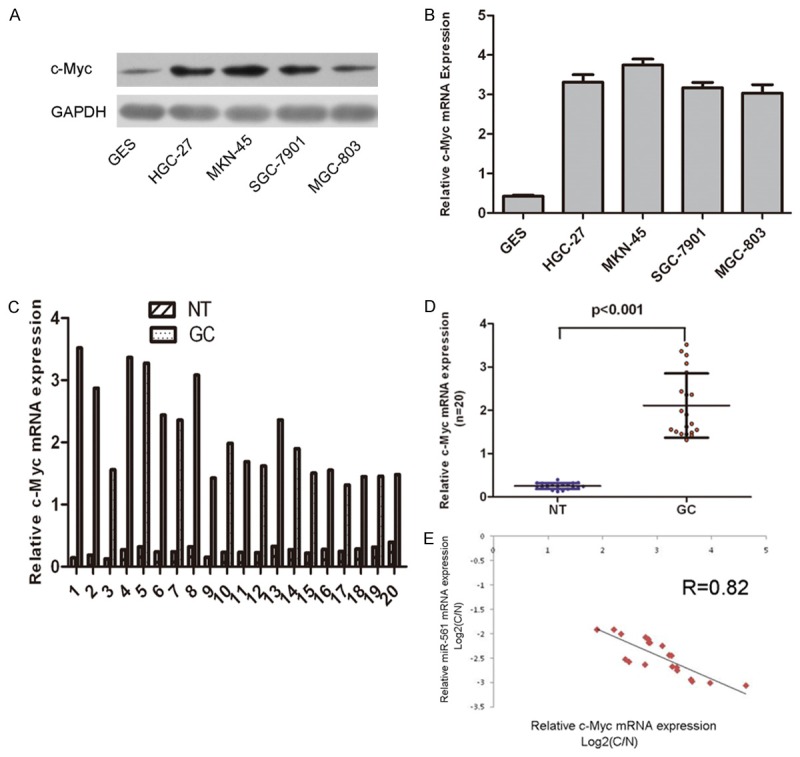
c-Myc was inversely expressed with miR-561 in GC. A. The relative c-Myc mRNA expression levels were determined by qRT-PCR in GC cell lines (SGC-7901, HGC-27, MKN-45, and MGC-803) and one normal gastric epithelial-1 cells (GES). B. Western blot analysis of c-Myc expression in GC cell lines (SGC-7901, HGC-27, MKN-45 and MGC-803) and one normal gastric epithelial-1 cells (GES). GAPDH was also detected as a loading control. C. qRT-PCR analysis of c-Myc expression in 20 pairs GC tissues and their corresponding adjacent normal gastric tissues. The expression of c-Myc was normalized to GAPDH. D. The expression of c-Myc in GC tissues was significant higher than in the corresponding adjacent normal tissues (NT). E. Analysis of correlation of miR-561 and c-Myc expression in GC tissues. (Two-tailed Pearson’s correlation analysis, r = -0.82; p<0.01). Data was presented as log2 of fold change of GC tissues relative to non-tumor adjacent tissues.
Discussion
The majority of cancer-related deaths are caused by metastasis [22]. Therefore, effective intervention targeting cancer metastasis might decrease the mortality of cancer patients. Increasing evidences suggest an important role of miRNAs in cancer metastasis, such as miR-10b, miR-181a and miR-409 [12,23-25]. However, the role of miRNAs in GC metastasis is still largely unknown. In our study, miR-561 was frequently downregulated in human GC cell lines and tissues and the downregulated miR-561 was associated with advanced clinical stage and lymph node-metastasis. Further studies showed that overexpression of miR-561 inhibited GC cell proliferation and invasion in vitro. Furthermore, c-Myc was identified as a direct and functional target of miR-561. The data from this study suggested that miR-561 acted as a novel tumor suppressor in GC and that downregulated miR-561 contributed to lymph node-metastasis and tumor progression in GC patients.
Deregulation of miR-561 is a frequent event in various cancers, suggesting that miR-561 may play an important role in tumor progression [19,20]. Yu and his colleagues showed that miR-561 was down-regulated human colon cancer stem cells [21]. Previous study also showed that DAX-1 was a target of miR-561, which modulated acetaminophen-induced hepatotoxicity through DAX-1-mediated modulation of NR transactivation, indicating that miR-561 may represent a novel therapeutic target to manage acetaminophen overdose poisoning [26]. However, the expression and function of miR-561 in GC development remainuninvestigated. Our results showed that miR-561 was down-regulated in 44 (44/50, 88%) GC tissues compared with the adjacent tissues and the expression of miR-561 in GC tissues was lower than in adjacent tissues. We also found that lower expression of miR-561 in GC specimens was correlated with pTNM stage. Forced expression of miR-561 inhibited cell proliferationand invasion in GC cell lines, indicating that repression of miR-561 might promote tumor progression in gastric carcinogenesis. All of this evidence indicated that miR-561 might contribute toGC development. miRNAdysregulation might result from various molecular mechanisms,, such as genetic mutation, epigenetic aberration and deregulated transcriptional activity [27,28]. Among them, epigenetic aberration may play critical roles in the transcriptional silencing of tumor suppressor genes or suppressor miRNAs by specific DNA methylation and histone modification in cancers [29]. Therefore, further experiments, such as DNA methylation and histone modification assays, might be necessary to pinpoint the molecular mechanisms of miR-561 dysregulation in GC.
We next try to identify the molecular mechanism by which miR-561 acts as a metastasis suppressor in GC. Luciferase assay and Western blot analysis showed that miR-561 inhibited the expression of c-Myc by directly targeting its 3’UTR. C-Myc is a key basic helix-loop-helix leucine zipper transcription factor that acts as an important regulator of several cellular processes, including cell migration, growth and proliferation in various cells [30-32]. It is overexpressed in a number of human cancers, and its overexpression contributes to malignant transformation by regulating the expression of a number of genes participating in multiple aspects of tumorigenesis, such as angiogenesis, cell cycle progression, cell invasion, migration, metastasis, and angiogenesis [33-35]. Recent studies have shown that c-Myc has important roles in GC development and progression [36]. Its high expression is correlated with advanced disease stage, lymph-node metastasis and poor survival rates [37,38]. Knockdown of c-Myc inhibited the invasion and metastasis of GC cell lines in vitro [39]. Consistent with that, we found that c-Myc was enriched in the primary GC tissues andinversely correlated to miR-561 levels. However, further studies with large number of primary GC samples were needed to confirm this interaction. These results demonstrate that miR-561 may act as a metastasis suppressor in gastric cancer by targeting c-Myc. Nevertheless, it should be noticed thatbesides c-Myc, lots of other genes could be regulated by miR-561. A more comprehensive profiling of gene dysregulation by microarray is needed to enhance the selection of target gene candidates for further functional analysis.
In conclusion, our study show that miR-561 is downregulated in GC cell lines and that low expression of miR-561 is associated with lymph-node metastasis, poor pTNM stage. Moreover, enforced expression of miR-561 suppressed GC cell proliferation and invasion through directly targeting c-Myc. miR-561 may act as an important tumor suppressor in GC.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Yang L. Incidence and mortality of gastric cancer in China. World J Gastroenterol. 2006;12:17–20. doi: 10.3748/wjg.v12.i1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao X, Yang L, Hu J. Down-regulation of miR-27a might inhibit proliferation and drug resistance of gastric cancer cells. J Exp Clin Cancer Res. 2011;30:55. doi: 10.1186/1756-9966-30-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X, Nie Y, Du Y, Cao J, Shen B, Li Y. MicroRNA-181a promotes gastric cancer by negatively regulating tumor suppressor KLF6. Tumour Biol. 2012;33:1589–1597. doi: 10.1007/s13277-012-0414-3. [DOI] [PubMed] [Google Scholar]
- 4.Zhang BG, Li JF, Yu BQ, Zhu ZG, Liu BY, Yan M. microRNA-21 promotes tumor proliferation and invasion in gastric cancer by targeting PTEN. Oncol Rep. 2012;27:1019–1026. doi: 10.3892/or.2012.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thiel A, Ristimaki A. Gastric cancer: basic aspects. Helicobacter. 2012;17(Suppl 1):26–29. doi: 10.1111/j.1523-5378.2012.00979.x. [DOI] [PubMed] [Google Scholar]
- 6.Gigek CO, Chen ES, Calcagno DQ, Wisnieski F, Burbano RR, Smith MA. Epigenetic mechanisms in gastric cancer. Epigenomics. 2012;4:279–294. doi: 10.2217/epi.12.22. [DOI] [PubMed] [Google Scholar]
- 7.Feng R, Chen X, Yu Y, Su L, Yu B, Li J, Cai Q, Yan M, Liu B, Zhu Z. miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett. 2010;298:50–63. doi: 10.1016/j.canlet.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Yu X, Li Z, Shen J, Wu WK, Liang J, Weng X, Qiu G. MicroRNA-10b Promotes Nucleus Pulposus Cell Proliferation through RhoC-Akt Pathway by Targeting HOXD10 in Intervetebral Disc Degeneration. PLoS One. 2013;8:e83080. doi: 10.1371/journal.pone.0083080. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Zheng F, Liao YJ, Cai MY, Liu YH, Liu TH, Chen SP, Bian XW, Guan XY, Lin MC, Zeng YX, Kung HF, Xie D. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut. 2012;61:278–289. doi: 10.1136/gut.2011.239145. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, Zhu W, Zhang J, Huo S, Zhou L, Gu Z, Zhang M. MicroRNA-650 targets ING4 to promote gastric cancer tumorigenicity. Biochem Biophys Res Commun. 2010;395:275–280. doi: 10.1016/j.bbrc.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z, Liu S, Shi R, Zhao G. miR-27 promotes human gastric cancer cell metastasis by inducing epithelial-to-mesenchymal transition. Cancer Genet. 2011;204:486–491. doi: 10.1016/j.cancergen.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Zheng B, Liang L, Huang S, Zha R, Liu L, Jia D, Tian Q, Wang Q, Wang C, Long Z, Zhou Y, Cao X, Du C, Shi Y, He X. MicroRNA-409 suppresses tumour cell invasion and metastasis by directly targeting radixin in gastric cancers. Oncogene. 2012;31:4509–4516. doi: 10.1038/onc.2011.581. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Lei H, Luo M, Wang Y, Dong L, Ma Y, Liu C, Song W, Wang F, Zhang J, Shen J, Yu J. DNA methylation downregulated mir-10b acts as a tumor suppressor in gastric cancer. Gastric Cancer. 2015;18:43–54. doi: 10.1007/s10120-014-0340-8. [DOI] [PubMed] [Google Scholar]
- 14.Zhang S, Shan C, Kong G, Du Y, Ye L, Zhang X. MicroRNA-520e suppresses growth of hepatoma cells by targeting the NF-kappaB-inducing kinase (NIK) Oncogene. 2012;31:3607–3620. doi: 10.1038/onc.2011.523. [DOI] [PubMed] [Google Scholar]
- 15.Zhou F, Zhu H, Luo D, Wang M, Dong X, Hong Y, Lu B, Zhou Y, Zhou J, Zhang Z, Gong W. A functional polymorphism in Pre-miR-146a is associated with susceptibility to gastric cancer in a Chinese population. DNA Cell Biol. 2012;31:1290–1295. doi: 10.1089/dna.2011.1596. [DOI] [PubMed] [Google Scholar]
- 16.Zhu LH, Liu T, Tang H, Tian RQ, Su C, Liu M, Li X. MicroRNA-23a promotes the growth of gastric adenocarcinoma cell line MGC803 and downregulates interleukin-6 receptor. FEBS J. 2010;277:3726–3734. doi: 10.1111/j.1742-4658.2010.07773.x. [DOI] [PubMed] [Google Scholar]
- 17.Yang X, Yu J, Yin J, Xiang Q, Tang H, Lei X. MiR-195 regulates cell apoptosis of human hepatocellular carcinoma cells by targeting LATS2. Pharmazie. 2012;67:645–651. [PubMed] [Google Scholar]
- 18.Yang Q, Jie Z, Cao H, Greenlee AR, Yang C, Zou F, Jiang Y. Low-level expression of let-7a in gastric cancer and its involvement in tumorigenesis by targeting RAB40C. Carcinogenesis. 2011;32:713–722. doi: 10.1093/carcin/bgr035. [DOI] [PubMed] [Google Scholar]
- 19.Kheirelseid EA, Miller N, Chang KH, Curran C, Hennessey E, Sheehan M, Newell J, Lemetre C, Balls G, Kerin MJ. miRNA expressions in rectal cancer as predictors of response to neoadjuvant chemoradiation therapy. Int J Colorectal Dis. 2013;28:247–260. doi: 10.1007/s00384-012-1549-9. [DOI] [PubMed] [Google Scholar]
- 20.Han Y, Staab-Weijnitz CA, Xiong G, Maser E. Identification of microRNAs as a potential novel regulatory mechanism in HSD11B1 expression. J Steroid Biochem Mol Biol. 2013;133:129–139. doi: 10.1016/j.jsbmb.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 21.Yu XF, Zou J, Bao ZJ, Dong J. miR-93 suppresses proliferation and colony formation of human colon cancer stem cells. World J Gastroenterol. 2011;17:4711–4717. doi: 10.3748/wjg.v17.i42.4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y, Zhao F, Wang Z, Song Y, Luo Y, Zhang X, Jiang L, Sun Z, Miao Z, Xu H. MicroRNA-335 acts as a metastasis suppressor in gastric cancer by targeting Bcl-w and specificity protein 1. Oncogene. 2012;31:1398–1407. doi: 10.1038/onc.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakayama I, Shibazaki M, Yashima-Abo A, Miura F, Sugiyama T, Masuda T, Maesawa C. Loss of HOXD10 expression induced by upregulation of miR-10b accelerates the migration and invasion activities of ovarian cancer cells. Int J Oncol. 2013;43:63–71. doi: 10.3892/ijo.2013.1935. [DOI] [PubMed] [Google Scholar]
- 24.Ouyang H, Gore J, Deitz S, Korc M. microRNA-10b enhances pancreatic cancer cell invasion by suppressing TIP30 expression and promoting EGF and TGF-beta actions. Oncogene. 2014;33:4664–74. doi: 10.1038/onc.2013.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pichler M, Winter E, Ress AL, Bauernhofer T, Gerger A, Kiesslich T, Lax S, Samonigg H, Hoefler G. miR-181a is associated with poor clinical outcome in patients with colorectal cancer treated with EGFR inhibitor. J Clin Pathol. 2014;67:198–203. doi: 10.1136/jclinpath-2013-201904. [DOI] [PubMed] [Google Scholar]
- 26.Li M, Yang Y, He ZX, Zhou ZW, Yang T, Guo P, Zhang X, Zhou SF. MicroRNA-561 promotes acetaminophen-induced hepatotoxicity in HepG2 cells and primary human hepatocytes through downregulation of the nuclear receptor corepressor dosage-sensitive sex-reversal adrenal hypoplasia congenital critical region on the X chromosome, gene 1 (DAX-1) Drug Metab Dispos. 2014;42:44–61. doi: 10.1124/dmd.113.052670. [DOI] [PubMed] [Google Scholar]
- 27.Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J, Prosper F, Garcia-Foncillas J. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125:2737–2743. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- 28.Breving K, Esquela-Kerscher A. The complexities of microRNA regulation: mirandering around the rules. Int J Biochem Cell Biol. 2010;42:1316–1329. doi: 10.1016/j.biocel.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 29.Ando T, Yoshida T, Enomoto S, Asada K, Tatematsu M, Ichinose M, Sugiyama T, Ushijima T. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer. 2009;124:2367–2374. doi: 10.1002/ijc.24219. [DOI] [PubMed] [Google Scholar]
- 30.Gamberi G, Benassi MS, Bohling T, Ragazzini P, Molendini L, Sollazzo MR, Pompetti F, Merli M, Magagnoli G, Balladelli A, Picci P. C-myc and c-fos in human osteosarcoma: prognostic value of mRNA and protein expression. Oncology. 1998;55:556–563. doi: 10.1159/000011912. [DOI] [PubMed] [Google Scholar]
- 31.Ikeda S, Sumii H, Akiyama K, Watanabe S, Ito S, Inoue H, Takechi H, Tanabe G, Oda T. Amplification of both c-myc and c-raf-1 oncogenes in a human osteosarcoma. Jpn J Cancer Res. 1989;80:6–9. doi: 10.1111/j.1349-7006.1989.tb02236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frick KK, Scher CD. Platelet-derived growth factor-stimulated c-myc RNA accumulation in MG-63 human osteosarcoma cells is independent of both protein kinase A and protein kinase C. Mol Cell Biol. 1990;10:184–192. doi: 10.1128/mcb.10.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun T, Wang C, Xing J, Wu D. miR-429 modulates the expression of c-myc in human gastric carcinoma cells. Eur J Cancer. 2011;47:2552–2559. doi: 10.1016/j.ejca.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 34.Shi JW, Liu W, Zhang TT, Wang SC, Lin XL, Li J, Jia JS, Sheng HF, Yao ZF, Zhao WT, Zhao ZL, Xie RY, Yang S, Gao F, Fan QR, Zhang MY, Yue M, Yuan J, Gu WW, Yao KT, Xiao D. The enforced expression of c-Myc in pig fibroblasts triggers mesenchymal-epithelial transition (MET) via F-actin reorganization and RhoA/Rock pathway inactivation. Cell Cycle. 2013;12:1119–27. doi: 10.4161/cc.24164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie X, Ye Z, Yang D, Tao H. Effects of combined c-myc and Bmi-1 siRNAs on the growth and chemosensitivity of MG-63 osteosarcoma cells. Mol Med Rep. 2013;8:168–172. doi: 10.3892/mmr.2013.1484. [DOI] [PubMed] [Google Scholar]
- 36.Calcagno DQ, Leal MF, Assumpcao PP, Smith MA, Burbano RR. MYC and gastric adenocarcinoma carcinogenesis. World J Gastroenterol. 2008;14:5962–5968. doi: 10.3748/wjg.14.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han S, Kim HY, Park K, Cho HJ, Lee MS, Kim HJ, Kim YD. c-Myc expression is related with cell proliferation and associated with poor clinical outcome in human gastric cancer. J Korean Med Sci. 1999;14:526–530. doi: 10.3346/jkms.1999.14.5.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khanna A, Bockelman C, Hemmes A, Junttila MR, Wiksten JP, Lundin M, Junnila S, Murphy DJ, Evan GI, Haglund C, Westermarck J, Ristimaki A. MYC-dependent regulation and prognostic role of CIP2A in gastric cancer. J Natl Cancer Inst. 2009;101:793–805. doi: 10.1093/jnci/djp103. [DOI] [PubMed] [Google Scholar]
- 39.Labisso WL, Wirth M, Stojanovic N, Stauber RH, Schnieke A, Schmid RM, Kramer OH, Saur D, Schneider G. MYC directs transcription of MCL1 and eIF4E genes to control sensitivity of gastric cancer cells toward HDAC inhibitors. Cell Cycle. 2012;11:1593–1602. doi: 10.4161/cc.20008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


