Abstract
The RNA-guided clustered regularly interspaced short palindromic (CRISPR) in combination with a CRISPR-associated nuclease 9 (Cas9) nuclease system is a new rapid and precise technology for genome editing. In the present study, we applied the CRISPR/Cas9 system to target ABCB1 (also named MDR1) gene which encodes a 170 kDa transmembrane glycoprotein (P-glycoprotein/P-gp) transporting multiple types of chemotherapeutic drugs including taxanes, epipodophyllotoxins, vinca alkaloids and anthracyclines out of cells to contribute multidrug resistance (MDR) in cancer cells. Our data showed that knockout of ABCB1 by CRISPR/Cas9 system was succesfully archieved with two target sgRNAs in two MDR cancer cells due to the alteration of genome sequences. Knockout of ABCB1 by CRISPR/Cas9 system significantly enhances the sensitivity of ABCB1 substrate chemotherapeutic agents and the intracellular accumulation of rhodamine 123 and doxorubicin in MDR cancer cells. Although now there are lots of limitations to the application of CRISPR/Cas9 for editing cancer genes in human patients, our study provides valuable clues for the use of the CRISPR/Cas9 technology in the investigation and conquest of cancer MDR.
Keywords: ABCB1, tumor, multidrug resistance, CRISPR/Cas9
Introduction
Genome engineering is a highly desirable technology in biological research and biomedical applications, allowing us to change genomic sequences at will to control the fundamental genetic information within a cell [1]. It has been implemented in creating model cell lines or organisms for the study of gene functions or genetics of human diseases, and in gene therapy to correct deleterious genomic changes or confer beneficial ones [2]. Genome engineering using engineered nucleases has been one of the most designable and scalable paths to achieve precise editing of genomic sequences. A number of genome editing technologies have emerged in recent years, including zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and the RNA-guided clustered regularly interspaced short palindromic (CRISPR) in combination with a CRISPR-associated nuclease 9 (Cas9) nuclease system [3]. The first two technologies use a strategy of tethering endonuclease catalytic domains to modular DNA binding proteins for inducing targeted DNA double-stranded breaks (DSBs) at specific genomic loci. By contrast, CRISPR/Cas9 is an adaptive immune system that exists in a variety of microbes which was used for eukaryotic genome engineering [4]. CRISPR/Cas was first discovered as a bacterial defense mechanism against foreign DNA [5]. The CRISPR locus in microbes typically consists of a set of noncoding RNA elements and enzymes that harbors the ability to recognize and cleave foreign nucleic acids based on their sequence signature [6,7]. Three known CRISPR systems, types I, II, and III, have been described so far. Among these different CRISPR systems, type II CRISPR systems usually only require a single protein Cas9 to perform the target cleavage [8]. The main endonucleases Cas9 in the type II CRISPR system has been used to obtain DNA deletion and insertion, gene mutation, as well as transcriptional activation and repression, with multiplex targeting ability, just leading by a 20-nt guide RNA component and cutting at the third nucleotide before protospacer adjacent motif (PAM) “NGG” [9]. The rapid development of CRISPR-Cas system results in an explosion of interest in the use of it in microbes, plants, animals and humans.
In the present study, we applied the CRISPR/Cas9 system to target ABCB1 (also named MDR1) gene which encodes a 170 kDa transmembrane glycoprotein (P-glycoprotein/P-gp) transporting multiple types of chemotherapeutic drugs including taxanes, epipodophyllotoxins, vinca alkaloids and anthracyclines out of cells to contribute multidrug resistance (MDR) in cancer cells.
Materials and methods
Cell culture and reagents
The two ABCB1-overexpressing MDR cell lines KBV200 and HCT-8/V were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/ml) and streptomycin (100 ng/ml) in a humidified incubator at 37°C with 5% CO2. Restriction endonuclease BsmBI was from New England Biolabs. Polyetherimide (PEI) was from Ploysciences. Vincristine, doxorubicin and cisplatin were ordered from LC Laboratories. Puromycin was from Selleck Chemicals. Methylthiazolyldiphenyl-tetrazolium bromide (MTT) was from ApexBio Technology. Rhodamine 123 and other chemicals were from Sigma-Aldrich. Anti-ABCB1 (SC-13131) and anti-14-3-3 (SC-629) antibodies were from Santa Cruz Biotechnology.
Vector generation, lentivirus production and transduction
LentiCRISPRv2 vector from Addgene (#52961) was digested with BsmBI and ligated with annealed oligonucleotides (ABCB1-sg1 and ABCB1-sg1; Table 1). Human embryonic kidney (HEK) cell line 293T were transfected using PEI at 70% confluency with LentiCRISPRv2 and packaging vectors pMD2G and psPAX2. Viral supernatant was harvested 48 hours after transfection and stored at -80°C. After transduction with viral supernatant containing 10 μg/ml polybrene, KBV200 and HCT-8/V cells were selected with 50 μg/ml puromycin to generate the stable cell lines.
Table 1.
Summary of the sequences of ABCB1 sgRNAs and primers
| Primer | Sequence (5’ to 3’) |
|---|---|
| Sg 1-F | CACCGTTGGACTGTCAGCTGCTGTC |
| Sg 1-R | AAACGACAGCAGCTGACAGTCCAAC |
| Sg 2-F | CACCGTGGACTTCCTCTCATGATGC |
| Sg 2-R | AAACGCATCATGAGAGGAAGTCCAC |
| Detection 1-F | TAGGTTGAATACTTCTTGTGTACAC |
| Detection 1-R | AAATAATCACATGGCCAAGAACAGC |
| Detection 2-F | TATATACCATTAAATACTTTTACAG |
| Detection 2-R | CTAATTAAGTTCCTATATTTCTTCT |
Western blot
Cells were harvested and lysed in RIPA buffer (1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 10 ng/ml PMSF, 0.03% aprotinin, 1 μM sodium orthovanadate) at 4°C for 30 minutes. After centrifuged for 10 minutes at 14,000×g, supernatants were collected. Protein concentration was quantified using with Bradford assay. Proteins were separated on 12% SDS-PAGE gels and transferred to polyvinylidene difluoride membranes. Membranes were blocked with 5% BSA and incubated with the indicated primary antibodies. Corresponding horseradish peroxidase-conjugated secondary antibodies were used against each primary antibody. Proteins were detected using the chemiluminescent detection reagents and films [10,11].
Genomic PCR and sequencing analysis
The genomic DNA of cells was extracted with the QuickExtract DNA extraction kit following the manufacturer’s protocol and amplified with a pair of primers (ABCB1-Detection 1 and ABCB1-Detection 2; Table 1) designed for the target region of interest using a Pfu DNA polymerase. Followed by agarose gel electrophoresis and ethidium bromide staining, the purified PCR products were sequencing with an ABI 3131xl Genetic analyzer.
Cell viability assay
Cells were harvested with trypsin and resuspended in fresh culture medium, then seeded into a 96-well plate at a density of 5000 cells/well. After incubation for 72 hours, added 10 µl of MTT solution (0.5 mg/ml) to each well, and then incubation for 4 hours in a 37°C incubator containing 5% CO2, subsequently the medium was discarded and 50 μl of dimethylsulfoxide (DMSO) were added into each well to dissolve the formazan crystals, and absorbance at 570 nm was measured by plate reader. The concentrations required to inhibit growth by 50% (IC50) were calculated from survival curves using the Bliss method [12,13].
Rhodamine 123 and doxorubicin accumulation
The intracellular accumulation of rhodamine 123 and doxorubicin in cells were performed as previously described. The cell was seeded into a 6-well plate at a density of 2.5×105 cells/well. After adhered, the cells were then incubated with 10 μM rhodamine 123 or doxorubicin for 2 hours at 37°C. After washing three times with ice-cold PBS, cells were analyzed with FCM as previously described [14,15].
Statistical analysis
A student’s t-test was used to compare individual data points among each group. A P-value of <0.05 was set as the criterion for statistical significance.
Results
Generate CRISPR/Cas9 vector to target ABCB1
To target ABCB1 with CRISPR/Cas9 system, we used LentiCRISPRv2 vector which expresses both hSpCas9 and the chimeric guide RNA (Figure 1A) [16]. The two targeting sequences from exon 5 and 8 of ABCB1 end with a 5’NGG3’ PAM (protospacer-adjacent motif) sequence were cloned into LentiCRISPRv2 vector respectively (Figure 1B). After identified by sequencing, the two vectors expressed sgRNA1 (sg1) or sgRNA2 (sg2) to target ABCB1 were successfully generated.
Figure 1.
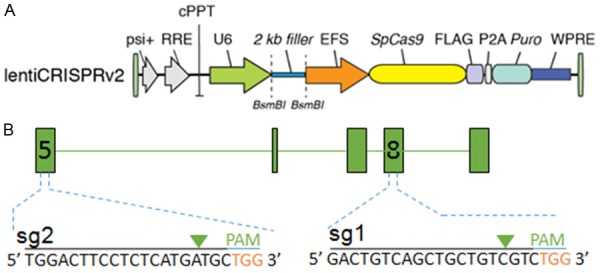
Generate CRISPR/Cas9 vector to target ABCB1. The map of LentiCRISPRv2 vector (A) and the locations and sequences of two sgRNAs for targeting ABCB1 (B) are shown.
Stable knockout of ABCB1 by CRISPR/Cas9 system
To establish cell lines stably expressed sgRNA to target ABCB1, the two ABCB1-overexpressing MDR cell lines KBV200 and HCT-8/V cells were selected with puromycin after transduction with LentiCRISPRv2 viral supernatant. The proteins of stable cell lines were isolated and analyzed by western blotting. As shown in Figure 2A, the protein levels of ABCB1 were undetectable in both KBV200 and HCT-8/V cells stably expressed either sg1 or sg2. To further identify the genomic change of targeting ABCB1 by CRISPR/Cas9 system, the genomic DNA of cells was extracted and amplified using the designed primers by PCR reaction. The sequencing results of PCR productions showed that there were an insert “T” before 4 nuclotides of PAM in the genomes of both KBV200 and HCT-8/V cells stably expressed sg1, and an mutant from “CT” to “GC” before 1 nuclotides of PAM in the target genomes of both KBV200 and HCT-8/V cells stably expressed sg2 (Figure 2B and 2C). These data suggest that cells with stable knockout of ABCB1 by CRISPR-Cas9 system were successfully established.
Figure 2.
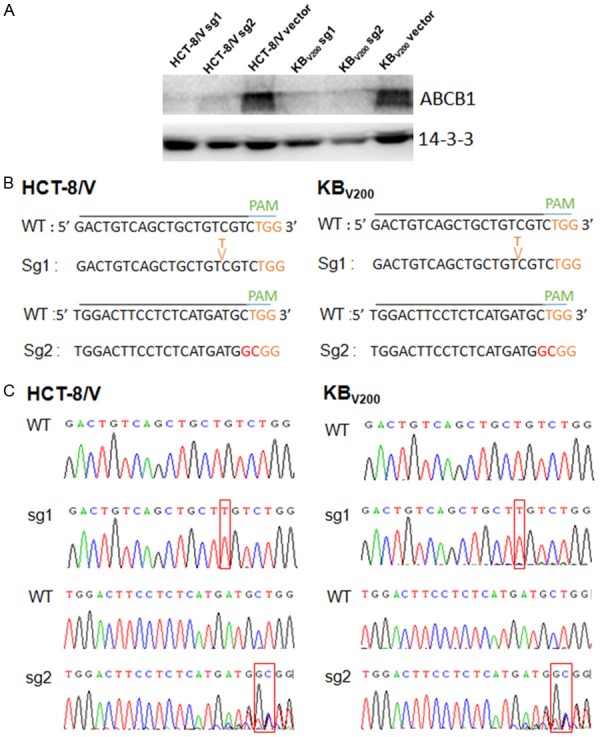
Stable knockout of ABCB1 by CRISPR/Cas9 system. KBV200 and HCT-8/V cells were selected with puromycin after transduction with LentiCRISPRv2 viral supernatant. The protein expression was examined by Western blot after lysing cells, and 14-3-3 was used as loading control. The genomic DNA of cells was amplified and sequenced by the designed primers. The representative Western blot results (A), sequencing comparison (B) and original data (C) are shown.
Knockout of ABCB1 by CRISPR/Cas9 system enhances the sensitivity of ABCB1 substrate chemotherapeutic agents in MDR cancer cells
To investigate the effects of knockout of ABCB1 by CRISPR/Cas9 system on the drug sensitivity in MDR cancer cells, we tested the cytotoxicity of two ABCB1 substrates vincristine and doxorubicin and one non-ABCB1 substrate cisplatin at the various concentrations in KBV200 and HCT-8/V cells with or without knockout of ABCB1. As shown in Figure 3, both KBV200 and HCT-8/V cells stably expressed either sg1 or sg2 were more sensitive to vincristine and doxorubicin but not to cisplatin. These results suggest that knockout of ABCB1 by CRISPR/Cas9 system could enhance the sensitivity of ABCB1 substrate chemotherapeutic agents in MDR cancer cells.
Figure 3.
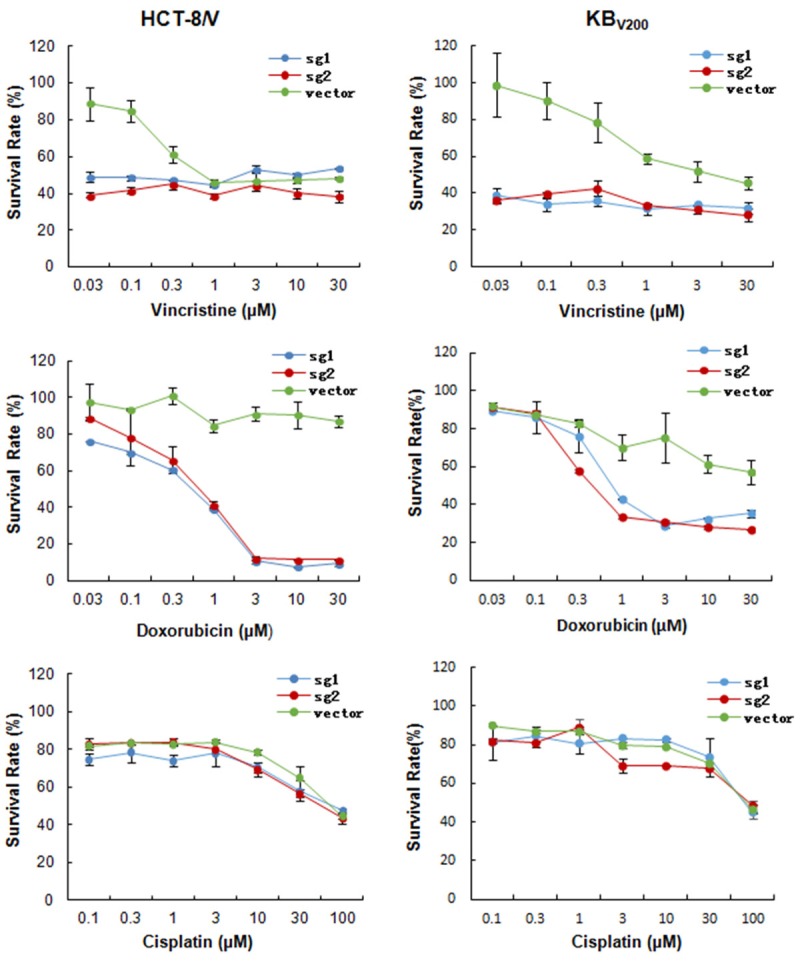
Knockout of ABCB1 by CRISPR/Cas9 system enhances the sensitivity of ABCB1 substrate chemotherapeutic agents in MDR cancer cells. Cells were treated with the indicated concentrations of vincristine, doxorubicin or cisplatin for 72 hours, and cell survival was measured by MTT assay. The representative growth curve of KBV200 and HCT-8/V treated with vincristine, doxorubicin and cisplatin are shown.
Knockout of ABCB1 by CRISPR/Cas9 system increases the intracellular accumulation of rhodamine 123 and doxorubicin in MDR cancer cells
To examine the effects of knockout of ABCB1 by CRISPR/Cas9 system on the intracellular drug accumulation in MDR cancer cells, we detected the intracellular accumulation of two ABCB1 substrates rhodamine 123 and doxorubicin in KBV200 and HCT-8/V cells with or without knockout of ABCB1. As shown in Figure 4A-C, both KBV200 and HCT-8/V cells stably expressed either sg1 or sg2 had more intracellular accumulation of rhodamine 123 and doxorubicin. These data suggest that knockout of ABCB1 by CRISPR/Cas9 system could increase the intracellular accumulation of rhodamine 123 and doxorubicin in MDR cancer cells.
Figure 4.
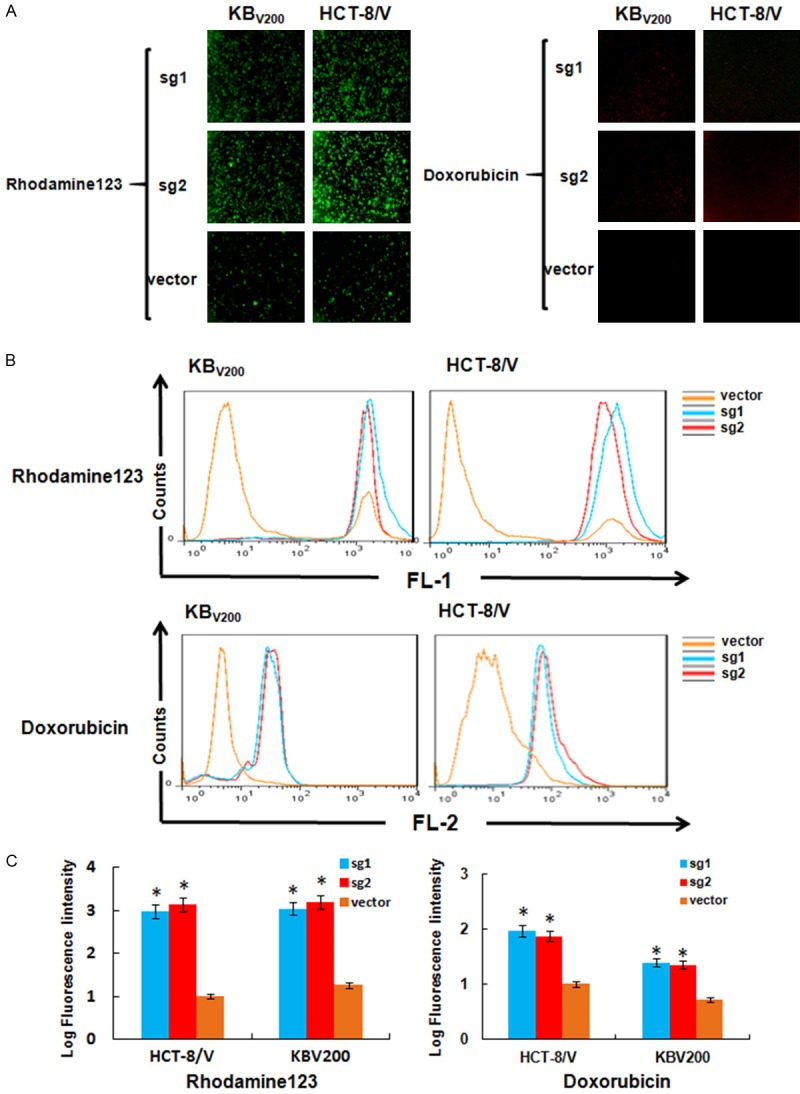
Knockout of ABCB1 by CRISPR/Cas9 system increases the intracellular accumulation of rhodamine 123 and doxorubicin in MDR cancer cells. Cells were incubated with 10 μM rhodamine 123 or doxorubicin for 2 hours at 37°C, measured by FCM and photographed by fluorescent microscope. The representative graphs (A), charts (B) and quantified data (C) are shown. *P<0.05 vs. corresponding control.
Discussion
MDR is a phenomenon wherein patients do not respond to chemotherapy from multiple anticancer drugs with diverse structures and mechanisms of action, and is one of the main reasons of failure of cancer chemotherapy [17]. Overexpressing the adenine triphosphate (ATP)-binding cassette (ABC) transporters, especially ABCB1, is one of common reasons to contribute MDR in cancer cells [18,19]. Human ABCB1 gene encodes a 170 kDa transmembrane glycoprotein which expresses in a variety of normal tissues such as the apical membrane of blood-brain barrier, kidney proximal tubule epithelial cells and small intestine to protect the body from xenobiotics and regulate drug absorption and disposition [20]. ABCB1 can transport multiple types of chemotherapeutic drugs out of cells, such as the taxanes, epipodophyllotoxins, vinca alkaloids and anthracyclines, and this process is coupled to the energy of ATP hydrolysis on the ATPase domain [21,22]. Therefore, inhibition of ABCB1 expression and activity may be an effective approach to overcome MDR in cancer chemotherapy. We and others have developed a lot of inhibitors to suppress the expression and activity of ABCB1, including small molecular compounds wallichinine [23], sipholenol A [24], UMMS-4 [25], sildenafil [26], trametinib [14], erlotinib [27], AG1478 [28], and biotechnological strategies antibodies [29], antisense oligonucleotides [30], RNA interference [15], etc. However, there was no inhibitors of ABCB1 succesfully applied in clinical today. Consequently, it is necessary to develop new approaches to target ABCB1 and reverse cancer MDR.
In the current study, we used the novel genome editing technology CRISPR/Cas system to target ABCB1. Our data showed that knockout of ABCB1 by CRISPR/Cas9 system was succesfully archieved with two target sgRNAs in two MDR cancer cells due to the alteration of genome sequences. Knockout of ABCB1 by CRISPR/Cas9 system significantly enhances the sensitivity of ABCB1 substrate chemotherapeutic agents and the intracellular accumulation of rhodamine 123 and doxorubicin in MDR cancer cells. Recently, Simoff et al also reported complete knockout of endogenous ABCB1 in MDCK cells by CRISPR/Cas9 system [31]. Many groups have used CRISPR/Cas9 system for precisely and rapidly engineering both loss-of-function and gain-of-function mutations in tumor-suppressor genes, oncogenes and other regulator genes of cellular transformation or drug response. For example, CRISPR/Cas9 system has been applied to model colorectal cancer from normal human intestinal epithelium organoids by introducing mutations in the tumor suppressor genes APC, SMAD4 and TP53, and oncogenes KRAS and/or PIK3CA [32,33]. Xue et al have generated liver tumors in mice by using hydrodynamic injection to deliver a CRISPR/Cas9 plasmid and sgRNAs to the liver that directly target the tumour suppressor genes PTEN and p53 [34]. The Cre-dependent CRISPR/Cas9 knockin mouse has lung adenocarcinoma formation by delivery of a single AAV vector expressing sgRNAs in the lung to target KRAS, p53, and LKB1 [35]. A genome-wide CRISPR screen finds that loss-of-function mutations of a small set of genes can drive tumor growth and metastasis after in a mouse model [36]. Another genome-wide CRISPR screen identifies CDC25A as a determinant of sensitivity to ATR Inhibitors [37]. CRISPR/Cas9 system can mediate efficient PD-1 disruption on human primary T cells from cancer patients [38]. Although now there are lots of limitations to the application of CRISPR/Cas9 for editing cancer genes in human patients, the use of CRISPR/Cas9 system provides numerous great opportunities for better understanding and treating cancer.
In summary, our results demonstrated that targeting ABCB1 by CRISPR/Cas9-based genome editing causes knockout of ABCB1 and reverses ABCB1-mediated MDR in cancer cells, resulting in the increase of the sensitivity and intracellular accumulation of the anti-cancer drugs. Our study provides valuable clues for the use of the CRISPR/Cas9 technology in the investigation and conquest of cancer MDR.
Acknowledgements
This work was supported by funds from the National Natural Science Foundation of China No. 31271444 and No. 81201726 (Z. S.), the Guangdong Natural Science Funds for Distinguished Young Scholar No. 2014A030306001 (Z. S.), the Guangdong Special Support Program for Young Talent No. 2015TQ01R350 (Z. S.) and the Science and Technology Program of Guangdong No. 2016A050502027 (Z. S.).
Disclosure of conflict of interest
None.
References
- 1.Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fujii M, Matano M, Nanki K, Sato T. Efficient genetic engineering of human intestinal organoids using electroporation. Nat Protoc. 2015;10:1474–1485. doi: 10.1038/nprot.2015.088. [DOI] [PubMed] [Google Scholar]
- 3.Kumar V, Jain M. The CRISPR-Cas system for plant genome editing: advances and opportunities. J Exp Bot. 2015;66:47–57. doi: 10.1093/jxb/eru429. [DOI] [PubMed] [Google Scholar]
- 4.Xie S, Shen B, Zhang C, Huang X, Zhang Y. sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS One. 2014;9:e100448. doi: 10.1371/journal.pone.0100448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 6.Sakuma T, Nishikawa A, Kume S, Chayama K, Yamamoto T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci Rep. 2014;4:5400. doi: 10.1038/srep05400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charpentier E, Richter H, van der Oost J, White MF. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol Rev. 2015;39:428–441. doi: 10.1093/femsre/fuv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Z, Park HR, Du Y, Li Z, Cheng K, Sun SY, Fu H, Khuri FR. Cables1 complex couples survival signaling to the cell death machinery. Cancer Res. 2015;75:147–158. doi: 10.1158/0008-5472.CAN-14-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Z, Li Z, Li ZJ, Cheng K, Du Y, Fu H, Khuri FR. Cables1 controls p21/Cip1 protein stability by antagonizing proteasome subunit alpha type 3. Oncogene. 2015;34:2538–2545. doi: 10.1038/onc.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mei XL, Yang Y, Zhang YJ, Li Y, Zhao JM, Qiu JG, Zhang WJ, Xue YQ, Zheng DW, Chen Y, Qin WM, Wei MN, Shi Z. Sildenafil inhibits the growth of human colorectal cancer in vitro and in vivo . Am J Cancer Res. 2015;5:3311–3324. [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang QW, Cheng KJ, Mei XL, Qiu JG, Zhang WJ, Xue YQ, Qin WM, Yang Y, Zheng DW, Chen Y, Wei MN, Zhang X, Lv M, Chen MW, Wei X, Shi Z. Synergistic anticancer effects of triptolide and celastrol, two main compounds from thunder god vine. Oncotarget. 2015;6:32790–32804. doi: 10.18632/oncotarget.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu JG, Zhang YJ, Li Y, Zhao JM, Zhang WJ, Jiang QW, Mei XL, Xue YQ, Qin WM, Yang Y, Zheng DW, Chen Y, Wei MN, Shi Z. Trametinib modulates cancer multidrug resistance by targeting ABCB1 transporter. Oncotarget. 2015;6:15494–15509. doi: 10.18632/oncotarget.3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi Z, Liang YJ, Chen ZS, Wang XW, Wang XH, Ding Y, Chen LM, Yang XP, Fu LW. Reversal of MDR1/P-glycoprotein-mediated multidrug resistance by vector-based RNA interference in vitro and in vivo. Cancer Biol Ther. 2006;5:39–47. doi: 10.4161/cbt.5.1.2236. [DOI] [PubMed] [Google Scholar]
- 16.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baguley BC. Multiple drug resistance mechanisms in cancer. Mol Biotechnol. 2010;46:308–316. doi: 10.1007/s12033-010-9321-2. [DOI] [PubMed] [Google Scholar]
- 18.Juliano RL, Ling V. A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim Biophys Acta. 1976;455:152–162. doi: 10.1016/0005-2736(76)90160-7. [DOI] [PubMed] [Google Scholar]
- 19.Gottesman MM, Ling V. The molecular basis of multidrug resistance in cancer: the early years of P-glycoprotein research. FEBS Lett. 2006;580:998–1009. doi: 10.1016/j.febslet.2005.12.060. [DOI] [PubMed] [Google Scholar]
- 20.International Transporter C, Giacomini KM, Huang SM, Tweedie DJ, Benet LZ, Brouwer KL, Chu X, Dahlin A, Evers R, Fischer V, Hillgren KM, Hoffmaster KA, Ishikawa T, Keppler D, Kim RB, Lee CA, Niemi M, Polli JW, Sugiyama Y, Swaan PW, Ware JA, Wright SH, Yee SW, Zamek-Gliszczynski MJ, Zhang L. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;9:215–236. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schinkel AH, Jonker JW. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family: an overview. Adv Drug Deliv Rev. 2003;55:3–29. doi: 10.1016/s0169-409x(02)00169-2. [DOI] [PubMed] [Google Scholar]
- 22.Ambudkar SV, Dey S, Hrycyna CA, Ramachandra M, Pastan I, Gottesman MM. Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu Rev Pharmacol Toxicol. 1999;39:361–398. doi: 10.1146/annurev.pharmtox.39.1.361. [DOI] [PubMed] [Google Scholar]
- 23.Lv M, Qiu JG, Zhang WJ, Jiang QW, Qin WM, Yang Y, Zheng DW, Chen Y, Huang JR, Wang K, Wei MN, Cheng KJ, Shi Z. Wallichinine reverses ABCB1-mediated cancer multidrug resistance. Am J Trans Res. 2016;8:2969–80. [PMC free article] [PubMed] [Google Scholar]
- 24.Shi Z, Jain S, Kim IW, Peng XX, Abraham I, Youssef DT, Fu LW, El Sayed K, Ambudkar SV, Chen ZS. Sipholenol A, a marine-derived sipholane triterpene, potently reverses P-glycoprotein (ABCB1)-mediated multidrug resistance in cancer cells. Cancer Sci. 2007;98:1373–1380. doi: 10.1111/j.1349-7006.2007.00554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiao D, Tang S, Aslam S, Ahmad M, To KK, Wang F, Huang Z, Cai J, Fu L. UMMS-4 enhanced sensitivity of chemotherapeutic agents to ABCB1-overexpressing cells via inhibiting function of ABCB1 transporter. Am J Cancer Res. 2014;4:148–160. [PMC free article] [PubMed] [Google Scholar]
- 26.Shi Z, Tiwari AK, Shukla S, Robey RW, Singh S, Kim IW, Bates SE, Peng X, Abraham I, Ambudkar SV, Talele TT, Fu LW, Chen ZS. Sildenafil reverses ABCB1- and ABCG2-mediated chemotherapeutic drug resistance. Cancer Res. 2011;71:3029–3041. doi: 10.1158/0008-5472.CAN-10-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi Z, Peng XX, Kim IW, Shukla S, Si QS, Robey RW, Bates SE, Shen T, Ashby CR Jr, Fu LW, Ambudkar SV, Chen ZS. Erlotinib (Tarceva, OSI-774) antagonizes ATP-binding cassette subfamily B member 1 and ATP-binding cassette subfamily G member 2-mediated drug resistance. Cancer Res. 2007;67:11012–11020. doi: 10.1158/0008-5472.CAN-07-2686. [DOI] [PubMed] [Google Scholar]
- 28.Shi Z, Tiwari AK, Shukla S, Robey RW, Kim IW, Parmar S, Bates SE, Si QS, Goldblatt CS, Abraham I, Fu LW, Ambudkar SV, Chen ZS. Inhibiting the function of ABCB1 and ABCG2 by the EGFR tyrosine kinase inhibitor AG1478. Biochem Pharmacol. 2009;77:781–793. doi: 10.1016/j.bcp.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fowers KD, Kopecek J. Targeting of multidrug-resistant human ovarian carcinoma cells with anti-P-glycoprotein antibody conjugates. Macromol Biosci. 2012;12:502–514. doi: 10.1002/mabi.201100350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nadali F, Pourfathollah AA, Alimoghaddam K, Nikougoftar M, Rostami S, Dizaji A, Azizi E, Zomorodipour A, Ghavamzadeh A. Multidrug resistance inhibition by antisense oligonucleotide against MDR1/mRNA in P-glycoprotein expressing leukemic cells. Hematology. 2007;12:393–401. doi: 10.1080/10245330701283991. [DOI] [PubMed] [Google Scholar]
- 31.Simoff I, Karlgren M, Backlund M, Lindstrom AC, Gaugaz FZ, Matsson P, Artursson P. Complete Knockout of Endogenous Mdr1 (Abcb1) in MDCK Cells by CRISPR-Cas9. J Pharm Sci. 2016;105:1017–1021. doi: 10.1016/S0022-3549(15)00171-9. [DOI] [PubMed] [Google Scholar]
- 32.Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 33.Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, Sachs N, Overmeer RM, Offerhaus GJ, Begthel H, Korving J, van de Wetering M, Schwank G, Logtenberg M, Cuppen E, Snippert HJ, Medema JP, Kops GJ, Clevers H. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 34.Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, Cai W, Yang G, Bronson R, Crowley DG, Zhang F, Anderson DG, Sharp PA, Jacks T. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, Scott DA, Song J, Pan JQ, Weissleder R, Lee H, Zhang F, Sharp PA. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruiz S, Mayor-Ruiz C, Lafarga V, Murga M, Vega-Sendino M, Ortega S, Fernandez-Capetillo O. A Genome-wide CRISPR Screen Identifies CDC25A as a Determinant of Sensitivity to ATR Inhibitors. Mol Cell. 2016;62:307–313. doi: 10.1016/j.molcel.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su S, Hu B, Shao J, Shen B, Du J, Du Y, Zhou J, Yu L, Zhang L, Chen F, Sha H, Cheng L, Meng F, Zou Z, Huang X, Liu B. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci Rep. 2016;6:20070. doi: 10.1038/srep20070. [DOI] [PMC free article] [PubMed] [Google Scholar]


