Abstract
Background
Esophageal cancer is a public health concern around the world; this cancer is the sixth leading cause of death of cancer in the world with about 386,000 deaths per year. Its risk factors include environmental factors such as tobacco smoke, gastroesophageal reflux and genetic changes. iNOS is stated by the effect of various inflammatory factors and is thus called inducible NOS. Investigating iNOS expression is a powerful tool for understanding effective molecular parameters at tissue and cellular responses to external factors. In this research work, iNOS expression in patients with esophageal cancer was studied in Iran.
Materials and Methods
15 formalin-fixed and paraffin-embedded (FFPE) esophageal cancer tissue samples and 15 normal FFPE samples were collected from various medical centers (Zabol, Zahedan, Kashan) to measure iNOS expression by real-time reverse transcriptase polymerase chain reaction (real-time RT-PCR). All PCR reactions were conducted by three replicates for iNOS and internal control (β-actin) by 2-ΔΔCT (Livak) method. Differences were measured in target gene expression in patients and control group using the t test. All statistical analyses were done using the SPSS software.
Results
The results showed that there was no significant difference between iNOS expression in the case and control groups (p > 0.05); however, there was an increase in iNOS expression in the case group. On the other hand, there was a significant difference between iNOS expression in males and females in the two groups of healthy subjects and patients, and it was higher in women than in men.
Conclusion
Further studies need to be conducted with larger sample sizes and in other populations to validate these findings.
Key Words: Esophageal cancer, iNOS, Quantitative real-time reverse transcriptase polymerase chain reaction
Introduction
Cancer is the third leading cause of death in Iran. About 30,000 of Iranians die of cancer annually [1]. Esophageal cancer is a common malignant neoplasm worldwide, with a varied geographical distribution which could be due to genetic or environmental risk factors such as smoking, alcohol or diet containing nitrosamines [2,3]. A recent study from esophageal squamous cell carcinoma showed that such systemic risk factors can produce nitric oxide (NO), which leads to tumor progression [4]. NO is a short-lived endogenous gas which acts as a signaling molecule in the body [5,6]. NO is a free radical gas that mediates various biological activities such as angiogenesis, transmission of nerve impulses and cytotoxic reactions [7]. It is synthesized from L-arginine by NOS [8]. There are two types of NOS, hybrid (neural and endothelial-cell) NOS and inducible NOS. Generally, neuronal NOS and endothelial-cell NOS produce NO in low concentrations, and they can be induced in neurons and endocrine cells. Their activity depends on high levels of cytoplasmic calcium/calmodulin. On the other hand, iNOS can be created by neutrophils, macrophages, endothelial cells and other cell types, and its induction depends on bacterial production or cytokines independent of calcium/calmodulin concentration [8,9]. Many recent studies confirm that iNOS can be found in different types of cancer and is correlated to clinical pathological factors such as histological grade, vascular invasion, high grade and correlation forecast [10,11,12,13,14]. In the present study we evaluated for the first time the expression of iNOS in tumoral and non-tumoral esophageal cancer tissue samples collected from two provinces of Iran by using quantitative real-time reverse transcriptase polymerase chain reaction (quantitative real-time RT-PCR).
Materials and Methods
In our study, formalin-fixed and paraffin-embedded (FFPE) samples were gathered from 15 cancerous tissue (case) samples as well as 15 healthy (control) samples. All samples were prepared in the pathology wards of Kashan, Zahedan and Zabol hospitals. RNA extraction from paraffin-embedded tissues was done by paraffin removal from samples and their surrounding tissues. In order to perform slicing (sectioning) appropriately, all blocks were placed at −20°C for 24 h, then slicing was done by a microtome tool in a sterile 1.5-µl tube for each block depending on the tissue contents. RNA extraction was performed in three stages as follows: Paraffin was removed by using xylene and ethanol. First, 1,000 µl of xylene was added to each tube. Tubes were vortexed for 10 s and placed in a 37°C water bath for 15 min, then they were centrifuged at 12,000 rpm for 2 min. Xylene was removed without damage to the deposit; these steps were repeated two or three times according to the samples' paraffin contents. To remove xylene, 1,000 µl of ethanol was added to the tube, then the tube was vortexed for 10 s. After that it was centrifuged at 12,000 rpm for 2 min. Finally, in order to dry the samples and remove residual ethanol, the tubes were left open and the samples were incubated at room temperature or a temperature >37°C for 15 min to completely remove residual ethanol. The third step of RNA extraction was done using the RNeasy® FFPE kit (Qiagen) according to its protocol, and extracted RNA was kept in a freezer at −80°C [15]. The quality of the extracted RNA was evaluated by uploading the extracted sample on 1% agarose gel; for higher reliability, the concentration and optical density of samples were measured using optical spectroscopy (ScanDrop®, Analytic Jena). The next step after RNA extraction was DNA synthesis. This reaction was performed by using a two-step RT-PCR kit (Vivantis). The first mixture was prepared on ice and contained the components listed in tables 1 and 2[16].
Table 1.
cDNA synthesis – first step
| Template | 5 µl |
| Primer oligo dt | 1 µl |
| Primer random hexamers | 1 µl |
| 10 mm dNTps mix | 1 µl |
| Nuclease-free water | up to 10 µl |
Table 2.
cDNA synthesis – second step
| Buffer M – MulV 10X | 2 µl |
| M-MulV | 0.5 µl |
| Nuclease-free water | up to 10 µl |
After first mixture synthesis, it was spun (to make a uniform mixture) and incubated at 65°C for 5 min, and then the mixture was placed on ice for 2 min and spun briefly afterward. Then the second mixture was synthesized and centrifuged briefly (10 s, 10,000 rpm) and incubated at room temperature (due to presence of random hexamers and to prevent primer separation), and the mixture was consecutively incubated for 60 min at 42°C, then the reaction continued by incubating the tube for 5 min at 85°C, then the tubes were cooled down by placing them on ice and then centrifuged briefly (10 s, 10,000 rpm); the resulting cDNA was stored at −20°C. Primer designing was done using CLC Main Work Bench 5 software and Allele ID 7 software, and the evaluation and analysis in terms of adhesion site and optimum properties were performed by online Beacon Designer Software [17] (tables 3, 4).
Table 3.
Characteristics of iNOS primers
| Reverse primer | Forward primer |
|---|---|
| ACATTCTGCTTCTGGAAACTA | CGCTACAACATCCTGGAG |
| GC = 38.1% | GC = 55.56% |
| Temperature = 55.5° C | Temperature = 56.1° C |
Table 4.
Characteristics of β-actin expression primers
| Reverse primer | Forward primer |
|---|---|
| CTAAGTCATAGTCCGCCTAG | GAAGATCAAGATCATTGCTCC |
| Temperature = 57.4° C | Temperature = 58.4° C |
PCR
Master gradient PCR was performed by a thermocycler machine (Mastercycler gradient, Eppendorf). A temperature of 52-60°C was considered to confirm the cDNA synthesis by iNOS primers and reference gene. For PCR, a two-step RT-PCR kit (Sinagene) was used. Preparation steps of major PCR solution were performed on ice and all tubes were placed on ice. In each reaction, the amount of material of PCR was determined empirically. Table 5 shows the materials and their concentration in the PCR. The PCR thermal gradient was programmed by the PCR thermocycler machine for each pair of primers to find the best connection temperature (table 6).
Table 5.
Optimum amount of PCR reaction component
| Master Mix Red | 12.5 µl |
| Forward primer | 1 µl |
| Reverse primer | 1 µl |
| cDNA template | 0.5 µl |
| Taq DNA polymerase | 1 µl |
| MgCl2 | 1 µl |
| Nuclease-free water | up to 25 µl |
Table 6.
PCR temperature cycle
| Cycle number | Time | Temperature | Step |
|---|---|---|---|
| 1 | 5 min | 95° C | initial denaturing |
| 35 | 30 s | 95° C | denaturing |
| 35 | 45 s | 56–57° C | annealing |
| 35 | 55 s | 72° C | extension |
| 1 | 8 min | 72° C | final extension |
Real-time PCR reaction was performed to analyze iNOS expression. β-Actin was used as an internal control gene. HotTaq EvaGreen qPCR Master Mix (Sinagene) was used for performing real-time PCR reaction. In tables 7 and 8, the composition and amount of reaction components of real-time PCR are shown. After placing samples in the system for real-time PCR reaction, the system's software was programmed as shown in table 9.
Table 7.
Amount of required material for real-time PCR reaction for iNOS primer
| Master Mix | 12.5 µl |
| Forward primer | 1 µl |
| Reverse primer | 1 µl |
| cDNA | 2 Ml |
| Nuclease-free water | up to 20 Ml |
Table 8.
Amount of required material for real-time PCR reaction for β-actin primer
| Master Mix | 12.5 Ml |
| Forward primer | 1 Ml |
| Reverse primer | 1 Ml |
| cDNA | 2 Ml |
| Nuclease-free water | up to 20 Ml |
Table 9.
Time and temperature conditions for real-time PCR reaction
| Time and temperature for initial enzyme activation: 95° C for 15 s 40 cycles involve the following denaturation steps: |
| Primer connection: 95° C for 30 s |
| Extended mix: 56.6° C for 45 s |
| Melting curve: 72° C for 30 s |
| Raising the temperature from 60° C to 99° C every 5 s by 1° C |
A fluorescent reporter was used for this method. These reporters are designed in a manner that if they have DNA replication, they should produce light, so increasing recorded light intensity in the device is directly proportional to the amount of obtained product. By continuing PCR, the fluorescent intensity will increase. The first cycle in which the fluorescent is greater than the baseline threshold is called threshold cycle (CT), in other words, in the initial phase of progressive steps, the amount of fluorescent increases to reach a threshold, and it is higher than the background level with a known amount, and this cycle is called CT. The CT amount was recorded based on drawing a threshold for all samples. For each sample, three replications were performed and the average CT was calculated for all samples. Information processing in real-time PCR is important. To investigate, there are two ways: (1) the Pfaffl method [18] and (2) the ΔΔCT method [19,20].
To reduce errors and correct variations in primer materials in RT-PCR, the approved method is simultaneous RNA amplification, which is used as an internal standard. In this study, the comparisons of the patient and control groups were tested by using the t test in SPSS software.
Results
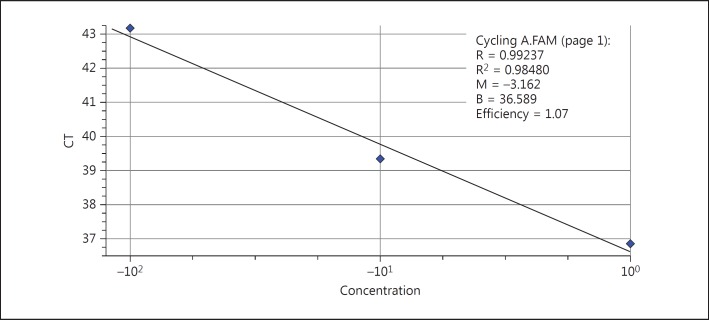
Samples' CTs were calculated based on the proliferation diagrams after performing reaction on all samples of any concentration using β-actin protein and their curvature drawn (based on the CT on the y-axis and the log of the copy amount on the x-axis). The efficiency of the reaction for β-actin was 1.07 and R2 was 98% after drawing the standard curvature (fig. 1).
Fig. 1.
β-Actin standard curvature.
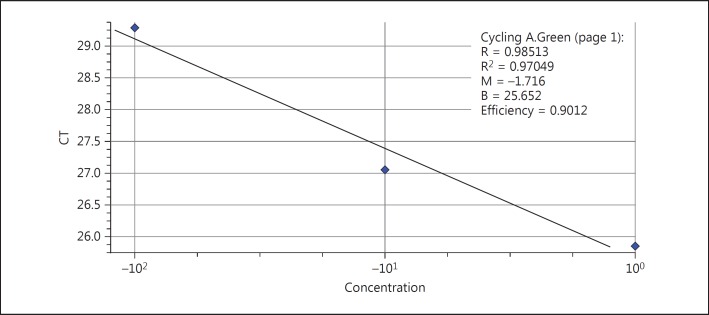
The same steps performed for the β-actin standard curvature were performed for the iNOS standard curvature as well. The efficiency of the reaction for iNOS was 90% and R2 was 97% after drawing the standard curvature (fig. 2).
Fig. 2.
iNOS standard curvature.
Results of Real-Time PCR Reaction
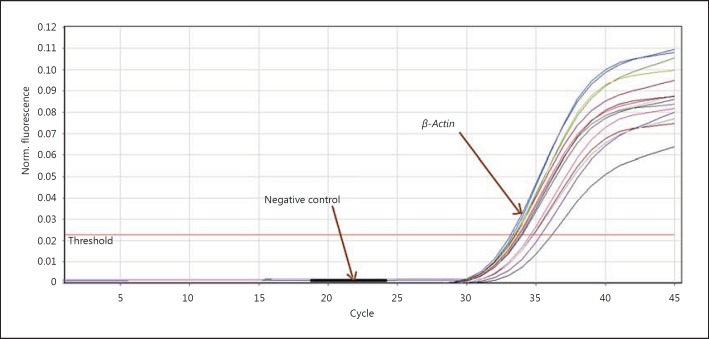
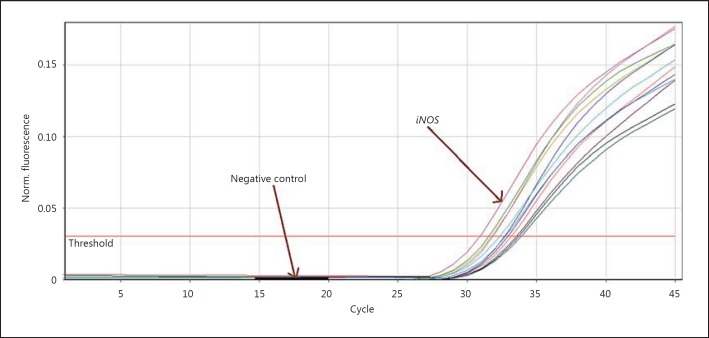
In figure 3 the β-actin proliferation progression in every cycle is shown. Figure 4 presents the progression of iNOS proliferation curvature in every cycle.
Fig. 3.
β-Actin proliferation curvature. The intensity of fluorescence is shown on the y-axis and the count of iNOS proliferation cycles on the x-axis.
Fig. 4.
iNOS proliferation curvature.
Results of Fusion Curvature Analysis
One of the most important advantages of real-time PCR is fusion curvature drawing by which we determined the diversity of products in the PCR reaction. In order to draw the curvature the instrument changes the samples temperature in specified durations, then the rate of change is shown on the y-axis and the instrument temperature is shown on the x-axis.
The Fusion Changes Curvature in Terms of β-Actin Gene Temperature
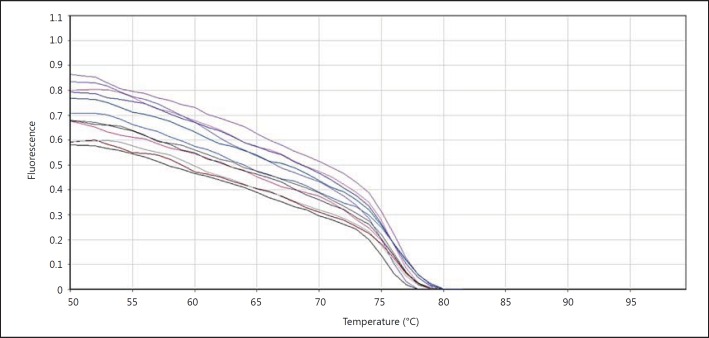
In order to draw the curvature, samples temperature increase and the light which was emitted from the samples were measured. In figure 5 it can be seen that there was a sudden drop of florescent light intensity at about 75°C.
Fig. 5.
Fluorescence changes in terms of temperature for β-actin. The temperature is shown on the x-axis and fluorescent light intensity on the y-axis.
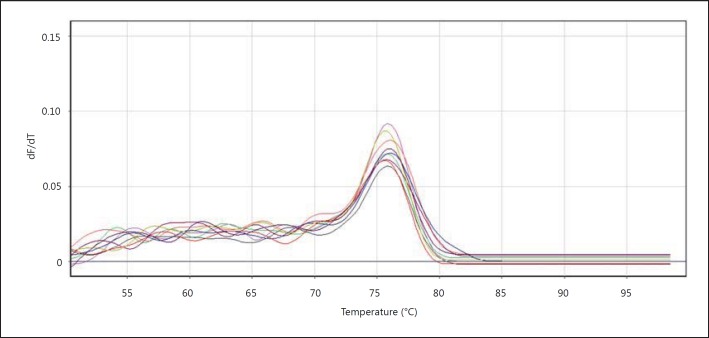
In the fusion curvature, every peak is indicative of one of the PCR products (fig. 6). The fusion peak includes a peak at 78°C.
Fig. 6.
β-Actin gene fusion curvature.
Fusion Changes Curvature in Terms of iNOS Temperature
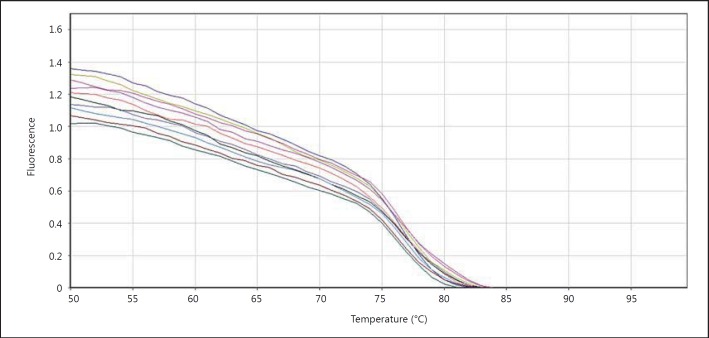
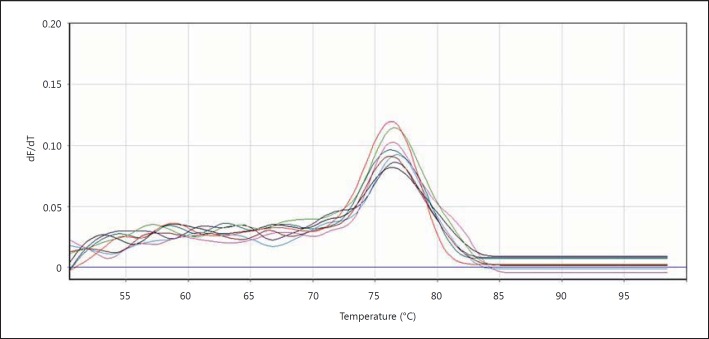
We then evaluated the fusion curvature in order to see whether PCR worked well or not. In this curvature the samples' temperature increases gradually and the emitted light is measured. The PCR product of iNOS begins to open in becoming single-strand following increasing the temperature of double-strand DNA and dye molecule separate from them. The intensity of fluorescence decreases following increasing the temperature until the temperature reaches around 74°C, which is the temperature of the PCR reaction product, then the intensity of fluorescence decreases and breaking will appear in the fusion curvature (fig. 7, 8).
Fig. 7.
Fluorescence changes in terms of temperature for iNOS. The temperature is shown on the x-axis and the light intensity on the y-axis.
Fig. 8.
iNOS fusion curvature. The temperature of 77°C was the temperature of iNOS product.
Statistical Results Obtained from the Analysis
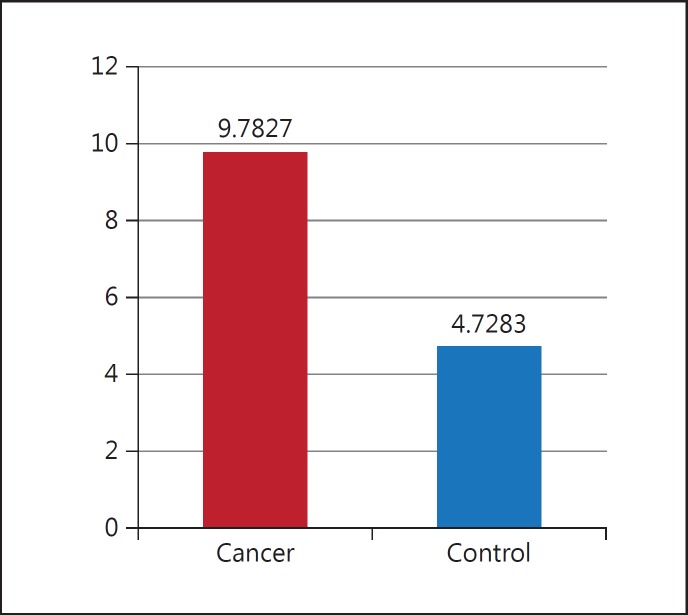
There was not statistically significant difference between iNOS expression among controls and patients. However, iNOS expression was higher in patients compared with controls as shown in figure 9. Differences between ΔCT averages were obtained from the control and patient groups for iNOS.
Fig. 9.
Comparing average ΔCT between the cancer and control group.
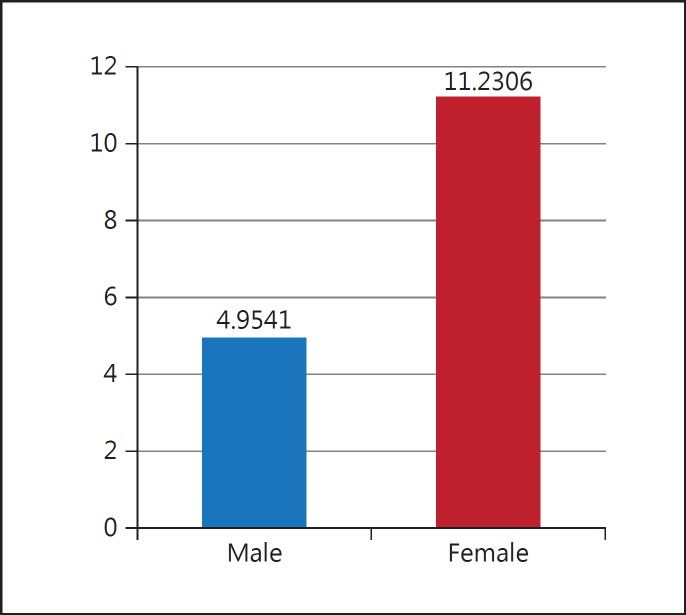
Thus, the results show that gene expression among patient is 2.068 times more than healthy subjects. In this study, two groups (controls and patients) were investigated in terms of gender. The results showed that there is a significant difference between men and women, iNOS expression increasing ore in women than men (fig. 10).
Fig. 10.
Comparing average ΔCT between men and women.
Checking the Uniqueness of Real-Time PCR
To ensure amplification of the gene, the samples resulted from PCR amplification, two amplified samples of each gene were randomly chosen and were put on 2% agarose gel for 60 min with the voltage of 100 V and the results obtained from agarose gel image confirmed the correct amplification of iNOS and β-actin (fig. 11).
Fig. 11.
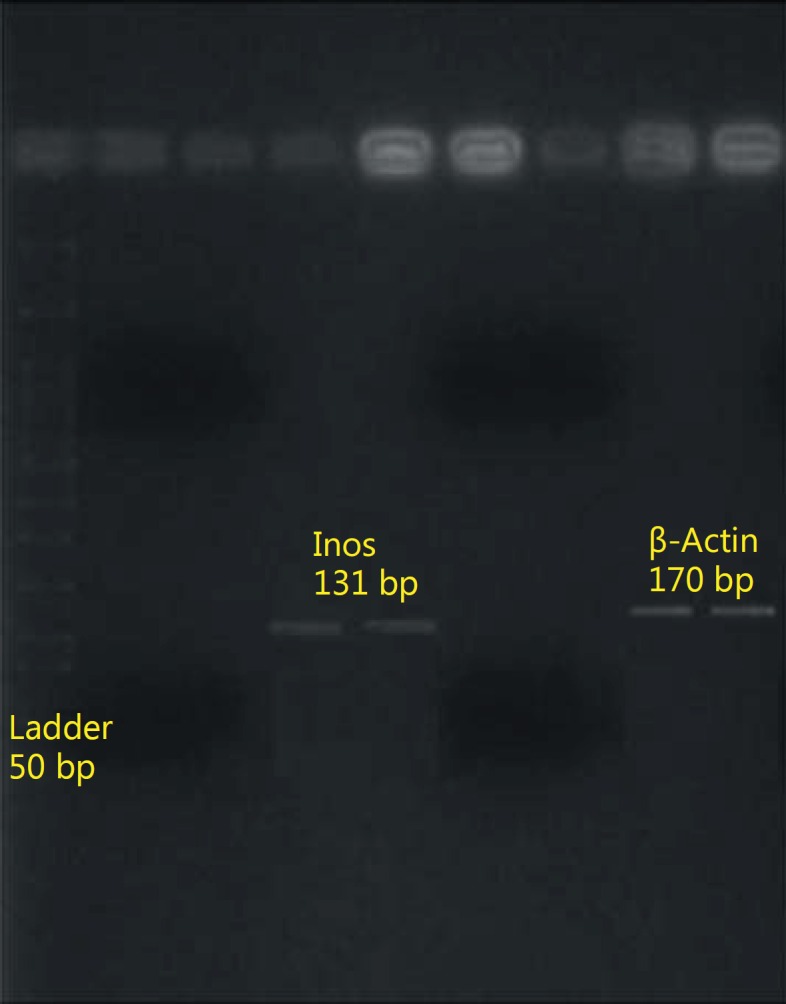
Amplified fragments in the real-time PCR reaction.
Discussion
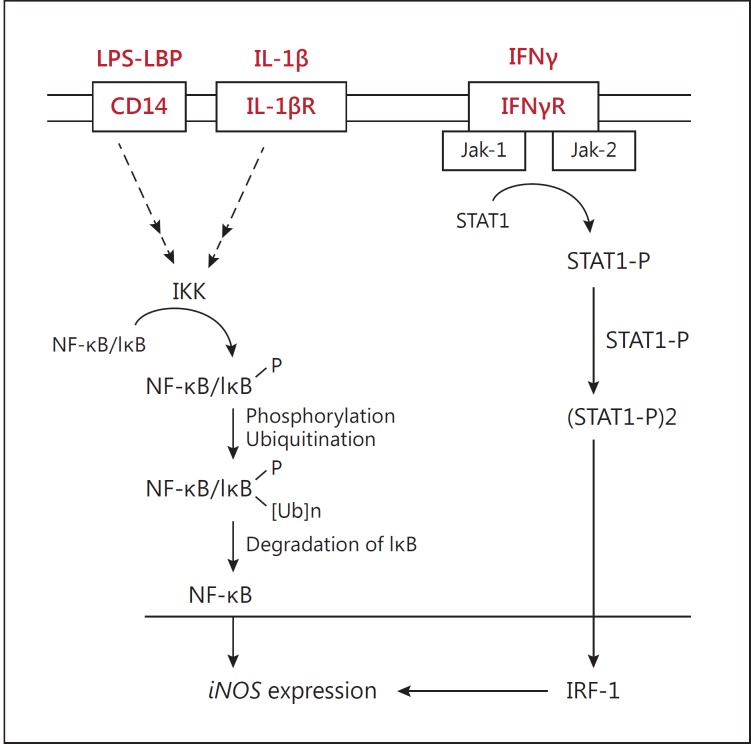
Esophageal cancer is a significant health problem throughout the world [21]. The effects of NO in tumor biology are broad, spanning its involvement in cellular transformation, formation of neoplastic lesions, and initiation and regulation of the metastatic cascade. NO participates in genotoxic events [22]. It may mediate DNA lesions through formation of toxic and mutagenic species, by direct modification of DNA, or by inhibition of DNA repair mechanisms [23]. NO and NOS are known to have an important role in many biological processes, including inflammation and cancer [6]. However, it seems that NO and iNOS are associated with antitumor effect and tumor progression [24]. Increased iNOS protein expression and NO level have been found in many chronic inflammatory and autoimmune diseases such as rheumatoid arthritis [25,26], multiple sclerosis [27], asthma [28] and type 1 diabetes [29]. An important inflammatory mediator linking chronic inflammation and cancer is NO, which is produced endogenously [8]. During inflammation, induced expression of iNOS in macrophages and epithelial cells leads to production of NO. The expression of iNOS and the level of NO have been shown to be elevated in various precancerous lesions and carcinomas [30,31]. The role of iNOS during tumor development is highly complex. Both promoting and deterring actions have been described, presumably depending upon the local concentration of iNOS within the tumor microenvironment [32]. The overexpression in Barrett's esophagus, and in adenocarcinomas arising from it, suggests that iNOS plays a role in the multistage development of esophageal cancer [33]. iNOS activity in macrophages is first regulated and modulated by cellular receptor molecules such as Toll-like receptors and CD14. CD14 is the receptor for lipopolysaccharide (LPS) and plays an essential role in pro-inflammatory responses in monocytes and macrophages via activation of the nuclear factor kappaB (NF-κB) pathway [34]. CD14 has two distinct forms: mCD14 (GPI-anchored form) and sCD14 (soluble CD14). mCD14 is believed to directly affect LPS simulation via interaction with Toll-like receptor 4 [35]. Interferon gamma (IFNγ) induces iNOS via the JAK-STAT signaling pathway. Activated JAK-STAT signaling pathway increases iNOS induction and NO production (fig. 12) [36].
Fig. 12.
Activation of NF-κB and STAT signaling pathway. LPS-binding protein (LBP) transfers LPS to the CD14 receptor on the cell membrane. Binding LPS activates NF-κB or AP-1. The IκB-NF-κB complex is inactive in the cytosol. After activation of the IκB/NF-κB complex, free NF-κB transfers into the nucleus and induces the expression of iNOS. IFNγ binds to IFNγR and phosphorylates STAT1. Activated STAT1 translocates into the nucleus and increases iNOS induction and NO production [37].
The regulation of NO synthesis by iNOS differs according to the strain and species of animals and depends on the inducers. Bovine and murine macrophages generate considerable amounts of iNOS in response to cytokine stimulation, but human and pig macrophages are resistant [38]. Recent developments are reviewed and show that NO biosynthesis is regulated by a variety of mechanisms at the transcriptional and posttranslational levels in activated macrophages and other cells. The level of NO produced by iNOS is mainly regulated at the transcriptional level. The molecular mechanisms for transcription of iNOS have been studied in different cells. Depending on the stimulator or cell type, different signaling pathways activate transcriptional factors, activators such as protein kinase C, tyrosine kinase, Janus kinases, Raf-1 protein kinase, mitogen-activated protein kinases and inhibitors such as protein tyrosine phosphatases and phosphoinositide 3-kinase [39,40,41,42,43]. The promoter region of the mouse iNOS contains several binding sites for transcription factors like NF-κB as well as Jun/Fos heterodimers, some C/EBP, CREB and the STAT family of transcription factors, within its proximal and distal [40,44,45,46]. The human iNOS promoter contains sequences homologous to mouse proximal and distal regions [36]. Transcription factors like NF-κB and activating protein-1 (AP-1) mediate the expression of iNOS and other inducible genes such as cyclooxygenase-2, vascular cell adhesion molecule-1 and intercellular cell adhesion molecule-1 in immune and inflammatory responses [47]. NF-κB is present in the cytosol as an inactive complex IκB-NF-κB (fig. 12). The IκB-NF-κB complex is phosphorylated by IκB kinase (IKK) through activation by stimulator(s) such as potent exogenous inducer (LPS) and endogenous inducers (cytokines, IFNγ and tumor necrosis factor alpha [TNFα]), which facilitate the translocation of free NF-κB from cytosol to the nucleus and the induction of iNOS expression [40,48,49]. LPS downregulates the DNA-binding activity of the inducible transcription factors NF-κB, CREB and AP-1, whereas it upregulates the DNA-binding activity of constitutive transcription factors promoter-selective transcription factor (Sp1) and activating protein-2 (AP-2) in a time-dependent manner [50]. The regulation of iNOS via the NF-κB pathway is an important mechanism in inflammatory processes and a potential site for intervention in inflammatory disease, the reversal effect of gypenosides, a herbal medicine, on LPS-induced iNOS expression through NF-κB activity, suggesting its use in the treatment of inflammatory diseases [51]. In addition, some compounds suppress iNOS induction via degradation of IκB. H2O2 increases the degradation of IκB and activation of the NF-κB pathway, whereas lactacystin prevents iNOS induction by the blocking degradation of IκB [52,53]. Therefore, the degradation of IκB, phosphorylation/dephosphorylation of the NF-κB-IκB complex and the translocation of free NF-κB to the nucleus are important in regulating NO production by iNOS. Activation of the C/EBP family transcription factors by protein kinases increases intracellular cAMP concentrations, inducing iNOS in rat vascular smooth muscle cells [44]. NF-κB and C/EBP can also cooperate in a synergistic manner to induce iNOS[54]. Combinations of cytokines or LPS with IFNγ induce iNOS expression synergistically. Suppressor of cytokine signaling-1 (SOCS-1) is a negative regulator of the JAK-STAT signaling pathway activated by cytokines and interferons. Exposure of cells to IFNγ activates Janus kinases, Jak-1 and Jak-2, which phosphorylate and activate STAT1. The activated STAT1 translocates into the nucleus, increases interferon regulatory factor-1 (IRF-1) levels and induces iNOS in murine macrophages, mouse and human islets, hepatocytes and renal mesangial cells (fig. 12) [55,56]. The overproduction of SOCS-1 inhibits cytokine signals that activate this pathway [36]. NO can regulate its own production. It has biphasic effects on NO synthesis by modulating iNOS mRNA expression [56]. The positive feedback regulation of iNOS expression in renal mesangial cells and vascular smooth muscle cells is mediated indirectly through increased intracellular cAMP levels via inhibition of phosphodiesterase III [57]. The negative feedback regulation of iNOS expression by NO in macrophages and hepatocytes is mediated by the inhibition of NF-κB activation. In contrast to rat renal mesangial cells, in rat aortic vascular smooth muscle cells, NO overproduction is upregulated by negative feedback regulation as well [44]. NO-releasing agents and cGMP analogs can modulate the transcription factors AP-1 and NF-κB in a positive or negative manner, depending on the tissue type [58]. On the other hand, H2O2 and ROS activate NF-κB with enhanced iNOS expression and this is suppressed by antioxidant compounds and enzymes. Thus, H2O2 production by LPS in macrophages participates in the upregulation of iNOS [52,53,54,55,56,57,58,59]. Some neuropeptides such as vasoactive intestinal peptide and the pituitary adenylate cyclase-activating polypeptide (PACAP) can function as macrophage-deactivating factors. Vasoactive intestinal peptide and PACAP use the cAMP-dependent pathway to stimulate CREB phosphorylation, causing a change in the CRE binding complex, a binding site for IL-10 as a macrophage-deactivating factor, and inhibiting IFNγ induced IRF-1 expression. A cAMP-independent pathway is also involved in the inhibition of IKK activity and NF-κB binding, thus reducing iNOS expression [60]. cAMP induction of iNOS has been documented for macrophages, vascular smooth muscle cells and renal mesangial cells [44]. iNOS expression is related to peroxisome proliferator-activated receptors (PPARs), which may antagonize the activities of transcriptional factors including AP-1, STAT and NF-κB. TNFα and IFNγ may modify PPAR activity, leading to a modified activity on the p65/RelA subunit of the NF-κB signaling pathway [61]. Also important may be the posttranscriptional regulation of iNOS expression. Steady-state iNOS mRNA levels depend on its synthesis, stability and degradation. The 3′-untranslated region of iNOS mRNA contains AU-rich sequences, known to destabilize iNOS mRNA [62,63]. The exposure of cells to transforming growth factor β (TGFβ) inhibits iNOS induction because it causes iNOS mRNA destabilization [64]. The posttranscriptional effects of TGFβ might be mediated via a modification of IRF-1 activation through the activation of nucleases acting on the AU-rich sequence [65]. In addition, protein kinase C alpha has been shown to play an essential role in iNOS mRNA stabilization in pancreatic β cells [66]. Conversely, iNOS mRNA stability is enhanced by elevation of intracellular cAMP levels, but reduced by increases in intracellular Ca2+[67]. Additionally, NO and/or cGMP can modulate iNOS expression by posttranscriptional mechanisms [57]. iNOS mRNA destabilization by cGMP limits iNOS induction during late treatment times of cells supplemented with NO or cGMP donors, and NO and/or cGMP generation and cGMP appear to control iNOS expression by positive and negative feedback mechanisms [57].
In the present study, the statistical data obtained by iNOS expression yielded that the difference of gene expression between the control and patient groups was not significant, but iNOS expression increased in the patient group. On the other hand, the difference between the two groups is significant in terms of gender, and the increase in gene expression is higher among women than men. In 2005, Chen et al. [68] concluded that iNOS induction with B[α]P can be a mechanism which is consistent with this sentence that B[α]P creates carcinogenesis effects on human esophageal. NO is a free radical and causes nitrogen radical creation which can damage DNA and cause cancer. Further studies indicated that induction of iNOS by B[a]P in the cells is mediated through both MEK/ERKs- and IKK/IκBa/NF-κB-dependent pathways. Also, a connection has been found between inflammation, cancer and NO composition. NO is produced when the body's immune system reacts to infectious bacteria and can cause cancer. The present study concludes that iNOS expression is more increased among women than men. In fact, the women's samples in this research included more inflammation and infection than those of the males. This increase in inflammation and infection is due to increased iNOS expression. Finally, high induction of iNOS leads to overproduction of NO, which is a key factor in cancers and inflammation that is involved in almost all stages of tumor development [69]. iNOS is on chromosome 17q11.2-12 and includes 27 exons [70]. In addition, in this study, to verify the real-time PCR operation, the amplified product was run for both genes by this method on 2% agarose gel and the band of amplified fragments was observed, and it was proved that the gene amplification action through real-time PCR had been correctly and appropriately performed. The real-time PCR method is the most accurate method to investigate changes in gene expression. In this method, only the increase or decrease in gene expression is of importance. This comparison of gene expression level change is done with a standard which is the internal control. The reference gene should be expressed in all samples at the same level; otherwise, the normalization process may cause errors in results/be inaccurate. In the present study, β-actin was used as the reference gene. Having shown more stability in esophageal tissue and more stability compared to 18s rRNA and GAPDH genes, β-actin is more reliable for the normalization of the data [71].
Conclusion
No significant difference was observed between iNOS expression in the two groups. However, in the patient group, iNOS expression was increased. There was a significant difference between iNOS expression in males and females in the two groups, and it was shown to be higher in women than in men.
References
- 1.Motalleb G, Gholipour N, Samaei NM. Association of the human astrocyte elevated gene-1 promoter variants with susceptibility to hepatocellular carcinoma. Med Oncol. 2014;31:916. doi: 10.1007/s12032-014-0916-5. [DOI] [PubMed] [Google Scholar]
- 2.Blot WJ. Esophageal cancer trends and risk factors. Semin Oncol. 1994;21:403–410. [PubMed] [Google Scholar]
- 3.Ribeiro U, Jr, Posner MC, Safatle-Ribeiro AV, Reynolds JC. Risk factors for squamous cell carcinoma of the oesophagus. Br J Surg. 1996;83:1174–1185. [PubMed] [Google Scholar]
- 4.Knowles RG, Moncada S. Nitric oxide synthases in mammals. Biochem J. 1994;298:249–258. doi: 10.1042/bj2980249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ignarro LJ, Buga GM, Wood KS, Byrns RE, Chaudhuri G. Endothelium-derived relaxing factor produced and released from artery and vein is nitric oxide. Proc Natl Acad Sci USA. 1987;84:9265–9269. doi: 10.1073/pnas.84.24.9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmer RM, Ferrige AG, Moncada S. Nitric oxide release accounts for the biological activity of endothelium-derived relaxing factor. Nature. 1987;327:524–526. doi: 10.1038/327524a0. [DOI] [PubMed] [Google Scholar]
- 7.Scott DJ, Hull MA, Cartwright EJ, Lam WK, Tisbury A, Poulsom R, Markham AF, Bonifer C, Coletta PL. Lack of inducible nitric oxide synthase promotes intestinal tumorigenesis in the Apc(Min/+) mouse. Gastroenterology. 2001;121:889–899. doi: 10.1053/gast.2001.27994. [DOI] [PubMed] [Google Scholar]
- 8.Moncada S, Palmer RMJ, Higgs EA. Nitric oxide: physiology, pathophysiology, and pharmacology. Pharmacol Rev. 1991;43:109–142. [PubMed] [Google Scholar]
- 9.Nathan C, Xie QW. Regulation of biosynthesis of nitric oxide. J Biol Chem. 1994;269:13725–13728. [PubMed] [Google Scholar]
- 10.Yagihashi N, Kasajima H, Sugai S, Matsumoto K, Ebina Y, Morita T, Murakami T, Yagihashi S. Increased in situ expression of nitric oxide synthase in human colorectal cancer. Virchows Arch. 2000;436:109–114. doi: 10.1007/pl00008208. [DOI] [PubMed] [Google Scholar]
- 11.Vakkala M, Kahlos K, Lakari E, Paakko P, Kinnula V, Soini Y. Inducible nitric oxide synthase expression, apoptosis, and angiogenesis in in situ and invasive breast carcinomas. Clin Cancer Res. 2000;6:2408–2416. [PubMed] [Google Scholar]
- 12.Wolf H, Haeckel C, Roessner A. Inducible nitric oxide synthase expression in human urinary bladder cancer. Virchows Arch. 2000;437:662–666. doi: 10.1007/s004280000296. [DOI] [PubMed] [Google Scholar]
- 13.Thomsen LL, Miles DW, Happerfield L, Bobrow LG, Knowles RG, Moncada S. Nitric oxide synthase activity in human breast cancer. Br J Cancer. 1995;72:41–44. doi: 10.1038/bjc.1995.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aaltomaa SH, Lipponen PK, Viitanen J, Kankkunen JP, Ala-Opas MY, Kosma VM. The prognostic value of inducible nitric oxide synthase in local prostate cancer. BJU Int. 2000;86:234–239. doi: 10.1046/j.1464-410x.2000.00787.x. [DOI] [PubMed] [Google Scholar]
- 15.Abramovitz M, Ordanic-Kodani M, Wang Y, Li Z, Catzavelos C, Bouzyk M, Sledge GW, Jr, Moreno CS, Leyland-Jones B. Optimization of RNA extraction from FFPE tissues for expression profiling in the DASL assay. Biotechniques. 2008;44:417–423. doi: 10.2144/000112703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taherian A, Mostafavi Zadeh SM, Ghani H, Khamechian T. ST6Gal1, Cox-2 and HB-EGF mRNA expression in breast cancer samples from Kashan, Iran. Middle East J Cancer. 2015;6:43–50. [Google Scholar]
- 17.Soleimani M, Zolfaghari MR, Morovvati A. Development and comparison of conventional PCR and SYBR green real time PCR for detection of Aggregatibacter actinomycetemcomitans and Tannerella forsythensis. Jundishapur J Microbiol. 2013;6:e6757. [Google Scholar]
- 18.Pfaffl MW, Hageleit M. Validities of mRNA quantification using recombinant RNA and recombinant DNA external calibration curves in real-time RT-PCR. Biotechnol Lett. 2001;23:275–282. [Google Scholar]
- 19.Morse DL, Carroll D, Weberg L, Borgstrom MC, Ranger-Moore J, Gillies RJ. Determining suitable internal standards for mRNA quantification of increasing cancer progression in human breast cells by real-time reverse transcriptase polymerase chain reaction. Anal Biochem. 2005;342:69–77. doi: 10.1016/j.ab.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ. Cancer statistics for Hispanics. CA Cancer J Clin. 2003;53:5–26. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- 22.Gal A, Wogan GN. Mutagenesis associated with nitric oxide production in transgenic SJL mice. Proc Natl Acad Sci USA. 1996;93:15102–15107. doi: 10.1073/pnas.93.26.15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wink DA, Vodovotz Y, Laval J, Laval F, Dewhirst MW, Mitchell JB. The multifaceted roles of nitric oxide in cancer. Carcinogenesis. 1998;19:711–721. doi: 10.1093/carcin/19.5.711. [DOI] [PubMed] [Google Scholar]
- 24.Vakkala M, Paakko P, Soini Y. eNOS expression is associated with the estrogen and progesterone receptor status in invasive breast carcinoma. Int J Oncol. 2000;17:667–671. doi: 10.3892/ijo.17.4.667. [DOI] [PubMed] [Google Scholar]
- 25.McInnes IB, Leung BP, Field M, Wei XQ, Huang FP, Sturrock RD, Kinninmonth A, Weidner J, Mumford R, Liew FY. Production of nitric oxide in the synovial membrane of rheumatoid and osteoarthritis patients. J Exp Med. 1996;184:1519–1524. doi: 10.1084/jem.184.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grabowski PS, Wright PK, Van 't Hof RJ, Helfrich MH, Ohshima H, Ralston SH. Immunolocalization of inducible nitric oxide synthase in synovium and cartilage in rheumatoid arthritis and osteoarthritis. Br J Rheumatol. 1997;36:651–655. doi: 10.1093/rheumatology/36.6.651. [DOI] [PubMed] [Google Scholar]
- 27.Bagasra O, Michaels FH, Zheng YM, Bobroski LE, Spitsin SV, Fu ZF, Tawadros R, Koprowski H. Activation of the inducible form of nitric oxide synthase in the brains of patients with multiple sclerosis. Proc Natl Acad Sci USA. 1995;92:12041–12045. doi: 10.1073/pnas.92.26.12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamid Q, Springall DR, Riveros-Moreno V, Chanez P, Howarth P, Redington A, Bousquet J, Godard P, Holgate S, Polak JM. Induction of nitric oxide synthase in asthma. Lancet. 1993;342:1510–1513. doi: 10.1016/s0140-6736(05)80083-2. [DOI] [PubMed] [Google Scholar]
- 29.Kroncke KD, Kolb-Bachofen V, Berschick B, Burkart V, Kolb H. Activated macrophages kill pancreatic syngeneic islet cells via arginine-dependent nitric oxide generation. Biochem Biophys Res Commun. 1991;175:752–758. doi: 10.1016/0006-291x(91)91630-u. [DOI] [PubMed] [Google Scholar]
- 30.Chen T, Rose ME, Hwang H, Nines RG, Stoner GD. Black raspberries inhibit N-nitrosomethylbenzylamine (NMBA)-induced angiogenesis in rat esophagus parallel to the suppression of COX-2 and iNOS. Carcinogenesis. 2006;27:2301–2307. doi: 10.1093/carcin/bgl109. [DOI] [PubMed] [Google Scholar]
- 31.Jaiswal M, LaRusso NF, Gores GJ. Nitric oxide in gastrointestinal epithelial cell carcinogenesis: linking inflammation to oncogenesis. Am J Physiol Gastrointest Liver Physiol. 2001;281:G626–G634. doi: 10.1152/ajpgi.2001.281.3.G626. [DOI] [PubMed] [Google Scholar]
- 32.Lechner M, Lirk P, Rieder J. Inducible nitric oxide synthase (iNOS) in tumor biology: the two sides of the same coin. Semin Cancer Biol. 2005;15:277–289. doi: 10.1016/j.semcancer.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Wilson KT, Fu S, Ramanujam KS, Meltzer SJ. Increased expression of inducible nitric oxide synthase and cyclooxygenase-2 in Barrett's esophagus and associated adenocarcinomas. Cancer Res. 1998;58:2929–2934. [PubMed] [Google Scholar]
- 34.Schroder NW, Opitz B, Lamping N, Michelsen KS, Zahringer U, Gobel UB, Schumann RR. Involvement of lipopolysaccharide binding protein, CD14, and Toll-like receptors in the initiation of innate immune responses by Treponema glycolipids. J Immunol. 2000;165:2683–2693. doi: 10.4049/jimmunol.165.5.2683. [DOI] [PubMed] [Google Scholar]
- 35.Du X, Low MG. Down-regulation of glycosylphosphatidylinositol-specific phospholipase D induced by lipopolysaccharide and oxidative stress in the murine monocyte-macrophage cell line RAW 264.7. Infect Immun. 2001;69:3214–3223. doi: 10.1128/IAI.69.5.3214-3223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rao KM. Molecular mechanisms regulating iNOS expression in various cell types. J Toxicol Environ Health B Crit Rev. 2000;3:27–58. doi: 10.1080/109374000281131. [DOI] [PubMed] [Google Scholar]
- 37.Aktan F. iNOS-mediated nitric oxide production and its regulation. Life Sci. 2004;75:639–653. doi: 10.1016/j.lfs.2003.10.042. [DOI] [PubMed] [Google Scholar]
- 38.Jungi TW, Adler H, Adler B, Thony M, Krampe M, Peterhans E. Inducible nitric oxide synthase of macrophages. Present knowledge and evidence for species-specific regulation. Vet Immunol Immunopathol. 1996;54:323–330. doi: 10.1016/s0165-2427(96)05690-5. [DOI] [PubMed] [Google Scholar]
- 39.Lahti A, Kankaanranta H, Moilanen E. p38 mitogen-activated protein kinase inhibitor SB203580 has a bi-directional effect on iNOS expression and NO production. Eur J Pharmacol. 2002;454:115–123. doi: 10.1016/s0014-2999(02)02490-1. [DOI] [PubMed] [Google Scholar]
- 40.Marks-Konczalik J, Chu SC, Moss J. Cytokine-mediated transcriptional induction of the human inducible nitric oxide synthase gene requires both activator protein 1 and nuclear factor kappaB-binding sites. J Biol Chem. 1998;273:22201–22208. doi: 10.1074/jbc.273.35.22201. [DOI] [PubMed] [Google Scholar]
- 41.Momose I, Terashima M, Nakashima Y, Sakamoto M, Ishino H, Nabika T, Hosokawa Y, Tanigawa Y. Phorbol ester synergistically increases interferon regulatory factor-1 and inducible nitric oxide synthase induction in interferon-gamma-treated RAW 264.7 cells. Biochim Biophys Acta. 2000;1498:19–31. doi: 10.1016/s0167-4889(00)00072-0. [DOI] [PubMed] [Google Scholar]
- 42.Muller MR, Pfannes SD, Ayoub M, Hoffmann P, Bessler WG, Mittenbuhler K. Immunostimulation by the synthetic lipopeptide P3CSK4: TLR4-independent activation of the ERK1/2 signal transduction pathway in macrophages. Immunology. 2001;103:49–60. doi: 10.1046/j.1365-2567.2001.01203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang BS, Lin JK, Lin-Shiau SY. Role of tyrosine kinase activity in 2,2′,2′′-tripyridine-induced nitric oxide generation in macrophages. Biochem Pharmacol. 1999;57:1367–1373. doi: 10.1016/s0006-2952(99)00047-7. [DOI] [PubMed] [Google Scholar]
- 44.Hecker M, Cattaruzza M, Wagner AH. Regulation of inducible nitric oxide synthase gene expression in vascular smooth muscle cells. Gen Pharmacol. 1999;32:9–16. doi: 10.1016/s0306-3623(98)00082-2. [DOI] [PubMed] [Google Scholar]
- 45.Kleinert H, Euchenhofer C, Ihrig-Biedert I, Forstermann U. In murine 3T3 fibroblasts, different second messenger pathways resulting in the induction of NO synthase II (iNOS) converge in the activation of transcription factor NF-kappaB. J Biol Chem. 1996;271:6039–6044. doi: 10.1074/jbc.271.11.6039. [DOI] [PubMed] [Google Scholar]
- 46.Kleinert H, Wallerath T, Fritz G, Ihrig-Biedert I, Rodriguez-Pascual F, Geller DA, Forstermann U. Cytokine induction of NO synthase II in human DLD-1 cells: roles of the JAK-STAT, AP-1 and NF-kappaB-signaling pathways. Br J Pharmacol. 1998;125:193–201. doi: 10.1038/sj.bjp.0702039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xia YF, Liu LP, Zhong CP, Geng JG. NF-kappaB activation for constitutive expression of VCAM-1 and ICAM-1 on B lymphocytes and plasma cells. Biochem Biophys Res Commun. 2001;289:851–856. doi: 10.1006/bbrc.2001.6067. [DOI] [PubMed] [Google Scholar]
- 48.Huang KT, Kuo L, Liao JC. Lipopolysaccharide activates endothelial nitric oxide synthase through protein tyrosine kinase. Biochem Biophys Res Commun. 1998;245:33–37. doi: 10.1006/bbrc.1998.8384. [DOI] [PubMed] [Google Scholar]
- 49.Obermeier F, Gross V, Scholmerich J, Falk W. Interleukin-1 production by mouse macrophages is regulated in a feedback fashion by nitric oxide. J Leukoc Biol. 1999;66:829–836. doi: 10.1002/jlb.66.5.829. [DOI] [PubMed] [Google Scholar]
- 50.Ye X, Liu SF. Lipopolysaccharide regulates constitutive and inducible transcription factor activities differentially in vivo in the rat. Biochem Biophys Res Commun. 2001;288:927–932. doi: 10.1006/bbrc.2001.5883. [DOI] [PubMed] [Google Scholar]
- 51.Aktan F, Henness S, Roufogalis BD, Ammit AJ. Gypenosides derived from Gynostemma pentaphyllum suppress NO synthesis in murine macrophages by inhibiting iNOS enzymatic activity and attenuating NF-kappaB-mediated iNOS protein expression. Nitric Oxide. 2003;8:235–242. doi: 10.1016/s1089-8603(03)00032-6. [DOI] [PubMed] [Google Scholar]
- 52.Bowie A, O'Neill LA. Oxidative stress and nuclear factor-kappaB activation: a reassessment of the evidence in the light of recent discoveries. Biochem Pharmacol. 2000;59:13–23. doi: 10.1016/s0006-2952(99)00296-8. [DOI] [PubMed] [Google Scholar]
- 53.Musial A, Eissa NT. Inducible nitric-oxide synthase is regulated by the proteasome degradation pathway. J Biol Chem. 2001;276:24268–24273. doi: 10.1074/jbc.M100725200. [DOI] [PubMed] [Google Scholar]
- 54.Hecker M, Preiss C, Schini-Kerth VB. Induction by staurosporine of nitric oxide synthase expression in vascular smooth muscle cells: role of NF-kappa B, CREB and C/EBP beta. Br J Pharmacol. 1997;120:1067–1074. doi: 10.1038/sj.bjp.0701026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chong MM, Thomas HE, Kay TW. Suppressor of cytokine signaling-1 regulates the sensitivity of pancreatic beta cells to tumor necrosis factor. J Biol Chem. 2002;277:27945–27952. doi: 10.1074/jbc.M110214200. [DOI] [PubMed] [Google Scholar]
- 56.Connelly L, Palacios-Callender M, Ameixa C, Moncada S, Hobbs AJ. Biphasic regulation of NF-kappa B activity underlies the pro-and anti-inflammatory actions of nitric oxide. J Immunol. 2001;166:3873–3881. doi: 10.4049/jimmunol.166.6.3873. [DOI] [PubMed] [Google Scholar]
- 57.Perez-Sala D, Cernuda-Morollon E, Diaz-Cazorla M, Rodriguez-Pascual F, Lamas S. Posttranscriptional regulation of human iNOS by the NO/cGMP pathway. Am J Physiol Renal Physiol. 2001;280:F466–F473. doi: 10.1152/ajprenal.2001.280.3.F466. [DOI] [PubMed] [Google Scholar]
- 58.Lee SK, Kim JH, Yang WS, Kim SB, Park SK, Park JS. Exogenous nitric oxide inhibits VCAM-1 expression in human peritoneal mesothelial cells. Role of cyclic GMP and NF-kappaB. Nephron. 2002;90:447–454. doi: 10.1159/000054733. [DOI] [PubMed] [Google Scholar]
- 59.Han YJ, Kwon YG, Chung HT, Lee SK, Simmons RL, Billiar TR, Kim YM. Antioxidant enzymes suppress nitric oxide production through the inhibition of NF-kappa B activation: role of H2O2 and nitric oxide in inducible nitric oxide synthase expression in macrophages. Nitric Oxide. 2001;5:504–513. doi: 10.1006/niox.2001.0367. [DOI] [PubMed] [Google Scholar]
- 60.Leceta J, Gomariz RP, Martinez C, Abad C, Ganea D, Delgado M. Receptors and transcriptional factors involved in the anti-inflammatory activity of VIP and PACAP. Ann NY Acad Sci. 2000;921:92–102. doi: 10.1111/j.1749-6632.2000.tb06954.x. [DOI] [PubMed] [Google Scholar]
- 61.Chinetti G, Griglio S, Antonucci M, Torra IP, Delerive P, Majd Z, Fruchart JC, Chapman J, Najib J, Staels B. Activation of proliferator-activated receptors alpha and gamma induces apoptosis of human monocyte-derived macrophages. J Biol Chem. 1998;273:25573–25580. doi: 10.1074/jbc.273.40.25573. [DOI] [PubMed] [Google Scholar]
- 62.Rodriguez-Pascual F, Hausding M, Ihrig-Biedert I, Furneaux H, Levy AP, Forstermann U, Kleinert H. Complex contribution of the 3′-untranslated region to the expressional regulation of the human inducible nitric-oxide synthase gene. Involvement of the RNA-binding protein HuR. J Biol Chem. 2000;275:26040–26049. doi: 10.1074/jbc.M910460199. [DOI] [PubMed] [Google Scholar]
- 63.Taylor BS, de Vera ME, Ganster RW, Wang Q, Shapiro RA, Morris SM, Jr, Billiar TR, Geller DA. Multiple NF-kappaB enhancer elements regulate cytokine induction of the human inducible nitric oxide synthase gene. J Biol Chem. 1998;273:15148–15156. doi: 10.1074/jbc.273.24.15148. [DOI] [PubMed] [Google Scholar]
- 64.Perrella MA, Yoshizumi M, Fen Z, Tsai JC, Hsieh CM, Kourembanas S, Lee ME. Transforming growth factor-beta 1, but not dexamethasone, down-regulates nitric-oxide synthase mRNA after its induction by interleukin-1 beta in rat smooth muscle cells. J Biol Chem. 1994;269:14595–14600. [PubMed] [Google Scholar]
- 65.Vodovotz Y. Control of nitric oxide production by transforming growth factor-beta1: mechanistic insights and potential relevance to human disease. Nitric Oxide. 1997;1:3–17. doi: 10.1006/niox.1996.0105. [DOI] [PubMed] [Google Scholar]
- 66.Carpenter L, Cordery D, Biden TJ. Protein kinase Cdelta activation by interleukin-1beta stabilizes inducible nitric-oxide synthase mRNA in pancreatic beta-cells. J Biol Chem. 2001;276:5368–5374. doi: 10.1074/jbc.M010036200. [DOI] [PubMed] [Google Scholar]
- 67.Oddis CV, Simmons RL, Hattler BG, Finkel MS. cAMP enhances inducible nitric oxide synthase mRNA stability in cardiac myocytes. Am J Physiol. 1995;269:H2044–H2050. doi: 10.1152/ajpheart.1995.269.6.H2044. [DOI] [PubMed] [Google Scholar]
- 68.Chen J, Yan Y, Li J, Ma Q, Stoner GD, Ye J, Huang C. Differential requirement of signal pathways for benzo[a]pyrene (B[a]P)-induced nitric oxide synthase (iNOS) in rat esophageal epithelial cells. Carcinogenesis. 2005;26:1035–1043. doi: 10.1093/carcin/bgi052. [DOI] [PubMed] [Google Scholar]
- 69.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu W, Lizhi L, Loizidou M, Ahmed M, Taylor I, Moncada S, Charles IG. Nitric oxide synthase in human breast cancer, a doubled-edged sword? Nitric Oxide. 2002;6:490. [Google Scholar]
- 71.Loeb KR, Loeb LA. Significance of multiple mutations in cancer. Carcinogenesis. 2000;21:379–385. doi: 10.1093/carcin/21.3.379. [DOI] [PubMed] [Google Scholar]