Abstract
Myeloid cell leukemia sequence 1 (MCL1), an anti-apoptotic BCL2 family protein, is a key regulator of intrinsic apoptosis. Normal cells require strict control over MCL1 expression with aberrant MCL1 expression linked to the emergence of various diseases and chemoresistance. Previous studies have detailed how MCL1 expression is regulated by multiple mechanisms both transcriptionally and translationally. However, characterization of the post-transcriptional regulators of MCL1 mRNA is limited. Polypyrimidine tract binding protein 1 (PTBP1) is a known regulator of post-transcriptional gene expression that can control mRNA splicing, translation, stability and localization. Here we demonstrate that PTBP1 binds to MCL1 mRNA and that knockdown of PTBP1 upregulates MCL1 expression in cancer cells by stabilizing MCL1 mRNA and increasing MCL1 mRNA accumulation in cytoplasm. Further, we show that depletion of PTBP1 protects cancer cells from antitubulin agent-induced apoptosis in a MCL1-dependent manner. Taken together, our findings suggest that PTBP1 is a novel regulator of MCL1 mRNA by which it controls apoptotic response to antitubulin chemotherapeutics.
Apoptosis, a highly regulated process of programmed cell death, is essential for maintaining tissue homeostasis and development. Dysregulation of apoptosis leads to a number of pathophysiologies including neurodegenerative diseases, inflammatory disorders, oncogenesis, and chemoresistance of certain malignancies.1 The B-cell lymphoma 2 BCL2 family of both pro- and anti-apoptotic proteins are regulators of intrinsic apoptosis which is responsible for cellular response to DNA damage, mitotic failure and homeostatic stress.2 The importance of the BCL2 family was recognized soon after the initial identification of the BCL2 gene wherein an aberrant chromosomal translocation leads to its upregulation in human follicular lymphoma.3, 4 More than 30 proteins sharing common sequence motifs with BCL2, most importantly a BH3 (BCL2 homology 3) helix, have subsequently been identified and are grouped together as the BCL2 family.5 The family is divided into three sub-families: anti-apoptotic, pro-apoptotic and pro-apoptotic BH3-only. BCL2 family regulation centers on the ability of the six anti-apoptotic BCL2 family members (BCL2, BCLXL, BCLW, MCL1 BCLB, and BFL1/A1) to bind the BH3 helices of the pro-apoptotic BCL2 proteins, BAX and BAK (BCL2 homologous antagonist/killer), thereby sequestering them in an inactive state. Multiple cellular stress pathways induce expression of one or a set of the pro-apoptotic BH3-only family members. These BH3-only proteins negatively regulate the anti-apoptotic BCL2 family members by competitively binding their BH3 binding pockets. A subset of BH3-only proteins has also been shown to directly activate BAX or BAK oligomerization.2, 5 Upregulation of the anti-apoptotic BCL2 family members has been observed across the cancer landscape6, 7 and is a recognized method for tumors to evade chemotherapeutic induced intrinsic apoptotic signaling.8, 9
MCL1, one of the anti-apoptotic BCL2 proteins and a homolog of BCL2, was first identified as a gene upregulated during differentiation of a myeloid leukemia cell line.10 It is unique among the BCL2 family due to its short half-life at both the mRNA and protein levels.1, 11 MCL1 is essential for normal homeostasis, as its knockdown in mice has been shown to impair neuronal differentiation12 and blood cell maturation.13 Further, MCL1 exhibits high expression in solid tumors6 and its overexpression has been shown to suppress cell death during extended mitotic arrest induced by microtubule-targeting agents such as taxanes and Vinca alkaloids.8, 9 MCL1 is strictly regulated by multiple mechanisms. Recent studies of the transcriptional control of MCL1 have tied its expression to multiple transcription factors (e.g., STAT3, STAT5, PU.1, HIF1 and E2F1).14, 15, 16, 17, 18 Other studies have identified multiple proteins that regulate the translation of MCL1 (CUGBP2, eIF2α and mTORC1).19, 20, 21 Further research has shown that MCL1 protein stability is regulated through binding by the BH3-only protein NOXA8, 22 and ubiquitin-mediated degradation through the E3-ligase Mule.23, 24 Together, this body of research has provided key insight into the importance of MCL1 regulation in normal and cancer biology. Yet, while these studies have established the importance of identifying regulators of MCL1 expression, few details regarding the protein regulators of MCL1 on post-transcriptional levels such as mRNA stability and subcellular distribution have been described.
The MCL1 mRNA contains a long 3′-untranslated region (3′UTR) of ~2800 bases. 3′UTRs often contain regulatory regions that mediate protein–mRNA interactions that affect mRNA stability, translation, and localization. A number of micro-RNAs (miRNAs) have been proven to target the MCL1 3′UTR for degradation.25, 26, 27 Although identification of proteins that modulate MCL1 mRNA stability and localization has been limited. One key regulator of mRNA is polypyrimidine tract binding protein 1 (PTBP1; also known as PTB or HNRNPI). PTBP1 is a RNA binding protein that regulates RNA splicing, IRES (internal ribosome entry segment)-mediated translation initiation, 3′-end processing, mRNA turnover, localization, and transportation.28 PTBP1 has been shown to be involved in distinct cellular processes including T-cell activation,29 HCV replication and infection,30 insulin secretion,31 and apoptosis32, 33, 34 through its post-transcriptional control of RNA. Recent studies have highlighted that PTBP1 expression is often dysregulated in human cancer, though its effect on malignancy appears to be cell-type dependent.35, 36, 37 In addition, it has been shown that PTBP1 knockdown can protect cancer cells from TRAIL (tumor necrosis factor-related apoptosis-inducing ligand)-induced apoptosis.32, 33 Our initial analysis of available cross-linking immunoprecipitation sequencing (CLIP-SEQ) data (GSM104818638) identified a strong association between PTBP1 and MCL1 mRNA with extensive interactions at the MCL1 3′UTR (Figure 1a). Therefore, the objective of the current study was to determine the effect that PTBP1 binding has on MCL1 mRNA and how this effect may influence cancer cell response to chemotherapeutics.
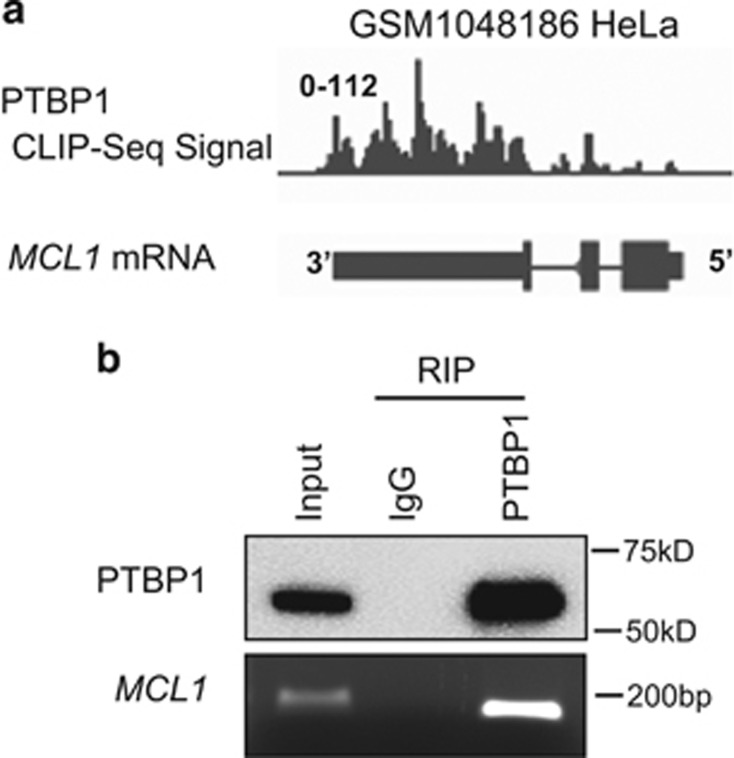
Figure 1.
PTBP1 binds to MCL1 mRNA. (a) The CLIP-SEQ mapped PTBP1 binding events are shown on the MCL1 gene. The CLIP-SEQ data GSM1048186 was previously described.38 (b) MCL1 RNA IP with PTBP1 antibody in PC3 cells. PTBP1 protein was immunoprecipitated by PTBP1 antibody and the co-immunoprecipitated MCL1 RNA was amplified by RT-PCR
Results
PTBP1 binds to MCL1 mRNA
To investigate the possible interaction between PTBP1 and MCL1 mRNA, we analyzed publicly available CLIP-SEQ data38 on genome-wide PTBP1–RNA interactions in HeLa cells. This analysis identified multiple sites of association between PTBP1 and MCL1 mRNA especially in its 3′UTR (Figure 1a). The interaction between PTBP1 and MCL1 mRNA was subsequently confirmed in the PC3 cell line (Figure 1b) using RNA immunoprecipitation (RIP).
Knockdown of PTBP1 upregulates MCL1 expression
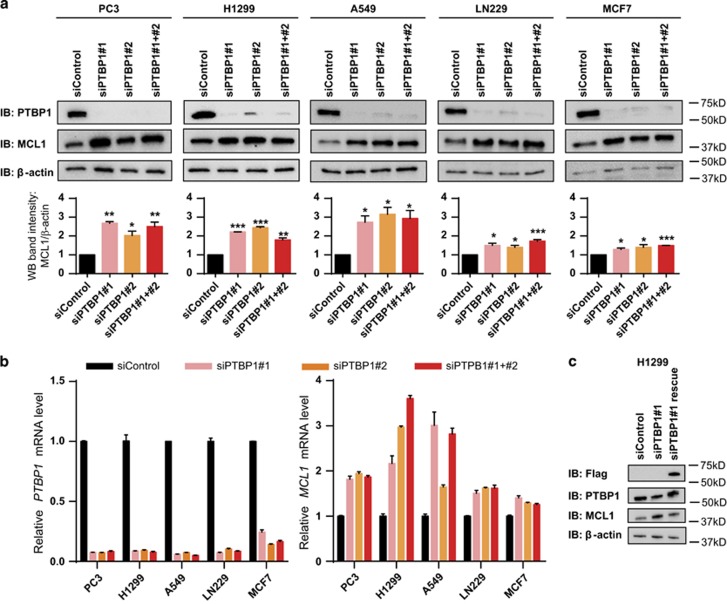
As a first step to understanding the effect that PTBP1 has on MCL1 mRNA, we assessed how changes in PTBP1 expression regulate MCL1 expression. For these studies we chose five human cancer cell lines from different tissue types—PC3 (prostate), H1299 (lung), A549 (lung), LN229 (CNS) and MCF7 (breast). We transfected each cell line with either negative control siRNA (siControl); one of two individual PTBP1-specific siRNA (siPTBP1#1 or siPTBP1#2); or a mixture of the two siPTBP1 and analyzed MCL1 protein levels by western blotting (Figure 2a) and MCL1 mRNA levels by real-time quantitative PCR (RT-qPCR; Figure 2b) 48 h after introduction of the siRNA. The mixture of siPTBP1#1 and siPTBP1#2 was used for all subsequent PTBP1 knockdowns and that mixture is hereafter referred to as siPTBP1. Knockdown of PTBP1 induced a significant upregulation of both MCL1 protein and mRNA in each of these cell lines. Overexpression of a Flag-PTBP1 construct in either PC3 cells or H1299 cells did not induce a significant change in MCL1 expression (Supplementary Figure S1).
Figure 2.
Upregulation of MCL1 in response to PTBP1 knockdown. PC3, H1299, A549, LN229 and MCF7 cells were transfected with either negative control siRNA (siControl), one of two individual siRNAs targeting PTBP1 (siPTBP1#1 or siPTBP1#2) or a mixture of both siPTBP1 for 48 h. (a) MCL1 protein level was detected by western blotting. Relative MCL1 band intensity was normalized to β-actin. (b) MCL1 and PTBP1– mRNA level was measured by RT-qPCR. Data was normalized against GAPDH. (c) H1299 cells were transfected with Flag-control vector or Flag-PTBP1 vector to rescue the upregulation of MCL1 by siPTBP1#1. Data are shown as mean±S.E.M., n=3 or more. The statistical significance was determined by unpaired student t-test where *P<0.05; **P<0.01; ***P<0.001
To demonstrate that the increase in MCL1 expression was due to PTBP1 knockdown and not an off-target effect of the siRNAs, we performed a rescue experiment using the siPTBP1#1, which targets the 3′UTR of PTBP1, in H1299 cells. For this experiment we first transfected H1299 cells with Flag-PTBP1 or Flag control. After 24 h we then treated the cells with siPTBP1#1 and measured both PTBP1 and MCL1 levels an additional 24 h later (Figure 2c). This alternate siPTBP1#1 treatment resulted in a less complete knockdown of PTBP1, but still resulted in increased expression of MCL1. Further, we observed that overexpression of Flag-PTBP1, lacking the PTBP1 3′UTR, in H1299 cells returned MCL1 expression to normal levels following siPTBP1#1 treatment (Figure 2c).
MCL1 mRNA can be alternatively spliced to produce a shortened form of MCL1, MCL1S. Unlike MCL1, MCL1S is pro-apoptotic.39 As PTBP1 is a well-known splicing regulator, we designed MCL1S specific primers to determine if knockdown of PTBP1 could also regulate MCL1S expression. However, in all five human cancer cell lines, we observed no evidence of MCL1S protein by western blotting nor mRNA by RT-qPCR in either siControl or siPTBP1-treated cells.
Knockdown of PTBP1 increases MCL1 mRNA stability
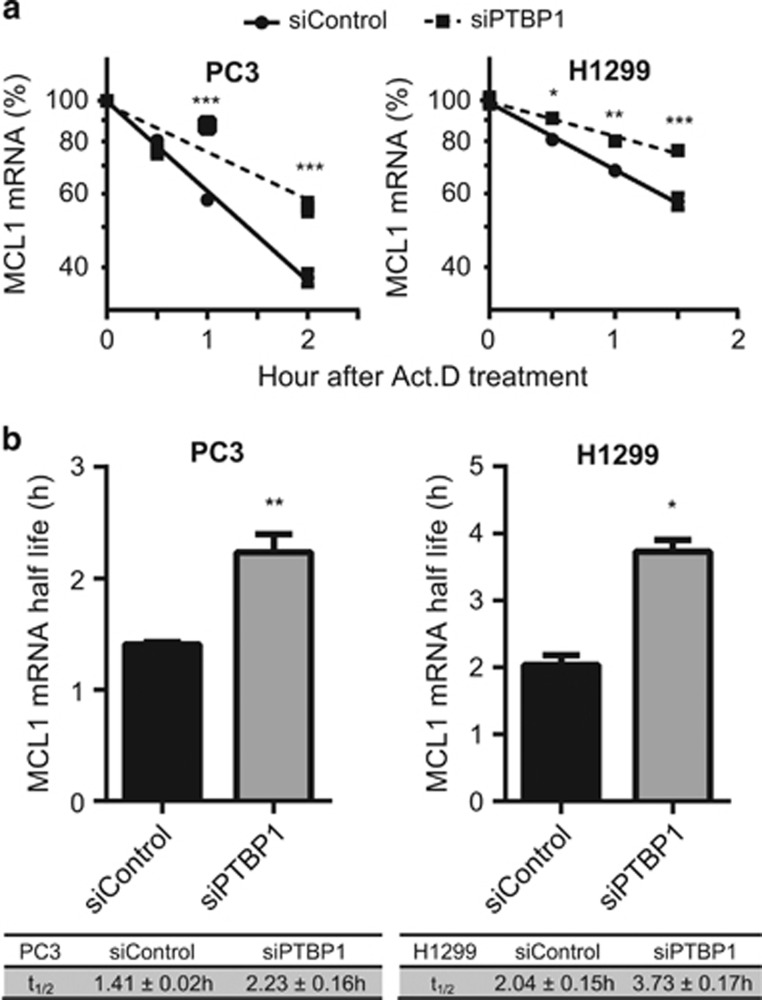
PTBP1 binding to mRNA has been reported in many cases to regulate mRNA turnover.26, 38 On the basis of the observed PTBP1 binding to MCL1 mRNA (Figure 1) and increase in MCL1 mRNA levels following PTBP1 knockdown (Figure 2b), we hypothesized that knockdown of PTBP1 stabilized MCL1 mRNA. To test this, siControl and siPTBP1 cells were treated with actinomycin D (Act. D) to block transcription and incremental RNA samples were harvested to assess MCL1 mRNA half-life. Prior studies have observed40 and our data confirm that GAPDH mRNA levels remain constant over the 8 h observation window in both PC3 and H1299 cells. MCL1 mRNA levels were therefore normalized to GAPDH levels in RT-qPCR and the relative MCL1 mRNA levels at 0 h were defined to be 100%. Linear regression decay curves of MCL1 mRNA (Figure 3) demonstrated a 1.6- and 1.8-fold increase in half-life for siPTBP1 treated PC3 and H1299 cells, respectively.
Figure 3.
Knockdown of PTBP1 stabilizes MCL1 mRNA. PC3 and H1299 cells transfected with either siControl or a mixture of two siPTBP1 for 48 h were treated with Act. D for indicated intervals. MCL1 mRNA decay was assessed by RT-qPCR using GAPDH as the internal control. (a) Data are represented as a log10 plot of the percentage of MCL1 mRNA remaining versus the time of incubation with Act. D from one representative RT-qPCR experiment. (b) MCL1 mRNA half-life (t1/2) was calculated from three independent experiments. All data are shown as mean±S.E.M., n=3 or more. The statistical significance was determined by unpaired student t-test where *P<0.05; **P<0.01; ***P<0.001
PTBP1 knockdown does not affect MCL1 protein stability
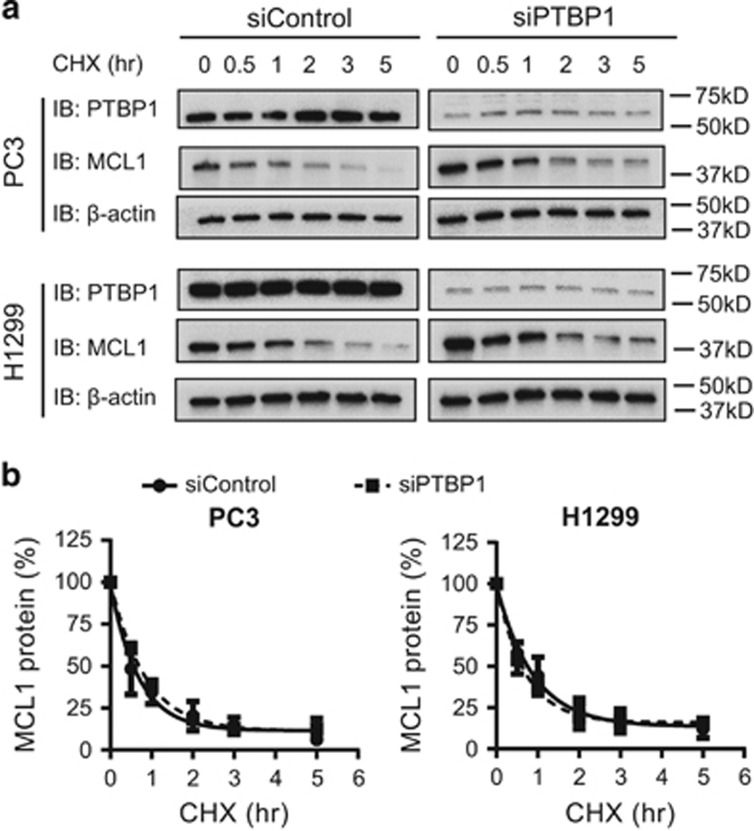
In addition to increasing MCL1 mRNA, knockdown of PTBP1 also increased MCL1 protein levels (Figure 2a). Although prior studies have not identified PTBP1 regulation of protein stability directly, we next sought to determine if PTBP1 regulation of proteins known to affect MCL1 protein stability affected MCL1 expression. We used cycloheximide (CHX) to block protein synthesis and determined the MCL1 protein decay curve by collecting and analyzing protein lysates at different time points after CHX treatment by western blotting. In each cell line, siControl or siPTBP1 treated, MCL1 half-life was unchanged at 30 and 45 min for PC3 and H1299 cells, respectively (Figure 4). These data show that PTBP1 knockdown does not affect MCL1 protein half-life in either PC3 or H1299 cells. This suggests that PTBP1 regulation of MCL1 expression occurs at the mRNA level and does not function through upstream regulation of proteins involved in MCL1 protein turnover.
Figure 4.
PTBP1 does not regulate MCL1 protein stability. PC3 and H1299 cells transfected with either siControl or a mixture of two siPTBP1 for 48 h were treated with CHX for time periods as indicated. (a) MCL1 protein levels were assessed by western blotting. (b) Relative MCL1 band intensity in a was quantified and normalized to β-actin. Data were plotted as the percentage of MCL1 protein remaining versus the times of incubation with CHX. The MCL1 protein decay curves were generated using one-phase exponential decay in Graphpad Prism. Each point was presented as mean±S.E.M., n=3
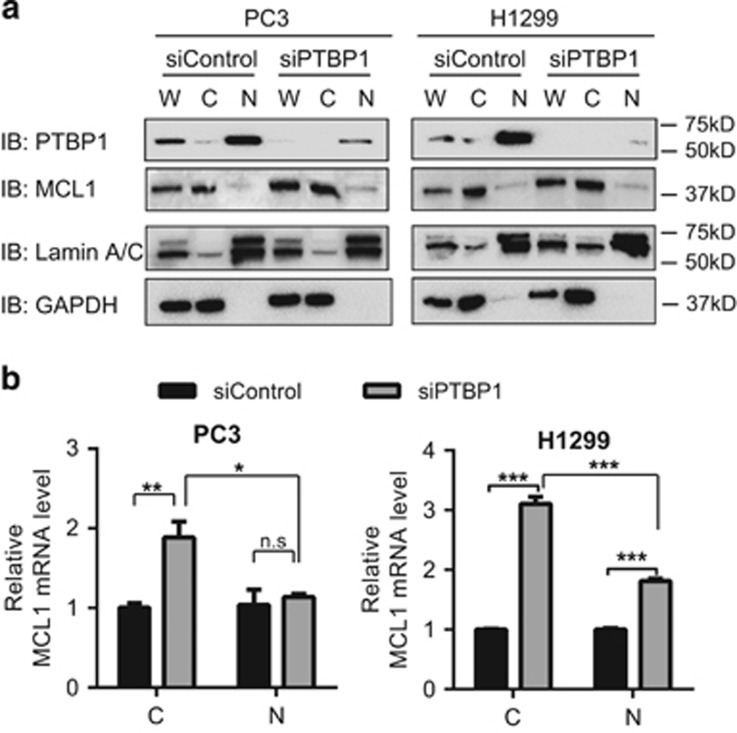
PTBP1 depletion increases MCL1 mRNA accumulation in cytoplasm
In addition to its role in regulating mRNA stability, PTBP1 is also known to regulate mRNA localization in cells. For instance, PTBP1 is required for correct Vg1 RNA localization in Xenopus oocytes during cytoplasmic RNA transport.41 Similar studies have demonstrated PTBP1's ability to regulate subcellular distribution of STX1B, VAMP2, SV2A, KIF5 and CD40L mRNA.29, 42 To determine whether PTBP1 influenced MCL1 mRNA cellular distribution, we performed nucleo-cytoplasmic fractionation of both protein and RNA in siControl and siPTBP1 cells. Both protein lysates and RNA extracts from whole-cell, cytoplasmic and nuclear fractions were collected. To confirm PTBP1 depletion efficiency and fractionation quality, equal volumes of the protein lysates were then separated on SDS-PAGE and immunoblotted for PTBP1, MCL1, Lamin A/C (nuclear control), and GAPDH (cytoplasmic control). Knockdown of PTBP1 did not influence MCL1 protein subcellular localization (Figure 5a). To assess the impact on MCL1 mRNA, we analyzed equal amounts of cytoplasmic or nuclear RNA extracts by RT-qPCR using GAPDH as an internal control. In each cell line, cytoplasmic total RNA was approximately four times that of nuclear RNA following subcellular fractionation. To assess fold change of MCL1 mRNA levels in both cytoplasmic and nuclear fractions following siPTBP1 treatment, both relative cytoplasmic and nuclear MCL1 mRNA levels in siControl cells were set as one. Knockdown of PTBP1 significantly increased MCL1 mRNA levels in the cytoplasm of both PC3 and H1299 cell lines (Figure 5b). In PC3 cells, MCL1 mRNA levels in cytoplasm increased markedly but the nuclear levels did not change (Figure 5b, left). In H1299 cells, although both the cytoplasmic and nuclear MCL1 mRNA levels increased, the increase in cytoplasm was significantly higher than that in nucleus (Figure 5b, right). These data suggest that in addition to increasing MCL1 mRNA levels, knockdown of PTBP1 also affects cellular localization of MCL1 mRNA.
Figure 5.
Downregulation of PTBP1 increases the cytoplasmic MCL1 mRNA levels more significantly. Cytoplasmic (C) and nuclear (N) fractions were prepared from PC3 and H1299 cells transfected with either siControl or a mixture of two siPTBP1 for 48 h. Whole-cell protein and RNA extracts (W) were also collected. (a) Protein fractions were analyzed by western blotting to show both the subcellular fractionation quality and that PTBP1 knockdown had no effect on MCL1 protein distribution in cells. GAPDH and Lamin A/C were used as the cytoplasmic and nuclear markers, respectively. (b) RNA fractions were quantified by RT-qPCR using GAPDH as the normalization control. Both relative cytoplasmic and nuclear MCL1 mRNA levels in siControl cells were set as one. Data are shown as mean±S.E.M., n=4. The statistical significance was determined by unpaired student t-test where *P<0.05; **P<0.01; ***P<0.001
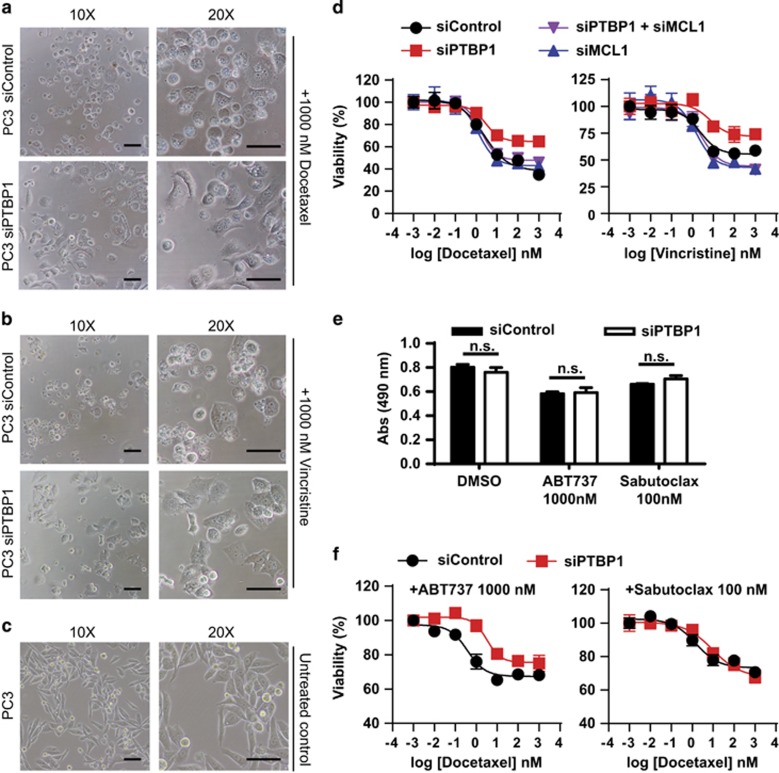
PTBP1 downregulation protects cancer cells from antitubulin agents in a MCL1-dependent manner
Prior studies have demonstrated that PTBP1 knockdown suppresses TRAIL-induced apoptosis in human cancer cells.32, 33 TRAIL has previously been identified as an extrinsic apoptotic activator.43 Further, PTBP1 has been reported to be involved in intrinsic apoptosis.34, 44 On the basis of our data demonstrating that PTBP1 knockdown increases MCL1 expression we firstly sought to determine if PTBP1 knockdown is pro-survival due to its upregulation of MCL1. For these studies we used antitubulin agents, docetaxel and vincristine, as apoptotic inducers. Antitubulin chemotherapeutics are widely used for many solid tumor types including breast cancer, prostate cancer and non-small cell lung cancer.45 The established antitubulin agents are mostly from two mechanistically different classes, taxanes (e.g., docetaxel) and Vinca alkaloids (e.g., vincristine). They cause mitotic arrest by stabilizing microtubules, which induces a cell cycle block and apoptosis.46 After treating siControl and siPTBP1 PC3 cells with 1000 nM docetaxel or 1000 nM vincristine for 48 h, siControl cells rounded up and underwent cell death while roughly half of PTBP1-depleted cells maintained a normal morphology (Figure 6a–c). To quantify this effect, we measured the viability of PC3 cells using the CellTiter 96 AQueous One Solution Cell Proliferation (MTS) Assay (Promega, Madison, WI, USA) following siControl or siPTBP1 treatment. We found that PTBP1-depleted cells were less sensitive to both docetaxel and vincristine treatment exhibiting increased cell viability (Figure 6d).
Figure 6.
Knockdown of PTBP1 increases cell viability through MCL1 under mitotic stress. PC3 cells transfected with either siControl or a mixture of two siPTBP1 were treated with 1000 nM docetaxel (a) or vinscristine (b) for 48 h. Cell morphology was checked by phase contrast microscope (scale bar, 100 μm). The morphology of untreated PC3 cells (c) were shown for comparison (scale bar, 100 μm). (d) PC3 cells were transfected with siControl; a mixture of two siPTBP1; a mixture of two siPTBP1 and two siMCL1; or a mixture of two siMCL1. Cell viability was assessed by MTS assay with various concentrations of docetaxel or vincristine for 48 h. The viability of cells treated with DMSO was set as 100%. (e) Abs (490 nm) was measured by MTS assay in siControl or siPTBP1 treated PC3 cells with DMSO, 1000 nM ABT737, or 100 nM Sabutoclax for 48 h, showing no significant change in cell viability following siPTBP1 treatment in DMSO control, ABT737, or Sabutoclax treated cells. (f) PC3 cells transfected with siControl or a mixture of two siPTBP1 were treated with 1000 nM ABT737 or 100 nM Sabutoclax combined with various doses of docetaxel for 48 h and cell viability was assessed by MTS assay. The viability of cells with 1000 nM ABT737 or 100 nM Sabutoclax alone was set as 100%. All data are presented as mean±S.E.M., n=3. The statistical significance was determined by unpaired student t-test where *P<0.05; **P<0.01; ***P<0.001
To test our hypothesis that this protective effect of PTBP1 knockdown is MCL1 dependent, we then knocked down both MCL1 and PTBP1 using siRNA and observed that siMCL1 (a mixture of two MCL1 targeted siRNAs) in PTBP1-depleted cells restored their sensitivity to both docetaxel and vincristine (Figure 6d). In addition to silencing MCL1 expression by siRNAs, we selectively blocked the anti-apoptotic BCL2 family proteins using the BH3 mimetics ABT73747 or Sabutoclax.48 ABT737 displays high binding affinity to BCL2, BCLW, and BCLXL but not to MCL1.47 Thus its efficacy is restricted to tumors with low levels of MCL1 and neutralization of MCL1 can re-sensitize resistant cancer cells to ABT737.49 Due to this limitation in ABT737, Sabutoclax (BI-97C1) was recently identified with pan-BCL2 family inhibitory potency.48 We firstly identified that 1000 nM ABT737 or 100 nM Sabutoclax could induce a small decrease in cell viability (shown as Abs 490 nm by MTS assay) which is not significantly different between cells treated with siControl and siPTBP1 (Figure 6e). We then treated siControl or siPTBP1 PC3 cells with 1000 nM ABT737 or 100 nM Sabutoclax and measured dose-dependent cell viability under docetaxel-induced mitotic stress. Following BCL2, BCLXL, and BCLW inhibition by ABT737, knockdown of PTBP1 retained its protective role on cell death (Figure 6f, left). However, when all the anti-apoptotic BCL2 family members including MCL1 were inhibited using Satutoclax, the pro-survival effect of siPTBP1 was negated (Figure 6f, right). These data support that the pro-survival effect of PTBP1 knockdown to antitubulin agents is MCL1 dependent.
In order to determine if the protective mechanism of PTBP1 knockdown can be more broadly applied to other classes of apoptotic inducers, we also assessed the viabilities of siControl or siPTBP1 treated PC3 cells with increasing doses of the DNA damage inducer cisplatin by MTS assay. In this case we did not observe a siPTBP1 induced pro-survival effect (Supplementary Figure S2).
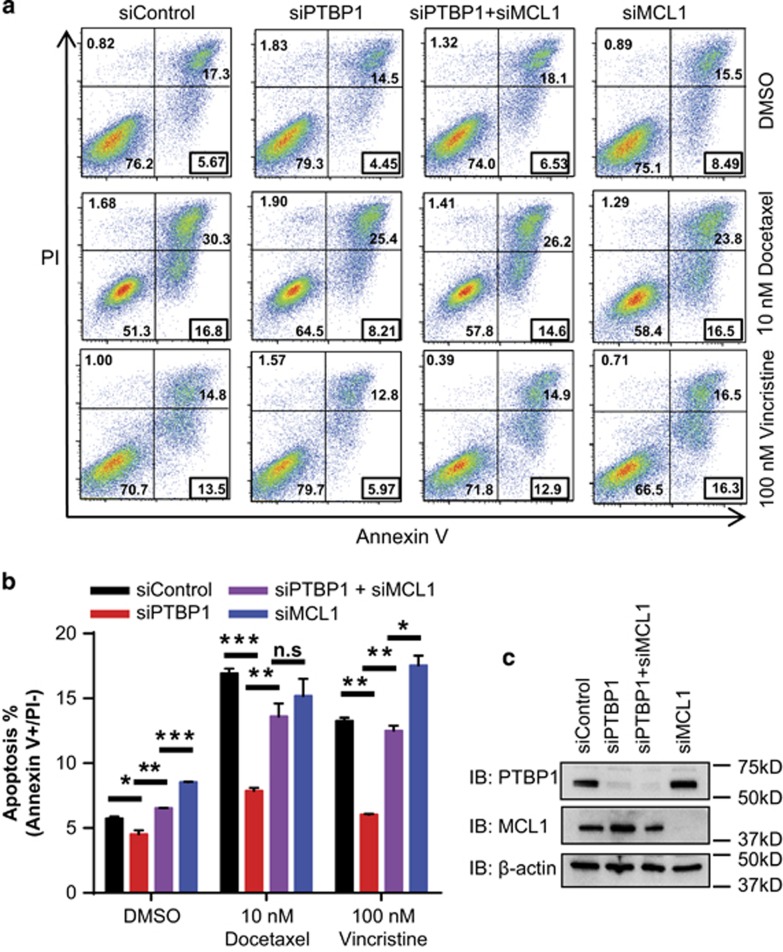
Knockdown of PTBP1 decreases apoptosis by antitubulin agents in a MCL1-dependent manner
To confirm that the PTBP1 knockdown induced pro-survival effect was due to changes in the population of apoptotic cells, PC3 cells were treated with 10 nM docetaxel or 100 nM vincristine for 48 h, harvested and analyzed by Annexin V/PI (propidium iodide) staining followed by flow cytometry to measure the degree of apoptosis. Annexin V−/PI− cells (lower left quadrant) are considered viable while Annexin V+/PI− cells (lower right quadrant) are considered apoptotic (Figure 7a).50 We observed that the viability data from the Annexin V/PI staining was in agreement with the MTS data in that depletion of PTBP1 increased cell viability and depletion of PTBP1 and MCL1 together reversed siPTBP1's effects (Figures 6d and 7a). Further, siPTBP1 caused a 50% decrease in both docetaxel and vincristine induced apoptosis compared with siControl. This reduction in the apoptotic population following siPTBP1 treatment of cells effectively returned the percent apoptosis to basal levels (DMSO treatment) suggesting siPTBP1 treatment almost completely blocked the apoptotic effects of antitubulin chemotherapeutics (Figure 7a and b). Although siMCL1+siPTBP1 together restored the apoptotic population (Figure 7a and b). The knockdown efficiency of each siRNA in both the viability and apoptosis assays was confirmed by western blotting (Figure 7c). The western blots revealed that siMCL1 in combination with siPTBP1 did not fully deplete MCL1 from the cell, as is seen during treatment with siMCL1 alone, but rather returned MCL1 levels to a basal (siControl) expression level in PC3 cells (Figure 7c). We observed similar results in H1299 cells that knockdown of PTBP1 decreased apoptosis compared with siControl (Supplementary Figure S3a and b). In H1299 cells where siMCL1+siPTBP1 treatment reduced MCL1 expression to levels comparable to siMCL1 treatment alone (Supplementary Figure S3c), there is no significant difference in apoptotic rates between siMCL1/siPTBP1 and siMCL1 alone. All data indicate that the anti-apoptotic effect of PTBP1 knockdown to antitubulin chemotherapeutics depends largely on its upregulation of MCL1.
Figure 7.
PTBP1 downregulation protects cells from antitubulin drug-induced apoptosis through MCL1. (a) Annexin V/PI staining followed by Flow Cytometry analysis was performed in PC3 cell transfected with siControl; a mixture of two siPTBP1; a mixture of two siPTBP1 and two siMCL1 or a mixture of two siMCL1 with 0.01% DMSO, 10 nM docetaxel or 100 nM vincristine for 48 h. Annexin V+/PI− gate frequencies are highlighted in the black box. (b) Statistical significance of the apoptosis rate (%, Annexin V+/PI−) was determined by unpaired student t-test. (c) MCL1 and PTBP1 protein levels were analyzed by western blotting in siControl, siPTBP1, siPTBP1+siMCL1, and siMCL1 PC3 cells. Data was shown as mean±S.E.M., n=3 where *P<0.05; **P<0.01; ***P<0.001
Discussion
The influence of PTBP1 on apoptosis continues to emerge. Thus far, PTBP1 has been demonstrated to influence TRAIL-induced extrinsic apoptosis in cancer cells32, 33 and caspase-dependent apoptosis during ischemia in cardiomyocytes.44 Studies focused on the impact of PTBP1 on apoptotic regulatory genes have shown that changes in PTBP1 expression modulate: the expression of CASPASE-3, CASPASE-9, APAF1, BAX, and BID;44 alternative splicing of BCLX into its pro-apoptotic (BCLXL) and anti-apoptotic (BCLXS) isoforms;34 and activation of IRES-mediated translation during apoptosis.32, 33 Our studies support the positioning of PTBP1 as a major regulator of apoptosis. We provide the first example that PTBP1 downregulation protects cancer cells from apoptosis induced by mitotic stress (docetaxel and vincrinstine) and further that PTBP1 is not a general apoptosis suppressor as there is no similar effect following cisplatin-induced DNA damage.
In regard to MCL1 mRNA regulation, these studies provide the first example of a protein that binds to MCL1 mRNA and regulates its mRNA stability. In addition, our findings show that PTBP1 downregulation changes the subcellular distribution of MCL1 mRNA. We observed both an increase in cytoplasmic MCL1 mRNAs and an increase in the cytoplasmic: nuclear ratio of MCL1 mRNA in PTBP1-depleted cells. The transcriptional and translational regulation of MCL1 has been extensively studied. Yet, thus far on the post-transcriptional level only miRNAs have been identified to regulate the half-life of MCL1 mRNA.25, 26, 27 Further, the identification of protein regulators of MCL1 mRNA localization has been largely ignored. PTBP1 has previously been shown to influence miRNA association with mRNAs.38 Further, a number of the regions identified in the published CLIP-SEQ study of PTBP1 in HeLa cells overlap with or neighbor known and putative miRNA sites in MCL1. Based on these studies and our results, detailed analysis of how different miRNAs that target MCL1 are influenced by PTBP1 and how overexpression or loss of one or a set of these may impact PTBP1 regulation of MCL1 mRNA could provide significant insight into the intricate control over MCL1 expression in normal homeostasis and cancer biology. Yet, the present study establishes that the regulation of MCL1 mRNA by proteins has significant impact on chemotherapeutic response and merits further study.
Recent studies have highlighted the critical positioning of MCL1 as a mitotic clock to balance cell death and survival during extended mitosis.8, 9 Thus, in periods of prolonged mitosis induced by microtubule stabilizing compounds, increased MCL1 expression enables the cells to survive. Conversely, inhibition or depletion of MCL1 during a prolonged mitotic period has been shown to induce apoptosis and to synergistically combine with compounds that induce prolonged mitosis.51 Our studies agree with these findings as we observed that PTBP1 depletion increased MCL1 expression, which protected cancer cells from apoptosis induced by antitubulin agents. Our analysis of both the prostate derived and lung derived cancer cell lines, PC3 and H1299, respectively shows that knockdown of MCL1 in PTBP1-depleted cells re-sensitizes the cells to docetaxel or vincristine. In addition, we show that when MCL1 is blocked by the pan-BCL2 family inhibitor Sabutoclax, the pro-survival effect of PTBP1 depletion is impaired completely. However, the BH3 mimetic inhibitor ABT737 which targets BCL2, BCLW, and BCLXL has no influence on the pro-survival role of PTBP1 knockdown to antitubulin chemotherapeutics. This effect may have significant implications in understanding PTBP1's roles during cancer progression and chemoresistance. In addition to its role in apoptosis regulation, MCL1 is also a key regulator of mitochondrial dynamics and metabolism.52, 53, 54, 55 Our report that PTBP1 modulates MCL1 expression thus implicates a possible involvement of PTBP1 in regulating mitochondrial dynamics. Further examination of such an effect may prove to be of interest to the field. Finally, as decreased PTBP1 has been observed as a key event during neuron,38 muscle56 and cardiomyocyte44 differentiation, its impact on MCL1 expression may serve as an important event during normal cellular development.
In summary, we have identified that downregulation of PTBP1 upregulates MCL1 expression both by increasing the half-life and cytoplasmic localization of MCL1 mRNA. Our data demonstrate that the chemo-protective effect of PTBP1 knockdown to antitubulin chemotherapeutics is due largely to this upregulation of MCL1.
Materials and Methods
Cell culture, siRNA and plasmid transfection
PC3, H1299, A549, LN229, and MCF7 cells were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum, 2.05 mM l-glutamine, 100 units per ml each of penicillin and streptomycin, and 0.25 μg/ml of Fungizone antimycotic (Life Technologies, Carlsbad, CA, USA) in a humidified atmosphere with 5% CO2.
Silencer Select validated siRNAs and negative control siRNAs were purchased from Ambion (Carlsbad, CA, USA). The siRNA sequences are: siPTBP1#1 (s11435) CAGUUUACCUGUUUUUAAAtt, siPTBP1#2 (s11436) GCAUCACGCUCUCGAAGCAtt, siMCL1#1 (s8583) CCAGUAUACUUCUUAGAAAtt, and siMCL1#2 (s8585) GTAATTAGGAACCTGTTTCtt. As a negative control, Silencer Select Negative Control #1 siRNA was used with no significant sequence similarity to human gene sequences. cDNA for PTBP1 (clone ID: 3863892) was obtained from Life Technologies and the coding sequence was cloned into the pcDNA3.1 plasmid with a Flag tag at the N terminus using a pcDNA 3.1 TOPO TA Expression Kit (Invitrogen, Carlsbad, CA, USA) for exogenous expression. siRNA and plasmid transfections were carried out using Lipofectamine RNA iMAX reagents and Lipofectamine 3000 reagents (Life Technolgies), respectively. Cells were harvested for analysis after 48 or 72 h transfection as designated in text.
Chemicals
Docetaxel, Vincrisitine, Cisplatin, ABT737, and Sabutoclax were purchased from Selleck Chemicals (Houston, TX, USA).
RT-qPCR
Total RNA was isolated from cells by Trizol reagent (Life Technologies) and purified by PureLink RNA Mini Kit (Ambion). Genomic DNA was removed using RNAse-free DNAse treatment. Final RNA concentrations were measured by absorbance at 260 nm and quality was confirmed using a A260/280 ratio of ~2.0. cDNA was prepared using 1 μg of total RNA in 20 μl reverse transcription reaction with qScript cDNA SuperMix (Quanta Biosciences, Gaithersburg, MD, USA) according to manufacturer's protocol.
RT-qPCR reactions were performed in a 10 μl reaction containing 4 μl of the diluted cDNA, 5 μl PerfeCta SYBR green FastMix, Low ROX (Quanta Biosciences), and 0.5 μl each of forward and reverse primers at final concentrations of 250 nM. All qPCR reactions were run in quadruplicate on MicroAmp Optical 384-well plates on a ViiA7 instrument (Applied Biosystems, Carlsbad, CA, USA). Amplification conditions consisted of the initial denaturation step at 95 °C for 3 min, followed by 40 cycles of 10 s at 95 °C and 30 s at 60 °C. Afterwards, melting curves were generated to confirm presence of a single uniform peak. Results were analyzed in ViiA 7 software by comparative CT (ΔΔCT) method using GAPDH as a normalization control and exported for analysis and presentation in GraphPad Prism (GraphPad Software, La Jolla, CA, USA).
The following primers (5′–3′) were used:
MCL1: (F) GGACATCAAAAACGAAGACG and (R) GCAGCTTTCTTGGTTTATGG; PTBP1: (F) ATTGTCCCAGATATAGCCGTTG and (R) GCTGTCATTTCCGTTTGCTG; GAPDH: (F) CCACATCGCTCAGACACCAT and (R) CCAGGCGCCCAATACG; MCL1S: (F) GCCTTCCAAGGATGGGTTTGTG and (R) CAAACCAGCTCCTACTCCAGCA.
Protein extraction and western blot
Whole-cell lysates were prepared by lysating cells on ice with lysis buffer (Pierce, Rockford, IL, USA): 25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP40, and 5% glycerol supplemented with protease inhibitors (1 mM AEBSF, 0.8 μM aprotinin, 0.05 mM bestatin, 0.015 mM E-64, 0.02 mM leupeptin, 0.01 mM pepstatin A).
Protein lysates were resolved by SDS-PAGE and transferred to PVDF membrane in a wet transfer system for 1 h at 100 V. Membranes were incubated with anti-MCL1 Abs (D35A5, Cell Signaling, Danvers, MA, USA), anti-PTBP1 Abs (RN011P, MBL International, Woburn, MA, USA), anti-β-actin Abs (PA1-21167, Pierce, Rockford, IL, USA), and anti-Lamin A/C Abs (H-110, Santa Cruz, Santa Cruz, CA, USA) at a dilution of 1 : 1000, and anti-GAPDH Abs (MAB373, Millipore, Billerica, MA, USA) at a dilution of 1 : 10 000 at 4 °C overnight. HPR-conjugated secondary Abs were used for detection using ECL2 reagent (Pierce). Immunoblots were visualized on Bio-Rad ChemiDoc MP imaging system. Band intensities were quantified using Bio-Rad Image Lab software.
Subcellular fractionation
Subcellular fractionation was performed by firstly suspending cells in hypotonic buffer A (10 mM Hepes pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, 0.15% Triton X-100, protease inhibitor) and swollen on ice for 8 min. After spinning down for 30 s at 12 000 × g, supernatant was saved as the cytoplasmic fraction. Cytoplasmic RNA was isolated by Trizol and cytoplasmic protein lysates can be used immediately. The nuclear pellet was rinsed with cold PBS once. For the nuclear protein extraction, pellets were lysed with equal volume of buffer B (20 mM Hepes pH 7.9, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 0.5% Triton X-100, protease inhibitor) on ice for 15 min. For nuclear RNA extraction, pellets were directly lysed by Trizol. Equal volumes of cytoplasmic and nuclear protein fractions were analyzed by western blot. Cytoplasmic and nuclear RNA fractions were analyzed by RT-qPCR using GAPDH as the normalization control.
RNA immunoprecipitation
RNA IP was done in PC3 cells using the RiboCluster Profiler RIP-Assay Kit (MBL International). In brief, cell extracts were incubated with anti-PTBP1-Abs (RN011P, MBL International) immobilized with protein A magnetic beads (Dynabeads, Thermo Fisher, Waltham, MA, USA), and the RNA–protein complex was then eluted. RNA was isolated and target RNA was analyzed by RT-PCR as outlined above.
mRNA and protein decay analysis
RNA stability was measured by incubating cells with 10 μg/ml Act. D and extracting total RNA at different time point. RT-qPCR was carried out to get relative RNA levels remaining (%) after Act.D treatment using GAPDH as the internal control. mRNA half-life was calculated by transforming the percentage of RNA levels remaining to log scale and linear regression of the mRNA decay in Graphpad Prism.
Protein stability was measured by treating cells with 5 μg/ml of cycloheximide and extracting total protein lysates at different time point. Total cell lysates (20 μg) were analyzed by western blotting and MCL1 and β-actin band intensities were quantified by Bio-Rad Image Lab. Percent MCL1 protein level remaining was calculated by setting the band intensity at 0 h as one and using β-actin band intensity as the internal control.
MTS assay
CellTiter 96 AQueous One Solution Cell Proliferation (MTS) Assay (Promega) was performed according to manufacturer's protocol. Viability (%) was calculated from the absorbance (490 nm) at different docetaxel/vincristine doses. Medium control absorbance (490 nm) was subtracted as the background read.
Flow cytometry
PC3 cells (2 × 105) were transfected with control siRNA or siPTBP1 alone or siMCL1 alone or siPTBP1 plus siMCL1. After 24 h, cells were treated with 0.1% DMSO as control or 10 nM docetaxel/100 nM vincristine in 0.1% DMSO for 48 h. Cells were trypsinized and 1 × 105 cells were stained with Annexin V-FITC and PI (BD Biosciences, San Jose, CA, USA) at room temperature for 15 min. Then cells were subjected to flow cytometry on BD LSRFortessa FACS, 50 000 events were collected for each sample and data was analyzed by FlowJo V10.
Statistical analysis
Statistical analysis was performed in Prism, Graphpad or Microsoft Excel. Unpaired Student t-test was used for all the statistical analysis. *P<0.05, **P<0.01, ***P<0.001.
Acknowledgments
We thank Hairui Su (UAB) for his help in FACS. The research was funded by grants from the UAB Comprehensive Cancer Center Young Supporters Board (WJP); UAB HHMI Med into Grad Fellowship (JC).
Glossary
- PTBP1
polypyrimidine tract binding protein 1
- BCL2
B-cell lymphoma 2
- BH3
BCL2 homology 3
- MCL1
myeloid cell leukemia sequence 1
- BAK
BCL2 homologous antagonist/killer
- 3′UTR
3′-untranslated region
- miRNA
micro-RNA
- CLIP-SEQ
cross-linking immunoprecipitation sequencing
- RIP
RNA immunoprecipitation
- Act. D
actinomycin D
- RT-qPCR
real-time quantitative PCR
- CHX
cycloheximide
- PI
propidium iodide
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by C Borner
Supplementary Material
References
- Thomas LW, Lam C, Edwards SW. Mcl-1; the molecular regulation of protein function. FEBS Lett 2010; 584: 2981–2989. [DOI] [PubMed] [Google Scholar]
- Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 2008; 9: 47–59. [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y, Cossman J, Jaffe E, Croce CM. Involvement of the bcl-2 gene in human follicular lymphoma. Science 1985; 228: 1440–1443. [DOI] [PubMed] [Google Scholar]
- Vaux DL, Cory S, Adams JM. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature 1988; 335: 440–442. [DOI] [PubMed] [Google Scholar]
- Moldoveanu T, Follis AV, Kriwacki RW, Green DR. Many players in BCL-2 family affairs. Trends Biochem Sci 2014; 39: 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placzek WJ, Wei J, Kitada S, Zhai D, Reed JC, Pellecchia M. A survey of the anti-apoptotic Bcl-2 subfamily expression in cancer types provides a platform to predict the efficacy of Bcl-2 antagonists in cancer therapy. Cell Death Dis 2010; 1: e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010; 463: 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haschka MD, Soratroi C, Kirschnek S, Hacker G, Hilbe R, Geley S et al. The NOXA-MCL1-BIM axis defines lifespan on extended mitotic arrest. Nat Commun 2015; 6: 6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature 2011; 471: 110–114. [DOI] [PubMed] [Google Scholar]
- Kozopas KM YT, Buchan HL, Zhou P, Craig RW. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc Natl Acad Sci USA 1993; 90: 3516–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Buchan HL, Townsend KJ, Craig RW. MCL-1, a member of the BLC-2 family, is induced rapidly in response to signals for cell differentiation or death, but not to signals for cell proliferation. J Cell Physiol 1996; 166: 532–536. [DOI] [PubMed] [Google Scholar]
- Arbour N, Vanderluit JL, Le Grand JN, Jahani-Asl A, Ruzhynsky VA, Cheung EC et al. Mcl-1 is a key regulator of apoptosis during CNS development and after DNA damage. J Neurosci 2008; 28: 6068–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opferman JT, Iwasaki H, Ong CC, Suh H, Mizuno S, Akashi K et al. Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science 2005; 307: 1101–1104. [DOI] [PubMed] [Google Scholar]
- Liu H, Ma Y, Cole SM, Zander C, Chen KH, Karras J et al. Serine phosphorylation of STAT3 is essential for Mcl-1 expression and macrophage survival. Blood 2003; 101: 344–352. [DOI] [PubMed] [Google Scholar]
- Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P et al. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol 2010; 11: 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JM, Lai MZ, Yang-Yen HF. Interleukin-3 stimulation of mcl-1 gene transcription involves activation of the PU.1 transcription factor through a p38 mitogen-activated protein kinase-dependent pathway. Mol Cell Biol 2003; 23: 1896–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piret JP, Minet E, Cosse JP, Ninane N, Debacq C, Raes M et al. Hypoxia-inducible factor-1-dependent overexpression of myeloid cell factor-1 protects hypoxic cells against tert-butyl hydroperoxide-induced apoptosis. J Biol Chem 2005; 280: 9336–9344. [DOI] [PubMed] [Google Scholar]
- Croxton R, Ma Y, Song L, Haura EB, Cress WD. Direct repression of the Mcl-1 promoter by E2F1. Oncogene 2002; 21: 1359–1369. [DOI] [PubMed] [Google Scholar]
- Subramaniam D, Natarajan G, Ramalingam S, Ramachandran I, May R, Queimado L et al. Translation inhibition during cell cycle arrest and apoptosis: Mcl-1 is a novel target for RNA binding protein CUGBP2. Am J Physiol Gastrointest Liver Physiol 2008; 294: G1025–G1032. [DOI] [PubMed] [Google Scholar]
- Fritsch RM, Schneider G, Saur D, Scheibel M, Schmid RM. Translational repression of MCL-1 couples stress-induced eIF2 alpha phosphorylation to mitochondrial apoptosis initiation. J Biol Chem 2007; 282: 22551–22562. [DOI] [PubMed] [Google Scholar]
- Mills JR, Hippo Y, Robert F, Chen SM, Malina A, Lin CJ et al. mTORC1 promotes survival through translational control of Mcl-1. Proc Natl Acad Sci USA 2008; 105: 10853–10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang X, Zhang J, Lopez H, Wang Y, Li W, O'Neill KL et al. The carboxyl-terminal tail of Noxa protein regulates the stability of Noxa and Mcl-1. J Biol Chem 2014; 289: 17802–17811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Bougie P, Menoret E, Juin P, Dousset C, Pellat-Deceunynck C, Amiot M. Noxa controls Mule-dependent Mcl-1 ubiquitination through the regulation of the Mcl-1/USP9X interaction. Biochem Biophys Res Commun 2011; 413: 460–464. [DOI] [PubMed] [Google Scholar]
- Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell 2005; 121: 1085–1095. [DOI] [PubMed] [Google Scholar]
- Garzon R, Heaphy CE, Havelange V, Fabbri M, Volinia S, Tsao T et al. MicroRNA 29b functions in acute myeloid leukemia. Blood 2009; 114: 5331–5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Zhang JP, Li B, Zeng C, You K, Chen MX et al. MicroRNA-125b promotes apoptosis by regulating the expression of Mcl-1, Bcl-w and IL-6R. Oncogene 2013; 32: 3071–3079. [DOI] [PubMed] [Google Scholar]
- Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y et al. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res 2009; 69: 1135–1142. [DOI] [PubMed] [Google Scholar]
- Sawicka K, Bushell M, Spriggs KA, Willis AE. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem Soc Trans 2008; 36: 641–647. [DOI] [PubMed] [Google Scholar]
- Matus-Nicodemos R, Vavassori S, Castro-Faix M, Valentin-Acevedo A, Singh K, Marcelli V et al. Polypyrimidine tract-binding protein is critical for the turnover and subcellular distribution of CD40 ligand mRNA in CD4+ T cells. J Immunol 2011; 186: 2164–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischendorf JJ, Begar C, Korf M, Manns MP, Krüger M. Polypyrimidine tract-binding protein (PTB) inhibits Hepatitis C virus internal ribosome entry site (HCV IRES)-mediated translation, but does not affect HCV replication. Arch Virol 2004; 149: 1955–1970. [DOI] [PubMed] [Google Scholar]
- Knoch KP, Bergert H, Borgonovo B, Saeger HD, Altkruger A, Verkade P et al. Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nat Cell Biol 2004; 6: 207–214. [DOI] [PubMed] [Google Scholar]
- Bushell M, Stoneley M, Kong YW, Hamilton TL, Spriggs KA, Dobbyn HC et al. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol Cell 2006; 23: 401–412. [DOI] [PubMed] [Google Scholar]
- King HA, Cobbold LC, Pichon X, Poyry T, Wilson LA, Booden H et al. Remodelling of a polypyrimidine tract-binding protein complex during apoptosis activates cellular IRESs. Cell Death Differ 2014; 21: 161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielli P, Bordi M, Di Biasio V, Sette C. Regulation of BCL-X splicing reveals a role for the polypyrimidine tract binding protein (PTBP1/hnRNP I) in alternative 5' splice site selection. Nucleic Acids Res 2014; 42: 12070–12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Norton JT, Ghosh S, Kim J, Fushimi K, Wu JY et al. Polypyrimidine tract-binding protein (PTB) differentially affects malignancy in a cell line-dependent manner. J Biol Chem 2008; 283: 20277–20287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Arslan AD, Ho TT, Yuan C, Stampfer MR, Beck WT. Involvement of polypyrimidine tract-binding protein (PTBP1) in maintaining breast cancer cell growth and malignant properties. Oncogenesis 2014; 3: e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Pool M, Darcy KM, Lim SB, Auersperg N, Coon JS et al. Knockdown of polypyrimidine tract-binding protein suppresses ovarian tumor cell growth and invasiveness in vitro. Oncogene 2007; 26: 4961–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 2013; 152: 82–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae J, Leo CP, Hsu SY, Hsueh AJ. MCL-1S, a splicing variant of the antiapoptotic BCL-2 family member MCL-1, encodes a proapoptotic protein possessing only the BH3 domain. J Biol Chem 2000; 275: 25255–25261. [DOI] [PubMed] [Google Scholar]
- Lam LT, Pickeral OK, Peng AC, Rosenwald A, Hurt EM, Giltnane JM et al. Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol 2001; 2: RESEARCH0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RA, Gagnon JA, Mowry KL. PTB/hnRNP I is required for RNP remodeling during RNA localization in Xenopus oocytes. Mol Cell Biol 2008; 28: 678–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K, Lim ZQ, Khandelia P, Friedman B, Makeyev EV. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev 2012; 26: 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, El-Deiry WS. TRAIL and apoptosis induction by TNF-family death receptors. Oncogene 2003; 22: 8628–8633. [DOI] [PubMed] [Google Scholar]
- Zhang J, Bahi N, Llovera M, Comella JX, Sanchis D. Polypyrimidine tract binding proteins (PTB) regulate the expression of apoptotic genes and susceptibility to caspase-dependent apoptosis in differentiating cardiomyocytes. Cell Death Differ 2009; 16: 1460–1468. [DOI] [PubMed] [Google Scholar]
- Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer 2004; 4: 253–265. [DOI] [PubMed] [Google Scholar]
- Topham CH, Taylor SS. Mitosis and apoptosis: how is the balance set? Curr Opin Cell Biol 2013; 25: 780–785. [DOI] [PubMed] [Google Scholar]
- Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 2005; 435: 677–681. [DOI] [PubMed] [Google Scholar]
- Wei J, Stebbins JL, Kitada S, Dash R, Placzek W, Rega MF et al. BI-97C1, an optically pure Apogossypol derivative as pan-active inhibitor of antiapoptotic B-cell lymphoma/leukemia-2 (Bcl-2) family proteins. J Med Chem 2010; 53: 4166–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Delft MF, Wei AH, Mason KD, Vandenberg CJ, Chen L, Czabotar PE et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell 2006; 10: 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger CP. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J Immunol Methods 1995; 184: 39–51. [DOI] [PubMed] [Google Scholar]
- Jackson RS 2nd, Placzek W, Fernandez A, Ziaee S, Chu CY, Wei J et al. Sabutoclax, a Mcl-1 antagonist, inhibits tumorigenesis in transgenic mouse and human xenograft models of prostate cancer. Neoplasia 2012; 14: 656–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morciano G, Giorgi C, Balestra D, Marchi S, Perrone D, Pinotti M et al. Mcl-1 involvement in mitochondrial dynamics is associated with apoptotic cell death. Mol Biol Cell 2016; 27: 20–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perciavalle RM, Stewart DP, Koss B, Lynch J, Milasta S, Bathina M et al. Anti-apoptotic MCL-1 localizes to the mitochondrial matrix and couples mitochondrial fusion to respiration. Nat Cell Biol 2012; 14: 575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadarajan S, Butterworth M, Wei J, Pellecchia M, Dinsdale D, Cohen GM. Sabutoclax (BI97C1) and BI112D1, putative inhibitors of MCL-1, induce mitochondrial fragmentation either upstream of or independent of apoptosis. Neoplasia 2013; 15: 568–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Shah K, Bradbury NA, Li C, White C. Mcl-1 promotes lung cancer cell migration by directly interacting with VDAC to increase mitochondrial Ca2+ uptake and reactive oxygen species generation. Cell Death Dis 2014; 5: e1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MP, Nagel RJ, Fagg WS, Shiue L, Cline MS, Perriman RJ et al. Quaking and PTB control overlapping splicing regulatory networks during muscle cell differentiation. RNA 2013; 19: 627–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.