Abstract
Caspases are a family of proteases found in all metazoans, including a dozen in humans, that drive the terminal stages of apoptosis as well as other cellular remodeling and inflammatory events. Caspases are named because they are cysteine class enzymes shown to cleave after aspartate residues. In the past decade, we and others have developed unbiased proteomic methods that collectively identified ~2000 native proteins cleaved during apoptosis after the signature aspartate residues. Here, we explore non-aspartate cleavage events and identify 100s of substrates cleaved after glutamate in both human and murine apoptotic samples. The extended consensus sequence patterns are virtually identical for the aspartate and glutamate cleavage sites suggesting they are cleaved by the same caspases. Detailed kinetic analyses of the dominant apoptotic executioner caspases-3 and -7 show that synthetic substrates containing DEVD↓ are cleaved only twofold faster than DEVE↓, which is well within the 500-fold range of rates that natural proteins are cut. X-ray crystallography studies confirm that the two acidic substrates bind in virtually the same way to either caspases-3 or -7 with minimal adjustments to accommodate the larger glutamate. Lastly, during apoptosis we found 121 proteins cleaved after serine residues that have been previously annotated to be phosphorylation sites. We found that caspase-3, but not caspase-7, can cleave peptides containing DEVpS↓ at only threefold slower rate than DEVD↓, but does not cleave the unphosphorylated serine peptide. There are only a handful of previously reported examples of proteins cleaved after glutamate and none after phosphorserine. Our studies reveal a much greater promiscuity for cleaving after acidic residues and the name 'cacidase' could aptly reflect this broader specificity.
Human caspases are a family of 12 homologous intracellular proteases known for driving cellular state changes such as apoptosis and differentiation, as well as inflammatory responses. Caspases are cysteine-class proteases named for their signature ability to cleave after aspartate residues, or P1 is aspartate using the Schetchter and Berger notation.1, 2 Synthetic peptide profiling for purified caspases show distinctive subsite preferences extending from P1 to P4 or P5.3, 4, 5 Sequence conservation and signal pathway analyses have further grouped the proteases into apoptotic initiators (−2, −8, −9 and −10), apoptotic executioners (−3, −6 and −7), regulators of inflammation (−1, −4, −5 and −12) and keratinocyte differentiation −14).
The past decade has seen a significant advancement in the use of LC-MS to define the spectrum of natural proteins cleaved by caspases in cells.6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 In addition to providing unbiased information about which proteins are cleaved, in some cases, these experiments locate the precise sites and quantify the rates of cleavage.13, 14, 15, 16, 17 Using the subtiligase-based N-terminomics approach, we identified more than 1700 aspartate cleavage events distributed among about 1200 different protein substrates upon induction of apoptosis across seven human cell lines (http://wellslab.ucsf.edu/degrabase/).18 These findings and others revealed structural preferences for cleavage in loops>helices>sheets.14, 19, 20 This large database of aspartate-cleaved proteins and their conservation in metazoans has helped to reveal the pathways and nodes that drive apoptosis.
Here, we extend this analysis to proteins cleaved at non-aspartate sites during apoptosis. Surprisingly, we find enrichment of proteins that are cut after glutamate in apoptotic cells. We find these glutamate sites have similar subsite specificities, degrees of conservation, and Gene Ontology (GO) term enrichments as seen for aspartate cleavages. Remarkably, the catalytic efficiency for cleaving a glutamate substrate is only twofold less than for cleaving the matched aspartate substrate by caspase-3 or caspase-7. Structural studies show that both acidic residues are accommodated in the binding sites of the two enzymes. Finally, we identified 121 P1 serine sites that are literature annotated phosphorylation sites and cut in apoptosis, and show caspase-3 can cleave after phosphoserine. Previously, there have been a handful of studies reporting proteins cut after glutamate by caspases.17, 22, 23, 24, 25, 26 Our studies reveal a surprising promiscuity for caspases to cleave P1 acidic residues suggesting a more expanded range of substrates than previously appreciated.
Results
Human and mouse apoptotic cells are similarly enriched for proteolysis after aspartate and glutamate
The DegraBase (http://wellslab.ucsf.edu/degrabase/) is a database comprising about 8000 unique proteolytic cuts identified by the subtiligase-based N-terminomics labeling technology from 33 apoptotic and 11 healthy cellular experiments.18 From this resource we ranked the percentage of instances that the 20 amino acids appear in the P1 position in apoptotic compared with healthy cells (Table 1). Consistent with major activation of caspases, the strongest enrichment ratio for substrates cut in apoptosis versus healthy cells is for aspartate residues (3.7-fold enriched). However, we were intrigued to find the second most enriched P1 residue during apoptosis is glutamate (3.1-fold enriched). From the Degrabase, we identified a total of 1706 P1 aspartate cleavage sites in human cells and 253 P1 glutamate sites (Table 2a). The 1706 P1 aspartate cleavages are distributed among 1268 proteins, which represent 1.3 cleavages per protein. Similarly, 253 P1 glutamate cleavages are found in 226 proteins, or 1.1 cuts per protein. We had previously generated a smaller apoptotic data set for mouse cells.27 From this data set we find 38 unique P1 glutamate cleavages compared with 259 P1 aspartate cleavages (Table 2a). We find a similar number of cut sites per protein from the mouse data: 38 cuts in 36 unique proteins for P1 glutamate compared with the 259 peptides in 221 proteins for P1 aspartate, reflecting 1.1 and 1.2 cuts per protein, respectively. These data suggest that cleavage after glutamate is quite frequent in both human and mouse cells and represents the same fraction of total cuts.
Table 1. Enrichment for each of the 20 possible amino acids observed at the P1 position in apoptotic samples compared with healthy cells.
| P1 | Apoptotic | % | Healthy | % | Apoptotic/healthy |
|---|---|---|---|---|---|
| K | 659 | 9.43% | 459 | 21.41% | 0.44 |
| R | 475 | 6.80% | 284 | 13.25% | 0.51 |
| F | 200 | 2.86% | 115 | 5.36% | 0.53 |
| — | 135 | 1.93% | 76 | 3.54% | 0.54 |
| Y | 187 | 2.68% | 100 | 4.66% | 0.57 |
| M | 356 | 5.09% | 174 | 8.12% | 0.63 |
| V | 96 | 1.37% | 35 | 1.63% | 0.84 |
| L | 351 | 5.02% | 122 | 5.69% | 0.88 |
| H | 97 | 1.39% | 30 | 1.40% | 0.99 |
| N | 449 | 6.42% | 129 | 6.02% | 1.07 |
| Q | 195 | 2.79% | 54 | 2.52% | 1.11 |
| A | 501 | 7.17% | 138 | 6.44% | 1.11 |
| C | 94 | 1.34% | 25 | 1.17% | 1.15 |
| I | 42 | 0.60% | 11 | 0.51% | 1.17 |
| W | 24 | 0.34% | 6 | 0.28% | 1.23 |
| S | 386 | 5.52% | 77 | 3.59% | 1.54 |
| G | 378 | 5.41% | 75 | 3.50% | 1.55 |
| P | 199 | 2.85% | 35 | 1.63% | 1.74 |
| T | 206 | 2.95% | 34 | 1.59% | 1.86 |
| E | 253 | 3.62% | 25 | 1.17% | 3.10 |
| D | 1706 | 24.41% | 140 | 6.53% | 3.74 |
| U | 1 | 0.01% | 0 | 0.00% | - |
| Total | 6990 | 100% | 2144 | 100% |
The fold enrichment of each residue is calculated from the ratio of the percent of peptides found with each P1 residue in apoptosis to the percent found in healthy cells. The fold enrichment values (final column) is ranked from low (green) to high (red) reflecting higher prevalence in apoptotic cells. Note the high enrichment in apoptotic cells for substrates cleaved with P1 aspartate, glutamate and serine. There is one cleavage in apoptotic cells after selenocysteine (U)
Table 2. Similar ratios of P1 Glu/Asp cuts in human and mouse apoptotic cells (a), and similar conservation of sites (b).
|
a | ||||
|---|---|---|---|---|
|
Human |
Mouse |
|||
| Count | Acidic %Glu | Count | Acidic %Glu | |
| Asp N-termini | 1706 | 12.9% | 259 | 12.8% |
| Glu N-termini | 253 | 38 | ||
| Asp Proteins | 1268 | 15.1% | 221 | 14.0% |
| Glu Proteins | 226 | 36 | ||
|
b | ||||
|---|---|---|---|---|
| Asp | Glu | Asp | Glu | |
| Human | ||||
| Asp | 1268 | |||
| Glu | 100 | 226 | ||
| Mouse | ||||
| Asp | 179 | 31 | 221 | |
| Glu | 14 | 18 | 10 | 36 |
(a) The number of unique N-termini and protein targets are shown for aspartate and glutamate sites for human and mouse. The percent of P1 glutamate sites is essentially the same between human and mouse data sets. (b) There is a significant overlap of aspartate and glutamate proteins within human and between human and mouse. Almost half the proteins cleaved after glutamate are also cleaved at an aspartate site
We next compared the conservation of P1 glutamate and aspartate apoptotic sites between mouse and human (Table 2b). Out of the 221 P1 aspartate-cleaved mouse proteins, 179 human orthologs were also cleaved.27 Here we find that of the 36 glutamate-cleaved mouse proteins identified, 18 proteins have human orthologs that are also cut. For the 179 targets that are cut at aspartate in both mouse and human, 149 are cleaved at precisely the same site (83% site conservation). Similarly for the 18 glutamate cut targets in common between human and mouse, 15 of the sites are conserved (83% site conservation). Thus, aspartate and glutamate sites appear equally conserved suggesting they are under roughly equivalent evolutionary pressure.
Proteins cleaved at P1 glutamate frequently have P1 aspartate cleavages
During apoptosis, some proteins are cut more than once by caspases. Thus, we compared the frequency that proteins are cut both after glutamate and aspartate (Table 2b). Out of 1268 aspartate-cleaved proteins, we find 100 are also cut at a P1 glutamate site, which is 8% of the aspartate-cleaved proteins. In contrast, of 226 proteins that cut after glutamate, 100 are also cut after aspartate representing 45% of the total glutamate-cleaved proteins. Similarly in mouse, for the 221 total aspartate-cleaved targets only 10 had an additional P1 glutamate cut site (~4%), but for 36 glutamate targets 10 had an additional P1 aspartate cut site (~30%). Thus, proteins targeted for glutamate cleavage have a higher propensity to contain an additional aspartate cut site. Moreover, there are two populations of dual acid cut proteins. About 40% of human proteins cut at both aspartate and glutamate sites (112 proteins total) are cleaved within 20 amino acids of each other, suggesting the presence of hot spots, and another 40% cut over 100 residues apart, suggesting unique cleavage effects (Supplementary Figure 1a). In contrast, out of the 726 cleavages from 287 proteins with multiple aspartate cleavages, only 11% are within the 20 amino acid hot spot and 70% are cut over 100 residues apart, suggesting aspartate sites are optimized for unique cleavage effects (Supplementary Figure 1b). We found no difference in the location of sites within the substrate for cutting after either aspartate or glutamate (Supplementary Figure 1c) as most cleavages appear after 60 residues of the protein. The glutamate cuts follow the same pattern of structural preference as aspartate cuts, loops>helices>sheets (Supplementary Figure 1d), and almost all of the glutamate substrates GO terms overlap with the aspartate substrates (Supplementary Figure 1e). Thus, from a secondary structural and functional bioinformatics perspective we cannot distinguish P1 glutamate from P1 aspartate-cleaved targets.
Caspases can cleave after aspartate or glutamate residues in cells and in vitro
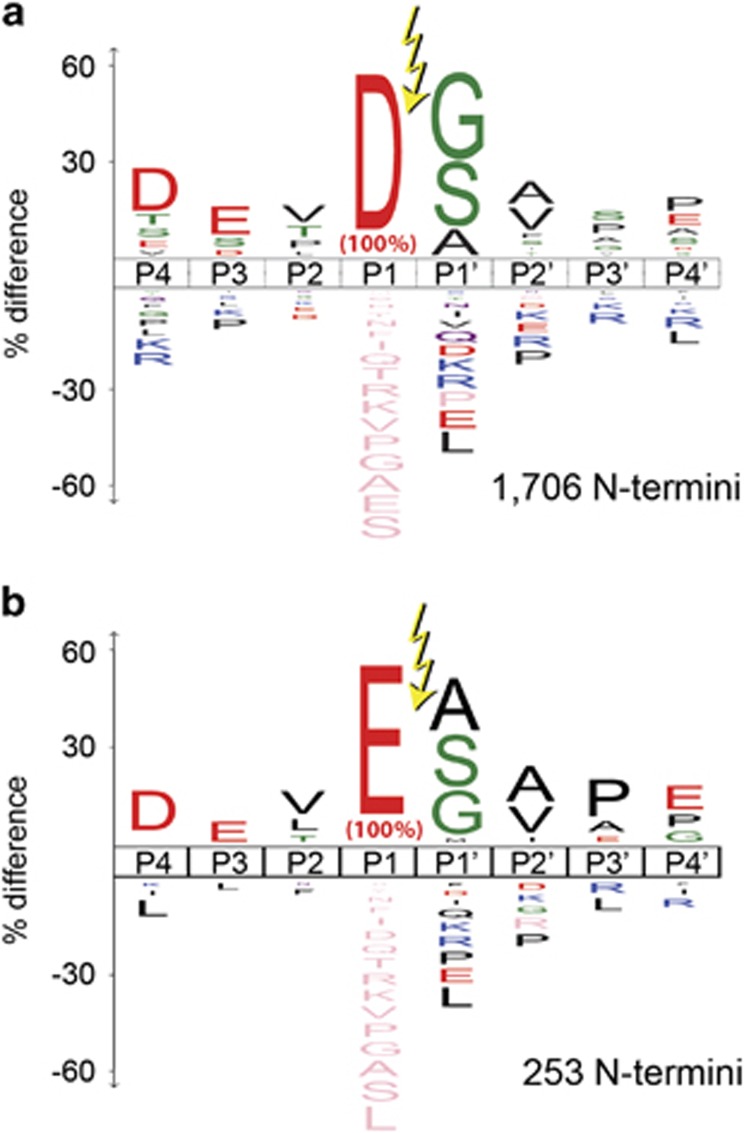
To determine the protease(s) responsible for glutamate cleavage, we compared the consensus cut-site motifs with those surrounding aspartate cuts using iceLogos. There are 2144 unique proteolytic events in healthy human cells, and the cleavage patterns are predominantly tryptic or chymotryptic-like with corresponding preference for lysine, arginine or large hydrophobic P1 side chains, and small side chains at P1'.18 The iceLogo changes dramatically for the apoptotic data set. From the nearly 7000 unique proteolytic events detected, there is an emergence of aspartate as the predominant P1 residue. Focusing on the 1706 unique aspartate cut sites (Figure 1a), the DEVD↓ represents the highest cleavage consensus motif and matches the apoptotic caspases-3 and −7 consensus motif based on synthetic substrates.28, 29 Remarkably, the iceLogo for the 253 unique P1 glutamate-cleaved sites (DEVE)↓ (Figure 1b) is virtually identical to that seen for the P1 aspartate-cleaved sites from P4 to P2'. The mouse data set shows essentially the same patterns (Supplementary Figure 2).27
Figure 1.
There is strong similarity between iceLogos for substrates cleaved after (a) P1 aspartate and (b) glutamate in apoptotic human samples. The iceLogos for P1 aspartate and P1 glutamate are virtually identical in the extended P4-P4' sequence around the cleavage site. (a) Reproduced with permission from Crawford et al.18
These data strongly suggest the same protease(s) are responsible for cleaving after aspartate and glutamate, likely the executioner caspases-3 and/or -7. Therefore, we quantified the rates that these two caspases cleave standard synthetic substrates containing the fluorogenic 7-amino-4-timethylfluoro-coumarin (AFC) reporter attached to the peptide, Ac-DEVx-AFC, where x is either aspartate or glutamate (Supplementary Figures 3a-b). These data allowed calculation of the kinetic constants (kcat and KM) for hydrolysis (Table 3). Remarkably, the catalytic efficiency values (kcat/KM) for cleaving after aspartate are only about twofold higher than for cleaving after glutamate by either caspase-3 or caspase−7.
Table 3. Summary of kinetic values for cleavage of P1 aspartate and glutamate by caspases-3 and -7.
|
D/E ratio |
||||||||
|---|---|---|---|---|---|---|---|---|
| Caspase | Substrate | Vmax (uM/s) | KM (uM) | kcat (1/s) | kcat/KM (1/(Ms)) | KM (uM) | kcat (1/s) | kcat/KM (1/(Ms)) |
| 3 | D | 5.3 (0.16) | 57.9 (5.6) | 355.73 | 6.14E+06 | 3.55 | 8.15 | 2.29 |
| 3 | E | 0.65 (0.04) | 16.3 (3.8) | 43.65 | 2.68E+06 | |||
| 7 | D | 2.6 (0.05) | 49.4 (3.1) | 173.80 | 3.52E+06 | 0.37 | 0.79 | 2.14 |
| 7 | E | 3.3 (0.06) | 133.8 (6.3) | 219.93 | 1.64E+06 | |||
The values for Vmax and KM with standard errors are reported from triplicate measurements on three biological replicates. The ratio of kcat/KM for P1 aspartate/P1 glutamate is just over twofold for both caspases-3 and -7
There are, however, counterbalancing differences seen in the specific kcat and KM values between the caspases. For example, the kcat values for caspase-3 are eightfold higher for cleaving aspartate, but the KM values are nearly fourfold higher. This suggests binding is actually weaker for P1 aspartate, but the catalytic step is faster. Caspase-7 shows the opposite trend, where the kcat is about the same for the P1 aspartate and glutamate, but the KM value is about twofold lower for the P1 aspartate. Commercial preparations of caspases-3 and -7 also had rate constants virtually identical to our recombinant forms (Supplementary Figures 3c-d). The rate constants we obtained with the P1 aspartate substrate are within threefold of published values for the.28, 30, 31 We extended the studies to mouse caspases-3 and -7 (Supplementary Figures 3e-f). The catalytic efficiency values for the P1 aspartate substrate were consistently only twofold above the P1 glutamate substrate, and mouse caspases-3 and -7 (Supplementary Table 1) similarly differed in their relative kcat and KM values as seen in the human homologs.
Since Ac-DEVE-AFC is cleaved only twofold slower than Ac-DEVD-AFC by caspases-3 and -7, we wondered if it is possible to substitute a P1 glutamate site for a known P1 aspartate cut site in cells and observe it being cleaved during apoptosis. PARP is a classic caspase substrate cut in apoptosis at a DEVD↓ sequence ending at aspartate-214.32 We introduced three human expression constructs into HEK293 cells: PARP harboring the wild-type P1 aspartate site (DEVD), a P1 glutamate site (DEVE) or a P1 alanine control site (DEVA) (Supplementary Figure 4). We induced apoptosis by treatment with staurosporine, and monitored cleavage of PARP by western blot. As expected, the native DEVD construct was rapidly cleaved, the DEVE was cut but more slowly, while the DEVA construct remained uncleaved. Thus, a P1 glutamate can substitute for P1 aspartate for cleavage in apoptotic cells, albeit more slowly.
Crystal structures show minimal differences in binding for P1 aspartate and glutamate peptides
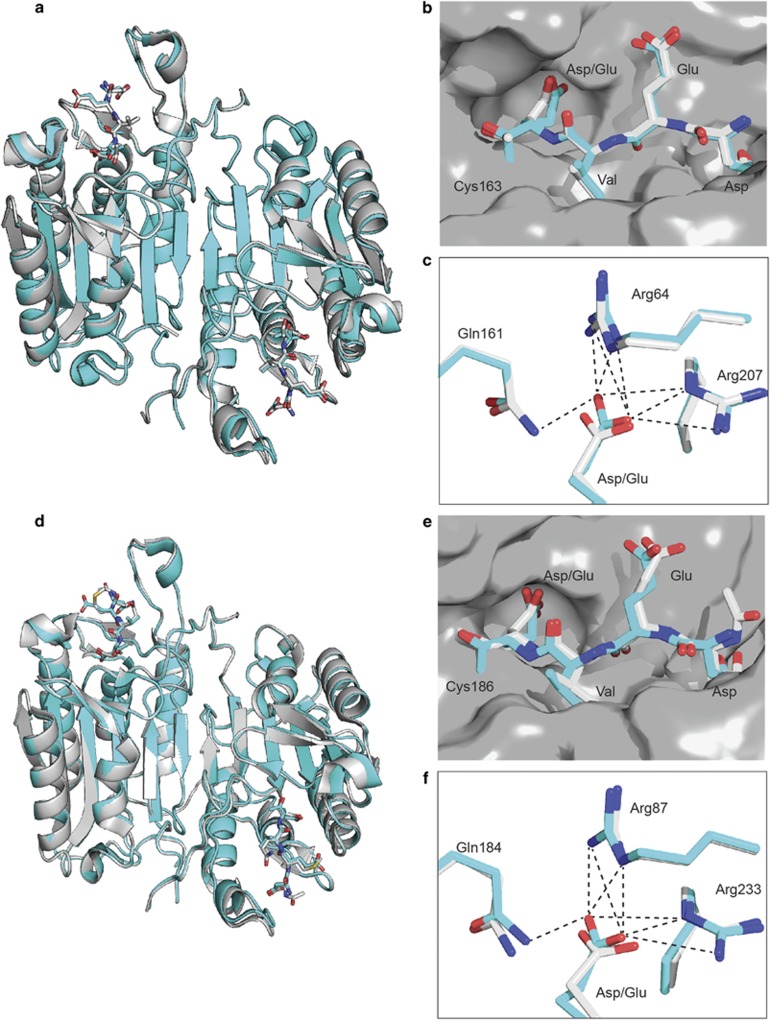
To understand the structural basis for the interactions of the P1 glutamate with caspases-3 and -7, we determined the crystal structures of the two caspases covalently labeled with DEVE-cmk (chloromethylketone) each at ~2.7 Å resolution (Figure 2,Supplementary Table 2 and Supplementary Figure 5). The asymmetric unit of the caspase-3-DEVE complex (PDB 5IC4) contains two structurally similar copies of the biological dimer that align well with a caspase-3-DEVD structure (Cα RMSD 0.4, PDB 2DKO31). The caspase-7-DEVE complex (PDB 5IC6) contains one biological dimer in the asymmetric unit, which is virtually identical to a caspase-7-DEVD structure (Cα RMSD 0.4, PDB 3H1P33). Importantly, the backbone and side chains of the DEVE and DEVD peptide inhibitors as well as the positions of the caspase peptide-binding residues align closely for both caspases-3 and -7 (Figures 2c and f). Although the P1 glutamate contains an additional methylene group compared with aspartate, both P1 acidic side chains interact with caspases-3 and -7 in virtually identical binding modes.
Figure 2.
Overlap of P1 glutamate and aspartate substrates in the caspases-3 and -7 pockets show minimal structural rearrangements to accommodate the extra methylene in P1 glutamate. (a–c) Human caspase-3 was labeled with Ac-DEVE-cmk, crystallized and X-ray structure solved to 2.65 Å (PDB: 5IC4). (d–f) Human caspase-7 was labeled with Ac-DEVE-cmk, crystallize and X-ray structure solved to 2.7 Å (PDB: 5IC6). The P1 glutamate labeled protein (blue) has minimal changes when overlayed with the P1 aspartate labeled protein caspase-3 (2DKO, gray) or caspase-7 (3H1P, gray). The differences between the glutamate and aspartate structures are localized to the binding pocket and substrate itself
Caspase-3 can cleave after phosphorylated serine
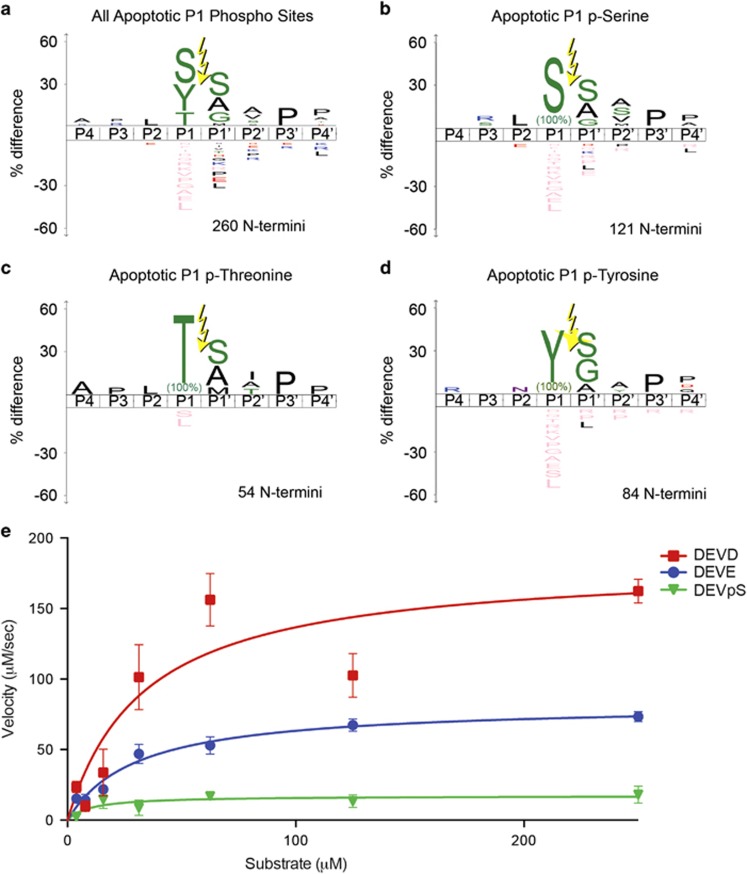
Given the significant promiscuity for apoptotic caspases to cleave after glutamate, we wondered if it was possible that they cleave directly after phosphorylated residues. It was possible to model a P1 phosphoserine into both caspases-3 and -7 with only small perturbations in the active site, though the bulkier phosphothreonine and phosphotyrosine predicted steric clashes. We explored our Degrabase to see if cleavages occur after serine, threonine and tyrosine residues known to be phosphorylated from the PhosphoSite Database (Table 4).34 Remarkably, we found 260 cuts after annotated phosphorylated sites during apoptosis compared with only 69 in healthy cells. A list of the annotated P1 phosphorylation substrates is shown in Supplementary Table 3. We were intrigued to find that P1 serine and threonine-annotated phosphosites are among the most abundantly enriched in the apoptotic set by 2.2- and 1.8-fold, respectively. In contrast, the much larger phosphotyrosine is significantly de-enriched in the apoptotic set. We analyzed the iceLogos for the phosphosites as well as the specific phosphorylated residues (Figure 3). There is little consensus from the iceLogo when grouping all phosphosites together or for the phosphotyrosine set. There is somewhat more defined consensus sequences for cleavages at phosphothreonine and phosphoserine. GO term analysis did not reveal any remarkably unique biology for the phosphoserine and phosphothreonine groups relative to each other or other P1 acid-cleaved substrates.
Table 4. Enrichment of annotated phosphorylation sites observed cleaved at the P1 position in apoptotic samples compared with healthy cells.
| P1 | Apoptotic | % | Healthy | % | Apoptotic/healthy |
|---|---|---|---|---|---|
| All phospho | 260 | 3.72% | 69 | 3.22% | 1.16 |
| p-Tyr | 84 | 1.20% | 43 | 2.01% | 0.60 |
| p-Thr | 54 | 0.77% | 9 | 0.42% | 1.84 |
| p-Ser | 121 | 1.73% | 17 | 0.79% | 2.18 |
The fold enrichment of each residue is calculated from the ratio of the percent of peptides found with each P1 residue in apoptosis to the percent found in healthy cells compared with the total peptides (6990 apoptotic and 2144 healthy). The fold enrichment values (final column) for phosphoserine and phospho-threonine are highly enriched in the apoptotic data set, whereas phospho-tyrosine is de-enriched
Figure 3.
Cleavage of P1-phosphoserine sites are frequently found in the Degrabase and caspase-3 can cleave synthetic substrate containing a P1 phosphoserine, biotin-WDEV(pS)↓SGVDEK(DNP). (a–d) The iceLogos for the combined group of three phospho-sites (a), phosphoserine (b), phospho-threonine (c) and phospho-tyrosine (d), do not match the typical caspase cleavage motif. (e) Caspase-3 can cleave the biotin-WDEV(pS)↓SGVDEK(DNP) substrate only threefold slower than the biotin-WDEV(D)↓SGVDEK(DNP) substrate. Error bars represent S.E.M
We next directly measured the kinetics of proteolysis of P1 phosphoserine or phosphothreonine substrates by caspases-3 and -7. We designed FRET substrates based on the PARP caspase cleavage sequence, DEV(x)↓GVDE containing a P1 aspartate, glutamate, phosphoserine or phosphothreonine and control substrates of P1 alanine and serine. As expected, both the P1 aspartate and glutamate FRET peptides were cleaved rapidly (Figure 3e). We observed quantitative cleavage of P1 phosphoserine by caspase-3 but not caspase-7, and no cleavage of P1 phosphothreonine by either caspases-3 or -7. There was no observable activity for the control alanine or serine substrates. The caspase-3 catalytic efficiency of the FRET peptides for aspartate was twofold more active than glutamate and only threefold more active than phosphoserine (Table 5). The caspase-3 phosphoserine kinetic constants kcat and KM are both lower than glutamate, and further reduced compared with aspartate. Thus, caspase-3 is more promiscuous than caspase-7 and able to cleave all three acidic functionalities at rates that are within an order of magnitude of each other.
Table 5. Summary of kinetic values for cleavage after aspartate, glutamate or phosphoserine FRET peptides and comparison of the ratios of k cat /K M for three FRET peptides shown in the last column.
| Caspase | Substrate | Vmax (mM/s) | KM (mM) | kcat (1/s) | kcat/KM (1/(Ms)) | kcat/KM ratio | |
|---|---|---|---|---|---|---|---|
| 3 | Asp | 182 (21.4) | 33.4 (11.9) | 1.8E−01 | 5.5E+03 | Asp/Glu | 2.00 |
| 3 | Glu | 82.4 (3.9) | 30.2 (4.6) | 8.2E−02 | 2.7E+03 | Asp/pSer | 3.14 |
| 3 | Phospho-ser | 17.3 (2.4) | 9.95 (5.6) | 1.7E−02 | 1.7E+03 | Glu/pSet | 1.57 |
P1 aspartate and P1 glutamate sites are well conserved between mouse and human, but not for distant metazoans
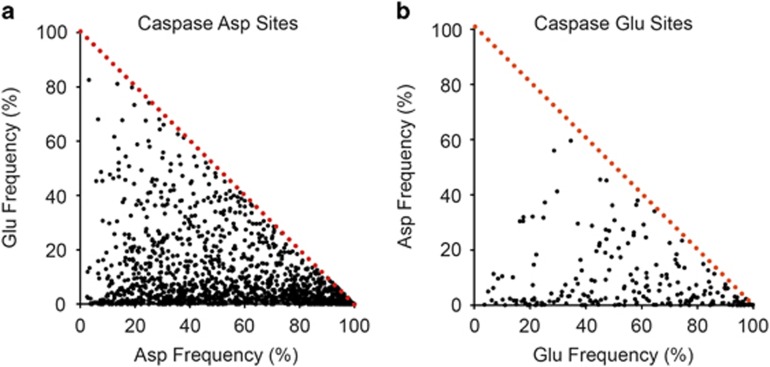
We took the 1706 observed P1 aspartate-cleaved human sites and determined the frequency these were conserved in homologs by sequence alignments and compared this with the frequency they were found altered to glutamate. There is a weak tendency for the 1706 aspartate sites to switch to glutamate (Figure 4a), and a similarly weak tendency for the 226 human glutamate sites to switch to aspartate (Figure 4b). However, both residues were more likely to remain unchanged than switch to the other, and almost as likely to switch to another amino acid as to its acidic relative.
Figure 4.
P1 aspartate and glutamate cleavage sites are not strongly conserved throughout the broader metazoan evolutionary record. Human caspase substrates with a P1 aspartate (a) and glutamate sites (b) are compared with all metazoan homologs for site conservation of a P1 aspartate or P1 glutamate in the sequence. The dashed red line indicates maximal conservation of 100% acidic residue (Asp+Glu). For both aspartate and glutamate sites, there is a tendency to remain itself as the data is skewed to the bottom left of the chart and minimal activity switching to the other acidic residue
We analyzed the relationship between evolutionary conservation of the acidic cleavage sites relative to recently measured rates of proteolysis of protein substrates from a subset of rate data compiled from 246 substrates cleaved by caspases-2, −3, −6, −7 and −8.21 We find a very weak trend where the faster the substrate is cleaved the stronger the acidic residue conservation, but only for caspases-3 and -7, and not caspases-2, −6 and −8 (Supplementary Figure 6). In addition, there is no significant relationship between conservation of acidic sites among the 84 substrates that are cleaved by more than one caspase within our kinetic data set of 246 substrates (Supplementary Figure 7). It is likely that these acidic cleavage sites evolve so quickly that only very mild trends are apparent in the evolutionary record.
Discussion
These studies highlight the use of proteomics coupled with detailed kinetic and structural analysis to reveal a greater prevalence and promiscuity for cleaving after glutamate residues in apoptosis. We found no remarkable differences in target proteins cleaved after aspartate or glutamate based upon iceLogo, secondary structure, evolution or cellular functions. The structural studies show that P1 glutamate can be accommodated in the substrate binding sites of both caspases-3 and -7 with minimal perturbations. Moreover, detailed kinetic analysis for matched synthetic substrates showed that aspartate is cleaved only twofold faster than glutamate by either caspases-3 or −7; this difference is roughly the same in cells undergoing apoptosis for the protein PARP containing the wild-type DEVD or replacement DEVE site. The twofold kinetic difference we see does not fully account for the eightfold difference from N-terminomics of aspartate over glutamate cleavages but prevalence and intrinsic rate measurements may not scale linearly. It is also possible that cellular cleavage activity of P1 glutamate substrate may rely on more optimized residues than is required for cleavage of P1 aspartate substrates. The rates of individual caspase substrates vary over 500-fold so this two- to eightfold rate differences between aspartate and glutamate cleavages is almost biologically indistiguishable.14, 21 Our studies indicate that P1 glutamate cleavages are biologically relevant and isofunctional with P1 aspartate cleavages.
There have been perhaps even isolated reports of single human proteins being cleaved after glutamate by human caspases-1, −3, −5 and −9[refs 17, 22, 23, 24, 25, 26] and a P1 glutamate synthetic substrate cleaved by caspase-14.35 Indeed, initial in vitro tests of human and other species' apoptotic and non-apoptotic caspase activities for Ac-DEVE-AFC have indicated broad glutamic acid activity (Supplementary Table 4).36 In addition, in silico caspase substrate algorithms have predicted, but not empirically confirmed, the ability for caspases to cleave P1 glutamate substrates.37, 38 Probably the best-characterized functional example is for DRONC, a Drosophila caspase, that cleaves itself during activation after glutamate.39, 40, 41 The active-site sequence around the catalytic cysteine of DRONC is quite different (PFCR versus QACQ, where C is the catalytic cysteine) from that of other fly caspases, such as DCP-1, which is shown to prefer DEVD.42 Hawkins et al.39 proposed this difference could facilitate more promiscuity for glutamate binding. However, the structures we present for inhibitors bound to caspases-3 and -7 show the P1 glutamate can be readily accommodated in the binding site.
Co-regulation of between kinases and caspases has long been recognized.43 Most caspases are known to be phosphorylated, and nearly half of the 518 human protein kinases can be cut during apoptosis.12, 18, 44 Phosphorylation on caspase substrates can also directly inhibit or activate cleavage rates by caspases.12 Our data here show that caspase cleavage may also occur directly following a phosphorylated serine site during apoptosis, suggesting kinase directed targeting of proteolysis. Although we infer the P1 residue from the gene sequence due to the method capturing the N-terminus of the P1' cleavage fragment, we do not know the explicit phosphylation state of the residue. Nonetheless we know these P1 serines can be phosphorylated at these sites, and have shown caspase-3 can directly cleave after a synthetic phosphoserine substrate. The synthetic substrates were not optimized to reflect the (R/P)LpS motif from apoptotic substrates. If these phosphoserine substrates were further optimized, it is possible these enzymes may indeed have higher activity for other P1 phosphoserine substrates and perhaps even activity for phosphothreonine. Further experiments will be needed to pursue optimization and kinetic determination of other phosphopeptides. To our knowledge this is the first example of any protease that cleaves immediately after a phosphorylated residue.
It is well-known that substitution of acidic residues at sites of phosphorylation can function as constitutive mimics,45 so perhaps it is not surprising we see capsase-3 cleave after phosphoserine. Ferrell and co-workers have systematically studied the evolutionary interplay between acidic and phosphorylation sites in kinases and estimate that upwards of 5% of the phosphorylation sites are derived from acidic precursors.46 They suggest phosphorylation sites tend to develop later in evolution as regulatory complexity develops. Phosphorylation sites are known to evolve rapidly in the sequence record, but they noted that phosphoserine sites most commonly mutate to threonine, glutamate and aspartate by enrichment factors of 1.5, 1.3 and 1.2, respectively. We have similarly found that caspase sites evolve very rapidly, with target being more conserved and pathway most conserved.27 With the direct interplay between kinases and caspases in apoptosis, we wondered if there was a common evolutionary pattern that may mimic or connect the two pathways. Indeed there is a trend for caspase sites to remain as an acidic amino acid, but it is not as strong as phosphorylation and is only mildly evident for caspase-3 and −7 substrates.
The data set presented here on nearly 2000 natural caspase substrates suggest human caspases have a much greater promiscuity for cleaving acidic functionalities than has been reported. Why was this overlooked? Before the application of LC-MS, proteomic studies only focused on tens of candidate-based protein substrate identifications using SDS-PAGE to estimate where a candidate protein was cleaved, and for only some of these did protein sequencing and mutagenesis identify the precise sites of cleavage. With such a small data set, it is not surprising P1 glutamate cuts would be missed systematically. Furthermore, specificity studies using positional-scanning libraries of synthetic substrates were focused on optimizing assays and finding strong inhibitors. These studies fixed on P1 aspartate as the sole P1 substrate for greatest activity levels and then thoroughly analyzed preferences in P4-P2 to identify optimal assay substrates and inhibitor sequences based on P1 aspartate.3 Subsequent proteomic studies, including our own, only focused on P1 aspartate cleavages as a traditional caspase signature. The more in-depth and unbiased bioinfomatic analysis of the DegraBase revealed the P1 glutamate and P1 phosphoserine signatures. We suggest P1 acidic residue promiscuity is general throughout the caspase family in all metazoans, and the term 'cacidases' aptly reflects this broader specificity.
Materials and Methods
Database analysis
The DegraBase (http://wellslab.ucsf.edu/degrabase/) compiles 8090 unique N-termini from 3206 proteins identified in subtiligase-based positive enrichment mass spectrometry experiments in 11 healthy and apoptotic human cell lines.18 The database was split into multiple subsets for analysis: N-termini from healthy cells, N-termini from apoptotic cells, apoptotic N-termini following an aspartate cleavage (Apop Asp) and apoptotic N-termini following a glutamate cleavage (Apop Glu). Sequence logos were made using iceLogo47 with the whole-human SwissProt library as background control. All cleavage iceLogo images were made with the percent difference scoring system and significance was determined by X2 analyses. The secondary structure was calculated using the NetSurfP server for the whole protein.48 The P4-P4' iceLogo was made using the filled logo option in iceLogo.
GO term enrichment was determined using the GO::TermFinder software.49 A list of unique proteins for each data set was created and uploaded to the database and tested for enrichment against the human SwissProt background using all evidence codes except ND (no biological data available) and IEA (inferred from electronic annotation). Enriched terms were defined using a corrected P-value cutoff of less than 0.01. To compare terms between data sets, a pairwise X2-test was performed using the Benjamini–Hochberg multiple testing correction procedure.
Enzyme purification and kinetics
Caspases-3 and -7 were purified using a C-terminal 6xHis tag in Escherichia coli as previously described.50 Enzyme activity was assayed using a 7-Amino-4-trifluoromethylcoumarin (AFC) substrate (excitation 395 nm and emission 505 nm) or a tryptophan/lysine-DNP FRET peptide substrate (excitation 280 nm and emission 355 nm) in caspase activity buffer (50 mM KCl, 50 mM HEPES pH 7.4, 0.1 mM EDTA, 0.1% CHAPS, 10 mM DTT) at room temperature. The AFC substrates, Ac-DEVD-AFC and Ac-DEVE-AFC (SMBiochemicals, Anaheim, CA, USA), were used with final caspase concentrations ranging from 10 to 50 nM. The FRET substrates, biotin-WDEV(x)GVDEK(dinitrophenol) where x is D, E, S, T, A, pS (phosphoserine) (Chinapeptides, Shanghai, China) or pT (phosphothreonine) (Shengnuo Peptide, Menlo Park, CA, USA), were used with enzyme concentrations ranging from 500 nM to 2 μM for optimized signal timing and intensity. FRET substrates were used instead of AFC reporter substrates for ease of synthesis of specified phospho-P1 residues. All enzymes were assayed at specified concentrations as determined by active-site titrations using z-VAD-fmk (Bachem Americas, Torrance, CA, USA). The final kinetics determined using triplicate biological replicates and each substrate concentration was assayed in triplicate (nine assays per substrate concentration total). Commercial sources of human caspase-3 (BD Biosciences, San Jose, CA, USA) and caspase-7 (G-Biosciences, St. Louis, MO, USA) were tested with the same kinetic protocols. All substrates showed regular parabolic Michaelis–Menten kinetic curves consistent with normal steady state kinetics and no evidence of product inhibition over the initial rate measurements.
Cellular assays
HEK293T (ATCC CLR-3216, Manassas, VA, USA) cells were cultured in DMEM high-glucose medium supplemented with sodium pyruvate, nonessential amino acids, penicillin/streptomycin, and 10% FBS at 37 oC. The cells were transiently transfected with pcDNA3.1 containing PARP (Uniprot P09874) with endogenous sequence (Asp214), or mutated to glutamate (Asp214Glu) or alanine (Asp214Ala), using Lipofectamine 2000 (ThermoFisher Scientific, Waltham, MA, USA) in OptiMEM media. After 6 h, the cells were supplemented with full DMEM media. The cells were treated with 500 nM staurosporine 24 h after transfections and harvested at various time-points up to 24 h later in M-PER Mammalian Protein Extract Reagent (Pierce, Rockford, IL, USA) with protease inhibitors (cOmplete, Mini, EDTA-free, Roche, Indianapolis, IN, USA). Extracts were run on SDS-PAGE gels and PVDF western blots. The antibodies used were anti-PARP (9542, Cell Signaling, Danvers, MA, USA) and anti-V5 (V8012, Sigma-Aldrich, St. Louis, MO, USA).
Crystallization, data collection, structure determination and refinement
Purified caspases-3 and -7 were treated with 1:8 and 1:32 ratio excess of z-DEVE-cmk (chloromethylketone) (American Peptide Company, Sunnyvale, CA, USA), respectively, in 50 mM HEPES pH 7.4, 50 mM KCl, 0.1 mM EDTA and 1 mM DTT for 2 h at 37 oC. Complete and single site covalent labeling was confirmed by mass spectrometry. The covalent caspase-DEVE complexes were dialyzed into 10 mM Tris pH 8.0, 50 mM NaCl, 10 mM DTT (caspase-3) or 2 mM DTT (caspase-7) and concentrated to 10 mg/ml. The caspase-3-DEVE and caspase-7-DEVE complexes were initially screened for crystallization using the Index Suite (Hampton Research, Aliso Viejo, CA, USA), PACT Suite, PEGs Suites and PEGs 2 suite (Qiagen, Valencia, CA, USA) screens. Caspase-3-DEVE crystal hits were optimized by mixing 1 μl of protein (10 mg/ml) with 1 μl of mother liquor (60% (v/v) tacsimate pH 7.0) at room temperature and crystals appeared in 14 days. Caspase-7-DEVE crystal hits were optimized by mixing 1 μl of protein (10 mg/ml) with 1 μl of mother liquor (0.2 M ammonium fluoride, 0.1 M sodium acetate pH 4.6, 20% (w/v) PEG 10 000) at room temperature and crystals appeared in 14 days. The crystals were cryoprotected with their respective mother liquors supplemented with 10% glycerol, flash cooled and stored in liquid nitrogen until data collection. Diffraction data were collected on the 8.3.1 beamline at the Advanced Light Source (ALS) and were processed in space groups F222 and P212121 for the caspase-3-DEVE and caspase-7-DEVE complexes, respectively, using the XDS package.51 The structures were determined by molecular replacement with Phaser52 using PDB codes 2DKO31 for caspase-3 and PDB code 2QL5,53 chains A and B, for caspase-7. The search models were prepared by removing the active-site peptide ligands. The complexes were iteratively built using Coot54 and refined in PHENIX.55 The covalent DEVE ligand was manually built into electron density. The restraint parameter file for the DEVE-cmk labeled protein was prepared with JLigand.56 The caspase peptide linkages were modeled as a covalent bond between the caspase catalytic cysteine residue and the ketomethylene group of the DEVE-cmk peptides. Refinement parameters included an initial round of simulated annealing, rigid body refinement and restrained refinement including TLS, NCS and weight optimization. Ramachandran statistics were calculated using MolProbity.57 Coordinates and structure factors have been deposited in the RCSB Protein Data Bank under accession codes 5IC4 (caspase-3-DEVE) and 5IC6 (caspase-7-DEVE).
Evolutionary analysis
All human apoptotic proteins cleaved after aspartate and glutamate were used as the input list. The identification of homologs (52–14 000 per protein substrate) were generated using a stand-alone protein BLAST (blastp)58 against the non-redundant 'nr' database with a E-value cutoff ≤10−16, as described previously.46 For each set of proteins and homologs, multiple sequence alignments were generated using Clustal Omega (clustalo) with default options.59 The output alignments were run through Perl codes to isolate the experimentally observed cleavage site conservation and all other non-cleaved background amino acids of the same type using only the human sequence as reference.
Acknowledgments
We thank Guy Salvesen and Scott Snipas at the Sanford Burnham Medical Research Institute for their helpful discussions and repeating some of kinetics with our AFC subtrates. We thank Emily Crawford, Zachary Hill (University of California, San Francisco), Dennis Wolan and Robyn Stanfield (The Scripps Research Institute) for helpful discussions and the staff of the ALS beamline 8.3.1 for support. This project was supported in part by Grants from National Institutes of Health R21 CA186077 (C.S.C.), R01 GM081051, R01 GM097316, and RO1 CA154802 (J.A.W.). It was also supported in part by NIH Training Grant T32 GM007175 (J.E.S.), the Global Healthy Living Foundation (J.E.S), NIH F31 CA180378 (T.J.R.) and the postdoctoral fellowships of a Banting Postdoctoral Fellowship from the Canadian Institutes of Health Research, Government of Canada (O.J.) and the Damon Runyon Cancer Research Foundation 2082-11 (N.D.T.).
Glossary
- AFC
7-amino-4-timethylfluoro-coumarin
- Ac
acetyl
- Cmk
chloromethylketone
- DNP
dinitrophenol
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- HEPES
2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid
- FBS
fetal bovine serum
- Fmk
fluormethylketone
- GO
gene ontology
- RMSD
root mean square deviation
- SDS
sodium dodecyl sulfate
- PBS
phosphate-buffered saline
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Salvesen
The authors declare no conflicts of interests.
Supplementary Material
References
- Alnemri ES, Livingston DJ, Nicholson DW, Salvesen G, Thornberry NA, Wong WW et al. Human ICE/CED-3 protease nomenclature. Cell 1996; 87: 171. [DOI] [PubMed] [Google Scholar]
- Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun 1967; 27: 157–162. [DOI] [PubMed] [Google Scholar]
- Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem 1997; 272: 17907–17911. [DOI] [PubMed] [Google Scholar]
- Poreba M, Strozyk A, Salvesen GS, Drag M. Caspase substrates and inhibitors. Cold Spring Harb Perspect Biol 2013; 5: a008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rano TA, Timkey T, Peterson EP, Rotonda J, Nicholson DW, Becker JW et al. A combinatorial approach for determining protease specificities: application to interleukin-1beta converting enzyme (ICE). Chem Biol 1997; 4: 149–155. [DOI] [PubMed] [Google Scholar]
- Pham VC, Anania VG, Phung QT, Lill JR. Complementary methods for the identification of substrates of proteolysis. Methods Enzymol 2014; 544: 359–380. [DOI] [PubMed] [Google Scholar]
- van den Berg BH, Tholey A. Mass spectrometry-based proteomics strategies for protease cleavage site identification. Proteomics 2012; 12: 516–529. [DOI] [PubMed] [Google Scholar]
- Staes A, Impens F, Van Damme P, Ruttens B, Goethals M, Demol H et al. Selecting protein N-terminal peptides by combined fractional diagonal chromatography. Nat Protoc 2011; 6: 1130–1141. [DOI] [PubMed] [Google Scholar]
- Impens F, Colaert N, Helsens K, Ghesquiere B, Timmerman E, De Bock PJ et al. A quantitative proteomics design for systematic identification of protease cleavage events. Mol Cell Proteomics 2010; 9: 2327–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drag M, Bogyo M, Ellman JA, Salvesen GS. Aminopeptidase fingerprints, an integrated approach for identification of good substrates and optimal inhibitors. J Biol Chem 2010; 285: 3310–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wejda M, Impens F, Takahashi N, Van Damme P, Gevaert K, Vandenabeele P. Degradomics reveals that cleavage specificity profiles of caspase-2 and effector caspases are alike. J Biol Chem 2012; 287: 33983–33995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turowec JP, Zukowski SA, Knight JD, Smalley DM, Graves LM, Johnson GL et al. An unbiased proteomic screen reveals caspase cleavage is positively and negatively regulated by substrate phosphorylation. Mol Cell Proteomics 2014; 13: 1184–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix MM, Simon GM, Cravatt BF. Global identification of caspase substrates using PROTOMAP (protein topography and migration analysis platform). Methods Mol Biol 2014; 1133: 61–70. [DOI] [PubMed] [Google Scholar]
- Julien O, Zhuang M, Wiita AP, O'Donoghue AJ, Knudsen GM, Craik CS et al. Quantitative MS-based enzymology of caspases reveals distinct protein substrate specificities, hierarchies, and cellular roles. Proc Natl Acad Sci USA 2016; 113: E2001–E2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoehr G, Schaab C, Graumann J, Mann M. A SILAC-based approach identifies substrates of caspase-dependent cleavage upon TRAIL-induced apoptosis. Mol Cell Proteomics 2013; 12: 1436–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimbo K, Hsu GW, Nguyen H, Mahrus S, Trinidad JC, Burlingame AL et al. Quantitative profiling of caspase-cleaved substrates reveals different drug-induced and cell-type patterns in apoptosis. Proc Natl Acad Sci USA 2012; 109: 12432–12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M, Kanneganti TD, Van Damme P, Vanden Berghe T, Vanoverberghe I, Vandekerckhove J et al. Targeted peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol Cell Proteomics 2008; 7: 2350–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford ED, Seaman JE, Agard N, Hsu GW, Julien O, Mahrus S et al. The DegraBase: a database of proteolysis in healthy and apoptotic human cells. Mol Cell Proteomics 2013; 12: 813–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell 2008; 134: 866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmer JC, Zhu W, Pop C, Regan T, Snipas SJ, Eroshkin AM et al. Structural and kinetic determinants of protease substrates. Nat Struct Mol Biol 2009; 16: 1101–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agard NJ, Mahrus S, Trinidad JC, Lynn A, Burlingame AL, Wells JA. Global kinetic analysis of proteolysis via quantitative targeted proteomics. Proc Natl Acad Sci USA 2012; 109: 1913–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasula SM, Hegde R, Saleh A, Datta P, Shiozaki E, Chai J et al. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature 2001; 410: 112–116. [DOI] [PubMed] [Google Scholar]
- Soares J, Lowe MM, Jarstfer MB. The catalytic subunit of human telomerase is a unique caspase-6 and caspase-7 substrate. Biochemistry 2011; 50: 9046–9055. [DOI] [PubMed] [Google Scholar]
- Moretti A, Weig HJ, Ott T, Seyfarth M, Holthoff HP, Grewe D et al. Essential myosin light chain as a target for caspase-3 in failing myocardium. Proc Natl Acad Sci USA 2002; 99: 11860–11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krippner-Heidenreich A, Talanian RV, Sekul R, Kraft R, Thole H, Ottleben H et al. Targeting of the transcription factor Max during apoptosis: phosphorylation-regulated cleavage by caspase-5 at an unusual glutamic acid residue in position P1. Biochem J 2001; 358: 705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checinska A, Giaccone G, Rodriguez JA, Kruyt FA, Jimenez CR. Comparative proteomics analysis of caspase-9-protein complexes in untreated and cytochrome c/dATP stimulated lysates of NSCLC cells. J Proteomics 2009; 72: 575–585. [DOI] [PubMed] [Google Scholar]
- Crawford ED, Seaman JE, Barber AE 2nd, David DC, Babbitt PC, Burlingame AL et al. Conservation of caspase substrates across metazoans suggests hierarchical importance of signaling pathways over specific targets and cleavage site motifs in apoptosis. Cell Death Differ 2012; 19: 2040–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talanian RV, Quinlan C, Trautz S, Hackett MC, Mankovich JA, Banach D et al. Substrate specificities of caspase family proteases. J Biol Chem 1997; 272: 9677–9682. [DOI] [PubMed] [Google Scholar]
- Pop C, Salvesen GS. Human caspases: activation, specificity, and regulation. J Biol Chem 2009; 284: 21777–21781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen ND, Koerber JT, Wells JA. Structural snapshots reveal distinct mechanisms of procaspase-3 and -7 activation. Proc Natl Acad Sci USA 2013; 110: 8477–8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan R, Mittl PR, Jelakovic S, Grutter MG. Extended substrate recognition in caspase-3 revealed by high resolution X-ray structure analysis. J Mol Biol 2006; 359: 1378–1388. [DOI] [PubMed] [Google Scholar]
- Lazebnik YA, Kaufmann SH, Desnoyers S, Poirier GG, Earnshaw WC. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature 1994; 371: 346–347. [DOI] [PubMed] [Google Scholar]
- Witkowski WA, Hardy JA. L2' loop is critical for caspase-7 active site formation. Protein Sci 2009; 18: 1459–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: a bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics 2004; 4: 1551–1561. [DOI] [PubMed] [Google Scholar]
- Park K, Kuechle MK, Choe Y, Craik CS, Lawrence OT, Presland RB. Expression and characterization of constitutively active human caspase-14. Biochem Biophys Res Commun 2006; 347: 941–948. [DOI] [PubMed] [Google Scholar]
- Quistad SD, Stotland A, Barott KL, Smurthwaite CA, Hilton BJ, Grasis JA et al. Evolution of TNF-induced apoptosis reveals 550 My of functional conservation. Proc Natl Acad Sci USA 2014; 111: 9567–9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, van Raam BJ, Salvesen GS, Cieplak P. Caspase cleavage sites in the human proteome: CaspDB, a database of predicted substrates. PLoS One 2014; 9: e110539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee LJ, Tan TW, Ranganathan S. CASVM: web server for SVM-based prediction of caspase substrates cleavage sites. Bioinformatics 2007; 23: 3241–3243. [DOI] [PubMed] [Google Scholar]
- Hawkins CJ, Yoo SJ, Peterson EP, Wang SL, Vernooy SY, Hay BA. The Drosophila caspase DRONC cleaves following glutamate or aspartate and is regulated by DIAP1, HID, and GRIM. J Biol Chem 2000; 275: 27084–27093. [DOI] [PubMed] [Google Scholar]
- Kumar S, Doumanis J. The fly caspases. Cell Death Differ 2000; 7: 1039–1044. [DOI] [PubMed] [Google Scholar]
- Snipas SJ, Drag M, Stennicke HR, Salvesen GS. Activation mechanism and substrate specificity of the Drosophila initiator caspase DRONC. Cell Death Differ 2008; 15: 938–945. [DOI] [PubMed] [Google Scholar]
- Song Z, Guan B, Bergman A, Nicholson DW, Thornberry NA, Peterson EP et al. Biochemical and genetic interactions between Drosophila caspases and the proapoptotic genes rpr, hid, and grim. Mol Cell Biol 2000; 20: 2907–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa M, Kornbluth S. Caspases and kinases in a death grip. Cell 2009; 138: 838–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix MM, Simon GM, Wang C, Okerberg E, Patricelli MP, Cravatt BF. Functional interplay between caspase cleavage and phosphorylation sculpts the apoptotic proteome. Cell 2012; 150: 426–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness PE, Koshland DE Jr. Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by the negative charge of the phosphate. J Biol Chem 1987; 262: 10422–10425. [PubMed] [Google Scholar]
- Pearlman SM, Serber Z, Ferrell JE Jr. A mechanism for the evolution of phosphorylation sites. Cell 2011; 147: 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. Improved visualization of protein consensus sequences by iceLogo. Nat Methods 2009; 6: 786–787. [DOI] [PubMed] [Google Scholar]
- Petersen B, Petersen TN, Andersen P, Nielsen M, Lundegaard C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct Biol 2009; 9: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM et al. GO::TermFinder—open source software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics 2004; 20: 3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolan DW, Zorn JA, Gray DC, Wells JA. Small-molecule activators of a proenzyme. Science 2009; 326: 853–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr 2010; 66: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr 2007; 40: 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agniswamy J, Fang B, Weber IT. Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis. FEBS J 2007; 274: 4752–4765. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 2010; 66: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N et al PHENIX: a comprehensive Python-based system for macromolecular structure solution Acta Crystallogr D Biol Crystallogr 2010; 66: 213–221. [DOI] [PMC free article] [PubMed]
- Lebedev AA, Young P, Isupov MN, Moroz OV, Vagin AA, Murshudov GN. JLigand: a graphical tool for the CCP4 template-restraint library. Acta Crystallogr D Biol Crystallogr 2012; 68: 431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 2010; 66: 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res 2008; 36: W5–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011; 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.