Abstract
Background
Since the discovery of specific histocompatibility, literature has associated genes involved in the immune response, like the Human Leucocyte Antigen (HLA), with a better prognosis in transplantation. However, other non-HLA genes may also influence the immune process, such as the genes encoding the immunoglobulin-like receptors of natural killer cells (KIRs). The discovery that NK cell KIR receptors interact with conservative epitopes (C1, C2, Bw4) presented in HLA class I molecules that are genetically polymorphic, also observed in KIR genes, led to the investigation of the relevance of the KIR system to hematopoietic stem cell transplant. The cure of patients with leukemias and other hematological malignancies after bone marrow transplantation (BMT) has been attributed in part to the ability of the donor immune cells, present in the graft, to recognize and eliminate neoplastic cells of the patient. The cytotoxic activity of NK cells is mediated by the absence of HLA class I-specific ligands on the target cell surface to inhibitory KIR receptors (hypothesis of “missing-self”).
Methods
We analyzed, by PCR typing-SSOP technique, the presence or absence of 16 KIR genes and haplotypes of 39 patients with hematopoietic disorders and 136 healthy individuals from Paraná State. The comparisons made between the patient and control group were performed using χ2 test or Fisher exact test (bilateral p-value), as appropriated. Significance level was considered when p-value ≤ 0.05.
Results
Framework genes KIR3DL3, KIR3DP1, KIR2DL4 and KIR3DL2 were positive in all samples. The comparison between KIR repertoire of patients and healthy individuals revealed significant differences (p < 0.05) in inhibitors genes KIR2DL2 (p = 0.0005) and KIR2DL5 (p = 0.0067) and activating genes KIR2DS1 (p = 0.0013), KIR2DS2 (p = 0.0038), KIR2DS3 (p = 0.0153) that are more frequent in controls than in patients. The KIR2DS3 was significantly more frequent (p = 0.0031) in patients with acute myeloid leukemia (AML) when compared to patients with acute lymphoblastic leukemia (ALL). We observed a higher frequency of haplotype A (59 %) in the patients.
Conclusion
Our data suggests that susceptibility to leukemia can be influenced, at least, partly byKIR receptors.
Electronic supplementary material
The online version of this article (doi:10.1186/s12878-016-0064-6) contains supplementary material, which is available to authorized users.
Keywords: KIR, HLA, Leukemia
Background
Natural killer (NK) cells play a pivotal role in innate immunity providing immediate protection against infections as well as in the early steps of neoplastic cellular transformation [1]. The antileukemic role of Natural Killer (NK) cells has been brought into focus in recent years.
Killer cell immunoglobulin-like receptors (KIRs) interactions with their ligands regulate the cytotoxicity activity of NK cells. HLA-Cw is the primary ligand for a significant number of inhibitory KIRs. HLA-Cw allotypes are categorized into C1 and C2 groups based on a polymorphism at residue 80 in the HLA-Cw molecule. Inhibitory KIR2DL2 and KIR2DL3 are specific for the C1 ligand group, and inhibitory KIR2DL1 is specific for the C2 ligand group. The inhibitory KIR3DL1 receptor is specific for HLA molecules with the HLA-Bw4 epitope (HLA-B, HLA-A3 e HLA-A11) [2]. When inhibitory KIRs encounter self-HLA class I ligands on target cells, they signal inhibition and establish tolerance.
KIR genotypes are organized into two main broad haplotypes termed A and B, according to KIR inhibitorys and activators genes content. Both A and B haplotype share four framework genes: KIR2DL4, KIR3DL2, KIR3DL3 and KIR3DP1. Haplotype A gene organization includes up to eight genes, those of the framework content together with KIR2DL1, KIR2DL3, KIR2DS4 and KIR3DL1. The activating KIR genes, KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS5, and KIR3DS1, as well as the genes encoding inhibitory KIRs, KIR2DL5A/B and KIR2DL2, are the principal representants of the Group B haplotypes [3].
The “missing-self” concept presented by Kärre and colleagues in the 1980s, paved the way for the understanding of NK-derived allorecognition mechanism [1, 4]. In brief, NK cells, through the expressing of cognate inhibitory receptor learn to detect and kill cells with reduced or “missing” expression of “self” MHC class I ligands [5].
Since the early 70s hematopoietic progenitor cells transplantation from different sources, have been used as a therapeutic alternative for a broad spectrum of cancers and hematological diseases [6].
The past 10 years have witnessed dramatic progress in our understanding of possible exploitation of NK cells in cancer therapy. Haplo-Hematopoietic Stem Cell Transplantation (haplo-HSCT) outcoming showed a positive effect related to NK cells in adults with AML and also in children with high-risk ALL [7, 8]. The increased activity of NK cells after transplanting, even when the donor and recipient are HLA- identical, suggest that the cytotoxicity of these receptor cells from the donor can be an additional component previously unrecognized in the rejection process.
Several studies indicate an association with disease role in interactions between these KIRs and HLA loci and infectious diseases, autoimmune/inflammatory diseases, cancer and reproduction [9].
Only a few studies have investigated the association between the genetic diversity of activating and inhibitory KIR genes in humans and the susceptibility and resistance to leukemia [10]. Some results pointed out association between a group of activating and inhibitory KIR genes with relapse, overall survival and relative risk [11].
Some results pointed out the association between a group of activating and inhibitory KIR genes with relapse, overall survival and relative risk [11].
More studies on KIR allelic diversity are needed in order to clarify the role of NK cells in hematopoietic diseases. Among the three already well- known gene families which encode for NK cell receptors, we aimed to characterize the KIR genes repertoire in patients from Paraná State with hematopoietic disorders.
Methods
Samples
The sample consisted of 39 patients (as shown in Table 1) with HSCT indication from the Erasto Gaertner Hospital (Curitiba, Paraná State, Brazil) and 136 healthy family members from LIGH-UFPR (Laboratory of Immunogenetics and Histocompatibility, Federal University of Paraná) database, in the period of July 2007 to May 2008 to carry out pre-transplant histocompatibility testing. All participants signed an informed consent document. The study was approved by the Ethics Committee from UFPR-CEP-HC number 037ext.019/2001-07.
Table 1.
General characteristics of patients (N = 39)
| Gender | Age | Ethnic group | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Disease | N | (%) | M | F | Average | Std. Dev. | White | Mulatto | Black |
| Bone Marrow Aplasia | 1 | 0.03 | 0 | 1 | 30.0 | 0.00 | 1 | 0 | 0 |
| Paroxysmal Nocturna Hemoglobinuria | 2 | 0.05 | 2 | 0 | 31.5 | 2.10 | 2 | 0 | 0 |
| NK Leukemia Cells | 1 | 0.03 | 1 | 0 | 13.0 | 0.00 | 1 | 0 | 0 |
| Non Hodgkin Lymphoma - Follicular | 1 | 0.03 | 0 | 1 | 39.0 | 0.00 | 1 | 0 | 0 |
| Acute Lymphoid Leukemia | 13 | 0.33 | 7 | 6 | 15.2 | 16.70 | 11 | 2 | 0 |
| Acute Myeloid Leukemia | 11 | 0.28 | 7 | 4 | 35.7 | 17.80 | 3 | 0 | 0 |
| Chronic Myeloid Leukemia | 3 | 0.08 | 3 | 0 | 31.7 | 9.60 | 1 | 0 | 0 |
| Myelodysplasia | 1 | 0.03 | 1 | 0 | 23.0 | 0.00 | 2 | 0 | 0 |
| Myelofibrosis | 2 | 0.05 | 1 | 1 | 50.5 | 12.00 | 1 | 0 | 0 |
| Multiple Myeloma | 1 | 0.03 | 1 | 0 | 40.0 | 0.00 | 1 | 1 | 0 |
| MyelodysplasticMyeloproliferativeSyndrome | 2 | 0.05 | 1 | 1 | 42.0 | 1.40 | 10 | 1 | 0 |
| Not Informed | 1 | 0.03 | 1 | 0 | NI | NI | 1 | 0 | 0 |
| Total | 39 | 1 | 25 | 14 | 35 | 4 | 0 | ||
M male, F female, Std. Dev. standard deviation; IBGE ethnic classification
Extraction of genomic DNA
Two tubes with 10 milliliters (10 ml) of peripheral blood were collected from each individual by venous puncture into sterile tubes containing EDTA vacutainer type. These samples were centrifuged to obtain leucocyte layer from which the DNA was extracted by salting-out technique [12]. The DNA concentration of the samples was measured by reading optical density, using the spectrophotometer Gene Quantpro RNA/DNA calculator.
Typing of KIR genes
The KIR gene typing was performed by PCR-SSOP (Polymerase Chain Reaction - Sequence Specific Oligonucleotide Probes), amplifying the exons 3, 5 and 7–9, using the kit “LabtypeKIR SSO Genotyping Test” (One Lambda Inc). The data analysis was performed using the HLA VISUAL version 2.0 software (One Lamda Inc.) that analyzes the combinations of probes in the microbeads detected by the instrument and consults an internal database that suggest what are the loci present.
Statistical analysis
Phenothypic frequencies regarding presence/absence of KIR genes for all samples (patient group and healthy control group) along with the haplotype frequencies were obtained by direct counting.
The frequencies of the 16 KIR genes obtained for ALL and AML patients group were compared using the χ2 test, and the comparisons made between the patient and control group were performed using Fisher exact test (bilateral p-value), with the aid of BioEstat 5.0 software. When the sample size of the analyzed group was very small (less than 10), which can decrease the accuracy of the test χ2, Yates correction was applied. The frequencies of haplotypes A and B were obtained by direct counting. The comparisons made between patient and controls were performed using χ2test. Significance level was considered when p-value ≤ 0.05.
Results
The epidemiology of the 39 patients with HSCT indication was analyzed according to disease, gender, age and ethnic/racial group (Table 1). Age analysis of the patients (N = 39) indicated that 64 % of the patients were male (25) and 36 % were female (14). The average age of the patients was 28.50 + 17.74 years. According to the racial group suggested by the Brazilian Institute of Geography and Statistics (IBGE), 89 % of patients were classified as White and 11 % Mulattos (mixed-descendent of White vs Black).
The frequencies of the presence/absence of 16 KIR genes in the patients are presented in Table 2. These data were compared with 136 healthy family members and it was observed that the KIR2DL2 (p = 0.0005); KIR2DL5 (p = 0.0067); KIR2DS1 (p = 0.0013); KIR2DS2 (p = 0.0038) and KIR2DS3 genes (p = 0.0153) were statistically more frequent in the healthy individuals than in the patients as can be seen in Table 3. The comparison between patients with acute lymphoid leukemia (ALL) and acute myeloid leukemia (AML) showed that the KIR2DS3 gene (p = 0.0013) was more frequent in AML patients (Table 4).
Table 2.
Distribution of KIR genes frequenciesin patients (N = 39)
| KIR genes | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2DL1 | 2DL2 | 2DL3 | 2DL4 | 2DL5 | 2DP1 | 2DS1 | 2DS2 | 2DS3 | 2DS4 | 2DS5 | 3DL1 | 3DL2 | 3DL3 | 3DP1 | 3DS1 | ||
| Patients | absolutefrequency | 35 | 9 | 36 | 39 | 11 | 37 | 6 | 12 | 5 | 36 | 11 | 37 | 39 | 39 | 39 | 10 |
| N = 39 | relativefrequency | 0,90 | 0,23 | 0,92 | 1,00 | 0,28 | 0,95 | 0,15 | 0,31 | 0,13 | 0,92 | 0,28 | 0,95 | 1,00 | 1,00 | 1,00 | 0,26 |
| genefrequency | 0,68 | 0,12 | 0,72 | 1,00 | 0,15 | 0,77 | 0,08 | 0,17 | 0,07 | 0,72 | 0,15 | 0,77 | 1,00 | 1,00 | 1,00 | 0,14 | |
| BoneMarrow Aplasia | absolutefrequency | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| N = 1 | relativefrequency | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 | 0,00 | 1,00 | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 1,00 |
| genefrequency | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 | 0,00 | 1,00 | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 1,00 | |
| ParoxysmalNocturnaHemoglobinuria | absolutefrequency | 2 | 0 | 2 | 2 | 0 | 2 | 0 | 1 | 0 | 2 | 0 | 2 | 2 | 2 | 2 | 0 |
| N = 2 | relativefrequency | 1,00 | 0,00 | 1,00 | 1,00 | 0,00 | 1,00 | 0,00 | 0,50 | 0,00 | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 |
| genefrequency | 1,00 | 0,00 | 1,00 | 1,00 | 0,00 | 1,00 | 0,00 | 0,29 | 0,00 | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 | |
| NK LeukemiaCells | absolutefrequency | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 |
| N = 1 | relativefrequency | 1,00 | 0,00 | 1,00 | 1,00 | 0,00 | 1,00 | 0,00 | 0,00 | 0,00 | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 |
| genefrequency | 1,00 | 0,00 | 1,00 | 1,00 | 0,00 | 1,00 | 0,00 | 0,00 | 0,00 | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 | |
| Non Hodgkin Lymphoma - Follicular | absolutefrequency | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 |
| N = 1 | relativefrequency | 1,00 | 0,00 | 1,00 | 1,00 | 0,00 | 1,00 | 0,00 | 0,00 | 0,00 | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 |
| genefrequency | 1,00 | 0,00 | 1,00 | 1,00 | 0,00 | 1,00 | 0,00 | 0,00 | 0,00 | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 | |
| AcuteLymphoidLeukemia | absolutefrequency | 12 | 3 | 12 | 13 | 4 | 13 | 2 | 3 | 2 | 12 | 5 | 13 | 13 | 13 | 13 | 2 |
| N = 13 | relativefrequency | 0,92 | 0,23 | 0,92 | 1,00 | 0,31 | 1,00 | 0,15 | 0,23 | 0,15 | 0,92 | 0,38 | 1,00 | 1,00 | 1,00 | 1,00 | 0,15 |
| genefrequency | 0,72 | 0,12 | 0,72 | 1,00 | 0,17 | 1,00 | 0,08 | 0,12 | 0,08 | 0,72 | 0,22 | 1,00 | 1,00 | 1,00 | 1,00 | 0,08 | |
| AcuteMyeloidLeukemia | absolutefrequency | 8 | 3 | 9 | 11 | 4 | 9 | 2 | 5 | 2 | 9 | 3 | 10 | 11 | 11 | 11 | 5 |
| N = 11 | relativefrequency | 0,73 | 0,27 | 0,82 | 1,00 | 0,36 | 0,82 | 0,18 | 0,45 | 0,18 | 0,82 | 0,27 | 0,91 | 1,00 | 1,00 | 1,00 | 0,45 |
| genefrequency | 0,48 | 0,15 | 0,57 | 1,00 | 0,20 | 0,57 | 0,10 | 0,26 | 0,10 | 0,57 | 0,15 | 0,70 | 1,00 | 1,00 | 1,00 | 0,26 | |
| ChronicMyeloidLeukemia | absolutefrequency | 3 | 0 | 3 | 3 | 0 | 3 | 0 | 0 | 0 | 3 | 1 | 3 | 3 | 3 | 3 | 0 |
| N = 3 | relativefrequency | 1,00 | 0,00 | 1,00 | 1,00 | 0,00 | 1,00 | 0,00 | 0,00 | 0,00 | 1,00 | 0,33 | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 |
| genefrequency | 1,00 | 0,00 | 1,00 | 1,00 | 0,00 | 1,00 | 0,00 | 0,00 | 0,00 | 1,00 | 0,18 | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 | |
| Myelodysplasia | absolutefrequency | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| N = 1 | relativefrequency | 1,00 | 1,00 | 1,00 | 1,00 | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 1,00 | 1,00 | 1,00 | 1,00 |
| genefrequency | 1,00 | 1,00 | 1,00 | 1,00 | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 1,00 | 1,00 | 1,00 | 1,00 | |
| Myelofibrosis | absolutefrequency | 2 | 0 | 2 | 2 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 2 | 2 | 2 | 2 | 0 |
| N = 2 | relativefrequency | 1,00 | 0,00 | 1,00 | 1,00 | 0,00 | 1,00 | 0,00 | 0,00 | 0,00 | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 |
| genefrequency | 1,00 | 0,00 | 1,00 | 1,00 | 0,00 | 1,00 | 0,00 | 0,00 | 0,00 | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 | |
| MultipleMyeloma | absolutefrequency | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 |
| N = 1 | relativefrequency | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 | 1,00 | 0,00 | 1,00 | 0,00 | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 |
| genefrequency | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 | 1,00 | 0,00 | 1,00 | 0,00 | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 | |
| MyelodysplasticMyeloproliferativeSyndrome | absolutefrequency | 2 | 0 | 2 | 2 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 2 | 2 | 2 | 2 | 1 |
| N = 2 | relativefrequency | 1,00 | 0,00 | 1,00 | 1,00 | 0,00 | 1,00 | 0,00 | 0,00 | 0,00 | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 1,00 | 0,50 |
| genefrequency | 1,00 | 0,00 | 1,00 | 1,00 | 0,00 | 1,00 | 0,00 | 0,00 | 0,00 | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 1,00 | 0,29 | |
| NotInformed | absolutefrequency | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 |
| N = 1 | relativefrequency | 1,00 | 1,00 | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 |
| genefrequency | 1,00 | 1,00 | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 0,00 | 1,00 | 1,00 | 1,00 | 1,00 | 0,00 | |
Genes in bold are those genes called “framewok genes
Table 3.
Frequency of each KIR gene in patients (N = 39) and controls (N = 136)
| KIR gene | Patient | Control | p-value | |
|---|---|---|---|---|
| (n = 39) | (n = 136) | |||
| Framework | KIR2DL4 | 39 (100 %) | 136 (100 %) | 1.0000 |
| KIR3DL2 | 39 (100 %) | 136 (100 %) | 1.0000 | |
| KIR3DL3 | 39 (100 %) | 136 (100 %) | 1.0000 | |
| KIR3DP1 | 39 (100 %) | 136 (100 %) | 1.0000 | |
| Haplotype A | KIR2DS4 | 36 (92 %) | 129 (95 %) | 0.6936 |
| KIR2DL1 | 35 (90 %) | 130 (96 %) | 0.2329 | |
| KIR2DL3 | 36 (92 %) | 118 (87 %) | 0.4173 | |
| KIR3DL1 | 37 (95 %) | 129 (95 %) | 1.0000 | |
| Haplotype B | KIR2DS1 | 6 (15 %) | 59 (43 %) | 0.0013 ( a ) |
| KIR2DS2 | 12 (31 %) | 78 (57 %) | 0.0038 ( a ) | |
| KIR2DS3 | 5 (13 %) | 45 (33 %) | 0.0153 ( a ) | |
| KIR2DS5 | 11 (28 %) | 45 (33 %) | 0.6976 | |
| KIR3DS1 | 10 (26 %) | 53 (39 %) | 0.1356 | |
| KIR2DL2 | 9 (23 %) | 75 (55 %) | 0.0005 ( a ) | |
| KIR2DL5 | 11 (28 %) | 72 (53 %) | 0.0067 ( a ) | |
| Haplotype A/B | KIR2DP1 | 37 (95 %) | 131 (96 %) | 0.6465 |
Genes in bold showed significance for the statistical test (a)
Table 4.
Frequency of each KIR gene from patients with ALL (N = 12) and AML (N = 11)
| All | AML | p-value | ||
|---|---|---|---|---|
| KIR GENE | (n = 12) | (n = 11) | ||
| Framework | KIR2DL4 | 12 (100 %) | 11 (100 %) | 1.0000 |
| KIR3DL2 | 12 (100 %) | 11 (100 %) | 1.0000 | |
| KIR3DL3 | 12 (100 %) | 11 (100 %) | 1.0000 | |
| KIR3DP1 | 12 (100 %) | 11 (100 %) | 1.0000 | |
| Haplotype A | KIR2DS4 | 11 (92 %) | 9 (82 %) | 0.5921 |
| KIR2DL1 | 11 (92 %) | 8 (73 %) | 0.3168 | |
| KIR2DL3 | 11 (92 %) | 9 (82 %) | 0.5901 | |
| KIR3DL1 | 12 (100 %) | 10 (91 %) | 0.4783 | |
| Haplotype B | KIR2DS1 | 10 (83 %) | 9 (82 %) | 1.0000 |
| KIR2DS2 | 3 (25 %) | 3 (27 %) | 1.0000 | |
| KIR2DS3 | 2 (17 %) | 9 (82 %) | 0.0031 ( a ) | |
| KIR2DS5 | 5 (42 %) | 3 (27 %) | 0.6668 | |
| KIR3DS1 | 2 (17 %) | 5 (45 %) | 0.1930 | |
| KIR2DL2 | 3 (25 %) | 3 (27 %) | 1.0000 | |
| KIR2DL5 | 4 (33 %) | 4 (36 %) | 1.0000 | |
| Haplotype A/B | KIR2DP1 | 12 (100 %) | 9 (82 %) | 0.2174 |
Genes in bold showed significance for the statistical test (a)
We also observed a higher frequency of haplotype A in patients (59 % of haplotype A and 41 % of haplotype B).
Discussion
Fourteen KIR genes plus two pseudogenes are joined in the leukocyte receptor complex (LCR) on chromosome 19q13.4 and display a high degree of genetic diversity concerning gene content and allelic polymorphism [13]. This genomic structure drives to non-allelic homologous recombination events, which potentially generates considerable genetic diversity in KIR gene repertoire among individuals and populations [14].
HLA and KIR gene clusters are functionally linked but segregate independently creating a genetic diversity that could have a different impact on transplantation outcome. Hence, studies regarding KIR and HLA genes in different populations can provide valuable information to several scientific fields [15, 16].
In our study, we evaluated the repertoire of KIR genes in patients with hematopoietic disorders. Overall, the frequencies of the presence/absence of KIR genes were similar to the frequencies observed in European populations, which would be expected considering the predominance of White Euro-descendants in southern Brazil.
The framework genes KIR3DL3, KIR3DP1, KIR2DL4 and KIR3DL2 were observed in all samples, in accordance to their presence in all known KIR haplotypes. The remaining inhibitorys and activators KIR genes showed frequencies that varied between individuals. The distribution of KIR gene frequencies among patients showed low frequencies as follows: KIR2DL2 (23 %), KIR2DL5 (28 %), KIR2DS1 (15 %), KIR2DS3 (13 %), KIR2DS5 (28 %) and KIR3DS1 (26 %). Despite the small sample size, the results are in agreement with those observed for populations of Caucasian origin. The KIR genes that showed higher frequencies in the patient group were: KIR2DL1 (90 %), KIR2DL3 (92 %), KIR2DP1 (95 %), KIR2DS4 (92 %) and KIR3DL1 (93 %).
A recent study of the association between polymorphisms in KIR and HLA genes and pediatric ALL in Hispanic and non-Hispanic children provided additional evidence about the contribution of genetic variation in ALL incidence. When the incidence and survival were evaluated between the two ethnic groups, a high incidence of ALL and a significantly worse survival was found in Hispanic children compared to non-Hispanic Whites. The genotypes diversity related to KIR and HLA ligands are very suggestive that these two loci may determine a different susceptibility effect depending on the ethnic groups. Such observed differences are probably multifactorial due to an interaction between KIR and environmental factors, e.g. patterns of infection, rather than merely allele frequencies differences between ethnic groups [14].
In a study carried out in Italian population Bontadini and colleagues reported the same general KIR gene patterns distribution observed in other Caucasian and non-Caucasian populations. Australian Aborigine, Chinese Han, and Japanese showed the most markedly different patterns, with significant differences from Italian population and other Caucasian populations, in particular for inhibitory gene KIR2DL2 and non inhibitorys KIR2DS1, KIR2DS2, KIR2DS3, KIR3DS1 [17–20]. The findings with respect to KIR gene diversity in different populations could provide relevant genomic diversity data for further studies on viral infection, autoimmune diseases, and reproductive fitness.
Inhibitorys genes KIR2DL2 and KIR2DL5B were also found at lower frequencies in the Italian population, as well as the activating genes KIR2DS3 and KIR2DS4 [15]. Notably, as to KIR2DS4 alleles, Han Chinese showed an inverse pattern compared to the Italian population [19].
The activating KIR2DS4 gene is unique in the haplotype A, whereas haplotype B contains up to five activating KIRs. Haplotypes A and B have been preserved in the human population (about 25 and 75 % in Caucasian), thus suggesting the occurrence of a balancing selection [13, 15].
Linkage disequilibrium or the non-random associations between alleles at two loci are also present in KIR genes repertoire. A high positive linkage disequilibrium between KIR2DL1 and KIR2DL3 has been observed in Caucasian and non-Caucasian populations [21]. Our data are consistent with this hypothesis since the frequencies of these two genes were the highest observed in both control and patient groups.
KIR repertoire comparisons between patients and healthy family members (Fig. 1) showed that the inhibitory genes KIR2DL2 (p = 0.0005) and KIR2DL5 (p = 0.0067), as well as the activating genes KIR2DS1 (p = 0.0013), KIR2DS2 (p = 0.0038), KIR2DS3 (p = 0.0153) were more frequently found (p <0.05) in healthy individuals than in patients.
Fig. 1.
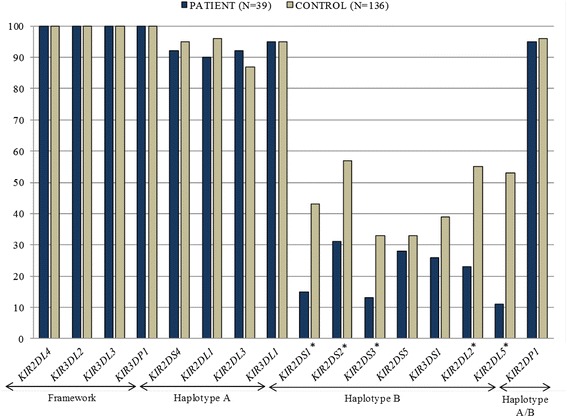
Phenotypic frequency related to the presence/absence of KIR genes in patients (N = 39) and healthy controls (N = 136). Genes marked with (*) showed significance for the stastical test
HLA-KIR genotypes have been associated with susceptibility to a variety of diseases such as psoriatic arthritis, type I diabetes, infectious diseases, cancer, and reproduction. Just a few of these studies revealed an influence of HLA-KIR gene interactions on disease outcome [9]. Others studies have investigated the frequency of KIR genes in patients with hematologic malignancies [22]. Most of these investigations have been performed in patients with different diseases such as AML, CML and MDS [23–27].
There are several studies that investigate the association between KIR haplotypes distribution and diseases; it is observed that Haplogroups A and B vary considerably between ethnic groups [15, 28–30]. The association of KIR gene/haplotypes has been investigated in patients with indication for hematopoietic stem cell transplantation, highlighting the role of KIR genes in the transplant outcome [7].
The stratification of the patients according to AML vs ALL groups, revealed that the KIR2DS3 gene presented higher frequencies (p = 0.0031) in AML when compared to ALL patients (Fig. 2). Although the sample size was relatively small, recent publications suggest important roles for specific KIR genes that may influence allogeneic hematopoietic stem cell transplantation (HSCT) outcome in HLA-compatible siblings, GvHD, relapse and other complications related to transplantation [31] in small and heterogeneous samples [32].
Fig. 2.
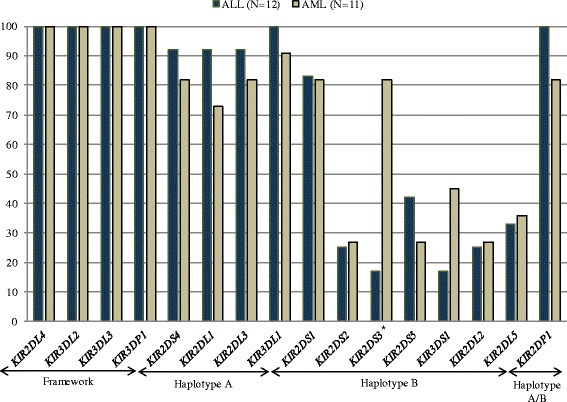
Phenotypic frequency related to the presence/absence of KIR genes in ALL patients (N = 12) and AML patients (N = 11). Genes marked with (*) showed significance for the stastical test
A study in China investigated KIR genotypes in 54 patients with hematopoietic malignancies, classified into two risk groups: standard and high. The frequency of activating KIR genes in standard-risk group was higher when compared to the high-risk group, specifically for KIR2DS1, KIR2DS2 and KIR3DS1. A secondary analysis of this study, comparing standard-risk group vs high-risk group in AML patients, revealed higher frequencies of activating KIR genes in the standard-risk group, particularly for KIR2DS1, KIR2DS2, and KIR2DS3 genes, the latter one in agreement with our findings in AML patients group [22].
In the same line of investigation, Kim and colleagues reported the influence of KIR genes in AML patients and HLA compatible donor siblings after HSCT. All the activating KIR genes in the donors showed an important role in transplant outcome and in the occurrence of acute graft-versus-host disease (GvHD) in HSCT in AML patients. Particularly, the KIR2DS2 gene and the allele KIR2DS4*003 were correlated with acute GvHD. This evidence suggests an immunogenic specificity in the Korean population compared to Caucasians since the frequency of KIR2DL2 and KIR2DS2 genes are comparatively lower in Koreans than in other countries. Long-term survival was noted even if the KIR2DS1 gene was only present in the donor and not in the recipient. The presence of both genes KIR2DS3-KIR2DS5 was more frequently found in a variety of complications related to transplant [31].
Mancusi and colleagues reported that donors, possessing KIR2DS1, KIR3DS1 or both activating genes, showed reduced infection rates and mortality, and a better event-free survival (EFS) [33].
Donor cells that express KIR haplotype B have been reported to contribute to relapse protection and improved survival after myeloablative allogeneic transplantation. Haplotype B/x donor cells have also been associated with a higher incidence of chronic GvHD. In HLA-haploidentical transplant setting, adonor KIR B haplotype has been associated with lower risk of relapse for patients with hematologic malignancies [2].
NK cells receptors of the family KIR may confer specific protector effect for different diseases. In a Turkish study carried out in a heterogeneous group of leukemia patients and controls, a protective effect was observed associated with KIR2DL2 and/or KIR2DS2 against CML [32].
From an evolutionary perspective, activating KIRs arose more recently from inhibitorys homologous genes [34]. A wide variation in KIR activators gene frequencies have been reported for different populational groups [35]. Nevertheless, the allelic diversity related to inhibitory receptors genes is limited when compared to activators genes [36]. The strong negative correlation observed between certain activating KIR and its ligands across populations, in contrast to weak positive correlations between several KIR inhibitory genes and their ligands, put forward a hypothesis that a pressure selection mechanism involving autoimmune disease is acting on the maintenance of lower frequencies of activator KIR receptor and their ligands [35].
KIR phenotypes analysis in Belgian leukemia patients indicated significantly higher frequencies of inhibitory KIR2DL1, KIR2DL2, and KIR2DL3 genes, suggesting their contribution for the lack of antitumor responses of NK cells [10]. On another study, conducted in 35 patients with a lymphoproliferative NK cell disease, inhibitory genes KIR2DL5A and KIR2DL5B were more frequently found in patients compared to healthy controls [37].
Epstein-Barr virus has been associated to the development of Hodgkin’s disease in some pathological conditions. An important review summarises current knowledge of the pathogenesis of Hodgkin’s disease with particular emphasis on the association with EBV. Besson and colleagues identified in a family study a stronger protector effect related to KIR2DS1/KIR3DS1 in patients with Hodgkin’s lymphoma [38].
A total of 50 Han Chinese patients were studied to explore the correlation between KIR genes and susceptibility to leukemia. The comparison made between patient and control groups showed lower frequencies of KIR3DL1 and KIR2DL1 genes amongst patients. Additionally, the results highlighted a negative correlation between the pathogenesis of leukemia and KIR3DL1, KIR3DS1, KIR2DL1, and KIR2DL5 genes [39], a very suggestive finding that KIR polymorphisms are associated with susceptibility to leukemia in Hans.
In the present study, patients had lower frequencies of KIR3DS1 (26 % versus 39 %) and KIR2DL5 (28 % versus 53 %) as compared to healthy family members group.
McQueen and collaborators analyzed KIR genes repertoire in donors, and found that KIR2DS3 conferred a protective effect against chronic GvHD in transplantation with HLA-compatible unrelated donor [40] and with donors who have more than four activating KIR in haploidentical transplants.
NK cells were components previously not recognized in HSCT rejection process and GvHD. More recently, investigation of human health/disease effects associated with KIR receptors have been reported and the majority of the described associations have been with activators KIR genes. Our data suggest that susceptibility to leukemia can be influenced, at least, partly by KIR receptors and an increased sample can confirm these findings for further investigations.
Conclusion
Our study with KIR receptors in patients with hematologic diseases showed that inhibitory genes KIR2DL2 and KIR2DL5 and activating genes KIR2DS1, KIR2DS2, KIR2DS3 were more frequently found in healthy family members than in patients. Also, the KIR2DS3 was significantly more frequent in patients with AML when compared to patients with ALL. We observed a higher frequency of haplotype A in the patients group. All these data are crucial to understanding the mechanisms underlying the dysfunction of NK in leukemia. The characterization of the genetic profile of patients brings together relevant information to enable robust investigations of KIR genes and their influence on important diseases such as Acute Myeloid Leukemia, Acute Lymphoblastic Leukemia, and others.
Acknowledgements
We are very grateful to the patients who generously accorded to provide samples for this study. We also thank the LIGH staff for technical support.
Funding
This study was supported by research funding from FUNPAR-LIGH and CAPES.
Availability of data and materials
The data sets supporting the results of this article are included in the article as Additional files 1 and 2.
Authors’ contributions
DKS: participated in the experimental design, performed DNA extraction and KIR typing, data collection, analysis and interpretation of data and writing of the manuscript. CEIG: participated in the writing of the manuscript, drawing up the tables and revision of the manuscript; MGB: participated in the experimental design and discussion of experiments, interpretation of data and writing of the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Consent for publication of raw data was obtained from all study participants.
Ethics approval and consent to participate
The study was approved by the Ethics Committee from UFPR-CEP-HC number 037ext.019/2001-07. All participants signed an informed consent document.
Abbreviations
- ALL
Acute lymphoblastic leukemia
- AML
Acute myeloid leukemia
- BMT
Bone marrow transplantation
- CML
Chronic myeloid leucemia
- DNTPs
Deoxynucleotide
- EDTA
Ethylene tetraacetic acid
- EFS
Event-free survival
- GvHD
Graft-versus-host disease
- HCT
Hematopoietic cell transplantation
- HLA
Human leukocyte antigen
- HSCT
Hematopoietic stem cell transplantation
- IBGE
InstitutoBrasileiro de Geografia e Estatística
- ILTs
Immunoglobulin-like transcript
- KIR
Immunoglobulin-like receptors of natural killer
- LCR
Leukocyte receptor complex
- LIGH
Laboratório de imunogenética e histocompatibilidade
- MDS
Myelodysplasia
- MHC
Major histocompatibility complex
- NK
Natural killer
- PCR-SSOP
Polymerase chain reaction - sequence specific oligonucleotide probes
Additional files
Patients – Supporting Information. (XLS 43 kb)
Healthy Family Members – Supporting Information. (XLS 69 kb)
Contributor Information
Daniele Kazue Sugioka, Email: danikazue@yahoo.com.br.
Carlos Eduardo Ibaldo Gonçalves, Email: carlosibaldo@gmail.com.
Maria da Graça Bicalho, Phone: +55 41 3361 1729, Email: ligh@ufpr.br.
References
- 1.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–74. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 2.Sobecks RM, Wang T, Askar M, Gallagher MM, Haagenson M, Spellman S, et al. Impact of KIR and HLA genotypes on outcomes after reduced-intensity conditioning hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2015;21(9):1589–96. doi: 10.1016/j.bbmt.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Middleton D, Gonzelez F. The extensive polymorphism of KIR genes. Immunology. 2009;129:8–19. doi: 10.1111/j.1365-2567.2009.03208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319(6055):675–8. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 5.Middleton D, Williams F, Halfpenny IA. KIR genes. Transpl Immunol. 2005;14(3-4):135–42. doi: 10.1016/j.trim.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Holowiecki J. Indications for hematopoietic stem cell transplantation. Pol Arch Med Wewn. 2008;118(11):658–63. [PubMed] [Google Scholar]
- 7.Ruggeri L, Capanni M, Urbani E, Perruccio K, Hlomchik WD, Tosti A, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295(5562):2097–100. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- 8.Marcenaro E, Carlomagn S, Pesce S, Chiesa MD, Moretta A, Sivor S. Role of alloreactive KIR2DS1(+) NK cells in haploidentical hematopoietic stem cell transplantation. J Leukoc Biol. 2011;90(4):661–7. doi: 10.1189/jlb.0311137. [DOI] [PubMed] [Google Scholar]
- 9.Kulkarni S, Martin MP, Carrington M. The Yin and Yang of HLA and KIR in human disease. Semin Immunol. 2008;20(6):343–52. doi: 10.1016/j.smim.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verheyden S, Bernier M, Demanet C. Identification of natural killer cell receptor phenotypes associated with leukemia. Leukemia. 2004;18(12):2002–7. doi: 10.1038/sj.leu.2403525. [DOI] [PubMed] [Google Scholar]
- 11.Babor F, Fischer JC, Uhrberg M. The role of KIR genes and ligands in leukemia surveillance. Front Immunol. 2013;4:27. doi: 10.3389/fimmu.2013.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.John SW, Weitzner G, Rozen R, Scriver CR. A rapid procedure for extracting genomic DNA from leukocyte. Nucleic Acids Res. 1991;19(2):408. doi: 10.1093/nar/19.2.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moretta L, Montaldo E, Vacca P, Zotto GD, Moretta F, Merli P, Locatelli F, Mingari MC. Human natural killer cells: origin, receptors, function, and clinical applications. Int Arch Allergy Immunol. 2014;164(4):253–64. doi: 10.1159/000365632. [DOI] [PubMed] [Google Scholar]
- 14.de Smith AJ, Walsh KM, Ladner MB, Zhang S, Xiao C, Cohen F, et al. The role of KIR genes and their cognate HLA class I ligands in childhood acute lymphoblastic leukemia. Blood. 2014;123(16):2497–503. doi: 10.1182/blood-2013-11-540625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uhrberg M, Parham P, Wernet P. Definition of content for nine commom group B haplotypes of the Caucasoid population: KIR haplotypes contain between seven and eleven KIR genes. Immunogenetics. 2002;54(4):221–9. doi: 10.1007/s00251-002-0463-7. [DOI] [PubMed] [Google Scholar]
- 16.Toneva M, Lepage V, Lafay G, Dulphy N, Busson M, Lester S, et al. Genomic diversity of natural killer cell receptor genes in three populations. Tissue Antigens. 2001;57(4):358–62. doi: 10.1034/j.1399-0039.2001.057004358.x. [DOI] [PubMed] [Google Scholar]
- 17.Bontadini A, Testi M, Cuccia MC, Martinetti M, Carcassi C, Chiesa A, et al. Distribution of killer cell immunoglobulin-like receptors genes in the Italian Caucasian population. J Transl Med. 2006;4:44. doi: 10.1186/1479-5876-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Witt CS, Dewing C, Sayer DC, Uhrberg M, Parham P, Christiansen FT. Population frequencies and putative haplotypes of the killer cell immunoglobulin-like receptor sequences and evidence for recombination. Transplantation. 1999;68(11):1784–9. doi: 10.1097/00007890-199912150-00024. [DOI] [PubMed] [Google Scholar]
- 19.Jiang K, Zhu FM, Lv QF, Yan LX. Distribution of killer cell immunoglobulin-like receptor genes in the Chinese Han population. Tissue Antigens. 2005;65(6):556–63. doi: 10.1111/j.1399-0039.2005.00412.x. [DOI] [PubMed] [Google Scholar]
- 20.Yawata M, Yawata N, McQueen KL, Cheng NW, Guethlein LA, Rajalingam R, Shilling HG, Parham P. Predominance of group A KIR haplotypes in Japanese associated with diverse NK cell repertories of KIR expression. Immunogenetics. 2002;54(8):543–50. doi: 10.1007/s00251-002-0497-x. [DOI] [PubMed] [Google Scholar]
- 21.Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale J. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A. 2000;97(9):4778–83. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao XY, Chang YJ, Huang XJ. Differential expression levels of killer immunoglobin-like receptor genotype in patients with hematological malignancies between high-risk and standard-risk groups. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2008;16(4):746–9. [PubMed] [Google Scholar]
- 23.Ruggeri L, Capanni M, Casucci M, Volpi I, Tosti A, Perruccio K, Urbani E, Negrin RS, Martelli MF, Velardi A. Role of natural killer cells alloreactivity in HLA-mismatched hematopoietic stem cell transplantation. Blood. 1999;94(1):333–9. [PubMed] [Google Scholar]
- 24.Elmaagacli AH, Ottinger H, Koldehoff M, Peceny R, Steckel NK, Trenschel R, Biersack H, Grosse-Wilde H, Beelen DW. Reduced risk for molecular disease in patients with chronic myeloid leukemia after transplantation from a KIR-mismatched donor. Transplantation. 2005;79(12):1741–7. doi: 10.1097/01.TP.0000164500.16052.3C. [DOI] [PubMed] [Google Scholar]
- 25.Sconocchia G, Lau M, Provenzano M, Rezvani K, Wongsena W, Fujiwara H, Hensel N, Melenhorst J, Li J, Ferrone S, Barrett AJ. The antileukemia effect of HLA-matched NK and NK-T cells in chronic myelogenous leukemia involves NKG2D-target-cell interactions. Blood. 2005;106(10):3666–72. doi: 10.1182/blood-2005-02-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khakoo SI, Carrington M. KIR and disease: a model system or system of models? Immunol Rev. 2006;214:186–201. doi: 10.1111/j.1600-065X.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- 27.Schellekens J, Rozemuller EH, Petersen EJ, Van Den Tweel JG, Verdonck LF, Tilanus MG. Activating KIRs exert a crucial role on relapse and overall survival after HLA-identical sibling transplantation. Mol Immunol. 2008;45(8):2255–61. doi: 10.1016/j.molimm.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 28.Shilling HG, Guethlein LA, Cheng NW, Gardiner CM, Rodriguez R, Tyan D, Parhan P. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol. 2002;168(5):2307–15. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]
- 29.Hsu KC, Chida S, Dupont B, Geragthy DE. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polimorphism. Immunol Rev. 2002;190:40–52. doi: 10.1034/j.1600-065X.2002.19004.x. [DOI] [PubMed] [Google Scholar]
- 30.Hsu KC, Liu XR, Selvakumar A, Mickelson E, O’Relly RJ, Dupont B. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol. 2002;169(9):5118–29. doi: 10.4049/jimmunol.169.9.5118. [DOI] [PubMed] [Google Scholar]
- 31.Kim HJ, Choi Y, Min WS, Kim TG, Sho BS, Kim SY, Eom KS, Lee S, Min CK, Cho SG, Kim DW, Lee JW, Kim CC. The activating killer cell immunoglobulin-like receptors as important determinants of acute graft-versus host disease in hematopoietic stem cell transplantation for acute myelogenous leukemia. Transplantation. 2007;84(9):1082–91. doi: 10.1097/01.tp.0000285918.72930.35. [DOI] [PubMed] [Google Scholar]
- 32.Middleton D, Diler AS, Meenagh A, Sleator C, Gourrau PA. Killer immunoglobulin-like receptors (KIR2DL2 and/or KIR2DS2) in presence of their ligand (HLA-C1 group) protect against chronic myeloid leukaemia. Tissue Antigens. 2009;73(6):553–60. doi: 10.1111/j.1399-0039.2009.01235.x. [DOI] [PubMed] [Google Scholar]
- 33.Mancusi A, Ruggeri L, Urbani E, Pierini A, Massei MS, Carotti A, et al. Haploidentical hematopoietic transplantation from KIR ligand-mismatched donors with activating KIRs reduces nonrelapse mortality. Blood. 2015;125(20):3173–82. doi: 10.1182/blood-2014-09-599993. [DOI] [PubMed] [Google Scholar]
- 34.Abi-Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J Exp Med. 2005;201(8):1319–32. doi: 10.1084/jem.20042558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Single RM, Martin MP, Gao X, Meyer D, Yeager M, Kidd JR, et al. Global diversity and evidence for coevolution of KIR and HLA. Nat Genet. 2007;39(9):1114–9. doi: 10.1038/ng2077. [DOI] [PubMed] [Google Scholar]
- 36.Hou L, Steiner NK, Chen M, Belle I, Kubit AL, et al. Limited allelic diversity of stimulatory two-domain killer cell immunoglobulin-like receptors. Hum Immunol. 2008;69(3):174–8. doi: 10.1016/j.humimm.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Scquizzato E, Teramo A, Miorin M, Facco M, Piazza F, Noventa F, Trentin L, Agostini C, Zambello R, Semenzato G. Genotypic evaluation of killer immunoglobulin-like receptors in NK-type lymphoproliferative disease of granular lymphocytes. Leukemia. 2007;21(5):1060–9. doi: 10.1038/sj.leu.2404634. [DOI] [PubMed] [Google Scholar]
- 38.Besson C, Roetynck S, Williams F, Orsi L, Amiel C, Lependeven C, et al. Association of killer cell immunoglobulin-like receptor genes with Hodgkin’s lymphoma in a familial study. PLoS One. 2007;2(5):e406. doi: 10.1371/journal.pone.0000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen AM, Guo XM, Yan WY, Xie SM, Zhu N, Wang XD, Xu R, Liu QP. Polymorphism of killer cell immunoglobulin-like receptor gene and its correlation with leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2007;15(1):35–8. [PubMed] [Google Scholar]
- 40.McQueen KL, Dorighi KM, Guethlein LA, Wong R, Sanjanwala B, Parham P. Donor-recipient combinations of group A and B KIR haplotypes and HLA class I ligand affect the outcome of HLA-matched, sibling donor hematopoietic cell transplantation. Hum Immunol. 2007;68(5):309–23. doi: 10.1016/j.humimm.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets supporting the results of this article are included in the article as Additional files 1 and 2.


