Abstract
Purpose
DNA repair defects have been previously reported in myeloproliferative neoplasms (MPNs). Inhibitors of poly(ADP-ribose) polymerase (PARP) have shown activity in solid tumors with defects in homologous recombination (HR). The present study was performed to assess MPN sensitivity to PARP inhibitors ex vivo.
Experimental Design
HR pathway integrity in circulating myeloid cells was evaluated by assessing formation of RAD51 foci after treatment with ionizing radiation or PARP inhibitors. Sensitivity of MPN erythroid and myeloid progenitors to PARP inhibitors was evaluated using colony formation assays.
Results
Six of 14 MPN primary samples had reduced formation of RAD51 foci after exposure to ionizing radiation, suggesting impaired HR. This phenotype was not associated with a specific MPN subtype, JAK2 mutation status or karyotype. MPN samples showed increased sensitivity to the PARP inhibitors veliparib and olaparib compared to normal myeloid progenitors. This hypersensitivity, which was most pronounced in samples deficient in DNA damage-induced RAD51 foci, was observed predominantly in samples from patients with diagnoses of chronic myelogenous leukemia, chronic myelomonocytic leukemia or unspecified myelodysplastic/MPN overlap syndromes.
Conclusions
Like other neoplasms with HR defects, MPNs exhibit PARP inhibitor hypersensitivity compared to normal marrow. These results suggest that further preclinical and possibly clinical study of PARP inhibitors in MPNs is warranted.
Keywords: Myeloproliferative neoplasm, poly(ADP-ribose) polymerase, targeted therapy, DNA repair, chronic myelomonocytic leukemia
Introduction
Myeloproliferative neoplasms (MPNs) represent a heterogeneous group of clonal diseases with a common propensity to progress to acute leukemia (1–4). Essential Thrombocythemia (ET), Polycythemia Vera (PV), Primary Myelofibrosis (MF), and mixed myelodysplastic/myeloproliferative neoplasms such as Chronic Myelomonocytic Leukemia (CMMoL) are all clonal neoplasms derived from aberrant early hematopoietic precursors but have varied clinical manifestations. Upon progression, they are uniformly refractory to standard acute leukemia therapies, with median survival less than six months.
One common mutation in MPNs, an activating V617F point mutation in the tyrosine kinase JAK2, is found in greater than 80% of cases of PV (5), 40% of cases of ET, and 30% of cases of MF (6). JAK2 mutation and overexpression have been associated with increased homologous recombination (HR) and genomic instability (7–9). Other alterations conferring an MPN-like phenotype, such as BCR/ABL translocations in CML, FIP1L1-PDGFR rearrangements in eosinophilic leukemias, and FLT3 mutations in acute myeloid leukemia, have also been associated with changes in the DNA repair pathways leading to increased genomic instability and drug resistance (9, 10). For example, even though early studies indicated that RAD51, a critical component of the homologous recombination (HR) pathway, is upregulated in BCR/ABL-positive CML cells (11), subsequent studies demonstrated that repair in these cells is error-prone and leads to mutations and large deletions or insertions (12). Further analysis traced this genomic instability to several changes, including (i) enhanced tyrosine phosphorylation of RAD51, leading to its aberrant function (13); (ii) downregulation of BRCA1 (14); (iii) stimulation of single-strand annealing, an error-prone DNA repair pathway (15); and (iv) other changes in the Fanconi anemia/BRCA pathway that can be reverted by ectopic BRCA1 expression (16).
Repair defects in BCR/ABL-negative MPNs are not as well characterized. Gross chromosomal lesions are common in MF and accelerating MPN; and SNP array karyotyping identified additional subcytogenetic abnormalities (17, 18). These types of changes are reminiscent of chromosomal aberrations observed in HR-deficient solid tumors such as BRCA1- or BRCA2-mutant breast and ovarian cancer (19). Although the upstream source and pathological consequences of these extensive genomic rearrangements are not as well understood in MPN, previous studies have reported increased error prone double-strand break repair in MPNs as well as CML (15).
Poly(ADP-ribose) polymerase (PARP) inhibitors are a class of antineoplastic agents being widely tested in solid tumors (20–25). These agents target PARP1, PARP2, and PARP3, three enzymes that contribute to various aspects of DNA repair (21, 23, 24, 26–28). PARP inhibition not only diminishes base excision repair, but also impairs alternative end-joining (29) and accelerates nonhomologous end-joining (30). In cells with diminished HR, these changes lead to error-prone DNA repair and cell death (21, 23, 31). In addition, PARP inhibition leads to trapping of PARP1 on DNA, preventing access of downstream repair proteins to sites of DNA damage (32–34) and providing a potential mechanism for PARP inhibitor-induced killing in HR proficient cells that contain high levels of PARP1 protein. Importantly, chromosome 1q (including the PARP1 locus at 1q42) is amplified in a subset of chronic phase MPNs and even more commonly in transformed MPNs (17, 35), providing a potential opportunity for PARP1 trapping even in MPNs without HR defects.
Consistent with the BCR/ABL-induced DNA repair abnormalities described above, PARP inhibitor hypersensitivity has been reported in CML (36). In view of the repair defects observed in other MPNs, as well as the copy number increases of the PARP1 locus in these disorders (17, 35), we have performed a survey comparing PARP inhibitor sensitivity of various MPNs, including CML, with that of normal hematopoietic progenitors. For this study, we have examined two PARP inhibitors that are undergoing extensive clinical testing. Veliparib, which is in National Cancer Institute-sponsored trials in a variety of neoplasms, including hematological neoplasms (www.clinicaltrials.gov), has recently been shown to exhibit favorable properties for combining with DNA damaging agents (34). Olaparib, which is roughly 10-fold more potent in vitro, has attained regulatory approval for the treatment of ovarian cancer (24, 25).
MATERIALS AND METHODS
Inhibitors
Veliparib (ABT-888, Enzo Life Sciences, Farmingdale, NY) and olaparib (ChemieTek, Indianapolis, IN) were dissolved in dimethyl sulfoxide (DMSO) at stock concentrations of 10 mM. Stocks were aliquoted in 10 μL volumes, stored at −80°C, and thawed once immediately before use. All samples in any given experiment contained identical concentrations of DMSO (0.1% vol/vol).
Clinical samples
After patients provided informed consent, specimens were obtained by purifying mononuclear cells from the peripheral blood of patients with MPN on Ficoll-Hypaque gradients. The MPN cohort included fresh samples from cases of BCR/ABL-positive CML; the classical BCR/ABL-negative MPNs ET, PV, primary and secondary MF; and mixed MDS/MPN diagnoses (CMMoL, aCML, MDS-MPN-U) (Table 1).
Table 1.
Clinical MPN Samples Studied
| MPN # | Clinical Diagnosisa | Karyotype | BRCA1 methylationc | JAK2 V617F Mutb,c | RAD51 foci Formationb,d | H2Ax foci formationb, | Veliparib IC50 (μM) | Olaparib IC50 (μM) | BROCA analysisb,e,f | |
|---|---|---|---|---|---|---|---|---|---|---|
| Deleterious alterations | Variants of unknown significance | |||||||||
| 1 | Post-ET MF | 46XY | No | − | Normal | Normal | 6.9 | b | CHEK2 c.1100delG | None |
| 2 | PMF | 46XY 11q- | + | 15 | ||||||
| 3 | PMFg | 7XY 13q- +14 | No | − | Impaired | Normal | 1.3 | 0.35 | None | LIG4 p.A857T PALB2 p.R414Q |
| 4 | PMF | 46XX 5q- | No | 48% | Normal | Normal | 8.6 | 0.83 | None | PRKDC p.M333I RAD51D p.R232Q |
| 5 | PV | 46XY inv9 | Yes | + | 1.0 | 0.56 | None | CDK12 p.P1275L, MSH2 p.R55G | ||
| 6 | CMMoL | 46XX | − | Impaired | Normal | 4.8 | 0.4 | |||
| 7 | MDS/MPN-U | 47XY +1p | Yes | − | Normal | Normal | 2.4 | 0.64 | None | PRKDC p.P695S TP53BP1 p.V1031A |
| 8 | Post-PV MF | 47XY +8 | 45% | Normal | Normal | 9.3 | 1.8 | |||
| 9 | MDS/MPN-U | 46XY | No | − | Impaired | Impaired | 0.8 | 0.14 | None | No alterations |
| 10 | CMMoL | 46XY | − | impaired | Normal | 0.7 | 1.0 | |||
| 11 | tPV | 46XY t(11;14) | + | Normal | Normal | >20 | 5.2 | |||
| 12 | CMMoL | 46XY | − | Normal | Normal | 2.6 | 0.30 | |||
| 13 | PMF | 46XY | 58% | Normal | Normal | 22 | 1.8 | |||
| 14 | PMF | 47XX +9 t(12;13) | + | Impaired | Normal | 7.0 | 1.1 | |||
| 15 | CML | 45X -Y t(9;22) | Impaired | Impaired | 2.2 | |||||
| 16 | PMF | 46XX 20q- | + | 2.7 | None | PRKDC p.A3904V | ||||
| 17 | ET | − | Normal | Normal | 2.9 | |||||
| 18 | ET | + | 0.3 | |||||||
| 19 | ET | + | 4.6 | |||||||
| 20 | CMMoL | 46XY | No | − | ovgrth | |||||
| 21 | tCMMoL | 46XY | No | 4.1 | None | No alterations | ||||
| 22 | CMMoL | 46,XY,del(2)(q) | No | − | 1.3 | XRCC4 c.24 delC |
XRCC4 c.24 delC DCLRE1C p.G38R |
|||
| 23 | CMMoL | 46XX | Yes | 2.5 | None | PIK3CA p.Y644H | ||||
| 24 | tPMF | 92XXYY der2 t(1;2) | + | 4.1 | ||||||
| 25 | CMMoL | 46XY | No | − | 3.2 | |||||
| 26 | CMMoL | 46XX | 5.5 | |||||||
| 27 | PMF | 45X -Y | Yes | + | 5.4 | None | SLX4 p.P385T, p.P957L, p.E942Q | |||
| 28 | CMMoL | 46XY | No | − | 1.9 | |||||
| 29 | CML | 46XX t(9;22) | No | 4.0 | None | MSH6p. I1054F PTEN p.H397R |
||||
| 30 | aCML eo | 46XX t(4;7) | No | 4.4 | None | CDK12 p.L988S, NBN p.I439M SLX4 p.R1372Q and p.A916S |
||||
| 31 | CMMoL | 46XX | No | 2.2 | BRCA1 5382insC (c.5263_5264insC) | RAD51B p.K243R TOPBP1 p.N1042S, |
||||
| 32 | PMF | 46XY t(1;7) +9 1q- | Yes | + | 4.2 | None | TOPBP1 p.R309C XRCC5 p.A550S |
|||
| 33 | PMF | 46 XY | No | + | 1.7 | None | LIG4 p.T9I | |||
| 34 | CMMoL | 48 XY +8 +14 | No | 0.8 | None | CDK12 p.L1189Q MLH1 p.H718Y, PALB2 p.D134N, PRKDC p.R1253H, SLX4 p.E701D, TOPBP1p.M293V |
||||
| 35 | CMMoL | 46 XX | No | 34% | 1.5 | None | XRCC5 p.R184H, | |||
| 36 | CMMoL | 46 XY | Yes | − | 4.3 | None | ATR L274F, RBBP8 C485 | |||
| 37 | PMF | 46 XY 20q- | No | + | 1.9 | None | BARD1 p.Q11H, BLM p.I366T, PIK3CA p.R524K | |||
| 38 | CMMoL | 46 XY | No | 4.3 | RAD50 c.3476delA | ATR p.H117R, CDK12 p.P645S, NBN p.N142S, SLX4 p.S1271F, UIMC1 p.Y564H | ||||
| 39 | CML | 46 t(9:22) | − | 4.8 | None | ATR p.I97F, MRE11 p.S334R, RBBP8 p.K357N, SLX4 p.K1635E, TP53BP1 p.H58R | ||||
| 40 | CMMoL | 46 XY | No | − | 3.2 | None | ATM p.S1691R, FAM175A p.T141I, TOPBP1 p.S817L, TP53BP1 p.E1019G | |||
| 41 | ET | 46 XY | 43% | 3.8 | .78 | |||||
Abbreviations used are: MF, myelofibrosis; CMMoL, chronic myelomonocytic leukemia; ET, essential thrombocythemia; PMF, primary myelofibrosis; PV, polycythemia vera; and t, transformed to AML at the time of study.
Blank cell indicates assay not performed.
+, JAK2 V617F mutation present; −, tested and mutation not present; numbers indicate quantitative allele burden where available.
Normal, foci form normally in response to ionizing radiation; impaired, foci form in fewer cells after ionizing radiation. See Fig. 1 for quantitation.
Sequence alterations deleterious to HR genes (bold) or nonhomologous end-joining (underlined) are shown here. Additional variants of unknown significance (previously reported allelic polymorphisms in the normal population at allele frequencies from 0.0005 to 0.24 and conservative substitutions) are listed in Table S2
All nomenclature is according to Human Genome Variation Society (HGVS) nomenclature except for the BRCA1 alteration, for which. Breast Cancer Information Core (BIC) nomenclature is provided along with the the HGVS nomenclature in parentheses.
Missing 1 copy of BRCA2 as a consequence of the 13q deletion.
RAD51 focus formation assay
Ten million Ficoll/Hypaque-purified mononuclear cells (MNCs) from normal controls and MPN patients were exposed to 10 Gy ionizing radiation (IR) from a Rad Source RS200 X-ray irradiator (Brentwood, TN), then allowed to recover for 6 h in a humidified 37 °C tissue culture incubator equilibrated at 5% (vol/vol) CO2. Leukocytes were pelleted by centrifugation at 200×g for 10 min and fixed in 2% (wt/vol) paraformaldehyde in Dulbecco’s calcium- and magnesium-free phosphate-buffered saline (PBS) for 10 min at 20–22 °C. Leukocytes were re-pelleted as above, washed with PBS, and stored in PBS at 4 °C. For analysis, 2.5 × 104 leukocytes were deposited onto glass coverslips by cytocentrifugation and processed as previously described (37). Briefly, coverslips were washed 3 times with PBS, permeabilized in PBS + 0.25 % (vol/vol) Triton X-100 for 10 min, washed an additional 3 times with PBS, then incubated for 1 h in blocking buffer [PBS, 1 % (vol/vol) glycerol, 0.1% (wt/vol) gelatin from cold-water fish, 0.1% (wt/vol) bovine serum albumin, 5% (vol/vol) goat serum and 0.4 % (wt/vol) sodium azide] for 1 h at room temperature. Coverslips were incubated with RAD51 rabbit polyclonal (Active Motif; Carlsbad, CA) and phospho-Ser139-H2AX mouse monoclonal (Millipore; Billerica, MA) antibodies diluted 1:500 in blocking buffer overnight at 4 °C. Coverslips were then washed 3 times with PBS, followed by incubation for 1 h in secondary Alexa Fluor 488-conjugated goat anti-mouse IgG and Alexa Fluor 568-tagged goat anti-rabbit IgG (Invitrogen; Carlsbad, CA) diluted 1:1000 in blocking buffer. Coverslips were further washed 3 times with PBS, counterstained with 1 μg/ml Hoechst 33258 in PBS, and mounted using UltraLong antifade reagent (Invitrogen, Carlsbad, CA). PEO1 and PEO4, cell lines with a truncating BRCA2 mutation and a reversion mutation, respectively, were utilized as negative and positive controls for radiation-induced RAD51 foci. Confocal images were captured on a Zeiss LSM 710 scanning confocal microscope with a 100×/1.4 N.A. oil immersion objective. For quantitation, 100 cells per sample from greater than 5 fields were manually scored for RAD51 and phoshopho-H2AX foci by an investigator blinded to clongenic assay results. Cells with >5 foci were graded as positive. Quantitation and image processing were performed with the Zeiss Zen software package and Adobe Photoshop CS3.
Colony forming assays
Colony forming assays were performed as previously described (38). Ficoll/Hypaque-purified MNCs were washed and resuspended in RPMI 1640 medium (Gibco). 0.5 ml aliquots were combined with 4.5 ml of MethoCult® medium (STEMCELL Technologies #H4435, Vancouver, BC) containing increasing concentrations of veliparib (0, 0.5, 1, 2.5, 5, 10, 20 μM) or olaparib (0, 0.25, 0.5, 1, 2 and 5 μM). Two or four 1 ml aliquots of each mixture were plated onto replicate 35 mm gridded culture plates to give final MNC concentrations of 1–5 × 105 cells per plate. After incubation at 37 °C in 5% CO2, most samples were scored for total colony number after 10–14 days; however, some CMMoL samples were scored earlier (5–7 days) due to hypersensitivity to growth factors. Colonies were defined as clusters of >40 cells. Drug concentrations at which total colony counts were inhibited to 50% of diluent treated controls (IC50) were determined using linear regression analysis of the dose response curves after linear transformation using an exponential model (CalcuSyn software, Biosoft, Inc., Cambridge, UK).
Genomic analysis
DNA was extracted from Ficoll/Hypaque-purified MNCs using a DNA midi kit (Qiagen, Valencia, CA). To search for somatic mutations in HR pathway proteins, aliquots of DNA (1 μg) were subjected to targeted capture and massively parallel sequencing using BROCA as previously described (39) for a panel of genes involved in HR (ATM, ATR, BABAM1, BARD1, BAP1, BLM, BRCA1, BRCA2, BRE, BRIP1, BRCC3, CHEK1, CHEK2, FAM175A, FANCC, FANCP, MRE11A, NBN, PALB2, RAD50, RAD51, RAD51B, RAD51C, RAD51D, RBBP8, TOPBP1, UIMC1, XRCC2, and XRCC3), regulation of HR (CDK12, C11orf30, ID4, TP53BP1, and USP28), or NHEJ (LIG4, XRCC4, XRCC5, XRCC6, PRKDC, DCLRE1C) and TP53.
Methylation-specific PCR (MSP), quantitative MSP (qMSP)
Genomic DNA was bisulfite-treated with the EZ DNA Methylation Kit (Zymo Research, Irvine, CA). The initial MSP screen was performed as previously described (40). qMSP for BRCA1 promoter methylation, normalized to β-actin levels, was carried out using the iTaq SYBR Green mix and 300 nM of each primer. qMSP, MSP primer sequences and annealing temperatures are listed in Supplemental Table 1. BRCA1 qMSP primers were adapted from Estellar et al. and cover the region −175 to +9 relative to the TSS (41). Cycling conditions are 95 °C for 5 min, followed by 40 cycles of 95 °C for 5 sec and 64 °C for 60 sec. Melt curve readings were recorded at 0.5 °C increments from 65 °C to 95 °C. The controls for unmethylated and methylated templates were bisulfite-treated samples from normal lymphocyte DNA and CpG methylated Jurkat genomic DNA (New England Biolabs, Ipswich, MA), respectively. Methylated samples are defined as amplicons with melting temperatures matching that of the methylated control.
RESULTS
Some MPN primary samples have an abnormal DNA damage response
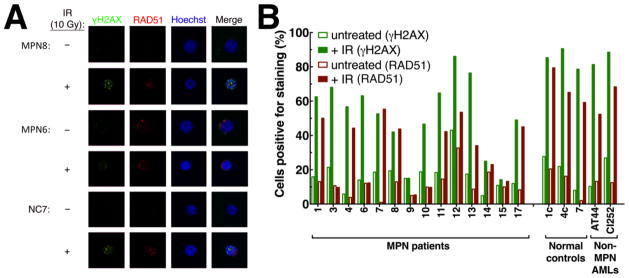
To assess whether PARP inhibitors might have any promise in non-CML MPNs, the present study examined primary samples from 41 patients with a variety of MPNs (Table 1). Following up on the observation that MPNs are associated with multiple genetic aberrations (17, 18), we examined a subset of 14 samples for integrity of the HR pathway. Upon exposure to ionizing radiation, cells with normal HR repair form foci of DNA repair complexes that can be detected by immunofluorescence after staining for RAD51, the recombinase responsible for pairing homologous sequences. Cells with impaired HR typically will not form RAD51 foci when exposed to ionizing radiation. Of 14 MPN samples examined using this assay, six (43%) displayed markedly diminished RAD51 foci formation (Figure 1 and Table 1). In contrast, 0 of 14 controls performed with these assays exhibited diminished RAD51 foci formation (p = 0.016, Fisher’s exact test).
Figure 1. Lack of radiation-induced RAD51 foci in a subset of MPNs.
A, RAD51 foci formation in circulating myeloid cells from the indicated samples before and after ionizing radiation. B, summary results from 14 MPN cases, 3 normal controls and 2 de novo acute myelogenous leukemia cases.
To search for potential explanations for this HR deficiency, we sequenced a panel of repair genes found to be mutated in a variety of malignancies, including BRCA1, BRCA2, RAD51, RAD51 paralogs and several Fanconi anemia pathway genes, in MPN samples from 24 of these patients. Results of this analysis are summarized in Table 1. Mutations observed included a well-established BRCA1 stop mutation (BRCA1 c.5382insC) and CHEK2 frameshift mutation (CHEK2 c.1100delG) as well as a deleterious mutation in the RAD50 gene (RAD50 c.3476delA). While a variety of rare normal alleles and conservative single nucleotide variants were also detected in other genes (Table 1), no deleterious homozygous or hemizygous mutations were observed that could by themselves account for the defect in RAD51 protein recruitment to sites of DNA damage. We also assessed BRCA1, BRCA2, FANCC, FANCF, and FANCL promoter CpG island methylation in these MPN samples. No hypermethylation was identified in the BRCA2, FANCC, FANCF, or FANCL promoters. However, six of 27 samples (22.2%) analyzed in the present study demonstrated hypermethylation of the BRCA1 promoter (Table 1).
Inhibition of colony formation by the PARP inhibitor veliparib
With the observation that a substantial fraction of MPN samples have a deficient DNA repair response, as manifested by lack of formation of radiation-induced RAD51 foci, we performed colony forming assays with continuous exposure to the PARP inhibitor veliparib using MNCs from 41 MPN patients and 13 normal controls. As shown in Figure S1, the sensitivities of granulocyte and erythroid colonies generally paralleled each other in both normal and MPN samples. Accordingly, for the remainder of this work we tabulated various colony types with each assay but combined them at the time of graphing. Assay reproducibility was established by comparing results of assays performed on samples from several patients at two points in time in the absence of changes in treatment (Figure S2).
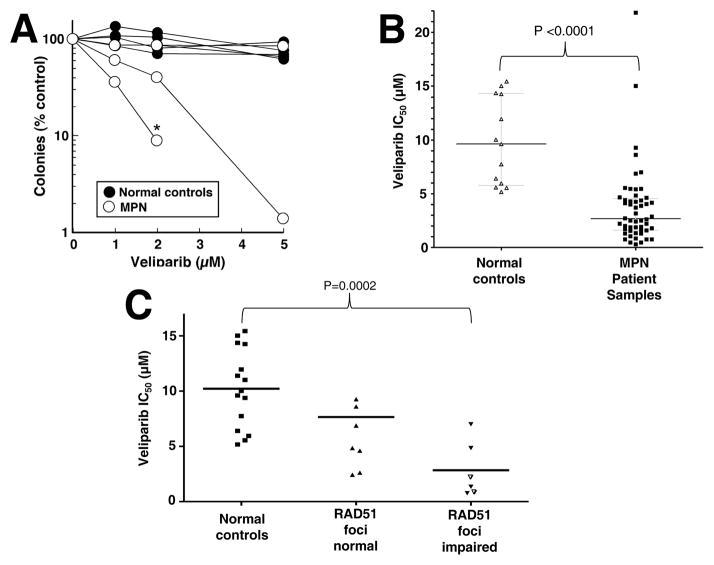
As shown in Figure 2A and B, the drug concentrations that inhibited colony formation by 50% (IC50) were an average of 4-fold lower in the MPN samples than in normal controls (2.5 vs 9.6 μM, P<0.0001). Focusing on the subset of samples that were subjected to assays for both RAD51 foci and colony formation (Figure 2C), PARP inhibitor sensitivity was greater (IC50 on average 3-fold lower) in MPN samples with impaired RAD51 foci formation compared to those without (mean 9.3 vs. 2.8 μM, p = 0.028). In contrast, BRCA1 promoter methylation status did not track with veliparib sensitivity (Figure S3).
Figure 2. Veliparib sensitivity in MPN samples and normal controls.
A, results of colony forming assays in three MPN samples and four normal controls. Each line and corresponding symbols represent the mean results of replicate plates from one assay performed as described in the Methods. * indicates sample with no colony growth at the next higher veliparib concentration. B, summary of IC50 values in MPN samples and normal controls from results in panel A and additional samples run at veliparib concentrations up to 20 μM. C, relationship between IC50 values and formation of RAD51 foci in MPN samples. Open triangles represent samples with impaired formation of both phospho-H2AX and RAD51 foci (Table 1).
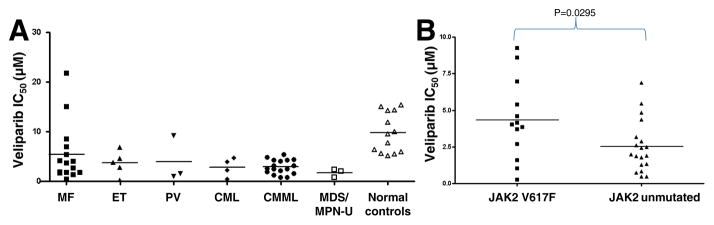
MPN primary samples demonstrate veliparib sensitivity across clinical subtypes
To determine whether PARP inhibitor sensitivity was linked to clinical characteristics, we analyzed colony-forming assay results based on clinical descriptions of disease at the time of sample acquisition. This analysis suggested broad sensitivity across clinical subtypes, with samples from MF patients demonstrating the greatest heterogeneity (Figure 3A). Although the number of samples in any MPN category was somewhat limited, CML, CMMoL and MDS/MPN-U samples were most sensitive, with IC50 values averaging 3, 3 and 1.5 μM, respectively.
Figure 3. Relationship between veliparib sensitivity and MPN biology.
A, IC50 values for veliparib in various MPN subsets. B, relationship between IC50 values for veliparib and JAK2 mutation status.
Activating JAK2 mutations correlate with decreased veliparib sensitivity
We also examined whether JAK2 mutation status, which was suggested as a surrogate marker for impaired DNA repair (15), correlated with PARP inhibitor sensitivity. In our sample set (Figure 3B), JAK2 non-mutated samples were significantly more sensitive to PARP inhibition than JAK2V617F mutated samples (mean IC50 2.8 vs. 5.7 μM, P = 0.04). Notably, however, 5 of 16 individual JAK2 mutated samples demonstrated veliparib IC50s below 2 μM despite the relative insensitivity of this subset as a whole.
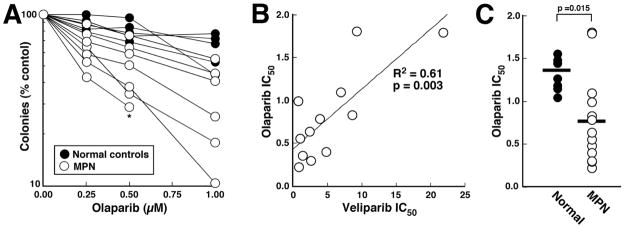
MPN hypersensitivity to the PARP inhibitor olaparib
While this work was in progress, it was reported that the PARP inhibitors PJ-34 (a preclinical tool compound) and veliparib might also inhibit certain kinases when assayed at high concentrations under cell-free conditions (42, 43). Importantly, the structurally dissimilar PARP inhibitor olaparib lacks this off-target kinase inhibitory activity (43). To provide assurance that the veliparib hypersensitivity observed in MPN samples was due to PARP inhibition, olaparib sensitivity was also examined using colony-forming assays (e.g., Figure 4A) in a subset of samples. For samples assayed using both veliparib and olaparib, there was a strong correlation (R2 = 0.61, p = 0.003) between sensitivity to the two agents (Figure 4B). Accordingly, as was the case with veliparib, a substantial fraction of these MPN samples were also hypersensitive to olaparib when compared to normal controls (Figure 4C).
Figure 4. Olaparib sensitivity in MPN samples and normal controls.
A, results of colony forming assays for 7 MPN samples and 4 normal controls. Each line and corresponding symbols represent the mean results of replicate plates from one assay performed as described in the Methods. * indicates sample with no colony growth at the next higher olaparib concentration. All samples assayed for olaparib sensitivity were also assayed using veliparib. B, correlation between IC50 for veliparib and olaparib in the 12 samples that overlapped. C, comparison of olaparib IC50 values in normal and MPN samples.
DISCUSSION
Because genomic instability is a hallmark of advanced MPNs, we examined whether clinical MPN specimens exhibit HR defects and whether they are hypersensitive to PARP inhibitors, which are known to be particularly effective in neoplasms with HR deficiency (20–24). Here we demonstrate PARP inhibitor hypersensitivity for the first time in a large subset of MPN samples.
An impaired DNA damage response, as evidenced by lack of development of RAD51 foci in response to ionizing radiation, was observed in ~40% of MPN samples assayed (Figure 1 and Table 1). This observation suggests that the HR pathway is impaired in those samples. These results are consistent with prior observations of extensive chromosomal and subchromosomal copy number changes in MPN (17, 18), which are another hallmark of HR deficiency (19). Because formation of RAD51 foci only provides an assessment of HR pathway integrity upstream of RAD51 and is unaffected by changes downstream, e.g., loss of RAD51C (44), it is important to emphasize that the present observations might underestimate the true frequency of HR repair defects in MPNs.
The causes of this HR pathway dysfunction in MPNs are incompletely understood at present. Some of the samples with impaired RAD51 foci formation also exhibited diminished formation of H2AX foci (Fig. 1B and Table 1). Because H2AX phosphorylation after double strand breaks typically reflects activation of ATM and phosphorylation of its substrate MDC1 (45), diminished formation of both phospho-H2AX and RAD51 foci might reflect a defect in ATM or its activation as described in other neoplasms. Other samples formed phospho-H2AX foci but nonetheless failed to form RAD51 foci, suggesting one or more defects between these two events in the DNA damage response. Consistent with this heterogeneity, we have previously examined 144 cases of MPN via SNP arrays and found that 26% have heterozygous deletions in genes encoding one or more DNA repair pathway proteins such as BRCA2, ATM, FANCC or FANCL (46). Because the other copy remains intact, however, it is unclear whether these heterozyogous deletions cause sufficient changes in protein expression to impact the HR pathway. Accordingly, we have examined a subset of MPN samples for methylation changes in FANC proteins, ATM, and BRCA2 but did not observe frequent changes in methylation that would confer increased genomic instability. BRCA1 promoter methylation was found in 6 of 27 samples analyzed (Table 1) but was not associated with increased sensitivity to PARP inhibition ex vivo (Figure S3). Given the results of drug sensitivity testing in the present study, further investigation of repair defects in MPNs appears warranted.
PARP inhibitors are currently undergoing extensive testing in other neoplasms with HR deficiencies (20–25). Here we found that MPN samples with impaired RAD51 foci formation exhibited increased sensitivity to the PARP inhibitor veliparib, suggesting that these cells depend on alternative repair pathways containing one or more veliparib-sensitive PARPs for their survival (Figure 2C). Further examination of MPN samples as a group revealed broad sensitivity to PARP inhibition across many subtypes of MPN (Figure 3A). This hypersensitivity relative to normal myeloid progenitors might reflect multiple alterations in MPNs. In addition to the sensitizing effects of HR deficiency, the presence of high concentrations of PARP1, e.g., as a consequence of chromosome 1q gains in copy number seen in a subset of MPNs (see Introduction), might also sensitize cells to the PARP trapping that accompanies treatment with PARP inhibitors (32, 37). Accordingly, there are multiple potential explanations for the observed hypersensitivity of MPNs relative to normal myeloid progenitors, just as there are in solid tumors (20–25); and further investigation is required to understand this hypersensitivity on a case by case basis in both instances.
While this work was in progress, it was reported that veliparib can inhibit purified kinases when applied under cell-free conditions at concentrations 100- to 1000-fold higher than those required to inhibit purified PARP1 and PARP2 (43). Importantly, the PARP inhibitor olaparib was shown to lack these off-target kinase inhibitor effects (43). In our study, there was a strong correlation between olaparib and veliparib sensitivity (Figure 4B), suggesting that the PARP inhibitor hypersensitivity of MPNs (Figures 2B and 4C) reflects PARP inhibition rather than an off-target effect.
To our knowledge the present study provides the first examination of PARP inhibitors in non-CML MPN. Because hypersensitivity of some MPN samples was apparent within the first few samples, we surveyed a broad range of MPN samples in an attempt to determine the types of diseases lumped under the categories of MPN and mixed MDS/MPN that might display this hypersensitivity. Further studies examining additional samples with each type of MPN or MDS/MPN syndrome at various stages of disease (e.g., initial diagnosis versus advanced disease requiring treatment versus recurrence after therapy or stem cell transplant) are required to assess whether PARP inhibitor hypersensitivity persists after current treatments, as appears to be the case in a subset of ovarian cancers, for example (23).
Our results indicate that MPN samples with the activating JAK2 V617F mutation are, on average, less sensitive than MPN samples without JAK2 mutations. Because the assays measured the sensitivity of proliferating cells, this difference is unlikely to reflect any difference in the rate of proliferation in JAK2 wildtype vs. JAK2 mutant samples. Instead, it is possible that the difference reflects the ability of JAK2 V617F to activate STAT-mediated transcription and inhibit apoptosis (47).
On the other hand, the therapeutic window for PARP inhibition in MPN, e.g., as measured by differences in mean IC50 of RAD51 foci-deficient MPNs and normal progenitors (Figure 2C), is somewhat narrower than the hundreds-fold difference in IC50 observed when comparing BRCA1- or BRCA2-deficient murine embryonic stem cells and their HR proficient counterparts (48). However, it is important to point out that the difference in sensitivity between BRCA2-deficient human ovarian cancer cells and their isogenic BRCA2-restored counterparts is also only 5- to 10-fold (30), yet clinical activity of PARP inhibitors is observed in ovarian cancer (20–25). The limited, albeit easily observed, sensitivity difference between normal and MPN progenitors suggests that activity of PARP inhibitors as single agents in MPN might merit further investigation, particularly in JAK2 wildtype MPNs, which sometimes lack therapeutic options (1–4). On the other hand, we have previously examined the impact of adding veliparib to topotecan and carboplatin (37), two agents that have exhibited some activity when combined to treat relapsed AML (49), and have observed synergistic cytotoxic effects with the topotecan/veliparib combination in AML lines in vitro (37). Moreover, because JAK2 mutations were associated with diminished PARP inhibitor sensitivity (Figure 3B), we have begun examining the effect of combining PARP inhibitors with other agents used in the treatment of MPNs, looking for for synergistic cytotoxic effects with PARP inhibitor/hydroxyurea and PARP inhibitor/ruxolitinib combinations. These observations will provide the impetus for further study of PARP inhibitors, alone and in combination, in MPNs.
Supplementary Material
Translational Relevance.
Myeloproliferative neoplasms (MPNs) are a heterogeneous group of clonal hematological disorders with limited current therapeutic options. Previous studies have shown that MPNs often have impaired DNA repair pathways. Poly(ADP-ribose) polymerase (PARP) inhibitors have shown promising activity in solid tumors with defects in homologous recombination repair. Here we compare the sensitivity of clinical isolates from several BCR/ABL-negative chronic myeloid neoplasms, including chronic myelomonocytic leukemia, essential thrombocythemia, and primary myelofibrosis, to normal controls using two different PARP inhibitors in colony forming assays ex vivo. Results of this analysis demonstrate that myeloid progenitors from many patients with JAK2 wildtype MPNs exhibit enhanced PARP inhibitor sensitivity, which is greatest in those with defective formation of RAD51 foci after DNA damage. These observations support the further study of PARP inhibitors, alone or in combination with other therapies, in certain MPNs.
Acknowledgments
Grant support: Procurement of specimens by the Sidney Kimmel Cancer Center at Johns Hopkins Tumor and Cell Procurement Bank (supported by NIH Grant P30 CA006973) is gratefully acknowledged by all authors. KW Pratz’s effort on these studies was supported as a co-investigator for translational science team at Johns Hopkins (NIH Grant UM1 CA186691). These studies were also supported by educational funds from the Mayo Foundation, including the M.D.-Ph.D. Program (A. Patel, NIH Grant T32 GM65841) and Clinical Pharmacology Training Program (B.D. Koh, NIH Grant T32 GM008685) and Clinician Investigator Training Program (B.D. Koh).
Footnotes
Conflict of Interest: No conflicts of interest exist for any authors of this manuscript.
References
- 1.Geyer HL, Mesa RA. Therapy for myeloproliferative neoplasms: when, which agent, and how? Blood. 2014;124:3529–37. doi: 10.1182/blood-2014-05-577635. [DOI] [PubMed] [Google Scholar]
- 2.Pemmaraju N, Moliterno AR. From Philadelphia-Negative to JAK2-Positive: Effect of Genetic Discovery on Risk Stratification and Management. Am Soc Clin Oncol Educ Book. 2015;35:139–45. doi: 10.14694/EdBook_AM.2015.35.139. [DOI] [PubMed] [Google Scholar]
- 3.Viny AD, Levine RL. Genetics of myeloproliferative neoplasms. Cancer J. 2014;20:61–5. doi: 10.1097/PPO.0000000000000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tefferi A, Barbui T. Polycythemia vera and essential thrombocythemia: 2015 update on diagnosis, risk-stratification and management. Am J Hematol. 2015;90:162–73. doi: 10.1002/ajh.23895. [DOI] [PubMed] [Google Scholar]
- 5.James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–8. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 6.Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–97. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 7.Plo I, Nakatake M, Malivert L, de Villartay JP, Giraudier S, Villeval JL, et al. JAK2 stimulates homologous recombination and genetic instability: potential implication in the heterogeneity of myeloproliferative disorders. Blood. 2008;112:1402–12. doi: 10.1182/blood-2008-01-134114. [DOI] [PubMed] [Google Scholar]
- 8.Nakatake M, Monte-Mor B, Debili N, Casadevall N, Ribrag V, Solary E, et al. JAK2(V617F) negatively regulates p53 stabilization by enhancing MDM2 via La expression in myeloproliferative neoplasms. Oncogene. 2012;31:1323–33. doi: 10.1038/onc.2011.313. [DOI] [PubMed] [Google Scholar]
- 9.Slupianek A, Hoser G, Majsterek I, Bronisz A, Malecki M, Blasiak J, et al. Fusion tyrosine kinases induce drug resistance by stimulation of homology-dependent recombination repair, prolongation of G(2)/M phase, and protection from apoptosis. Mol Cell Biol. 2002;22:4189–201. doi: 10.1128/MCB.22.12.4189-4201.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan J, Li L, Small D, Rassool F. Cells expressing FLT3/ITD mutations exhibit elevated repair errors generated through alternative NHEJ pathways: implications for genomic instability and therapy. Blood. 2010;116:5298–305. doi: 10.1182/blood-2010-03-272591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slupianek A, Schmutte C, Tombline G, Nieborowska-Skorska M, Hoser G, Nowicki MO, et al. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol Cell. 2001;8:795–806. doi: 10.1016/s1097-2765(01)00357-4. [DOI] [PubMed] [Google Scholar]
- 12.Slupianek A, Nowicki MO, Koptyra M, Skorski T. BCR/ABL modifies the kinetics and fidelity of DNA double-strand breaks repair in hematopoietic cells. DNA Repair (Amst) 2006;5:243–50. doi: 10.1016/j.dnarep.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slupianek A, Dasgupta Y, Ren SY, Gurdek E, Donlin M, Nieborowska-Skorska M, et al. Targeting RAD51 phosphotyrosine-315 to prevent unfaithful recombination repair in BCR-ABL1 leukemia. Blood. 2011;118:1062–8. doi: 10.1182/blood-2010-09-307256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deutsch E, Jarrousse S, Buet D, Dugray A, Bonnet ML, Vozenin-Brotons MC, et al. Down-regulation of BRCA1 in BCR-ABL-expressing hematopoietic cells. Blood. 2003;101:4583–8. doi: 10.1182/blood-2002-10-3011. [DOI] [PubMed] [Google Scholar]
- 15.Cramer K, Nieborowska-Skorska M, Koptyra M, Slupianek A, Penserga ET, Eaves CJ, et al. BCR/ABL and other kinases from chronic myeloproliferative disorders stimulate single-strand annealing, an unfaithful DNA double-strand break repair. Cancer Res. 2008;68:6884–8. doi: 10.1158/0008-5472.CAN-08-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valeri A, Alonso-Ferrero ME, Rio P, Pujol MR, Casado JA, Perez L, et al. Bcr/Abl interferes with the Fanconi anemia/BRCA pathway: implications in the chromosomal instability of chronic myeloid leukemia cells. PLoS One. 2010;5:e15525. doi: 10.1371/journal.pone.0015525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thoennissen NH, Krug UO, Lee DHT, Kawamata N, Iwanski GB, Lasho T, et al. Prevalence and prognostic impact of allelic imbalances associated with leukemic transformation of Philadelphia chromosome–negative myeloproliferative neoplasms. Blood. 2010;115:2882–90. doi: 10.1182/blood-2009-07-235119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brecqueville M, Rey J, Devillier R, Guille A, Gillet R, Adelaide J, et al. Array comparative genomic hybridization and sequencing of 23 genes in 80 patients with myelofibrosis at chronic or acute phase. Haematologica. 2014;99:37–45. doi: 10.3324/haematol.2013.091454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watkins JA, Irshad S, Grigoriadis A, Tutt AN. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res. 2014;16:211. doi: 10.1186/bcr3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yap TA, Sandhu SK, Carden CP, de Bono JS. Poly(ADP-ribose) polymerase (PARP) inhibitors: Exploiting a synthetic lethal strategy in the clinic. CA Cancer J Clin. 2011;61:31–49. doi: 10.3322/caac.20095. [DOI] [PubMed] [Google Scholar]
- 21.Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12:801–17. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 22.Lee JM, Ledermann JA, Kohn EC. PARP Inhibitors for BRCA1/2 mutation-associated and BRCA-like malignancies. Ann Oncol. 2014;25:32–40. doi: 10.1093/annonc/mdt384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott CL, Swisher EM, Kaufmann SH. Poly (ADP-Ribose) Polymerase Inhibitors: Recent Advances and Future Development. J Clin Oncol. 2015;33:1397–406. doi: 10.1200/JCO.2014.58.8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng FY, de Bono JS, Rubin MA, Knudsen KE. Chromatin to Clinic: The Molecular Rationale for PARP1 Inhibitor Function. Mol Cell. 2015;58:925–34. doi: 10.1016/j.molcel.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D’Andrea AD. Homologous Recombination Deficiency: Exploiting the Fundamental Vulnerability of Ovarian Cancer. Cancer Discov. 2015;5:1137–54. doi: 10.1158/2159-8290.CD-15-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–93. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 27.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–82. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 28.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ceccaldi R, Liu J, Amunugama R, Hajdi I, Primack B, Petalcorin MIR, et al. Homologous recombination-deficient tumors are hyperdependent on PolQ-mediated repair. Nature. 2015;518:258–62. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel A, Sarkaria J, Kaufmann SH. Nonhomologous end-joining drives PARP inhibitor synthetic lethality in homologous recombination-deficient cells. Proc Natl Acad Sci U S A. 2011;108:3406–11. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashworth A. A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J Clin Oncol. 2008;26:3785–90. doi: 10.1200/JCO.2008.16.0812. [DOI] [PubMed] [Google Scholar]
- 32.Satoh MS, Lindahl T. Role of Poly (ADP-ribose) Formation in DNA Repair. Nature. 1992;356:356–8. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 33.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–99. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hopkins TA, Shi Y, Rodriguez LE, Solomon LR, Donawho CK, Digiammarino EL, et al. Mechanistic Dissection of PARP1 Trapping and the Impact on in vivo Tolerability and Efficacy of PARP Inhibitors. Mol Cancer Res. 2015;13:1465–77. doi: 10.1158/1541-7786.MCR-15-0191-T. [DOI] [PubMed] [Google Scholar]
- 35.Chen JY, Wang CM, Lu SC, Chou YH, Luo SF. Association of apoptosis-related microsatellite polymorphisms on chromosome 1q in Taiwanese systemic lupus erythematosus patients. Clin & Exp Immunol. 2006;143:281–7. doi: 10.1111/j.1365-2249.2005.02984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tobin LA, Robert C, Rapoport AP, Gojo I, Baer MR, Tomkinson AE, et al. Targeting abnormal DNA double-strand break repair in tyrosine kinase inhibitor-resistant chronic myeloid leukemias. Oncogene. 2013;32:1784–93. doi: 10.1038/onc.2012.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel AG, Flatten KS, Schneider PA, Dai NT, McDonald JS, Poirier GG, et al. Enhanced killing of cancer cells by poly(ADP-ribose) polymerase inhibitors and topoisomerase I inhibitors reflects poisoning of both enzymes. J Biol Chem. 2012;287:4198–210. doi: 10.1074/jbc.M111.296475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mesa RA, Tefferi A, Gray LA, Reeder T, Schroeder G, Kaufmann SH. In vitro antiproliferative activity of the farnesyltransferase inhibitor R115777 in hematopoietic progenitors from patients with myelofibrosis with myeloid metaplasia. Leukemia. 2003;17:849–55. doi: 10.1038/sj.leu.2402901. [DOI] [PubMed] [Google Scholar]
- 39.Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:18032–7. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–9. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 42.Antolin AA, Jalencas X, Yelamos J, Mestres J. Identification of pim kinases as novel targets for PJ34 with confounding effects in PARP biology. ACS Chem Biol. 2012;7:1962–7. doi: 10.1021/cb300317y. [DOI] [PubMed] [Google Scholar]
- 43.Antolin AA, Mestres J. Linking off-target kinase pharmacology to the differential cellular effects observed among PARP inhibitors. Oncotarget. 2014;5:3023–8. doi: 10.18632/oncotarget.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sonoda E, Zhao GY, Kohzaki M, Dhar PK, Kikuchi K, Redon C, et al. Collaborative roles of gammaH2AX and the Rad51 paralog Xrcc3 in homologous recombinational repair. DNA Repair (Amst) 2007;6:280–92. doi: 10.1016/j.dnarep.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 45.Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 46.McDevitt MA, Koh BD, Patel AG, Moliterno AR, Poh W, Herman JG, et al. Genetic and epigenetic defects in DNA repar lead to synthetic lethality of poly(ADP-ribose) polyerase inhibitors in aggressive myeloproliferative disorders. Blood. 2011;118 Abstract 400. [Google Scholar]
- 47.Mesa RA, Tefferi A, Lasho TS, Loegering D, McClure RF, Powell HL, et al. Janus kinase 2 (V617F) mutation status, signal transducer and activator of transcription-3 phosphorylation and impaired neutrophil apoptosis in myelofibrosis with myeloid metaplasia. Leukemia. 2006;20:1800–8. doi: 10.1038/sj.leu.2404338. [DOI] [PubMed] [Google Scholar]
- 48.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 49.Kaufmann SH, Karp JE, Letendre L, Kottke TJ, Safgren S, Greer J, et al. Phase I and Pharmacological Study of Infusional Topotecan and Carboplatin in Relapsed and Refractory Leukemia. Clin Cancer Res. 2005;11:6641–9. doi: 10.1158/1078-0432.CCR-05-0817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.