Abstract
Differentiation of skeletal muscle cells, like most other cell types, requires a permanent exit from the cell cycle. The epigenetic programming underlying these distinct cellular states is not fully understood. In this study, we provide evidence that the lysine methyltransferase G9a functions as a central axis to regulate proliferation and differentiation of skeletal muscle cells. Transcriptome analysis of G9a knockdown cells revealed deregulation of many cell cycle regulatory genes. We demonstrate that G9a enhances cellular proliferation by two distinct mechanisms. G9a blocks cell cycle exit via methylation-dependent transcriptional repression of the MyoD target genes p21Cip/Waf1 and Rb1. In addition, it activates E2F1-target genes in a methyltransferase activity-independent manner. We show that G9a is present in the E2F1/PCAF complex, and enhances PCAF occupancy and histone acetylation marks at E2F1-target promoters. Interestingly, G9a preferentially associates with E2F1 at the G1/S phase and with MyoD at the G2/M phase. Our results provide evidence that G9a functions both as a co-activator and a co-repressor to enhance cellular proliferation and inhibit myogenic differentiation.
INTRODUCTION
During myogenic differentiation, proliferating myoblasts differentiate into multinucleated myotubes and mature to form adult muscle fibers. This involves two distinct stages: an irreversible withdrawal of proliferating myoblasts from the cell cycle; and subsequent expression of differentiation specific genes. In muscle cells, proliferation and differentiation are mutually exclusive events. Thus pathways driving proliferation have to be suppressed for induction of differentiation. The transcription factors E2F1 and MyoD as well as chromatin modifying and remodelling factors that associate with them play a major role in controlling these processes (1,2).
In proliferating myoblasts, E2F1-dependent cell cycle genes are activated whereas MyoD-dependent differentiation genes are switched off. Conversely during differentiation, MyoD-dependent myogenic genes are activated, and E2F1-dependent cell cycle genes are permanently silenced. This is achieved through differential association of E2F1 and MyoD with co-factors. In myoblasts, MyoD interacts with co-repressors HDAC1, G9a and Suv39h1 (3–7) which catalyse histone deacetylation and methylation marks resulting in repression of muscle gene promoters. In contrast, E2F1 activates S-phase genes (Cyclins) and DNA synthesis genes (DHFR, DNA Pol) by association with co-activators p300 and PCAF (8,9). Upon induction of differentiation, MyoD associates with PCAF and p300 (10), resulting in acetylation of histones and activation of muscle promoters, whereas the Rb1/E2F1 complex associates with HDAC1 and Suv39h1 resulting in permanent silencing of cell cycle genes (11–13). Corresponding with this differential recruitment of co-factors, in myoblasts, histone H3 lysine 9 di-methylation (H3K9me2), H3K9me3 and H3K27me3 repression marks catalysed by G9a, Suv39h1/2 and Ezh2 respectively are present at myogenin and muscle creatine kinase (MCK) promoters (7,14,15). On the other hand, H3K9me3 silences E2F1-dependent gene promoters in myotubes (13,16,17).
Upon induction of differentiation, MyoD is transcriptionally activated and switches on p21Cip1/Waf1 (p21) and Rb1 expression (18–20) for an irreversible exit from the cell cycle and maintenance of permanent arrested state of myotubes (21). Indeed, inactivation of p21 and Rb1 by E1A has been shown to induce DNA synthesis in myotubes (21). Conversely, high levels of p21 result in reduced Cyclin-CDK activity and Rb1 phosphorylation, leading to cell cycle arrest (22). During growth factor withdrawal and induction of differentiation, Rb1 is hypo-phosphorylated and recruited by E2F1 family members. The Rb1/E2F1 complex is required to repress E2F1-target genes involved in cell cycle progression and DNA synthesis (8,12). Apart from its role in regulating E2F1 activity, Rb1 is also involved in cell cycle exit and activation of differentiation genes (23). Rb−/− myocytes can differentiate into myotubes and express early differentiation genes such as p21 and myogenin, but exhibit defects in terminal differentiation with reduced expression of late markers such as myosin heavy chain (MHC) and MCK (24,25) and display DNA synthesis after re-addition of serum to the cultures (23,24).
We and others have shown that overexpression of G9a inhibits myogenic differentiation (5,6,14,26,27). However, whether or not G9a impacts proliferation and cell cycle exit of myoblasts has not been addressed. In the present study, we have globally identified G9a target genes in muscle cells. Interestingly, a number of genes involved in cell cycle control are differentially regulated in G9a knockdown cells. We demonstrate that G9a inhibits irreversible cell cycle exit by transcriptionally repressing p21 and Rb1 in a methyltransferase activity-dependent manner. Consequently, re-expression of p21 or Rb1 rescue the G9a-mediated block of myogenic differentiation. In addition, G9a positively regulates E2F1-target genes in a methylation-independent manner. Through protein interaction assays, we show that G9a associates with E2F1 during the G1/S phase of the cell cycle and results in increased PCAF occupancy at E2F1 target promoters. Our results provide evidence that G9a is a pivotal molecule that balances proliferation and differentiation of muscle cells. Our data suggest that deregulation of G9a expression may be important in muscle disorders characterized by a differentiation defect.
MATERIALS AND METHODS
Primary myoblast isolation, cell culture and differentiation assays
All animal procedures were approved by the Institutional Animal Care and Use Committee. Primary myoblasts were isolated from hind limb muscles of wild-type C57BL/6 mice as described previously (28). Cells were cultured in F10 media supplemented with 20% fetal bovine serum (FBS) and 5 ng/ml basic fibroblast growth factor and plated on collagen-coated dishes. To enrich myoblasts, cells were trypsinized and pre-plated onto culture dishes for 15–30 min to remove strongly adherent fibroblasts. The purity of myoblasts was confirmed by staining cells with anti-Pax7 antibody. More than 95% of cells stained positively for Pax7 and were used for experiments. C2C12 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Sigma, St Louis, MO, USA) supplemented with 20% FBS (Hyclone, Logan UT, USA). Proliferating C2C12 cells were cultured at 60% confluency. For differentiation, cells at 80–90% confluency were cultured in DMEM supplemented with 2% horse serum. To monitor differentiation, cells were immunostained with anti-MHC antibody (Sigma). Myogenic index was calculated by quantifying the ratio of total nuclei within myotubes to the total number of nuclei. At least 500 nuclei were counted. Phoenix cells and 293T cells were cultured in DMEM supplemented with 10% FBS. All cells were incubated at 37°C with 5% CO2 in a humidified incubator.
Plasmids, site-directed mutagenesis and luciferase assays
pBabe, pBabe-G9a and EGFP-G9a have been previously described (5), and were provided by Dr Martin J Walsh. Flag-PCAF was provided by Dr Yoshihiro Nakatani (29). E2F1 was provided by Dr Srikumar Chellappan (30). CyclinD1 promoter reporter (pD1luc) was provided by Dr Michael Strauss (31). The E2F1 binding site in pD1luc was mutated (TTTGGCGC to TTTGATGC) by site directed mutagenesis, using the QuikChange™ site-directed mutagenesis kit (Agilent, Santa Clara, CA, USA). The following primers were used: Forward 5′- CTCCCGGCGTTTGATGCCCGCGCC-3′ and Reverse 5′-GGCGCGGGCATCAAACGCCGGGAG-3′. The construct was sequenced to confirm the mutation. Mouse p21 and Rb1 cDNAs from C2C12 cells were cloned into pCMV 3× Flag vector flanking restriction sites HindIII and BamHI and confirmed by sequencing. Luciferase assays were performed using the Dual Luciferase Reporter System (Promega, Madison, WI, USA). pBabe and pBabe-G9a cells were transfected with 200 ng of wild-type or E2F1-mutant CyclinD1 reporters in a 24-well plate. A total of 3 ng of renilla reporter was co-transfected as a normalization control. Transfection was carried out in triplicates using Lipofectamine Plus. Luminescence was analysed with Varioskan plate reader using SkanIt software.
siRNA knockdown and retroviral infection
Knockdown experiments were performed using 100 nM siRNA specific for G9a (on-target plus smart pool, Mouse BAT 8; accession number: NM_147151; NM_145830) or siRNA specific for CyclinD1 (on-target plus smart pool, mouse ccnd1 Accession number: NM_007631;XM_011241977) from Dharmacon (Lafayette, CO, USA) using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA) according to manufacturer's instructions. Control cells were transfected with scrambled siRNA (on-target plus control pool). Transfection of siRNAs was done in growth medium for 48 hr. To overexpress G9a, Phoenix cells were transfected with pBabe or pBabe-G9a. Retroviral supernatants were used to transduce C2C12 cells. pBabe and pBabe-G9a cells were selected with 2 μg/ml puromycin for 48 hr and used for subsequent experiments. Where indicated, cells were differentiated for 12, 24 and 36 hr.
Microarray analysis and quantitative RT-PCR (qRT-PCR)
C2C12 cells were transfected with scrambled siRNA (control) or siG9a for 48 hr. Microarray was performed with RNA from two biological replicates of siRNA and siG9a cells at Day 0 (48 hr post-transfection in growth medium) and after culture in differentiation medium for 1 day (Day 1). RNA was cleaned using RNeasy MiniElute Cleanup Kit (Qiagen, Valencia CA, USA), and quantified using Nanodrop. For microarray analysis, RNA quality was checked using Bioanalyzer (Agilent Technologies). RNA was reverse transcribed and cRNA was synthesized using Total Prep RNA amplification kit (Ambion, Foster City, CA, USA). cRNA was labelled and subsequently hybridized to Illumina mouse WG-6 v2.0 array (Illumina Inc, CA, USA). Partek Genomics Suite version 6.5 (Partek Inc., MO, USA) was used for gene expression analysis. The microarray data are compliant with MIAME guidelines and have been submitted to the GEO repository [accession number GSE70039]. Analysis of Variance (ANOVA) was applied on the dataset and differentially expressed gene list was generated using P-values of <0.05 with 1.3-fold-change cut off. Unsupervised two-dimensional hierarchical clustering with complete linkage was performed on selected genes using Spearman's correlation as similarity matrix. To define biological networks among the differentially regulated genes, pathway analyses were carried out using Ingenuity Pathway Analysis software (IPA) (Ingenuity Systems, Redwood City, CA, USA). The significance of the association between the dataset and the canonical pathways were calculated by ratio and/or Fisher's exact. For qRT-PCR 2 μg of RNA was reverse transcribed using Super Script III first strand cDNA synthesis kit according to manufacturer's instruction (Invitrogen, Carlsbad, CA, USA). Quantitative real time polymerase chain reaction (PCR) reaction was carried out with 0.2 μM of primer in triplicates using LightCycler 480 SYBR green I master mix (Roche, Basel Switzerland). PCR amplification was performed under following conditions: pre-amplification at 95°C for 5 min, amplification including denaturation at 95°C for 10 s, annealing at 60°C for 10 s, followed by 72°C for 10 s for 45 cycles. Melting curves were generated and tested for a single product after amplification. The threshold cycle (Ct) values of the test primers were normalized to Ct values of GAPDH primers to derive normalized Ct (ΔCt). Gene expression was quantified using the 2−ΔCt method. The primer sequences for CyclinD1, CyclinE, p21, GAPDH (32,33), DHFR (34) and G9a (5) have been previously described. Rb1 primer sequences are: Forward 5′-ACGCTGCCCAGGAGACCTTT-3′ and Reverse 5′- AGGGCTTCGAGGAATGTGAGGT-3′
Cell proliferation assays
Proliferation was measured by BrdU incorporation assays (35). Cells were pulsed with 10 μM BrdU for 30 min. Cells were fixed and stained with anti-BrdU antibody according to manufacturer's protocol (Roche). For flow cytometry, cells were stained with propidium iodide mix (10 μg/ml propidium iodide solution with RNase 200 μg/ml) for 30 min at room temperature and strained using 40 μm filters. Cell cycle profiling was performed with at least 10 000 gated population using a flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA) and the data was analysed using FACSDIVA and Flowing software. Where indicated, cells were synchronized at G1/S boundary using 1 mM hydroxyurea (HU) for 14 hr and subsequently released in normal growth media. 300 ng/ml of nocodozole was used to synchronize cells at mitosis. G9a methyltransferase activity was blocked using 0.25 μM UNC0638 (Sigma).
Immunofluorescence
Cells were cultured on sterile cover slips placed in 6-well dishes, and fixed with 4% paraformaldehyde (Sigma). After blocking, cells were permeabilized using phosphate buffered saline (PBS) containing 10% horse serum (Gibco, Carlsbad, CA) and 0.1% Triton X-100 (Biorad, Hercules, CA). Cells were incubated with either anti-G9a (3306S, Cell Signaling) or anti-MHC (M4276, Sigma) in blocking solution (PBS with 10% horse serum) followed by secondary antibody tagged with fluorophore (Invitrogen). DNA was counter stained with DAPI and mounted using mounting agent (Vectashield, Burlingame, CA, USA). Images were obtained using Zeiss (Axioplan) or Olympus (DP72) microscope.
Co-immunoprecipitation (Co-IP) and chromatin immunoprecipitation (ChIP)
293T cells were transfected with EGFP-G9a and Flag-PCAF and E2F1 plasmids. About 24 h after transfection cells were lysed with NP40 buffer (20 mM Tris–HCl pH 7.4, 137 mM NaCl, 2 mM ethylenediaminetetraacetic acid and 1% NP-40 with protease inhibitor). Lysates were immunoprecipitated with anti-Flag agarose beads (Sigma, St Louis, MO, USA). After washes, samples were loaded on sodium dodecyl sulphate-polyacrylamide gel electrophoresis gels and immunoblotted with antibodies as indicated. For endogenous co-immunoprecipitation (Co-IP), nuclear extracts were prepared from C2C12 cells as described (36). Protein lysates from C2C12 cells were pre-cleared and incubated with desired antibodies overnight at 4°C using Protein A/G agarose beads (Santa Cruz Biotech Inc., Dallas, TX, USA). For sequential immunoprecipitation, the protein complex from washed beads (both from control and test antibody) was eluted using Tris–HCl pH 7.4 (65 mM), sodium dodecyl sulphate (1%) and dithiothreitol (5 mM). Samples were boiled for 5 min at 95°C and the supernatant (70 μl) was transferred to a new tube. A total of 20 μl of the elute was kept to check the first IP, and the remaining 50 μl was diluted 10 times with NP40 buffer. Samples were then incubated overnight with the second antibody. Antibodies used for Co-IP and western blotting are as follows: anti-G9a (3306S), pRbS780 (9300) from Cell Signaling (Danvers, MA, USA); anti-CyclinD1 (sc-753), anti-CyclinE (sc-481), anti-CyclinA (sc-239), anti-p21 (sc-397), anti-MyoD (5.8A) (sc-32758), anti-Myogenin (sc-576), anti-GFP (sc-9996), anti-E2F1 (sc-193), normal rabbit IgG (sc-2027) from SantaCruz Biotech Inc.; Rb1 (554136) from BD Biosciences (San Jose, CA, USA); anti-PCAF (ab12188), anti-E2F1 (ab179445) anti-G9a (ab40542) from Abcam (Cambridge, MA, USA); anti-H3S10p (06-570) from Millipore (Billerica, MA, USA); and anti-Troponin-T (T6277), anti-Flag (F3165), anti-β-actin (A1978) from Sigma. Chromatin immunoprecipitation (ChIP) assays were done as described (5). Briefly, 106 control and treated cells were fixed in 1% formaldehyde for 10 min at 37°C. Cells were sonicated using Bioruptor (Diagenode, Liege, Belgium). ChIP was carried out according to kit protocol (Millipore). Immunoprecipitates were reverse crosslinked and DNA was extracted using phenol–chloroform–isoamylalcohol (Sigma). Quantitative RT-PCR reaction was performed in triplicates as described above. DNA isolated from 10% input was used as control. Relative enrichment was calculated using 2−ΔCt method. Antibodies used for ChIP assays are as follows: anti-E2F1 (ab179445), anti-PCAF (ab12188), anti-G9a (ab40542), anti-H3K9ac (ab4441) from Abcam; and anti-H3K9me2 (17-681) from Millipore. Primers used for ChIP assays have been previously described: CyclinD1 (32), DHFR (37) and Rb1 (38). The following primer sequences were used: p21: Forward 5′-CCCCGCATGCCCAGTTTATGG-3′ Reverse 5′-CCGCGTCACATAGCAGGTCCC-3′; β-actin: Forward 5′ – GCTTCTTTGCAGCTCCTTCGTTG-3′ Reverse 5′- TTTGCACATGCCGGAGCCGTTGT-3′.
Statistical analysis
Significance was calculated using student's t-test (two-sided) and P-values of <0.05 were considered to be statistically significant (*P < 0.05, **P < 0.01, ***P < 0.001). Error bars represent the mean ± standard deviation (SD) unless specified otherwise.
RESULTS
Global gene expression analysis in G9a knockdown myoblasts
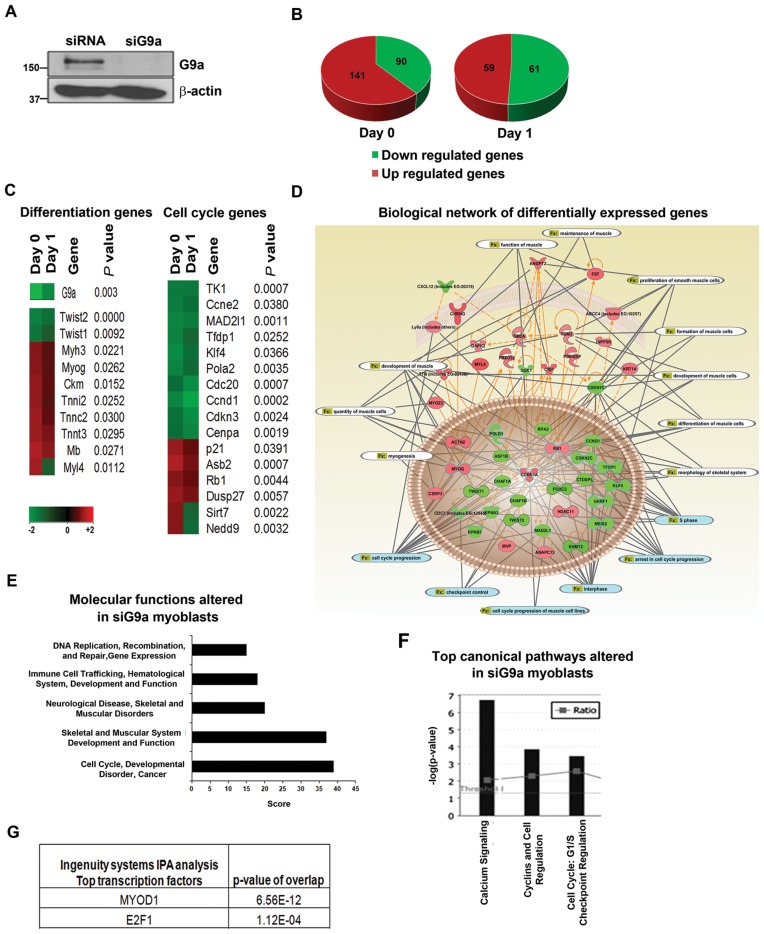
Several recent studies have established a role for G9a as an inhibitor of myogenic differentiation (5,6,14,26,27). However, genome-wide targets of G9a in myoblasts have not been identified. Moreover, whether G9a regulates cell cycle exit that is a pre-requisite for differentiation has not been investigated. To address these outstanding questions, we performed genome-wide transcriptomic analysis using microarrays. Endogenous G9a was depleted using siRNA specific for G9a (siG9a). Control cells were transfected with scrambled siRNA (siRNA). G9a knockdown in siG9a cells was confirmed by western blot (Figure 1A). RNA was isolated from proliferating (Day 0) and differentiating (Day 1) siG9a and siRNA cells and hybridized to mouse WG6v2.0 arrays (Illumina Inc, CA, USA). A total of 231 genes were differentially regulated in Day 0 siG9a cells (Figure 1B). Functional annotation revealed that the expression of several genes was upregulated in siG9a cells consistent with the role of G9a as a transcriptional repressor. Interestingly, many genes were also downregulated suggesting that G9a may directly activate their expression, or indirectly regulate them. Similarly during differentiation, 59 genes were upregulated and 61 genes were downregulated in siG9a cells. Consistent with our previous findings (5), G9a knockdown upregulated differentiation genes such as myogenin (Myog) and Troponin T (Tnnt3) whereas differentiation inhibitory genes Twist1 and Twist2 were downregulated (Figure 1C). Interestingly, in addition to differentiation genes, many cell cycle control genes were found to be positively and negatively regulated by G9a. For instance, MyoD target genes important for cell cycle exit such as p21 and Rb1 were upregulated in siG9a cells, whereas E2F1 target genes involved in proliferation namely CyclinD1 (Ccnd1), CyclinE (Ccne2) and Thymidine kinase (TK1) were downregulated. IPA of the differentially expressed genes revealed gene expression, cellular location, biological functions as well as biological networks (Figure 1D). Cell cycle progression and muscle differentiation were identified to be significantly altered functions (Figure 1D). Moreover, cell cycle regulation and muscle development were among the top molecular functions and canonical pathways that were significantly altered in G9a knockdown cells (Figure 1E and F). Consistent with the differential expression of genes, the transcriptional activity of MyoD and E2F1 was predicted to be deregulated upon G9a knockdown (Figure 1G).
Figure 1.
Global gene expression analysis in G9a knockdown C2C12 myoblasts. (A) C2C12 cells were transfected with scrambled siRNA (siRNA) or G9a specific siRNA (siG9a) for 48 hr. G9a expression was analysed in siRNA and siG9a cells by western blot. β-actin was used as loading control. (B) Microarray analysis was done with two biological replicates of siRNA and siG9a cells in proliferating (Day 0) and differentiating conditions (Day 1). Pie charts show the total number of genes whose expression is significantly upregulated and downregulated in Day 0 and Day 1 siG9a cells compared to control siRNA cells. Differential gene expression was determined by comparing the mean of siG9a biological replicates to control replicates. Green indicates downregulation and red indicates upregulation of gene expression. (C) Heatmap was generated with a partial list of G9a target genes involved in differentiation and cell cycle control in Day 0 and Day 1 cells with corresponding P-values and fold changes according to the colour scale at the bottom. Gene expression clustering was performed by the associate program Gene Cluster and viewed in TreeView software (D) Differentially expressed genes were used to create biological networks. Orange lines indicate direct regulation between genes whereas dotted lines indicate indirect regulation. Grey lines indicate functions (Fx) altered by the genes which are related to cell cycle progression (blue) and muscle differentiation (white). (E) Top molecular functions altered in siG9a myoblasts include cell cycle regulation, skeletal muscle development and function and muscle disorders. (F) Top canonical pathways altered upon G9a knockdown including Cyclins and cell cycle regulation. (G) IPA analysis revealed that the activity of the transcription factors MyoD and E2F1 is significantly altered in siG9a cells.
G9a promotes myoblast proliferation
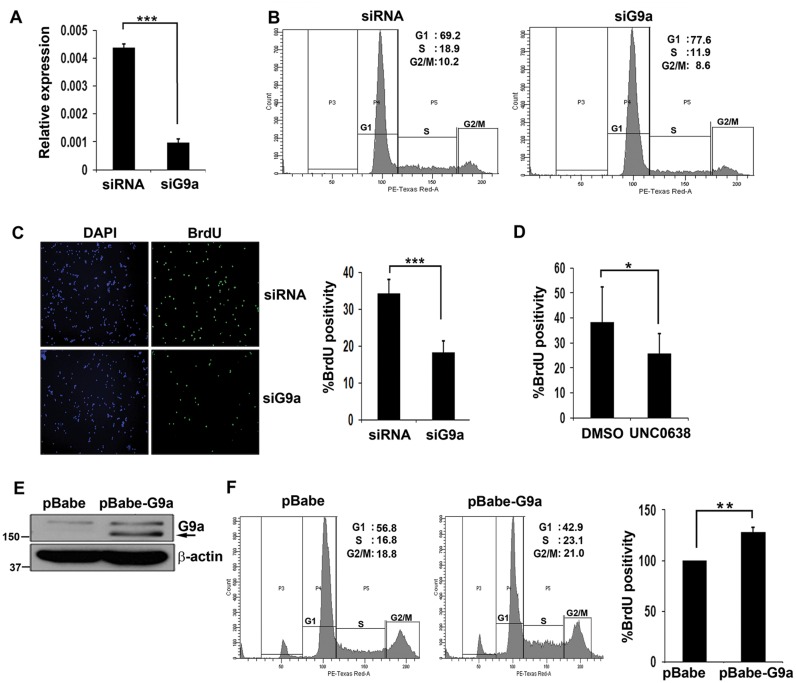
To examine whether G9a indeed has a role in regulating proliferation of myoblasts, we performed loss-of-function studies. To extend the relevance of our findings, we used primary myoblasts that were transfected with siRNA against G9a and the knockdown was confirmed by qRT-PCR (Figure 2A). Compared to control cells, a reduction in S-phase cells was apparent upon G9a depletion by flow cytometry (Figure 2B). To validate these findings, both siRNA control and siG9a cells were pulsed with BrdU and stained with anti-BrdU antibody. siG9a cells showed significantly lesser BrdU incorporation compared to control cells (Figure 2C). To examine whether G9a methyltansferase activity is needed for its impact on proliferation, cells were treated with UNC0638 (39). Consistent with G9a knockdown, UNC0638 treated cells displayed reduced proliferation compared to control cells (Figure 2D) albeit to a lesser extent than siG9a cells. Similar results were obtained in C2C12 cells (data not shown). To determine if G9a is sufficient to induce cellular proliferation, C2C12 cells were transduced with a retroviral vector expressing G9a (pBabe-G9a) or with vector (pBabe) alone (Figure 2E). Using an equivalent number of cells, an increase in S-phase cells was apparent upon G9a overexpression compared to control cells (Figure 2F), and correspondingly, pBabe-G9a cells displayed higher percentage of BrdU+ cells.
Figure 2.
G9a promotes myoblast proliferation. (A) Primary myoblasts were transfected with scrambled siRNA (siRNA) or G9a siRNA for 48 hr in growth medium. G9a expression in control (siRNA) and siG9a cells was analysed by qRT-PCR. (B) Control and siG9a primary myoblasts cultured in growth medium were stained with PI and analysed by flow cytometry. siG9a primary myoblasts had less S phase cells compared to control cells. At least 10 000 cells were counted. (C) BrdU incorporation was analysed in proliferating control and siG9a primary myoblasts. Nuclei were counter stained with DAPI. To quantify the percentage of BrdU+ cells, at least 500 cells were counted. (D) Primary myoblasts were treated with UNC0638 or DMSO as a vehicle control for 36 hr in growth medium. For each experiment, at least 500 nuclei in 5–6 random fields were counted and the percentage of BrdU+ cells was quantified. The results are representative of two independent experiments. Error bars indicate the mean of BrdU+ cells in each experiment ± SD. (E) pBabe and pBabe-G9a expressing C2C12 cells were analysed for G9a expression by western blot. β-actin was used as loading control. Arrow indicates G9a overexpression. Numbers indicate the molecular weight of proteins. (F) Equivalent number of pBabe and pBabe-G9a C2C12 cells were stained with PI and at least 10 000 gated population were analysed. Flow cytometric analysis showed higher S phase cells upon G9a overexpression. All results are representative of at least three independent experiments. pBabe and pBabe-G9a cells were pulsed with BrdU and the percentage of BrdU+ cells was quantified. At least 500 cells were counted in each experiment from 5–6 random fields. Error bars indicate the mean of BrdU+ cells in three independent experiments ± SEM (n = 3).
Proliferation and cell cycle exit genes are differentially regulated by G9a
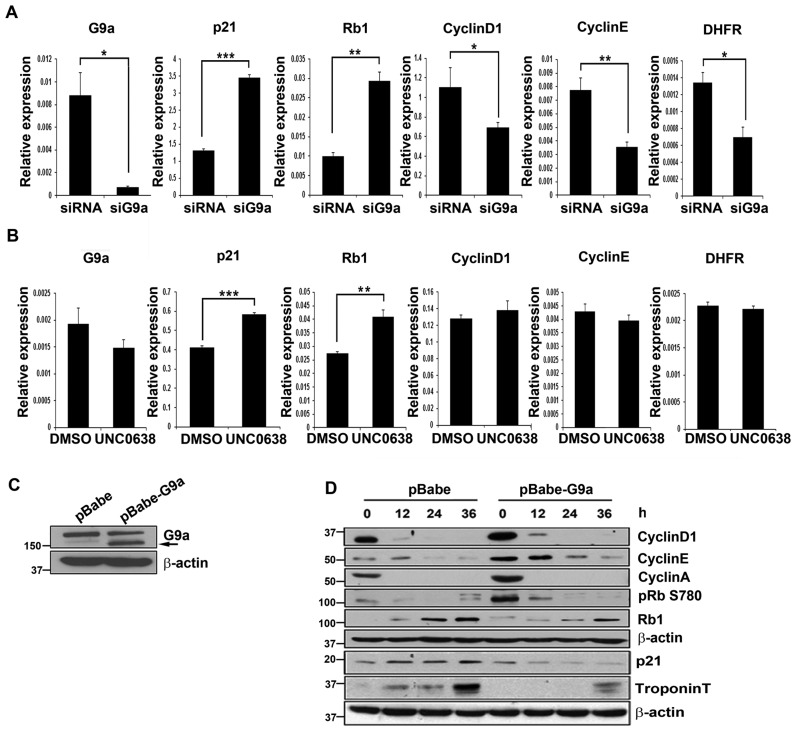
To examine the mechanisms by which G9a regulates proliferation, we first validated the expression of cell cycle genes identified in microarrays by q-RT-PCR in primary myoblasts. Consistent with the microarray data, siG9a cells showed significant upregulation of MyoD target genes p21 and Rb1 compared to control primary cells (Figure 3A), as well as in siG9a C2C12 cells (data not shown). On the other hand, E2F1 targets genes CyclinD1, CyclinE and DHFR were downregulated in siG9a cells. Interestingly, inhibition of endogenous G9a methyltransferase activity using UNC0638 resulted in upregulation of p21 and Rb1 expression, while no significant change in the expression of E2F1 target genes was evident (Figure 3B). These results suggest that G9a-mediated regulation of cell cycle exit genes is dependent on its methyltransferase activity, whereas E2F1 target genes are regulated in a methyltransferase-independent manner. To further validate these findings, pBabe and pBabe-G9a cells were analysed during differentiation (Figure 3C). The expression of CyclinD1, CyclinE and CyclinA was higher, and Rb1 phosphorylation was elevated in undifferentiated G9a overexpressing cells (Figure 3D). On the other hand, Rb1, p21 and Troponin-T levels were reduced during differentiation. Taken together, these results suggest that G9a may promote proliferation by transcriptionally repressing the cell cycle inhibitors p21 and Rb1, as well as directly or indirectly activating expression of E2F1-responsive genes.
Figure 3.
Proliferation and cell cycle exit genes are differentially regulated by G9a. (A) Primary myoblasts were transfected with scrambled siRNA and siG9a in growth medium. Forty-eight hours post-transfection, cells were analysed for expression of G9a, p21, Rb1, CyclinD1, CyclinE and DHFR by qRT-PCR. (B) Primary myoblasts were treated with DMSO (vehicle control) or with UNC0638 for 48 hr in growth medium. Expression of cell cycle genes was analysed by qRT-PCR. The results are representative of three independent experiments. Error bars indicate the mean of q-RT-PCR triplicates in each experiment ± SD. (C) C2C12 cells expressing pBabe and pBabe-G9a were analysed for G9a expression by western blot. β-actin was used as loading control. Arrow indicates G9a overexpression. (D) pBabe and pBabe-G9a cells were cultured in growth medium (0 hr) or induced to differentiate for 12–36 hr and analysed for CyclinD1, CyclinE, CyclinA, Rb1 phosphorylation at serine 780 (pRbS780), Rb1, p21 and Troponin-T expression. β-actin was used as an internal control. The results are representative of three independent experiments.
G9a mediates H3K9me2 on promoters of cell cycle exit genes but not on E2F1 target genes
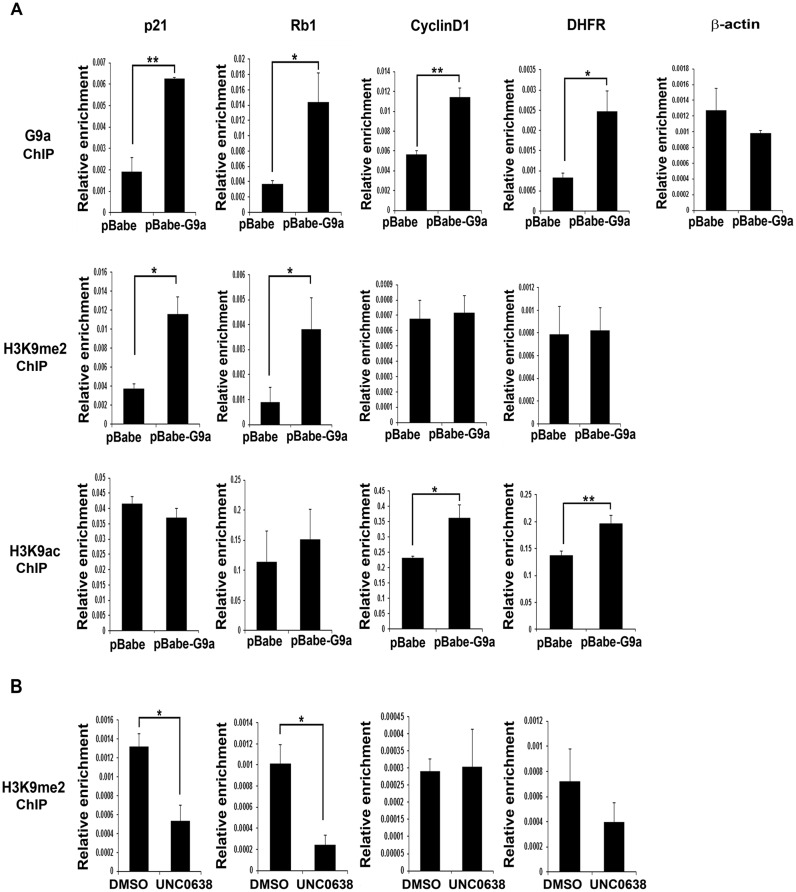
Previous studies have shown that in addition to its canonical role as a transcriptional repressor that is methyltransferase activity-dependent, G9a can activate expression of genes independent of its catalytic activity (40,41). To examine mechanisms by which G9a regulates cell cycle genes, we analysed its occupancy and H3K9me2 enrichment, a signature of G9a activity, at its target gene promoters in proliferating myoblasts. Compared to controls, G9a occupancy was increased at p21 and Rb1 promoters in pBabe-G9a cells and correlated with elevated H3K9me2 repression marks (Figure 4A). H3K9ac, a mark of transcriptional activation was however not altered at these promoters. In contrast, G9a occupancy was not correlated with H3K9me2 at E2F1 target gene promoters namely CyclinD1 and DHFR which instead showed H3K9ac enrichment. Moreover, in presence of UNC0638, a significant reduction of H3K9me2 levels was apparent at p21 and Rb1 promoters but not at CyclinD1 and DHFR promoters (Figure 4B). These results indicate that G9a represses p21 and Rb1 by mediating H3K9me2 marks, whereas it functions to regulate E2F1 target genes independent of its methylation activity.
Figure 4.
G9a mediates H3K9me2 on cell cycle exit genes but not on E2F1 target genes. (A) G9a occupancy, H3K9me2 and H3K9ac marks were analysed by ChIP assays at p21, Rb1, CyclinD1, DHFR and β-actin promoters in pBabe and pBabe-G9a cells cultured in growth medium. G9a occupancy was increased in G9a overexpressing cells at all these promoters except β-actin, which was used as a negative control. H3K9me2 enrichment was apparent at the p21 and Rb1 promoters in pBabe-G9a cells, but not at CyclinD1 and DHFR promoters. In contrast, H3K9ac was seen at CyclinD1 and DHFR promoters but not at p21 and Rb1 promoters. The results are representative of two independent experiments. Error bars indicate the mean of qRT-PCT triplicates in each experiment ± SD. (B) p21, Rb1, CyclinD1 and DHFR promoters were analysed for H3K9me2 in C2C12 cells treated with either DMSO or UNC0638 for 48 hr in growth medium. UNC0638 treatment reduced H3K9me2 at p21 and Rb1 promoters, but did not impact this mark at CyclinD1 and DHFR promoters. The results are representative of two independent experiments. Error bars indicate the mean of qRT-PCR triplicates in each experiment ± SD.
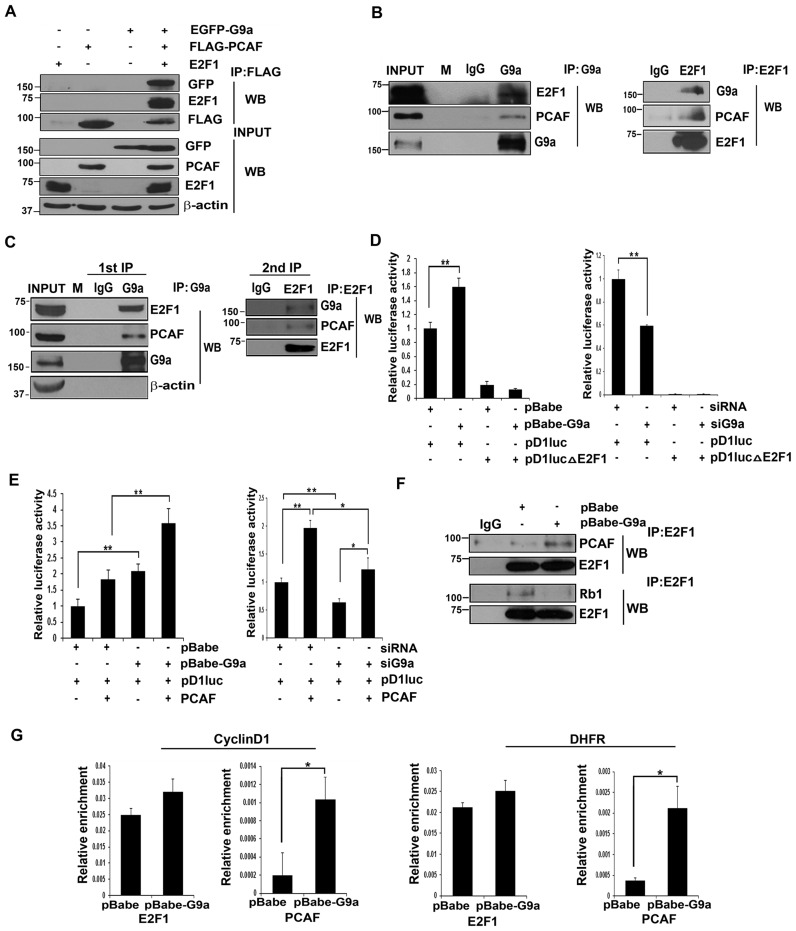
G9a associates with E2F1 and increases PCAF occupancy at E2F1 target promoters
Previous studies have shown that the role of G9a as an activator or repressor depends on its association with co-factors (40–42). Proliferation of cells is dependent on E2F1 which recruits co-activators PCAF and p300 to mediate the expression of its downstream target genes (8,9,43). Since E2F1 target genes were upregulated in presence of G9a, we tested if G9a associates with E2F1 and PCAF. Due to high transfection efficiencies in 293T cells, we initially tested interaction using exogenously expressed EGFP-G9a, Flag-PCAF and E2F1 constructs. Immunoprecipitation of PCAF showed that it is associated with E2F1 and G9a (Figure 5A). To test their interaction in myoblasts, endogenous G9a was immunoprecipitated. Consistent with the above results, E2F1 and PCAF associated with G9a in proliferating myoblasts (Figure 5B, left panel). Reciprocally, endogenous E2F1 immunoprecipitated with G9a and PCAF in myoblasts (Figure 5B, right panel). Furthermore, sequential immunoprecipitation of C2C12 myoblasts using anti-G9a antibody, followed by anti-E2F1 antibody confirmed that G9a, E2F1 and PCAF are present in the same complex (Figure 5C). To determine the significance of this association, we performed luciferase assays with the CyclinD1 promoter reporter (pD1luc) as a readout of E2F1 activity. Higher CyclinD1 promoter activity was evident in G9a over expressing cells compared to control cells. To determine if the elevated CyclinD1 activity in pBabe-G9a cells is E2F1-dependent, we generated point mutations at the E2F1 binding site (pD1lucΔE2F1) in the reporter construct. The basal promoter activity of the mutant construct was expectedly reduced in control cells. Interestingly, the mutant reporter activity was not elevated in pBabe-G9a cells (Figure 5D, left panel). Conversely, pD1luc activity was significantly reduced in siG9a cells, and pD1lucΔE2F1 activity was barely detectable in both control and siG9a cells (Figure 5D, right panel) confirming that G9a increases CyclinD1 expression in an E2F1-dependent manner.
Figure 5.
G9a associates with E2F1 and increases PCAF occupancy at E2F1 target promoters. (A) EGFP-G9a, Flag-PCAF and E2F1 were transfected in 293T cells. PCAF was immunoprecipitated using anti-Flag beads. Interaction with G9a, E2F1 and PCAF was analysed by western blot. Expression of G9a, PCAF and E2F1 was analysed in lysates (input). (B) Endogenous G9a was immunoprecipitated from C2C12 cells using nuclear extracts (left panel). Association with E2F1 and PCAF was tested by immunoblotting with anti-E2F1 and anti-PCAF antibodies. M denotes lane with molecular weight markers. In reverse IP experiments, endogenous E2F1 was immunoprecipitated and the association with G9a and PCAF was tested (right panel). (C) Immunoprecipitation was performed with anti-G9a antibody (first IP) and checked for association with E2F1 and PCAF. Elute from the first IP was re-immunoprecipitated (second IP) with anti-E2F1 antibody and immunoblotted with anti-G9a and anti-PCAF antibodies. The results are representative of at least two independent experiments. (D) pBabe and pBabe-G9a cells (left panel) as well as siRNA and siG9a C2C12 cells (right panel) were transfected in triplicates with 200 ng of a wild-type cyclin D1 reporter (pD1luc) or pD1lucΔE2F1 that harbours point mutations at E2F1 binding site. Luciferase activity was measured using dual luciferase reporter assays. (E) G9a overexpressed (left panel) and knockdown cells (right panel) were transfected in triplicates with 200 ng pD1luc in the absence and presence of PCAF (50 ng) and luciferase activity was measured. Results are representative of at least two independent experiments. Error bars indicate the mean luciferase activity from triplicate transfections in each experiment ± SD. (F) Endogenous E2F1 was immunoprecipitated from pBabe and pBabe-G9a cells grown in growth medium (Day 0). Interaction was examined by immunoblotting with anti-PCAF, anti-Rb1 and anti-E2F1 antibodies. (G) ChIP assays were performed with anti-E2F1 and anti-PCAF antibodies at CyclinD1 and DHFR promoters in proliferating pBabe and pBabe-G9a cells. Results are representative of two independent experiments. Error bars indicate the mean of triplicate q-RT-PCR in each assay ± SD.
PCAF is a co-factor that increases E2F1 transcriptional activity (9). We therefore examined whether G9a impacts PCAF-mediated activation of E2F1. Overexpression of PCAF augmented CyclinD1 promoter in both control and G9a overexpressing cells, although the overall reporter activity was higher in G9a overexpressing cells due to the increased basal activity (Figure 5E, left panel). Even in presence of PCAF, pD1luc activity was significantly lesser in siG9a cells compared to controls (Figure 5E, right panel). To define the mechanisms by which G9a augments PCAF-mediated E2F1 activity, we tested endogenous E2F1 association with PCAF and Rb1 in presence of G9a in proliferating cells. Interestingly, the interaction of E2F1 with PCAF was increased in presence of G9a, whereas the association with Rb1 was reduced (Figure 5F). We then examined E2F1 and PCAF recruitment at E2F1 target promoters in proliferating myoblasts. As expected, E2F1 was present at CyclinD1 and DHFR promoters in proliferating pBabe cells and the occupancy was not significantly altered in G9a overexpressing cells. However, increased PCAF occupancy was apparent at these promoters in G9a over expressing cells compared to control cells (Figure 5G).
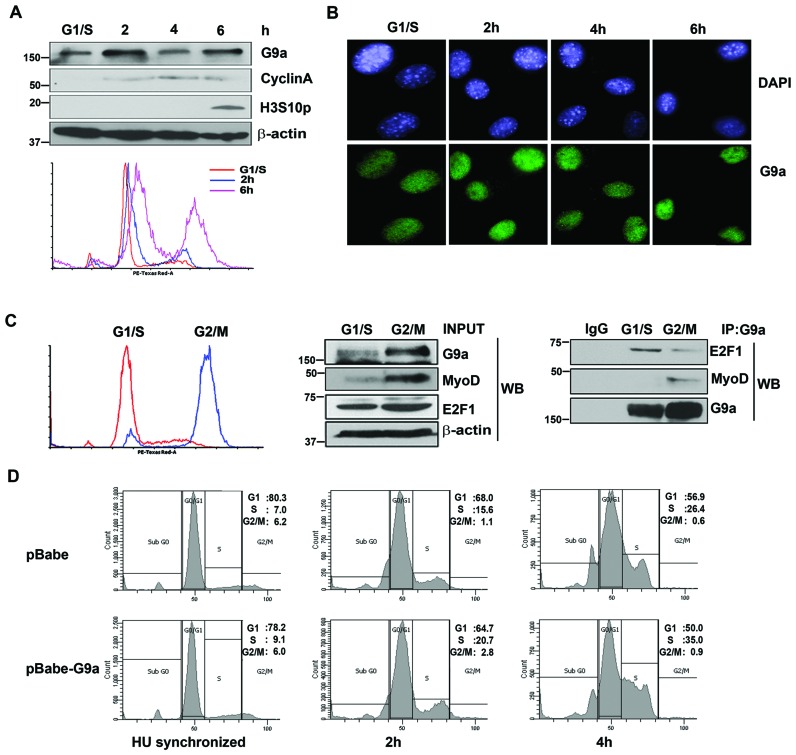
G9a associates with E2F1 and MyoD in a cell cycle-specific manner
We have previously reported that G9a associates with MyoD to inhibit its activity in myoblasts (5). Since our data showed that G9a also interacts with E2F1 and promotes its transcriptional activity, we hypothesized that G9a may associate with these two transcription factors at different phases of the cell cycle. To test this hypothesis, we first examined G9a expression in cycling myoblasts. Cells were synchronized at G1/S boundary using HU and subsequently released in growth media. The synchronization was monitored by western blot using cell cycle markers as well as by flow cytometry. Interestingly, G9a expression oscillated during different phases of cell cycle. The expression peaked during transition into the S phase and into mitosis (Figure 6A) but remained nuclear at all phases (Figure 6B). As E2F1 activity increases during transition into the S phase, we speculated that G9a interaction with E2F1 during S phase may be required for its impact on proliferation. To test the interaction with E2F1 and MyoD, myoblasts were synchronized at G1/S and G2/M boundary using hydroxyurea and nocodozole respectively. Synchronization was monitored by flow cytometry (Figure 6C) and lysates from synchronized cells were used for immunoprecipitation. Interestingly, the association of G9a with E2F1 was higher at G1/S phase, whereas the association with MyoD was higher at G2/M (Figure 6C, right panel). In addition, G1/S synchronized pBabe-G9a cells showed a higher percentage of cells entering the S-phase upon release into growth media compared to control cells (Figure 6D). These results suggest that in proliferating myoblasts, G9a exists in two distinct complexes at different phases of the cell cycle to regulate proliferation and myogenic differentiation.
Figure 6.
G9a associates with E2F1 and MyoD in distinct phases of the cell cycle. (A) C2C12 cells were synchronized at G1/S boundary using HU. Transition into the S phase (2 and 4 hr culture in growth media after HU removal) and mitosis (6 hr culture in growth media after HU removal) was checked using CyclinA, as a marker of the S phase, and Histone H3 serine 10 phosphorylation (H3S10p) as a mitotic marker. G9a expression was checked using western blot. β-actin was used as an internal control. Synchronization of cells with HU was also confirmed by flow cytometry using PI staining (lower panel). (B) HU synchronized C2C12 cells were released and fixed 2, 4 and 6 hr after HU removal and immunostained with anti-G9a antibody (green). Nuclei were counterstained with DAPI (blue). (C) C2C12 cells were synchronized at G1/S and G2/M boundary using HU and nocodozole respectively. Synchronization was examined by flow cytometry (left panel). The expression of G9a, MyoD and E2F1 in lysates is shown (middle panel). Endogenous G9a was immunoprecipitated from synchronized cells and tested for association with E2F1 and MyoD respectively (right panel). The interaction of G9a with E2F1 is higher at G1/S phase, and with MyoD at the G2/M phase. (D) Flow cytometric analysis of pBabe and pBabe-G9a cells synchronized at G1/S boundary using HU and released into growth media for 2hr and 4hr. The results are representative of at least two independent experiments.
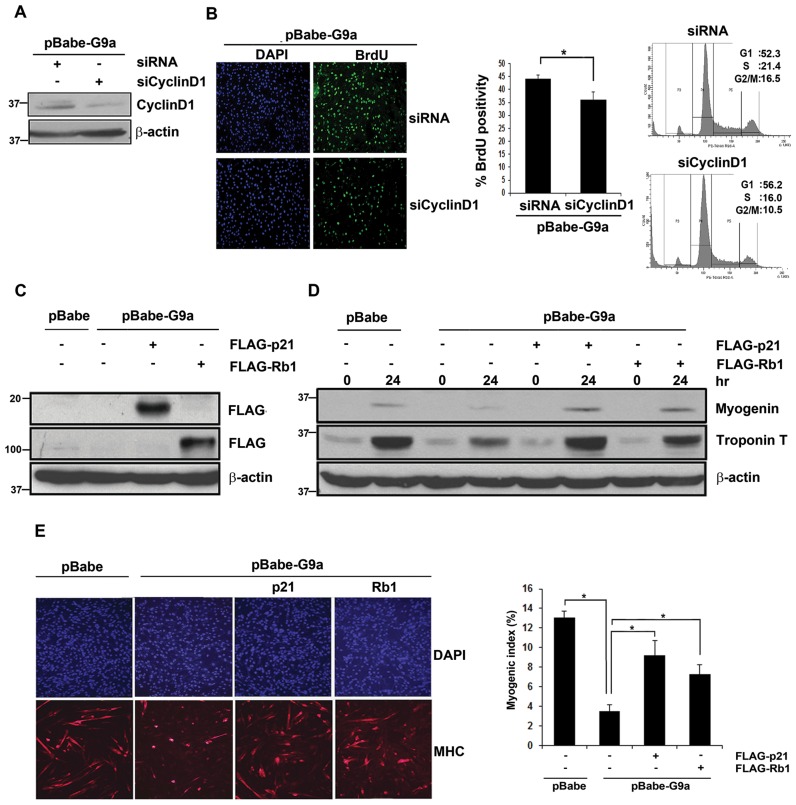
Modulation of cell cycle genes blocks proliferation and rescues myogenic differentiation in G9a overexpressing cells
Given that cessation of proliferation and irreversible cell cycle exit are essential for myogenic differentiation, we examined whether G9a-mediated inhibition of differentiation is mediated through its impact on the cell cycle genes (44). CyclinD1 knockdown was performed in G9a over expressing cells using control scrambled siRNA and CyclinD1 siRNA (Figure 7A). Downregulation of CyclinD1 resulted in reduced BrdU positivity compared to control siRNA cells, and a reduction in S-phase cells was also apparent by flow cytometry (Figure 7B). Rb1 and p21 play a central role in myogenic differentiation by promoting irreversible cell cycle exit. We therefore examined whether the repression of p21 and Rb1 by G9a is important for the block of myogenic differentiation observed in pBabe-G9a cells. Rb1 and p21 were individually transfected in pBabe-G9a cells (Figure 7C) and cells were analysed for differentiation. Consistent with our previous results (5), G9a over expression inhibited differentiation. Re-expression of p21 or Rb1 in G9a over expressing cells rescued expression of myogenin and Troponin-T (Figure 7D). In addition, the number of MHC+ cells was increased along with an increase in the myogenic index (Figure 7E).
Figure 7.
Modulation of cell cycle genes blocks proliferation and rescues myogenic differentiation in G9a overexpressing cells. (A) pBabe-G9a cells were transfected with control or CyclinD1 siRNA (siCyclinD1) in growth medium for 48 hr. The knockdown of CyclinD1 in pBabe-G9a cells was examined by western blot. β-actin was used as an internal control. (B) Control and siCyclinD1 pBabe-G9a cells were pulsed with BrdU and analysed by immunofluorescence with anti-BrdU antibody. The percentage of BrdU+ cells was quantified. Results are representative of two independent experiments. Error bars indicate mean BrdU+ cells obtained by counting at least 500 cells in each experiment ± SD. The reduction in S-phase population was also verified by PI staining of control and siCyclinD1 cells. (C) pBabe-G9a cells were transfected with Flag-p21 and Flag-Rb1 individually and their expression was analysed by western blot using anti-Flag antibody. (D) pBabe and pBabe-G9a cells transfected with p21 and Rb1 were differentiated for 24 hr and lysates were analysed for myogenin and Troponin-T by western blot. (E) Cells were immunostained with anti-MHC antibody (red) 36 hr after differentiation. Myogenic index was quantified and is shown in the right panel. The results are representative of two independent experiments. Error bars indicate the mean myogenic index ± SD.
DISCUSSION
G9a, a euchromatic lysine methyltransferase, mediates H3K9me2 that is associated with transcriptional repression (45). Recent studies have shown that G9a is overexpressed in various cancers (46–48) and its downregulation reduces cellular proliferation (49,50). However, genome wide targets of G9a, and the mechanisms by which it impacts cellular proliferation and cell cycle progression have not been defined. In this study, using a transcriptomic approach in skeletal myoblasts, we have identified G9a target genes and provide evidence for a dual role for G9a in regulating cell cycle progression. We demonstrate that G9a not only prevents cell cycle exit through repression of its down stream targets p21 and Rb1, but also promotes proliferation by activating expression of E2F1 targets such as CyclinD1 and DHFR in a methylation-independent manner. We demonstrate that regulation of cell cycle genes is critical for the ability of G9a to block myogenic differentiation.
While G9a has been predominantly studied as a transcriptional repressor, knockdown of G9a in skeletal myoblasts resulted in both upregulation as well as downregulation of a substantial number of genes. Consistent with its function as a co-repressor, our results show that G9a transcriptionally represses expression of cell cycle exit genes p21 and Rb1 by mediating H3K9me2 at their promoters. Consistently, treatment of cells with UNC0638 upregulates expression of these targets concomitant with a reduction in this repressive mark. These findings are in line with previous studies in other cell types that have demonstrated direct transcriptional regulation of p21 expression by G9a (51,52). It is interesting to note that while Rb1 expression is also repressed by G9a, the levels of phosphorylated Rb1 are enhanced by G9a overexpression. It is likely that the reduced p21 expression in G9a overexpressing cells results in increased CyclinD1/CDK activity, and consequently higher phosphorylated Rb1 levels. While reduced expression of Rb1 and p21 in G9a overexpressing cells could also account for the increased expression of E2F1 target genes such as CyclinD1, several lines of evidence suggest that E2F1 target genes are directly regulated by G9a, rather than a consequence of reduced p21 and Rb1 expression: (i) G9a occupancy is apparent on E2F1 target genes promoters. (ii) Treatment with UNC0638 does not impact expression of E2F1 target genes suggesting that the effect on their expression is not a consequence of reduced p21 and Rb1. (iii) H3K9me2 marks are not correlated with G9a occupancy on E2F1 target gene promoters. (iv) Transcriptional activation marks (H3K9ac) and increased PCAF enrichment is apparent on E2F1 promoters in G9a overexpressing cells. (v) G9a associates with E2F1 and PCAF, and enhances their association. (vi) The impact of G9a on CyclinD1 expression is E2F1-dependent. Overall, these observations are in line with recent studies which have shown that G9a can activate gene expression independent of its methyltransferase activity through association with co-activators (40–42). Consistent with a potential role for G9a in enhancing E2F1 activity, it preferentially associates with E2F1 during the G1/S transition of cells. Indeed, G1/S synchronized pBabe-G9a cells showed a higher number of cells in S phase, and conversely, G9a knockdown in primary myoblasts led to a reduced number of cells in S phase. These observations are in agreement with a recent study demonstrating that G9a−/− mouse embryonic fibroblasts (MEFs) show reduced cell numbers in the S phase compared to wild-type MEFs (53).
We have previously shown that G9a associates with MyoD resulting in its methylation that blocks its transcriptional activity (5). Interestingly, the association of G9a with MyoD is higher at G2/M phase which may be important for MyoD degradation at G2/M (54). Indeed, methylation of MyoD by G9a has been reported to be a signal for its degradation (6). Moreover, our observation that G9a and MyoD association is lower at the G1/S phase is consistent with enhanced MyoD transcriptional activity during differentiation which occurs at the G1 phase. The molecular switch that controls the association of G9a with E2F1 during the G1/S phase, and with MyoD during G2/M remains to be investigated. The stoichiometry of G9a, MyoD and E2F1 at different phases of the cell cycle, or post-translational modifications might dictate the association of G9a with the activating E2F1/PCAF complex or with the repressive MyoD/HDAC1 complex.
Given its role in proliferation of cells, it is not surprising that G9a overexpression is apparent in various cancers (55,56). Since G9a represses tumour suppressor genes, pharmacological compounds targeting its activity are an attractive tool for cancer therapy (57). However, since G9a also positively regulates the expression of genes which drive cellular proliferation, our findings imply that the methyltransferase-independent function of G9a may also contribute to tumorigenesis. Thus targeting G9a expression may have greater therapeutic value than just inhibition of its activity.
Acknowledgments
We thank Dr Martin J. Walsh, Dr Y Nakatani, Dr M Strauss and Dr S. Chellappan for the gift of various plasmid constructs.
FUNDING
National Medical Research Council [NMRC/CBRG/0063/2014 to R.T]; NUS Graduate School for Integrative Sciences and Engineering Scholarship (to J.R.O.). Funding for open access charge: National Medical Research Council Grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Tapscott S.J. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 2.Wang J., Helin K., Jin P., Nadal-Ginard B. Inhibition of in vitro myogenic differentiation by cellular transcription factor E2F1. Cell Growth Differ. 1995;6:1299–1306. [PubMed] [Google Scholar]
- 3.Puri P.L., Iezzi S., Stiegler P., Chen T.T., Schiltz R.L., Muscat G.E., Giordano A., Kedes L., Wang J.Y., Sartorelli V. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol. Cell. 2001;8:885–897. doi: 10.1016/s1097-2765(01)00373-2. [DOI] [PubMed] [Google Scholar]
- 4.Mal A., Sturniolo M., Schiltz R.L., Ghosh M.K., Harter M.L. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 2001;20:1739–1753. doi: 10.1093/emboj/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ling B.M.T., Bharathy N., Chung T.-K., Kok W.K., Li S., Tan Y.H., Rao V.K., Gopinadhan S., Sartorelli V., Walsh M.J., et al. Lysine methyltransferase G9a methylates the transcription factor MyoD and regulates skeletal muscle differentiation. Proc. Natl. Acad. Sci. U.S.A. 2012;109:841–846. doi: 10.1073/pnas.1111628109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung E.-S., Sim Y.-J., Jeong H.-S., Kim S.-J., Yun Y.-J., Song J.-H., Jeon S.-H., Choe C., Park K.-T., Kim C.-H., et al. Jmjd2C increases MyoD transcriptional activity through inhibiting G9a-dependent MyoD degradation. Biochim. Biophys. Acta. 2015;1849:1081–1094. doi: 10.1016/j.bbagrm.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Mal A.K. Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. EMBO J. 2006;25:3323–3334. doi: 10.1038/sj.emboj.7601229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Falco G., Comes F., Simone C. pRb: master of differentiation. Coupling irreversible cell cycle withdrawal with induction of muscle-specific transcription. Oncogene. 2006;25:5244–5249. doi: 10.1038/sj.onc.1209623. [DOI] [PubMed] [Google Scholar]
- 9.Martínez-Balbás M.A., Bauer U.-M., Nielsen S.J., Brehm A., Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19:662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puri P.L., Sartorelli V., Yang X.-J., Hamamori Y., Ogryzko V.V., Howard B.H., Kedes L., Wang J.Y.J., Graessmann A., Nakatani Y., et al. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 11.Robertson K.D., Ait-Si-Ali S., Yokochi T., Wade P.A., Jones P.L., Wolffe A.P. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 12.Frolov M.V., Dyson N.J. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J. Cell Sci. 2004;117:2173–2181. doi: 10.1242/jcs.01227. [DOI] [PubMed] [Google Scholar]
- 13.Ait-Si-Ali S., Guasconi V., Fritsch L., Yahi H., Sekhri R., Naguibneva I., Robin P., Cabon F., Polesskaya A., Harel-Bellan A. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 2004;23:605–615. doi: 10.1038/sj.emboj.7600074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ling B.M.T., Gopinadhan S., Kok W.K., Shankar S.R., Gopal P., Bharathy N., Wang Y., Taneja R. G9a mediates Sharp-1-dependent inhibition of skeletal muscle differentiation. Mol. Biol. Cell. 2012;23:4778–4785. doi: 10.1091/mbc.E12-04-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caretti G., Di Padova M., Micales B., Lyons G.E., Sartorelli V. The polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guasconi V., Pritchard L.-L., Fritsch L., Mesner L.D., Francastel C., Harel-Bellan A., Ait-Si-Ali S. Preferential association of irreversibly silenced E2F-target genes with pericentromeric heterochromatin in differentiated muscle cells. Epigenetics. 2010;5:704–709. doi: 10.4161/epi.5.8.13025. [DOI] [PubMed] [Google Scholar]
- 17.Blais A., van Oevelen C.J.C., Margueron R., Acosta-Alvear D., Dynlacht B.D. Retinoblastoma tumor suppressor protein–dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. J. Cell Biol. 2007;179:1399–1412. doi: 10.1083/jcb.200705051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halevy O., Novitch B.G., Spicer D.B., Skapek S.X., Rhee J., Hannon G.J., Beach D., Lassar A.B. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 19.Parker S.B., Eichele G., Zhang P., Rawls A., Sands A.T., Bradley A., Olson E.N., Harper J.W., Elledge S.J. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science. 1995;267:1024–1027. doi: 10.1126/science.7863329. [DOI] [PubMed] [Google Scholar]
- 20.Martelli F., Cenciarelli C., Santarelli G., Polikar B., Felsani A., Caruso M. MyoD induces retinoblastoma gene expression during myogenic differentiation. Oncogene. 1994;9:3579–3590. [PubMed] [Google Scholar]
- 21.Mal A., Chattopadhyay D., Ghosh M.K., Poon R.Y., Hunter T., Harter M.L. p21 and retinoblastoma protein control the absence of DNA replication in terminally differentiated muscle cells. J. Cell Biol. 2000;149:281–292. doi: 10.1083/jcb.149.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo K., Wang J., Andrés V., Smith R.C., Walsh K. MyoD-induced expression of p21 inhibits cyclin-dependent kinase activity upon myocyte terminal differentiation. Mol. Cell. Biol. 1995;15:3823–3829. doi: 10.1128/mcb.15.7.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu W., Schneider J.W., Condorelli G., Kaushal S., Mahdavi V., Nadal-Ginard B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 24.Novitch B.G., Mulligan G.J., Jacks T., Lassar A.B. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J. Cell Biol. 1996;135:441–456. doi: 10.1083/jcb.135.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zacksenhaus E., Jiang Z., Chung D., Marth J.D., Phillips R.A., Gallie B.L. pRb controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev. 1996;10:3051–3064. doi: 10.1101/gad.10.23.3051. [DOI] [PubMed] [Google Scholar]
- 26.Wang J., Abate-Shen C. The MSX1 homeoprotein recruits G9a methyltransferase to repressed target genes in myoblast cells. PLoS One. 2012;7:e37647. doi: 10.1371/journal.pone.0037647. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Choi J., Jang H., Kim H., Lee J.-H., Kim S.-T., Cho E.-J., Youn H.-D. Modulation of lysine methylation in myocyte enhancer factor 2 during skeletal muscle cell differentiation. Nucleic Acids Res. 2014;42:224–234. doi: 10.1093/nar/gkt873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun H., Li L., Vercherat C., Gulbagci N.T., Acharjee S., Li J., Chung T.-K., Thin T.H., Taneja R. Stra13 regulates satellite cell activation by antagonizing Notch signaling. J. Cell Biol. 2007;177:647–657. doi: 10.1083/jcb.200609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X.J., Ogryzko V.V., Nishikawa J., Howard B.H., Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 30.Wang S., Fusaro G., Padmanabhan J., Chellappan S.P. Prohibitin co-localizes with Rb in the nucleus and recruits N-CoR and HDAC1 for transcriptional repression. Oncogene. 2002;21:8388–8396. doi: 10.1038/sj.onc.1205944. [DOI] [PubMed] [Google Scholar]
- 31.Müller H., Lukas J., Schneider A., Warthoe P., Bartek J., Eilers M., Strauss M. Cyclin D1 expression is regulated by the retinoblastoma protein. Proc. Natl. Acad. Sci. U.S.A. 1994;91:2945–2949. doi: 10.1073/pnas.91.8.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y., Rao V.K., Kok W.K., Roy D.N., Sethi S., Ling B.M.T., Lee M.B.H., Taneja R. SUMO modification of Stra13 is required for repression of cyclin D1 expression and cellular growth arrest. PLoS One. 2012;7:e43137. doi: 10.1371/journal.pone.0043137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheedipudi S., Puri D., Saleh A., Gala H.P., Rumman M., Pillai M.S., Sreenivas P., Arora R., Sellathurai J., Schroeder H.D., et al. A fine balance: epigenetic control of cellular quiescence by the tumor suppressor PRDM2/RIZ at a bivalent domain in the cyclin a gene. Nucleic Acids Res. 2015;43:6236–6256. doi: 10.1093/nar/gkv567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandromme M., Chailleux C., Escaffit F., Trouche D. Binding of the retinoblastoma protein is not the determinant for stable repression of some E2F-regulated promoters in muscle cells. Mol. Cancer Res. 2008;6:418–425. doi: 10.1158/1541-7786.MCR-07-0381. [DOI] [PubMed] [Google Scholar]
- 35.Azmi S., Ozog A., Taneja R. Sharp-1/DEC2 inhibits skeletal muscle differentiation through repression of myogenic transcription factors. J. Biol. Chem. 2004;279:52643–52652. doi: 10.1074/jbc.M409188200. [DOI] [PubMed] [Google Scholar]
- 36.Dignam J.D., Lebovitz R.M., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferreira R., Naguibneva I., Mathieu M., Ait-Si-Ali S., Robin P., Pritchard L.L., Harel-Bellan A. Cell cycle-dependent recruitment of HDAC-1 correlates with deacetylation of histone H4 on an Rb-E2F target promoter. EMBO Rep. 2001;2:794–799. doi: 10.1093/embo-reports/kve173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berkes C.A., Bergstrom D.A., Penn B.H., Seaver K.J., Knoepfler P.S., Tapscott S.J. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell. 2004;14:465–477. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- 39.Vedadi M., Barsyte-Lovejoy D., Liu F., Rival-Gervier S., Allali-Hassani A., Labrie V., Wigle T.J., Dimaggio P.A., Wasney G.A., Siarheyeva A., et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat. Chem. Biol. 2011;7:566–574. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bittencourt D., Wu D.-Y., Jeong K.W., Gerke D.S., Herviou L., Ianculescu I., Chodankar R., Siegmund K.D., Stallcup M.R. G9a functions as a molecular scaffold for assembly of transcriptional coactivators on a subset of glucocorticoid receptor target genes. Proc. Natl. Acad. Sci. U.S.A. 2012;109:19673–19678. doi: 10.1073/pnas.1211803109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaturvedi C.-P., Hosey A.M., Palii C., Perez-Iratxeta C., Nakatani Y., Ranish J.A., Dilworth F.J., Brand M. Dual role for the methyltransferase G9a in the maintenance of beta-globin gene transcription in adult erythroid cells. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18303–18308. doi: 10.1073/pnas.0906769106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shankar S.R., Bahirvani A.G., Rao V.K., Bharathy N., Ow J.R., Taneja R. G9a, a multipotent regulator of gene expression. Epigenetics. 2013;8:16–22. doi: 10.4161/epi.23331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ait-Si-Ali S., Polesskaya A., Filleur S., Ferreira R., Duquet A., Robin P., Vervish A., Trouche D., Cabon F., Harel-Bellan A. CBP/p300 histone acetyl-transferase activity is important for the G1/S transition. Oncogene. 2000;19:2430–2437. doi: 10.1038/sj.onc.1203562. [DOI] [PubMed] [Google Scholar]
- 44.Skapek S.X., Rhee J., Spicer D.B., Lassar A.B. Inhibition of myogenic differentiation in proliferating myoblasts by cyclin D1-dependent kinase. Science. 1995;267:1022–1024. doi: 10.1126/science.7863328. [DOI] [PubMed] [Google Scholar]
- 45.Tachibana M., Sugimoto K., Fukushima T., Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 46.Casciello F., Windloch K., Gannon F., Lee J.S. Functional role of G9a histone methyltransferase in cancer. Front. Immunol. 2015;6:487. doi: 10.3389/fimmu.2015.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehnertz B., Pabst C., Su L., Miller M., Liu F., Yi L., Zhang R., Krosl J., Yung E., Kirschner J., et al. The methyltransferase G9a regulates HoxA9-dependent transcription in AML. Genes Dev. 2014;28:317–327. doi: 10.1101/gad.236794.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong C., Wu Y., Yao J., Wang Y., Yu Y., Rychahou P.G., Evers B.M., Zhou B.P. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J. Clin. Invest. 2012;122:1469–1486. doi: 10.1172/JCI57349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ueda J., Ho J.C., Lee K.L., Kitajima S., Yang H., Sun W., Fukuhara N., Zaiden N., Chan S.L., Tachibana M., et al. The hypoxia-inducible epigenetic regulators Jmjd1a and G9a provide a mechanistic link between angiogenesis and tumor growth. Mol. Cell. Biol. 2014;34:3702–3720. doi: 10.1128/MCB.00099-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding J., Li T., Wang X., Zhao E., Choi J.-H., Yang L., Zha Y., Dong Z., Huang S., Asara J.M., et al. The histone H3 methyltransferase G9A epigenetically activates the serine-glycine synthesis pathway to sustain cancer cell survival and proliferation. Cell Metab. 2013;18:896–907. doi: 10.1016/j.cmet.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishio H., Walsh M.J. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc. Natl. Acad. Sci. U.S.A. 2004;101:11257–11262. doi: 10.1073/pnas.0401343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim J.K., Estève P.-O., Jacobsen S.E., Pradhan S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res. 2009;37:493–505. doi: 10.1093/nar/gkn961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim K.-B., Son H.-J., Choi S., Hahm J.Y., Jung H., Baek H.J., Kook H., Hahn Y., Kook H., Seo S.-B. H3K9 methyltransferase G9a negatively regulates UHRF1 transcription during leukemia cell differentiation. Nucleic Acids Res. 2015;43:3509–3523. doi: 10.1093/nar/gkv183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kitzmann M., Carnac G., Vandromme M., Primig M., Lamb N.J., Fernandez A. The muscle regulatory factors MyoD and myf-5 undergo distinct cell cycle-specific expression in muscle cells. J. Cell Biol. 1998;142:1447–1459. doi: 10.1083/jcb.142.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hua K.-T., Wang M.-Y., Chen M.-W., Wei L.-H., Chen C.-K., Ko C.-H., Jeng Y.-M., Sung P.-L., Jan Y.-H., Hsiao M., et al. The H3K9 methyltransferase G9a is a marker of aggressive ovarian cancer that promotes peritoneal metastasis. Mol. Cancer. 2014;13:189. doi: 10.1186/1476-4598-13-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen M.-W., Hua K.-T., Kao H.-J., Chi C.-C., Wei L.-H., Johansson G., Shiah S.-G., Chen P.-S., Jeng Y.-M., Cheng T.-Y., et al. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res. 2010;70:7830–7840. doi: 10.1158/0008-5472.CAN-10-0833. [DOI] [PubMed] [Google Scholar]
- 57.Yuan Y., Wang Q., Paulk J., Kubicek S., Kemp M.M., Adams D.J., Shamji A.F., Wagner B.K., Schreiber S.L. A small-molecule probe of the histone methyltransferase G9a induces cellular senescence in pancreatic adenocarcinoma. ACS Chem. Biol. 2012;7:1152–1157. doi: 10.1021/cb300139y. [DOI] [PMC free article] [PubMed] [Google Scholar]