Abstract
ZFP57 is necessary for maintaining repressive epigenetic modifications at Imprinting control regions (ICRs). In mouse embryonic stem cells (ESCs), ZFP57 binds ICRs (ICRBS) and many other loci (non-ICRBS). To address the role of ZFP57 on all its target sites, we performed high-throughput and multi-locus analyses of inbred and hybrid mouse ESC lines carrying different gene knockouts. By using an allele-specific RNA-seq approach, we demonstrate that ZFP57 loss results in derepression of the imprinted allele of multiple genes in the imprinted clusters. We also find marked epigenetic differences between ICRBS and non-ICRBS suggesting that different cis-acting regulatory functions are repressed by ZFP57 at these two classes of target loci. Overall, these data demonstrate that ZFP57 is pivotal to maintain the allele-specific epigenetic modifications of ICRs that in turn are necessary for maintaining the imprinted expression over long distances. At non-ICRBS, ZFP57 inactivation results in acquisition of epigenetic features that are characteristic of poised enhancers, suggesting that another function of ZFP57 in early embryogenesis is to repress cis-acting regulatory elements whose activity is not yet required.
INTRODUCTION
Genomic imprinting is a phenomenon involving a small but developmentally important subset of mammalian genes that are transcribed only from the maternal or the paternal allele (1,2). Key elements for the establishment and maintenance of imprinted gene expression are imprinting control regions (ICRs). These are 0.6–10 kb-long CpG-rich sequences that acquire DNA methylation in the germline in a sex-specific manner (‘germline-differentially methylated regions’, gDMRs) and control in cis the gamete of origin-dependent expression of clusters of imprinted genes through several distinct mechanisms (3). ICR methylation is erased in the primordial germ cells, re-established in developing gametes around meiosis and faithfully maintained on only one parental chromosome in the zygote and somatic cells during development. In early embryogenesis, imprints are maintained despite a genome-wide CpG demethylation occurring after fertilization and de novo methylation during implantation (4,5). ICRs are protected from the genome-wide waves of DNA demethylation by specific DNA binding factors (6). These include the zinc-finger protein ZFP57 and its corepressor KRAB-A-interacting protein (KAP1) (7,8). The ZFP57–KAP1 complex interacts with ICRs in a parent-of-origin-specific manner, by recognizing the methylated [TG]GCCGC motif that is highly enriched at the ICRs (9–13). ZFP57-KAP1 bind to both parental alleles at many other loci that are not ICRs in mouse embryonic stem cells (ESCs) (9,12,13).
In addition to differential DNA methylation, ICRs also display different histone modifications on their maternal and paternal alleles (14). In particular, H3K9me3 is associated with the DNA methylated allele and H3K4me3 is associated with the non-methylated allele. A functional link between mCpG and histone modifications at ICRs is evoked by several observations. These include the demonstration that the ZFP57–KAP1 complex recruits both the histone H3K9 methyltransferase SET Domain Bifurcated 1 (SETDB1) and the DNA methyltransferases DNMT1, DNMT3A and DNMT3B, and that ZFP57 inactivation results in loss of mCpG and histone H3K9me3 at ICRs, in mouse ESCs (9,15).
Despite the above cited studies, the role of ZFP57 in controlling imprinted and non-imprinted gene expression remains largely undefined. By employing genome-wide and locus-specific approaches in multiple knockout ESC systems, here we show that despite its restricted binding to ICRs, ZFP57 controls the parent of origin-dependent epigenetic features and gene expression of extensive genomic regions at imprinted gene loci. In addition, we demonstrate that ZFP57 binding prevents the acquisition of enhancer-specific histone marks at many other regions through the recruitment of heterochromatin marks. These results provide new insights into the mechanisms of imprinting maintenance in somatic cells and disclose a possible novel role of ZFP57 in the epigenetic control of early embryogenesis.
MATERIALS AND METHODS
Cell lines and culture conditions
WT and Zfp57 −/− (clone A9) A3 ESCs (16,17), WT and Zfp57 −/− E14 and JB1 ESCs and WT J1 and triple Dnmt1, Dnmt3a and Dnmt3b mutation (TKO, 18) ESCs were cultured under standard feeder-free conditions on gelatinized tissue culture dishes with media containing DMEM (EuroClone ECM0101L) supplemented with 100 μM 2-mercaptoethanol (Sigma), 1× non-essential amino acids (only for A3), 1 mM sodium pyruvate, 2 mM l-glutamine, 1x penicillin–streptomycin, 10% (15% for E14 and JB1) foetal calf serum (HyClone) and 103 U/ml leukemia inhibitory factor (LIF, Millipore) at 37°C under an atmosphere of 5% CO2.
CRISPR-Cas9-mediated Zfp57 knockout
The expression vector pX459 (Addgene) and pX461 (Addgene), pX462 (Addgene) were used to express Cas9 (Nuclease), Cas9n (Nickase) and sgRNAs. Targeting sequences of sgRNAs were designed using CRISPR Design Tool (http://tools.genome-engineering.org) available at the Feng Zhang Laboratory website (19). sgRNAs of Zfp57 were synthesized, annealed, and ligated to pX459, pX461 and pX462 plasmids that were digested with BbsI (New England Biolabs) according to Ran et al. (19). 4 × 106 WT E14 and JB1 ESCs were transfected with 4 μg of pX459 expressing both Cas9 and sgRNA targeting Zfp57 exon 4 (UCSC transcript ID uc012atb.1) or plasmids pX461 and pX462 (2 μg each) expressing Cas9n and sgRNAs targeting Zfp57 exon 5 using Nucleofector (Amaxa) according to the manufacturer's protocol. Forty eight hours after transfection, the cells expressing GFP were enriched with flow cytometry (FACS Aria, Becton Dickinson) and plated at low density. Cells were incubated with 1 μg/ml puromycin for 72 h. Five to six days after plating, single colonies were picked and screened for the expected genotype by amplifying the target sites. The sequences for the target sites and primers for validation are reported in Supplementary Table S1. The western blot performed for validation of the knockout cell lines was obtained with a polyclonal anti-ZFP57 antibody (Abcam ab45341) that was raised against a synthetic peptide derived from residues 150–250 of mouse ZFP57.
Chromatin immunoprecipitation (ChIP)
ChIP of ZFP57, KAP1 and H3K9me3 was performed as previously described (13). The same protocol was used for H3K4me1 (7 μg of Abcam ab8580 for 100 μg of chromatin in 600 μl of shearing buffer), H3K4me3 (7 μg of Abcam ab8895 for 100 μg of chromatin in 600 μl of shearing buffer), H3K27me3 (10 μg of Millipore 07-449 for 100 μg of chromatin in 600 μl of shearing buffer) and H3K27ac (10 μg of Abcam ab4729 for 100 μg of chromatin in 600 μl of shearing buffer). The rabbit control IgG ChIP-grade antibody (5 μg of Abcam ab46540-1) was used as a control in the ChIP. Q-PCR assays of locus-specific ChIPs were performed in triplicate. Primers for locus-specific ChIP are listed in Supplementary Table S1. Conditions were: one cycle at 95.0°C for 3 min, 40 cycles at 95.0°C for 10 s, 60.0°C for 30 s, 75.0°C for 30 s, followed by melt curve at 60°C to 90°C increment 0.5°C for 5 s, in the presence of 1x iTaq™ Universal SYBR® Green Supermix (Biorad) on a CFX Connect™ Real-Time PCR Detection System (Biorad). Two nanogram of DNA from immunoprecipitated and input chromatin were used for Illumina library preparation. Chromatin immunoprecipitation sequencing (ChIP-seq) libraries were generated by using the NuGen Ovation Ultralow Library System Kit and sequenced 50 bp single-end at IGA Technology Services (Italy) using an Illumina HiSeq2500 platform. One ChIP-seq library per cell line/Ab was sequenced and the results validated by Q-PCR.
DNA methylation analysis
CpG methylation was studied with three methods: locus-specific direct bisulfite sequencing, whole genome-bisulfite sequencing (WGBS-seq) and Reduced Representation Bisulfite Sequencing (RRBS). For locus-specific bisulfite sequencing, 2 μg of genomic DNA was bisulfite-converted with the EpiTect Bisulfite Kit (QIAGEN) and the regions of interest were amplified by PCR and directly sequenced. DNA sequences were analysed by using the Chromas Lite 2.1.1 (Technelysium) program. Primers are listed in Supplementary Table S1. For WGBS-seq and RRBS library preparation, 100–200 ng of genomic DNA was used with the NuGEN Ovation Ultralow Methyl-Seq Library System. Libraries were sequenced 100 bp paired-end (WGBS-seq) or 50bp single-end (RRBS) at IGA Technology Services (Italy) using the Illumina HiSeq2500 platform.
RNA analysis
RNA was extracted from cultured ESCs by lysing the cells on the culture dish with the addition of the TRI reagent (Sigma-Aldrich) and following the protocol of the manufacturer. For qRT-PCR analysis, 1 μg of total RNA was retrotranscribed by using the QuantiTect Reverse Transcription Kit (Qiagen), according to the protocol of the manufacturer. cDNA was amplified by real-time PCR using SYBR Green PCR Master Mix (Bio-Rad) on a C1000 Thermal Cycler (Bio-Rad). Primers are listed in Supplementary Table S1. RNA sequencing (RNA-seq) libraries were generated from 200 ng of RNA by using the Illumina TruSeq Stranded mRNA Sample Prep Kit and sequenced 50bp single-end at IGA Technology Services (Italy) using the Illumina HiSeq2500 platform.
NGS data analysis
Reads were processed by adaptor trimming (Illumina Pipeline Casava v1.8.2 or Cutadapt for RNA-seq), filtered for low quality reads (Trim Galore v0.2.8) and subjected to quality control (FastQC). Reads were aligned to the Mus musculus genome (assembly NCBI37/mm9) and results were displayed on the UCSC Genome Browser (ChIP-seq and global and allele-specific RNA-seq tracks have normalized genome coverage as reads per million; WGBS and RRBS tracks displayed methylation levels of symmetric CpGs covered by at least five reads). ChIP-seq reads were aligned using the Bowtie v0.12.7 with uniquely mapping option (20) and duplicate reads were removed. Peaks were defined using the MACS algorithm (21) implemented in SeqMonk v0.32.1 (Simon Andrews, Babraham Institute, UK) with 1.0e−5 P-values cut-off; to reduce false positives, we considered only ZFP57 peaks overlapping KAP1 binding sites. Hierarchical cluster analysis was performed using package heatmap.2 of the R v3.0.1 gplots library (Gregory R. Warnes). WGBS/RRBS-seq reads were aligned using Bismark v0.7.9 (22) and CpG methylation calls were extracted using the Bismark methylation extractor v0.7.9. The methylation level of a DNA region was defined as the average methylation level of all its symmetric CpGs (at least 2 CpGs covered by at least five reads). Hypomethylated DNA regions in Zfp57 −/− compared to WT ESCs were identified by searching for sequences with common symmetric CpGs (at least 5 CpGs covered by at least 5 reads that were less than 1 kb apart) with a decrease in methylation of >25%.
H3K4me1 and H3K4me3 ChIP-seq peaks regions data sets for mouse ESC and differentiated tissues were downloaded from the UCSC mm9 annotation database (GEO GSE29184, 23). The number of ICRBS and non-ICRBS (E14 ESC) overlapping with H3K4me1 and H3K4me3 peaks were calculated by using the BEDTools suite v2.14.3-1 (24). The significance of such co-bindings was assessed using permutations. An analogous number of random genomic locations of both ICRBS and non-ICRBS were generated preserving region sizes, chromosome distributions and not allowing overlaps among them. The process was repeated 1000 times in order to estimate the P-value (Python custom script and BEDTools suite v2.14.3-1).
RNA-seq reads were aligned using the TopHat v2.0.8b (25). Aligned RNA-seq reads were analysed using SeqMonk v0.32.1; for gene expression, the RNA-seq quantification pipeline was used with default parameters returning log2-transformed counts of reads falling within the exons of each gene (Ensembl Database modified by adding uc012fxx.1, uc007wb0.1, uc009hep.2 and uc007iem.1 genes from UCSC Genes) corrected for the total number of sequences in the dataset. The differentially expressed genes between WT and knockout samples were identified using the intensity difference method (without multiple testing correction and with 0.05 P-values cut-off) on the genes with a minimum of 10 reads. To reduce the false positives, we considered only the genes that were concordantly deregulated in both knockout cell lines. The distance of the deregulated genes from the ZFP57 binding sites was calculated as the shortest distance between the ZFP57 ChIP-seq peaks and the gene coordinates. The distance between overlapping peaks and genes was set to 0. ZFP57 and KAP1 ChIP-seq datasets on JB1 hybrid mouse ESC were downloaded from GEO (GSE74757). The allele-specific mapping of ChIP-seq and RNA-seq data in hybrid JB1 mouse ESC was performed as previously described (13). Allele-specific ZFP57/KAP1 binding sites were identified as described by Anvar et al. (13), with range of bi-allelicity Y = j- ± 2σ and considering only the ZFP57/KAP1 JB1 peaks (monoallelic = xJB1 ∈ YJB1). Allele-specific expression was calculated as for global gene expression (see above), but considering only allele-specific reads without log2 transformation (values were rounded to the nearest integer number). We considered only the genes with at least five SNPs in the exons and at least 10 reads in one allele of either the WT or the knockout. To find the differences in the JF1/B6 allelic expression ratio, we applied the Fisher-exact test with Benjamini–Hochberg multiple-testing correction (P-value < 0.05).
Data access
All NGS data produced for this manuscript have been deposited in the Gene Expression Omnibus (GEO) Data Sets with the accession number GSE77444.
RESULTS
Extensive mCpG loss at ICRs in Zfp57 −/− ESCs
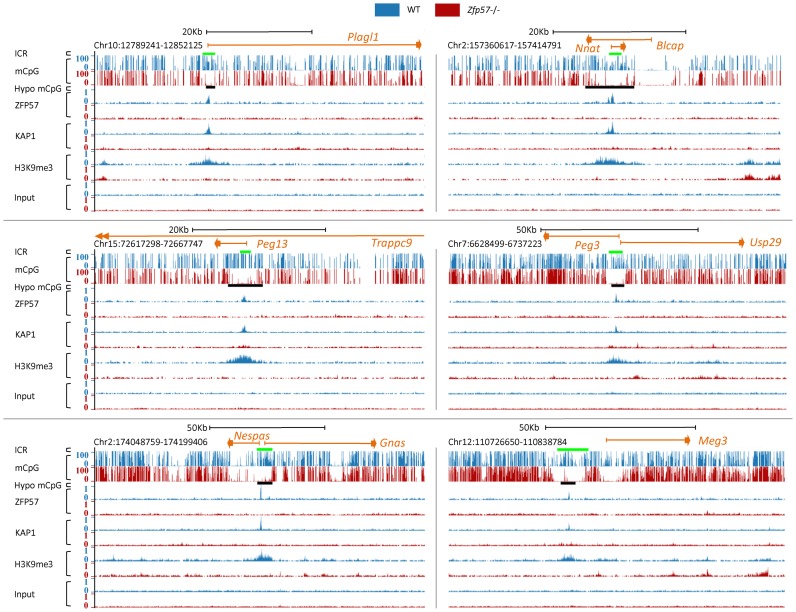
To explore the mechanism by which ZFP57 maintains repressive chromatin modifications, we investigated the effect of ZFP57 loss on the entire mouse genome by using high-throughput methods. For the analysis of DNA methylation, we produced Whole-Genome Bisulfite Sequencing (WGBS-seq) profiles; ZFP57 and KAP1 binding and H3K9me3 enrichment were determined by Chromatin Immunoprecipitation-Sequencing (ChIP-seq) in wildtype (WT) and Zfp57 −/− A3 ESCs (16). We found 63 ZFP57-specific ChIP-seq peaks in WT-A3 ESCs (Supplementary Table S2). Eleven of these colocalized with the ICRs maintaining differential DNA methylation in ESCs (26,27; see also Supplementary Table S3). All these target sites showed pronounced loss of mCpG, KAP1 binding and H3K9me3 in Zfp57 −/− A3 ESCs (Figure 1 and Supplementary Figure S1 and Table S4). While ZFP57 and KAP1 binding was concentrated at the center of the ICRs, the mCpG and H3K9me3 loss generally corresponded to the entire extension of the ICRs. At Meg3, mCpG loss also included the promoter of the Meg3 gene (Figure 1, bottom-right panel) that is known to correspond to a somatic (secondary) DMR (28), but like the other secondary DMRs, it does not bind ZFP57 (9,12).
Figure 1.
Extensive loss of mCpG and H3K9me3 at ICRs in Zfp57 −/− A3 ESCs. Screen shots from the UCSC Genome Browser of examples of mCpG and H3K9me3 profiles together with ZFP57 and KAP1 binding profiles at ICRs in WT (in blue) and Zfp57 −/− (in red) A3 ESCs, as determined by WGBS-seq and ChIP-seq. ICRs are indicated by green bars, and hypomethylated regions (hypo mCpG) in Zfp57 −/− A3 ESCs that overlap with ZFP57 peaks by black bars. Genes are indicated by orange lines at the top of each panels. Note that in all cases ZFP57 and KAP1 bind within the ICR sequences that are characterised by ZFP57-dependent mCpG and H3K9me3.
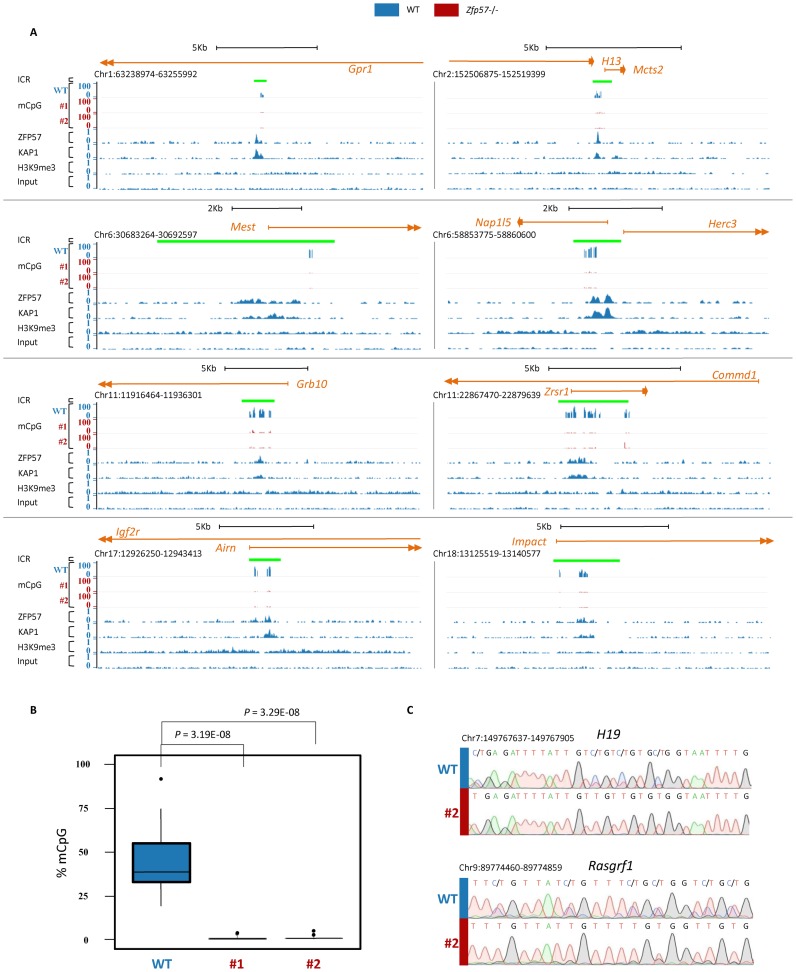
We were unable to determine the effect of ZFP57 loss at all imprinted loci in Zfp57 −/− A3 ESCs, because 9 ICRs were not bound by ZFP57 and undermethylated in the parental cell line which was used for generating the Zfp57 knockout (Supplementary Figure S2 and Table S4). This finding is consistent with what was previously found in other ESC lines (29). To overcome this problem, we identified an ESC line (WT-E14) that showed 161 ZFP57 binding sites and consistently maintained ZFP57 binding at all ICRs, as determined by ChIP-seq (Figure 2A, Supplementary Figure S3 and Table S5). 39 of these ZFP57 binding sites correspond to sites identified also in WT A3 ESCs, 54 correspond to previously reported sites in hybrid M. m. castaneus × M. m. domesticus ESC lines (12) and 96 were uniquely found in WT E14 ESCs (Supplementary Table S5). We used the CRISPR-Cas9 methodology (19) to generate two new Zfp57 knockout lines from this ESC strain (Supplementary Figure S4). DNA methylation was analysed by Reduced Representation Bisulfite Sequencing (RRBS) in these cell lines. Significantly, the mCpG profiles demonstrated complete loss of methylation at 18 ICRs detected by RRBS in the E14 knockout lines compared with the parental line (Figure 2A, B, Supplementary Figure S3 and Table S6). Although the H19 and Rasgrf1 ICRs were not detectable by RRBS, complete demethylation at the Rasgrf1 and H19 ICRs was demonstrated by direct bisulfite sequencing (Figure 2C). In addition, there was complete demethylation of the H19 promoter (Supplementary Figure S3), which is a somatic DMR close to the H19 ICR (28). Overall, these combined approaches show that ZFP57 maintains DNA methylation at all ICRs in mouse ESCs, and that the control of mCpG at these regions extends further than the ZFP57 binding sites to include the entire ICRs and close-by somatic DMRs.
Figure 2.
ZFP57 binding maintains mCpG at all ICRs in mouse E14 ESCs. A) Screen shots from the UCSC Genome Browser showing the mCpG profiles at ICRs determined by RRBS in WT (in blue) and two Zfp57 -/- (#1 and #2, in red) E14 ESC lines, together with ZFP57 and KAP1 binding and H3K9me3 profiles determined by ChIP-seq in WT E14 ESCs (in blue). The ICRs are indicated as green bars. (B) Box plot showing the distribution of mCpG levels at 18 ICRs determined by RRBS in WT (in blue) and Zfp57 −/− (line 1 and line 2, in red; see also Supplementary Table S6) E14 ESC lines. In the box-plot, upper and lower hinges correspond to the first and third quartiles, the upper whisker extends from the hinge to the highest value that is within 1.5 × IQR of the hinge, where IQR is the inter-quartile range, or distance between the first and third quartiles, the lower whisker extends from the hinge to the lowest value within 1.5 × IQR of the hinge. Data beyond the end of the whiskers are outliers and plotted as points. The difference between the methylation level of 18 ICRs in WT and two Zfp57 −/− E14 ESC lines is statistically significant (P < 0.001 calculated with two-tailed Student's t-test assuming unequal variances). (C) DNA methylation analysis of H19 and Rasgrf1 ICRs determined by direct bisulfite sequencing in WT (indicated by vertical blue bar) and Zfp57 −/− line 1 E14 (red bar) ESCs. Comparable results have been obtained in the Zfp57 −/− E14 ESC line 2. Note ZFP57 binding and mCpG maintenance in WT and complete mCpG loss in Zfp57 −/− E14 ESCs at all ICRs.
Incomplete loss of mCpG at ZFP57 binding sites not overlapping ICRs
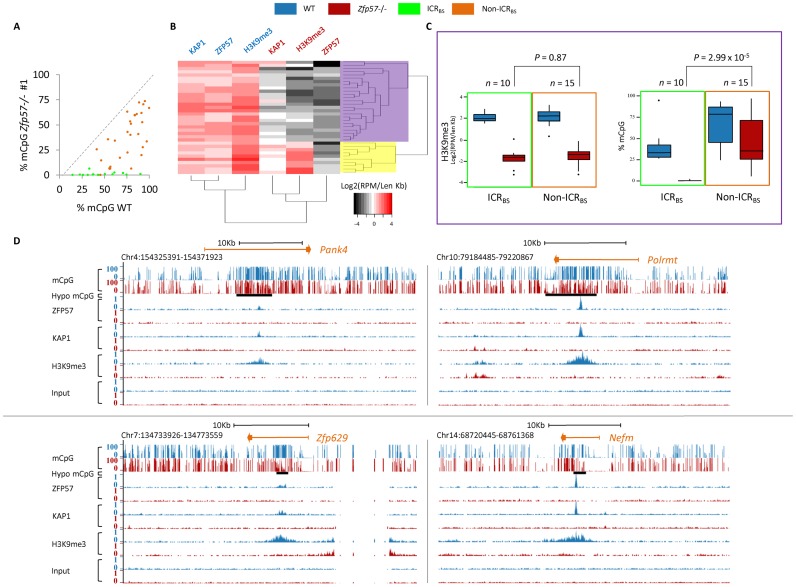
In addition to the ICRs, ZFP57 shows binding to many other genomic loci as well (9,12,13). We sought to investigate what role ZFP57 plays in the control of epigenetic modifications at these sites. In contrast to the ICR ZFP57 binding sites (ICRBS), non-ICR bound sites (non-ICRBS) generally underwent moderate or incomplete loss of DNA methylation in both the Zfp57 −/− E14 ESC lines (Figure 3A and Supplementary Table S7). To investigate if the different behavior of the non-ICRBS was due to incomplete KAP1 and/or H3K9me3 loss, we classified by hierarchical clustering ZFP57-, KAP1- and H3K9me3-specific read counts within 35 ZFP57 ChIP-seq peak regions (reported as reads per million and normalized by length), which were determined in WT-A3 and Zfp57 −/− A3 ESCs and for which methylation level was assessed (Figure 3B and Supplementary Table S8). This method allowed the identification of a large cluster of ZFP57 binding sites showing intense loss of KAP1 and H3K9me3 in Zfp57 -/- A3 ESCs (Figure 3B, highlighted in purple). The results were validated by locus-specific ChIP (Supplementary Figure S5). This cluster included many non-ICRBS and all but one (Peg10) ICRBS. Interestingly, although H3K9me3 peak intensities were similar (P-value = 0.87), mCpG levels differed significantly (P-value = 2.99 × 10−5) between ICRBS (n = 10) and non-ICRBS (n = 15) within this cluster, in Zfp57 −/− A3 ESCs (Figure 3C and Supplementary Table S8). This differential behavior was confirmed by locus-specific ChIP and direct bisulfite sequencing in WT and Zfp57 −/− E14 ESCs (Supplementary Figure S6). A minor cluster of ZFP57 binding sites showed weaker loss of KAP1 and H3K9me3 enrichment in Zfp57 −/− A3 ESCs (Figure 3B, highlighted in yellow). Significantly, most of these loci overlapped or were close to long terminal repeats (LTRs) of endogenous retroviral elements (Supplementary Figure S7). In summary, loss of ZFP57 binding has little effect on repressive epigenetic marks at loci overlapping with retroviral elements, but generally leads to extensive H3K9me3 loss at both ICRBS and non-ICRBS, and intense mCpG loss only at ICRBS (Figures 1 and 3D and Supplementary Figure S1).
Figure 3.
ZFP57 controls repressive epigenetic modifications at ICRBS and non-ICRBS. (A) Scatter plot showing mCpG levels under ZFP57 peaks in WT and Zfp57 -/- E14 ESC line 1, as determined by RRBS. ICRBS are in green and non-ICRBS in orange. Comparable results were obtained in Zfp57 −/− E14 ESC line 2 (see Supplementary Table S7). (B) Hierarchical clustering of ZFP57, KAP1 and H3K9me3 ChIP-seq peak intensities in WT (indicated in blue) and Zfp57 −/− (indicated in red) A3 ESCs. A larger cluster of ZFP57 binding sites (shaded in purple) loses H3K9me3 and KAP1, while a minor cluster (shaded in yellow) shows similar or slightly reduced levels in Zfp57 −/− A3 ESCs. (C) Box plots showing distributions of H3K9me3 peak intensities and mCpG levels of the ZFP57 binding sites highlighted in purple in (B) divided in ICRBS (included in green frame) and non-ICRBS (included in orange frame), in WT (blue) and Zfp57 −/− (red) A3 ESCs. The distributions of H3K9me3 of ICRBS and non-ICRBS in Zfp57 −/− ESCs do not differ (P > 0.05 calculated with two-tailed Student's t-test assuming unequal variances) while those of methylation levels differ significantly (P < 0.001). (D) Screen shots from the UCSC Genome Browser describing examples of mCpG and H3K9me3 profiles together with ZFP57 and KAP1 binding profiles at non-ICRBS in WT and Zfp57 −/− A3 ESCs, reported as in Figure 1. Note that loss of ZFP57 and KAP1 binding leads to extensive loss of H3K9me3 with little effect on mCpG at non-ICRBS.
Different histone marks are acquired at ICRBS and non-ICRBS in Zfp57 −/− ESCs
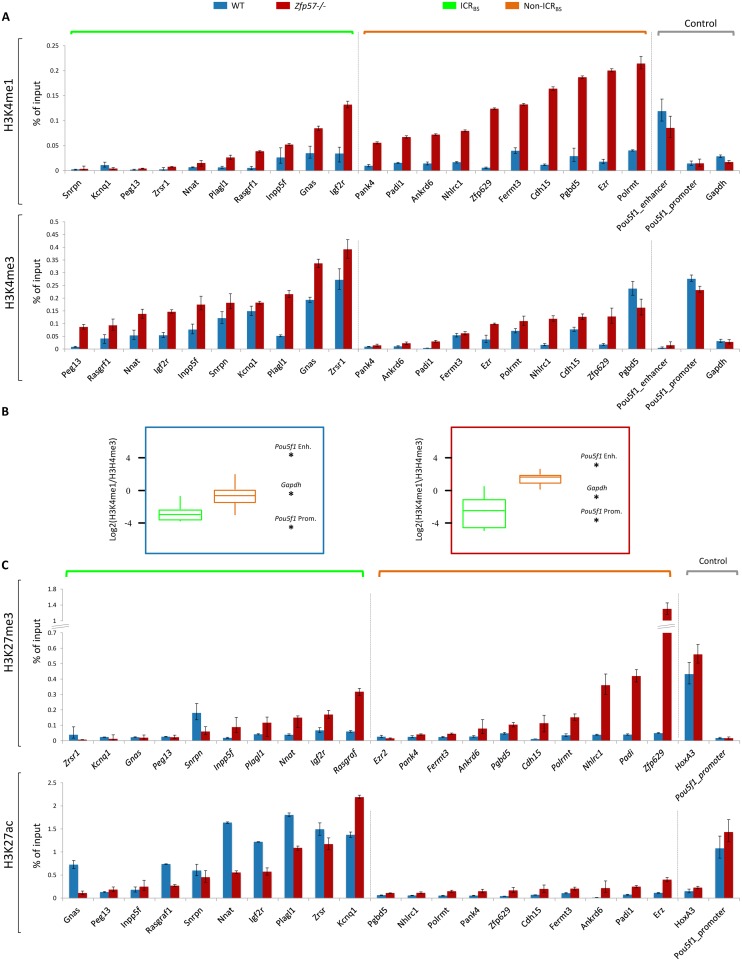
The different response of DNA methylation to Zfp57 inactivation suggests that the ICRBS and non-ICRBS have different functional properties. To investigate this issue further, we looked at the enrichment of several other histone mofifications at these two groups of ZFP57 binding sites that do not overlap with LTRs. First, we measured the level of H3K4me1 and H3K4me3 enrichment in Zfp57 −/− relative to WT E14 ESCs by locus-specific ChIP. We found that H3K4me1 was generally increased and reached levels comparable or higher than the ESC-specific Pou5f1 enhancer at all non-ICRBS investigated, but was less frequently increased and generally present at lower levels at ICRBS in Zfp57 −/− E14 ESCs (Figure 4A, top panel). H3K4me3 showed the opposite behavior. It was generally increased reaching in several cases levels comparable to the ESC-specific Pou5f1 promoter at ICRBS, but was less frequently increased and with lower levels of enrichment at non-ICRBS (Figure 4A, lower panel). Therefore, in absence of ZFP57, H3K4me1 is frequently gained at non-ICRBS while H3K4me3 is generally increased at ICRBS. Indeed, the H3K4me1/H3K4me3 ratio was generally lower at ICRBS than at non-ICRBS in WT E14 ESCs, and was further increased at non-ICRBS approaching that of the Pou5f1 enhancer in Zfp57 −/− E14 ESCs (Figure 4B). To analyse if the differential increase of H3K4me3 and H3K4me1 at ICRBS and non-ICRBS in Zfp57 −/− E14 ESCs was a consequence of the residual DNA methylation level or was due to the structural characteristics of these loci, we measured the enrichment of these two marks in ESCs that had very low level of mCpG in the entire genome because of the triple knockout of Dnmt1, Dnmt3a and Dnmt3b (TKO, 18). We found that loss of DNA methylation caused changes of H3K4me1 and H3K4me3 at the ZFP57 binding sites similar to those observed in the Zfp57 −/− ESCs. H3K4me1 enrichment was increased only at non-ICRBS (Supplementary Figure S8, top panel), while H3K4me3 was increased only at ICRBS (Supplementary Figure S8, bottom panel). Therefore, the low H3K4me3 level of the non-ICRBS is not caused by maintenance of mCpG in Zfp57 −/− ESCs, but rather to their DNA sequence characteristics.
Figure 4.
Loss of ZFP57 affects histone modifications at ZFP57 binding sites. (A) H3K4me1 (top) and H3K4me3 (bottom) enrichment at ICRBS (green) and non-ICRBS (orange) in WT (blue) and Zfp57 −/− (red) E14 ESCs, as determined by ChIP. The Pou5f1 promoter and enhancer and Gapdh were analysed as controls. Precipitated target loci with non-immune IgG were in the range of 0.00007–0.02% of input. (B) Box-plot (represented as in Figure 2B) showing the ratio of H3K4me1 to H3K4me3 enrichment at ICRBS and non-ICRBS and control loci in WT and Zfp57 −/− E14 ESCs. The H3K4me1/H3K4me3 ratios of the Pou5f1 enhancer, Gapdh and Pou5f1 promoter, used as controls, have been indicated on the right as asterisks. (C) H3K27me3 (top) and H3K27ac (bottom) enrichment at ICRBS and non-ICRBS in WT and Zfp57 −/− E14 ESCs, as determined by ChIP. The Pou5f1 and Hoxa3 promoter were analysed as controls. Precipitated target loci with non-immune IgG were in the range of 0.002–0.02% of input. Note that ZFP57 inactivation differentially affects the enrichment of histone marks at ICRBS and non-ICRBS.
We then analysed H3K27me3 and H3-lysine-27-acetylation (H3K27ac). The enrichment of H3K27me3 was increased at several ICRBS and most non-ICRBS, in Zfp57 −/− E14 ESCs (Figure 4C, top panel). Concerning H3K27ac, we found that the enrichment of this mark decreased at ICRBS in Zfp57 −/− E14 ESCs. At non-ICRBS, the enrichment of H3K27ac was generally lower than at ICRBS in WT-E14 ESCs, and its level was slightly increased or not affected in Zfp57 −/− E14 ESCs (Figure 4C, bottom panel).
Overall, these data indicate that according to the histone marks, which were acquired upon Zfp57 inactivation in ESCs, the ZFP57 binding sites that do not overlap with ERVs may be divided in two broad groups. The first group, including most ICRBS, is characterized by a general increase of H3K4me3, a low H3K4me1/H3K4me3 ratio, loss of H3K27ac and often increase of H3K27me3. The second group, corresponding to most non-ICRBS, is characterized by acquisition of H3K4me1 and H3K27me3, and a high H3K4me1/H3K4me3 ratio.
Differential H3K4me1 and H3K4me3 enrichment at ICRBS and non-ICRBS in differentiated mouse tissues
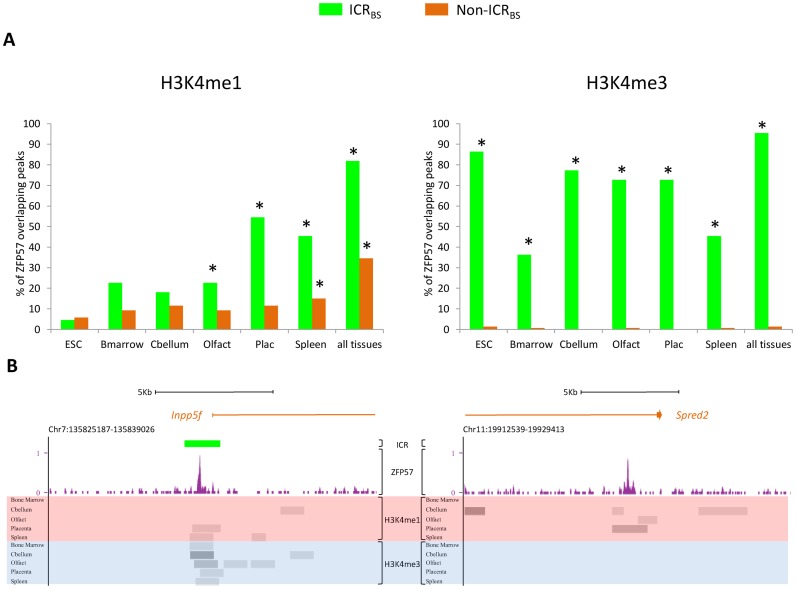
Zfp57 is poorly expressed in most adult mouse tissues (7). We therefore asked if the differential enrichment of H3K4me1 and H3K4me3 that we had observed in Zfp57 −/− ESCs between ICRBS and non-ICRBS was also present in adult tissues. To address this question, we took advantage of the genomic profiles of histone marks determined by the ENCODE project in mouse ESCs and differentiated adult tissues (23). We calculated the number of ICRBS and non-ICRBS (E14 ESCs) overlapping with H3K4me1 and H3K4me3 peaks and the probability to find this overlap. The result of this analysis is shown in Figure 5 and in Supplementary Table S9. We found significant (estimated P-value < 0.01) overlap of H3K4me1 immunoprecipitation peaks and both ICRBS and non-ICRBS in at least one differentiated tissue (Figure 5A). The percentage of overlapping peaks increased when five tissues were considered together, indicating that ZFP57 binding sites acquire H3K4me1 in a tissue-specific manner (Figure 5B). In contrast, H3K4me3 peaks consistently overlapped ICRBS, but not non-ICRBS, in all tested tissues. Therefore, ICRBS and non-ICRBS display different epigenetic marks also in differentiated adult tissues. As in undifferentiated ESCs lacking ZFP57, H3K4me1 is present at both ICRBS and non-ICRBS, while H3K4me3 is characteristic of only ICRBS (Figure 5B).
Figure 5.
ZFP57 binding sites and H3K4me1- and H3K4me3-enriched regions overlap in undifferentiated ESCs and differentiated tissues, as determined by in silico analysis. (A) Percentage of ICRBS and non-ICRBS ChIP-seq peaks overlapping genomic regions enriched with H3K4me1 and H3K4me3 in undifferentiated ESCs and differentiated tissues. *estimated P-value < 0.01. (B) Screen shots from the UCSC Genome Browser showing examples of overlaps between ICRBS (left panel) and non-ICRBS (right panel) and H3K4me1- and H3K4me3-enriched regions. Note that ICRBS overlap with both H3K4me1 and H3K4me3 while non-ICRBS generally overlap only with H3K4me1, in differentiated tissues.
Alteration of allele-specific gene expression at ZFP57-bound imprinted loci in Zfp57 −/− ESCs
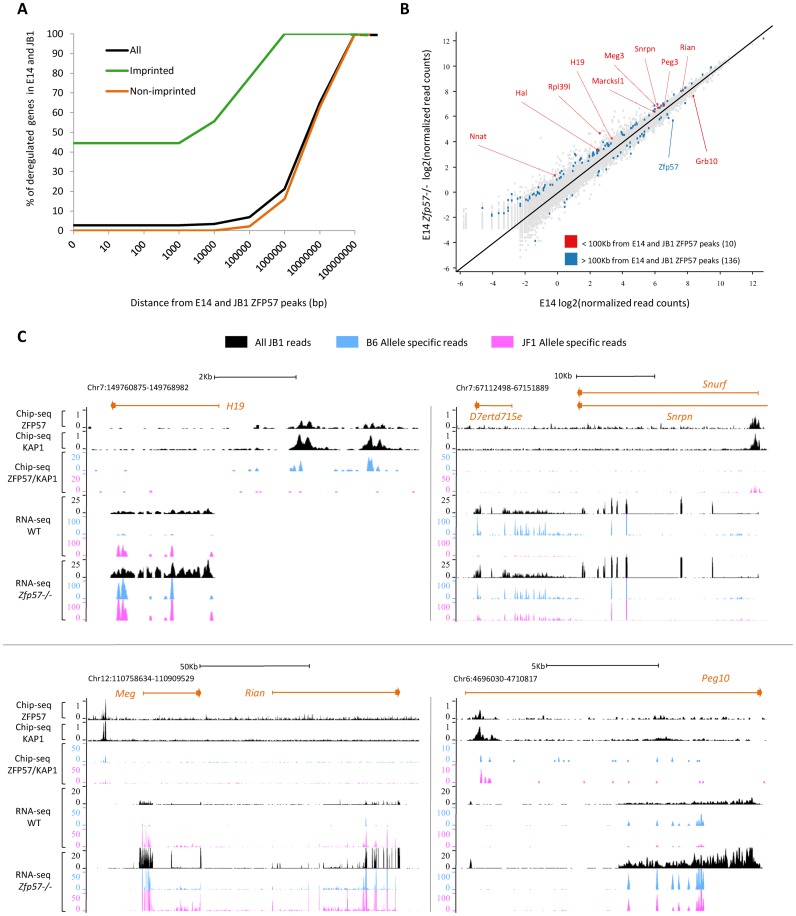
In our previous study, we demonstrated that only a few imprinted genes were deregulated in Zfp57 −/− A3 ESCs (9). In that study, however, only the global RNA level was measured, without distinguishing the two parental alleles of each gene. For this reason, we generated a further Zfp57 knockout (Supplementary Figure S4) in the mouse intra-specific hybrid ESC line JB1 (JF1 × B6; 30) that allows to discriminate the maternal from the paternal alleles of most genes. We first looked for concordantly deregulated genes in both Zfp57 −/− E14 and JB1 ESCs by global (non-allele-specific) RNA-seq analysis. By using this approach, 146 genes were identified (Supplementary Table S10). To further spot the genes whose deregulation more likely results directly from ZFP57 binding, we measured the distance of the deregulated genes from the overlapping E14/JB1 ZFP57 binding sites (N = 120 and Supplementary Table S5). We observed that only a few deregulated genes are located close to ZFP57 peaks and these correspond mostly to imprinted genes (Figure 6A and Supplementary Table S10). Indeed, we found only 10 deregulated genes (nine up- and one down-regulated) within 100 kb from the ZFP57 peaks, of which seven are imprinted (Figure 6B and Supplementary Table S10). These results were consistent with the previous study (9). We then analysed gene expression in WT and Zfp57 −/− JB1 ESCs by allele-specific RNA-seq mapping. By using this approach, the relative expression of the maternal and paternal alleles of 8127 genes overlapping the SNPs (see Materials and Methods) that are present between the B6 and JF1 genomes were determined. We found significant (corrected Fisher-exact test P-value < 0.05) differences in the JF1/B6 allelic expression ratio of 37 genes between WT and Zfp57 −/− JB1 ESCs (Supplementary Table S11). Ten of these deregulated genes were located closer than 100 kb from the mono-allelic ZFP57 binding sites determined by allele-specific ChIP-seq in the WT JB1 ESCs (Supplementary Table S12). These sites include regions with parent-of-origin-specific ZFP57 binding corresponding to the ICRBS and regions with strain-specific binding corresponding to the non-ICRBS (13). Strikingly, we found that 9 of the 10 genes were imprinted. All these imprinted genes but Igf2r had ZFP57 binding on the repressed allele in WT JB1 ESCs and this allele was relatively activated in Zfp57 −/− JB1 ESCs (Figure 6C and Supplementary Table S11). Interestingly, the imprinted expression of multiple genes (Meg3 and Rian; Snrpn and D7Ertd715e) within the Meg3 and Snrpn clusters was lost, demonstrating that ZFP57 exerts a long-range regulatory function on genomic imprinting (Figure 6C and Supplementary Table S13). Overall, the allele-specific RNA-seq analysis allowed to demonstrate that nine imprinted genes within seven clusters were transcriptionally deregulated in Zfp57 −/− ESCs (Supplementary Table S13). In contrast, close to the non-ICRBS, the relative expression of the JF1 and B6 alleles was found deregulated in only one gene (Eprs) and in this case the allele (JF1) not bound by ZFP57 was found slightly (2-fold) repressed (Supplementary Table S11). Overall, these results demonstrate that upon ZFP57 inactivation the imprinted gene expression is generally lost while the expression of non-imprinted target genes is mostly unaffected in undifferentiated ESCs.
Figure 6.
Effect of ZFP57 loss on gene expression and genomic imprinting in inbred and hybrid ESCs, determined by global and allele-specific RNA-seq analyses. (A) Empirical cumulative distribution of the distances (bp) of deregulated genes (all genes, black; imprinted genes, green; non imprinted genes, orange) from ZFP57 binding sites in both Zfp57 −/− E14 and JB1 ESCs. (B) Scatter plot showing deregulated genes in both Zfp57 −/− E14 and JB1 ESCs. Deregulated genes are indicated by red dots if distant <100 kb and blue dots if distant >100 kb from ZFP57 binding sites. (C) Screen shots from the UCSC Genome Browser showing the results of global and allele-specific RNA-seq analysis (bottom of each panel) in WT and Zfp57 −/− JB1 ESCs, and global and allele-specific ZFP57 and KAP1 ChIP-seq (top of each panel) in WT JB1 ESCs at the Snrpn, H19, Meg3 and Peg10 ICRBS. Global mapping is indicated in black, while allele-specific mapping is in blue (B6 allele) and pink (JF1 allele). Note that the imprinted alleles bound by ZFP57 become derepressed in Zfp57 −/− JB1 ESCs. Multiple genes are deregulated at Snrpn (Snrpn and D7Ertd715e) and Meg3 (Meg3 and Rian) ICRBS, indicating that ZFP57-bound ICRs control the imprinted expression of the entire clusters.
DISCUSSION
Zfp57 is a maternal-zygotic effect gene whose deficiency results in highly penetrant embryonic lethality around midgestation (7,31). In humans, ZFP57 mutation causes a complex form of Transient Neonatal Diabetes Mellitus-type 1 with multiple hypomethylation at imprinted loci (32). The function of this gene has not been investigated genome-wide so far. By using mouse ESCs as model, we demonstrate in this study that ZFP57 controls the expression and epigenetic features of large domains in the mouse genome. The binding of the ZFP57/KAP1 complex appears necessary for maintaining H3K9me3 at a large number of loci not overlapping with LTRs. However, the consequence of ZFP57 inactivation on acquisition of active histone marks and gene expression is dependent on the sequence context. ICRBS are characterized by dramatic mCpG loss, H3K4me3 acquirement and activation of imprinted alleles over long distances. Instead, non-ICRBS are characterized by moderate mCpG loss, H3K4me1 and H3K27me3 acquirement, possibly corresponding to poised enhancers that can be activated only in specific differentiated cell types.
Concerning genomic imprinting, we previously demonstrated that ZFP57 is necessary to maintain mCpG and H3K9me3 at several ICRs (9). The Zfp57 knockout system (16) employed in that study did not allow us to determine the effect of ZFP57 on about half of the ICRs, because the parental cell line had lost DNA methylation and ZFP57 binding at many loci. By using newly derived knockout ESC lines, we now demonstrate that ZFP57 binding is necessary to maintain differential methylation at all ICRs in mouse ESCs. Although they do not bind ZFP57, several somatic DMRs, including the H19 and Meg3 promoters, also lost mCpG in Zfp57 −/− ESCs, consistent with their dependence on ICR methylation. In apparent contrast with the dramatic mCpG loss found in ESCs, differential methylation is maintained at some imprinted loci in mice lacking both maternal and zygotic ZFP57 and in ESC lines directly derived from the knockout mice (7,33). This discrepancy is possibly explained by the incomplete penetrance of the mouse phenotype due to the limited time frame during which ZFP57 works or to the partial redundancy of imprinting maintenance factors in early embryogenesis, or by intrinsic differences present in the different ES cell lines used for analyzing the function of ZFP57 at the ICRs (6). Consistent with this hypothesis, a hypomorphic KAP1 mutation results in fully penetrant loss of the imprinted expression of 9 genes in mouse embryos (34).
Zfp57 inactivation in hybrid ESCs allowed us to also demonstrate that mono-allelic ZFP57 binding is required for the parent-of-origin-specific expression of single or multiple genes located in imprinted clusters. To our knowledge, this is the first report showing that ZFP57 is necessary to maintain imprinted gene expression. By including also two genes (Grb10 and Nnat) that were identified only by the global RNA-seq analysis likely because of the paucity of SNPs, we found deregulated gene expression in 9 imprinted domains in Zfp57 −/− ESCs (Supplementary Table S13). For most genes, the ZFP57-bound allele was activated. In the case of Igf2r and Grb10, the RNA levels of the ZFP57-bound (maternal) alleles were repressed 4-fold and 26-fold, respectively, consistent with the activating role of ICR methylation demonstrated on the expression of these two genes (1; see also Supplementary Table S11). The unaffected expression of the remaining hypomethylated imprinted loci is probably due to their tissue- and/or developmental stage-specific expression as well.
DNA sequences enriched with H3K9me3 are resistant to the DNA demethylation induced by treatment with ERK1/2 and GSK3β inhibitors (2i) that decrease the level of DNA methyltransferases, indicating that H3K9me3 contributes to the maintenance of DNA methylation in ESCs (35,36). However, we found that although all ZFP57 binding sites not overlapping with LTRs lose H3K9me3, only ICRBS become completely demethylated in serum-cultured Zfp57 −/− ESCs. In addition, only ICRBS acquire H3K4me3, likely on the ZFP57-bound allele, that is added to that already present on the non-CpG-methylated allele (14). This is likely mediated by CxxC-zinc finger proteins (e.g. CFP1) or other factors recognizing non-methylated CpG-rich regions and promoters and recruiting the SET1A/B H3K4 methyltransferases (37). Since H3K4 methylation prevents de novo DNA methylation by inhibiting DNMT3L (38), it is likely that this histone mark contributes to the maintenance of the non-methylated status at the ICRs. Thus, H3K9me3 and H3K4me3 appear as guardians of the differential methylation of the two parental alleles of the ICRs. ZFP57/KAP1 binding approximately at the centre of these regions appears critical for the maintenance of these epigenetic marks. Consistent with these observations, SETDB1 inactivation results in reduced mCpG and increased 5hmC specifically at ERVs and ICRs (39). The H3K4me3-bearing allele of the ICRs also displays H3K27me3 in mouse ESCs and embryos (40). Consistent with this finding, we found increased enrichment of H3K27me3 and decreased enrichment of H3K27ac at several ICRBS upon ZFP57 inactivation.
Concerning non-imprinted loci, we found that mCpG is moderately reduced and H3K4me1 is particularly increased at non-ICRBS upon ZFP57 loss. Also, the H3K4me1/H3K4me3 ratio at most of these sites is higher than at ICRBS and is similar to that of enhancers (23). Furthermore, the results obtained in TKO ESCs indicate that this differential H3K4me1/H3K4me3 ratio is not a consequence of the different mCpG levels but rather of the sequence characteristics of ICRBS and non-ICRBS. Interestingly, in silico analysis indicates that these epigenetic differences are present also in differentiated tissues, suggesting that non-ICRBS correspond at least in part to enhancers that are activated in specific cell types. The binding of ATRX (recruiting histone H3.3 and maintaining heterochromatin marks through cell division) to the ICRs but not to most of non-ICRBS (41; and data not shown) is consistent with the differential role played by these two groups of ZFP57 target sites in mouse ESCs. Luo et al. (42) recently reported that the transcription elongation factor AFF3 interacts with ZFP57 and is required for enhancer activity. We found that 78 AFF3 ChIP-seq peaks, including all ICRBS (the Blcap ICR has two ZFP57 peaks both colocalizing with AFF3) and 57 non-ICRBS, colocalize with ZFP57 binding sites in WT-E14 ESCs (Supplementary Table S5). AFF3 occupancy is decreased at 97% of these loci in Zfp57 −/− ESCs (Supplementary Table S5 and 42). These data support the hypothesis that ZFP57 may be involved in recruiting AFF3 at both ICRBS and non-ICRBS, possibly contributing to the inhibition of enhancer activity. Both global and allele-specific RNA-seq analyses demonstrated infrequent activation of genes close to non-ICRBS in Zfp57 −/− ESCs. This may possibly be explained by acquisition of the repressive H3K27me3 mark that is characteristic of poised enhancers at most of these sites (43).
Consistent with previously described data (12,13,44), we found binding of ZFP57 to some gDMRs that maintain differential methylation only in pre-implantation embryos (Zfp787) or in a tissue-specific manner (Cdh15, Nhlrc1 and Fkbp6). These loci are biallelically methylated in WT ESCs, and therefore not included in the ICRBS for the purpose of this study. These sites lose mCpG, but their hypomethylation is either incomplete or less extended than that of the ICRBS, and some of them (Cdh15 and Nhlrc1) display the histone marks of poised enhancers as other non-ICRBS in Zfp57 −/− ESCs.
A number of ZFP57 peaks overlap LTRs and it has been suggested that ZFP57 targets KAP1, which, in turn, is necessary for silencing of these loci (12,45). Although KAP1 binding is reduced, we observe maintenance of H3K9me3 and no gene activation at these loci in Zfp57 −/− ESCs. It is therefore likely that KAP1- and H3K9me3-mediated silencing of endogenous retroviral elements works through multiple targeting proteins of which ZFP57 is only one member.
Previous genome-wide studies have demonstrated the binding of ZFP57 to the methylated allele of the ICRs, identified its recognition sequence motif and addressed the influence of genetic variations on ZFP57 binding and associated epigenetic modifications and gene expression (9,12,13). Our new study explores for the first time the role of ZFP57 in DNA methylation, H3K9me3 and allele-specific expression along the entire mouse genome. The results obtained identify ZFP57 as a key protein for the maintenance of repressive epigenetic marks at both imprinted and non-imprinted target sites in undifferentiated mouse ESCs (Figure 7). Its binding to the methylated allele of the ICRs is pivotal to maintain the parent-of-origin-specific epigenetic marks and expression of the imprinted gene clusters. In addition to ICRs, ZFP57 binding contribute to repress a number of other cis-acting regulatory elements with the characteristic of enhancers that likely need to be activated at later developmental stages.
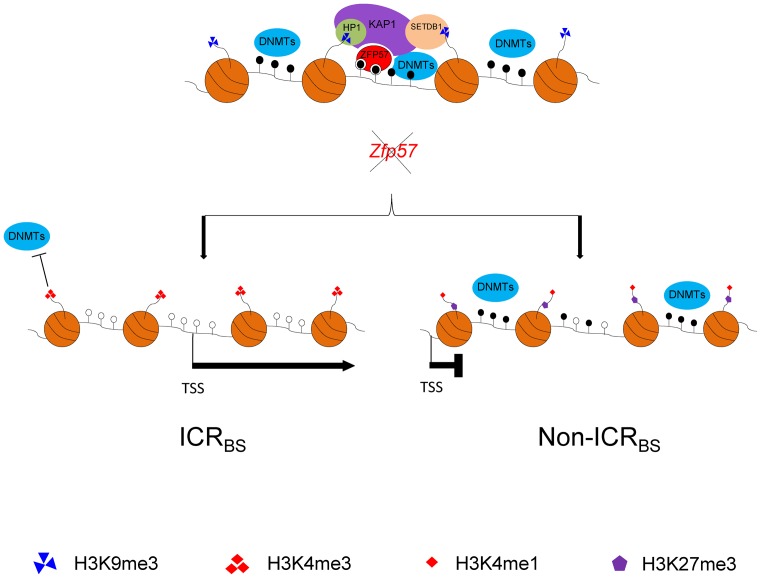
Figure 7.
Main epigenetic marks and gene expression changes at ICRBS and non-ICRBS (non-LTR) upon ZFP57 inactivation. For ICRBS, only the ZFP57-bound allele is shown. Note that KAP1 and H3K9me3 are lost at both ICRBS and non-ICRBS. However, ICRBS are characterized by dramatic mCpG loss, H3K4me3 acquirement and gene deregulation, while non-ICRBS are characterized by moderate mCpG loss, H3K4me1 and H3K27me3 acquirement and no consistent effect on gene expression.
Supplementary Material
Acknowledgments
We thank T. Yokota for the WT and Zfp57 −/− A3 ESC lines, Genesia Manganelli for help in ESC culturing and Andrea Oneglia, Flavia Cerrato and the other members of Andrea Riccio's lab for useful discussion on the manuscript.
Footnotes
Present addresses:
Zahra Anvar, Infertility Research Center, Department of Obstetrics and Gynecology, School of Medicine, Shiraz University of Medical Sciences, Shiraz, Iran.
Gaetano Verde, Programa de Recerca en Càncer, Institut Hospital del Mar d'Investigacions Mèdiques (IMIM), 08003 Barcelona, Spain.
Enza Lonardo, Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Baldiri Reixac, 10, 08028 Barcelona, Spain.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Telethon-Italia [GGP15131 to A.R.]; Associazione Italiana Ricerca sul Cancro [IG 2012 N.12815 to A.R.]; PON Ricerca e Competitività 2007–2013: Codice Progetto [PON02_ 00677–PON02_00619_3470457 to G.G.]; Progetto Bandiera MIUR-CNR Epigenomica (to A.R. and G.G.); EU-FP7-ITN INGENIUM [290123 to A.R., G.G., K.H. and R.F.]; Marie Curie fellowships from the EU-FP7-ITN INGENIUM (H.K., A.S. and S.L.); POR Campania [FSE 2007/2013 to V.R. and Z.A.]; Reti di eccellenza tra Università – Centri di Ricerca – Imprese, progetto ‘MODO – Model Organism’. Funding for open access charge: Fondazione Telethon.
Conflict of interest statement. None declared.
REFERENCES
- 1.Barlow D., Bartolomei M. Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 2014;6:a018382. doi: 10.1101/cshperspect.a018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weaver J.R., Bartolomei M.S. Chromatin regulators of genomic imprinting. Biochim. Biophys. Acta. 2014;1839:169–177. doi: 10.1016/j.bbagrm.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacDonald W.A., Mann M.R. Epigenetic regulation of genomic imprinting from germ line to preimplantation. Mol. Reprod. Dev. 2014;81:126–140. doi: 10.1002/mrd.22220. [DOI] [PubMed] [Google Scholar]
- 4.Smallwood S., Tomizawa S., Krueger F., Ruf N., Carli N., Segonds-Pichon A., Sato S., Hata K., Andrews S., Kelsey G. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 2011;43:811–814. doi: 10.1038/ng.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith Z., Chan M., Mikkelsen T., Gu H., Gnirke A., Regev A., Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelsey G., Feil R. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos. Trans. R Soc. Lond. B Biol. Sci. 2013;368:20110336. doi: 10.1098/rstb.2011.0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X., Ito M., Zhou F., Youngson N., Zuo X., Leder P., Ferguson-Smith A. A maternal-zygotic effect gene, ZFP57, maintains both maternal and paternal imprints. Dev. Cell. 2008;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Messerschmidt D., de Vries W., Ito M., Solter D., Ferguson-Smith A., Knowles B. Trim28 Is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335:1499–1502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- 9.Quenneville S., Verde G., Corsinotti A., Kapopoulou A., Jakobsson J., Offner S., Baglivo I., Pedone P.V., Grimaldi G., Riccio A., et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y., Toh H., Sasaki H., Zhang X., Cheng X. An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes Dev. 2012;26:2374–2379. doi: 10.1101/gad.202200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baglivo I., Esposito S., De Cesare L., Sparago A., Anvar Z., Riso V., Cammisa M., Fattorusso R., Grimaldi G., Riccio A., et al. Genetic and epigenetic mutations affect the DNA binding capability of human ZFP57 in transient neonatal diabetes type 1. FEBS Lett. 2013;587:1474–1481. doi: 10.1016/j.febslet.2013.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strogantsev R., Krueger F., Yamazawa K., Shi H., Gould P., Goldman-Roberts K., McEwen K., Sun B., Pedersen R., Ferguson-Smith A. Allele-specific binding of ZFP57 in the epigenetic regulation of imprinted and non-imprinted monoallelic expression. Genome Biol. 2015;16:112. doi: 10.1186/s13059-015-0672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anvar Z., Cammisa M., Riso V., Baglivo I., Kukreja H., Sparago A., Girardot M., Lad S., De Feis I., Cerrato F., et al. ZFP57 recognizes multiple and closely spaced sequence motif variants to maintain repressive epigenetic marks in mouse embryonic stem cells. Nucleic Acids Res. 2016;44:1118–1132. doi: 10.1093/nar/gkv1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanli I., Feil R. Chromatin mechanisms in the developmental control of imprinted gene expression. Int. J. Biochem. Cell Biol. 2015;67:139–147. doi: 10.1016/j.biocel.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Zuo X., Sheng J., Lau H., McDonald C., Andrade M., Cullen D., Bell F., Iacovino M., Kyba M., Xu G., et al. Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain. J Biol Chem. 2011;287:2107–2118. doi: 10.1074/jbc.M111.322644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akagi T., Usuda M., Matsuda T., Ko M., Niwa H., Asano M., Koide H., Yokota T. Identification of Zfp-57 as a downstream molecule of STAT3 and Oct-3/4 in embryonic stem cells. Biochem. Biophys. Res. Commun. 2005;331:23–30. doi: 10.1016/j.bbrc.2005.03.118. [DOI] [PubMed] [Google Scholar]
- 17.Azuma S., Toyoda Y. Production of a germ-line chimeric mouse derived from newly established embryonic stem cells. Jpn. J. Anim. Reprod. 1991;37:37–43. [Google Scholar]
- 18.Tsumura A., Hayakawa T., Kumaki Y., Takebayashi S., Sakaue M., Matsuoka C., Shimotohno K., Ishikawa F., Li E., Ueda H.R., et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- 19.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen Y., Yue F., McCleary D.F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V.V., et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quinlan A., Hall I. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie W., Barr C.L., Kim A., Yue F., Lee A.Y., Eubanks J., Dempster E.L., Ren B. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148:816–831. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song Q., Decato B., Hong E., Zhou M., Fang F., Qu J., Garvin T., Kessler M., Zhou J., Smith A.D. A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PLoS One. 2013;8:e81148. doi: 10.1371/journal.pone.0081148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edwards C.A., Ferguson-Smith A.C. Mechanisms regulating imprinted genes in clusters. Curr. Opin. Cell Biol. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 29.Takikawa S., Wang X., Ray C., Vakulenko M., Li X. Human and mouse ZFP57 proteins are functionally interchangeable in maintaining genomic imprinting at multiple imprintedregions in mouse embryonic stem cells. Epigenetics. 2013;8:1268–1279. doi: 10.4161/epi.26544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kota S., Llères D., Bouschet T., Hirasawa R., Marchand A., Begon-Pescia C., Sanli I., Arnaud P., Journot L., Girardot M., et al. ICR noncoding RNA expression controls imprinting and DNA replication at the Dlk1-Dio3 domain. Dev Cell. 2014;31:19–33. doi: 10.1016/j.devcel.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 31.Shamis Y., Cullen D.E., Liu L., Yang G., Ng S.F., Xiao L., Bell F.T., Ray C., Takikawa S., Moskowitz I.P., et al. Maternal and zygotic ZFP57 modulate NOTCH signaling in cardiac development. Proc Natl Acad Sci U.S.A. 2015;112:E2020–E2029. doi: 10.1073/pnas.1415541112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackay D.J., Callaway J.L., Marks S.M., White H.E., Acerini C.L., Boonen S.E., Dayanikli P., Firth H.V., Goodship J.A., Haemers A.P., et al. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat. Genet. 2008;40:949–951. doi: 10.1038/ng.187. [DOI] [PubMed] [Google Scholar]
- 33.Lau H.-T., Liu L., Li X. Zfp57 mutant ES cell lines directly derived from blastocysts. Stem Cell Res. 2016;16:282–286. doi: 10.1016/j.scr.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexander K.A., Wang X., Shibata M., Clark A.G., García-García M.J. TRIM28 Controls Genomic Imprinting through Distinct Mechanisms during and after Early Genome-wide Reprogramming. Cell Rep. 2015;13:1194–1205. doi: 10.1016/j.celrep.2015.09.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ficz G., Hore T.A., Santos F., Lee H.J., Dean W., Arand J., Krueger F., Oxley D., Paul Y.L., Walter J., et al. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell. 2013;13:351–359. doi: 10.1016/j.stem.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Habibi E., Brinkman A.B., Arand J., Kroeze L.I., Kerstens H.H., Matarese F., Lepikhov K., Gut M., Brun-Heath I., Hubner N.C., et al. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell. 2013;13:360–369. doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 37.Long H.K., Blackledge N.P., Klose R.J. ZF-CxxC domain-containing proteins, CpG islands and the chromatin connection. Biochem. Soc. Trans. 2013;41:727–740. doi: 10.1042/BST20130028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ooi S.K., Qiu C., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D., et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leung D., Du T., Wagner U., Xie W., Lee A.Y., Goyal P., Li Y., Szulwach K.E., Jin P., Lorincz M.C., et al. Regulation of DNA methylation turnover at LTR retrotransposons and imprinted loci by the histone methyltransferase SETDB1. Proc. Natl. Acad. Sci. U.S.A. 2014;111:6690–6695. doi: 10.1073/pnas.1322273111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maupetit-Méhouas S., Montibus B., Nury D., Tayama C., Wassef M., Kota S.K., Fogli A., Cerqueira Campos F., Hata K., Feil R., et al. Imprinting control regions (ICRs) are marked by mono-allelic bivalent chromatin when transcriptionally inactive. Nucleic Acids Res. 2016;44:621–635. doi: 10.1093/nar/gkv960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voon H.P., Hughes J.R., Rode C., De La Rosa-Velázquez I.A., Jenuwein T., Feil R., Higgs D.R., Gibbons R.J. ATRX Plays a Key Role in Maintaining Silencing at Interstitial Heterochromatic Loci and Imprinted Genes. Cell Rep. 2015;11:405–418. doi: 10.1016/j.celrep.2015.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo Z., Lin C., Woodfin A.R., Bartom E.T., Gao X., Smith E.R., Shilatifard A. Regulation of the imprinted Dlk1-Dio3 locus by allele-specific enhancer activity. Genes Dev. 2016;30:92–101. doi: 10.1101/gad.270413.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shlyueva D., Stampfel G., Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 44.Proudhon C., Duffié R., Ajjan S., Cowley M., Iranzo J., Carbajosa G., Saadeh H., Holland M.L., Oakey R.J., Rakyan V.K., et al. Protection against de novo methylation is instrumental in maintaining parent-of-origin methylation inherited from the gametes. Mol. Cell. 2012;47:909–920. doi: 10.1016/j.molcel.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rowe H.M., Jakobsson J., Mesnard D., Rougemont J., Reynard S., Aktas T., Maillard P.V., Layard-Liesching H., Verp S., Marquis J., et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.