Abstract
Retinoic acid-inducible gene I (RIG-I) recognizes specific molecular patterns of viral RNAs for inducing type I interferon. The C-terminal domain (CTD) of RIG-I binds to double-stranded RNA (dsRNA) with the 5′-triphosphate (5′-PPP), which induces a conformational change in RIG-I to an active form. It has been suggested that RIG-I detects infection of influenza A virus by recognizing the 5′-triphosphorylated panhandle structure of the viral RNA genome. Influenza panhandle RNA has a unique structure with a sharp helical bending. In spite of extensive studies of how viral RNAs activate RIG-I, whether the structural elements of the influenza panhandle RNA confer the ability to activate RIG-I signaling has been poorly explored. Here, we investigated the dynamics of the influenza panhandle RNA in complex with RIG-I CTD using NMR spectroscopy and showed that the bending structure of the panhandle RNA negates the requirement of a 5′-PPP moiety for RIG-I activation.
INTRODUCTION
To defend against viral infections, host immune systems employ a multitude of highly sophisticated pathways. As the first step in surveillance for viral infection, the pattern recognition receptors (PRRs) recognize molecular patterns of the viral pathogens, and then initiate a signaling cascade which culminates with induction of type I interferons (IFN) for antiviral activity (1). Retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), recognize double-stranded viral RNAs in the cytosol of cells (2). Three RLRs, RIG-I, melanoma differentiation associated factor 5 (MDA5) and laboratory of genetics and physiology 2 (LGP2), are known and belong to the DExD/H-box family of helicases. Among them, RIG-I has been reported as an important sensor of negative-sense RNA viruses such as influenza, hepatitis C, Sendai and vesicular stomatitis viruses, for induction of interferon beta (IFN-β) (3).
RIG-I consists of two N-terminal caspase recruitment domains (CARDs), a central RNA helicase domain, and a C-terminal RNA-binding domain. The C-terminal domain (CTD) of RIG-I recognizes the 5′-triphosphate (PPP) group of non-self RNAs and undergoes a conformational change to induce IFN-β production (4). Structural and biochemical studies have demonstrated that RIG-I CTD can bind to blunt-ended double stranded (ds) RNAs containing either PPP or a hydroxyl residue (OH) at the 5′ end (5,6). Although 5′-PPP dsRNA binds to the CTD more strongly and stimulates interferon production more effectively compared to 5′-OH dsRNA (7,8), it has been shown that RIG-I can be activated by non-triphosphorylated RNAs (9–12). Therefore, the mechanism underlying the discrimination of 5′-PPP or 5′-OH ends of dsRNAs by RIG-I is not fully understood.
The genome of the influenza A virus is composed of 8 segmented, single-stranded RNAs. Each RNA segment has a highly conserved sequence of 12 and 13 nucleotides at the 5′ and 3′ ends, respectively (Figure 1A) (13). The 5′- and 3′- end sequences form a partial duplex (referred to as panhandle RNA), and this panhandle region functions as a promoter for viral transcription and replication by interacting with RNA-dependent RNA polymerase (14,15). RIG-I was suggested to target the panhandle structure, leading to induction of interferons for antiviral activity (16–19). A recent study reported that defective interfering (DI) RNAs of the influenza virus, which were generated by deletion of genomic RNA and shorter genomic segments with the same ends, preferentially bound to RIG-I compared to the full-length genomic RNA (20). However, another study reported that 5′-PPP-containing full-length genomic RNAs were RIG-I agonists (17). Thus, it remains to be determined which type of RNA is a bona fide ligand for RIG-I activation during the course of physiological infection. Previously, we demonstrated that the panhandle structure had an internal loop and an unusual bending at the 5′-terminal stem (21). Whether this structural feature influences the recognition by RIG-I remains unexplored. Thus, it is of interest to determine which structural element—the 5′-PPP moiety or the fine structure at the terminal sequence of the RNA genome—is the more critical factor for recognition by the CTD of RIG-I.
Figure 1.
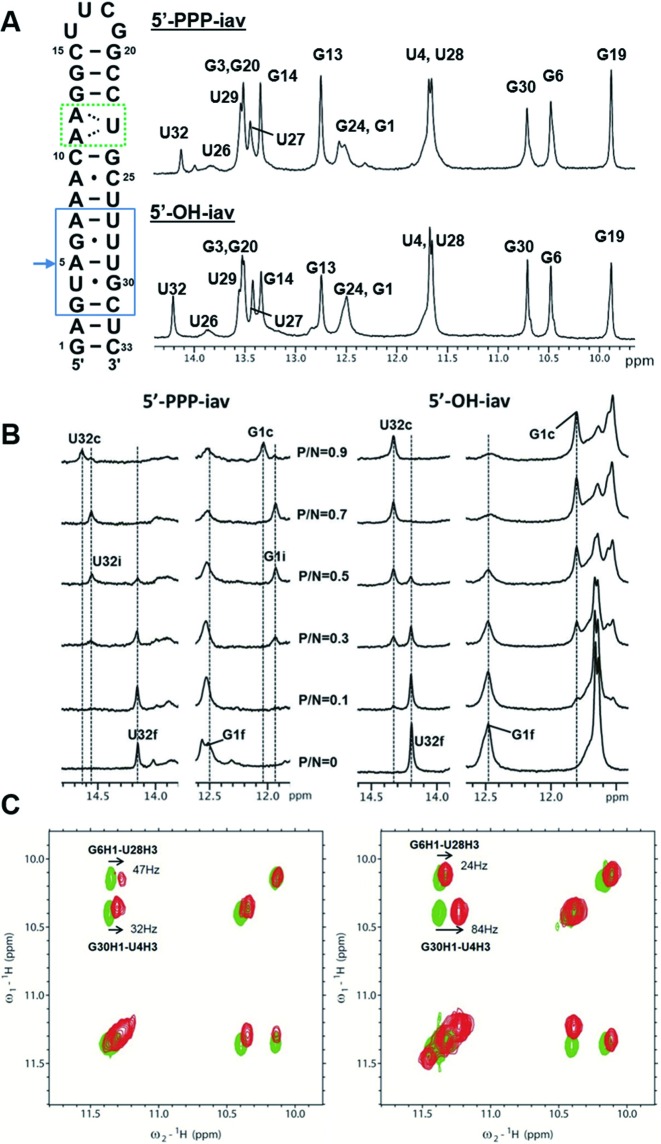
NMR experiments for influenza A virus RNA-RIG-I CTD complex. (A) The sequence and 1D proton NMR spectra of free influenza A virus (iav) panhandle RNA. The green dotted box indicates the (A11-A12)-U23 internal loop. The blue box indicates the region of the bent helix. The arrow indicates the pivot point of bending. (B) Titration of iav RNAs with RIG-I CTD. Spectral changes in the free and complexed forms of 5′-PPP-iav and 5′-OH-iav RNAs at various P/N ratios. Imino resonances are labeled as ‘f’ (free), ‘c’ (complex), and ‘i’ (intermediate). (C) Imino proton regions of 2D NOESYs of free and complexed RNAs. Left spectrum; free 5′-PPP-iav (green) and RIG-I CTD bound RNA (red). Right spectrum; free 5′-OH-iav (green) and RIG-I CTD bound RNA (red).
Using NMR spectroscopy, we investigated the structure and dynamics of the influenza promoter RNA complexed with RIG-I CTD with and without the 5′-PPP moiety. We showed that the RIG-I CTD formed a different complex with the RNA without the 5′-PPP moiety from that with the 5′-PPP moiety. Biophysical experiments suggested that the unique structure within the influenza genomic RNA could enhance binding to RIG-I in the absence of the 5′-PPP moiety. These structural features of the RNA also enabled the RIG-I-mediated immunogenic activity in cells without the 5′-PPP moiety. Our results suggest that RIG-I might be a dynamic protein that is capable of recognizing more complex structures within RNA than the 5′-PPP moiety.
MATERIALS AND METHODS
Preparation of RNAs and RIG-I CTD
RNAs with triphosphate (5′-PPP) were prepared by in vitro transcription using T7 RNA polymerase and plasmids containing T7 RNA polymerase promoter (Integrated DNA Technologies, Inc). The transcribed RNAs were purified by standard denaturing PAGE and dialyzed against NMR buffer (10 mM potassium phosphate pH 6.85, 50 mM NaCl and 1 mM DTT). Chemically synthesized 5′-OH RNAs (Integrated DNA Technologies, Inc) were purified by standard desalting and dialyzed against NMR buffer. The human RIG-I CTD (residues 802–925) was cloned into the bacterial expression vector pET-MBP (Novagen). The RIG-I CTD was expressed in E. coli BL21(DE3) by induction with 0.8 mM isopropyl-β-D-thiogalactoside, starting at OD600 of 0.6, overnight at 16°C. 15N-labeled or 14N-His6-MBP-RIG-I CTD was purified using nickel agarose resin (Qiagen) and then His6-MBP was removed by digestion with TEV protease. The protein was further purified using nickel agarose and HiTrapTM SP-sepharose columns (GE Healthcare).
NMR spectroscopy
Conventional NMR experiments used for spectral assignments were performed on INOVA 600 MHz and 700 MHz spectrometers (Agilent) with TXI-HCN cryogenic probes. 1D proton spectra, exchangeable 2D nuclear Overhauser effect spectroscopy (NOESY), and [15N,1H]-heteronuclear single quantum coherence (HSQC) spectra were recorded in 90% H2O/10% D2O, 10 mM potassium phosphate buffer (pH 6.85) with 50 mM NaCl and 1 mM DTT. The non-exchangeable 2D NOESY spectra were recorded in 100% D2O buffer at 298 K. Both exchangeable and non-exchangeable 2D NOESY spectra were collected at 150 ms and 300 ms mixing times to facilitate spectral assignments of the RNA proton resonances. The NMR data were processed and analyzed using NMRPipe (22) and Sparky (23).
Hydrogen exchange rate (kex) measurement
The apparent longitudinal relaxation rate constants (R1a = 1/T1a) of the imino protons of free 5′-PPP or 5′-OH RNAs and the RIG-I CTD bound RNAs at various P/N ratios were determined by semiselective inversion recovery 1D NMR experiments. The apparent relaxation rate constant of water (R1w) was determined by a selective inversion recovery experiment, using a DANTE sequence for selective water inversion (24). R1a and R1w were determined by curve fitting of the inversion recovery data to the appropriate single-exponential function. The hydrogen exchange rate constants (kex) of the imino protons were measured by a water magnetization transfer experiment. The intensities of each imino proton were measured with 20 different delay times. The kex values for the imino protons were determined by fitting the data to Equation (1), where I0 and I(t) are the peak intensities of the imino proton in the water magnetization transfer experiments at times zero and t, respectively, and R1a and R1w are the apparent longitudinal relaxation rate constants for the imino proton and water, respectively, measured in the semiselective inversion recovery 1D NMR experiments (25).
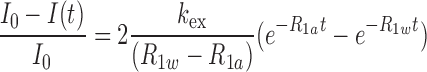 |
(1) |
RNA interference and quantitative real-time PCR (qRT-PCR)
hRIG-I and non-target control siRNAs were transfected into HEK 293T cells (3 × 106) with Lipofectamine LTX (Invitrogen). hRIG-I siRNA sequences are 5′-ACGGAUUAGCGACAAAUUUAA-3′ (sense) and 5′-UUAAAUUUGUCGCUAAUC CGU-3′ (antisense). Thirty six hours later, the cells were plated in 6 well plates and then stimulated with RNA ligands for 6 h. The cells were transfected with 1 μM RNA ligands (5′-PPP-iav, 5′-OH-iav, 5′-PPP-cont, 5′OH-cont, poly I:C, 5′-OH-NS1-80, 5′-OH-bend-GC) using Lipofectamine LTX. Total RNA was isolated from cells using TRI-reagent (Ambion). cDNA were synthesized using the SuperScript III enzyme (Invitrogen). Quantitative real-time PCR (qRT-PCR) analysis was performed using EvaGreen Dye (Biotium) and following primers: hIFN-β forward, 5′-GGAGGACGCCGCATTGAC-3′, hIFN-β reverse, 5′-CAATAGTCTCATTCCAGCCAGTGC-3′; hRIG-I forward, 5′-GCATTGCCCTCAACGACCAC-3′ hRIG-I reverse, 5′-GAGGCCATGTGGGCCATGAG-3′. For the hISG56 (human interferon-stimulated gene 56) expression test, qRT-PCR analysis was performed using EvaGreen Dye (Biotium), h-taq DNA polymerase (Solgent) and following primers: hISG56 forward, 5′- GCCTCCTTGGGTTCGTCTACAA-3′, hISG56 reverse, 5′-GAGGCCATGTGGGCCATGAG-3′, human glyceraldehyde 3-phosphate dehydrogenase (hGAPDH) forward, 5′-GCATTGCCCTCAACGACCAC-3′ hGAPDH reverse, 5′-GAGGCCATGTGGGCCATGAG-3′ hRIG-I forward, 5′-GCATTGCCCTCAACGACCAC-3′ hRIG-I reverse, 5′-GAGGCCATGTGGGCCATGAG-3′. hISG56 and hRIG-I expression data were normalized to hGAPDH.
Isothermal titration calorimetry experiment
Calorimetric titrations of RIG-I CTD into RNAs were performed on a VP-ITC microcalorimeter (MicroCal) at 25°C. Protein and RNA samples were dialyzed against reaction buffer containing 10 mM potassium phosphate (pH 6.85), 50 mM NaCl and 5 mM β-mercaptoethanol. The 1.4 mL sample cell was filled with a 5 μM solution of RNA and the 300 μl injection syringe with 50 μM of the titrating RIG-I CTD. Binding isotherms were generated by plotting heats of reaction normalized by the modes of injectant versus the ratio of total injectant to total RNA per injection. The data were fit using the one site binding site model embedded in Origin 7.0 (MicroCal).
RESULTS
Different binding modes of 5′-PPP-iav RNA and 5′-OH-iav RNA to RIG-I CTD
The influenza panhandle RNA bearing the 5′-PPP moiety (5′-PPP-iav) was designed by addition of a single G·C base pair to facilitate the synthesis of the RNA by in vitro transcription (Figure 1A). We also employed the UUCG tetraloop to help RNA to stably fold into a hairpin structure (Figure 1A). Previously, we determined the structure of the influenza panhandle RNA which was composed of two helical stems, the (A11-A12)-U23 internal loop, and the UUCG tetraloop (21). The structure of the panhandle RNA showed an A-form helix with a helical bend of ∼46° at the A5·U29 base pair. The 5′-PPP-iav and a chemically synthesized RNA without the 5′-PPP moiety (5′-OH-iav) showed similar 1D imino proton spectra (Figure 1A) with minor differences in the chemical shifts of the G1 and U32 between the two RNA species.
The changes in the 1D imino proton spectra of the 5′-PPP-iav and 5′-OH-iav upon binding of RIG-I CTD was examined at various RIG-I protein-to-RNA (P/N) molar ratios (Supplementary Figure S1). Most of the imino resonances from the iav RNAs showed chemical shift perturbations upon binding of RIG-I CTD. Notably, the imino proton peaks of terminal bases, G1 and U32, showed the dramatic chemical shift perturbations. Various residues between the terminal stem and the nearby (A11-A12)-U23 internal loop (G3, U4, G6, G24, U27, U28 and G30) also showed changes in chemical shifts or peak intensities upon addition of the RIG-I CTD, whereas the binding of RIG-I CTD had no effects on the resonances in the UUCG loop and its proximal helix (G13, G14, G19 and G20). This indicates that the RIG-I CTD predominantly associates with the terminal base pairs of the panhandle RNA, consistent with the complex structures of RIG-I CTD and duplex RNAs (5,6).
The 1D proton NMR spectra showed different complex formation with the 5′-PPP-iav and 5′-OH-iav (Figure 1B). As the molar ratio of the RIG-I CTD increased, G1 and U32 of the 5′-PPP-iav shifted from the free state (G1f and U32f) to the RIG-I-bound state (G1c and U32c). Notably, additional peaks (G1i and U32i) were observed during the titration of RIG-I CTD. However, G1 and U32 of the 5′-OH-iav shifted from the free state to the bound state without having additional peaks. (Figure 1B), suggesting that RIG-I CTD might differentiate the 5′-PPP moiety from a 5′-OH moiety through an intermediate state. In addition, the wobble base pairs (U4·G30 and G6·U28) adjacent to the helical bending at A5·U29 showed different 2D NOESY cross-peaks between the 5′-PPP-iav and the 5′-OH-iav (Figure 1C). The G30-H1↔U4-H3 cross-peak of 5′-OH-iav RNA showed a larger change in chemical shift (84 Hz) when transitioning from the free state to the RIG-I-bound state than that of the 5′-PPP-iav (32 Hz). The G6-H1↔U28-H3 cross-peak in 5′-OH-iav RNA shifted less (24 Hz) than that of 5′-PPP-iav RNA (47 Hz). These differences in the chemical shift perturbations between 5′-OH iav and 5′-PPP-iav suggested that RIG-I CTD might differentiate 5′-PPP-iav from 5′-OH-iav through a different binding mechanism.
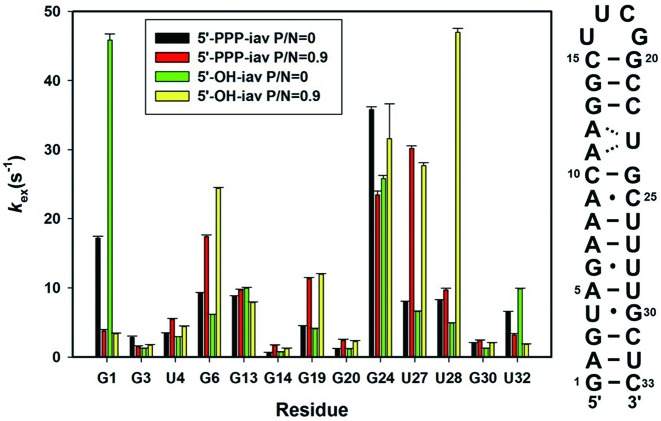
RIG-I-CTD binding stabilizes the terminal base-pairing of iav RNAs
It is well known that imino protons of RNA will exchange with solvent protons at a characteristic rate (24). Some imino protons in solvent-accessible or flexible region of RNA will exchange quickly. On the contrary, the imino protons in highly structured regions or in stable base-pairs will exchange slowly. To investigate the dynamics and the structures of the panhandle RNAs in the presence of RIG-I CTD, we measured the hydrogen exchange rate constants (kex) of imino protons in RNAs. The kex values for the imino protons of the free and RIG-I CTD-bound RNAs at various P/N ratios were determined at 25°C by the water magnetization transfer method (24). We observed the stabilization of the terminal base pairs by comparing the kex values of the free states and RIG-I-bound states of 5′-PPP-iav and 5′-OH-iav. The G1 resonances of both iav RNA species showed a dramatic decrease in kex upon binding of the RIG-I CTD (Figure 2 and Supplementary Figure S2). Similarly, the U32 resonances of both iav RNAs showed the reduced kex upon binding of RIG-I CTD, though the changes were smaller. The reduced kex values in G1 and U32 indicated that RIG-I CTD stabilized the two terminal base pairs (G1·C33 and A2·U32) of both 5′-PPP-iav and 5′-OH-iav. In contrast, the 4th to 7th base pairs from the terminus including the helical bend (U4, G6, U27, U28 and G30) and the trans-wobble base pair from the UUCG tetraloop (G19) showed the increased kex upon binding of RIG-I CTD, indicating the destabilization of the base pairs upon binding of RIG-I CTD (Figure 2 and Supplementary Figure S2) (26). Of note, the G6 and U27 of both iav RNAs showed significantly increased kex values. The increased kex in G19 suggested that the binding of RIG-I CTD destabilized the trans-wobble base pair (U16·G19) of the UUCG tetraloop. These results showed that RIG-I CTD stabilized the two terminal base pairs (G1·C33 and A2·U32) but destabilized the base pairs near the helical bend regardless of the presence of the 5′-PPP moiety. The kex of the G13, G14 and G20 in both iav RNAs did not significantly change, indicating that the proximal stem of the UUCG tetraloop was not greatly affected by RIG-I CTD binding. Thus, the destabilization of U16·G19 base pair upon binding of RIG-I CTD might not be significant and might be an indirect consequence of the binding event.
Figure 2.
Determination of hydrogen exchange rates in iav-RNAs. The sequence of iav RNA and kex values of free and complexed forms of influenza A virus RNAs.
We also observed differences in kex between the RIG-I-bound states of 5′-PPP-iav and 5′-OH-iav. The U28 located in the helical bend of the 5′-PPP-iav showed slightly increased kex compared to the free 5′-PPP-iav, whereas the kex of U28 in the RIG-I-bound 5′-OH-iav was dramatically increased from that of the free 5′-OH-iav (∼9.6 fold) (Figure 2). In the case of G24 which is located next to the (A11-A12)-U23 internal loop, the kex in the 5′-PPP-iav decreased upon binding of RIG-I CTD, whereas the kex of the 5′-OH-iav RNA increased. In line with the results of our NOESY spectra experiment (Figure 1C), these results suggest that RIG-I CTD interacts differently with 5′-PPP-iav and 5′-OH-iav RNA at the helical bend and internal loop structures.
The helical bend and internal loop in the panhandle RNA is critical for RIG-I CTD binding
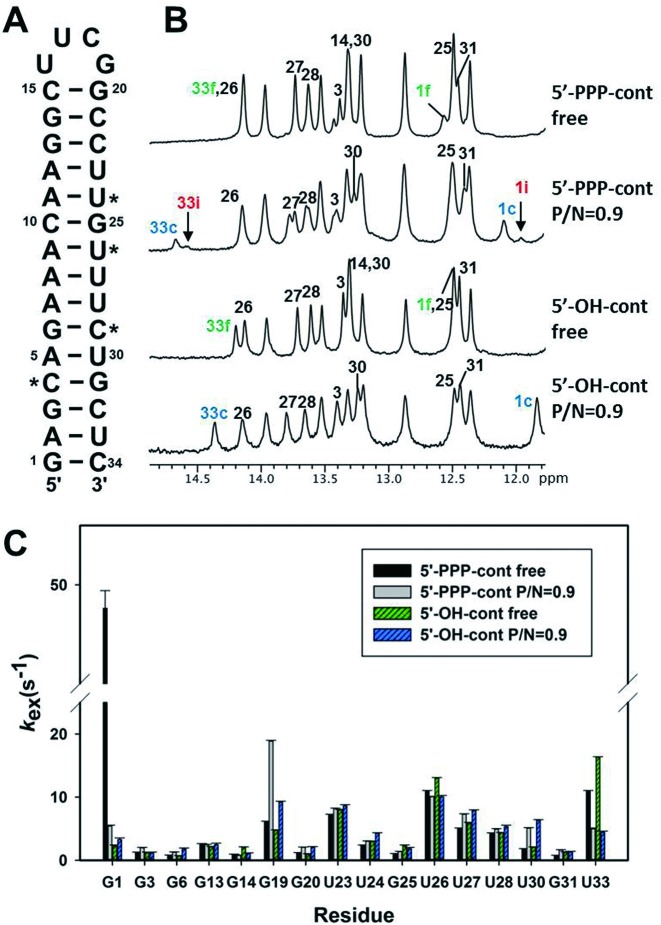
As shown in Figure 2, the kex values of residues in the bent helical stem (U4, G6, U27 and U28) or near the internal loop (G24) of iav RNAs bound to RIG-I CTD were significantly different between the 5′-PPP- or 5′-OH-iav RNAs. Thus, we investigated the effect of the bend and the loop structures by employing new hairpin RNA constructs (referred to as control RNAs) which eliminated the internal loop and the wobble base pairs by replacing them with the complementary Watson–Crick base pairs (Figure 3A). We designed these control RNAs with and without the 5′-PPP moiety (5′-PPP-cont and 5′-OH-cont, respectively). First, the imino resonances of the control RNAs were completely assigned by 2D NOESY. Free control RNAs produced different 1D proton NMR spectra than the iav RNAs (Figures 1A and 3B). The G1 and U33 resonances at the terminal end of both 5′-PPP-cont and 5′-OH-cont showed significant changes in chemical shifts upon binding of RIG-I CTD. Similar to the interaction between the iav RNA and RIG-I CTD (Figure 1B), only 5′-PPP-cont showed additional peaks (1i and 33i) of G1 and U33 during the titration of RIG-I CTD (Figure 3B). This confirmed that RIG-I CTD might employ different mechanisms to distinguish between 5′-PPP RNA and 5′-OH RNA.
Figure 3.
Determination of hydrogen exchange rates in cont RNAs. (A) The sequence of the cont RNA. The residues of iav RNA that were changed to C or U in cont RNA, for removal of helical bend and the internal loop, are starred. (B) 1D imino proton NMR spectra of free cont RNAs (5′-PPP-cont and 5′-OH-cont) and RIG-I CTD-bound cont RNAs. The residues that showed spectral changes upon RIG-I CTD binding are labeled with numbers. Intermediate peaks of G1 and U33 of 5′-PPP-cont RNA are labeled with ‘i’ in red color. As in Figure 1B, c (blue) and f (green) indicate the complex and the free form, respectively. (C) The kex values of free and bound cont RNA.
The G1 and U33 imino peaks of 5′-PPP-cont RNA showed large decreases in kex upon RIG-I CTD binding (Figure 3C). In particular, the kex of the G1 resonance of the bound 5′-PPP-cont RNA dramatically decreased to 11% of the free state. We could not accurately determine the kex of G1 in the 5′-OH-cont due to severe spectral overlap with G25 peak. However, the U33 peak of the 5′-OH-cont RNA showed a ∼73% decrease in kex upon binding of RIG-I, suggesting that the RIG-I CTD stabilized the terminal base pairs in both control RNAs. In contrast with the iav RNAs, most of the helical residues in the control RNAs showed relatively small changes in kex (1–3 s−1) upon binding of RIG-I. These results indicated that the RIG-I CTD binding to the iav RNA significantly affected the dynamics of the helical bend and internal loop structures, and these non-canonical structures might be important for interaction between iav RNA and RIG-I CTD.
RIG-I CTD uses alternative binding interfaces for the bent 5′-OH-iav RNA
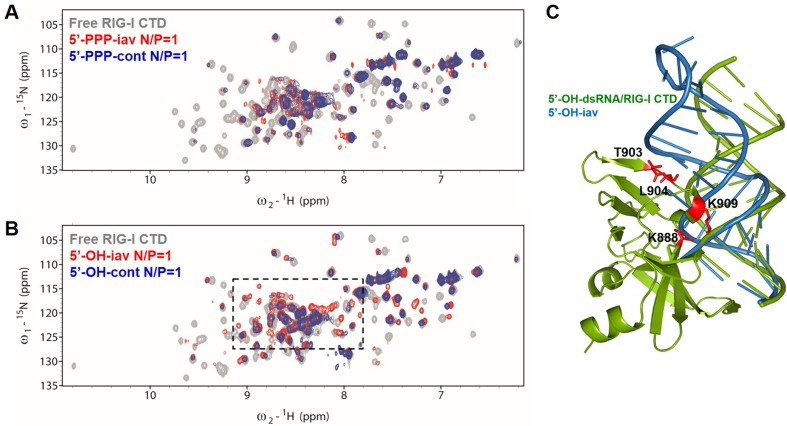
To investigate binding modes of RIG-I CTD against the iav RNA and the control RNA, we monitored the chemical shift perturbations in 15N-1H HSQCs of RIG-I CTD upon the addition of RNAs to the 15N-labeled protein. Amide proton peaks of RIG-I CTD significantly changed in intensity and chemical shift compared to the free protein (Supplementary Figures S3 and S4). First, all four RNAs (5′-PPP-iav, 5′-OH-iav, 5′-PPP-cont, and 5′-OH-cont) induced significant line-broadening upon binding to RIG-I CTD (Figure 4 and Supplementary Figures S3 and S4). The line-broadening might suggest that the interaction between the RIG-I CTD and the RNA is an intermediate exchange process on the NMR chemical shift timescale. The spectra of RIG-I CTD complexed with 5′-PPP-iav showed similar chemical shifts to the RIG-I CTD complexed with 5′-PPP-cont (Figure 4A), suggesting that the complex formation with the RIG-I CTD was similar between the 5′-PPP-iav and the 5′-PPP-cont. On the other hand, the HSQC spectra RIG-I CTD complexed with 5′-OH-iav and 5′OH-cont showed different chemical shift perturbations, as indicated by dotted boxes in the Figure 4B. In addition, NMR peaks of the RIG-I CTD complexed with 5′-OH-iav showed dramatic differences in chemical shifts from the RIG-I CTD complexed with 5′-PPP-iav (Supplementary Figure S4). These spectral differences between the RIG-CTDs complexed with 5′-OH-iav and 5′-PPP-iav show that RIG-I CTD might form a complex with 5′-OH-iav different from other three RNAs.
Figure 4.
NMR titration of 15N-labeled RIG-I CTD with iav RNAs and cont RNAs. (A) 15N-1H-HSQC spectra of free RIG-I CTD, 5′-PPP-iav/RIG-I CTD complex and 5′-PPP-cont/RIG-I CTD complex (gray, free protein; red, 1:1 complex with 5′-PPP-iav; blue, 1:1 complex with 5′-PPP-cont). N/P indicates RNA-to-Protein molar ratio. (B) 15N-1H-HSQC spectra of free RIG-I CTD, 5′-OH-cont/RIG-I CTD complex, and 5′-OH-iav/RIG-I CTD complex (gray, free protein; red, 1:1 complex with 5′-OH-iav; blue, 1:1 complex with 5′-OH-cont). Amide protons in the dotted boxes revealed dramatic changes in chemical shift upon RIG-I CTD binding to 5′-OH-iav. (C) Model of 5′-OH-iav/RIG-I CTD complex structure. Structure of iav RNA (blue, PDB 1JO7) was superimposed on 5′-OH-14bp-GC-dsRNA/RIG-I CTD complex (green, PDB 3OG8). Mutations at RIG-I CTD (K888, T903, L904 and K909) are indicated in red.
To explore binding interfaces between RIG-I CTD and RNAs, we first measured the binding affinities of RIG-I CTD against the RNA constructs using isothermal titration calorimetry (ITC; Table 1). Remarkably, 5′-OH-iav showed a stronger binding affinity (Kd = 10.6 ± 2.6 nM) than 5′-PPP-iav (96.1 ± 24.6 nM). In contrast, 5′-OH-cont showed weaker binding affinity (75.1 ± 26.2 nM) than 5′-PPP-cont (12 ± 3.8 nM), suggesting that the internal loop and/or the bent helix in the iav RNAs were critical for strong binding to RIG-I CTD without the 5′-PPP moiety. The wild type (WT) RIG-I CTD binding to the iav RNAs showed a favorable ΔG, (−9.57 to −10.88 kcal mol−1) with favorable enthalpy (ΔH = −10.29 to −17.71 kcal mol−1). Interestingly, only 5′-OH-iav showed favorable entropy (TΔS = +0.59 kcal mol−1), implying that 5′-OH-iav structure is energetically favorable for the complex formation with RIG-I CTD, comparable to the 5′-PPP RNAs.
Table 1. Thermodynamic parameters of ITC titrations.
| RIG-I CTD | Parameters | 5′-OH-iav | 5′-OH-cont | 5′-PPP-iav | 5′-PPP-cont |
|---|---|---|---|---|---|
| K888A | Kd (nM) | nda | nd | - | - |
| T903A | Kd (nM) | 79.4 ± 11.3 (∼7.5)b | 44.2 ± 4.5 (∼0.59) | - | - |
| L904A | Kd (nM) | 67.6 ± 22.9 (∼6.4) | 39.7 ± 13.9 (∼0.53) | - | - |
| K909A | Kd (nM) | 122 ± 2.3 (∼11.5) | 110 ± 13.1 (∼1.5) | - | - |
| WTc | Kd (nM) | 10.6 ± 2.6 | 75.1 ± 26.2 | 96.1 ± 24.6 | 12 ± 3.8 |
| WT | ΔH (kcal mol−1) | −10.29 ± 0.1 | −10.52 ± 0.3 | −17.71 ± 0.4 | −13.02 ± 0.2 |
| WT | TΔS (kcal mol−1) | 0.59 | −0.80 | −8.14 | −2.21 |
| WT | ΔG (kcal mol−1) | −10.88 ± 0.1 | −9.72 ± 0.3 | −9.57 ± 0.4 | −10.81 ± 0.2 |
aNot determined.
bThe number in the blank represents the ratio of Kd between the mutant and the WT.
cWild type RIG-I CTD.
To further evaluate the binding interface of RIG-I CTD against 5′-OH-iav, we designed RIG-I CTD mutants. Mutated residues of RIG-I CTD were selected based on previous mutagenesis studies and our model of RIG-I CTD complexed with the iav RNA (Figure 4C). We generated four mutant RIG-I CTD (K888A, T903A, L904A and K909A) and measured binding affinities of 5′-OH-iav and 5′-OH-cont against the mutant proteins. First, K888A mutant dramatically abolished binding affinities of both 5′-OH-RNAs. This was consistent with previous studies demonstrated that the mutation in K888 abolished binding with blunt-ended dsRNAs and 5′-PPP dsRNAs (6,27). We also showed that K909A mutant weakened the binding affinities of 5′-OH-iav (∼12-fold increase in Kd) and 5′-OH-cont (∼1.46-fold increase in Kd). This was also consistent with the previous studies that K909 forms a solvent-mediated hydrogen bond between its amine and the phosphodiester backbone from the dsRNA (27), and the mutation in K909 that disrupts the hydrogen bond reduced the binding affinity of the dsRNA against RIG-I (6). According to our model (Figure 4C), residue K909 was close to both 5′- and 3′-strand backbones of 5′-OH-iav due to the bent RNA helix, suggesting that the bent helix in 5′-OH-iav was responsible for stronger binding affinity with wild type RIG-I CTD (∼10.6 nM) than 5′-OH-cont (∼75.1 nM). As a result, K909A mutant might disrupt the interaction with both strands of 5′-OH-iav, resulting in larger increase in Kd for 5′-OH-iav than 5′-OH-cont. Of note, both T903A and L904A mutants inhibited binding of 5′-OH-iav to RIG-I CTD (∼ 7.5- and ∼6.4-fold increase in Kd, respectively), whereas the interaction between 5′-OH-cont and the mutant protein was not much affected. T903 and L904 are remarkably close to the helical stem of 5′-OH-iav in our model, whereas 5′-OH-cont is bound away from these residues. Taken together, our results implied that the helical bend of 5′-OH-iav might play a pivotal role for the recognition by RIG-I CTD.
The terminal helical bend of RNA activates the RIG-I dependent IFN-β production
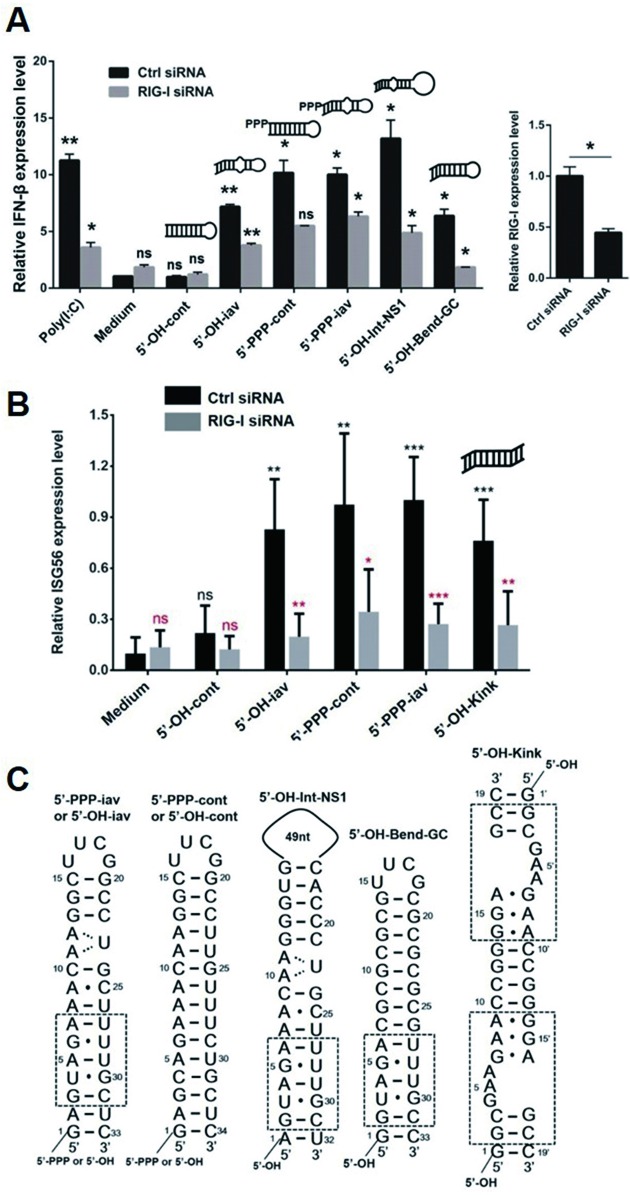
To elucidate whether the structural features of iav RNAs have any effect on stimulating RIG-I signaling, we performed a quantitative real-time PCR to measure the RIG-I-mediated immunogenic response upon treatment of iav RNAs. The 5′-PPP and 5′-OH forms of iav and cont RNA were transfected into cells to analyze their ability to activate RIG-I-dependent IFN-β induction. The activity of the triphosphorylated RNAs, 5′-PPP-iav and 5′-PPP-cont, were similar to each other and higher than 5′-OH-cont RNA as expected (Figure 5A and Supplementary Table S1). However, the blunt-ended 5′-OH-iav RNA induced IFN-β expression similarly to the 5′-PPP-iav and 5′-PPP-cont. This suggested that (i) the unique structure, such as the bend, within 5′-OH-iav RNA rendered it an effective agonist to activate RIG-I signaling pathway, comparable to the 5′-triphosphorylated RNAs, and (ii) the 5′-PPP of dsRNA was not essential for activating RIG-I-dependent signaling if the RNA could bind to RIG-I via other structural features. In addition, we tested IFN-β inducibility of 5′-OH panhandle RNA with intact NS1 gene sequences including terminal A-U base pair and 49-nucleotide-long loop (5′-OH-Int-NS1). 5′-OH-Int-NS1 induced IFN-β expression comparably to 5′-OH-iav RNAs (Figure 5A and Supplementary Table S1) indicating that neither the terminal G·C pair nor the length of the loop in iav RNA was critical for inducing IFN-β expression. In order to examine the effect of the bent helix on induction of IFN-β, we designed the G-C base pair rich dsRNA (5′-OH-Bend-GC) bearing the bent helix originated in influenza panhandle RNA (G3-C31 to A7-U27 base pairs), which represented the helical bend in the solution structure (Figure 5C and Supplementary Figure S5). Interestingly, 5′-OH-Bend-GC showed significant IFN-β induction without the 5′-PPP moiety (Figure 5A, Supplementary Table S1), suggesting that the bent helix of panhandle RNA, even without internal loop, might play a crucial role in RIG-I dependent IFN-β signaling.
Figure 5.
Quantitative real-time PCR (qRT-PCR). (A) The 5′-PPP and 5′-OH forms of panhandle RNAs (1 μM) were tested for their ability to induce RIG-I dependent IFN-β expression by a quantitative real-time PCR (qRT-PCR) (left). RIG-I siRNA was transfected into the cells for RIG-I knockdown (right). The transfection agent, PEI (medium) and a viral dsRNA mimetic, poly (I:C), were included as negative and positive controls, respectively. (B) The ISG56 expression test of iav RNAs, cont- RNAs, and 5′-OH-Kink. Representative data from three independent experiments are shown. *P < 0.05; **P <0.01; ns, not significant. Error bars, s.d. Black asterisks indicates the comparison to the medium only. Red asterisks indicates the comparison to the control siRNA. (C) Full sequences of RNAs used for the stimulation. The sequences in the dotted box represented the helical bend or kink structure.
To further confirm that the bent helix in the iav RNA induced the RIG-I-mediated immunogenic activity in the absence of the 5′-PPP moiety, we synthesized a RNA construct containing a kink-turn motif which contained two RNA helices connected by a trinucleotide bulge (5′-OH-Kink, PDB 4CS1). The high resolution structure of 5′-OH-Kink determined by X-ray crystallography showed a bent helix similar to that found in the iav RNA (Supplementary Figure S5) (28). Interestingly, 5′-OH-Kink induced the expression of ISG56 in a RIG-I-dependent manner (Figure 5B and C, Supplementary Table S2), suggesting that the helical bending in RNA might play a crucial role in RIG-I-dependent immunogenic activity.
DISCUSSION
According to the previously reported complex structures of RIG-I CTD and various RNAs, the 5′-PPP of dsRNA interacts with positively charged residues of RIG-I such as lysine or histidine (5,6). The dsRNA stabilizes the binding interface through electrostatic interactions between its backbone and RIG-I. The panhandle structure within the genomic RNA of influenza A virus, which also has a 5′-PPP and a blunt-ended double-stranded structure, is an appropriate agonist for RIG-I. It is well known that the binding affinity between RIG-I CTD and 5′-PPP-containing dsRNA is higher than that of 5′-OH-dsRNA (8). In this regard, we measured the binding affinity between the influenza panhandle RNAs, 5′-PPP-iav and 5′-OH-iav RNA, and RIG-I CTD. Surprisingly, the absence of 5′-PPP did not decrease but rather enhanced the binding affinity for RIG-I CTD. This implies that the helical bend in the terminal stem, which is different from the double-stranded structure of typical RIG-I agonists, is more important for interaction with the RIG-I CTD than the 5′-PPP moiety. Given that the Kd of 5′-PPP-iav RNA is similar to that of 5′-OH-cont RNA, but the former can induce IFN-β production whereas the latter cannot, the high binding affinity alone cannot fully explain the better stimulatory activity of the iav RNA.
The crystal structures of the human RIG-I CTD complexed with a dsRNA showed that the bound conformations of 5′-PPP dsRNA and the 5′-OH dsRNA are different in contact sites and orientation (27). The 5′-PPP moiety in the 5′-PPP dsRNA interacts with the positively charged Lys and His residues in the cleft of the CTD through electrostatic interactions (27). The first four nucleotides at the 5′-end, but not the complementary strand, are the key nucleotides for the 5′-PPP dsRNA to form a complex with the RIG-I CTD. On the other hand, the 5′-OH RNA makes alternative contacts with the RIG-I CTD which tilted the RNA helix toward the protein by ∼15° with respect to the 5′-PPP dsRNA. Specifically, R811 and H817 of the RIG-I CTD interact with the complementary 3′-strand of the dsRNA (27). To understand the significance of the bent RNA helix in the recognition of the 5′-OH-iav by the RIG-I CTD, we superimposed the structure of 5′-OH-iav with the 5′-OH dsRNA bound to the human RIG-I CTD (Supplementary Figure S6). The helix of the 5′-OH-iav was more tilted toward RIG-I CTD than the 5′-OH dsRNA (Supplementary Figure S6). Thus, the helical bending around the A5·U29 base pair in 5′-OH-iav might create additional contacts with the RIG-I CTD to facilitate the binding to the protein in the absence of the 5′-PPP moiety.
We confirmed the similar induction level of IFN-β between two types of iav RNAs as shown in the Figure 5. However, the binding affinity of 5′-PPP-iav RNA against the RIG-I CTD was weaker than that of 5′-OH-iav RNA (Table 1). It may be that the binding mode and affinity of 5′-PPP-iav RNA are set by the competition between binding of the 5′-PPP to the core cleft and binding of the phosphodiester backbone to the alternative binding site and that these two opposing forces reflect the dynamics of the influenza RNA. As shown in Figure 2 and Supplementary Figure S2, binding of the RIG-I CTD significantly decreased the kex values of the first two base pairs (G1·C33 and A2·U32) of both 5′-PPP- and 5′-OH-iav RNAs, indicating the stabilization of the base pairs. However, the kex values of G1 and U32 of 5′-OH-iav RNA decreased more than those of 5′-PPP-iav RNA (Supplementary Figure S2). In addition, the 6th and 7th base pairs within the bend (G6·U28 and A7·U27) and the 10th base pair (C10·G24) adjacent to the internal loop exhibited dramatically different dynamics changes between 5′-PPP-iav and 5′-OH-iav (Figure 2 and Supplementary Figure S2). These base pairs in the bent helix and internal loop are positioned close to the alternative RIG-I binding site for 5′-OH RNA, as shown in the RIG-I CTD/5′-OH dsRNA complex structure (Supplementary Figure S6). Indeed, we discovered two residues of RIG-I CTD, T903 and L904, which could potentially contribute to alternative binding with the bent 5′-OH-iav. Taken together, although the changes in dynamics of the two influenza RNA species are distinct, RIG-I CTD does not seem to discriminate 5′-PPP-iav from 5′-OH-iav RNA as it does for standard duplex dsRNA.
In the dynamics study for the control RNAs, which are neither bent nor possess an internal loop, both 5′-PPP- and 5′-OH-cont RNAs showed relatively minor changes in the exchange rate upon protein binding (Figure 3C and Supplementary Figure S2). The binding of RIG-I CTD induced distinguishable decreases in the kex of terminal bases G1 and U33 in both types of the control RNAs. However, most of the other imino protons in the helix showed no significant change in their exchange rates, in contrast to the results from the iav RNAs. This remarkable difference in dynamics between iav RNAs and control RNAs suggests that the control RNAs are not bound closely enough to the RIG-I CTD to manifest dynamics changes in the backbone when interacting with RIG-I CTD and that, in those constructs, the modification at the 5′ end is the main determinant of the binding mode of duplex dsRNAs.
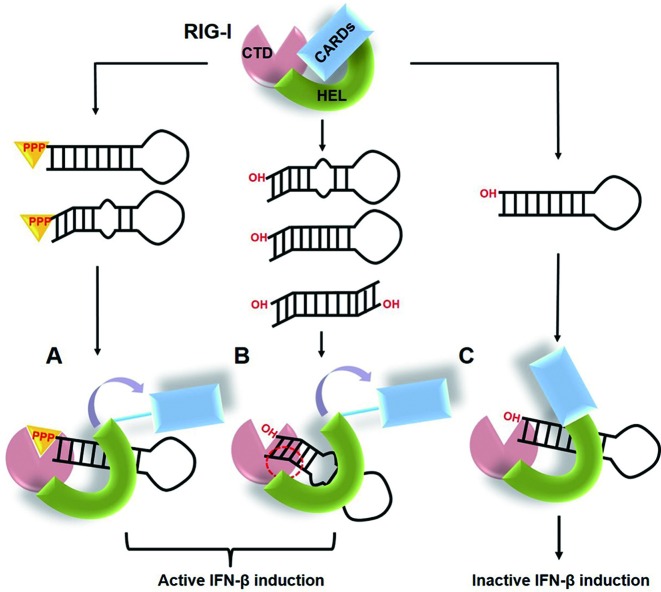
Based on these results, we propose a model for RNA recognition by RIG-I to stimulate IFN-β induction (Figure 6). When RIG-I binds to 5′-triphosphorylated dsRNAs, the protein implements conformational changes exposing the CARDs to activate downstream signaling (29,30). However, 5′-OH dsRNAs are not able to completely activate the conformational change in RIG-I because there is no strong interaction to compensate the lack of the 5′-PPP. If RIG-I associates with RNAs of unusual structure like influenza panhandle RNA, structural features such as the bent helix and internal loop may provide the necessary swing motion for the RNA to contact RIG-I tightly, probably through an additional site. In addition, the kinking the helical axis is inducible tight binding with RIG-I without the 5′-PPP. These non-canonical interaction may be sufficient to induce conformational changes in RIG-I that release CARDs to form lock-washer CARD tetramers (31), leading to IFN-β production. Recently, it was elucidated that the activation of RIG-I signaling is governed by ubiquitin binding and/or conjugation of CARDs and filament formation mediated by helicase activity, and the length of dsRNA has been suggested to be a key determinant to select the activation mechanism. Here, we demonstrated that the unique panhandle structure with a bend or kink-turn structures constitutes a type of RIG-I agonists without the 5′-PPP moiety, providing the first structural determinant for RIG-I activation in the absence of 5′-PPP. RIG-I recognizes RNA structures of negative-sense viruses including Hepatitis C virus (HCV) RNA. Despite the extensive studies on RNA agonists of RIG-I, a few structures of RNA were determined. In Hepatitis C virus, the viral RNA motif including poly-U/UC or X-region containing 5′-PPP binds with RIG-I. (32) The RIG-I binding to HCV RNA requires 5′-PPP, but the 5′-PPP moiety alone is not sufficient. The additional poly-U/UC RNA motif is necessary for stable activation of RIG-I, even though its structure might be flexible without a rigid conformation (33). Our RIG-I agonists with a bent RNA helix and the RIG-I agonists derived from HCV suggest that RIG-I might be a dynamic sensor that could detect different structural features of RNA to counter the evasion maneuvers employed by RNA viruses. The structures of RIG-I complexed with its agonist can provide deep insights into the mechanism of how RIG-I probes the structural features of RNA. We are currently investigating the complex structure of RIG-I and influenza panhandle RNA.
Figure 6.
A working model for RIG-I activation by panhandle RNAs. (A) The RIG-I protein recognizes the non-self dsRNAs through 5′-PPP and then activates the IFN-β induction pathway by a conformational change that exposes the CARDs domain. (B) The bent or kink-turn structure within the panhandle RNA may induce the active conformational change for IFN-β production, even without the 5′-PPP moiety. The red dotted circle indicates an alternative binding site between the bent 5′-OH panhandle RNAs and RIG-I CTD. (C) The 5′-OH standard dsRNAs can bind to RIG-I, but they are not able to induce the conformational change of RIG-I leading to IFN-β production.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge assistance from Dr Junsang Ko.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea (NRF), Korean Government [2011-0020322, 2015R1A2A1A15055329 and 2015R1A2A2A01005688 to B.-S.C., 2012M3A9B4027955 and 2011–0020334 to S.-J.K., 2013R1A2A2A05003837 to J.-H.L., and NRF-2015M3A9C4076320 to M.-K.L.]; Next-Generation BioGreen 21 Program [SSAC, No. PJ01117701 to J.-H.L.]; Korea Health Technology R&D Project through the Korea Health Industry Development Institute [HI14C0211 to S.-J.K.]; National Research Council of Science and Technology [DRC-14-2-KRISS to .S.-J.K.]. Funding for open access charge: 2015R1A2A2A01005688.
Conflict of interest statement. None declared.
REFERENCES
- 1.Goubau D., Deddouche S., Reis E.S.C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kato H., Takahasi K., Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol. Rev. 2011;243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 3.Loo Y.M., Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y., Ludwig J., Schuberth C., Goldeck M., Schlee M., Li H., Juranek S., Sheng G., Micura R., Tuschl T., et al. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat. Struct. Mol. Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu C., Xu H., Ranjith-Kumar C.T., Brooks M.T., Hou T.Y., Hu F., Herr A.B., Strong R.K., Kao C.C., Li P. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahasi K., Yoneyama M., Nishihori T., Hirai R., Kumeta H., Narita R., Gale M., Jr, Inagaki F., Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol. Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 7.Pichlmair A., Schulz O., Tan C.P., Naslund T.I., Liljestrom P., Weber F., Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 8.Vela A., Fedorova O., Ding S.C., Pyle A.M. The thermodynamic basis for viral RNA detection by the RIG-I innate immune sensor. J. Biol. Chem. 2012;287:42564–42573. doi: 10.1074/jbc.M112.385146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X., Ranjith-Kumar C.T., Brooks M.T., Dharmaiah S., Herr A.B., Kao C., Li P. The RIG-I-like receptor LGP2 recognizes the termini of double-stranded RNA. J. Biol. Chem. 2009;284:13881–13891. doi: 10.1074/jbc.M900818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marques J.T., Devosse T., Wang D., Zamanian-Daryoush M., Serbinowski P., Hartmann R., Fujita T., Behlke M.A., Williams B.R. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat. Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 11.Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T.S., Fujita T., Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malathi K., Dong B., Gale M., Jr, Silverman R.H. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desselberger U., Racaniello V.R., Zazra J.J., Palese P. The 3′ and 5′-terminal sequences of influenza A, B and C virus RNA segments are highly conserved and show partial inverted complementarity. Gene. 1980;8:315–328. doi: 10.1016/0378-1119(80)90007-4. [DOI] [PubMed] [Google Scholar]
- 14.Fodor E., Pritlove D.C., Brownlee G.G. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 1994;68:4092–4096. doi: 10.1128/jvi.68.6.4092-4096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coloma R., Valpuesta J.M., Arranz R., Carrascosa J.L., Ortin J., Martin-Benito J. The structure of a biologically active influenza virus ribonucleoprotein complex. PLoS Pathog. 2009;5:e1000491. doi: 10.1371/journal.ppat.1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlee M., Hartmann G. The chase for the RIG-I ligand–recent advances. Mol. Ther. 2010;18:1254–1262. doi: 10.1038/mt.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rehwinkel J., Tan C.P., Goubau D., Schulz O., Pichlmair A., Bier K., Robb N., Vreede F., Barclay W., Fodor E., et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 18.Schlee M., Roth A., Hornung V., Hagmann C.A., Wimmenauer V., Barchet W., Coch C., Janke M., Mihailovic A., Wardle G., et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt A., Schwerd T., Hamm W., Hellmuth J.C., Cui S., Wenzel M., Hoffmann F.S., Michallet M.C., Besch R., Hopfner K.P., et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baum A., Sachidanandam R., Garcia-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc. Natl. Acad. Sci. U.S.A. 2010;107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bae S.H., Cheong H.K., Lee J.H., Cheong C., Kainosho M., Choi B.S. Structural features of an influenza virus promoter and their implications for viral RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10602–10607. doi: 10.1073/pnas.191268798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 23.Goddard T.D., Kneller D.G. Sparky 3. San Francisco: University of California Press; 2008. [PubMed] [Google Scholar]
- 24.Lee J.H., Pardi A. Thermodynamics and kinetics for base-pair opening in the P1 duplex of the Tetrahymena group I ribozyme. Nucleic Acids Res. 2007;35:2965–2974. doi: 10.1093/nar/gkm184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee Y.M., Kim H.E., Park C.J., Lee A.R., Ahn H.C., Cho S.J., Choi K.H., Choi B.S., Lee J.H. NMR study on the B-Z junction formation of DNA duplexes induced by Z-DNA binding domain of human ADAR1. J. Am. Chem. Soc. 2012;134:5276–5283. doi: 10.1021/ja211581b. [DOI] [PubMed] [Google Scholar]
- 26.Nozinovic S., Fürtig B., Jonker H.R., Richter C., Schwalbe H. High-resolution NMR structure of an RNA model system: the 14-mer cUUCGg tetraloop hairpin RNA. Nucleic Acids Res. 2010;38:683–694. doi: 10.1093/nar/gkp956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu C., Ranjith-Kumar C.T., Hao L., Kao C.C., Li P. Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5′ triphosphate. Nucleic Acids Res. 2011;39:1565–1575. doi: 10.1093/nar/gkq974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McPhee S.A., Huang L., Lilley D.M. A critical base pair in k-turns that confers folding characteristics and correlates with biological function. Nat. Commun. 2014;5:5127. doi: 10.1038/ncomms6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo D., Ding S.C., Vela A., Kohlway A., Lindenbach B.D., Pyle A.M. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kowalinski E., Lunardi T., McCarthy A.A., Louber J., Brunel J., Grigorov B., Gerlier D., Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 31.Peisley A., Wu B., Xu H., Chen Z.J., Hur S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014;509:110–114. doi: 10.1038/nature13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito T., Owen D.M., Jiang F., Marcotrigiano J., Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seol Y., Skinner G., Visscher K. Elastic Properties of a Single-Stranded Charged Homopolymeric Ribonucleotide. Phys. Rev. Lett. 2004;93:118102. doi: 10.1103/PhysRevLett.93.118102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.