Abstract
Small interfering RNAs (siRNAs) defend the organism against harmful transcripts from exogenous (e.g. viral) or endogenous (e.g. transposons) sources. Recent publications describe the production of siRNAs induced by DNA double-strand breaks (DSB) in Neurospora crassa, Arabidopsis thaliana, Drosophila melanogaster and human cells, which suggests a conserved function. A current hypothesis is that break-induced small RNAs ensure efficient homologous recombination (HR). However, biogenesis of siRNAs is often intertwined with other small RNA species, such as microRNAs (miRNAs), which complicates interpretation of experimental results. In Drosophila, siRNAs are produced by Dcr-2 while miRNAs are processed by Dcr-1. Thus, it is possible to probe siRNA function without miRNA deregulation. We therefore examined DNA double-strand break repair after perturbation of siRNA biogenesis in cultured Drosophila cells as well as mutant flies. Our assays comprised reporters for the single-strand annealing pathway, homologous recombination and sensitivity to the DSB-inducing drug camptothecin. We could not detect any repair defects caused by the lack of siRNAs derived from the broken DNA locus. Since production of these siRNAs depends on local transcription, they may thus participate in RNA metabolism—an established function of siRNAs—rather than DNA repair.
INTRODUCTION
Genomes are constantly challenged by intrinsic and extrinsic damaging agents or processes; appropriate repair is thus essential. DNA double-strand breaks (DSBs) are particularly harmful lesions because they interrupt the continuity of genetic information. They arise physiologically due to replication fork stalling and collapse in S-phase of the mitotic cell cycle. Furthermore, the essential cross-overs during the meiotic cell cycle are initiated by deliberately induced DNA DSBs. Finally, the rearrangement of immunoglobulin and T-cell receptor variable regions is triggered by DSB induction. Broken ends of DNA can be re-ligated by the non-homologous end-joining (NHEJ) or the microhomology-mediated end-joining (MMEJ) pathways. Such events may lead to sequence losses and are thus mutagenic. Alternatively, homologous recombination (HR) can repair the break using the sister chromatid as template, but it is accordingly limited to the late S and G2 phases of the cell cycle.
Several pathways can mediate homologous recombination. These include gene conversion (GC), double Holliday junctions (dHJ), synthesis-dependent strand annealing (SDSA) and break-induced replication (BIR) (1). A special situation occurs if the DNA DSB arises between directly repeated sequences. In this case, the single-strand annealing (SSA) pathway can lead to the deletion of one copy of the direct repeat, thus shortening the genomic sequence. A common step in all HR events is the initial processing by the Mre11-Rad50-Nbs1 complex (MRN) together with the nuclease CtIP. This step is followed by further 3′-to-5′ resection via exonuclease-1 (Exo-1); the downstream genetic requirements then differ between the individual HR pathways.
In recent years, the discovery of DNA DSB-derived siRNAs in plants as well as cultured human and Drosophila cells has raised the question whether small RNA mediated processes may influence the repair of DNA DSBs. Assays that report the activity of the SSA pathway in plants and homologous recombination in human cells showed decreased, but not abolished, repair activity when small RNA biogenesis factors were depleted (2,3). In addition, a physical interaction between the human siRNA effector protein Ago2 and the Rad51 recombinase has been described (3). Repair of damaged DNA follows different kinetics in the absence of locus-derived small RNAs (4), since the repair-associated γ-H2Ax foci persist for a longer time. On the other hand, generation of DSB-derived siRNAs only occurred if the break was located within a transcribed region in Drosophila (5). Furthermore, the phenotype of flies with mutations in the siRNA pathway is rather mild (6,7), while miRNA factor mutant flies are not viable (6,7).
Since the biogenesis enzymes for DSB-derived siRNAs usually also process precursors for other small RNAs, in particular miRNAs, it is challenging to conceive experiments that are not at risk of indirect effects via the miRNA pathway. Indeed, a recently published study concludes that the DSB-repair defect detected in cultured human cells depleted of Dicer is due to secondary effects on cell cycle progression and can be rescued by transfection of a let-7 miRNA mimic (8). Results in Neurospora crassa, on the other hand, have demonstrated that mutants of the Argonaute-protein Qde2 are hypersensitive to DNA damaging agents (9) and that DNA damage is necessary to trigger the transposon-defense mechanism of quelling in this organism (10). This required the DNA-dependent and RNA-dependent RNA polymerase activities of Qde1, which lacks homologs in both Drosophila and vertebrates.
In Drosophila, Dicer-1 removes the loop of pre-miRNAs to generate the miRNA/miRNA* biogenesis intermediate. Double-stranded RNA, on the other hand, is nucleolytically processed into siRNAs by the paralog Dicer-2. Although Dicer-2 also processes endogenous precursors of the long hairpin derived endo-siRNAs, the Drosophila system allows examining the effects of siRNA removal without a major disturbance of the miRNA pathway. We therefore developed an SSAreporter as well as a homologous recombination assay based on a GFP knock-in strategy (11) for Drosophila. This allowed us to test the requirement of the siRNA pathway for repair efficiency. Neither in cultured cells depleted for siRNA biogenesis components nor in dcr-2 mutant flies, an influence of siRNA factors on HR efficiency was observed. Finally, the challenge of dcr-2 or ago2 mutant flies with the DSB-inducing drug camptothecin did not show any hypersensitivity. We therefore hypothesize that the transcription dependent, break-derived siRNAs of Drosophila are part of an mRNA surveillance phenomenon that is invoked as a consequence of the perturbation of mRNA biogenesis caused by a DNA DSB within the transcribed region.
MATERIALS AND METHODS
Molecular biology
Tagging of the act5C locus with GFP via polymerase chain reaction (PCR) products or a targeting vector was performed as previously described (11). The SSA assay stable reporter cell line as well as DSB induction by I-SceI transfection was also previously published (12). To generate a Δlig4 line in the SSA reporter background, cells were treated with a sgRNA against the lig4 ORF with the targeting sequence 5′-GCCAGCACGATCAAGTTCC-3′. Subsequently a clonal selection was performed and a clone recovered where a deletion of ∼0.8 kB removed the 5′ UTR and the first 30 nt of the lig4 ORF.
Cell culture and transfection
Cells were cultured in standard Schneider's medium supplemented with 10% fetal bovine serum (FBS) (both Bio&Sell, Nürnberg/Germany). Cells were seeded at a density of 0.5 (for the HR assay using the short homology HR donor) or 1 × 106 cells/ml (for all other experiments). Transfections were performed with Fugene HD (Promega) according to the manufacturer's instructions. For RNAi cells were treated with dsRNA (sequences published in (5,13–14) or described in Supplementary Table S1) at a final concentration of 2–10 ng/μl. For our SSA reporter cells were split 1:3 four days after induction of RNAi and transfected with pKF259 at a final concentration of 0.5 ng/ μl (for introduction of the I-SceI nuclease). To assess transfection efficiencies, wildtype S2-cells were treated as described above but transfected with pKF63 (15) that encodes GFP expressed from the ubiquitin promoter. Readout by flow cytometry was performed five days post transfection.
For the HR assay with a donor molecule in trans, wildtype cells with stable Cas9 expression were seeded at a concentration of 0.5–1 × 106 cells/ml and treated with dsRNA for RNAi. Two days after induction of RNAi, cells were transfected either with sgRNA/HR donor (0.75 ng/μl each) for tagging of act5C or with pKF63 (1.5 ng/μl) to assess transfection efficiencies. Cells were grown for an additional five days before measurement. For quantification of the percentage of GFP positive cells a Becton Dickinson FACSCalibur flow cytometer equipped with a 96-well plate autosampler was used. Data analysis was performed using flowing software version 2.5.1 (http://www.flowingsoftware.com/).
Deep sequencing and data analysis
RNA extraction, library preparation and data analysis were performed as described previously (16). The sequences have been deposited at the European Nucleotide Archive (ENA) under the accession number PRJEB12939.
Fly strains and husbandry
The fly strains carrying the dcr-2 null allele (w; dcr-2[L811fsX]/CyO), the dcr-2 catalytically inactive allele (w; dcr-2[G31R]/CyO) and the fly strain with the dcr-2 rescue transgene y,w,eyFLP; FRT42Ddcr-2[L811fsX]; P{w+,Dcr-2} are described in (6). Transgenic flies carrying the SSA reporter and the I-SceI transgene were generated by PhiC31 recombinase mediated integration into attP sites 19E7 and 2A3, respectively; injections were performed by Rainbow Transgenics (Camraillo/CA, USA). The transgenes and dcr-2 alleles were crossed together with standard genetic approaches.
For analysis of CPT sensitivity in absence of dcr-2 or ago2, mutant strains with the reporter background were crossed to yield either homozygous or trans-heterozygous mutants. In addition to the fly strains mentioned above, we tested also the dcr-2 mutant w[1118]; PBac{w[+mC] = WH}dcr-2[f06544]/CyO as well as the deficiency strain y[1] w[1118]/Dp(1;Y)y[+]; Df(2R)ED3385, P{w[+mW.Scer\FRT.hs3] = 3′.RS5+3.3′}ED3385/CyO.
We also investigated the CPT sensitivity in the absence of Ago2 by crossing the strain w[1118]; AGO2[321]/TM3, Sb[1], carrying an ago2 loss of function allele to the ago2 point mutant yw; ago2[V966M].
F0 females were kept on fly food coated with different concentrations of CPT (Sigma) or the respective DMSO control during four days after eclosion. The cross was performed on fly food treated with different concentrations of CPT (or the respective control). After 7 days, flies were transferred to freshly treated fly food for additional 7 days. dcr-2 proficient and dcr-2 deficient progeny was counted. For the rescue experiment, dcr-2 null mutants were crossed to flies carrying the dcr-2 rescue transgene (y,w,eyFLP; FRT42Ddcr-2[L811fsX];P{w+, Dcr-2}) and treated as described above. To evaluate the CPT sensitivity of a corresponding wildtype, flies carrying a dcr-2 mutant allele were first backcrossed with w1118. The resulting offspring with a wildtype second chromosome and the balancer CyO was then used for the cross and CPT sensitivity assay as described above.
Western blotting
A total of 3 × 106 cells were treated with 3 ng/μl dsRNA and harvested for protein isolation after five days. Harvested cells were washed twice with 1x phosphate buffered saline (PBS) and lysed by resuspension in 1x PBS supplemented with 1x Proteinase Inhibititor without EDTA (Roche) and 1% Tween. Resuspended cells were frozen in liquid nitrogen, incubated at room temperature until almost complete thawing and then transferred to ice for 30 min. After a centrifugation step (16000 x g, 4°C, 15 min) supernatant was mixed with SDS-loading buffer and comparable amounts of protein were applied to a 12% SDS-gel.
For fly extraction individual flies were anesthetized, ground in 75 μl lysis buffer (30 mM HEPES, 100 mM KOAc, 2 mM MgOAc, 1 mM DTT, 1% triton-X and 1x proteinase inhibitor cocktail (Roche)) and incubated on ice for 30 min. After centrifugation (16000 xg, 4°C, 15 min) the supernatant was mixed with SDS-loading buffer and 24 μg of total protein was applied to a 12% SDS-gel. For Western blotting, the monoclonal B-2 antibody was used (sc-9996, Santa Cruz) for detection of GFP. For loading control the monoclonal anti beta-Tubulin (E7) antibody from the Developmental Studies Hybridoma Bank (DSHB) was used. Chemiluminescence images were recorded with a Fuji LAS-3000 mini system and quantification of band intensity was performed using ImageJ.
Fluorescence microscopy
Fluorescence microscopy was performed on a Leica MZ16FA fluorescence stereomicroscope. Images were taken with a Leica DFC 320 camera at a fixed magnification (99.8x). Acquisition was done using Leica DFC Twain camera software with maximum gain setting and an exposure time of 2.9 s.
RESULTS
A reporter system to analyze DNA double-strand break repair in Drosophila
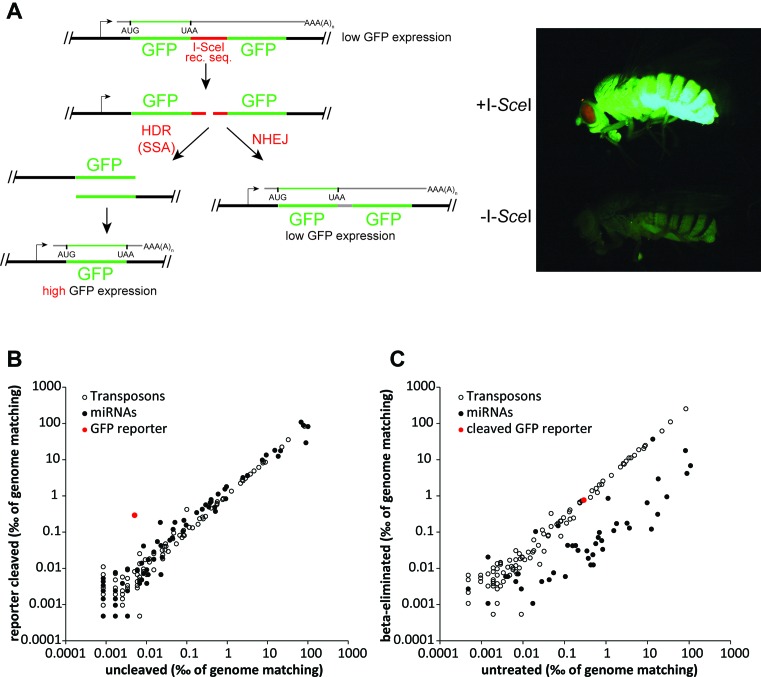
In Drosophila, the generation of small RNAs from a DNA end has only been described upon transfection of linearized plasmids. We thus established a reporter system that makes use of the highly specific homing-endonuclease I-SceI (17) in flies and S2-cells. Similar to a previously used reporter system in Arabidopsis (2), we separated two stretches of full-length GFP coding sequence by an I-SceI recognition sequence. In the original state, only the first GFP open reading frame is translated, while the second GFP sequence is part of the 3′-UTR due to the presence of a stop-codon upstream of the I-SceI site. Prior to cleavage and repair, the construct yields only low amounts of GFP (Figure 1A). In Drosophila, the nonsense-mediated mRNA decay pathway can be triggered by long 3′-untranslated regions (18) and we suspected that the long 3′-UTR (comprised of a GFP sequence and the SV40 polyadenylation signal) renders the transcript susceptible to degradation via mRNA surveillance pathways (19,20). Consistent with this hypothesis, the uncleaved reporter construct can be de-repressed in S2-cells by knock-down of the nonsense mediated decay (NMD) factor upf1 (Supplementary Figure S1). Upon introduction of the I-SceI nuclease, the construct is cleaved between the two GFP coding sequences. Subsequent repair can either proceed via the NHEJ, resulting in no change of GFP expression, or via the SSA pathway, a form of homology directed repair which in this case removes one GFP coding sequence and thus shortens the 3′-UTR. The resulting substantial increase in GFP fluorescence can be visualized in flies with the help of a fluorescence stereomicroscope (Figure 1A, right) and in cells via flow cytometry (12). We verified the rearrangements in individual offspring animals from a fly cross that combined the double-GFP reporter with the I-SceI nuclease. The locus can be PCR-amplified and yields a smaller product if a rearrangement via the SSA pathway has occurred. Uncleaved or faithfully repaired loci can be distinguished from those repaired via via mutagenic NHEJ: In vitro digestion of the PCR product with I-SceI produces two fragments of equal size if a non-modified locus was amplified, while products of mutagenic NHEJ events are resistant to digestion (Supplementary Figure S2).
Figure 1.
siRNAs are generated against the SSA reporter upon introduction of a DSB. (A) Schematic overview over the SSA reporter strategy. Two copies of GFP are separated by a recognition site for the I-SceI nuclease. The original construct results in only little GFP expression, since the reporter transcript is targeted by NMD due to its long 3′-UTR. Upon introduction of a DSB, cells repair the break either by NHEJ or SSA. Only the latter will lead to an increase in GFP expression, which can be observed in vivo (compare flies with and without I-SceI nuclease expression). (B) The comparison of small RNA libraries after control treatment or transfection of the I-SceI nuclease shows that introduction of a break in the reporter leads to generation of siRNAs against the break. Generation of microRNAs and siRNAs against transposons is unaffected by the break. (C) Comparison of ß-eliminated versus untreated control of small RNA libraries after introduction of the break reveals loading of break-derived siRNAs into Ago2, as is the case for transposon mapping reads. Most miRNAs on the other hand, are loaded into Ago1 and thus susceptible to β-elimination.
Chromosomally integrated reporters generate Ago2-loaded siRNAs upon induction of a DNA double-strand break
Transfected linearized DNA will generate siRNAs if a transcribed region is interrupted by the cut (5). Does this reflect a response that occurs at a chromosomal DNA break? We established stable S2 cell lines carrying our GFP-I-SceI reporter construct and introduced the I-SceI nuclease via transfection. Three days later, we isolated small RNAs and subjected them to deep sequencing. Induction of the chromosomally located DNA break via I-SceI strongly stimulated the generation of siRNAs (Figure 1B). The pattern of siRNA generation from the chromosomally located break is similar to the one we observed previously upon transient transfection (Supplementary Figure S3). Their abundance was roughly 10-fold lower, presumably due to the reduced copy number of specific DNA breaks in the stable cell line compared with transient plasmid transfection.
Are the break-derived siRNAs incorporated into Ago2 effector complexes? We subjected the RNA to periodate oxidation and beta-elimination prior to sequencing. This treatment renders unmodified RNA 3′-OH ends incompetent for adapter ligation and thus increases the abundance of 2′-O-methyl modified small RNAs among the sequencing reads (21). Since Drosophila siRNAs are 2′-modified only after loading into Ago2 (22), enrichment in the sequencing reads obtained from treated RNA demonstrates their incorporation into Ago2 RNAi effector complexes. Break-derived small RNAs were resistant to oxidation and are thus loaded into Ago2 (Figure 1C), consistent with the observation of an Ago2-dependent repressive response at DNA breaks (5). In summary, chromosomally located DNA DSBs in a transcriptionally active region can lead to the production and Ago2-loading of small RNAs with corresponding sequence.
The siRNA pathway components have no effect on SSA efficiency
Using RNAi, we depleted our SSA reporter cell lines of factors with a known role in DNA repair, transfected the I-SceI expression plasmid to induce the break and assayed the extent of repair via homologous recombination by flow cytometry. Depletion of the essential HR protein RPA resulted in reduced HR activity as measured by our reporter. Consistent with the notion that SSA does not depend on Rad51 in budding yeast (23), knock-down of Rad51 had no effect on SSA efficiency. On the other hand, depletion of the NHEJ factors Ku80 and Lig4 resulted in increased HR activity, presumably because the competing NHEJ pathway had been attenuated (Figure 2A). Our reporter system thus allows us to detect both increases and decreases in the relative efficiency of HR by SSA.
Figure 2.
HDR efficiencies of the SSA reporter are unaffected by knock-down of siRNA factors. (A and B) HDR efficiencies were determined by flow cytometry after knock-down of individual factors and subsequent transfection with a plasmid encoding the I-SceI nuclease. Knock-down of Renilla luciferase (RLuc) served as knock-down control and was used for normalization. Depicted in (A) are HDR efficiencies for the parental SSA reporter cell line, while (B) shows HDR efficiencies for the Δlig4 SSA reporter cell line. (C) To assess the influence of knockdown of individual factors on the transfection efficiency, we transfected wildtype (S2) cells with a plasmid encoding GFP following the respective knock-down. Five days later, GFP-expressing cells were quantified via flow cytometry. Again, knock-down of RLuc was used as control for normalization. Error bars represent the standard deviation (SD); for Rad51, three independent dsRNA designs were tested, two designs for Ago2. Significant changes compared to the Firefly Luciferase (FLuc) are indicated by an asterisk (*) (P < 0.05; two-sided t-Test equal variance, n ≥ 3 biological replicates).
Having successfully validated our reporter assay, we tested the effect of depleting miRNA pathway components (Ago1, Dcr-1) and siRNA pathway components (Ago2, Dcr-2) (13,15) on DNA repair. Inactivation of miRNA but not siRNA pathway components resulted in an apparent reduction of homologous recombination (Figure 2A and B). However, since induction of the DNA break requires transfection of the I-SceI expression vector, we also measured how knock-down of DNA repair proteins (Nbs1, Rad51, RPA, Lig4, Ku80, mus308), miRNA factors (Ago1, Dcr-1) and siRNA factors (Ago2, Dcr-2) affected the transfection (Figure 2C). We found that part of the Ago1 knock-down effect can be explained by a reduced transfection efficiency. Furthermore, as reported by Liu et al., miRNA factor depletion can lead to cell cycle abnormalities in human cells which in turn influence HR repair (8). Indeed, knock-down of the miRNA pathway components Drosha and Ago1, but not the siRNA pathway components Dcr-2 or Ago2, resulted in a pronounced increase of the G1 population in cultured Drosophila cells (Supplementary Figure S4). It is thus likely that the apparent reduction in HR repair events was the consequence of changes in the cell cycle distribution, augmented by a lower transfection rate in the case of Ago1. This certainly does not exclude a further contribution of other miRNA-related regulatory events.
The siRNA pathway also processes hairpin-RNAs (24), a class of small RNAs that originate from genomically encoded long RNA hairpins. An abundant small RNA derived from the CG4068 hairpin represses the mus308 gene, which encodes the Drosophila homolog of the mammalian DNA polymerase theta (24–26). This polymerase mediates an alternative end-joining mechanism that relies on microhomologies (27). It is therefore formally possible that depletion of siRNA factors leads to up-regulation of mus308, and that this compensates for changes in HR pathway activity because our SSA reporter is analogously re-arranged via the hyper-activated MMEJ system. We reasoned that upon genetic ablation of lig4, mus308 should always be the predominant end-joining activity. In other words, although the balance between HR and end-joining is overall shifted toward HR in lig4 deletion cells, the potentially compensating effect of MMEJ after RNAi factor knock-down should be abolished. We used cas9-CRISPR to mutagenize the lig4 gene in our reporter cell line, then re-cloned the cells and analyzed the lig4 locus by PCR. We recovered one clone that harbored a deletion of ∼0.8 kB in the vicinity of the lig4 gene, removing its 5′ UTR and the first 10 aminoacids of the N-terminus (Supplementary Figure S5). We then measured the efficiency of SSA after knock-down of various factors and subsequent transfection of I-SceI in the Δlig4 mutant reporter cell line (Figure 2B). Similar to wildtype cells, we observed decreased SSA efficiency after knock-down of RPA, but not after depletion of Rad51. Knock-down of Ku80 resulted in increased SSA efficiency while, as predicted, depletion of lig4 by RNAi had no effect in the lig4 mutant cells (Figure 2B). Importantly, also in the lig4 mutant cell line depletion of RNAi factors had no effect, while knock-down of miRNA factors led to an apparent reduction of SSA efficiencies that may be explained by indirect effects (see above). We thus conclude that homology directed repair via the SSA pathway functions independently of break-derived siRNAs in cultured Drosophila cells.
dcr-2 mutant flies have normal levels of homology directed repair
We repeated our analysis in vivo by crossing dcr-2 mutant alleles to flies that transgenically harbored our SSA reporter construct. We crossed trans-heterozygous dcr-2L811fsX/dcr-2G31R or dcr-2L811fsX/dcr-2G31Rfemales carrying the DSB reporter to dcr-2L811fsX/CyO or dcr-2G31R/CyO mutant males carrying a ubiquitously expressed I-SceI nuclease transgene. Upon activation of the zygotic genome, the nuclease will thus cleave the GFP reporter and repair by SSA will lead to patches of high GFP fluorescence in the F1 adults (individual patches are not discernible in Figure 1A, right). We quantified the GFP fluorescence in the eye as a proxy for the balance between HR and end joining events in mutant and control animals. Consistent with the cell culture data, there was no difference in the rate of homology-directed repair between maternal and zygotic dcr-2 mutant animals and heterozygous controls (Figure 3A and B). To obtain an independent measure of GFP expression, we also extracted protein from individual whole flies and quantified the total amount of GFP by Western blotting (Figure 3C). Again, no difference between dcr-2 mutant and control animals was evident. Taken together, our results obtained in vivo with mutant flies do not suggest any requirement of Dcr-2 for SSA-mediated repair.
Figure 3.
HDR efficiency of the SSA reporter is unaffected in maternal and zygotic dcr-2 mutants. The crossing scheme shows the process to generate maternal and zygotic dcr-2 mutants. First trans-heterozygous females that carry two dcr-2 mutant alleles are generated to diminish the influence of potential background mutations. Trans-heterozygous females were then used in the consecutive cross with heterozygous males that carry the I-SceI transgene. Progeny (F1) was then subjected to (A and B) fluorescence microscopy and (C) Western Blotting. Heterozygous F1 animals were used as control. (A) Example pictures of fly eyes in adult F1 flies. dcr-2 proficient flies (Dcr2 +/-) were compared to dcr-2 deficient flies (Dcr2 -/-). (B) Quantification of HDR efficiency of the SSA reporter was measured via the mean fluorescence within the fly eye using ImageJ. Black bars show the mean GFP fluorescence in dcr-2 proficient controls, grey bars the fluorescence in dcr-2 deficient flies. The genotype of F0 females and males is indicated, as are the number of eyes measured, the number of flies (n) and the number of independent crosses. Error bars show the SD. C) Quantification of Western Blot experiments to assess the relative GFP expression (normalized to tubulin as loading control) within the whole fly. Again, black bars show the GFP protein levels in dcr-2 proficient animals, while the results for dcr-2 deficient flies are depicted in grey; error bars represent the SD of three biological replicates. Depicted are also the blots used for quantification. For each lane, whole fly protein extract of a single dcr-2 proficient (+) or dcr-2 deficient (-) animal was applied.
Homologous recombination is unaffected by depletion of siRNA factors
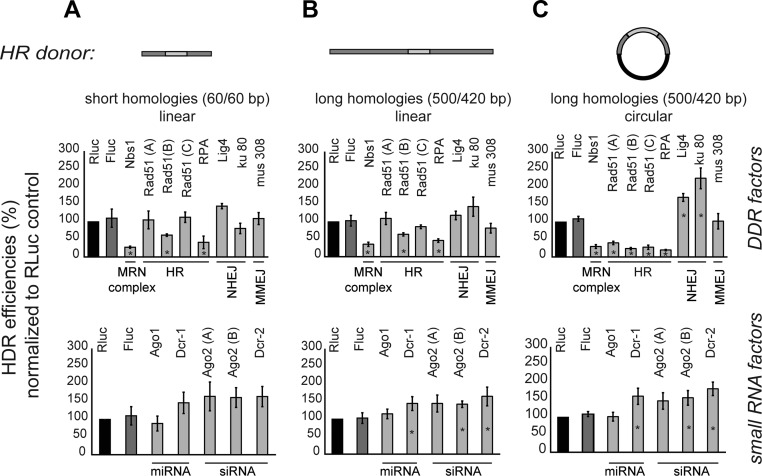
SSA is a rather specific case of homology-directed repair and may not be fully representative of homologous recombination. In order to test whether homologous recombination repair with a template in trans is affected by depletion of siRNA biogenesis factors, we exploited our approach to tag proteins at their genomic locus via the CRISPR/Cas9 system (12). For our assay, we programmed Cas9 to introduce a DSB adjacent to the stop codon of the act5C gene. We provided not only the guide RNA, but also a template for HR repair by transfection. To enable readout via flow cytometry, the two homology regions are flanking a GFP coding sequence. Upon integration of our HR donor at the act5C gene, a strong fluorescence signal from the fusion protein can be observed in individual cells and quantified by flow cytometry (11). Since there is evidence that Rad51 independent HR repair might be favored for short homology regions (<100 bp) (28) we varied the HR donor length upstream and downstream of the DSB (60/60 bp and 500/420 bp). Additionally, we tested the role of HR donor topology (linear and circular).
We compared the efficiency of the GFP knock-in after knock-down of the MRN-complex member Nbs1, known HR factors (RPA, Rad51), NHEJ factors (Lig4, Ku80), miRNA pathway components (Ago1, Dcr-1), as well as siRNA factors (Ago2, Dcr-2) (Figure 4). As before, we observed no decrease in the efficiency of HR upon depletion of siRNA factors, while absence of Nbs1 and RPA clearly reduces HR efficiency. Our results provide insight into the HR donor requirements for engagement of Rad51: Upon knock-down of Rad51 with three independent Rad51 dsRNA designs, we found no strong effect with linear donor molecules bearing either short or long homology arms; the relative HR efficiencies did not fall below 62.2 ± 3.4% (short homologies, Figure 4A) and 64.01 ± 4.5% (long homologies, Figure 4B) of the control treatment. In contrast, when cells were provided with a circular HR donor with long homology arms for repair, Rad51 depletion had a more pronounced effect and HR efficiencies dropped to 23.8 ± 3.1% of the control treatment (Figure 4C). Most importantly, though, we did not observe any reduction in HR with either linear or circular donors upon depletion of the siRNA factors Dcr-2 and Ago2; if anything, the HR efficiency appeared increased after siRNA pathway inhibition (Figure 4, bottom panel). However, this is likely due to technical reasons since detection of Act5C-GFP expression by FACS is facilitated by the absence of repressive siRNAs from the locus (note that the initial recombination frequencies were measured without marker selection).
Figure 4.
HDR efficiencies of genome editing are unaffected by knock-down of siRNA factors. Top: schematic representation of the HR donors employed; homology arms are not drawn to scale. S2-cells stably expressing the Cas9 protein were transfected with an sgRNA expression construct and the indicated homology donor to append a GFP moiety to the Act5C gene at its chromosomal locus. Seven days later, cells were analyzed by flow cytometry and the Act5C-GFP positive population was quantified (absolute values were 3–8% for the Rluc control). Knock-down of Renilla luciferase (RLuc) served as control for normalization. We tested three independent knock-down constructs for Rad51, two for Ago2. The asterisk (*) indicates significant changes (P < 0.05; two-sided t-Test equal variance, n ≥ 3 biological replicates). (A) PCR-based HR donor with 60 nt of homologous sequence at either end; although the MRN complex and RPA appear to be required for homology-directed repair with this donor, the role of Rad51 is limited. Only one knock-down construct resulted in a significant reduction; for the small RNA biogenesis factors no significant change was observed. (B) Extension of the homology arms in the linear configuration (targeting vector pIS1 linearized) results in values largely comparable with the ones observed in (A). For the small RNA factors, a slight apparent increase of HR efficiency is detected; we assume this to be a technical artefact because detection of Act5C-GFP is facilitated in the absence of repressive siRNAs generated at the break. (C) If the HR donor is supplied as a circular targeting vector, the dependency on Rad51 is far more pronounced. In this case, all three knock-down constructs resulted in a highly significant reduction of HR efficiency. For the small RNA biogenesis factors, however, the results are essentially unchanged.
Response to DSB stress is independent of Dcr-2
Given that faithful HR repair both in cis and in trans is independent of break derived siRNAs, we investigated whether Dcr-2 might be important for a general DSB stress response. To this end, we treated dcr-2 mutant flies with the topoisomerase I inhibitor camptothecin (CPT). CPT treatment results in the formation of a covalent link between the nicked DNA and topoisomerase I, thus preventing re-ligation. Replication forks that run into this complex collapse, which ultimately leads to the formation of DSBs at numerous sites throughout the genome. This stands in contrast to the single, directed DSBs we introduced for our HR assays. Flies with a mutation in the brca2 gene, which encodes an important factor for Rad51-dependent homologous recombination, are hypersensitive to camptothecin (29).
We reasoned that in the case of an essential role for Dcr-2 in a DSB stress response, dcr-2 mutant animals should show impaired development to adult flies in the presence of CPT. Thus, a decrease in the proportion of dcr-2 mutant flies recovered from a cross of heterozygous parents should be indicative of CPT hypersensitivity (Table 1, see Supplementary Figure S7A for a graphical representation). We did not observe any increased CPT sensitivity with control animals that carried a wildtype chromosome 2 in combination with a CyO balancer (Table 1, cross 1). In contrast, treatment with CPT severely reduced hatching of dcr-2 deficient animals (Table 1, cross 2 and 3). However, we were not able to rescue this phenotype by an ectopically placed and functionally validated dcr-2 wildtype rescue transgene (6) (Table 1, cross 4). Since both alleles used in our experiments (individually and the trans-heterozygous combination) were derived from the same genetic screen (6), the observed hypersensitivity is most likely due to a background mutation present in the strain that was mutagenized. We therefore attempted to combine the alleles with independent dcr-2 mutant chromosomes. The deficiency Df(2R)ED3385 comprises ∼118 kb surrounding the dcr-2 locus. Trans-heterozygous flies with the dcr-2l811fsX allele remained hypersensitive to CPT (Table 1, cross 5) but again the rescue with a wildtype dcr-2 transgene was not successful (Table 1, cross 6). In contrast, a trans-heterozygous combination of the piggyBAC transposon insertion dcr2f06544 with either the deficiency Df(2R)ED3385 or dcr-2L811fsX had no increased CPT sensitivity (Table 1, cross 7 and 8). This transposon insertion is located 29 nt downstream of the annotated transcription start site and the allele is dysfunctional in transgenic RNA interference (Supplementary Figure S6). Finally, we found no CPT sensitivity conferred by the catalytically inactive ago2V966M mutation (30) in trans-heterozygous animals with the ago2321 P-element insertion (31) (Table 1, cross 9). We conclude that an impaired RNAi pathway does not lead to hypersensitivity to the DSB inducing agent camptothecin. The simplest interpretation of our results is that the background mutation responsible for CPT-sensitivity in certain dcr-2 alleles is either also present on or resides within the region deleted in Df(2R)ED3385.
Table 1. Camptothecin-challenge of dcr2 and ago2 mutant flies (CyO and TM3 are homozygous lethal balancer chromosomes with functional Dcr-2 or Ago2 alleles and a visible marker).
| cross | genotypes | phenotype | expected ratio | control | DMSO | 0.01 mM | 0.03 mM | 0.1 mM | 0.3 mM |
|---|---|---|---|---|---|---|---|---|---|
| 1 | +/CyO | Cy | 2 | 75 | 43 | 112 | 142 | 69 | 84 |
| x | non-Cy | 1 | 54 | 38 | 94 | 70 | 38 | 52 | |
| +/CyO | # crosses | (1) | (2) | (3) | (3) | (2) | (3) | ||
| 2 | dcr2G31R/CyO | Cy | 2 | 234 | 236 | 105 | 74 | 220 | 131 |
| x | non-Cy | 1 | 100 | 141 | 62 | 41 | 21 | 0 | |
| dcr2L811fsX/CyO | # crosses | (3) | (2) | (2) | (2) | (3)* | (3)(*) | ||
| 3 | dcr2L811fsX/CyO | Cy | 2 | 155 | 157 | 8 | 100 | 219 | 135 |
| x | non-Cy | 1 | 61 | 82 | 2 | 5* | 2 | 0 | |
| dcr2L811fsX/CyO | # crosses | (3) | (2) | (1) | (2) | (3)* | (3)(*) | ||
| 4 | dcr2L811fsX/CyO | Cy | 1 | 219 | 186 | 183 | 117 | 31 | 20 |
| x | non-Cy | 1 | 191 | 163 | 196 | 68 | 1 | 0 | |
| dcr2L811fsX; P{Dcr2} | # crosses | (4) | (3) | (5) | (4) | (1)* | (1)(*) | ||
| 5 | dcr2L811fsX/CyO | Cy | 2 | 62 | 55 | 45 | 108 | 126 | 23 |
| x | non-Cy | 1 | 41 | 36 | 19 | 37 | 4 | 0 | |
| dcr2Df(2R)ED3385/CyO | # crosses | (3) | (3) | (2) | (3) | (3)* | (2)(*) | ||
| 6 | dcr2Df(2R)ED3385/CyO | Cy | 1 | 68 | 49 | 77 | 112 | 95 | 117 |
| x | non-Cy | 1 | 81 | 87 | 112 | 114 | 8 | 0 | |
| >dcr2L811fsX; P{Dcr2} | # crosses | (2) | (3) | (3) | (3) | (3)* | (3)(*) | ||
| 7 | dcr2f06544c/CyO | Cy | 2 | 93 | 81 | 66 | 18 | 12 | 62 |
| x | non-Cy | 1 | 51 | 43 | 37 | 23 | 9 | 25 | |
| dcr2Df(2R)ED3385/CyO | # crosses | (2) | (2) | (2) | (2) | (2) | (2) | ||
| 8 | dcr2f06544c/CyO | Cy | 2 | 175 | 92 | 100 | 230 | 146 | 137 |
| x | non-Cy | 1 | 83 | 52 | 69 | 135 | 89 | 66 | |
| dcr2L811fsX/CyO | # crosses | (4) | (3) | (3) | (5) | (3) | (4) | ||
| 9 | ago2321/ TM3, Sb | Sb | 1 | 51 | 85 | 49 | 72 | 60 | 58 |
| x | non-Sb | 1 | 64 | 89 | 56 | 111 | 91 | 52 | |
| ago2V966M | # crosses | (2) | (2) | (2) | (3) | (3) | (3) | ||
*distribution different from expected, P < 10−5, χ2 test.
(*)no exact χ2 test calculation possible because of 0 observed events in one category.
A recent publication described frequent variants of the cyp6d2 gene that either impair splicing (SD allele) or transcript accumulation (NT allele) and, in both cases, lead to camptothecin hypersensitivity (29). We thus used the published PCR-based assay to test the stocks from our camptothecin experiments. We detected the presence of the SD allele in the dcr-2L811fsX and dcr-2G31R strains (including those employed for the transgenic Dcr-2 rescue cross). Furthermore, we found the NT allele in the Df(2R)ED3385 and ago2V966M stocks. In contrast, the dcr-2f06544 piggyBAC allele and the ago2321 allele did not carry any of the described cyp6d2 alleles (Supplementary Figure S7B). Thus, all genetic combinations that showed hypersensitivity to camptothecin were also homozygous for cyp6d2 mutations, while none of the combinations with a functional cyp6d2 copy was hypersensitive for this drug. Taken together, our analysis demonstrates that the camptothecin hypersensitivity in certain RNAi mutant stocks is caused by cyp6d2 background mutations.
DISCUSSION
In this manuscript we examined the requirement of dcr-2 for efficient DNA double-strand break (DSB) repair in Drosophila melanogaster. We present a GFP-based system that reports on the balance of homology-directed repair versus non-homologous end joining upon induction of a single break with the I-SceI nuclease. The reporter is established both in a stable, clonally derived S2-cell line and in transgenic flies. With this reporter, we demonstrate that a break located in the chromosome induces an siRNA response that is very similar to the one we had observed previously with transfected linearized plasmids (5). This is in accordance with reports in Neurospora crassa, Arabidopsis thaliana and human cell culture, demonstrating the conservation of the siRNA response to a DSB within a transcribed region. Cells of Neurospora are indeed hypersensitive to DNA-damaging conditions if important RNAi-factors are mutated (9). Furthermore, there is a genetic interdependence between homologous recombination and the generation of the damaged-induced qiRNAs from repetitive sequences in Neurospora (10).
In Arabidopsis and human cells, a link between DSB induced small RNA biogenesis and efficient DSB repair was also proposed (2) and a physical interaction between mammalian Rad51 and Ago2 was recently described (3). A potential caveat of work with A. thaliana and mammalian cells is that the small RNA biogenesis pathways are intertwined. In Arabidopsis, several classes of small RNAs with distinct functions have been described. Although they may be preferentially produced by one of the four Dicer-like proteins and loaded into a specific Argonaute effector, none of the Dicer-like proteins produces exclusively a single class of siRNAs (32); genetic ablation may therefore affect more than one pathway. In mammalian cells, a single Dicer enzyme produces both siRNAs and miRNAs. Consequently, Dicer mutant mice are not viable. Analogously, although there are four distinct Argonaute proteins in mammals, only Ago2 is catalytically active and the major acceptor of both, miRNAs and siRNAs. Depletion of Ago2 and Drosha, as reported by Wei et al. (2), could thus indirectly affect how DNA damage is dealt with since miRNAs target DNA repair factors and regulate cell cycle progression. Indeed, a recent publication demonstrated that overexpression of the miRNA let-7 or related miRNAs rescues the observed HR deficiency in Dicer knock-down cells (8).
We have previously demonstrated that the Drosophila DNA DSB induced siRNAs depend on Dcr-2 and Ago2 for their function, while depletion of the miRNA factors Drosha and Dcr-1 had no detectable influence on their generation (5). This was now confirmed by the demonstration that the break-derived siRNAs are fully resistant to periodate oxidation and beta-elimination (Figure 1C), indicating that they all carry the 2′-O-methyl modification at their 3′-end that is characteristic of Ago2-loaded small RNAs. In Drosophila, the biogenesis of most miRNAs can be separated from siRNAs because Dcr-1 is dedicated to the processing of miRNAs, while Dcr-2 produces siRNAs from long double-stranded RNA (6,15,33). Similarly, the Ago1 protein is the essential miRNA acceptor while Ago2 is, like Dcr-2, non-essential. Nonetheless, male flies with a mutation in ago2 or dcr-2 show impaired fertility due to reduced expression of hairpin RNAs (34). In addition, zygotic defects in chromatin structure and mitosis were reported for ago2 mutant embryos (35). Thus, Drosophila offers the possibility to study the effect of siRNA ablation with most likely marginal side-effects on the miRNA pathway. We therefore developed a SSA reporter analogous to the ones employed in Arabidopsis to probe HR efficiency between two repeats of GFP coding sequence. Both in dcr-2 mutant flies and in cells depleted for siRNA factors, we found that SSA repair was unaffected. Knock-down of miRNA factors, however, led to an apparent reduction of SSA efficiencies; a likely reason for this is a shift in cell cycle distribution and proliferation as well as a decrease in transfection efficiency (see Figure 2 and Supplementary Figure S5).
SSA is a special subtype of homology directed repair; we also tested whether break-induced siRNAs influence the efficiency of repair when homologous recombination occurs with a donor in trans. Using a GFP knock-in approach as reporter for HR repair, we were able to confirm the observation that efficient HR repair is independent of an siRNA response at the DSB. We noticed that the requirement for Rad51 was particularly strong in case of circular HR donors, while the siRNA pathway components showed no change for any of the HR donors employed. Finally, a general DSB stress response to camptothecin treatment, which results in the creation of many DSBs scattered throughout the genome, appeared unaffected by mutation of dcr-2 in vivo. This experiment was complicated by a background mutation of the cyp6d2 gene in certain strains that renders the flies hypersensitive to CPT in a dcr-2 independent manner as demonstrated with an ectopic rescue transgene (Table 1). We confirmed that the sensitivity to CTP was not caused by the dcr-2 mutations with the help of a number of different dcr-2 alleles in trans-heterozygous combinations and PCR-based genotyping for known cyp6d2 alleles (Supplementary Figure S7). In addition, trans-heterozygous ago2 mutant flies also showed no CPT hypersensitivity. Taken together, we conclude that in Drosophila melanogaster the outcome of DNA DSB repair does not depend on break-derived siRNAs. It is a formal possibility that alternative backup mechanisms take over if siRNAs cannot be generated. This might result in changes that can only be observed during a time-window that is below the resolution of our assays (readout occurs days after induction of the DNA DSB). Nonetheless, any putative role of siRNAs should be of accessory nature because they are only generated when the break occurs within a transcribed region (5), yet chromosomes must also be repaired when they break in between genes. This is corroborated by the fact that dcr-2 and ago2 are non-essential in flies. In contrast, mutant alleles of core DSB repair factors are generally either lethal or sterile due to problems with meiotic crossover resolution.
Studies with Dicer knock-out mice also allow some conclusions on other DNA repair pathways: During vertebrate B-cell development, specific DNA DSBs are created by the Rag1/2 proteins to initiate rearrangement to a recombined V(D)J locus; these somewhat atypical DNA DSBs are exclusively repaired by NHEJ. Conditional ablation of murine dicer in developing B-cells did not affect repair pathway choice, indicating that the NHEJ pathway is unperturbed in the absence of damage-induced siRNAs. There were, however, indirect effects because miRNAs no longer repressed pro-apoptotic genes and altered transcriptional control of the enzyme terminal deoxynucleotidyl transferase (36). Similar processes may take place in flies, where the MMEJ-factor DNA polymerase theta (mus308) may be repressed by a hairpin-derived endogenous siRNA in a dcr-2 dependent manner (24,26). We have not examined in detail whether this changes the nature of the end-joining products in the absence of dcr-2 because the focus of our manuscript is on the role of the siRNAs generated at the site of the DNA DSB. We do note, however, that depletion of lig4 has a stronger influence on the ratio of homology-directed versus end-joining repair than depletion of mus308 (e.g. Figure 4).
The findings presented in our current study, as well as the previous observation that generation of DSB induced siRNAs depends on transcription, are in line with a role of break-derived siRNAs in RNA quality control rather than DNA repair. Furthermore, the presence of break-derived siRNAs could slow down the kinetics of re-expression after the break has been mended. It is also currently unclear whether in vivo a DNA DSB within a transcriptional unit will lead to run-off or rather stalling of RNA polymerase II. In the latter case, the generation of break-derived siRNAs bears at least conceptual similarity to the recently described identification of transposon transcripts through stalled spliceosomes in Cryptococcus neoformans (37). Since Neurospora qiRNAs are primarily directed against repetitive DNA and thus are potential players in transposon defense, they might also function in a self versus non-self recognition system. Further studies will reveal the precise role of break-derived siRNAs in mRNA quality control as well as the mechanism of dsRNA synthesis at the break.
Supplementary Material
Acknowledgments
The anti-tubulin antibody E7 was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (DFG) [SFB646]. Funding for open access charge: Deutsche Forschungsgemeinschaft [SFB 646]; Ludwig-Maximilians-Universität München.
Conflict of interest statement. None declared.
REFERENCES
- 1.Mehta A., Haber J.E. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2014;6:a016428. doi: 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei W., Ba Z., Gao M., Wu Y., Ma Y., Amiard S., White C.I., Danielsen J.M., Yang Y.G., Qi Y. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Gao M., Wei W., Li M.M., Wu Y.S., Ba Z., Jin K.X., Li M.M., Liao Y.Q., Adhikari S., Chong Z., et al. Ago2 facilitates Rad51 recruitment and DNA double-strand break repair by homologous recombination. Cell Res. 2014;24:532–541. doi: 10.1038/cr.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francia S., Michelini F., Saxena A., Tang D., de Hoon M., Anelli V., Mione M., Carnici P., d'Adda di Fagagna F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michalik K.M., Bottcher R., Forstemann K. A small RNA response at DNA ends in Drosophila. Nucleic Acids Res. 2012;40:9596–9603. doi: 10.1093/nar/gks711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y.S., Nakahara K., Pham J.W., Kim K., He Z., Sontheimer E.J., Carthew R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 7.Okamura K., Ishizuka A., Siomi H., Siomi M.C. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu B., Liu M., Wang J., Zhang X., Wang X., Wang P., Wang H., Li W., Wang Y. DICER-dependent biogenesis of let-7 miRNAs affects human cell response to DNA damage via targeting p21/p27. Nucleic Acids Res. 2015;43:1626–1636. doi: 10.1093/nar/gku1368. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Lee H.C., Chang S.S., Choudhary S., Aalto A.P., Maiti M., Bamford D.H., Liu Y. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature. 2009;459:274–277. doi: 10.1038/nature08041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Q., Ye Q.A., Liu Y. Mechanism of siRNA production from repetitive DNA. Genes Dev. 2015;29:526–537. doi: 10.1101/gad.255828.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunzelmann S., Böttcher R., Schmidts I., Förstemann K. A comprehensive toolbox for genome editing in cultured Drosophila cells. G3: Genes|Genomes|Genetics. 2016;6:1777–1785. doi: 10.1534/g3.116.028241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bottcher R., Hollmann M., Merk K., Nitschko V., Obermaier C., Philippou-Massier J., Wieland I., Gaul U., Forstemann K. Efficient chromosomal gene modification with CRISPR/cas9 and PCR-based homologous recombination donors in cultured Drosophila cells. Nucleic Acids Res. 2014;42:e89. doi: 10.1093/nar/gku289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartig J.V., Esslinger S., Bottcher R., Saito K., Forstemann K. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. EMBO J. 2009;28:2932–2944. doi: 10.1038/emboj.2009.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartig J.V., Forstemann K. Loqs-PD and R2D2 define independent pathways for RISC generation in Drosophila. Nucleic Acids Res. 2011;39:3836–3851. doi: 10.1093/nar/gkq1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forstemann K., Horwich M.D., Wee L., Tomari Y., Zamore P.D. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by Dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elmer K., Helfer S., Mirkovic-Hosle M., Forstemann K. Analysis of endo-siRNAs in Drosophila. Methods Mol. Biol. 2014;1173:33–49. doi: 10.1007/978-1-4939-0931-5_4. [DOI] [PubMed] [Google Scholar]
- 17.Pierce A.J., Johnson R.D., Thompson L.H., Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Behm-Ansmant I., Gatfield D., Rehwinkel J., Hilgers V., Izaurralde E. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 2007;26:1591–1601. doi: 10.1038/sj.emboj.7601588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giuliani G., Giuliani F., Volk T., Rabouille C. The Drosophila RNA-binding protein HOW controls the stability of dgrasp mRNA in the follicular epithelium. Nucleic Acids Res. 2014;42:1970–1986. doi: 10.1093/nar/gkt1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giuliani F., Giuliani G., Bauer R., Rabouille C. Innexin 3, a new gene required for dorsal closure in Drosophila embryo. PLoS One. 2013;8:e69212. doi: 10.1371/journal.pone.0069212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vagin V.V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 22.Horwich M.D., Li C., Matranga C., Vagin V., Farley G., Wang P., Zamore P.D. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 23.Ivanov E.L., S N., Fishman-Lobell J., Haber J.E. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces Cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okamura K., Chung W.J., Ruby J.G., Guo H., Bartel D.P., Lai E.C. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan S.H., Yu A.M., McVey M. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 2010;6:e1001005. doi: 10.1371/journal.pgen.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Czech B., Malone C.D., Zhou R., Stark A., Schlingeheyde C., Dus M., Perrimon N., Kellis M., Wohlschlegel J.A., Sachidanandam R., et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kent T., Chandramouly G., McDevitt S.M., Ozdemir A.Y., Pomerantz R.T. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nat. Struct. Mol. Biol. 2015;22:230–237. doi: 10.1038/nsmb.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ira G., Haber J.E. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 2002;22:6384–6392. doi: 10.1128/MCB.22.18.6384-6392.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas A.M., Hui C., South A., McVey M. Common variants of Drosophila melanogaster Cyp6d2 cause camptothecin sensitivity and synergize with loss of Brca2. G3 (Bethesda) 2013;3:91–99. doi: 10.1534/g3.112.003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim K., Lee Y.S., Carthew R.W. Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes. RNA. 2007;13:22–29. doi: 10.1261/rna.283207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hain D., Bettencourt B.R., Okamura K., Csorba T., Meyer W., Jin Z., Biggerstaff J., Siomi H., Hutvagner G., Lai E.C., et al. Natural variation of the amino-terminal glutamine-rich domain in Drosophila argonaute2 is not associated with developmental defects. PLoS One. 2010;5:e15264. doi: 10.1371/journal.pone.0015264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gasciolli V., Mallory A.C., Bartel D.P., Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 33.Tomari Y., Du T., Zamore P.D. Sorting of Drosophila Small Silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen J., Duan H., Bejarano F., Okamura K., Fabian L., Brill J.A., Bortolamiol-Becet D., Martin R., Ruby J.G., Lai E.C. Adaptive regulation of testis gene expression and control of male fertility by the Drosophila harpin RNA pathway. Mol. Cell. 2015;57:165–178. doi: 10.1016/j.molcel.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deshpande G., Calhoun G., Schedl P. Drosophila argonaute-2 is required early in embryogenesis for the assembly of centric/centromeric heterochromatin, nuclear division, nuclear migration, and germ-cell formation. Genes Dev. 2005;19:1680–1685. doi: 10.1101/gad.1316805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koralov S.B., Mulijo S.A., Galler G.R., Krek A., Chakraborty T., Kanellopoulou C., Jensen K., Cobb B.S., Merkenschlager M., Rajewsky N. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 37.Dumesic P.A., Natarajan P., Chen C., Drinnenberg I.A., Schiller B.J., Thompson J., Moresco J.J., Yates J.R., 3rd, Bartel D.P., Madhani H.D. Stalled spliceosomes are a signal for RNAi-mediated genome defense. Cell. 2013;152:957–968. doi: 10.1016/j.cell.2013.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.