Abstract
Alternative DNA structures that deviate from B-form double-stranded DNA such as G-quadruplex (G4) DNA can be formed by G-rich sequences that are widely distributed throughout the human genome. We have previously shown that Pif1p not only unfolds G4, but also unwinds the downstream duplex DNA in a G4-stimulated manner. In the present study, we further characterized the G4-stimulated duplex DNA unwinding phenomenon by means of single-molecule fluorescence resonance energy transfer. It was found that Pif1p did not unwind the partial duplex DNA immediately after unfolding the upstream G4 structure, but rather, it would dwell at the ss/dsDNA junction with a ‘waiting time’. Further studies revealed that the waiting time was in fact related to a protein dimerization process that was sensitive to ssDNA sequence and would become rapid if the sequence is G-rich. Furthermore, we identified that the G-rich sequence, as the G4 structure, equally stimulates duplex DNA unwinding. The present work sheds new light on the molecular mechanism by which G4-unwinding helicase Pif1p resolves physiological G4/duplex DNA structures in cells.
INTRODUCTION
G-quadruplex (G4) DNA is a four-stranded non-canonical structure held together by Hoogsteen base pairs and further stabilized by monovalent cations K+ or Na+ (1–3). Stable G4 structures were found in sub-telomeres, intron-extron splicing junction sites, untranslated regions, gene bodies and gene expression regulatory regions such as promoters (4). The existence of G4 in living cells has been confirmed using an engineered antibody that can recognize G4 structure with high affinity and specificity (5,6). Initial computational analyses have revealed that there are >375 000 G4 motifs in the human genome (7,8). Recent high-resolution sequencing based method has identified >2 times higher than the previous prediction (9), highlighting the importance of G4 in genome integrity. Indeed, G4s have been shown to be implicated in critical cellular processes including initiation of DNA replication at the origin, restart of the collapsed replication fork, DNA recombination and telomere maintenance (10).
At each cell division in human, 30 000–50 000 DNA replication origins are activated, and it remains unclear how they are selected and recognized by replication factors (11). The recent advances have shown that G-rich repeated elements are present in 67–90% of the DNA replication origins from Drosophila to human cells (12). More importantly, it appears that it is G4 and its orientation that determine the precise position of the replication start site (13). These observations raise the question: how G4 influences its downstream duplex DNA unwinding for providing a platform for replicon assembly?
Similarly, the same question may also be raised for G4-induced replication fork stalling and restart of the collapsed replication fork after G4 unfolding. During DNA replication, replicative helicases separate the two strands to form a two-pronged replication fork. The synthesis along the leading-strand template is continuous and that along the lagging strand is discontinuous, leading to the formation of long single-stranded regions termed as Okazaki fragments (14). If these regions contain G-rich sequences, formation of G4s along the lagging-strand template will slow down replication and increase the likelihood of chromosomal breakage and genomic rearrangement (15,16). Similarly, G4 may also be formed along the leading-strand template in regions of G-rich single-stranded DNA due to transient discordance between the replicative helicase and leading-strand DNA polymerase (17). The cell must face the serious problem of handling these obstacles for an ongoing synthesis of lagging or leading strand by unfolding G4 (18). In this regard, there are at least two fundamental questions: (i) how these G4s are resolved by special helicase? (ii) After unfolding a G4, the helicase immediately meets the downstream duplex DNA, then how the helicase toggles its function or activity from G4 unfolding to duplex unwinding?
Concerning the first question, there are many studies showing that as formation of such stable G4 structures may threaten genomic stability, cells have evolved a special family of helicases to unfold G4 and remove those obstacles. A number of G4-helicases have been identified, illustrated by, but not limited to, RecQ family (19), Pif1 family DNA helicases (20,21) and adenosine triphosphate (ATP)-dependent DExH/D family RNA helicases (22). The Saccharomyces cerevisiae Pif1 helicase (Pif1p), has been shown to suppress genome instability at G4 motifs by means of its potent G4 unwinding activity and to keep cells from replication fork impairment, unusual epigenetic silencing and gross chromosomal rearrangement (23–25), which were otherwise observed in a Pif1p deficient strain (Pif1Δ) (23,26). We and others have studied the molecular mechanism of Pif1p-mediated G4 unfolding (27,28) and found that Pif1p unfolds G4 in two large steps, and then halts at the ss/dsDNA junction, followed by rapid reformation of G4 and re-initiation of unfolding by the same monomer (27). As to the second question, we have previously studied, using stopped-flow method, how the duplex DNA downstream of G4 was unwound by Pif1p helicase and found that G4 greatly stimulates Pif1p-catalyzed duplex DNA unwinding (29). However, the molecular details about Pif1p toggling between G4 unfolding and duplex DNA unwinding cannot be observed in the bulk assay. Therefore, the molecular mechanism of G4-stimulated duplex DNA unwinding still remains to be revealed.
The various single-molecule techniques developed recently have opened a new window to monitor transient events from individual molecules that are otherwise hidden in ensemble-averaged experiments. By taking this advantage, we further characterized the G4-stimulated duplex DNA unwinding using single-molecule fluorescence resonance energy transfer (smFRET) (30). Here we have found that, while a partial duplex DNA could be unwound significantly only at high protein concentrations with a remarkable ‘waiting time’ from protein binding to duplex DNA unwinding, G4 greatly reduces the waiting time and enhances the unwinding fraction. We also found a novel toggling occurs between G4 unfolding and duplex unwinding at a G4/duplex junction, due to Pif1p oligomeric state adjustment. Fluorescence anisotropy studies indicated the ‘waiting time’ was in fact related to Pif1p dimerization process which is required for duplex DNA unwinding. These observations imply that dimerization may play an important role in regulation of helicase activities in resolving different DNA structures. In addition, G-rich sequence was found to similarly stimulate duplex unwinding as well as folded G4, and we think that both G-rich sequence and folded G4 are responsible for the stimulation of downstream duplex DNA unwinding. Altogether, these new findings may deepen our understanding of the unwinding mechanisms of Pif1p helicase on different DNA substrates and provide insights into the molecular mechanism behind G4-stimulated duplex DNA unwinding by Pif1p.
MATERIALS AND METHODS
Buffers
Pif1p reaction buffer contains 5 mM MgCl2, 50 mM NaCl in 25 mM Tris–HCl, pH 7.5 with 2 mM dithiothreitol (DTT), unless otherwise specified. For single-molecule measurements, 0.8% D-glucose, 1 mg/ml glucose oxidase (266 600 units/g, Sigma), 0.4 mg/ml catalase (2000–5000 units/mg, Sigma) and 1 mM Trolox were added to the reaction buffer (30).
DNA constructs
All oligonucleotides required to make the DNA substrates were purchased from Sangon Biotech (Shanghai, China). Sequences and labeling positions of all the oligonucleotides were listed in Supplementary Table S1. For DNA constructs used in single-molecule measurements, DNA was annealed with 1:3 mixture of the stem and ssDNA or G4 strands by incubating the mixture at 95°C for 5 min, then slowly cooling down to room temperature in about 7 h. The strand without biotin was used in excess to reduce the possibility of having non-annealed strand anchored at the coverslip surface. The concentration of stem strand was 2.5 μM and all annealing were carried out in annealing buffer containing 50 mM NaCl, 25 mM Tris-HCl, pH 7.5, unless otherwise specified.
Protein purification
The plasmid encoding yeast Pif1p gene was a gift from Dr Zakian. The protein expression and purification were performed essentially according to Boule and Zakian with minor modifications (31). Truncated BLM642-1290 was expressed and purified as previously described (32).
Single-molecule fluorescence data acquisition
Single-molecule FRET study was carried out with a home-built objective-type total-internal-reflection microscopy. Cy3 was excited by a 532-nm Sapphire laser (Coherent Inc., USA). An oil immersion objective (100×, N.A.1.49) was used to generate an evanescent field of illumination. Fluorescence signal from Cy3 and Cy5 were split by a dichroic mirror, and finally collected by an electron-multiplying charge-coupled device camera (iXON, Andor Technology, South Windsor, CT, USA). Fluorescence imaging processes were controlled and recorded by MetaMorph (Molecular Device, California). The coverslips (Fisher Scientific, USA) and slides were cleaned thoroughly by a mixture of sulfuric acid and hydrogen peroxide, acetone and sodium ethoxide, then the surfaces of coverslip were coated with a mixture of 99% mPEG (m-PEG-5000, Laysan Bio, Inc.) and 1% of biotin-PEG (biotin-PEG-5000, Laysan Bio, Inc.). Streptavidin (10 μg/ml) in buffer containing 50 mM NaCl, 25 mM Tris–HCl, pH 7.5, were added to the microfluidic chamber made of the polyethylene glycol (PEG) coated coverslip, and incubated for 10 min. After washing, 50 pM DNA were added to the chamber and allowed to be immobilized for 10 min. Then free DNA was removed by washing with the reaction buffer. After that, chamber was filled with the reaction buffer with an oxygen scavenging system (0.8% D-glucose, 1 mg/ml glucose oxidase, 0.4 mg/ml catalase and 1 mM Trolox). Imaging was initiated before Pif1p and ATP was flowed into the chamber. We used an exposure time of 100 ms for all single-molecule measurements at a constant temperature of 22°C. To obtain the fraction of DNA unwinding with time, a series of movies were recorded with 1-s duration at different times, and the acceptor spots were counted to represent the number of remaining DNA molecules.
FRET data analyses
The FRET efficiency was calculated using IA/(ID+IA), where ID and IA represent the intensity of donor and acceptor, respectively. The FRET value above 1 is due to background subtraction from very low intensity in the donor channel, giving rise to negative donor intensity. Basic data analysis was carried out by scripts written in Matlab, and all data fitting were generated by Origin 8.0. Histograms were fitted by multi-peak Gaussian distributions, with the peak position unstrained. Waiting time was measured manually from individual FRET traces, and the resulting histograms were fitted with a single-exponential decay. An auto-mated step-finding method (from http://bio.physics.illinois.edu/HaMMy.asp) was employed to characterize the association and dissociation of Pif1p in Supplementary Figure S6A, and the time ton and toff were determined accordingly.
Equilibrium DNA-binding assay with Pif1p
Binding of Pif1p to DNA was analyzed by fluorescence polarization assay using Infinite F200 PRO (TECAN) at a constant temperature of 25°C. DNA labeled only with carboxy-fluorescein (FAM) was used in the present study (Supplementary Table S1). Various amounts of protein were added to a 150 μl aliquot of binding buffer (25 mM Tris–HCl, pH 7.5, 50 mM NaCl, 5 mM MgCl2 and 2 mM DTT) containing 5 nM DNA. Each sample was allowed to equilibrate in solution for 5 min, after which fluorescence polarization was measured. A second reading was taken after 10 min, in order to ensure that the mixture was well equilibrated. Less than 5% change was observed between the 5- and 10-min measurements, indicating that equilibrium was reached in 5 min. If the binding curve is in sigmoidal shape, the curve is fitted by Hill equation: y = [Pif1p]n/(KDn+[Pif1p]n). y is binding fraction, KD is the equilibrium dissociation constant and n is Hill coefficient, describing cooperativity. n > 1 indicates more than one Pif1p can bind to one DNA.
Kinetics of Pif1p binding to DNA substrates
Binding of Pif1p to DNA was analyzed by fluorescence polarization assay (33,34) using Infinite F200 PRO (TECAN). DNA labeled only with FAM was used in this study (Supplementary Table S1). Binding kinetics were measured at a constant temperature of 25°C in a two-syringe mode, where Pif1p helicase was in one syringe while DNA substrate was in another syringe. Each syringe contained buffer of 5 mM MgCl2, 50 mM NaCl in 25 mM Tris–HCl, pH 7.5 with 2 mM DTT and the measurement was initiated by rapid mixing. The final concentrations of DNA and Pif1p were 40 and 40–200 nM, respectively. The binding time constant t1 and t2 were obtained by fitting the data with y = y0-A1exp(-t/ t1)-A2exp(-t/t2) where t represents the time, y represents the fluorescence anisotropy and y0 is a constant.
Circular dichroism spectropolarimetry
Circular dichroism (CD) experiments were performed with a Bio-Logic MOS450/AF-CD optical system (BioLogic Science Instruments, France) equipped with a temperature-controlled cell holder, using a quartz cell with 1-mm path length. A 2.5 μM solution of G-rich DNA or G4 DNA was prepared in 25 mM Tris–HCl, pH 7.5, with different cation. CD spectra were recorded in the UV (220–320 nm) regions in 0.75 nm increments with an averaging time of 2 s at 25°C.
RESULTS
Pif1p-mediated duplex DNA unwinding is characterized by a waiting time
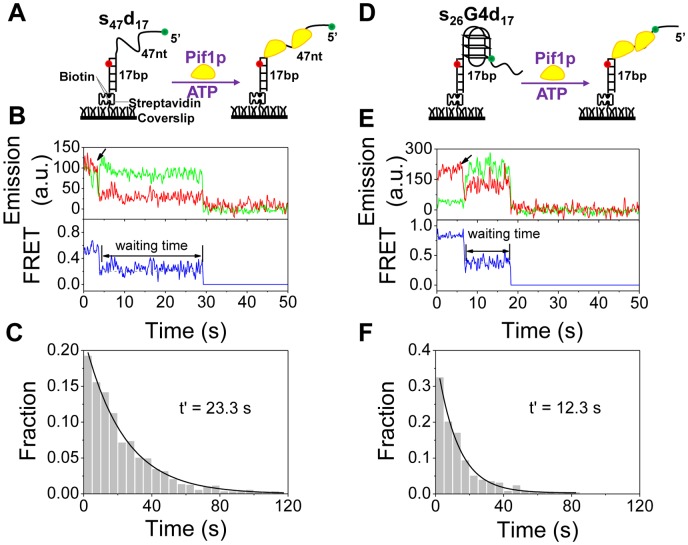
To understand how G4 stimulates Pif1p-catalyzed duplex DNA unwinding, it is necessary to determine the unwinding properties of Pif1p helicase with a partial duplex DNA alone. In order to use an appropriate labeled partial duplex DNA which allows us to monitor FRET signal without potential influence of inhibitory effect for protein translocation due to dye labeling at ss/dsDNA junction which may stabilize dsDNA (35,36), we screened a series of partial duplex DNAs labeled at different relative position of Cy3 and Cy5. Finally, the substrate s47d17, which was labeled with Cy3 at 5′-end of 64 nt-ssDNA and Cy5 at fifth nucleotide from the 3′ terminal of biotinylated-ssDNA, was used for monitoring DNA unwinding (Figure 1A).
Figure 1.
G4 structure reduces the waiting time for downstream duplex DNA unwinding by Pif1p. (A) Schematic representation of experimental procedures for partial duplex DNA unwinding. (B) Representative fluorescence emission and FRET traces for s47d17 unwinding by 80 nM Pif1p and 2 mM ATP. Upon addition of Pif1p and ATP as indicated by the arrow, FRET signal decreases abruptly, indicating the binding of Pif1p. It then remains at a constant value for ∼25 s and then the Cy3 and Cy5 signals disappear simultaneously as the duplex is separated. The waiting time is defined as the time between Pif1p binding and duplex separation. (C) Histogram of the waiting time follows an exponential decay with a time constant of 23.3 s. (D) Schematic representation of experimental procedures for G4/ds DNA unwinding. (E) Representative fluorescence emission and FRET traces for Pif1p-catalyzed unwinding of s26G4d17. Upon Pif1p and ATP addition as indicated by the arrow, FRET signal decreases rapidly, then keeps at constant value for ∼12 s, until both Cy3 and Cy5 signals disappear abruptly. (F) Histogram of the waiting time follows an exponential decay with a time constant of 12.3 s.
By inspecting the individual smFRET traces obtained at protein concentration ranges between 2–60 nM, we found that Pif1p switches between binding and dissociate status (Supplementary Figure S1). However, above 80 nM Pif1p, upon addition of 2 mM ATP, smFRET value decreased from 0.5 to 0.2, then lasted for ∼25 s at stable FRET value and suddenly disappeared (Figure 1B). The characters of smFRET traces at higher protein concentration can be interpreted as that after forming a stable protein–DNA complex, Pif1p translocates to ss/dsDNA junction, lasts there for ∼25 s and finally separates the two DNA strands, as judged from the simultaneous disappearance of Cy3 and Cy5 signal (Figure 1B). We designated the time from initial Pif1p binding to the final duplex unwinding as waiting time. The histogram of the waiting time can be well fitted by a single exponential decay with a time constant of 23.3 s (Figure 1C), indicating that an average waiting time of about 23.3 s was required before a 17-bp duplex DNA was abruptly unwound. Taken together, the above results indicate that a partial duplex DNA was unwound through multiple Pif1p acting in concert and requires a long waiting time to adjust the protein conformation for effective DNA unwinding.
G4 structure significantly reduces the waiting time for duplex DNA unwinding
Having characterized the unwinding properties of Pif1p for duplex DNA alone, we then set about investigating how G4 structure stimulates the duplex DNA unwinding as we have previously revealed in bulk assays (29). For this purpose, we designed a DNA substrate which contains a three-layered G4 with a 5′ tail of 26-nt ssDNA and a 3′ tail of 17-bp dsDNA (Figure 1D, named s26G4d17), which mimics an ongoing synthesis of lagging-strand stalled by G4 and is similar to that we used for bulk assays (29). s26G4d17 was labeled with Cy3 at the 26th nucleotide from the 5′ end in the G4 strand and with Cy5 at the fifth nucleotide from the 3′ end in the complementary strand. The dyes are spaced in this way so that FRET change between the two fluorophores can report the unfolding of a single G4, and the simultaneous disappearance of Cy3 and Cy5 signals would reflect the escape of the unwound DNA from the coverslip.
smFRET trajectories recorded with s26G4d17 significantly differed from that obtained with the s47d17 substrate. After addition of 80 nM Pif1p and 2 mM ATP, the FRET value abruptly dropped to ∼0.4 and then remained in this state for about 10 s before suddenly disappearing completely (Figure 1E). This phenomenon was similarly interpreted as above that, upon addition of Pif1p and ATP, Pif1p unfolds G4 immediately, then translocates to the ss/dsDNA junction and waits there for about 10 s, and finally unwinds the duplex DNA abruptly. To confirm the above interpretation, we plotted the FRET distribution before duplex DNA unwinding from 97 traces and by Gaussian fitting, we obtained two peaks at 0.37 and 0.91, respectively (Supplementary Figure S2). These values are quite close to the two peaks at 0.39 and 0.90 in the FRET distribution of G4 under a dilute buffer condition (20 mM NaCl) that correspond to the unfolded (EFRET = 0.39) and completely folded G4 structure (EFRET = 0.90), respectively (27). This confirms that Pif1p does not immediately unwind the downstream duplex DNA after unfolding G4. We then measured the waiting time for each individual trace and constructed a histogram as shown in Figure 1F. The waiting time follows a single exponential decay quite well as we observed before, but with an average time constant of 12.3 s, which was significantly reduced as compared to that determined with the duplex DNA without G4 insertion (s47d17, Figure 1C). The potential possibility that the internal labeled Cy3 could inhibit protein translocation was ruled out by studying the same substrate in which Cy3 was attached at the 5′-teminal of the ssDNA tail (Supplementary Figure S3).
One may query the above phenomenon in comparing with the previous smFRET studies in which Pif1p was observed to repeatedly unwind G4 DNA as monomer, but without waiting time (27). In fact, this discrepancy comes from the different experimental condition used. In our previous study, the repetitive unfolding/folding of G4 phenomenon was observed with lower protein and ATP concentrations (5 nM and 25 μM, respectively). In the present study, according to our experimentally determined condition, higher concentrations of protein (80 nM) and ATP (2 mM) were used to study G4-stimulated duplex DNA unwinding. Higher ATP concentration stabilize Pif1p binding on the substrate (27) and the unfolded G4 kept in such linear status should be coated by high protein concentration Pif1p, impeding G4 refolding.
To probe whether the stimulating effect was only associated with three-layered G4, a substrate s264-G4d17 which contains a four-layered G4 was studied. Supplementary Figure S4A shows the FRET trace recorded with this new substrate, and the waiting time distribution also follows an exponential decay with a time constant of 11.0 s (Supplementary Figure S4B), indicating that the acceleration of waiting process is not restricted with a specific G4 structure.
To rule out the possibility that any G4-unfolding helicase can display the waiting time phenomenon with G4/ds substrate structure, we measured BLM642-1290-mediated G4/dsDNA unwinding and found that G4/ds was unwound continuously without any visible waiting step (Supplementary Figure S4C), indicating that G4-stimulated duplex DNA unwinding by reducing the waiting time is an intrinsic property of Pif1p rather than G4/ds substrate caused artifact. Overall, these results provide a mechanistic insight into how G4 structure stimulates its downstream duplex DNA unwinding: the G4 structure greatly reduces the waiting time that is otherwise a definite rate-limiting step for duplex DNA unwinding.
High Pif1p concentrations reduce the waiting time through accelerating protein dimerization
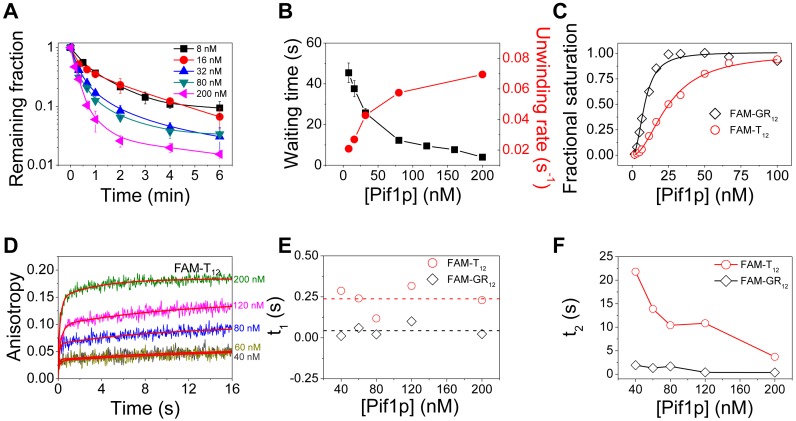
To determine whether G4-induced reduction in waiting time was influenced by protein concentration, we firstly determined the unwinding fraction by counting the decrease number of Cy5 spots over time with Pif1p concentration ranging from 8 to 200 nM (28). The unwinding fractions increase as the protein concentration increasing (Figure 2A). In consistent with the above results, the unwinding rate at each protein concentration, determined by fitting the unwinding kinetic curves with single exponential equation, increases with increasing protein concentrations (Figure 2B, circle). More importantly, the determined waiting time from smFRET traces is decreased as Pif1p concentration increased, and becomes as low as 4 s at 200 nM Pif1p (Figure 2B). Altogether, the fact that the striking anti-correlation between waiting time and unwinding rate as a function of protein concentration indicates that higher protein concentration resulted in oligomerization which reduces significantly the waiting time and enhances the unwinding amplitude and unwinding rate.
Figure 2.
Pif1p concentration influences the waiting time before duplex DNA unwinding. (A) Fractions of remaining s26G4d17 molecules on coverslip versus time after addition of 2 mM ATP and different concentrations of Pif1p. (B) The average waiting time for s26G4d17 unwinding decreases significantly with increasing Pif1p concentration, while the unwinding rate obtained from single exponential fitting of curves in (A) increases with increasing Pif1p concentration. (C) DNA binding curve determined with fluorescence anisotropy assay using 5 nM FAM-T12 or FAM-GR12 and increasing Pif1p concentration under equilibrium conditions. The titration curves were fitted by the Hill equation y = [Pif1p]n/(KDn+ [Pif1p]n) with KD of 25.2 nM, Hill coefficient of 1.97 ± 0.08 for FAM-T12 and KD of 8.7 nM, Hill coefficient of 2.20 ± 0.27 for FAM-GR12, y represents the binding fraction. (D) Stopped-flow kinetic time-courses for Pif1p binding to a FAM-T12. The final concentration of FAM-labeled DNA was 40 nM and that of Pif1p was from 40 to 200 nM. Fluorescence signal was recorded upon addition of Pif1p. The initial binding time course was fitted to a double-exponential process: y = y0 − A1exp(−t/t1) − A2exp(−t/t2), where t represents the time, y represents the fluorescence anisotropy and y0 is a constant. The two binding time constants t1 and t2 are shown in (E) and (F). Lines in (F) are the simple connections of the individual data points.
The above observations lead us to think that the so called ‘waiting time’ may be in fact related to the time needed for Pif1p to assemble into an oligomer that is required for the duplex DNA unwinding. To determine the nature of the oligomer, we further investigated the DNA binding properties of Pif1p under equilibrium conditions at a series of Pif1p concentrations by fluorescence anisotropy measurements. The binding of Pif1p with 12-nt ssDNA FAM-T12 exhibited a sigmoid curve pattern which was best fitted by the Hill equation with a Hill coefficient of 1.97 and an equilibrium dissociation constant KD of 25.2 nM (Figure 2C). Because the ssDNA is too short to accommodate two independent monomers side by side (37), this result suggests that ssDNA binding may induce dimerization of Pif1p, being consistent with the previous observations (37).
Since the binding kinetics may help to clarify the nature of dimer formation in more detail, we then studied the Pif1p binding kinetic of FAM-T12 with increasing protein concentration. The binding processes can be best fitted by a two-step kinetic model (Figure 2D), suggesting that Pif1p successively binds to DNA first as a monomer, and then forms a dimer. We assigned the first step to the binding of the first monomer to the ssDNA with binding time constant t1, and the second step to binding of the second monomer with binding time constant t2. While t1 keeps almost constant (t1 = 0.24 s, Figure 2E), t2 decreases from 22 to 5 s as the protein concentration increases from 40 to 200 nM (Figure 2F). These results can be interpreted as that the ssDNA was quickly coated with monomeric Pif1p and the higher protein concentration enhances the dimer formation. Interestingly, there is a correlation between the increase in dimer formation rate (decrease in t2) and reduction in the waiting time (compare Figure 2B with F). Thus, higher protein concentration accelerates protein dimerization and reduces the waiting time, and consequently initiates duplex DNA unwinding.
G4 stimulating effect is stronger with the substrates mimicking DNA replication forks
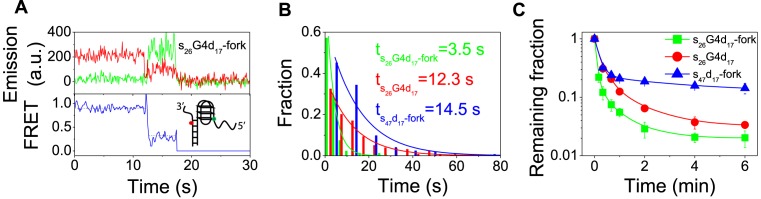
Growing evidence indicates that G4 structures indeed form in cell (5), probably in DNA replication origin foci and replication forks area where G4 exists at one side of forked ss/ds DNA junction. To further study G4 stimulation effect under the conditions which may be relevant biological context, we modified s26G4d17 substrate by adding a 10-nt at 3′ end of ss/dsDNA junction (s26G4d17-fork).
Similarly as before, the traces of these smFRET display the previously observed waiting time characters (Figure 3A). However, the waiting time was significantly reduced from 12.3 to 3.5 s due to the addition of the 10-nt 3′ end in the s26G4d17-fork substrate (Figure 3B). Meanwhile, the kinetic of unwinding of each substrate was determined under the same experimental conditions (Figure 3C). From the results shown in Figure 3B and C, it can be seen that the presence of G4 structure at forked duplex DNA greatly enhanced the duplex unwinding efficiency, which is correlated with the reduction of waiting time. We also determined the waiting time with a forked duplex DNA (s47d17-fork) under the same experimental condition. The obtained value (14.5 s) is much longer than the 3.5 s for s26G4d17-fork, further confirming the stimulation effect of G4. Altogether, the above results indicate that G4 displays maximal stimulating effects for duplex DNA unwinding in the context of the substrates with structures resembling DNA replication forks.
Figure 3.
G4 stimulating effect is stronger in the presence of a fork structure. (A) Representative fluorescence emission and FRET traces for s26G4d17-fork unwinding at 80 nM Pif1p and 2 mM ATP. (B) For s26G4d17-fork and s26G4d17, the waiting time distributions follow a single-exponential decay with different time constants. The distribution for s26G4d17 is replotted from Figure 1E. The Pif1p and ATP concentrations are 80 nM and 2 mM, respectively. (C) Fractions of remaining molecules on the coverslip decrease with time after addition of 80 nM Pif1p and 2 mM ATP. The unwinding rates for s26G4d17-fork and s26G4d17 are 0.167 s−1 and 0.057 s−1 by single-exponential fitting.
G-rich sequence stimulates Pif1p-catalyzed duplex DNA unwinding
We noted that after quickly unfolding, G4 remains in unfolded state, as judged from the smFRET value at ∼0.4 (Figure 1E). Therefore, during the so called ‘waiting time’, Pif1p is in fact kept in G-rich linear state. Thus we were wondering whether G-rich sequence alone stimulates duplex DNA unwinding and is enough to induce Pif1p dimerization and enhance the duplex DNA unwinding.
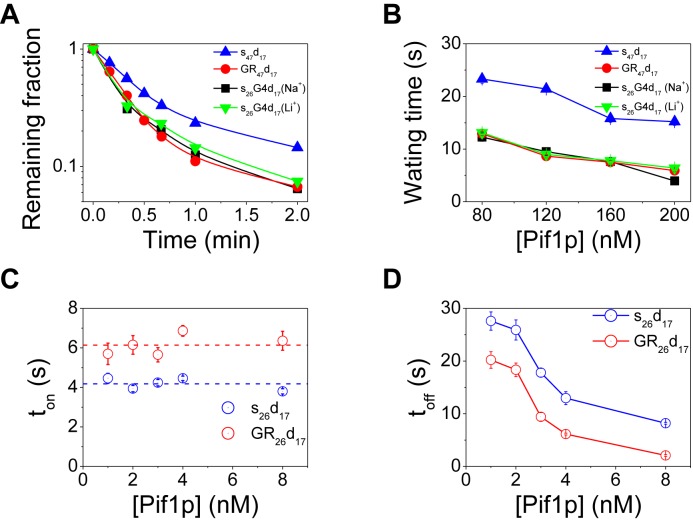
To address this question we prepared an s26G4d17 substrate derivative, substrate GR47d17 that contains, instead of a G4 motif, a G-rich sequence which cannot fold into G4 structure as confirmed by CD spectra (Supplementary Figure S5A). We studied the Pif1p-catalyzed duplex unwinding with this new substrate together with s26G4d17. The fraction of DNA unwound was determined by counting the fluorescent spots as before and the unwinding rate was obtained from a single-exponential fitting of the data (Figure 4A). The results show that Pif1p unwinds both s26G4d17 and GR47d17 almost in the same efficiency, as judged from unwinding fractions in two minutes (both are ∼ 94%) and unwinding rates (0.057 s−1 and 0.054 s−1). This finding suggests that G-rich sequence can stimulate duplex DNA unwinding even without the G4 structure.
Figure 4.
Effects of G-rich sequence on Pif1p binding and unwinding of downstream duplex DNA. (A) Fractions of remaining DNA molecules on coverslip decrease with time after addition of 80 nM Pif1p and 2 mM ATP. The unwinding rates for s26G4d17, s26G4d17 (Li+), GR47d17 and s47d17 are 0.057 s−1, 0.054 s−1, 0.054 s−1 and 0.036 s−1 by single-exponential fittings. (B) The average waiting times for the unwinding of s47d17, GR47d17, s26G4d17 and s26G4d17 (Li+) decrease with increasing Pif1p concentration. (C and D) The dwell time ton for Pif1p to remain bound to ss/dsDNA with a random sequence or G-rich sequence overhang, and the time interval of two successive binding at protein concentration from 1–8 nM. ton and toff were obtained from single-molecule fluorescence traces in Supplementary Figure S6. Lines in (D) are the simple connections of the individual data points.
To further confirm the stimulation effect of G-rich sequence without G4 structure, we performed unwinding reactions with its cognate DNA substrate s26G4d17 prepared in the presence of LiCl (s26G4d17 (Li+)), which does not support G4 formation even with the existence of 10 mM Mg2+ (27), as confirmed by CD (Supplementary Figure S5B). The unwinding reactions were carried out with the buffer containing 50 mM LiCl and 2 mM MgCl2. The results shown in Figure 4A indicate that Pif1p-catalyzed DNA unwinding of s26G4d17 (Li+) is similarly as with substrates s26G4d17 (Na+) and GR47d17.
We then further analyzed smFRET traces recorded with the above substrates and determined the corresponding waiting times. A comparison of the waiting times deduced from GR47d17, s26G4d17 and s26G4d17 (Li+) indicates that they are almost identical at each protein concentration, but significantly smaller than that for the s47d17 substrate (Figure 4B). This observation suggests that G-rich sequence indeed can stimulate duplex DNA unwinding by reducing the waiting time even without forming G4 structure. Taken together, the above findings clearly suggest that like the G4 DNA structure, the G-rich sequence has the activator role on the duplex DNA unwinding activity of Pif1p.
To clarify the mechanism by which G-rich sequence could stimulate downstream duplex DNA unwinding, we performed three additional experiments: (i) titration of DNA binding with the G-rich DNA (FAM-GR12, Figure 2C). Similarly to the above determined titration curve obtained with FAM-T12, the binding isotherm can be best fitted by the Hill equation with a Hill coefficient of 2.20, but with KD value (8.70 nM) which is smaller than that determined with FAM-T12 (25.2 nM), indicating that Pif1p binds tightly to G-rich sequence and induces a dimer formation. (ii) Characterizing the binding properties of Pif1p toward both substrates s26d17 (random ssDNA at 5′ end) and GR26d17 (G-rich ssDNA at 5′ end) under the same experimental conditions by single-molecule protein-induced fluorescence enhancement (PIFE, Supplementary Figures S6 and 7) (38). When Cy3 was labeled at the junction of the partial duplex DNA instead of the 5′ end, we observed that, at lower protein concentrations, Cy5 intensity jumped randomly between two levels, while the Cy3 intensity remained constant (Supplementary Figure S7), suggesting the repetitive association with and dissociation from the DNA substrate of Pif1p without duplex unwinding. The association and dissociation kinetics between Pif1p and the both substrates were determined by recording a series of fluorescence emission trajectories at protein concentration from 1–8 nM (Supplementary Figure S6). We defined two parameters, the dwell time ton for the helicase to remain bound to the DNA substrate, and the time interval toff between two successive binding events (Supplementary Figure S6A). Typical distribution of ton and toff are given in Supplementary Figure S6B, C, E and F. ton values are independent of protein concentrations, and that obtained with GR26d17 is ∼ 1.5 times higher than s26d17 (6.14 versus 4.18 s, Figure 4C). toff decreased as protein concentration increased, however toff values of s26d17 are systematically higher than GR26d17 (Figure 4D). As expected for binary reaction, koff (1/ton) is independent of Pif1p concentration while kon (1/toff) has a linear dependence on it (Supplementary Figure S6D and G) (38). These results altogether indicate that Pif1p binds GR26d17 more readily and tightly than s26d17. (iii) Finally, we measured the binding kinetics of Pif1p with fluorescent-labeled 12-nt G-rich sequence (FAM-GR12) with a bulk anisotropy assay (33,34) as we performed above. Similarly, the binding processes can be best fitted by a two-step kinetic model (Supplementary Figure S8), as we found for FAM-T12. The t1 value for FAM-T12 is 6-fold higher than that for FAM-GR12 (0.24 versus to 0.04), indicating that binding rates of the first monomer with the G-rich sequence are 6 folds higher than that with the poly-T (Figure 2E). Interestingly, the binding constant t2 (represent for dimer formation rate) is systematically higher than for FAM-GR12 (Figure 2F). The differences between them are 20- and 3-folds at protein concentration 40 and 200 nM, respectively, indicating that Pif1p forms an active dimer on FAM-GR12 at least 20 times rapidly than on FAM-T12 at lower protein concentration.
Altogether, these results revealed the molecular mechanism by which G4/G-rich sequence stimulate duplex DNA unwinding: Pif1p binds more rapidly and quickly to form a stable dimer with G-rich sequence than with poly-T, thus reducing the waiting time and enhancing the duplex DNA unwinding.
DISCUSSION
The mechanisms of Pif1p-mediated G4 DNA unfolding have been investigated previously by two smFRET studies, and both studies revealed Pif1p monomers to be a poor helicase to unwind duplex DNA (27,28). Interestingly, we have recently found that the duplex DNA unwinding activity could be significantly stimulated by the presence of G4 DNA upstream of the duplex DNA. Here, we used smFRET method to characterize Pif1p-catalyzed duplex unwinding with different substrates containing G4/G-rich at ss/dsDNA junctions, which Pif1p may encounter in the biological context. The present work provides a comprehensive assessment of G4-stimulated duplex DNA unwinding at the single-molecule level.
The existence of a ‘waiting time’ between G4 and duplex DNA unwinding revealed that Pif1p used different mechanisms to unwind G4 and duplex DNA, respectively
We have previously found that the presence of G4 structure may greatly stimulate the Pif1p helicase to unwind duplex DNA in our stopped-flow study, and proposed that this stimulation results from G4-enhanced Pif1p dimerization (29). However, the underlying mechanism by which Pif1p sequentially unwind G4/duplex DNA was still obscure. In the current study, we took the advantage of smFRET to directly visualize the molecular detail of G4-stimulated duplex DNA unwinding. In particular, the covalently linked G4/duplex DNA which mimic the physiological substrate allows us to compare side by side the helicase activities of Pif1p with G4 and duplex DNA. The most striking feature is that Pif1p unfolds rapidly G4 DNA, but it does not successively perform the immediate unwinding of the downstream duplex DNA. Instead, Pif1p dwells at the ss/dsDNA junction with a waiting time. The fact that G4 DNA could greatly reduce the waiting time suggests that Pif1p unwinds G4 and duplex DNA through different mechanisms. Indeed, we and others have shown that Pif1p repetitively unfolds G4 as a monomer (27,28), while dimerization might be required for duplex DNA unwinding as evidenced by: (i) analytical sedimentation velocity analysis shows that binding to ssDNA induces Pif1p dimerization, and formation of dimer prevents an additional oligomerization (37); (ii) our recent stopped-flow study suggests Pif1p unwinds duplex DNA as a dimer, which is characterized by both equilibrium DNA binding assay and dynamic light scattering measurement (29). Importantly and interestingly, we captured snapshots of the toggling of Pif1p during the transition from a state for G4 unfolding to that for duplex unwinding, which is characterized by a waiting time. This feature appears unique for Pif1p family helicase, since Bloom syndrome protein (BLM) (39), Werner and Escherichia coli RecQ do not exhibit such properties (unpublished observation).
The mechanism of G4-stimulated duplex DNA unwinding
Two possible scenarios may be envisioned to explain the G4-induced duplex DNA unwinding phenomenon: (i) the special topology of G4 may convert the monomeric Pif1p from an inactive to an active conformation for unwinding duplex DNA; (ii) G4 induces an oligomerization of Pif1p, which is required for efficient duplex DNA unwinding. As our previous bulk assay (29), the present study also supports the latter scenario. In addition, we further verified the link between Pif1p dimerization and efficient duplex DNA unwinding by that: (i) binding between Pif1p and DNA under equilibrium conditions exhibited a sigmoid curve pattern which was fitted with a Hill coefficient ∼2, suggesting that ssDNA binding may induce dimerization of Pif1p (Figure 2C); (ii) the binding kinetics between Pif1p and ssDNA exhibit two steps, assigned to Pif1p binding to DNA and Pif1p dimerization, respectively (Figure 2D and Supplementary Figure S8); (iii) the waiting time before duplex unwinding exhibits a single-exponential decay (Figures 1C, F and 3B). As the distribution of duration time can provide information about the number of intermediates or rate limiting steps, even those steps cannot be observed directly (40), the single-exponential decay suggests that there is only one rate-limiting step, which might be an assembly-state adjustment for Pif1p before unwinding the duplex DNA. Therefore, we propose that the stimulation of duplex DNA unwinding by G4 should result from enhanced helicase dimerization.
More importantly, smFRET studies not only further confirm our previously proposed G4-induced dimerization, but also reveal the origin of G4-stimulated duplex DNA unwinding that is otherwise impossible to explore in our previous bulk assay. As the G4 structure is rapidly disrupted upon Pif1p addition (Figure 1E) and the unfolded G-rich ssDNA is then always occupied by Pif1p during the following waiting step, the G4 sequence cannot refold back to a folded structure as revealed by the recorded smFRET trajectories (Figure 1E). Therefore, we may deduce that it should be the G-rich sequence rather than the G4 structure that is responsible for reducing the waiting time, and consequently stimulating the duplex DNA unwinding. This inference is further verified by the observations that a G-rich sequence which cannot fold into a higher order structure can also stimulate duplex DNA unwinding, and in a Li+ buffer in which G4 cannot be formed, the stimulation effect still exists for the DNA substrate with a G4 motif (Figure 4A and B).
Previous studies have shown that Pif1p binds to telomeric or other G-rich DNA with a much higher affinity compared to random sequence DNA (23,41). Our current study reveals that G4 motif or G-rich sequence may enhance Pif1p-catalyzed duplex unwinding. In addition, we further revealed the mechanism behind the stimulation effects of G-rich DNA. From binding kinetics study, equilibrium binding measurements and single-molecule PIFE study, we confirmed that Pif1p has a much higher binding affinity for G-rich sequence than for other random sequences, therefore G-rich sequence enhances the protein dimerization process.
Potential significances of G4/G-rich stimulated duplex DNA unwinding in vivo
Many studies from different laboratories with different approaches have revealed that Pif1p unwinds duplex DNA with very low activity (Supplementary Table S2). This intrinsic biochemical property of Pif1p appears to be inconsistent with its diverse cellular functions in telomere lengthening, processing Okazaki fragments, promoting break-induced replication and suppressing DNA damages at G4 sites (25,26,42,43). Our discoveries in the previous (29) and present work provide hints for understanding how Pif1p achieves its biological functions by means of G4/G-rich stimulating duplex DNA unwinding. In model systems such as S. cerevisiae, while most cells use telomerases to elongate telomeres, some cells use recombination-dependent DNA replication mechanism for maintenance of eroding telomeres, known as alternative lengthening of telomeres (ALT) pathway (44). It is well documented that Pif1p plays an important role in ALT (43,45,46). In this process, the 3′-G-rich single-strand tail invades an intact donor molecule and results in a structure known as D-loop where the invading strand is replicated (44,45). Pif1p protein plays two essential roles during the bubble migration (Supplementary Figure S9): (i) Pif1p in combination with Polδ, participates in template strand separation to promote DNA synthesis; and (ii) Pif1p displaces the newly synthesized strand from template by unwinding duplex DNA (42). The two processes must be coordinated in the sense that the long duplex DNA formed between the template and the newly synthesized DNA will be rapidly accumulated if Pif1p cannot efficiently unwind the newly synthesized strand. In this regard and according to our findings, the G-rich telomere sequence will confer Pif1p a high unwinding activity to enhance the bubble migration and telomere elongation, otherwise, the telomere replication will fail. Due to the space limitation, we will not discuss how Pif1p participates in other biological processes by G4/G-rich stimulating duplex DNA unwinding in great detail. Providing in vivo evidences to support our in vitro findings is a current undertaking project.
Supplementary Material
Acknowledgments
We thank Xi's laboratory members and Dr. Malcolm Buckle for insightful discussions. The Research was conducted within the context of the International Associated Laboratory ‘Helicase-mediated G-quadruplex DNA unwinding and Genome Stability’.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31370798, 11304252, 11574252, 31301632]; Northwest A&F University Startup Funding [Z101021102 to X.-G.X., Z111021205 to X.-M.H.]. Funding for open access charge: College of Life Sciences, Northwest A&F University.
Conflict of interest statement. None declared.
REFERENECS
- 1.Lane A.N., Chaires J.B., Gray R.D., Trent J.O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bochman M.L., Paeschke K., Zakian V.A. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maizels N., Gray L.T. The G4 genome. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murat P., Balasubramanian S. Existence and consequences of G-quadruplex structures in DNA. Curr. Opin. Genet. Dev. 2014;25:22–29. doi: 10.1016/j.gde.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 5.Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biffi G., Di Antonio M., Tannahill D., Balasubramanian S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014;6:75–80. doi: 10.1038/nchem.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Todd A.K., Johnston M., Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huppert J.L., Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015;33:877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- 10.Balasubramanian S., Neidle S. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009;13:345–353. doi: 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mechali M. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat. Rev. Mol. Cell Biol. 2010;11:728–738. doi: 10.1038/nrm2976. [DOI] [PubMed] [Google Scholar]
- 12.Cayrou C., Coulombe P., Puy A., Rialle S., Kaplan N., Segal E., Mechali M. New insights into replication origin characteristics in metazoans. Cell Cycle. 2012;11:658–667. doi: 10.4161/cc.11.4.19097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valton A.-L., Hassan-Zadeh V., Lema I., Boggetto N., Alberti P., Saintomé C., Riou J.F., Prioleau M.-N. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J. 2014;33:732–746. doi: 10.1002/embj.201387506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balakrishnan L., Bambara R.A. Okazaki fragment metabolism. Cold Spring Harb. Perspect. Biol. 2013;5:a010173. doi: 10.1101/cshperspect.a010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung I., Schertzer M., Rose A., Lansdorp P.M. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat. Genet. 2002;31:405–409. doi: 10.1038/ng928. [DOI] [PubMed] [Google Scholar]
- 16.Mirkin E.V., Mirkin S.M. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopes J., Piazza A., Bermejo R., Kriegsman B., Colosio A., Teulade-Fichou M.P., Foiani M., Nicolas A. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011;30:4033–4046. doi: 10.1038/emboj.2011.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Kregten M., Tijsterman M. The repair of G-quadruplex-induced DNA damage. Exp. Cell Res. 2014;329:178–183. doi: 10.1016/j.yexcr.2014.08.038. [DOI] [PubMed] [Google Scholar]
- 19.Huber M.D., Duquette M.L., Shiels J.C., Maizels N. A conserved G4 DNA binding domain in RecQ family helicases. J. Mol. Biol. 2006;358:1071–1080. doi: 10.1016/j.jmb.2006.01.077. [DOI] [PubMed] [Google Scholar]
- 20.Bochman M.L., Sabouri N., Zakian V.A. Unwinding the functions of the Pif1 family helicases. DNA Repair. 2010;9:237–249. doi: 10.1016/j.dnarep.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanders C.M. Human Pif1 helicase is a G-quadruplex DNA-binding protein with G-quadruplex DNA-unwinding activity. Biochem. J. 2010;430:119–128. doi: 10.1042/BJ20100612. [DOI] [PubMed] [Google Scholar]
- 22.Booy E.P., Meier M., Okun N., Novakowski S.K., Xiong S., Stetefeld J., McKenna S.A. The RNA helicase RHAU (DHX36) unwinds a G4-quadruplex in human telomerase RNA and promotes the formation of the P1 helix template boundary. Nucleic Acids Res. 2012;40:4110–4124. doi: 10.1093/nar/gkr1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paeschke K., Bochman M.L., Garcia P.D., Cejka P., Friedman K.L., Kowalczykowski S.C., Zakian V.A. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ribeyre C., Lopes J., Boulé J.-B., Piazza A., Guédin A., Zakian V.A., Mergny J.-L., Nicolas A. The Yeast Pif1 Helicase Prevents Genomic Instability Caused by G-Quadruplex-Forming CEB1 Sequences In Vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paeschke K., Capra J.A., Zakian V.A. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boulé J.-B., Zakian V.A. Roles of Pif1-like helicases in the maintenance of genomic stability. Nucleic Acids Res. 2006;34:4147–4153. doi: 10.1093/nar/gkl561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou X.-M., Wu W.-Q., Duan X.-L., Liu N.-N., Li H.-H., Fu J., Dou S.-X., Li M., Xi X.-G. Molecular mechanism of G-quadruplex unwinding helicase: sequential and repetitive unfolding of G-quadruplex by Pif1 helicase. Biochem. J. 2015;466:189–199. doi: 10.1042/BJ20140997. [DOI] [PubMed] [Google Scholar]
- 28.Zhou R., Zhang J., Bochman M.L., Zakian V.A., Ha T. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. Elife. 2014;3:e02190. doi: 10.7554/eLife.02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duan X.-L., Liu N.-N., Yang Y.-T., Li H.-H., Li M., Dou S.-X., Xi X.-G. G-quadruplexes Significantly Stimulate Pif1 Helicase-catalyzed Duplex DNA Unwinding. J. Biol. Chem. 2015;290:7722–7735. doi: 10.1074/jbc.M114.628008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy R., Hohng S., Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boule J.-B., Zakian V.A. The yeast Pif1p DNA helicase preferentially unwinds RNA-DNA substrates. Nucleic Acids Res. 2007;35:5809–5818. doi: 10.1093/nar/gkm613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun H., Karow J.K., Hickson I.D., Maizels N. The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 33.Dou S.-X., Wang P.-Y., Xu H.Q., Xi X.G. The DNA binding properties of the Escherichia coli RecQ Helicase. J. Biol. Chem. 2004;279:6354–6363. doi: 10.1074/jbc.M311272200. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X.-D., Dou S.-X., Xie P., Hu J.-S., Wang P.-Y., Xi X.G. Escherichia coli RecQ is a rapid, efficient, and monomeric helicase. J. Biol. Chem. 2006;281:12655–12663. doi: 10.1074/jbc.M513089200. [DOI] [PubMed] [Google Scholar]
- 35.Moreira B.G., You Y., Owczarzy R. Cy3 and Cy5 dyes attached to oligonucleotide terminus stabilize DNA duplexes: Predictive thermodynamic model. Biophys. Chem. 2015;198:36–44. doi: 10.1016/j.bpc.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 36.Iqbal A., Wang L., Thompson K.C., Lilley D.M.J., Norman D.G. The structure of cyanine 5 terminally attached to double-stranded DNA: Implications for FRET studies. Biochemistry. 2008;47:7857–7862. doi: 10.1021/bi800773f. [DOI] [PubMed] [Google Scholar]
- 37.Barranco-Medina S., Galletto R. DNA binding induces dimerization of Saccharomyces cerevisiae Pif1. Biochemistry. 2010;49:8445–8454. doi: 10.1021/bi100984j. [DOI] [PubMed] [Google Scholar]
- 38.Markiewicz R.P., Vrtis K.B., Rueda D., Romano L.J. Single-molecule microscopy reveals new insights into nucleotide selection by DNA polymerase I. Nucleic Acids Res. 2012;40:7975–7984. doi: 10.1093/nar/gks523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu W.-Q., Hou X.-M., Li M., Dou S.-X., Xi X.-G. BLM unfolds G-quadruplexes in different structural environments through different mechanisms. Nucleic Acids Res. 2015;43:4614–4626. doi: 10.1093/nar/gkv361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie S. Single-molecule approach to enzymology. Single Mol. 2001;2:229–236. [Google Scholar]
- 41.Zhang D.H., Zhou B., Huang Y., Xu L.X., Zhou J.Q. The human Pif1 helicase, a potential Escherichia coli RecD homologue, inhibits telomerase activity. Nucleic Acids Res. 2006;34:1393–1404. doi: 10.1093/nar/gkl029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson M.A., Kwon Y., Xu Y., Chung W.-H., Chi P., Niu H., Mayle R., Chen X., Malkova A., Sung P., et al. Pif1 helicase and Polδ promote recombination-coupled DNA synthesis via bubble migration. Nature. 2013;502:393–396. doi: 10.1038/nature12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geronimo C.L., Zakian V.A. Getting it done at the ends: Pif1 family DNA helicases and telomeres. DNA Repair (Amst) 2016;44:151–158. doi: 10.1016/j.dnarep.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McEachern M.J., Haber J.E. Break-induced replication and recombinationaltelomere elongation in yeast. Annu. Rev. Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 45.Vasianovich Y., Harrington L.A., Makovets S. Break-induced replication requires DNA damage-induced phosphorylation of Pif1 and leads to telomere lengthening. PLoS Genet. 2014;10:e1004679. doi: 10.1371/journal.pgen.1004679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu Y., Tang H.-B., Liu N.-N., Tong X.-J., Dang W., Duan Y.-M., Fu X.-H., Zhang Y., Peng J., Meng F.-L., et al. Telomerase-null survivor screening identifies novel telomere recombination regulators. PLoS Genet. 2013;9:e1003208. doi: 10.1371/journal.pgen.1003208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.