Abstract
The superfamily of 3′-5′ polymerases synthesize RNA in the opposite direction to all other DNA/RNA polymerases, and its members include eukaryotic tRNAHis guanylyltransferase (Thg1), as well as Thg1-like proteins (TLPs) of unknown function that are broadly distributed, with family members in all three domains of life. Dictyostelium discoideum encodes one Thg1 and three TLPs (DdiTLP2, DdiTLP3 and DdiTLP4). Here, we demonstrate that depletion of each of the genes results in a significant growth defect, and that each protein catalyzes a unique biological reaction, taking advantage of specialized biochemical properties. DdiTLP2 catalyzes a mitochondria-specific tRNAHis maturation reaction, which is distinct from the tRNAHis maturation reaction typically catalyzed by Thg1 enzymes on cytosolic tRNA. DdiTLP3 catalyzes tRNA repair during mitochondrial tRNA 5′-editing in vivo and in vitro, establishing template-dependent 3′-5′ polymerase activity of TLPs as a bona fide biological activity for the first time since its unexpected discovery more than a decade ago. DdiTLP4 is cytosolic and, surprisingly, catalyzes robust 3′-5′ polymerase activity on non-tRNA substrates, strongly implying further roles for TLP 3′-5′ polymerases in eukaryotes.
The discovery of a superfamily of 3′-5′ polymerases challenged the dogma that synthesis of DNA and RNA always occurs in the 5′-3′ direction. The founding member of this family, the tRNAHis guanylyltransferase (Thg1), was identified in Saccharomyces cerevisiae and named after its essential function in tRNAHis maturation (1–3). However, initial characterization of S. cerevisiae Thg1 (SceThg1) revealed that it catalyzed an unexpected template-dependent 3′-5′ polymerase activity in vitro, and subsequent structures uncovered a striking similarity between Thg1 and canonical 5′-3′ polymerases (4–8). Proteins related to Thg1 were subsequently identified in all three domains of life, and are broadly classified into two groups, as either bona fide Thg1 enzymes (so far only found in Eukarya) or Thg1-like proteins (TLPs, found in all three domains), based on characteristic conserved sequence differences and likely distinct functions (9–11).
To date, the only physiological activity unequivocally demonstrated for any Thg1/TLP enzyme is the original function of Thg1 in S. cerevisiae (and likely most eukaryotes). Thg1 uses its 3′-5′ addition activity to add a G-nucleotide (known as G-1) to the 5′-end of cytosolic tRNAHis, thus incorporating the required identity element for recognition by histidyl-tRNA synthetase (HisRS) (1,2,12–15). For this reaction, Thg1 adds only a single G residue in a non-Watson–Crick (WC) base pair (G-1 is added opposite A73). Therefore, the biological relevance of the 3′-5′ polymerase activity that adds multiple WC base-paired nucleotides to various substrates remained unclear (6,16). It has been speculated that some archaeal TLPs also catalyze post-transcriptional G-1-addition to tRNAHis (10,16,17). However, many archaea and bacteria encode the G-1 residue in their tRNAHis genes and predictably retain G-1 during 5′-end processing (18,19). Thus, many organisms with genes encoding TLPs do not apparently require addition of the tRNAHis G-1 identity element and physiological substrates for these enzymes remain uncharacterized.
In vitro characterization revealed three biochemical properties that distinguish TLPs from their Thg1 enzyme counterparts. First, TLPs selectively catalyze WC base pair-dependent addition reactions, and do not efficiently catalyze the non-WC (G-1 opposite A73) reaction associated with Thg1 (11,16). Second, TLPs prefer to repair 5′-truncated tRNAs instead of adding nucleotides to the full-length tRNAHis that is the substrate for Thg1 (11). Third, the repair activities of TLPs are not tRNA species-specific, in contrast to the strict tRNAHis substrate selectivity exhibited by Thg1 (9,11,20–22). Thus, we hypothesized that TLPs take advantage of their distinct biochemical properties to catalyze different biological reactions from Thg1-type enzymes.
The goal of this study was to determine physiological functions for these unusual TLP 3′-5′ polymerases using the eukaryotic slime mold, Dictyostelium discoideum as a model system. While most metazoa and fungi encode one unique (Thg1) member of the 3′-5′ polymerase family (3,10), many protozoa encode multiple Thg1-related genes. Indeed, four members of the enzyme family (DdiThg1, DdiTLP2, DdiTLP3 and DdiTLP4) were identified in D. discoideum (9). The function of DdiThg1 was readily predicted based on homology to S. cerevisiae Thg1, and its involvement in G-1 addition to tRNAHis was supported by biochemical studies and the ability of DdiThg1 to complement the lethality of S. cerevisiae thg1Δ mutants (9). Thg1/TLP enzymes have been suggested to play a role in mitochondrial-tRNA (mt-tRNA) 5′-editing, which is widespread among protozoa and replaces genomically-encoded 5′-mismatches from mt-tRNA with exact matches (Supplementary Figure S1) (10,23–25). The repair step of this reaction was speculated to require a 3′-5′ polymerase, and DdiTLP3 and DdiTLP4 both exhibited robust in vitro activity with mt-tRNA 5′-editing substrates, suggesting that either or both proteins might catalyze this reaction in vivo (9,20). However, the role of TLPs in 5′-editing has not been addressed in any in vivo system. Moreover, no robust in vitro activity has ever been detected with the fourth enzyme (DdiTLP2), and its role remained a mystery.
Here, we use subcellular localization, biochemical assays, and analysis of genetically depleted strains to demonstrate distinct biologically important functions for each of the 3′-5′ polymerases in D. discoideum. We found growth defects upon depletion of each individual 3′-5′ polymerase, demonstrating their non-redundant functions. DdiThg1 was found to be cytosolic, consistent with its previously predicted role in tRNAHis maturation, and we proved this role by analysis of tRNAHis from RNAi-depletion strains. The mitochondrial localization of DdiTLP2 and DdiTLP3 led to the findings that DdiTLP2 added G-1 to mitochondrial tRNAHis in vivo, with uniquely different biochemical specificity, while DdiTLP3 was the physiologically relevant 5′-editing enzyme, establishing for the first time a firm biological role for TLPs in vivo, in this unusual editing reaction. Finally, and most surprisingly, the cytosolic protein DdiTLP4 was shown to act on several small non-coding RNAs in vitro and in vivo, and its depletion was shown to cause a severe growth defect, suggesting for the first time that a TLP might have a critical role in non-tRNA-related activity associated with this enzyme family. These observations thus demonstrate important and distinct biological roles for 3′-5′ polymerases, demonstrate unexpected breadth in their functions, and open the door to subsequent genetic and biochemical analysis of additional activities catalyzed by this unique family of proteins.
MATERIALS AND METHODS
D. discoideum genetics and in vivo characterization
Strains, plasmids and cloning strategies are described in Supplementary Material (see also Supplementary Tables S1 and S2); standard methods for axenic growth, transformation and construction of deletion strains by homologous recombination were followed (26,27). GFP-fusion proteins and DAPI-stained cells were visualized using a Ti-E inverted fluorescence microscope after fixation. RNAi targeting each 3′-5′ polymerase gene was performed using the MB35/MB38 tet-repressible system according to previously published protocols, and RNA levels were measured by northern blots of total RNA (28–30). See methods in Supplementary Material for detailed protocol information.
Phosphatase protection assay for 3′-5′ addition
Substrate RNAs were produced by in vitro transcription with T7 RNA polymerase, followed by 5′-32P labeling as previously described (21) and assays were performed according to (9). 5S rRNA and Class I ncRNA transcripts were transcribed from 5′-hammerhead ribozyme constructs to generate the desired 5′-U start site, as in (9). Briefly, reactions containing 5′-32P-labeled RNA, 0.1 mM ATP and 1.0 mM GTP were initiated by addition of purified enzymes, as indicated. After RNase A digestion and phosphatase treatment, reactions were resolved by silica TLC and visualized using a Typhoon trio imager (9).
Primer extension assay for G-1 addition
Individual clones of [RNAi]thg1 and vector control strains were grown with and without tetracycline for 4 days and total RNA was isolated with Tri Reagent (Sigma). Primer extension to detect G-1 addition was performed as in (3) with 5′-32P labeled primer complementary to tRNAHis (5′- GCCACAATCTGATGTACTACCACT).
Mitochondrial tRNA sequencing
Total RNA was isolated from D. discoideum with Tri Reagent, followed by circularization by ligation with T4 RNA ligase, RT-PCR with mt-tRNA-specific primers, and restriction cloning of the amplified products into digested pUC19 vectors for sequencing (Supplementary Table S3), all performed as described previously (20,22).
α-NTP labeling and RNase H assays for DdiTLP4 substrates
[RNAi]tlp4 strains were selected and grown as above in media with and without tetracycline and used for total RNA isolation with Tri Reagent. Total RNA (8 μg each) was incubated with 13 μM purified DdiTLP4 enzyme and 4 μl of the indicated [α-32P]NTP (3000 Ci/mmol) in the same buffer used for the phosphatase protection assay at a final volume of 20 μl reaction. Reactions were quenched after 2 h at room temperature by addition of formamide loading dye, then resolved by 4M urea-10% PAGE, dried and visualized with a Typhoon trio. For detection of substrates by RNase H, reactions were performed with α-32P-NTP as described above, but prior to gel resolution, DNA oligonucleotides complementary to 5S rRNA and Class I ncRNA were added to each reaction and annealed by heating to 95°C for 5 min followed by slow cooling to room temperature for 30 min. After annealing, 0.1 U RNase H (Promega) was added to reactions and incubated at 37°C for 30 min.
RESULTS
Subcellular localization of 3′-5′ polymerases in Dictyostelium discoideum
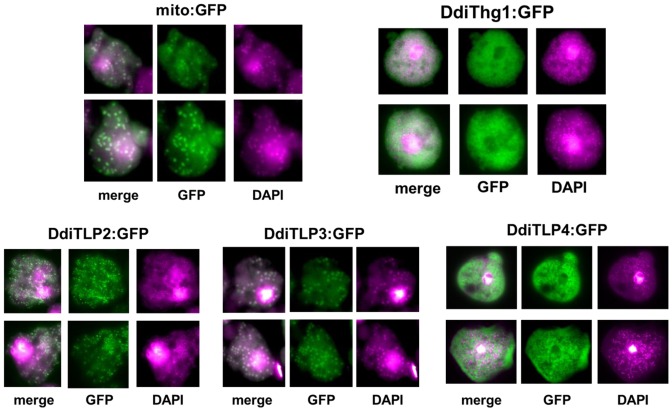
Since the few previously predicted substrates for 3′-5′ polymerases exist in distinct cellular locations, the primary localization of each of the four D. discoideum proteins (DdiThg1 and DdiTLP2-4) was determined. We visualized each protein after expression as a fusion with a C-terminal green fluorescent protein (GFP) (Supplementary Table S1) (31), and compared the results with known mitochondrial and DNA images. The mitochondrial targeting sequence of the TopA gene was added to the N-terminus of GFP (mito:GFP) (32), resulting in typical mitochondrial morphology in D. discoideum, characterized by small spherical structures (33) that were coincident with the structures arising from 4′,6-diamidino-2-phenylindole (DAPI) fluorescence (Figure 1). Consistent with the expected role of DdiThg1 in maturation of nucleus-encoded tRNAHis and cytosolic localization of Thg1 in S. cerevisiae (34), DdiThg1:GFP was largely observed as diffuse fluorescence throughout the cytosol, distinct from DAPI-stained mitochondria (Figure 1).
Figure 1.
Localization of Thg1/TLP enzymes in D. discoideum. DdiThg1/DdiTLP enzymes were expressed as a C-terminal GFP fusions and imaged for GFP (green) and DAPI (purple) fluorescence. The left-most panel in each group shows merged images of GFP and DAPI fluorescence signals from the same cell. Mito:GFP expresses GFP fused with the TopA N-terminal mitochondrial targeting peptide. Images of two representative cells are shown for each strain.
Analysis of the remaining DdiTLP:GFP expressing strains revealed distinct localization patterns. DdiTLP2:GFP and DdiTLP3:GFP are mitochondrial, based on clearly exhibited fluorescence in spherical particles that co-localize with the DAPI-stained mitochondria, while DdiTLP4:GFP is cytoplasmic, based on diffuse cytosolic localization similar to that observed with DdiThg1 (Figure 1). These results are consistent with predicted localizations based on MitoProt sequence analysis (35) (Supplementary Figure S2).
DdiTLP2 catalyzes G-1 addition to mitochondrial tRNAHisin vitro
In numerous in vitro assays, DdiTLP2 has never been associated with any robust biochemical activity, including repair of any 5′-truncated tRNA substrates proposed to occur during mitochondrial editing (9,20). However, the mitochondrial localization of DdiTLP2 led us to consider a possible function for this enzyme with mitochondrial tRNAHis (mt-tRNAHis), which does not undergo 5′-editing to repair mismatches, but may require addition of G-1 (19,20). In vertebrates, Thg1 is targeted to both cytosol and mitochondria through the use of alternative translation initiation sites, and the mitochondrial isoform is presumably responsible for introducing the G-1 residue to mt-tRNAHis (3,36,37). In D. discoideum, however, where DdiThg1 is not localized to mitochondria (Figure 1), two possible scenarios are suggested. Either mt-tRNAHis lacks a G-1 residue, as recently observed for some other protozoan eukaryotes (22,38), or another enzyme such as DdiTLP2 is responsible for the mt-tRNAHis G-1 addition reaction.
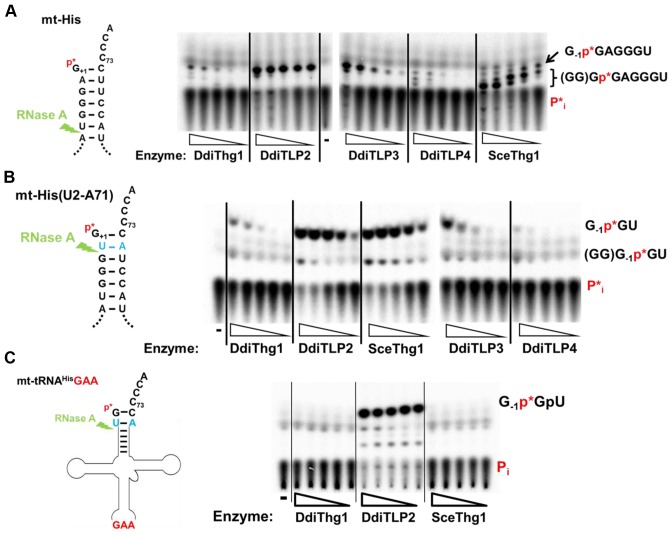
To test post-transcriptional addition of G-1, we performed in vitro assays with 5′-32P-labeled D. discoideum mt-tRNAHis using the previously described phosphatase protection assay (9,20). Addition of G-1 is reflected by production of a 7-mer oligonucleotide (G-1p*GAGGGU, with p* indicating the position of the labeled phosphate), in contrast to inorganic phosphate (P*i) produced from unreacted substrate (Figure 2A). With this assay robust activity was observed with purified DdiTLP2, and a lesser amount of activity was observed with DdiTLP3, while DdiThg1 and DdiTLP4 had only minimal activity. This is in stark contrast to the robust in vitro 5′-tRNA repair activities that were demonstrated for the same recombinant DdiTLP3 and DdiTLP4 enzymes in earlier work (9), suggesting that the purified enzyme is folded well enough to exhibit activity with other substrates. To more clearly resolve the reaction products, we created a modified mt-tRNAHis variant in which the orientation of the 2–71 base pair was reversed to yield mt-tRNAHis(U2-A71). Digestion of this tRNA with RNase A produced a more easily resolved trinucleotide (G-1p*GU), and spots corresponding to the G-1 addition product were also readily observed by silica TLC (Figure 2B).
Figure 2.
Addition of G-1 to D. discoideum mitochondrial tRNAHis. Each of the four purified DdiThg1/DdiTLP enzymes and SceThg1 were used in assays with 5′-32P-labeled mt-tRNAHis (either (A) wild-type or (B) U2-A71 variant) and analyzed for G-1 addition by phosphatase protection (21). Labeled RNA was incubated with 1 mM GTP and 0.1 mM ATP and 5-fold serial dilutions of each enzyme, as indicated (DdiThg1, 7-0.01 μM; DdiTLP2, 44-0.07 μM; DdiTLP3, 5-0.008 μM; DdiTLP4, 25-0.04 μM; SceThg1 75-0.12 μM), or with no enzyme (lanes indicated by -). Reactions were digested with RNase A (jagged arrow) and treated with phosphatase, releasing Pi from unreacted substrate and protected oligonucleotide fragments, whose identities are shown on the right of each panel. (C) DdiThg1 and DdiTLP2 exhibit distinct dependence on the GUG anticodon. Phosphatase protection assays were performed with a variant of the mt-tRNAHisU2-A71 substrate that contains a GAA anticodon in place of the wild-type GUG. Reactions contain 5-fold serial dilutions of the indicated enzymes (or no enzyme, -).
With this improved assay, we observed a similar result. DdiTLP2, and to a lesser extent DdiTLP3, catalyzed the most efficient G-1 addition to the modified mt-tRNAHis substrate, as judged by the relative intensities of products observed at each concentration. Since both DdiTLP2 and DdiTLP3 exhibit predominantly mitochondrial localization (Figure 1) and significant in vitro activity with mt-RNAHis (Figure 2), these data suggest that either enzyme (or a complex of both) could catalyze this reaction in vivo.
Surprisingly, DdiThg1 only weakly adds G-1 to mt-tRNAHis (Figure 1), while previous results with the nucleus-encoded tRNAHis suggested an opposite pattern, with DdiThg1 able to readily add G-1 and DDiTLP2 exhibiting little detectable activity with this substrate (9). This suggests that there are distinct specificities of DdiThg1 and DdiTLP2 for two different tRNAHis species (cytosolic versus mitochondrial, respectively), raising questions about the mechanism of recognition employed by each enzyme. To test this, we created a variant of the mt-tRNAHis(U2-A71) substrate, in which the His-specific GUG anticodon was replaced with Phe-specific GAA (Figure 2C). Consistent with previous characterization of SceThg1 (21), alteration of GUG to GAA completely abolished the ability of both SceThg1 and DdiThg1 to add G-1 (compare Figure 2B versus C). In contrast, the G-1 addition activity of DdiTLP2 was not detectably affected by the GAA alteration. Thus, the identity of mt-tRNAHis for G-1 addition by DdiTLP2 is specified by a different mechanism from the canonical GUG-dependent G-1 addition pathway utilized in the cytosol.
DdiTLP2 is the sole mt-tRNAHis-specific G-1 addition enzyme in D. discoideum
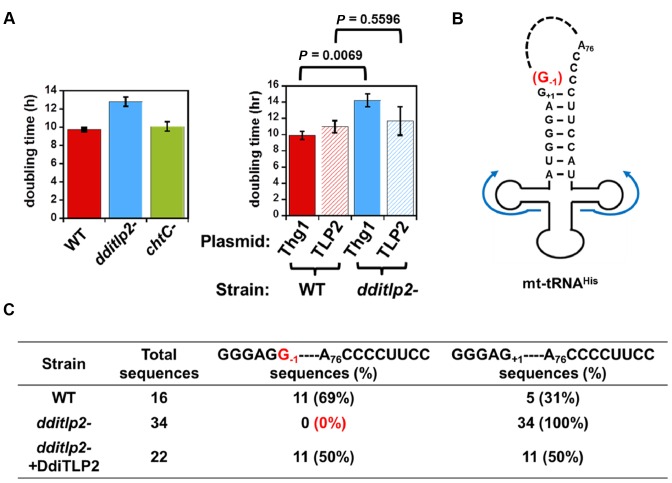
To conclusively assess in vivo function, we sought to obtain deletion strains for each of the four genes in D. discoideum by homologous recombination of a blasticidin S-resistance (bsR) cassette into each chromosomal locus (39,40) (Supplementary Results and Supplementary Figure S3A). A dditlp2- strain was readily obtained (Supplementary Figure S3B), and although this strain was viable, doubling time measurements indicated a reproducible and statistically significant vegetative growth defect associated with the deletion strain (Figure 3A). The increased doubling time was complemented by transformation with a plasmid expressing DdiTLP2 (but not DdiThg1) under control of the act15 promoter (Figure 3A). These results indicated that the growth defect was directly attributable to loss of DdiTLP2 function and not likely due to a secondary mutation acquired during generation of the dditlp2- strain.
Figure 3.
DdiTLP2 catalyzes G-1 addition to mt-tRNAHisin vivo. (A) Deletion of DdiTLP2 causes an increase in doubling time. Average doubling times are reported for 2–3 independent cultures of each indicated strain, with P-values determined by T-test. (B) Circularization and RT-PCR to determine G-1-status of mt-tRNAHis. After circularization of total RNA by T4 RNA ligase (indicated by dotted line), RT-PCR was performed with the two primers indicated by the blue lines on the diagram, and amplified inserts were cloned by restriction ligation into appropriately-digested vectors. (C) mt-tRNAHis sequences. For each strain, the number of sequences either containing or lacking G-1 out of the total sequences obtained is indicated, with the corresponding percent of total sequences indicated in parentheses.
Since the dditlp2- strain was viable, the role for this enzyme in mt-tRNAHis maturation was investigated further. Total RNA was extracted from parental AX2 (WT) and dditlp2- strains, and analyzed by circularization (to join the 5′- and 3′-ends of the tRNA) followed by RT-PCR of the junction and analysis of the amplified reaction products after ligation into a plasmid for sequencing of independent clones (Figure 3B) (22). We observed a complete loss of G-1 from the 5′-end of mt-tRNAHis in the dditlp2- strain, which was restored upon expression of DdiTLP2 in the complemented strain (Figure 3C). Taken together with the in vitro activity and localization results, these data conclusively demonstrate that DdiTLP2 is the exclusive G-1 addition enzyme for mt-tRNAHis in D. discoideum. Although DdiTLP3 exhibited some in vitro G-1 addition activity with mt-tRNAHis (albeit ∼100-fold less efficiently, as judged by the products observed with each enzyme in the endpoint assay (Figure 2)) and also localizes to mitochondria, this residual activity did not apparently complement the function of DdiTLP2 in vivo.
Depletion of thg1, dditlp3 or dditlp4 by RNAi causes a significant growth defect
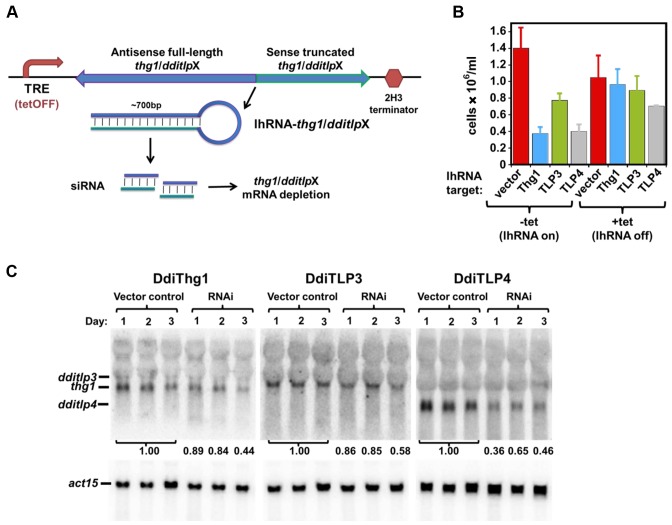
Despite repeated attempts and successful deletion of a control non-essential gene (chtC) (41) in addition to dditlp2 (Supplementary Figure S3B), viable deletion stains for any of the other three genes (thg1, dditlp3 or dditlp4) could not be isolated, suggesting that they are essential. The inability to obtain viable thg1 deletion strains in D. discoideum is consistent with previous studies in yeast and human cells (2,42–44) and the failure to delete DdiTLP3 and DdiTLP4 may suggest similarly critical and non-redundant roles for these enzymes in D. discoideum. Therefore, RNA interference (RNAi) was used to target each gene (Figure 4A), using a tet-repressible system previously applied to silence other genes in D. discoideum (29,30,45). To quantify the effects of RNAi on growth, selected cells were diluted to uniform density in the presence or absence of tet and counted after 8 days. Expression of lhRNA targeting thg1, dditlp3 or dditlp4 was associated with a severe growth defect compared to cells transformed with the pMB38 empty vector (Figure 4B). Slightly impaired growth under tet-repressed conditions is likely due to incomplete lhRNA repression. Decreased mRNA levels of the targeted genes were verified by northern analysis (Figure 4C). To quantify expression, mRNA signals observed for each targeted gene were normalized to the actin mRNA signal from the same RNA sample; these normalized expression levels were compared to the average actin-normalized signal observed in vector control samples. In each case, mRNA levels of the targeted gene decreased upon induction of RNAi, with ∼50% of their wild-type levels observed by the third day of growth under lhRNA-inducing conditions, confirming partial depletion of the correct target mRNA in the RNAi strains (Figure 4C).
Figure 4.
RNAi depletion of DdiThg1, DdiTLP3 or DdiTLP4 causes growth defects in D. discoideum. (A) RNAi strategy. Plasmids express lh-RNA targeting each indicated mRNA under control of a tetracycline-inducible repressor (tetOFF) system. (B) Growth defect associated with expression of lhRNA targeting DdiThg1/TLP genes. Cultures of cells expressing the indicated lhRNA construct were inoculated at the same density and grown +/- tet; average cell counts after 8 days of growth are reported from 2–3 independently grown cultures. (C) Decreased levels of each target mRNA. Northern analysis was performed on RNA isolated after 1–3 days of growth in the absence of tet; bands corresponding to expected sizes of mRNA for act15, thg1, tlp3 and tlp4 are indicated. The intensity of each target band was normalized to the intensity of the act15 signal in the same RNA sample. Ratios of normalized band intensity for each RNAi sample to the average normalized band intensity for the three vector control RNA samples are reported.
DdiThg1 is the sole G-1 addition enzyme for nucleus-encoded tRNAHis
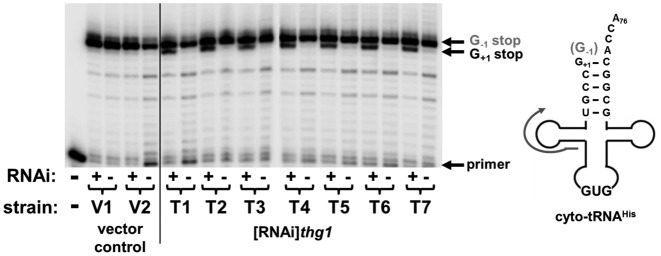
The availability of the [RNAi]thg1 strain enabled a direct validation of the biological function of Thg1 for the first time in any organism outside of S. cerevisiae. Primer extension was used to determine the status of G-1 on tRNAHis derived from vector control and [RNAi]thg1 strains. Premature termination of the primer extension products at G+1 due to loss of the G-1 residue was observed in 7 independent [RNAi]thg1 clones under RNAi inducing conditions (Figure 5). These experiments further confirmed the partial nature of the RNAi depletion, since there was not a complete loss of the G-1 residue under RNAi-inducing conditions (Figure 5). Due to the importance of G-1 for tRNAHis aminoacylation and translation fidelity, the decrease in G-1 addition likely explains the growth defect associated with RNAi targeting DdiThg1 (2,42).
Figure 5.
Partial depletion of DdiThg1 by RNAi decreases the levels of G-1 on nucleus-encoded tRNAHis. Primer extension analysis of cytosolic tRNAHis (cyto-tRNAHis) was performed on RNA isolated from independent vector control (V1-V2) or [RNAi]thg1 (T1-T7) cells grown ± tet. The position of the unextended primer is seen in the lane with no added RNA (-); primer extension products that contain G-1 (G-1 stop) or lack G-1 (G+1 stop) are indicated.
DdiTLP3 is the 3′-5′ polymerase that catalyzes mt-tRNA editing in D. discoideum
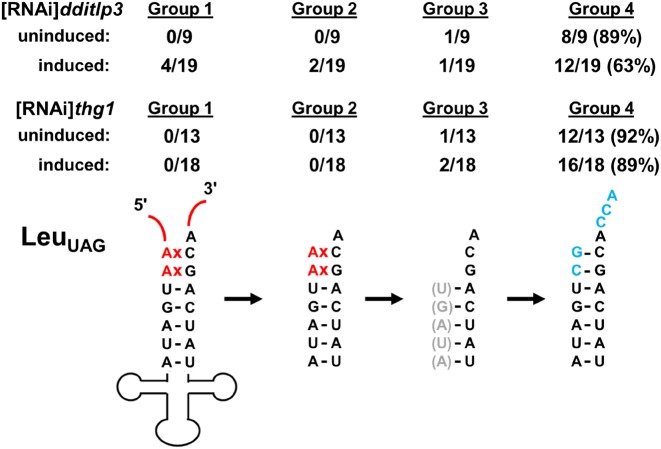
We directly investigated the role of DdiTLP3 in 5′-editing of mt-tRNA by analyzing the editing status of RNA isolated from the [RNAi]tlp3 strain. To do this, we used the circularization/RT-PCR approach (Figure 3B) to amplify the 5′ and 3′-ends of mt-tRNALeuUAG (Figure 6) and mt-tRNAIleCAU (Supplementary Figure S4), both of which undergo 5′-editing in D. discoideum (Supplementary Figure S1) and were previously demonstrated to be in vitro substrates for 5′-repair by DdiTLP3 (9,20). Because editing intermediates are observed even among sequences of RNA isolated from wild-type cells (20), sequences of independently-isolated clones were categorized into 4 major groups reflecting different stages of 5′-editing (Figure 6, Supplementary Figure S4 and Table S3). Group 1 reflects unprocessed mt-tRNA transcripts (containing extensive 5′-leader and 3′-trailer sequences). Groups 2 and 3 represent various partially-processed intermediates, with the major distinction being whether the RNA still retains any 5′-mismatched nucleotides (Group 2) or whether the mismatched nucleotides (and in some cases other nucleotides within the 5′-acceptor stem) have been removed by the unidentified editing nuclease (Group 3). As we observed previously (20), completion of 5′-editing is not required for 3′-end processing (including addition of the 3′-CCA), and therefore sequences obtained for Groups 2 and 3 include RNAs at various stages of 3′-end processing (Supplementary Table S3). Group 4 comprises fully-edited mt-tRNA, with correct 5′- and 3′-ends.
Figure 6.
Depletion of DdiTLP3 causes decreased mt-tRNA 5′-editing. The editing status of mt-tRNALeuUAG was determined from [RNAi]tlp3 or [RNAi]thg1 cells, as indicated, with RNAi induced (-tet) or uninduced (+tet). Sequences obtained from independent clones (Supplementary Table S3) were categorized into four groups according to their processing status. In Group 1, red lines indicate unprocessed 5′-leader or 3′-trailer sequences; 5′-mismatched nucleotides to be repaired by 5′-editing are indicated in red. For clarity, only aminoacyl-acceptor stem sequences are shown for Groups 2–4. Since 3′-end processing can occur prior to 5′-editing, sequences in Group 2 and Group 3 can contain either unprocessed 3′-ends or 3′-CCA. Some sequences in Group 3 reveal removal of additional normally-paired 5′-nucleotides beyond the 5′-mismatched residues; such nucleotides are indicated in gray parentheses. Group 4 corresponds to fully 5′-edited mt-tRNA; 5′-edited/3′-processed nucleotides are indicated in blue.
For both mt-tRNA, we observed a decrease in the overall percent of Group 4 (fully-edited) mt-tRNA upon induction of RNAi against DdiTLP3, with a corresponding increase in the proportion of unedited or partially-edited intermediates (Figure 6 and Supplementary Figure S4). Sequencing of mt-tRNALeuUAG from the RNAi strain targeting thg1 revealed no change in the level of editing upon induction of RNAi, demonstrating that the effects on 5′-editing are specific to DdiTLP3 (Figure 6). Due to the partial nature of the RNAi depletion (Figure 4C), complete loss of mt-tRNA 5′-editing was not observed for either substrate. Interestingly, consistent with previous investigations of 5′-editing (20,46), the percent of fully-edited tRNA is variable and no investigation of the possible impact of the presence of unedited or partially-edited tRNA has been reported. These data nonetheless confirm that DdiTLP3 is a bona fide part of the long-sought 5′-editing enzyme in D. discoideum, likely using its WC-dependent 3′-5′ polymerase activity to repair the 5′-ends of mt-tRNA in which incorrectly base paired 5′-nucleotides have been removed by an as of yet unidentified nuclease (Supplementary Figure S1).
DdiTLP4 is a 3′-5′ polymerase that acts on non-tRNA substrates
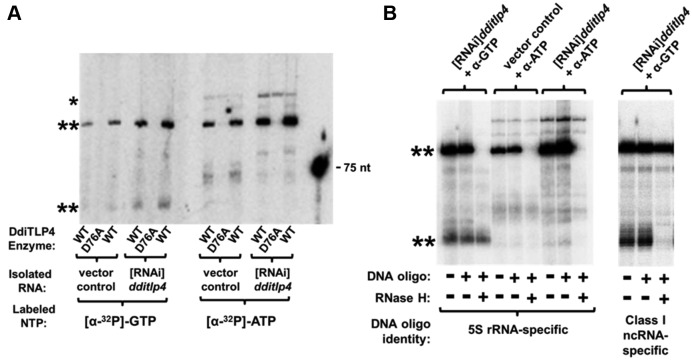
The growth defect due to depletion of DdiTLP4 suggested that its activity was important for the function of one or more D. discoideum RNA(s). To identify these, we isolated total RNA from [RNAi]dditlp4 cells, reasoning that substrates for DdiTLP4 might accumulate in a hypomodified state. To detect substrate RNAs, we used a labeling approach in which equal amounts of isolated RNA from [RNAi]dditlp4 and vector control cells were treated with purified DdiTLP4 and [α-32P]-NTP, followed by resolution of resulting products by urea-PAGE. Reactions performed in the presence of [α-32P]-GTP yielded two prominent bands, both of which were labeled to higher intensity in RNA from DdiTLP4-depleted cells (Figure 7A, two stars). Reactions performed with [α-32P]-ATP generated one major band that migrated similarly to the longer GTP-labeled RNA, and several less intensely labeled products. Most of the labeled species were absolutely dependent on DdiTLP4 activity, since they were not generated in reactions with a catalytically inactive (D76A) DdiTLP4 variant (7) (Figure 7A). However, one higher migrating ATP-labeled band (Figure 7A, one star) was not DdiTLP4-activity dependent, indicating that it is likely the product of some labeling activity that co-purifies with the bacterially-expressed DdiTLP4 used in these assays.
Figure 7.
Non-tRNA substrates for DdiTLP4 in D. discoideum RNA. (A) Labeling in vivo isolated RNA with α-NTP. Total RNA from [RNAi]tlp4 and vector control strains was incubated with either wild-type (WT) or catalytically inactive (D76A) DdiTLP4, and either [α-32P]-GTP or [α-32P]-ATP. The two WT lanes represent independent replicates for each sample. Labeled products were resolved by urea-PAGE; a 75 nt labeled tRNA transcript was included as a size marker. Double stars mark DdiTLP4 activity-dependent labeled bands; single star marks product of a co-purifying activity. (B) Identification of DdiTLP4-labeled RNAs. Labeling reactions were performed as described in (A), and then treated with DNA oligonucleotides complementary to either 5S rRNA or Class I ncRNA and RNase H, as indicated. Products were resolved by urea-PAGE; double stars highlight the same bands identified in (A).
An RNase H-based assay was used to identify the labeled RNAs. Labeling reactions were performed as above, but prior to gel resolution, RNAs were purified by phenol extraction and incubated with DNA oligonucleotides complementary to known D. discoideum non-coding RNAs. Two specific non-coding RNAs were chosen to be tested because they represent abundant cytosolic RNAs of the appropriate size for the observed bands. In this assay, RNase H will degrade the labeled RNA if it hybridizes with the specific DNA oligonucleotide. The larger band labeled by both GTP and ATP was consequently identified as 5S rRNA because it was specifically degraded in reactions that contain both the 5S-specific DNA oligonucleotide and RNase H (Figure 7B). Notably, migration of this band is consistent with the predicted size (120 nt) of 5S rRNA in D. discoideum. Likewise, the smaller GTP-labeled band was identified as a recently-discovered Dictyostelium-specific ncRNA known as Class I ncRNA. A DNA oligonucleotide complementary to the conserved 5′-end of these RNAs caused specific degradation of the lower band in the presence of RNase H (Figure 7B). Class I ncRNA are a group of abundant small cytosolic RNAs transcribed from at least 14 unique genes in D. discoideum (47–50). These RNAs range in size from 55–65 nt, consistent with the observed migration of the lower band.
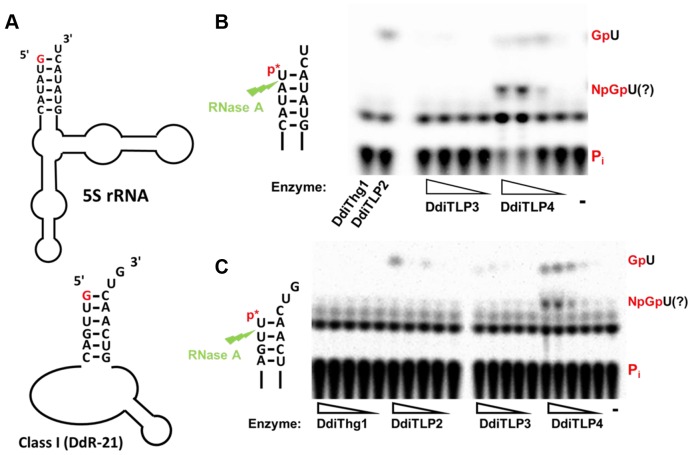
To further evaluate 5S rRNA and Class I ncRNA as potential substrates for DdiTLP4, we determined whether DdiTLP4 can act on either of these RNAs in an in vitro assay. The labeling assay suggested that DdiTLP4 could add either GTP or ATP to 5S rRNA, but does not indicate where each nucleotide might be incorporated. However, since the annotated 5′-nucleotide of both substrates is a G (indicated in red in Figure 8A), we tested whether DdiTLP4 could act on substrates engineered to lack this 5′-nucleotide. Indeed, an in vitro transcript of an otherwise mature 5S rRNA that lacks the 5′-terminal G is a substrate for DdiTLP4, as demonstrated using the 5′-32P-labeled transcript in a phosphatase protection assay (Figure 8B). Upon incubation of the labeled RNA with GTP and ATP (both nucleotides were included since either could be used to activate the 5′-monophosphate for 3′-5′ addition (5)), products due to the activity of DdiTLP4 were readily observed. The upper-most fragment observed in the reactions with DdiTLP4 (and to a lesser extent with DdiTLP2) is Gp*U generated by addition of the missing 5′-G, based on its similar migration to previously characterized standards (11). At the highest enzyme concentrations, additional products that likely correspond to further nucleotide addition (based on the lower migration of these products) also occur (Figure 8B), but the lack of defined standards prevents conclusive identification of these. For Class I ncRNA, which also contains a 5′-G, we similarly observed that DdiTLP4 catalyzes addition of at least one GTP to a 5′-truncated transcript corresponding to one of the Class I ncRNAs (DdR-22), also missing its 5′-terminal G residue (Figure 8C). As with 5S rRNA, the ability to act on the Class I ncRNA is predominantly associated with DdiTLP4.
Figure 8.
DdiTLP4 acts on non-tRNA substrates in vitro. (A) Secondary structure diagrams of annotated 5S rRNA and Class I ncRNA. The 5′-terminal G removed from each RNA for in vitro assay is underlined. (B and C) Phosphatase protection assays with (B) 5S rRNA and (C) Class I ncRNA transcripts lacking 5′-terminal G nucleotides. 5′-32P-labeled RNA was incubated with the indicated enzymes (or no enzyme, lane -) and 1.0 mM GTP/0.1 mM ATP, digested with RNase A (jagged arrow) and then treated with phosphatase. Reactions contained 5-fold serial dilutions of the indicated enzymes (3–0.024 μM), or 3 μM DdiThg1 or DdiTLP2 only in (B). Identification of the G-addition product (GpU) is based on comparison to a previously characterized standard; lower migrating products likely correspond to further nucleotide additions observed at the highest enzyme concentrations of DdiTLP4.
DISCUSSION
In this paper, we demonstrate that each of the four 3′-5′ polymerase family members in D. discoideum has a distinct and important function in the cell. Prior to this study, the only biological activity established for any of these enzymes was the G-1 addition reaction required for maturation of tRNAHis in S. cerevisiae (2,42). Yet, biochemical studies suggested that the Thg1/TLP enzyme family was composed of 3′-5′ polymerases capable of a much broader spectrum of biological activities (6,9,11,16,20). Here, we provide biochemical, cellular and genetic evidence confirming that the templated 3′-5′ polymerase activity of Thg1/TLP enzymes has a bona fide place in the cellular arsenal of nucleic acid processing enzymes, being used in this case by DdiTLP3 to repair 5′-truncated tRNA during 5′-editing (Figures 6, Supplementary Figures S1 and S4). Moreover, the apparently essential nature of three of the four enzymes (and small, but measurable growth defect with the fourth) indicates that these functions are non-redundant and biologically important (Figures 3 and 4). Finally, our demonstration of the first activity of any 3′-5′ polymerase family member on non-tRNA substrates such as 5S rRNA and Class I ncRNA (Figures 7 and 8) paves the way for the discovery of additional roles for 3′-5′ polymerases in biology.
Mitochondrial tRNA maturation requires unusual enzyme activities
Many types of tRNA editing, defined as alterations to the encoded sequence of a tRNA that result in alteration to another standard nucleotide, are now known to occur in a broad range of eukaryotic species (51,52). Editing events that involve removal of encoded tRNA residues and replacement with alternative sequences have been shown to act at both the 5′- and 3′-ends of tRNA (10,53). Yet, despite the apparently widespread occurrence of these events, many of which were identified decades ago, this work is the first to conclusively identify any enzyme that catalyzes one of these unusual reactions. This slow progress is undoubtedly related to the non-canonical nature of many of the enzymatic activities associated with these processes. For example, although standard 5′-3′ polymerases such as the CCA-adding enzyme are known to act on tRNA 3′-ends, 3′-editing reactions have been identified that require addition of specific nucleotides using the 5′-half of the tRNA as a template (54,55). This type of templated addition is not traditionally associated with CCA-adding enzyme, or any other known components of the 3′-end maturation machinery, and thus the specific enzymes that catalyze these 3′-end editing reactions remain unidentified. Likewise, the 5′-editing reaction that was first identified more than 20 years ago consists of a two-step process in which first, 5′-mismatched nucleotides are removed by a nuclease (still unidentified), and second, the resulting 5′-truncated tRNA is repaired by the activity of a 3′-5′ polymerase (Supplementary Figure S1) (23–25). However, no enzyme capable of catalyzing such an unusual 3′-5′ polymerase activity was known at that time. Our discovery of the function of DdiTLP3 in repair of 5′-truncated tRNA during the second step of the 5′-editing reaction (Figure 6 and Supplementary Figure S4) finally answers the long-standing question of the identity of one component of the 5′-editing enzyme machinery, and is to our knowledge the first conclusive demonstration of any enzyme that participates in mt-tRNA 5′- or 3′-end editing in any organism. Future identification of the remaining players in 5′-editing, especially the missing nuclease(s), and elucidation of functions for the many TLPs widely observed in other organisms (10) will be important to answer the many outstanding questions about these unusual RNA editing pathways.
A unique mitochondrial function for DdiTLP2: addition of G-1 to mt-tRNAHis
In this work, we also provide the first report, to our knowledge, of any activity catalyzed by DdiTLP2, whose function remained a mystery even after it was identified as a member of the 3′-5′ polymerase superfamily several years ago (9). Although DdiTLP2 is more similar in sequence to other TLP enzymes than to the Thg1 enzymes that are the traditional G-1 addition enzymes in other eukaryotes (10), DdiTLP2 is unlike all other TLPs investigated to date in that it does not catalyze detectable 5′-end repair of any tested tRNA. The similarity of DdiTLP2 to other TLP enzymes is, however, consistent with our observation that unlike Thg1, DdiTLP2 does not depend on the GUG anticodon for mt-tRNAHis recognition (Figure 2C), and suggests that G-1 addition activity may have arisen multiple independent times during evolution from a common ancestral polymerase. Interestingly, the mt-tRNAHis substrate for DdiTLP2 contains a C73 discriminator nucleotide, as opposed to the universally conserved A73 found in all cytosolic tRNAHis (19). Addition of G-1 to the C73-containing mt-tRNA allows DdiTLP2 to utilize the WC-dependent activity that is biochemically preferred by TLPs, but raises new questions about how DdiTLP2 prevents addition of multiple G-nucleotides opposite C74 and C75, as has been observed with other Thg1/TLP family enzymes acting on similar C73-containing substrates (6,16). Determination of the molecular signals that allow TLPs to recognize a correct tRNA 5′-end is an important future step for this enzyme family.
The viability of the dditlp2- strain also brings into question the biological role of the additional G-1 on tRNAHis in mitochondria. A modest growth defect was observed due to the loss of DdiTLP2 activity (Figure 3A), but this is much less severe than the defect associated with loss of the G-1 residue from cytosolic tRNAHis in D. discoideum and other eukaryotes (2,3,44). D. discoideum are strictly aerobic both in vegetatively-growing and developing cells, indicating that mitochondria in the dditlp2- cells must be at least minimally functional (56). Thus, unlike for the cytosolic tRNA, D. discoideum mt-tRNAHis may not require G-1 for its proper function in mitochondrial translation. In D. discoideum, cytosolic and mitochondrial HisRS are encoded by two separate genes. It is possible that D. discoideum mitochondrial HisRS can catalyze sufficient aminoacylation of mt-tRNAHis in the absence of G-1, as already observed for some HisRS enzymes, including the mitochondrial isoform of S. cerevisiae HisRS (22,38,57). Interestingly, a precedent for incomplete modification of mt-tRNAHis with G-1 has already been established in potato mitochondria, where despite the presence of an encoded G-1, a substantial fraction of the mt-tRNAHis lacks G-1 due to an apparent miscleavage by mitochondrial RNase P (58). In this case, however, the HisRS activity in mitochondrial extracts retains its dependence on the G-1 for recognition (58), and the function of the substantial pool of mt-tRNAHis that lacks G-1, similar to the significant fraction observed even in wild-type AX2 mitochondria, is unknown. Evaluating these possibilities, as well as the potential for import of cytosolic tRNAHis containing G-1 into mitochondria, will provide additional insight into the diverse pathways for specifying tRNAHis identity that are now known to exist among eukaryotes.
Predicting biological functions for 3′-5′ polymerases: challenges and opportunities
In this and previous work, we have observed several instances of overlapping biochemical functions associated with the four D. discoideum 3′-5′ polymerases, yet there appears to be little, if any, overlap in function between the four enzymes in vivo. In the case of 5′-editing, although DdiTLP3 and DdiTLP4 both catalyze robust tRNA repair in vitro, making either enzyme initially a candidate for the 5′-editing enzyme (9,20), the cytosolic localization of DdiTLP4 clearly prevents it from carrying out this function in vivo. For G-1 addition to mt-tRNAHis, the situation is even more complex. None of the 34 sequenced mt-tRNAHis clones contained the G-1 residue in the dditlp2- strain (Figure 3C), despite the presence of a fully functional DdiTLP3 enzyme in this strain, and the reduced, but still significant in vitro G-1 addition activity exhibited by this enzyme with mt-tRNAHis (Figure 2). It is possible that the reduced amount of activity of DdiTLP3 measured in vitro is insufficient for G-1 addition in the milieu of the mitochondria, or it is possible that DdiTLP3 forms a complex with other components of the 5′-editing enzyme (such as the unknown 5′-editing nuclease) in vivo that restricts its activity to the 5′-truncated mt-tRNA that undergo 5′-editing. Evaluation of this and other hypotheses will require identification of the complete 5′-editing enzyme. Regardless of the mechanisms involved, it is clear that simply determining in vitro substrate specificities of 3′-5′ polymerase enzymes is not sufficient to determine biological function in many cases.
Finally, although the data presented here suggest that DdiTLP4 has the biochemical capacity to act on at least two non-tRNA substrates, whether the essential phenotype of the DdiTLP4 mutant (Figure 4) is caused by loss of its activity on either 5S rRNA or Class I ncRNA processing remains to be demonstrated. The abundant nature of 5S rRNA and Class I ncRNA likely enabled detection of these RNAs in the labeling experiments (Figure 7), but important functions for other less abundant DdiTLP4 substrates that were also observed to be labeled but not yet identified (Figure 7) can not be excluded. It is also possible that DdiTLP4 could act more generally in RNA surveillance or quality control, using its 3′-5′ polymerization activity to repair RNAs that are incidentally damaged or improperly processed at their 5′-ends. The recent observation that many tRNAs in yeast appear to be under constant surveillance by the rapid tRNA decay pathway opens the door to this possibility (59). Notably, no biological functions for any TLPs in Bacteria and Archaea have been established (11,16,17,60), and our results demonstrate that alternative non-tRNA substrates for TLP enzymes in other domains of life are a distinct possibility. Regardless of the physiological function of DdiTLP4, this ability of 3′-5′ polymerases to act on RNAs that do not share the overall three-dimensional structure of tRNA raises important questions about substrate recognition and catalysis.
Through this work, we also demonstrate that D. discoideum is a powerful model system in which to investigate functions for 3′-5′ polymerase family enzymes. Most animals and fungi appear to have lost the ancient TLP enzymes and instead have retained the single eukaryotic Thg1 that was the founding member of the enzyme family. However, D. discoideum is representative of many TLP-containing organisms that are widespread among the eukaryotic family tree, which also includes some species that lack Thg1 entirely and only encode TLP orthologs with as yet undetermined functions (10,38). Therefore, studies in D. discoideum, where both types of enzymes are present and are biologically relevant, promise to provide fertile ground for a deeper understanding of the evolutionary history and biological capabilities of these unusual enzymes.
Supplementary Material
Acknowledgments
The authors are extremely grateful to Dr. Michael Gray for his ongoing intellectual contributions to these studies and to Dr. Eric Phizicky and Dr. Anita Hopper for their generous advice provided during preparation of this manuscript. The Dicty stock center is acknowledged for advice, strains and plasmids and we thank Dr. Jian-Qiu Wu, Ning Wang and Yihua Zhu for the technical support in the microscopy experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM-087543 to J.E.J]; Pelotonia Fellowship [to Y.L]; NSF REU support [to E.Y.C.]. Funding for open access charge: NIH [GM-087543].
Conflict of interest statement. None declared.
REFERENCES
- 1.Cooley L., Appel B., Soll D. Post-transcriptional nucleotide addition is responsible for the formation of the 5′ terminus of histidine tRNA. Proc. Natl. Acad. Sci. U.S.A. 1982;79:6475–6479. doi: 10.1073/pnas.79.21.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gu W., Hurto R.L., Hopper A.K., Grayhack E.J., Phizicky E.M. Depletion of Saccharomyces cerevisiae tRNA(His) guanylyltransferase Thg1p leads to uncharged tRNAHis with additional m(5)C. Mol. Cell. Biol. 2005;25:8191–8201. doi: 10.1128/MCB.25.18.8191-8201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu W., Jackman J.E., Lohan A.J., Gray M.W., Phizicky E.M. tRNAHis maturation: an essential yeast protein catalyzes addition of a guanine nucleotide to the 5′ end of tRNAHis. Genes Dev. 2003;17:2889–2901. doi: 10.1101/gad.1148603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hyde S.J., Eckenroth B.E., Smith B.A., Eberley W.A., Heintz N.H., Jackman J.E., Doublie S. tRNA(His) guanylyltransferase (THG1), a unique 3′-5′ nucleotidyl transferase, shares unexpected structural homology with canonical 5′-3′ DNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20305–20310. doi: 10.1073/pnas.1010436107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyde S.J., Rao B.S., Eckenroth B.E., Jackman J.E., Doublie S. Structural Studies of a Bacterial tRNA(HIS) Guanylyltransferase (Thg1)-Like Protein, with Nucleotide in the Activation and Nucleotidyl Transfer Sites. PLoS One. 2013;8:e67465. doi: 10.1371/journal.pone.0067465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackman J.E., Phizicky E.M. tRNAHis guanylyltransferase catalyzes a 3′-5′ polymerization reaction that is distinct from G-1 addition. Proc. Natl. Acad. Sci. U.S.A. 2006;103:8640–8645. doi: 10.1073/pnas.0603068103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith B.A., Jackman J.E. Kinetic analysis of 3′-5′ nucleotide addition catalyzed by eukaryotic tRNA(His) guanylyltransferase. Biochemistry. 2012;51:453–465. doi: 10.1021/bi201397f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamura A., Nemoto T., Heinemann I.U., Yamashita K., Sonoda T., Komoda K., Tanaka I., Soll D., Yao M. Structural basis of reverse nucleotide polymerization. Proc. Natl. Acad. Sci. U.S.A. 2013;110:20970–20975. doi: 10.1073/pnas.1321312111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abad M.G., Long Y., Willcox A., Gott J.M., Gray M.W., Jackman J.E. A role for tRNA(His) guanylyltransferase (Thg1)-like proteins from Dictyostelium discoideum in mitochondrial 5′-tRNA editing. RNA. 2011;17:613–623. doi: 10.1261/rna.2517111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackman J.E., Gott J.M., Gray M.W. Doing it in reverse: 3′-to-5′ polymerization by the Thg1 superfamily. RNA. 2012;18:886–899. doi: 10.1261/rna.032300.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao B.S., Maris E.L., Jackman J.E. tRNA 5′-end repair activities of tRNAHis guanylyltransferase (Thg1)-like proteins from Bacteria and Archaea. Nucleic Acids Res. 2011;39:1833–1842. doi: 10.1093/nar/gkq976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connolly S.A., Rosen A.E., Musier-Forsyth K., Francklyn C.S. G-1:C73 recognition by an arginine cluster in the active site of Escherichia coli histidyl-tRNA synthetase. Biochemistry. 2004;43:962–969. doi: 10.1021/bi035708f. [DOI] [PubMed] [Google Scholar]
- 13.Himeno H., Hasegawa T., Ueda T., Watanabe K., Miura K., Shimizu M. Role of the extra G-C pair at the end of the acceptor stem of tRNA(His) in aminoacylation. Nucleic Acids Res. 1989;17:7855–7863. doi: 10.1093/nar/17.19.7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosen A.E., Brooks B.S., Guth E., Francklyn C.S., Musier-Forsyth K. Evolutionary conservation of a functionally important backbone phosphate group critical for aminoacylation of histidine tRNAs. RNA. 2006;12:1315–1322. doi: 10.1261/rna.78606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudinger J., Florentz C., Giege R. Histidylation by yeast HisRS of tRNA or tRNA-like structure relies on residues -1 and 73 but is dependent on the RNA context. Nucleic Acids Res. 1994;22:5031–5037. doi: 10.1093/nar/22.23.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abad M.G., Rao B.S., Jackman J.E. Template-dependent 3′-5′ nucleotide addition is a shared feature of tRNAHis guanylyltransferase enzymes from multiple domains of life. Proc. Natl. Acad. Sci. U.S.A. 2010;107:674–679. doi: 10.1073/pnas.0910961107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinemann I.U., O'Donoghue P., Madinger C., Benner J., Randau L., Noren C.J., Soll D. The appearance of pyrrolysine in tRNAHis guanylyltransferase by neutral evolution. Proc. Natl. Acad. Sci. U.S.A. 2009;106:21103–21108. doi: 10.1073/pnas.0912072106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orellana O., Cooley L., Soll D. The additional guanylate at the 5′ terminus of Escherichia coli tRNAHis is the result of unusual processing by RNase P. Mol. Cell. Biol. 1986;6:525–529. doi: 10.1128/mcb.6.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juhling F., Morl M., Hartmann R.K., Sprinzl M., Stadler P.F., Putz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abad M.G., Long Y., Kinchen R.D., Schindel E.T., Gray M.W., Jackman J.E. Mitochondrial tRNA 5′-editing in Dictyostelium discoideum and Polysphondylium pallidum. J. Biol. Chem. 2014;289:15155–15165. doi: 10.1074/jbc.M114.561514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackman J.E., Phizicky E.M. tRNAHis guanylyltransferase adds G-1 to the 5′ end of tRNAHis by recognition of the anticodon, one of several features unexpectedly shared with tRNA synthetases. RNA. 2006;12:1007–1014. doi: 10.1261/rna.54706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao B.S., Mohammad F., Gray M.W., Jackman J.E. Absence of a universal element for tRNAHis identity in Acanthamoeba castellanii. Nucleic Acids Res. 2013;41:1885–1894. doi: 10.1093/nar/gks1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lonergan K.M., Gray M.W. Editing of transfer RNAs in Acanthamoeba castellanii mitochondria. Science. 1993;259:812–816. doi: 10.1126/science.8430334. [DOI] [PubMed] [Google Scholar]
- 24.Lonergan K.M., Gray M.W. Predicted editing of additional transfer RNAs in Acanthamoeba castellanii mitochondria. Nucleic Acids Res. 1993;21:4402. doi: 10.1093/nar/21.18.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price D.H., Gray M.W. A novel nucleotide incorporation activity implicated in the editing of mitochondrial transfer RNAs in Acanthamoeba castellanii. RNA. 1999;5:302–317. doi: 10.1017/s1355838299981840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fey P., Kowal A.S., Gaudet P., Pilcher K.E., Chisholm R.L. Protocols for growth and development of Dictyostelium discoideum. Nat. Protoc. 2007;2:1307–1316. doi: 10.1038/nprot.2007.178. [DOI] [PubMed] [Google Scholar]
- 27.Gaudet P., Pilcher K.E., Fey P., Chisholm R.L. Transformation of Dictyostelium discoideum with plasmid DNA. Nat. Protoc. 2007;2:1317–1324. doi: 10.1038/nprot.2007.179. [DOI] [PubMed] [Google Scholar]
- 28.Blaauw M., Linskens M.H., van Haastert P.J. Efficient control of gene expression by a tetracycline-dependent transactivator in single Dictyostelium discoideum cells. Gene. 2000;252:71–82. doi: 10.1016/s0378-1119(00)00227-4. [DOI] [PubMed] [Google Scholar]
- 29.Martens H., Novotny J., Oberstrass J., Steck T.L., Postlethwait P., Nellen W. RNAi in Dictyostelium: the role of RNA-directed RNA polymerases and double-stranded RNase. Mol. Biol. Cell. 2002;13:445–453. doi: 10.1091/mbc.01-04-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosel D., Kimmel A.R. The COP9 signalosome regulates cell proliferation of Dictyostelium discoideum. Eur. J. Cell Biol. 2006;85:1023–1034. doi: 10.1016/j.ejcb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Levi S., Polyakov M., Egelhoff T.T. Green fluorescent protein and epitope tag fusion vectors for Dictyostelium discoideum. Plasmid. 2000;44:231–238. doi: 10.1006/plas.2000.1487. [DOI] [PubMed] [Google Scholar]
- 32.Komori K., Kuroe K., Yanagisawa K., Tanaka Y. Cloning and characterization of the gene encoding a mitochondrially localized DNA topoisomerase II in Dictyostelium discoideum. Western blot analysis. Biochim. Biophys. Acta. 1997;1352:63–72. doi: 10.1016/s0167-4781(96)00229-1. [DOI] [PubMed] [Google Scholar]
- 33.Schimmel B.G., Berbusse G.W., Naylor K. Mitochondrial fission and fusion in Dictyostelium discoideum: a search for proteins involved in membrane dynamics. BMC Res. Notes. 2012;5:505. doi: 10.1186/1756-0500-5-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breker M., Gymrek M., Schuldiner M. A novel single-cell screening platform reveals proteome plasticity during yeast stress responses. J. Cell Biol. 2013;200:839–850. doi: 10.1083/jcb.201301120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Claros M.G. MitoProt, a Macintosh application for studying mitochondrial proteins. Comput. Appl. Biosci. 1995;11:441–447. doi: 10.1093/bioinformatics/11.4.441. [DOI] [PubMed] [Google Scholar]
- 36.Corcoran J.B., McCarthy S., Griffin B., Gaffney A., Bhreathnach U., Borgeson E., Hickey F.B., Docherty N.G., Higgins D.F., Furlong F., et al. IHG-1 must be localised to mitochondria to decrease Smad7 expression and amplify TGF-beta1-induced fibrotic responses. Biochim. Biophys. Acta. 2013;1833:1969–1978. doi: 10.1016/j.bbamcr.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 37.Hickey F.B., Corcoran J.B., Docherty N.G., Griffin B., Bhreathnach U., Furlong F., Martin F., Godson C., Murphy M. IHG-1 promotes mitochondrial biogenesis by stabilizing PGC-1alpha. J. Am. Soc. Nephrol. 2011;22:1475–1485. doi: 10.1681/ASN.2010111154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao B.S., Jackman J.E. Life without post-transcriptional addition of G-1: two alternatives for tRNAHis identity in Eukarya. RNA. 2015;21:243–253. doi: 10.1261/rna.048389.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faix J., Kreppel L., Shaulsky G., Schleicher M., Kimmel A.R. A rapid and efficient method to generate multiple gene disruptions in Dictyostelium discoideum using a single selectable marker and the Cre-loxP system. Nucleic Acids Res. 2004;32:e143. doi: 10.1093/nar/gnh136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimmel A.R., Faix J. Generation of multiple knockout mutants using the Cre-loxP system. Methods Mol. Biol. 2006;346:187–199. doi: 10.1385/1-59745-144-4:187. [DOI] [PubMed] [Google Scholar]
- 41.Khare A., Shaulsky G. Cheating by exploitation of developmental prestalk patterning in Dictyostelium discoideum. PLoS Genet. 2010;6:e1000854. doi: 10.1371/journal.pgen.1000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Preston M.A., Phizicky E.M. The requirement for the highly conserved G-1 residue of Saccharomyces cerevisiae tRNAHis can be circumvented by overexpression of tRNAHis and its synthetase. RNA. 2010;16:1068–1077. doi: 10.1261/rna.2087510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rice T.S., Ding M., Pederson D.S., Heintz N.H. The highly conserved tRNAHis guanylyltransferase Thg1p interacts with the origin recognition complex and is required for the G2/M phase transition in the yeast Saccharomyces cerevisiae. Eukaryot. Cell. 2005;4:832–835. doi: 10.1128/EC.4.4.832-835.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo D., Hu K., Lei Y., Wang Y., Ma T., He D. Identification and characterization of a novel cytoplasm protein ICF45 that is involved in cell cycle regulation. J. Biol. Chem. 2004;279:53498–53505. doi: 10.1074/jbc.M406737200. [DOI] [PubMed] [Google Scholar]
- 45.Kuhlmann M., Popova B., Nellen W. RNA interference and antisense-mediated gene silencing in Dictyostelium. Methods Mol. Biol. 2006;346:211–226. doi: 10.1385/1-59745-144-4:211. [DOI] [PubMed] [Google Scholar]
- 46.Laforest M.J., Bullerwell C.E., Forget L., Lang B.F. Origin, evolution, and mechanism of 5′ tRNA editing in chytridiomycete fungi. RNA. 2004;10:1191–1199. doi: 10.1261/rna.7330504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aspegren A., Hinas A., Larsson P., Larsson A., Soderbom F. Novel non-coding RNAs in Dictyostelium discoideum and their expression during development. Nucleic Acids Res. 2004;32:4646–4656. doi: 10.1093/nar/gkh804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Avesson L., Reimegard J., Wagner E.G., Soderbom F. MicroRNAs in Amoebozoa: deep sequencing of the small RNA population in the social amoeba Dictyostelium discoideum reveals developmentally regulated microRNAs. RNA. 2012;18:1771–1782. doi: 10.1261/rna.033175.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Avesson L., Schumacher H.T., Fechter P., Romby P., Hellman U., Soderbom F. Abundant class of non-coding RNA regulates development in the social amoeba Dictyostelium discoideum. RNA Biol. 2011;8:1094–1104. doi: 10.4161/rna.8.6.17214. [DOI] [PubMed] [Google Scholar]
- 50.Hinas A., Soderbom F. Treasure hunt in an amoeba: non-coding RNAs in Dictyostelium discoideum. Curr. Genet. 2007;51:141–159. doi: 10.1007/s00294-006-0112-z. [DOI] [PubMed] [Google Scholar]
- 51.Grosjean H., Benne R. Modification and editing of RNA. Washington, D.C: ASM Press; 1998. [Google Scholar]
- 52.Gray M.W. Evolutionary Origin of RNA Editing. Biochemistry. 2012;51:5235–5242. doi: 10.1021/bi300419r. [DOI] [PubMed] [Google Scholar]
- 53.Betat H., Long Y., Jackman J.E., Morl M. From end to end: tRNA editing at 5′- and 3′-terminal positions. Int. J. Mol. Sci. 2014;15:23975–23998. doi: 10.3390/ijms151223975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lavrov D.V., Brown W.M., Boore J.L. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13738–13742. doi: 10.1073/pnas.250402997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leigh J., Lang B.F. Mitochondrial 3′ tRNA editing in the jakobid Seculamonas ecuadoriensis: a novel mechanism and implications for tRNA processing. RNA. 2004;10:615–621. doi: 10.1261/rna.5195504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Srinivas U.K., Katz E.R. Oxygen utilization in Dictyostelium discoideum. FEMS Microbiol. Lett. 1980;9:53–55. [Google Scholar]
- 57.Su D., Lieberman A., Lang B.F., Simonovic M., Soll D., Ling J. An unusual tRNAThr derived from tRNAHis reassigns in yeast mitochondria the CUN codons to threonine. Nucleic Acids Res. 2011;39:4866–4874. doi: 10.1093/nar/gkr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Placido A., Sieber F., Gobert A., Gallerani R., Giege P., Marechal-Drouard L. Plant mitochondria use two pathways for the biogenesis of tRNAHis. Nucleic Acids Res. 2010;38:7711–7717. doi: 10.1093/nar/gkq646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guy M.P., Young D.L., Payea M.J., Zhang X., Kon Y., Dean K.M., Grayhack E.J., Mathews D.H., Fields S., Phizicky E.M. Identification of the determinants of tRNA function and susceptibility to rapid tRNA decay by high-throughput in vivo analysis. Genes Dev. 2014;28:1721–1732. doi: 10.1101/gad.245936.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heinemann I.U., Randau L., Tomko R.J., Jr, Soll D. 3′-5′ tRNAHis guanylyltransferase in bacteria. FEBS Lett. 2010;584:3567–3572. doi: 10.1016/j.febslet.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.