Abstract
Genome transplantation (GT) allows the installation of purified chromosomes into recipient cells, causing the resulting organisms to adopt the genotype and the phenotype conferred by the donor cells. This key process remains a bottleneck in synthetic biology, especially for genome engineering strategies of intractable and economically important microbial species. So far, this process has only been reported using two closely related bacteria, Mycoplasma mycoides subsp. capri (Mmc) and Mycoplasma capricolum subsp. capricolum (Mcap), and the main factors driving the compatibility between a donor genome and a recipient cell are poorly understood. Here, we investigated the impact of the evolutionary distance between donor and recipient species on the efficiency of GT. Using Mcap as the recipient cell, we successfully transplanted the genome of six bacteria belonging to the Spiroplasma phylogenetic group but including species of two distinct genera. Our results demonstrate that GT efficiency is inversely correlated with the phylogenetic distance between donor and recipient bacteria but also suggest that other species-specific barriers to GT exist. This work constitutes an important step toward understanding the cellular factors governing the GT process in order to better define and eventually extend the existing genome compatibility limit.
INTRODUCTION
Synthetic biology encompasses a variety of disciplines that aim to design and create new systems either derived from natural organisms (top-down approaches) or built from scratch (bottom-up approaches). From a fundamental perspective, being able to design and build a biological system stands as a very practical measure of understanding of how this system operates and can help to identify molecular components essential for life. The engineering of new and well understood cellular chassis would provide platforms for innovative applications in biotechnology, agriculture or medicine. Different and complementary approaches are currently developed using bacteria that have lost much of their genomes through regressive evolution (1–3). Among them, Mollicutes are considered as the best representatives of natural minimal cells with some of the smallest genomes among free-living organisms (0.58 to ∼2.2 Mbp) that can grow in axenic medium (4). Innovative strategies toward the production of minimalist chassis rapidly emerged from global gene inactivation experiments (3,5) and from comparative genome analyses for a better comprehension of minimal cellular machineries such as the translation apparatus (6). A recent breakthrough was achieved with the design and synthesis of Syn3.0 (0.531 Mbp; 473 genes) (7), a reduced version of the 1.08 Mbp Mycoplasma mycoides subsp. capri (Mmc) natural genome. This work represents a technological milestone with the creation of the smallest genome ever carried by a microorganism capable of autonomous replication. It is also a step forward in defining the minimal set of genes essential for life and the possibility to customize bacterial genomes.
The drastic minimization of Mmc genome succeeded because of the emergence of cutting-edge technologies allowing the synthesis and manipulation of its genetic information on a genome-wide scale. Historically, mycoplasmas have long been considered intractable for genome engineering (4), just like a large majority of non-model microorganisms. Newly developed approaches now rely on the (i) cloning and (ii) engineering of the whole Mmc genome into an intermediate host possessing well-developed tools followed by (iii) the back-transplantation of the newly engineered genome into suitable bacterial recipient cells.
Because of its cloning capacities and large repertoire of available genetic tools, the yeast Saccharomyces cerevisiae was chosen as the host for whole bacterial genome manipulations and first studies have consisted in the cloning of mycoplasma genomes into S. cerevisiae as yeast centromeric plasmids (8,9). So far, seven complete bacterial genomes have been cloned in yeast, including micro-organisms that, unlike most Mollicutes (10), use the universal genetic code (11,12). Known key factors limiting in-yeast cloning are the genome size, high G + C %, the absence of ARS-like sequences (autonomous replication sequences) (13) and the presence of any toxic genes (11).
Following in-yeast cloning, new yeast molecular and editing tools have been developed to manipulate cloned bacterial genomes. For instance, the TREC system (Tandem Repeat Endonuclease Cleavage) (14) and TREC-IN derivatives (15) allow the deletion, insertion, or replacement of genes without any scars on bacterial chromosomes cloned in yeast. Such systems have notably been used in mycoplasma to assess gene function (8,16,17) and for genome minimization purposes (15). More recently, the CRISPR/Cas9 system, first developed for yeast mutagenesis purposes (18), has been also adapted for the engineering of bacterial genomes cloned in yeast (19).
Altogether, in-yeast cloning strategies associated with genome engineering tools now pave the way to high-throughput manipulation of natural or synthetic genomes in yeast, allowing the study of virtually any gene of interest on a bacterial chromosome. However, for this extremely powerful set of methods to be widely used, engineered bacterial chromosomes need to be isolated from yeast and transplanted back into a recipient bacterium that provides compatible cellular machineries able to replicate and express its genetic information. This critical whole genome transplantation (GT) step was first established with a whole genome isolated from bacteria (20) and then with natural and synthetic bacterial genomes cloned in yeast (7,8,21). These results demonstrated that a bacterial genome can be designed, synthetized, genetically manipulated on a large scale and transplanted into a recipient cell resulting in a living cell that is genetically programmed by the engineered genome. In these cases, two closely related species, sharing more than 99% identity on 16S rDNA as well as on their core proteome (6; also see Supplementary Table S2), Mmc and Mycoplasma capricolum subsp. capricolum (Mcap) were respectively used as donor genome and recipient cell. However, GT technology is currently limited to these two bacterial species and this limitation prevents the demonstration of the strategy for other non-model microorganisms with agricultural, medical or biotechnological interest. In particular, mechanisms underlying the GT process are still unclear and major technological difficulties might arise every time GT technology needs to be applied to other bacterial species.
In order to circumvent these difficulties, we explored the effect of the phylogenetic distance between donor genomes and the recipient cell on GT efficiency using seven species from the Spiroplasma phylogenetic group with increasing phylogenetic distance from the Mcap recipient cell. Our results demonstrate for the first time that GT can be achieved with several Mcap related genomes, including a Mollicutes belonging to a different genus. Overall, GT efficiency decreases as phylogenetic distance increases, suggesting that a compatibility limit is eventually reached between incoming genome and recipient cell enzymatic machineries. With the cloning of several Mollicutes genomes in yeast, this work offers efficient genetic engineering tools in these otherwise difficult-to-manipulate organisms.
MATERIALS AND METHODS
Bacterial and yeast strains, culture conditions
Competent Escherichia coli cells (Electromax DH10B from Invitrogen) [F−-mcrAΔ(mrr-hsdRMS-mcrBC) ф80dlacZ ΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galKλ- rpsL nupG] served as the host strain for cloning experiments and plasmid propagation. E. coli cells transformed with plasmids were grown at 37°C in Luria-Bertani (LB) broth medium or in LB agar supplemented with 100 μg/ml of ampicillin.
Saccharomyces cerevisiae strain W303a (MATa his3-11, 15 trp11 leu2-3,112 ura3-1 ade2-1 can1-100) VL6-48N (MATα trp1-Δ1 ura3-Δ1 ade2-101 his3-Δ200 lys2 met14 cir) (22) was cultured at 30°C in YPDA medium (Clontech) according to a standard protocol (23). Yeast transformed with mycoplasma genomes was grown in minimal SD Base medium (Clontech), complemented with—HIS DO supplement (Clontech) (SD-HIS medium).
Wild-type Mcap (wtMcap) strain California KidT (ATCC 27343) was used in this study as well as a restriction-free Mcap mutant (McapΔRE) obtained by inactivation of the CCATC-restriction enzyme in the wild-type strain (8). McapΔRE harbors (i) a puromycin resistance marker for selection in mycoplasma background and (ii) yeast elements (a centromere CEN6, an autonomously replicating sequence ARSH4 and the auxotrophic marker HIS) for selection and propagation of the Mcap genome as a centromeric plasmid in yeast. Wild-type Mycoplasma leachii strain PG50 (wtMlea) (24,25), wild-type Mmc strain GM12 (wtMmc) (24), wild-type Mycoplasma mycoides subsp. mycoides strain PG1 (wtMmm) (26), wild-type Mycoplasma putrefaciens strain 156 (wtMputr), wild-type Mesoplasma florum strain L1 (wtMflorum; ATCC 33453) (27) and wild-type Spiroplasma citri strain GII3 (wtScitri; ATCC 27556) (28) were transformed with the newly constructed vector pMT85tetM-PSlacZ-pRS313 (Supplementary Figure S1) as described by others (9,29), then selected as donor species for GT experiments.
All species were cultured in SP5 medium, deriving from the original SP4 medium (30). SP5 medium is composed of 3.5 g/l of Mycoplasma broth base (Fisher Scientific), 10 g/l of Bacto Tryptone (Fisher Scientific) and 5.3 g/l of Bacto Peptone (Fisher Scientific). The solution was adjusted to pH 7.5, autoclaved for 20 min at 120°C, then supplemented with 0.125% (w/v) glucose, 5% (v/v) CMRL 1066 10× (Invitrogen), 0.11% (w/v) sodium bicarbonate, 1 mM L-glutamine, 3.5% (v/v) yeast extract (Fisher Scientific), 0.2% (w/v) TC yeastolate, 17% (v/v) fetal bovine serum, 0.1 mg/ml ampicillin and 0.002% (w/v) phenol red.
Mycoplasma strains were all cultured at 37°C, whereas Mflorum and Scitri were cultured at 30 and 32°C, respectively. Tetracycline was added to the medium when needed at concentrations ranging from 2 to 15 μg/ml, depending on the species.
For transplantation experiments, McapΔRE recipient cells were grown at 30°C in super optimal broth (SOB) (31) supplemented with 17% (v/v) fetal bovine serum, glucose at 10 g/l, 0.002% (w/v) phenol red, and penicillin at 0.5 μg/ml (SOB (+) medium).
Phylogenetic reconstruction and similarity matrix
The phylogenetic tree was generated using concatenated multiple alignments of selected 79 orthologous proteins involved in translation as previously described (6). Multiple alignments were generated using MUSCLE (32), concatenated using Seaview (33) and curated from unreliable sites with GBlock (34). The final concatenated alignment contained 10 686 sites. The phylogenetic tree was constructed by the Maximum Likelihood method using PhyML (35) available on the web server Phylogeny.fr (36). The list of Mollicutes analyzed with some of their genomic characteristics is given in Supplementary Table S1. A similarity matrix was produced from the final concatenated alignment using BLASTP 2.4.0 (37)
Construction of oriC-based replicative plasmids
The oriC-based plasmids pMCO3, pMYCO1, pMYSO1 and pSD4, containing the origin of replication of Mcap, Mmc, Mmm and Scitri, respectively, have been previously described (38).
OriC plasmids pMleaOriC, pMputrOriC and pMflOriC containing the origin of replication of Mlea, Mputr and Mflorum were constructed for this study as described in Lartigue et al. (38). Primers used for their construction are summarized in Supplementary Table S4. OriC segments of these plasmids have a global organization identical to those previously constructed. Each region includes the dnaA gene of each species studied surrounded by natural intergenic regions on each side of the gene. Prior to being used for transformation into McapΔRE, the purified plasmids were verified by restriction analyses and the integrity of the dnaA gene sequences was checked by DNA sequencing. Transformations of McapΔRE were conducted using 1 or 10 μg of plasmids and transformants were grown on selective medium for 5–8 days depending on the species under study.
Plasmid construction used to mark donor Mollicutes genomes
A transposon-based plasmid (pMT85tetM-PSlacZ-pRS313, Supplementary Figure S1, accession number KX011460) designed for marking the genomes of all Mollicutes species selected as donor for GT experiments was constructed using the Gibson Assembly® method (39). Details for this construction are described in the Supplementary material. Ten to thirty micrograms of pMT85tetM-PSlacZ-pRS313 were used to transform wtMmm, wtMlea, wtMputr species by PEG-mediated transformation (40) and wtScitri by electroporation (41). A marked Mflorum donor genome was obtained by transformation of wtMflorum as described by Baby et al. (V. Baby, F. Labroussaa, J. Brodeur, D. Matteau, G. Gourgues, C. Lartigue and S. Rodrigue, in preparation).
Transformants were grown in SP5 medium containing suitable concentration of tetracycline, filter cloned three times to ascertain the clonality of the population and stored at −80°C. Genomic DNA from subclones was extracted with the Wizard Genomic DNA Purification Kit (Promega) and further analyzed by direct sequencing to localize the transposon insertion site. Primers used for direct sequencing and localization of insertions into Mollicutes genomes are summarized in Supplementary Table S5.
Isolation of intact Mollicutes genomes in agarose plugs
The protocol for whole, intact genomic DNA isolation from bacteria cultures was done as previously described by Lartigue et al. (20) with some modifications. First, the amount of bacterial cells present in each agarose plug was accurately measured using a newly developed quantitative real-time polymerase chain reaction (qPCR) assay, (see Supplementary Figure S2 and Supplementary Data for details). This allowed the preparation of a reproducible series of plugs (2-fold serial dilutions) ranging from 8 to 0.25 μg of genomic DNA. The resuspension buffer used for Mflorum cells preparation was also modified. Briefly, a 50-ml Mflorum culture was centrifuged at 5000 g for 15 min at 4°C and resuspended into 25 ml of a cold resuspension buffer (8 mM Hepes, 272 mM Sucrose; pH 7.4). After repeating the centrifugation step, cells were resuspended into 500 μl of the same resuspension buffer and then mixed with a 2% low-melting-point agarose solution. This modification prevented cell aggregation when Mflorum cells were resuspended before their inclusion into agarose plugs. This protocol was applied to all the others species used in this study.
Preparation of yeast agarose plugs was performed according to Lartigue et al. (8).
Genome transplantation into McapΔRE recipient cells
Prior to transplantation experiments, entrapped Mollicutes genomes (isolated from bacterial cultures or yeast cells) were released from the agarose plug with β-agarase I (3 units/plug) per the manufacturer's recommendations (New England Biolabs). All transplantation experiments in this study were performed with as previously developed using a 5% polyethylene glycol (PEG) mediated protocol (8,20), except for a few modifications found to improve transplantation/transformation efficiencies. McapΔRE recipient cells were cultivated at 30°C instead of 37°C in SOB(+) and this temperature was also used during the 90 min recovery period after incubation of cells with PEG. Also, SP5 medium was used instead of SP4. This protocol was also used for the transformation of plasmids carrying each oriC of the different bacterial species transplanted. Depending of the transformation efficiencies observed, 1 or 10 μg of plasmid was used instead of 20 or 100 μl of melted agarose plug solutions, respectively used for from-bacteria or from-yeast transplantations.
Whole genome sequencing
For Mcap, Mmc, Mmm, Mlea, Mputr and Mflorum, gDNA from the donor strains and two from-bacteria transplants was extracted using the Wizard genomic DNA purification kit (Promega) for mycoplasmas and the Puregene yeast/bact. kit B (QIAGEN) for Mflorum using the manufacturer's recommendations. For Mmc, Mlea and Mputr, four transplants obtained from-yeast GT assays were also processed. Illumina libraries were prepared as in Baby et al. (V. Baby, F. Labroussaa, J. Brodeur, D. Matteau, G. Gourgues, C. Lartigue and S. Rodrigue, in preparation) and sequenced on a HiSeq2000 instrument at the McGill University and Génome Québec Innovation Centre (Montreal, Canada). The resulting 50-bp paired-end reads were aligned to the available RefSeq genomes; Mcap NC_007633.1, Mmc NZ_CP001621.1, Mmm NC_005364.2, Mlea NC_014751.1, Mputr NC_015946.1. For Mflorum, the expected sequence containing the yeast elements and tetM gene was used and variant detection was performed as in Baby et al. (V. Baby, F. Labroussaa, J. Brodeur, D. Matteau, G. Gourgues, C. Lartigue and S. Rodrigue, in preparation). Briefly, the reads were trimmed using Trimmomatic (v. 0.32) (42) and aligned using BWA-mem (v. 0.7.10) (43). GATK (v. 3.3.0) (44) was used to detect variants, and VCFtools (v. 0.1.12b) (45) was used to compare the donor and transplant genomes
Statistics
Transformation rates are the mean of three independent transformation experiments performed in triplicates. Transplantation rates correspond to the average of four independent GT events obtained during the same GT experiment. Three independent GT experiments were conducted for each species. We used the Tukey's HSD test for all pair-wise comparisons among oriC-based plasmid transformation rates, from-bacteria and from-yeast GT experiments. From-bacteria and from-yeast GT data were treated independently for statistical calculations. We used a parametric paired t-test (46) to assess statistical difference during chloramphenicol treatment for each dilution point. qPCR data were log-transformed to meet model assumptions. The Benjamini–Hochberg (47) method was used to adjust the false discovery rate associated with the test.
RESULTS
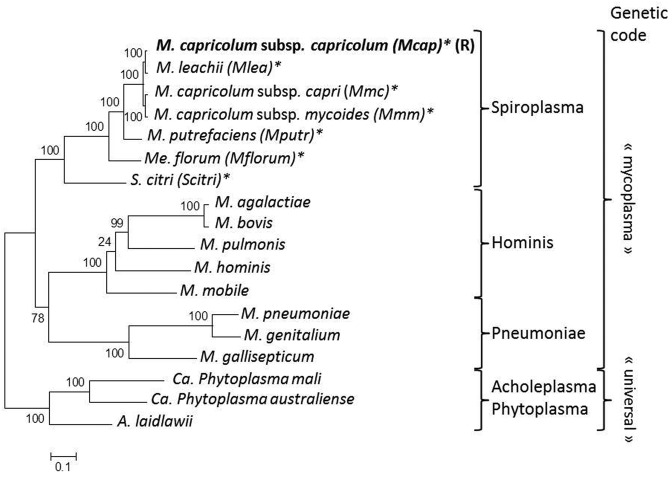
Mcap is currently the only recipient bacterium that has been used successfully in GT experiments. The McapΔRE strain, a derivative of Mcap that was obtained by inactivating the gene encoding the CCATC-restriction endonuclease, was chosen as the recipient cell since this deletion eliminates the need for a DNA methylation step before GT (8). Here, seven bacterial species of the Spiroplasma phylogenetic group were selected as donor-genome strains to investigate the importance of phylogenetic distance on GT (Figure 1). A first group comprising Mmc, Mmm as well as Mlea, all belonging to the Mycoides cluster that also includes Mcap, are more closely related to the recipient cell (>99% identity on the 79 core proteins; Supplementary Table S2). A second group of species, more phylogenetically distant from Mcap (between 85 and 96% identity on the 79 core proteins; Supplementary Table S2), included Mputr, Mflorum and Scitri. All species can be cultured under the same growth conditions and use the alternative mycoplasma genetic code for gene expression, thus constituting plausible candidates for GT given our experimental framework.
Figure 1.
Phylogenetic tree of Mollicutes. The phylogenetic tree was generated using concatenated multiple sequence alignments of selected 79 orthologous proteins involved in translation as described in Grosjean et al. (6). The genetic code used is indicated on the right side. Phylogenetic group: Spiroplasma; Pneumoniae; Hominis; Acholeplasma-Phytoplasma. Mcap used as the recipient cell is indicated with the (R) symbol. Species used as donor genomes, all belonging to the Spiroplasma phylogenetic group, are marked with an asterisk.
Heterologous transformation of Mcap with oriC plasmids
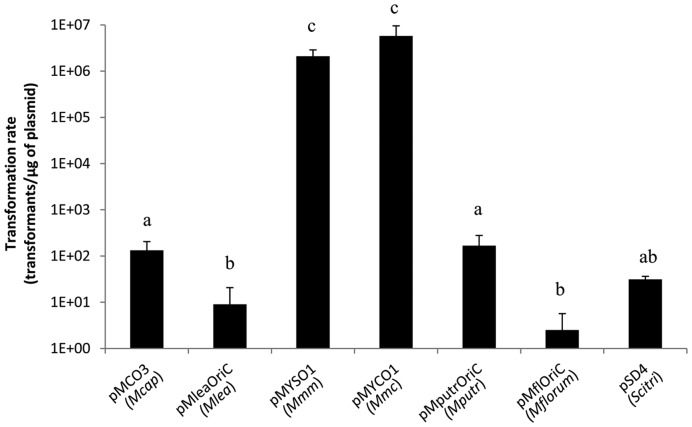
Considering that few critical cellular processes are likely to ensure the ‘boot up’ of the new chromosome by the recipient cell, we surmised that the cellular machinery of Mcap should allow the replication of plasmids carrying the origin of replication region (oriC region) of donor genomes. We thus sought to evaluate whether McapΔRE can be transformed with plasmids containing the oriC region of the seven individual potential donor genomes. Genomic oriC regions are highly syntenic among the considered species and all oriC plasmids were designed similarly to include the dnaA gene and the intergenic regions located on each side. Four oriC plasmids, pMCO3 (Mcap), pMYCO1 (Mmc), pMYSO1 (Mmm) and pSD4 (Scitri) were previously shown to contain all functional elements required for efficient replication (38). Three additional plasmids were built using the same backbone to contain the complete ∼1.9–2.0 kbp oriC regions of Mlea, Mputr or Mflorum. All oriC plasmids were found to replicate in McapΔRE suggesting that the enzymatic machinery of the bacteria should be able to initiate the replication of genomes with a diversity of oriC. However, significant differences in transformation rates were observed (Figure 2). Strikingly, no direct relationship was observed between the transformation efficiencies of oriC plasmids and their phylogenetic distances to the recipient cell. Plasmids pMYSO1 and pMYCO1 containing oriC regions of Mmm and Mmc were transformed with higher efficiencies (>106 transformants/μg of plasmid) than the pMCO3 plasmid (∼102 transformants/μg of plasmid) even if this last one was derived from the genome of the recipient cell. Overall, these data support the choice of McapΔRE as a suitable recipient cell for GT experiments within the Spiroplasma phylogenetic group.
Figure 2.
Transformation of McapΔRE with different oriC-based replicative plasmids. Plasmids containing complete oriC regions of all Mollicutes donor genomes used in GT experiments were tested. Letters indicate statistically different transformation rates (pair-wise comparisons).
Insertion of genetic markers in donor Mollicutes genomes
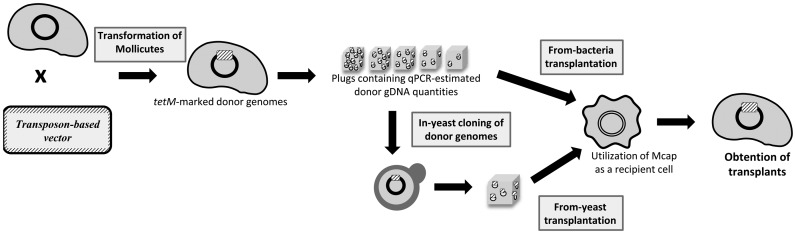
GT can be accomplished using chromosomes directly isolated from bacteria (from-bacteria GT) or from yeast containing the cloned bacterial genome (from-yeast GT) (Figure 3). However, isolated donor genomes must harbor a marker (tetM, conferring resistance to tetracycline) for selection of bacterial transplants. For genomes cloned in yeast, additional elements are important to allow maintenance and selection of the chromosome once inserted in its intermediate host (i.e. ARSH4, CEN6 and HIS3). Mcap, Mmc and Mflorum genomes containing these yeast and bacterial elements were obtained from other studies ((8); V. Baby, F. Labroussaa, J. Brodeur, D. Matteau, G. Gourgues, C. Lartigue and S. Rodrigue, in preparation). We then constructed a novel transposon-based plasmid, named pMT85tetM-PSlacZ-pRS313 (Supplementary Figure S1) to allow a single-step integration of all elements. This plasmid was used to similarly mark the genomes of Mmm, Mlea, Mputr and Scitri. For each of these species, a clone in which the transposon inserted into intergenic sequences or genes not likely to interfere with GT was used for the following experiments (Supplementary Table S5).
Figure 3.
Global scheme representing the main steps conducted in this study. Donor-genome cells were first transformed with an integrative transposon-based plasmid. Newly marked genomes were then gently isolated and either directly back-transplanted into restriction free Mcap recipient cells for from-bacteria transplantation experiments or transformed into yeast spheroplasts for further from-yeast transplantation experiments. All putative transplants originating from from-bacteria and from-yeast transplantation experiments were genotypically and phenotypically characterized systematically.
Impact of gDNA concentration on genome transplantation
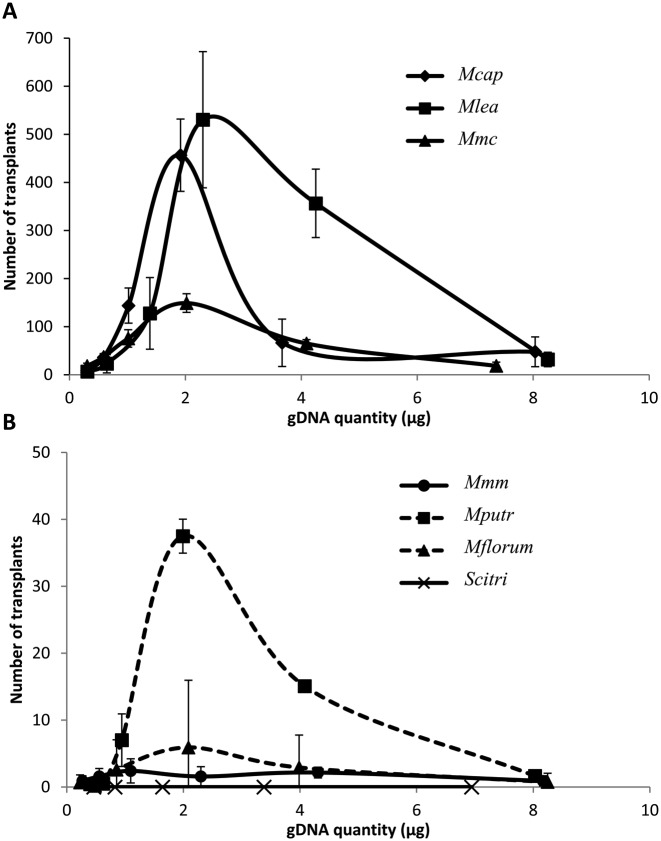
Bacterial cultures from the seven Mollicutes species, followed by a qPCR assay developed to accurately estimate cell concentration, were used to prepare 2-fold dilution series of DNA plugs with similar donor genome gDNA concentrations (∼400, 200, 100, 50, 25 and 12.5 ng/μl) (Supplementary Figure S2). Each plug of every series was then used in an independent GT experiment using McapΔRE as the recipient cell. Transplants were obtained for every species tested (Figure 4), except for Scitri, a species that remained refractory to transplantation in McapΔRE under all tested conditions (Figure 4B). A similar pattern was even observed between all species tested when transplantation rates obtained with all the gDNA concentrations used for GT were compared. Transplantation rates increased when gDNA concentrations were comprised between 0 and 2 μg of gDNA and then progressively decreased when higher gDNA concentrations were used, giving bell-shape curves for all species (Figure 4). Surprisingly, the same optimal gDNA amount (∼2 μg), corresponding to plugs containing ∼100 ng/μl of bacterial DNA, was clearly needed for successful GT for all transplanted species. We categorized these species in two sub-groups based on their respective transplantation rates. Mcap, Mlea and Mmc were considered as highly transplantable with around or over 100 transplants obtained when gDNA quantities were in the range of 1–4 μg (Figure 4A). On the other hand, Mputr, Mmm and Mflorum were considered as poorly transplantable species, especially for the two latter with <10 transplants obtained regardless of the gDNA quantity used (Figure 4B).
Figure 4.
Effect of gDNA quantity in from-bacteria GT assays. Different quantities of donor gDNA were tested in GT experiments using McapΔRE as the recipient cell. Numbers of transplants obtained in relation to the gDNA quantity used are reported for (A) highly transplantable species (Mcap, Mlea, Mmc) or (B) poorly transplantable species (Mmm, Mputr, Mflorum, Scitri).
Importance of the phylogenetic distance in the GT process
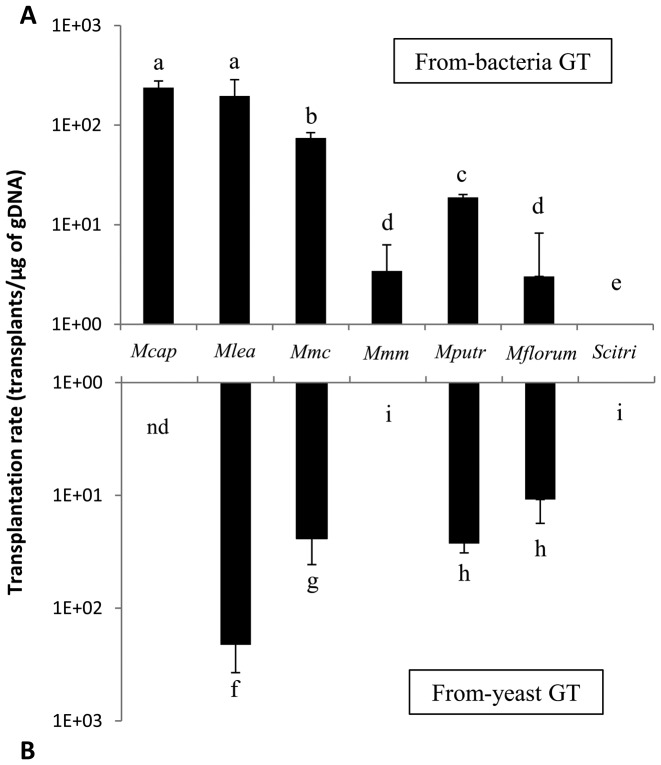
We next used the previously determined optimal gDNA quantity to accurately measure the transplantation rate for each of the seven targeted species (Figure 5A). All species considered, the transplantation of Mcap into its own genetic background resulted in the highest transplantation rate (Table 1). In contrast to results obtained for oriC plasmids transformation, the other genomes were successfully transplanted into McapΔRE and a negative correlation between transplantation efficiencies and phylogenetic distance was observed (Figure 5A). The only exception to this general trend was Mmm, for which transplants were only obtained at a rate of ∼3.43 transplants/μg of gDNA. This rate is significantly lower than its closest phylogenetic neighbors Mlea (∼1.96 × 102 transplants/μg of gDNA) and Mmc (∼7.44 × 101 transplants/μg of gDNA). Altogether, these results showed that we have extended GT technology to five new Mollicutes species, with Mflorum being the Mcap most-distant compatible genome in our experimental system (92.47% identity on the 79 core proteins).
Figure 5.
Transplantation rates obtained for (A) from-bacteria and (B) from-yeast GT assays. Optimal gDNA quantities were used to determine an optimal transplantation rate for all species tested. Nd, not determined. Letters indicate statistically different transformation rates (pair-wise comparisons).
Table 1. Optimal genomic DNA and transplantation rates obtained for each donor species tested, in either from-bacteria or from-yeast GT experiments.
| Species transplanted | From-bacteria transplantation | From-yeast transplantation | ||
|---|---|---|---|---|
| Optimal gDNA concentration (μg) | Transplantation rate (transplants/μg DNA ± SEM) | Optimal gDNA concentration (μg) | Transplantation rate (transplants/μg DNA ± SEM) | |
| M. capricolum subsp. capricolum CK | 1.92 | 2.38 × 102 ± 3.93 × 101 | nd | nd |
| M. leachii PG50 | 2.29 | 1.96 × 102 ± 8.94 × 101 | 0.66 | 2.11 × 102 ± 1.61 × 102 |
| M. mycoides subsp. capri GM12 | 2.02 | 7.44 × 101 ± 9.66 | 0.71 | 2.41 × 101 ± 1.69 × 101 |
| M. mycoides subsp. mycoides PG1 | 2.30 | 3.43 ± 2.86 | nd | 0 |
| M. putrefaciens strain 156 | 1.99 | 1.88 × 101 ± 1.27 | 0.57 | 2.67 × 101 ± 5.66 |
| M. florum L1 | 2.09 | 3.02 ± 5.23 | 0.52 | 2.14 × 101 ± 6.84 |
| S. citri GII3 | nd | 0 | nd | 0 |
SEM (standard error of the mean).
Transplantation assays using bacterial genomes cloned in yeast
We next tested the ability of these seven donor Mollicutes genomes to be transplanted when they are cloned and isolated from yeast. Mcap, Mmc and Mflorum genomes were already cloned in yeast at the beginning of this work (9); V. Baby, F. Labroussaa, J. Brodeur, D. Matteau, G. Gourgues, C. Lartigue and S. Rodrigue, in preparation). For other species (Mmm, Mlea, Mputr, Scitri), intact donor genomes were released from agarose plugs and transformed into yeast spheroplasts as previously described (8,9) (Figure 3). After transformation, the integrity of all four individual genomes was assessed by multiplex PCR and pulsed-field gel electrophoresis (PFGE) analysis (data not shown).
All seven Mollicutes genomes cloned in yeast were isolated in agarose plugs then transplanted into McapΔRE recipient cells (Figure 3). The Mcap genome cloned in yeast was previously transplanted at a rate equivalent to ∼103 transplants/μg of gDNA, and was not re-attempted in this study (17). From-yeast transplantation of Mlea, Mmc, Mputr and Mflorum whole genomes were also successful (Figure 5B). The general trend is in good agreement with the negative correlation previously observed for GT using chromosomes directly isolated from bacteria, with overall rates decreasing as the phylogenetic distance from Mcap increased (Figure 5A and B). No transplants were obtained for Scitri, but more surprisingly we did not observe any GT events for Mmm.
Confirmation of transplant phenotypes and genotypes
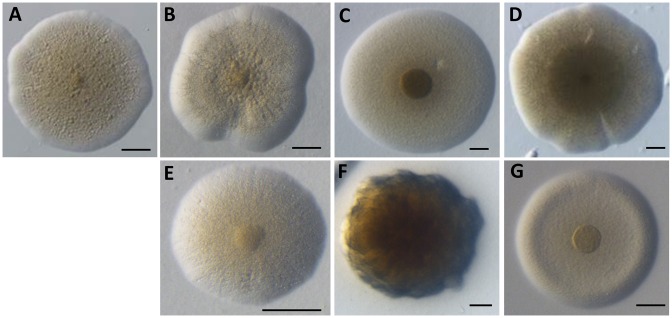
Putative transplants were first observed as colonies on plates to provide a first assessment of the nature of the species obtained (Figure 6). The colony morphology of each transplant was in all aspects similar to the donor species. The colonies of all species tested displayed the typical Mollicutes ‘fried-egg’ phenotype, except for Mputr colonies showing a fuzzier form, brown color and uneven margin. Phenotypic distinctions could also distinguish the different species tested. Colonies from Mcap and Mmc showed wavy margins in comparison to Mlea, Mmm and Mflorum that all had even and defined margins. Colony sizes were also different (Figure 6, 100 μm scale) with Mmc and Mlea showing very large colonies in comparison to Mmm. All colonies were convex except for Mflorum transplants that showed a rounded elevation near the margin associated with a flat center.
Figure 6.
Observed morphologies of transplant colonies obtained during GT experiments. (A) McapΔRE, (B) Mcap, (C) Mlea, (D) Mmc, (E) Mmm, (F) Mputr, (G) Mflorum. Scale: 100 μm.
Transplants were further analyzed using (i) species-specific PCR, (ii) PFGEs and (iii) whole genome sequencing to ensure that they had the expected genotype. For each bacterial species, ten transplants were initially investigated by PCR using species-specific primers (Supplementary Table S6). All tested transplants harbored the amplicons corresponding to the expected donor genome and no sign of the Mcap recipient genome was detected (Supplementary Figure S3). Two of these ten transplants per species were subjected to PFGE analysis to validate the proper global organization of the transplanted chromosome. Again, profiles, in accordance with each respective donor genotype, were obtained (Supplementary Figure S4 and Table S7). Finally, the genome of the original donor bacteria was sequenced along with the same two transplants subjected to PFGE analysis. With the exception of a few single nucleotide polymorphisms and indels, the transplants’ genomes fully matched the expected sequence and sequences from the recipient chromosome were not detected at significant rates in unmapped reads (Supplementary Table S3). Taken together, these data confirm that the recipient Mcap strain was effectively changed into five new species, adopting the phenotype and genotype conferred by the donor genome.
DISCUSSION
Many recent genome engineering techniques such as TREC, TREC-IN and CRISPR-Cas9 use S. cerevisiae as a ‘living workbench’ to allow on-demand editing of almost any genes or group of genes from bacterial genomes. However, the back-transplantation of such engineered genomes into a suitable recipient cell is an essential process to access and study this modified genetic information. Understanding the mechanisms governing GT is thus of major importance for the study of intractable microorganisms presenting interests in biotechnology, but also in biomedical applications with the development of innovative vaccines (48,49,50).
The success of a GT experiment depends on several factors, many remaining poorly understood or completely unknown. In order to learn more about the mechanisms underlying the GT process, we broached the question of the degree of relatedness necessary between donor and recipient cells for successful transplantation and pursued genetic factors that possibly govern this compatibility. We selected McapΔRE as the recipient cell for several reasons: (i) this species, Mcap, was used as recipient cell in the initial work showing the transplantation of whole bacterial genomes (8,20), (ii) it supports the replication of plasmids carrying the chromosomal origin of replication of different donor genomes (Figure 2) (38) and (iii) the mutant McapΔRE does not express any restriction enzymes, which removes a known barrier in GT experiments (8).
The genomes that were evaluated for GT all belong to the same phylogenetic group within the Mollicutes and represent three different genera (Figure 1). The seven selected genomes were first modified by simultaneous integration of bacterial (tetM) and yeast (ARSH4, CEN6 and HIS) genetic markers into their genomes. Following this marker integration, the genomes were successfully cloned as independent replicons in yeast. This study added four genomes (Mlea, Mmm, Mputr and Scitri) to the list of cloned genomes in yeast, confirming that this eukaryote host is extremely versatile for cloning bacterial genomes. The Scitri genome, with a size of 1.84 Mbp, is with the Haemophilus influenzae genome (51) the largest bacterial genome cloned into yeast. Although difficulties have been reported with genomes expressing toxic products for the yeast cell, such as nucleases (11), the genome cloning reported here was straightforward, which was probably facilitated by the alternative genetic code of this group of organisms (Figure 1). With the availability of these cloned genomes in yeast, we compared the efficiency of GT between donor genomes directly extracted from bacteria (from-bacteria GT) and donor genomes extracted from yeast (from-yeast GT) (Figure 3).
Using from-bacteria GT, we first evaluated the optimal concentration of donor gDNA necessary to obtain the highest transplantation rate. The development of a qPCR assay allowed us to determine precisely the amount of genomic DNA added to the recipient cells during the GT experiment. Using the same number of McapΔRE recipient cells (∼5 × 109), we found that optimal yields of transplantation were obtained using about 2 μg of donor gDNA; the only exception being Scitri, for which no transplant could be obtained whatever the gDNA quantity used (Figure 4 and Table 1). Beyond this optimal 2 μg-amount of donor gDNA, the number of transplants dropped dramatically. This result is reminiscent of standard bacterial transformation as it is known that above a given concentration of DNA resulting in the highest number of transformants, there is a drop in the transformation efficiency. This decrease has been attributed potentially to impurities in the reaction or to the saturation of DNA transporters at cell surface in other cases (52–54). Whole GT is an artificial process, limited to the laboratory, that occur concomitantly to PEG-mediated cell fusion events (8,51,55). Besides the possible requirement of cell syncytia for the reaction, it is yet unknown if specific DNA channels and/or transmembrane proton motive forces must be mobilized for DNA translocation.
Using an optimal quantity of gDNA, transplantation efficiencies were compared in-between donor species. For all genomes that were successfully transplanted into McapΔRE, a negative correlation between the transplantation efficiency and the phylogenetic distance was observed. Highest transplantation efficiencies were obtained with genomes closest to McapΔRE and proportionally diminished with the increase in phylogenetic distance (Mcap > Mlea > Mmc > Mputr > Mflorum). Very similar results were obtained for from-yeast GT with the Mmc, Mlea, Mputr and Mflorum donor genomes although a higher number of recipient cells was required for the reaction (1.5 × 1010 cells/transplantation). However, for Mmm and Scitri, no colonies were obtained in the from-yeast GT experiments despite numerous attempts. Regardless, the success obtained with the other four genomes implies that it is now possible to perform genome engineering for these species of interest and that GT can be obtained between bacteria from different genera. In this system, we established that the transplantation limit was located between Mflorum and spiroplasmas, since the Scitri genome was the only genome that could not be transplanted into McapΔRE. Although phylogenetic distance is probably the main factor in the establishment of this barrier, it is also possible that other factors such as genome size and growth rates of the donor cells might contribute to this phenomenon. Scitri, Mcap-most distant phylogenetic species, has the largest genome size (1.84 Mbp) of the tested donor species and has the slowest growth rate under selective pressure (doubling time > 4 h), it is currently difficult to draw any clear conclusions on the relative importance of these factors. However, we attempted to transplant the genome of another Spiroplasma species, Spiroplasma floricola (56), that has a genome size close to Mmc (∼1.3 versus ∼1.1 Mbp respectively) and is considered as a fast grower among Mollicutes (doubling time < 1 h). Despite these properties, no transplants were obtained using S. floricola as the donor genome (data not shown). This result reinforces the idea that the main constraint in compatibility between the donor genome and the recipient cell is indeed the phylogenetic distance. The current frontier stands between Mflorum (92.47% identity on the core proteome) and spiroplasmas species (<86% identity). The identification of Mflorum as the McapΔRE most-distant compatible GT organism remains extremely interesting and should greatly facilitate our search for genetic factors responsible for genome compatibility between the recipient cell and potential donor genomes. Investigating proteins involved in cellular processes supposed to play a key role during genome boot-up such as replication, transcription and translation but differently conserved between compatible and non-compatible genomes, should allow us to identify potential barriers for GT technology. To our knowledge, this is also the first time that a GT was successfully achieved between bacteria belonging to different genera.
Among the factors that probably play a role in the success of GT experiments, the compatibility between the origin of replication (oriC) from the donor genome and the cellular machinery in the recipient cell was tested using plasmids carrying the chromosomal oriC of genome donor species. Although McapΔRE was transformed by all oriC plasmids tested, differences in transformation efficiencies were observed and were not directly related to the phylogenetic distance between donor species and the recipient cell. In particular, no direct correlation was observed between transformation rates obtained with pMCO3 (Mcap), pMYSO1 (Mmm) and pSD4 (Scitri) and transplantation rates of their corresponding whole genomes. These results suggest that, even though the initiation of replication of the transplanted genome might be triggered by the recipient cell machinery, others cellular functions are likely to be required for a successful GT. We hypothesize that the compatibility between the transcription and translation apparatus of the recipient cell and incoming genome is essential. This is based on the requirements for the host machinery to be able to read the genetic information carried by the input genome, to direct protein synthesis and possibly replicate the donor genome, all likely critical processes in the initial boot-up of the transplanted genome. Once key genes from the donor genome are properly expressed by the machinery from the recipient cell, the expression of other genes should normally follow using the newly synthesized transcription-translation system.
All previously described factors, related to the phylogenetic distance between the donor cell and the recipient cell, cannot explain the low yield of transplantation obtained when Mmm was used as the donor genome (Figure 5 and Table 1). Indeed, Mmm is much more related to Mcap (∼99% core proteome similarity) than Mput and Mflorum but GT transplantation rates remained very low or null in comparison to the other species. These data suggest that some species-specific genetic factors, present or absent on Mmm genome could prevent GT. Genome comparison between Mmm and all other transplanted genomes highlighted the presence of few genes of unknown functions (hypothetical membrane proteins) but also some involved in capsular biosynthesis pathway. Indeed, Mmm has the capacity to produce capsular galactan (57,58) but also exopolysaccharides as secreted or attached molecules (59) that may contaminate the purified gDNA and present some toxicity for the recipient cells. However, a large majority of sequences were associated with insertion sequences (IS). Overall, ∼13% of the Mmm genome is composed of IS elements belonging to different families (26). We hypothesize that the transposition of these IS may be triggered in the recipient cell (Mcap) that lacks these elements and cellular mechanisms involved in their regulation (60). The stress caused by the GT process might also be responsible for IS movement since stress is known to induce transposition events (61). Such transpositions may result in lethal events for the recipient cell and therefore explain the drop in the transplantation efficiency observed for Mmm in our from-bacteria GT or our incapacity to transplant its genome in from-yeast GT. Inversely, no sequences common to all transplanted genomes but absent on Mmm were identified in silico.
This work provides a useful platform for the study of several mycoplasmas, ideal organisms toward the creation of minimal cells and essential biological units for our comprehension of life. As more and more bacterial genomes are now cloned in yeast and can be engineered using powerful genetic tools, one of the main bottlenecks of synthetic biology strategies remains our capacity to extent GT technics to a growing number of species. Our work shows that the transplantation of a bacterial genome into a recipient cell, that was until now limited to two very closely related ruminant mycoplasmas, can be extended to more distant species including species from a different genus. This achievement paves the way for future studies devoted to better understand and control the cellular factors that define the compatibility between a donor genome and a recipient cell. In addition, several genomes for which in-yeast cloning and back transplantation is now possible originate from mycoplasmas species pathogenic for ruminants, and our work could, thus, lead to significant breakthroughs in the biology of these pathogenic micro-organisms as well as in the development of new and innovative treatments.
ACCESSION NUMBER
GenBank KX011460.
Supplementary Material
Acknowledgments
We thank Enora Bodin for technical assistance on genotype confirmation of bacterial transplants. We thank Prof Alain Blanchard for helpful discussions and for reviewing of the manuscript. We also thank Dr Florence Tardy (Anses, Laboratoire de Lyon, UMR Mycoplasmoses des Ruminants, Lyon, France) for providing the wtMputr strain 156. Part of the experiments was performed at the Genomic and Transcriptomic Facility of Bordeaux. This work was also made possible with the support of M. Arieis and Prof E. Gnant.
Authors contributions: F.L., A.L., P.S.P. and C.L. designed the research. F.L., A.L., V.B., G.G. and C.L. performed the experiments. D.M., V.B and S.R. performed Mollicutes genomes sequencing and subsequent analyses. F.L., S.V., P.S.P., S.R. and C.L. wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
French National Funding Research Agency [ANR-13-JSV5-0004-01 ‘SYNBIOMOLL’]; National Science Foundation [IOS-1110151]; Conseil Régional d'Aquitaine [20030304002FA, 20040305003FA]; European Union, FEDER [2003227]; Investissements d'Avenir, Convention attributive d'aide [ANR-10-EQPX-16-01]. Funding for open access charge: French National Funding Research Agency [ANR-13-JSV5-0004-01 ‘SYNBIOMOLL’]; National Science Foundation [IOS-1110151].
Conflict of interest statement. None declared.
REFERENCES
- 1.Mushegian A.R., Koonin E. V. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. U.S.A. 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gil R., Sabater-Muñoz B., Latorre A., Silva F.J., Moya A. Extreme genome reduction in Buchnera spp.: toward the minimal genome needed for symbiotic life. Proc. Natl. Acad. Sci. U.S.A. 2002;99:4454–4458. doi: 10.1073/pnas.062067299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hutchison C.A., Peterson S.N., Gill S.R., Cline R.T., White O., Fraser C.M., Smith H.O., Venter J.C. Global transposon mutagenesis and a minimal Mycoplasma genome. Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- 4.May M., Balish M.F., Blanchard A. Mycoplasmatales. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes. Berlin Heidelberg: Springer-Verlag; 2014. pp. 515–550. [Google Scholar]
- 5.Glass J.I., Assad-Garcia N., Alperovich N., Yooseph S., Lewis M.R., Maruf M., Hutchison C.A., Smith H.O., Venter J.C. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. U.S.A. 2006;103:425–430. doi: 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grosjean H., Breton M., Sirand-Pugnet P., Tardy F., Thiaucourt F., Citti C., Barré A., Yoshizawa S., Fourmy D., de Crécy-Lagard V., et al. Predicting the minimal translation apparatus: lessons from the reductive evolution of mollicutes. PLoS Genet. 2014;10:e1004363. doi: 10.1371/journal.pgen.1004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hutchison C.A., Chuang R.-Y., Noskov V.N., Assad-Garcia N., Deerinck T.J., Ellisman M.H., Gill J., Kannan K., Karas B.J., Ma L., et al. Design and synthesis of a minimal bacterial genome. Science. 2016;351:aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- 8.Lartigue C., Vashee S., Algire M.A., Chuang R.-Y., Benders G.A., Ma L., Noskov V.N., Denisova E.A., Gibson D.G., Assad-Garcia N., et al. Creating bacterial strains from genomes that have been cloned and engineered in yeast. Science. 2009;325:1693–1696. doi: 10.1126/science.1173759. [DOI] [PubMed] [Google Scholar]
- 9.Benders G.A., Noskov V.N., Denisova E.A., Lartigue C., Gibson D.G., Assad-Garcia N., Chuang R.-Y., Carrera W., Moodie M., Algire M.A., et al. Cloning whole bacterial genomes in yeast. Nucleic Acids Res. 2010;38:2558–2569. doi: 10.1093/nar/gkq119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Citti C., Maréchal-Drouard L., Saillard C., Weil J.H., Bové J.M. Spiroplasma citri UGG and UGA tryptophan codons: sequence of the two tryptophanyl-tRNAs and organization of the corresponding genes. J. Bacteriol. 1992;174:6471–6478. doi: 10.1128/jb.174.20.6471-6478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karas B.J., Tagwerker C., Yonemoto I.T., Hutchison C.A., Smith H.O. Cloning the Acholeplasma laidlawii PG-8A genome in Saccharomyces cerevisiae as a yeast centromeric plasmid. ACS Synth. Biol. 2012;1:22–28. doi: 10.1021/sb200013j. [DOI] [PubMed] [Google Scholar]
- 12.Tagwerker C., Dupont C.L., Karas B.J., Ma L., Chuang R.-Y.Y., Benders G.A., Ramon A., Novotny M., Montague M.G., Venepally P., et al. Sequence analysis of a complete 1.66 Mb Prochlorococcus marinus MED4 genome cloned in yeast. Nucleic Acids Res. 2012;40:10375–10383. doi: 10.1093/nar/gks823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noskov V.N., Karas B.J., Young L., Chuang R.-Y., Gibson D.G., Lin Y.-C., Stam J., Yonemoto I.T., Suzuki Y., Andrews-Pfannkoch C., et al. Assembly of large, high G+C bacterial DNA fragments in yeast. ACS Synth. Biol. 2012;1:267–273. doi: 10.1021/sb3000194. [DOI] [PubMed] [Google Scholar]
- 14.Noskov V.N., Segall-Shapiro T.H., Chuang R.-Y. Tandem repeat coupled with endonuclease cleavage (TREC): a seamless modification tool for genome engineering in yeast. Nucleic Acids Res. 2010;38:2570–2576. doi: 10.1093/nar/gkq099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandran S., Noskov V.N., Segall-Shapiro T.H., Ma L., Whiteis C., Lartigue C., Jores J., Vashee S., Chuang R.-Y. TREC-IN: gene knock-in genetic tool for genomes cloned in yeast. BMC Genomics. 2014;15:1180–1190. doi: 10.1186/1471-2164-15-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schieck E., Lartigue C., Frey J., Vozza N., Hegermann J., Miller R.A., Valguarnera E., Muriuki C., Meens J., Nene V., et al. Galactofuranose in Mycoplasma mycoides is important for membrane integrity and conceals adhesins but does not contribute to serum resistance. Mol. Microbiol. 2016;99:55–70. doi: 10.1111/mmi.13213. [DOI] [PubMed] [Google Scholar]
- 17.Lartigue C., Lebaudy A., Blanchard A., El Yacoubi B., Rose S., Grosjean H., Douthwaite S. The flavoprotein Mcap0476 (RlmFO) catalyzes m5U1939 modification in Mycoplasma capricolum 23S rRNA. Nucleic Acids Res. 2014;42:8073–8082. doi: 10.1093/nar/gku518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiCarlo J.E., Norville J.E., Mali P., Rios X., Aach J., Church G.M. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsarmpopoulos I., Gourgues G., Blanchard A., Vashee S., Jores J., Lartigue C., Sirand-Pugnet P. In-yeast engineering of a bacterial genome using CRISPR/Cas9. ACS Synth. Biol. 2016;5:104–109. doi: 10.1021/acssynbio.5b00196. [DOI] [PubMed] [Google Scholar]
- 20.Lartigue C., Glass J.I., Alperovich N., Pieper R., Parmar P.P., Hutchison C.A., Smith H.O., Venter J.C. Genome transplantation in bacteria: changing one species to another. Science. 2007;317:632–638. doi: 10.1126/science.1144622. [DOI] [PubMed] [Google Scholar]
- 21.Gibson D.G., Glass J.I., Lartigue C., Noskov V.N., Chuang R.-Y., Algire M.A., Benders G.A., Montague M.G., Ma L., Moodie M.M., et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 22.Larionov V., Kouprina N., Solomon G., Barrett J.C., Resnick M.A. Direct isolation of human BRCA2 gene by transformation-associated recombination in yeast. Proc. Natl. Acad. Sci. U.S.A. 1997;94:7384–7387. doi: 10.1073/pnas.94.14.7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Treco D.A., Winston F. Growth and manipulation of yeast. Curr. Protoc. Mol. Biol. 2008 doi: 10.1002/0471142727.mb1302s19. doi:10.1002/0471142727.mb1302s19. [DOI] [PubMed] [Google Scholar]
- 24.Manso-Silván L., Vilei E.M., Sachse K., Djordjevic S.P., Thiaucourt F., Frey J. Mycoplasma leachii sp. nov. as a new species designation for Mycoplasma sp. bovine group 7 of Leach, and reclassification of Mycoplasma mycoides subsp. mycoides LC as a serovar of Mycoplasma mycoides subsp. capri. Int. J. Syst. Evol. Microbiol. 2009;59:1353–1358. doi: 10.1099/ijs.0.005546-0. [DOI] [PubMed] [Google Scholar]
- 25.Wise K.S., Calcutt M.J., Foecking M.F., Madupu R., DeBoy R.T., Röske K., Hvinden M.L., Martin T.R., Durkin A.S., Glass J.I., et al. Complete genome sequences of Mycoplasma leachii strain PG50T and the pathogenic Mycoplasma mycoides subsp. mycoides small colony biotype strain Gladysdale. J. Bacteriol. 2012;194:4448–4449. doi: 10.1128/JB.00761-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westberg J. The genome sequence of Mycoplasma mycoides subsp. mycoides SC type strain PG1T, the causative agent of Contagious Bovine Pleuropneumonia (CBPP) Genome Res. 2004;14:221–227. doi: 10.1101/gr.1673304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mccoy R.E., Basham H.G., Tully J.G., Rose D.L., Carle P., Bove J.M. Acholeplasma florum, a new species isolated from plants. Int. J. Syst. Bacteriol. 1984;34:11–15. [Google Scholar]
- 28.Vignault J.C., Bové J.M., Saillard C., Vogel R., Faro A., Venegas L., Stemmer W., Aoki S., McCoy R.E., Al-Beldawi A.S., et al. Mise en culture de spiroplasmes à partir de matériel végétal et d'insectes provenant de pays circum méditerranéens et du Proche Orient. C. R. Acad. Sci. III. 1980;290:775–780. [Google Scholar]
- 29.Kouprina N., Larionov V. Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat. Protoc. 2008;3:371–377. doi: 10.1038/nprot.2008.5. [DOI] [PubMed] [Google Scholar]
- 30.Tully J.G., Whitcomb R.F., Clark H.F., Williamson D.L. Pathogenic mycoplasmas: cultivation and vertebrate pathogenicity of a new spiroplasma. Science. 1977;195:892–894. doi: 10.1126/science.841314. [DOI] [PubMed] [Google Scholar]
- 31.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 32.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gouy M., Guindon S., Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 34.Talavera G., Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 35.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 36.Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.-F., Guindon S., Lefort V., Lescot M., et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lartigue C., Blanchard A., Renaudin J., Thiaucourt F., Sirand-Pugnet P. Host specificity of mollicutes oriC plasmids: functional analysis of replication origin. Nucleic Acids Res. 2003;31:6610–6618. doi: 10.1093/nar/gkg848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A., Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 40.King K.W., Dybvig K. Transformation of Mycoplasma capricolum and examination of DNA restriction modification in M. capricolum and Mycoplasma mycoides subsp. mycoides. Plasmid. 1994;31:308–311. doi: 10.1006/plas.1994.1033. [DOI] [PubMed] [Google Scholar]
- 41.Stamburski C., Renaudin J., Bove J.M. First step toward a virus-derived vector for gene cloning and expression in spiroplasmas, organisms which read UGA as a tryptophan codon: synthesis of chloramphenicol acetyltransferase in Spiroplasma citri. J. Bacteriol. 1991;173:2225–2230. doi: 10.1128/jb.173.7.2225-2230.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glover T., Mitchell K. An Introduction to Biostatistics. 2nd edn. Long Grove: Waveland Pr. Inc; 2008. [Google Scholar]
- 47.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 1995;57:289–300. [Google Scholar]
- 48.Ardiani A., Higgins J.P., Hodge J.W. Vaccines based on whole recombinant Saccharomyces cerevisiae cells. FEMS Yeast Res. 2010;10:1060–1069. doi: 10.1111/j.1567-1364.2010.00665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donati C., Rappuoli R. Reverse vaccinology in the 21st century: improvements over the original design. Ann N Y Acad Sci. 2013;1285:115–132. doi: 10.1111/nyas.12046. [DOI] [PubMed] [Google Scholar]
- 50.Kindsmüller K., Wagner R. Synthetic biology: impact on the design of innovative vaccines. Human Vaccines. 2011;7:658–662. doi: 10.4161/hv.7.6.14987. [DOI] [PubMed] [Google Scholar]
- 51.Karas B.J., Jablanovic J., Sun L., Ma L., Goldgof G.M., Stam J., Ramon A., Manary M.J., Winzeler E.A., Venter J.C., et al. Direct transfer of whole genomes from bacteria to yeast. Nat. Methods. 2013;10:410–412. doi: 10.1038/nmeth.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maier B., Chen I., Dubnau D., Sheetz M.P. DNA transport into Bacillus subtilis requires proton motive force to generate large molecular forces. Nat. Struct. Mol. Biol. 2004;11:643–649. doi: 10.1038/nsmb783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwarzenlander C., Averhoff B. Characterization of DNA transport in the thermophilic bacterium Thermus thermophilus HB27. FEBS J. 2006;273:4210–4218. doi: 10.1111/j.1742-4658.2006.05416.x. [DOI] [PubMed] [Google Scholar]
- 54.Burton B., Dubnau D. Membrane-associated DNA transport machines. Cold Spring Harb. Perspect. Biol. 2010;2:a000406. doi: 10.1101/cshperspect.a000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gyuris J., Duda E.G. High-efficiency transformation of Saccharomyces cerevisiae cells by bacterial minicell protoplast fusion. Mol. Cell. Biol. 1986;6:3295–3297. doi: 10.1128/mcb.6.9.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davis R.E., Lee I.-M., Worley J.F. Spiroplasma floricola, a new species isolated from surfaces of flowers of the tulip tree, Liriodendron tulipifera L. Int. J. Syst. Bacteriol. 1981;31:456–464. [Google Scholar]
- 57.Buttery S.H., Plackett P. A specific polysaccharide from Mycoplasma mycoides. J. Gen. Microbiol. 1960;23:357–368. doi: 10.1099/00221287-23-2-357. [DOI] [PubMed] [Google Scholar]
- 58.Plackett P., Buttery S.H. A galactan from Mycoplasma mycoides. Nature. 1958;182:1236–1237. doi: 10.1038/1821236a0. [DOI] [PubMed] [Google Scholar]
- 59.Bertin C., Pau-Roblot C., Courtois J., Manso-Silván L., Thiaucourt F., Tardy F., Le Grand D., Poumarat F., Gaurivaud P. Characterization of free exopolysaccharides secreted by Mycoplasma mycoides subsp. mycoides. PLoS One. 2013;8:e68373. doi: 10.1371/journal.pone.0068373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mahillon J., Chandler M. Insertion sequences. Microbiol. Mol. Biol. Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagy Z., Chandler M. Regulation of transposition in bacteria. Res. Microbiol. 2004;155:387–398. doi: 10.1016/j.resmic.2004.01.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.