Abstract
RNA-binding proteins (RBPs) play crucial roles in all organisms. The protein Gemin5 harbors two functional domains. The N-terminal domain binds to snRNAs targeting them for snRNPs assembly, while the C-terminal domain binds to IRES elements through a non-canonical RNA-binding site. Here we report a comprehensive view of the Gemin5 interactome; most partners copurified with the N-terminal domain via RNA bridges. Notably, Gemin5 sediments with the subcellular ribosome fraction, and His-Gemin5 binds to ribosome particles via its N-terminal domain. The interaction with the ribosome was lost in F381A and Y474A Gemin5 mutants, but not in W14A and Y15A. Moreover, the ribosomal proteins L3 and L4 bind directly with Gemin5, and conversely, Gemin5 mutants impairing the binding to the ribosome are defective in the interaction with L3 and L4. The overall polysome profile was affected by Gemin5 depletion or overexpression, concomitant to an increase or a decrease, respectively, of global protein synthesis. Gemin5, and G5-Nter as well, were detected on the polysome fractions. These results reveal the ribosome-binding capacity of the N-ter moiety, enabling Gemin5 to control global protein synthesis. Our study uncovers a crosstalk between this protein and the ribosome, and provides support for the view that Gemin5 may control translation elongation.
INTRODUCTION
RNA-binding proteins (RBPs) play a pivotal role in the regulation of gene expression due to their capacity to interact with different targets, either RNAs or other proteins (1,2). Additionally, studies on the conformational plasticity of many RBPs (3,4) together with the incessant discovery of novel RNA-binding motifs have increased the number of RBPs and, importantly, have shed light on new functions performed by these proteins within the cell (5,6). Recent studies have shown that certain RBPs can perform a dedicated function on the translation of selective mRNAs (7–9), and others sediment with the actively translating polyribosomes (10–12). Beyond the role of RBPs in controlling protein synthesis, ribosomal proteins can interact with non-ribosomal components to perform extra-ribosomal functions (13,14). Additionally, ribosomal proteins can regulate viral RNA functions. For instance, RACK1 enhances hepatitis C virus (HCV) internal ribosome entry site (IRES)-dependent translation (15), whereas P0 is associated to the Potato virus A membrane ribonucleoprotein complex, synergistically enhancing viral translation with the viral protein VPg and the eukaryotic initiation factor eIF(iso)4E (16). In contrast, L13a acts as an antiviral agent inhibiting translation by forming a complex with a hairpin of the respiratory syncytial virus M viral RNA (17).
Initiation of translation in eukaryotic mRNAs depends on the m7GTP residue (or cap) located at the 5′end of most mRNAs. In this process, translation initiation factors (eIFs) recruit the small ribosome subunit to the 5′end of the mRNA (18). However, a subset of viral mRNAs have evolved cap-independent mechanisms that allow to evade cap-dependent inhibition and to bypass the translation shut down induced in infected cells (19). This mechanism is based on IRES elements (20). Viral IRESs are RNA functional elements able to recruit the ribosomal subunits internally promoting translation initiation at internal start codons independent of the 5′end of the mRNA. IRES-dependent translation is modulated by a subset of eIFs and various RBPs (21–23), with the exception of the dicistrovirus intergenic region (24,25). Riboproteomic approaches conducted with two genetically distant viral IRESs, HCV and foot-and-mouth disease virus (FMDV), identified Gemin5 as a regulator of both cap-dependent and IRES-dependent translation (26), revealing a new role for this protein.
The RBP Gemin5 performs critical functions in evolutionary distant organisms. In humans, the highest expression of Gemin5 occurs in the gonads (27,28), and loss of Gemin5/Rigor mortis protein is lethal at the larva stage in Drosophila (29). Gemin5 is a peripheral protein of the survival of motor neuron (SMN) complex (30) found in metazoan cells. This multi-protein complex plays a critical role on the biogenesis of small nuclear ribonucleoproteins (snRNPs), the components of the splicing machinery. Gemin5 recognizes the Sm site of snRNAs, and delivers these molecules to the SMN complex (31). The Gemin5 residue involved in snRNA interaction was mapped to the 5th WD repeat within the N-terminal region (32). Independent studies of our laboratory showed that a polypeptide encompassing the C-terminal region of Gemin5 was able to interact directly with the IRES element to a similar extent than the full-length protein (33). In contrast, its N-terminal region had no IRES-binding capacity. Hence, separate regions of the protein are involved in the recognition of RNAs with different functions, distinct primary sequence and structural organization. This finding suggests the existence of multiple RNA targets recognized by specialized domains likely assembled in distinct functional complexes.
Since Gemin5 is mainly found in the cell cytoplasm outside of the SMN complex (34), it is plausible that Gemin5 may recruit (or interfere with) other factors that have RNA-binding capacity and thus, regulate translation. Understanding the complexity of Gemin5 function in translation control would greatly benefit from a global approach to identify novel regulators. Here, we have undertaken this challenge to identify the Gemin5-associated proteins in vivo. Mass spectrometry identification of TAP-tagged Gemin5 complexes showed that the protein associates preferentially through its N-terminal domain with many factors, among them members of the SMN complex, RBPs, eIFs and ribosomal proteins. Most associated factors were lost upon exhaustive ribonuclease (RNase) treatment of the TAP-purification complexes, uncovering the existence of a Gemin5 network guided by RNA bridges. Among the RNase-resistant factors, we identified several ribosomal proteins, besides a few SMN complex components. Here, we show that the protein Gemin5 sediments with the ribosome fractions, and that the purified protein interacts directly with purified 60S ribosomal subunits and 80S ribosomes via its N-terminal domain. Specific mutations within the N-terminal domain of the Gemin5 protein impair ribosome interaction. These mutations also abolish the direct interaction of the ribosomal proteins L3 and L4 with purified Gemin5 in pull-down assays using glutathione S-transferase (GST) fusion proteins. These data strongly suggest that Gemin5 may control global protein synthesis through its direct binding with the ribosome.
MATERIALS AND METHODS
Constructs
The plasmid pcDNA3-Xpress-G5, expressing the full-length Gemin5 protein was generated in two steps. The N-terminal fragment was excised from pRSETB-13WD (33) and inserted into pcDNA3-Xpress via BamHI-EcoRI. Next, the C-terminal fragment was fused in frame using a polymerase chain reaction (PCR) fragment via EcoR1-XbaI. The sequences encoding the N-terminal and C-terminal domains of Gemin5 present in pRSETBG5 were transferred to pcDNA3-NTAP or pcDNA3-CTAP (35) using standard procedures. Oligonucleotides (Sigma) used for PCR and the restriction enzyme sites used for cloning are described in Supplementary Table S1. Plasmids were transformed into E. coli DH5a. Mutagenesis of the plasmid pcDNA3-Xpress-G5 to obtain the F381A mutant (pcDNA3-Xpress-G5-F381A construct) was performed using the QuikChange mutagenesis procedure (Agilent Technologies) with the pair of primers G5_F381As and G5_F381Aas described in Supplementary Table S1. All plasmids were confirmed by DNA sequencing (Macrogen).
pGEXKG plasmids expressing GST fusions of HnRNP U (36), IGF2BP2 (37), SYNCRIP (ABM, ORF010243), S3A (ABM, PV036253), RACK1 (38), L3 (ABM, ORF008983), L4 (ABM, ORF009002), L5 (ABM, ORF009003) and P0 (39) were generated using standard procedures. See Supplementary Table S1 for oligonucleotides sequences used for PCR and the restriction enzyme sites used for cloning.
Protein complexes purification by tandem affinity purification (TAP)
HEK293 cells (4 × P100 plates), grown in Dulbecco's Modified Eagle Medium (DMEM), transfected with the TAP-Gemin5 plasmids, were harvested 24 h post-transfection. The protein complexes associated to TAP-Gemin5 were purified as described (40,41). An optional treatment with RNase A (75 μg/1.5 ml) 30 min at room temperature immediately following TEV protease (Life Technologies) digestion (41) eliminates the factors bound by RNA bridges; this concentration of RNase degrades all RNA probe in UV-crosslink assays. The supernatant of the first purification, treated or untreated with RNase A, was subsequently subjected to a second Calmodulin (Agilent Technologies) purification step. Purified proteins were precipitated with 10% trichloroacetic acid at 4°C overnight, pelleted at 14 000 g 15 min at 4°C, washed three times with 1 ml of acetone and, finally, dissolved in SDS-loading buffer. A small aliquot was analyzed on silver stained SDS-PAGE gels to visualize the purification of proteins associated to TAP-Gemin5.
Mass spectrometry identification
The samples obtained by TAP were applied onto a SDS-PAGE gel, stopping the run to concentrate the proteome in the stacking/resolving gel interface. Following digestion of the proteins with sequencing grade trypsin (Promega) (42), the protein digest was analyzed by reverse phase-liquid chromatography (RP-LC)-MS/MS in an Easy-nLC II system coupled to an ion trap LTQ-Orbitrap-Velos-Pro mass spectrometer (Thermo Scientific). The peptides were concentrated and then eluted using a 90-min gradient from 5 to 40% of 0.1% formic acid, 80% acetonitrile in water. Electrospray ionization (ESI) was done using a Nano-bore emitters Stainless Steel ID 30 μm (Proxeon) interface. The Orbitrap resolution was set at 30 000. The mass spectrometer was operated in the selected MS/MS ion-monitoring mode. The LTQ-Orbitrap-Velos-Pro detector was programmed to perform a continuous sequential operation in the MS/MS mode on the doubly or triply charged ions corresponding to the peptide/s selected previously from the theoretical prediction. The MS/MS peptide spectra was analyzed by assigning the fragments to the candidate sequence after calculating the series of theoretical fragmentations, according to the nomenclature of the series (43).
Two independent biological replicates were analyzed for G5-Nter, G5-Cter and the control TAP polypeptides; only factors identified in both replicates were considered for further studies (Supplementary Figure S1). The factors associated to the control TAP polypeptide were subtracted from the overlap identified with more than 2 peptides (FDR < 0.01), in G5-Nter or G5-Cter samples. After background subtraction, only proteins differing in four or more peptides were considered preferentially associated to either the N-ter for C-ter region of Gemin5. In a few cases where no peptides were identified in one protein, a difference of 2 peptides was considered as the cut-off.
Subcellular fractionation
HEK293 cells, grown to 90% confluence in 10–12 P100 dishes, were washed with ice cold phosphate buffered saline (PBS), and lysed in buffer 1 (15 mM Tris-HCl pH 7.4, 80 mM KCl, 5 mM MgCl2, 1% Triton-X-100, Protease inhibitors (Complete mini, Roche)). Cell debris was discarded by spinning at 14 000 g 10 min 4°C, twice. The supernatant of the second spinning is the S30 fraction. S30 centrifugation at 95 000 rpm during 1.5 h using the TLA100.3 rotor yielded the S100 fraction (supernatant), and the ribosomes plus associated factors (pellet - R fraction). To prepare the fraction containing ribosomes free of associated factors, the ribosomal pellet was resuspended in 200 μl of high salt buffer 2 (15 mM Tris-HCl pH 7.4, 500 mM KCl, 5 mM MgCl2, 2 mM DTT, 290 mM sucrose), loaded in a discontinuous sucrose gradient (1.5 ml buffer 40% (w/v) sucrose, 15 mM Tris-HCl pH 7.4, 500 mM KCl, 5 mM MgCl2, 2 mM DTT (bottom layer) and 1 ml buffer 20% (w/v) sucrose, 15 mM Tris-HCl pH 7.4, 500 mM KCl, 5 mM MgCl2, 2 mM DTT (top layer)), centrifuged at 4°C 95 000 rpm 2 h using a TLA100.3 rotor. The pure ribosomes pellet (RSW fraction) was resuspended in 100 μl of buffer 1. The total protein content in S30 and S100 fractions was measured by the Bradford assay; the ribosome concentration in R and RSW fractions was determined as 14 units A260 = 1 mg/ml.
Polysome profiles and analysis
Ribosome profiles were prepared from HEK293 cells (4 P100 dishes per gradient) as described (44). Briefly, cells were washed with ice-cold PBS containing 100 μg/ml cycloheximide (Sigma) to block ribosomes in the elongation step. Then, cells were lysed with buffer A (15 mM Tris-HCl pH 7.4, 80 mM KCl, 5 mM MgCl2, 100 μg/ml cycloheximide), supplemented with 1% (v/v) Triton X-100, 40 U/ml RNase OUT (Life Technologies), Protease inhibitors). Cytoplasmic extracts obtained by centrifugation at 14 000 g 10 min at 4°C, were loaded into a linear 10–50% (w/v) sucrose gradient in buffer A, centrifuged at 39 000 rpm in a SW40 Ti rotor 2 h 15 min at 4°C. Gradients were fractionated by upward displacement with 87% (v/v) glycerol using an ISCO density-gradient fractionator, monitoring A260 continuously (ISCO UA-5 UV monitor). Fractions (1 ml) were collected from gradients; the proteins of interest were analyzed in all the gradient fractions (15 μl) by Western blot (WB).
In ribosome-dissociation experiments, cycloheximide was omitted from all buffers, and the KCl concentration in the cytoplasmic extracts was increased to 800 mM (high salt). Ribosomal subunits were resolved by centrifugation at 39 000 rpm in a SW40 Ti rotor 3 h 30 min at 4°C on 10–30% (w/v) sucrose gradient containing buffer B (15 mM Tris-HCl pH 7.4, 800 mM KCl, 5 mM MgCl2).
Purification of 40S, 60S ribosomal subunits and 80S ribosome
Fractions corresponding to 80S peak from polysome profiles, and the fractions corresponding to the 40S and 60S peaks from a high salt dissociation gradient, were collected and pooled at 48 000 rpm using a T865 rotor 2 h at 4°C. 80S, 60S and 40S pellets were resuspended in 10 mM HEPES pH 7, 10 mM MgCl2, 50 mM KCl, 5 mM β-ME and stored at −70°C. Ribosome concentration was calculated as 1 A260 unit = 20 pmol/ml 80S ribosome (14).
Ribosome binding assay
His-tagged Gemin5 (His-G5), His-G51–739 wild type and the mutants W14A, Y15A, F381A and Y474A (a kind gift of Dr Chao Xu and Dr Jinrong Min) or His-G51287-1508 proteins (4 pmols) were coupled to Ni-agarose resin (25 μl of beads suspension) (Qiagen) during 1 h at 4°C in binding buffer (RBB) (50 mM TrisOAc pH 7.7, 50 mM KOAc, 5 mM Mg (OAc)2, 10 mM DTT, 30 μg/ml tRNA). Unbound protein was removed by 3 washings with RBB, spinning at 14 000 g 3 min at 4°C. Beads-protein complexes, resuspended in 100 μl of RBB, were incubated with 80S ribosomes, 40S or 60S ribosomal subunits (0.7 pmol) 1 h at 4°C. After three washes of the beads-complexes with RBB supplemented with NP40 0.05%, spinning at 14 000 g 3 min at 4°C, beads-bound proteins were resuspended in SDS-loading buffer, heated at 92°C 3 min, resolved by SDS-PAGE and detected by WB using anti-His (Gemin5), anti-RACK1 (40S) or 3BH5 (anti-P0, 60S and 80S) antibodies. Independent binding assays were conducted at least three times.
Expression and purification of proteins
Escherichia coli BL21 transformed with plasmids pET-G51287–1508 and pRSETBG5 grown at 37°C were induced with Isopropyl-D-1-thiogalactopyranoside (IPTG) 0.5 mM during 2 h. Bacterial cell lysates were prepared in binding buffer (20 mM NaH2PO4, 500 mM NaCl, 20 mM Imidazole) using a French press and cell debris was eliminated by centrifugation at 16 000 g 30 min at 4°C, twice. The lysate was loaded in His-GraviTrap columns (GE HealthCare) and the recombinant protein was eluted using Imidazole 500 mM (45). Proteins were dialyzed against phosphate buffer pH 6.8, 1 mM DTT and stored at −20°C in 50% glycerol.
siRNA interference
siRNAs targeting Gemin5 (CCUUAAUCAAGAAGAGAAAUU) or a control sequence (AUGUAUUGGCCUGUAUUAGUU) were purchased from Dharmacon. HEK293 cells grown to 70% confluent were treated with 100 nM siRNA using lipofectamine 2000 (Life Technologies) according to the manufacturer instructions. After 48 h, cell lysates were prepared in buffer A for polysome profile analysis.
Gemin5 expression in mammalian cells
HEK293 cells 70% confluent were transfected with the plasmid pcDNA3-Xpress-G5, pcDNA3-Xpress-G5-F381A, TAP-G5-Nter, TAP-G5-Cter or the empty vector pcDNA3.1 using lipofectamine (Life Technologies) according to the manufacturer instructions. Cell lysates were prepared 24 h post-transfection in buffer A for polysome profile analysis, or in buffer B for ribosome-dissociation experiments. Similarly, cells transfected with TAP-G5-Nter, TAP-G5-Cter and pcDNA3-CTAP, were lysed 24 h post-transfection to perform TAP procedure.
HEK293 cells transfected with pcDNA3-Xpress-G5, pcDNA3-Xpress-G5-F381A, TAP-G5-Nter, TAP-G5-Cter and siRNAG5-depleted cells, as well, were radiolabeled with [35S]-methionine at the end of the transfection during a 3 h pulse with 10 μCi / well (about 106 cells). In all cases, cells were kept for 1 h in methionine-free medium before labeling. Protein lysates were separated on SDS-polyacrylamide gels. Dried gels were used to determine incorporation of [35S]-methionine in newly synthesized proteins relative to control cells, set to 100%. The statistical significance of the efficiency of protein synthesis was determined by the unpaired two-tail Student's t-test.
Immunodetection
Gemin5 protein or its tagged-domains were immunodetected using anti-Gemin5 (Novus) antibody. P0 was detected with the monoclonal antibody 3BH5 (46). Commercial antibodies were used to detect eIF3b, eIF4B, DHX9 (Bethyl laboratories), His-tag, tubulin (Sigma), PTB (Acris), hnRNPA1, hnRNPU (Immuquest), CBP (Abcam), GST, RACK1, RPL3, RPL4 (Santa Cruz), eIF4E (Transduction laboratories). Appropriate secondary antibodies (Thermo Scientific) were used according to the manufacturer instructions. Protein signals were visualized with ECL plus (Millipore). Quantification of the signal detected was done in the linear range of the antibodies.
GST-pull down assay
The proteins of interest were prepared as GST-fusions as described (41). For binding, the GST-fusion protein (4 μg) bound to the glutathione resin (GE healthcare) was incubated with myc/DDK-Gemin5 (250 ng) (ORIGENE, TP317516), or His-G51–739 proteins, in 5 volumes of binding buffer (50 mM HEPES pH 7.4, 100 mM NaCl, 2 mM DTT, 2 mM MgCl2, 0.5% Igepal CA-630, 10% glycerol) 2 h at 4°C in a rotating wheel. Beads were pelleted at 3000 g 2 min at 4°C and washed three times with 5 volumes of binding buffer, rotating the reaction tube 5 min at 4°C. Finally, the beads were boiled in SDS-loading buffer and proteins resolved by SDS-PAGE. WB analysis was performed using anti-Gemin5 for Gemin5, and anti-GST to detect the GST-fusion protein. Independent pull-down assays were conducted at least three times.
RESULTS
Identification of Gemin5 partners by TAP
We have shown that Gemin5 downregulates translation via a novel RNA-binding domain placed at the C-terminal region (33,45). In contrast, the motif involved in snRNA interaction was mapped to 5th WD repeat within the N-terminal region (32). Thus, separate regions of the protein are involved in the recognition of RNAs with different functions. This finding suggests the existence of multiple RNA targets recognized by specialized domains likely assembled in distinct functional ribonucleoprotein (RNP) complexes.
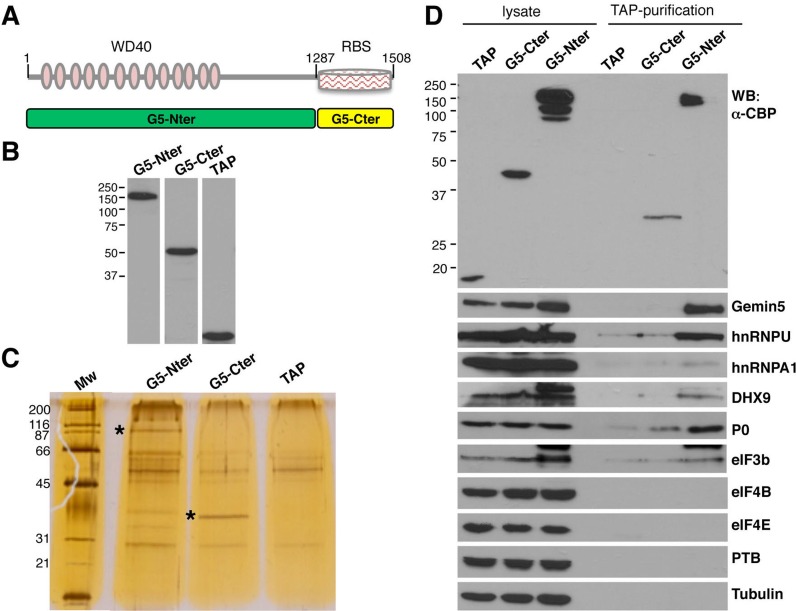
Here, we sought to define the Gemin5-associated protein network in vivo to elucidate the role of this protein in cellular processes. To identify the factors associated to Gemin5 within human cells we attempted their purification by TAP followed by mass spectrometry (LC-MS/MS) analysis. To this end, the TAP tag was fused to the Nter or the Cter open-reading-frame regions (Figure 1A). Plasmids expressing the TAP-tagged proteins G5-Nter and G5-Cter were transfected in HEK293 cells in parallel to the empty vector pcDNA3-CTAP; the expression of G5-Nter and G5-Cter was determined using anti-CBP antibody (Figure 1B). Following purification of the TAP-tagged proteins, soluble extracts containing the factors associated to G5-Nter, G5-Cter and TAP polypeptide were analyzed by SDS-PAGE. The Gemin5 TAP-tagged purified proteins were readily detected following silver staining (depicted by asterisks in Figure 1C). In addition, co-purifying factors, although observed in both G5-Nter and G5-Cter samples, were more abundant with G5-Nter.
Figure 1.
Purification of Gemin5 associated factors in HEK293 cells. (A) Schematic of TAP-tagged Gemin5 polypeptides. Numbers indicate the amino acid residues flanking each polypeptide. Grey-pink ovals depict the WD40 motifs located within the N-terminal domain; a striped rectangle depicts a non-canonical RNA-binding site (RBS) located at the C-terminal domain. (B) Expression of the TAP-tagged G5-Nter, G5-Cter and the control TAP polypeptide in total cell extracts of transfected HEK293 cells monitored by WB using anti-CBP recognizing the TAP polypeptide. (C) Analysis of the TAP complexes by SDS-PAGE and silver staining. The size (kDa) of the MW markers is indicated on the left. An asterisk depicts the mobility of the G5-Ner and G5-Cter. (D) Western blot of total cell extracts (lysate) and TAP complexes (TAP-purification) associated to the TAP, G5-Cter or G5-Nter polypeptides. The small TAP polypeptide resulting from the TEV-digested control TAP sample was lost in this gel. A lower mobility of the G5-Cter TAP-purified sample results from TEV cleavage.
To verify that the TAP procedure yielded samples enriched in Gemin5 partners, we analyzed the total lysate and the purified samples for the presence of hnRNP U, hnRNPA1 and DHX9, previously described to be associated to the SMN complex (30,47,48). These proteins were immunodetected in total lysates prepared from cells transfected with the empty vector, G5-Cter or G5-Nter (Figure 1D, top panel). Relative to the total lysate, the molecular weight of the TAP-purified G5-Cter protein was decreased after TEV protease treatment, as expected (Figure 1D, top panel). Following purification, hnRNP U, DHX9 and Gemin5 were enriched in the G5-Nter purification sample, relative to G5-Cter and control TAP samples (Figure 1D). Notwithstanding, the full-length endogenous Gemin5 protein was immunodetected in the G5-Nter TAP, but not in G5-Cter or control TAP samples, suggesting that this protein oligomerizes through its N-terminal domain. In addition, the ribosomal protein P0 and the initiation factor eIF3b were detected in the G5-Nter sample, while these proteins were faint or undetectable in the G5-Cter sample. Other proteins such as eIF4B, eIF4E, PTB and tubulin, were absent in all TAP samples. These results indicate that G5-Nter and G5-Cter TAP-purified complexes are differentially enriched in associated factors.
RNA bridges connect the network of Gemin5 partners identified by Mass Spectrometry
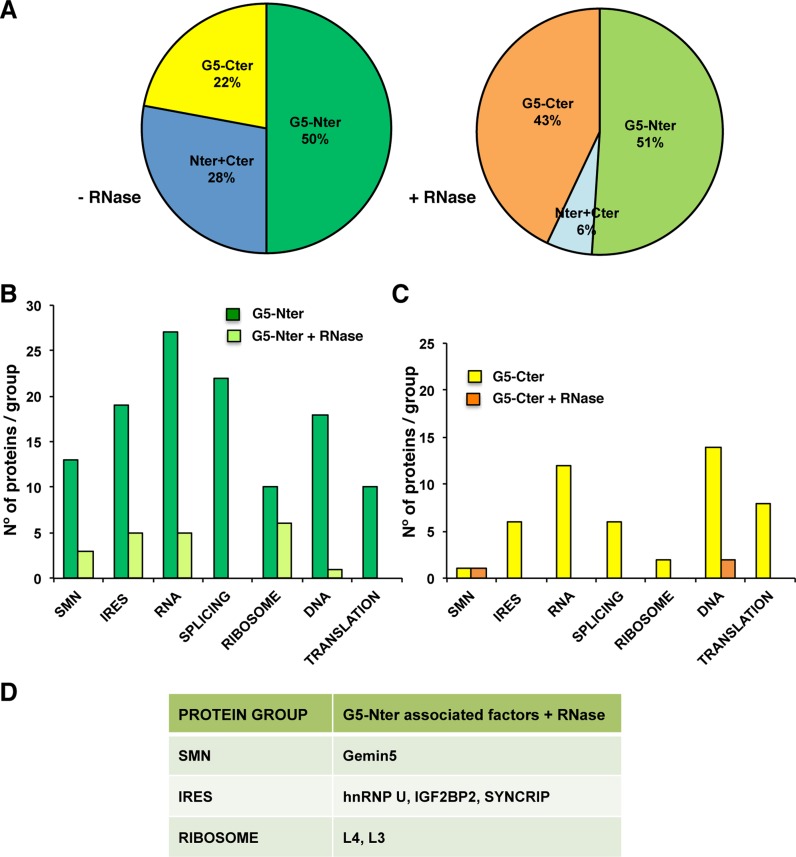
Given that Gemin5 and many of its associated factors were RBPs, we performed new rounds of TAP including an optional exhaustive RNase treatment prior to the second affinity purification step. The TAP purified proteins of two independent biological replicates were digested with trypsin and their tryptic fingerprints were identified by mass spectrometry. Then, the factors associated to the control polypeptide TAP were subtracted from the overlap of two independent biological replicates associated to either G5-Nter or G5-Cter. Of these proteins, 188 were identified with G5-Nter in the absence of RNase treatment while 133 were identified with G5-Cter. After RNase treatment, only 27 proteins were identified associated to G5-Nter and 22 with G5-Cter. Regarding the identity of the proteins, 50% copurified preferentially with the N-terminal domain, 22% with the C-terminal domain and 28% with both regions in the absence of RNase treatment (Figure 2A). The vast majority of associated factors were preferentially associated to the N-terminal domain of the protein, both in the presence (51%) and absence (50%) of RNase treatment. The number of factors associated to G5-Cter was also increased after RNase treatment (43%), while only 6% were associated to both regions. Together, these data indicated that the total number of proteins co-purifying with G5-Nter was generally higher than those associated to G5-Cter.
Figure 2.
Summary of proteins associated with Gemin5 complexes. (A) Pie chart plot depicting the percentage of proteins preferentially associated to Gemin5 domains, G5-Nter (dark or light green), G5-Cter (yellow or orange) and both regions (dark or light blue), in the absence or presence of RNase treatment, respectively. (B) Proteins associated to the N-terminal domain of Gemin5 identified by LC-MS/MS with more than two peptides in two independent biological replicate assays (about 500 proteins were unequivocally detected in each replica, data are available upon request) were grouped according to their gene function. The histogram depicts the number of proteins in each group versus the mean score (average of the score determined in the proteomic analysis for the proteins within each functional group) of the group associated to the G5-Nter in the absence (dark green bars) or the presence of RNase A treatment during the second TAP step (light green bars). (C) Representation of the proteins associated to the C-terminal domain of Gemin5 identified without (yellow bars) or with (orange bars) RNase A treatment during the second TAP step. (D) Proteins associated to G5-Nter resistant to RNase treatment analyzed in further detail in this work.
MS/MS data obtained in the presence or absence of RNase treatment, were classified by protein function. Next, due to the focus of this work, only groups of proteins related with RNA functions were considered for further studies. Representation of the number of proteins identified in each group (Figure 2B and C) readily showed that factors associated to G5-Nter was higher in the groups including the SMN complex (SMN), the RBPs involved in internal initiation of translation (designated IRES), RNA metabolism in general (RNA), mRNA splicing (splicing) and the ribosomal proteins (ribosome). Notably, the total number of proteins associated to the functional domains of Gemin5 was significantly reduced after RNase treatment in most groups (Figure 2B and C). In marked difference, 50% of the ribosomal proteins remained associated to G5-Nter (Figure 2B).
Diversity of RNA-binding proteins among Gemin5-associated factors
Since Gemin5 function is involved in at least two RNA biology processes, snRNPs biogenesis and translation control (26,32), we focused our attention on four groups of proteins, (i) the SMN complex (SMN, mean score 48.6), (ii) factors known to modulate IRES-driven translation initiation (IRES, mean score 21.7), (iii) factors involved in protein synthesis (Translation, mean score 11.6) and (iv) the ribosomal proteins (Ribosome, mean score 12.4). Interestingly, many components of the SMN complex (30,48,49) were exclusively identified associated to the N-terminal domain of Gemin5 (Supplementary Figure S2). Specifically, the factors identified with largest number of peptides and highest score were Gemin5, Gemin4, Gemin3, snRNP70, SNUT1, UNRIP and SMN (ranging between 53 to 7 peptides, score 266 to 27). Other components of the SMN complex, such as Gemin6, and Gemin2, were also identified, confirming that this procedure recruited bona fide Gemin5 partners. Upon RNase treatment, only Gemin5, Gemin4 and SNRP70 were unequivocally identified; the number and identity of Gemin5 peptides identified was similar in the presence or absence of RNase treatment, suggesting that the protein oligomerizes through its N-terminal domain, consistent with results shown in Figure 1D.
The group of RBPs modulating IRES-dependent translation initiation is shown in Supplementary Figure S3. Among the factors identified with G5-Nter, the highest number of peptides and highest score (16 and 50.37, respectively) was observed for hnRNP U, a factor known to be associated with the SMN complex (48). Other proteins reported as IRES-binding factors NCL, IGF2BP2, STAU1, LARP1, LAR4B, YBX1 and SYNCRIP were also preferentially associated to G5-Nter. RNase digestion of the TAP-purified complexes revealed that hnRNP U, IGF2BP2, YBX1, SYNCRIP and FUS remained bound to G5-Nter (Figure 2D, Supplementary Figure S3). Confirming the validity of our approach, the physical and functional interactions among FUS, SMN and Gemins has been recently reported (50). Proteins exclusively identified with G5-Cter, such as RAVER1 and PCBP2, did not resist the RNase treatment.
A similar analysis of the group of factors related to general translation control and protein synthesis revealed that the proteins associated to G5-Nter, identified with higher number of peptides and highest score, were EPRS and eIF4ENIF1, but none of them were resistant to RNase treatment suggesting that RNA bridges held Gemin5-eIFs and Gemin5-eEF2 association (Supplementary Figure S4). Similarly, none of the translation control factors associated to G5-Cter (eIF4ENIF1, eIF3b, etc.) remained bound after RNase treatment.
Finally, the ribosomal proteins identified in the G5-Nter TAP-purified complexes belong to both ribosomal subunits (Supplementary Figure S5), with L4 and L3 having the largest score and number of identified peptides even upon RNase treatment (Figure 2D). The number of peptides identified in the ribosomal proteins was generally lower (number of peptides ranging between 9 and 2, score ranging between 21.7 for L4 to 4.2 for L36A) than the groups mentioned earlier, but the preferential association to the N-terminal domain was similar to the SMN complex.
In spite of the diversity of the Gemin5-partners composition, we conclude that most of the proteins identified in the selected groups were preferentially associated to the G5-Nter domain, as they were not identified with G5-Cter. Furthermore, the differential association of most proteins to Gemin5 upon RNase treatment (Figure 2B and C) suggests that only a small number of factors bind to Gemin5 via direct protein–protein interaction.
Gemin5 associates with the ribosomal particles in cell lysates
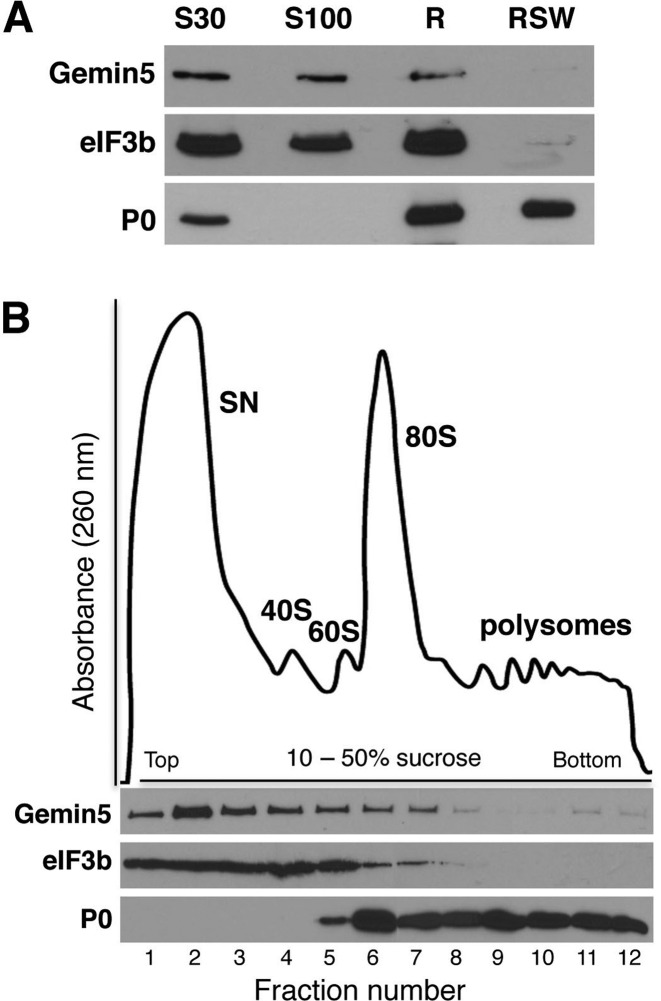
We have shown that Gemin5 has the ability to regulate translation (26,45). Additionally, we show here that the network of Gemin5 partners included a subset of ribosomal proteins, irrespectively of the RNase treatment during the affinity purification (Figure 2B and C; Supplementary Figure S5). Subsequently, to determine its presence in subcellular fractions, Gemin5 was immunodetected in S30 and S100 fractions as well as in the total ribosomes (R), but not in the ribosome salt wash (RSW) (Figure 3A). As expected, a subunit of a diagnostic initiation factor eIF3b was detected in S30, S100 and R fractions, but not in RSW. In contrast, the ribosomal protein P0 was detected in S30, R and RSW fractions, confirming the composition of the subcellular fractions. Together, these results demonstrate that Gemin5 sediments with the total ribosome fraction, although it is not an integral component of the particle.
Figure 3.
Gemin5 associates with the ribosomal particles in cell lysates. (A) HEK293 fractions corresponding to S30 and S100 (100 μg of total protein), ribosomes (R) and pure ribosomes (ribosome salt wash, RSW) (30 μg of total protein) were analyzed by Western blot to detect the presence of Gemin5, the translation initiation factor eIF3b and the ribosomal protein P0. (B) Polysome profile from a total lysate of HEK293 cells separated on a 10–50% sucrose gradient. The fractions corresponding to the supernatant (SN), 40S and 60S ribosomal subunits, the 80S ribosomes and the polysomes are indicated. Gemin5, eIF3b and the ribosomal protein P0 were analyzed in the fractions (15 μl) of the gradient by WB, using specific antibodies.
Next, to determine the type of ribosomal particle (free subunits or ribosomes) interacting with Gemin5 we conducted a ribosome profile analysis (Figure 3B). To this end, HEK293 cytoplasmic lysate was loaded on a linear 10–50% sucrose gradient, and fractions were collected. Then, WB analysis of equal volume (15 μl) of all the fractions revealed that Gemin5 was found in the supernatant, but interestingly, it was also detected all along the gradient including fractions corresponding to the 40S, 60S, 80S and although weakly, the polysomes. Immunodetection of eIF3b, a subunit of the eIF3 factor associated to the translation initiation complex, indicated the sedimentation of free eIFs and preinitiation complexes, while immunodetection of P0 was used to visualize the position of ribosomal 60S subunit, 80S ribosomes and the polyribosomes along the gradient (Figure 3B).
Gemin5 interacts directly with the 60S ribosomal subunit through its N-terminal domain
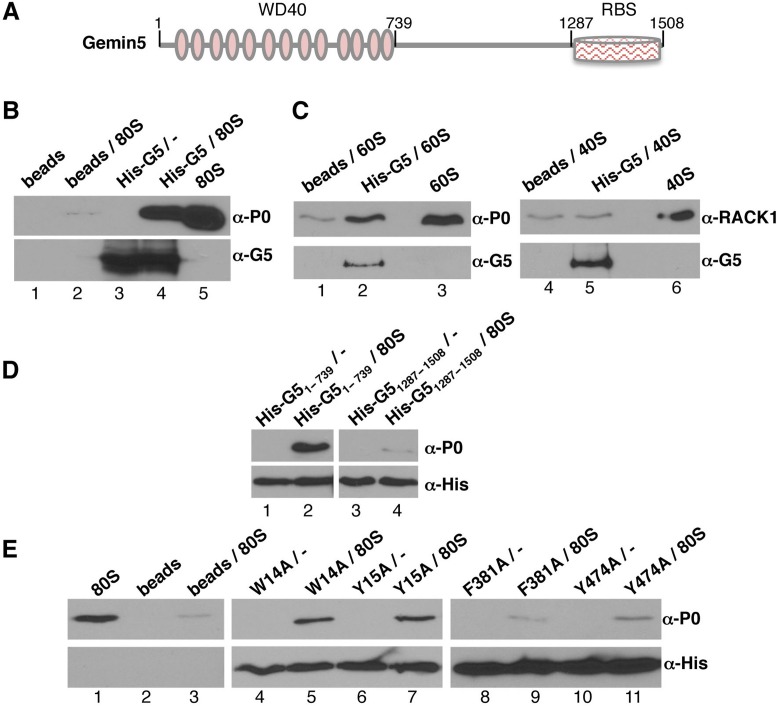
The observation that Gemin5 sedimented with the ribosomal particles in the polysome profile fractions, together with its role in translation control (26), suggested to us that the protein could interact directly with the ribosome. To test this hypothesis, we prepared N-terminal His-tagged constructs of the full-length Gemin5 (His-G5), the N-terminal fragment His-G51–739 and the C-terminal fragment His-G51287–1508 (Figure 4A). The N-terminal region (residues 1–739) includes the WD domain repeats (51), whereas the C-terminal region harbors a non-canonical RBS (45).
Figure 4.
Purified Gemin5 binds directly to the ribosome. (A) Diagram showing His-tagged Gemin5 polypeptides used in the ribosome-binding assays. Numbers indicate the amino acid residues flanking each polypeptide. (B) Binding assay of 80S ribosomes with His-Gemin5. 80S ribosomes (0.7 pmol) were incubated with the full length His-G5 (4 pmol) previously bound to Ni-agarose beads. After extensive washings, His-G5 was immunodetected using anti-G5 antibody. WB anti-P0 was used to determine the presence of ribosomes bound to Gemin5. (C) Binding assay of 40S and 60S ribosome subunits (0.7 pmol) with His-tagged Gemin5 (4 pmol). WB anti-P0 was used to determine the presence of 60S subunits bound to Gemin5, while anti-RACK1 was used to monitor the presence of 40S subunits. (D) Binding assay of 80S ribosomes with His-G51–739 or His-G51287–1508. Both, His-G51–739 or His-G51287–1508 were immunodetected using anti-His. WB anti-P0 was used to determine the presence of ribosomes bound to Gemin5. (E) Binding assay of 80S ribosomes carried out with the indicated mutants of His-G51–739. All binding assays were conducted independently at least three times.
A binding assay was then conducted using purified His-G5 and 80S ribosomes, corresponding to the pellet of 80S peak fractions (Supplementary Figure S6A). The 80S pellet did not contain detectable amounts of eIFs, as shown by WB of eIF3b or eIF4E (Supplementary Figure S6B). The His-G5 protein bound to prewashed Ni-agarose beads was incubated with 80S ribosomes. Following extensive washing of the beads-bound complexes, the presence of Gemin5 was determined by WB using anti-G5 antibody while ribosomes were detected using anti-P0 antibody. Control assays carried out in parallel omitted the His-G5 protein, the 80S particles or both. Samples containing only beads (Figure 4B, lane 1) were negative for both, Gemin5 and P0, as expected. Beads incubated only with 80S ribosomes (Figure 4B, lane 2) were also negative, demonstrating that 80S particles were not unspecificaly bound to the Ni-agarose beads. Beads incubated with His-G5 alone (Figure 4B, lane 3) were positive for Gemin5, but not for P0. Interestingly, His-G5 coated beads incubated with 80S pellet (Figure 4B, lane 4) were positive for both, Gemin5 and P0, demonstrating that the purified His-G5 can recruit 80S ribosomes in a very efficient manner. Indeed, the intensity of the 80S particles, monitored by immunodetection of P0, was 89.7-fold higher than the signal observed in the control (lane 2). The 80S pellet alone was loaded in lane 5 as a control of the specificity of P0 antibody.
These results prompted us to determine the ribosomal subunit responsible for Gemin5 interaction using free 40S and 60S ribosomal subunits isolated from a high salt dissociation sucrose gradients (Supplementary Figure S6C) that contained the expected composition of ribosomal proteins (Supplementary Figure S6D). The results indicated that only the 60S, but not the 40S ribosomal subunits, retained the ability to bind His-G5 (Figure 4C). Indeed, the binding of the 60S subunit (lane 2), monitored by P0 immunodetection, revealed 11.4-fold higher intensity than the signal observed in the control (lane 1). In contrast, the binding of the 40S subunit (lane 5), identified using anti-RACK1 antibody, was similar to the control (lane 4, 1.3-fold). Hence, these results show that Gemin5-ribosome interaction takes place through the 60S subunit.
Next, in order to identify the regions of Gemin5 responsible for the binding with the ribosomes we used the polypeptides His-G51–739, and His-G51287–1508 (Figure 4D). The binding assay performed using beads-His-G51–739 (lane 2) showed an intense interaction with the 80S ribosomes (38.8-fold relative to the signal of the control (Figure 4B, lane 2). In contrast, the C-terminal fragment, His-G51287–1508 (Figure 4D, lane 4) failed to recruit 80S particles (0.5-fold relative to the control). We conclude that the region of Gemin5 interacting with the ribosomal particles is placed at the N-terminal domain of the protein.
Given the direct interaction between the N-terminal region of Gemin5 protein and 80S ribosomes in vitro, we sought to investigate this region in further detail. Since the WD domain of Gemin5 was reported to be responsible for its binding to a methylguanosine cap-resin (52), in addition to snRNAs (32), we made use of four different amino acid mutations in positions W14, Y15, F381 and Y474 to alanine within His-G51–739. The first two, W14, Y15, are located at the very end of the N-terminus, while F381 and Y474 are placed on the 7th and 9th WD repeat domain of Gemin5. Relative to the wild-type His-G51–739, the proteins harboring the substitution W14A or Y15A did not modify the binding of the His-tagged protein to 80S particles (Figure 4E, lanes 5 and 7) (13.8- and 14.4-fold, respectively, compared to the beads-ribosome background, Figure 4E, lane 3). In contrast, the protein harboring the substitution F381A did not interact with the 80S particles (1.1-fold over the background, Figure 4E, lane 9), and the protein Y474A showed a very weak binding (3.0-fold over the background) (Figure 4E, lane 11). These results revealed that the interaction of the N-terminal domain of Gemin5 with the ribosome involves the residues F381 and Y474 within the 7th and 9th WD repeats.
Gemin5 interacts directly with the ribosomal proteins L3 and L4
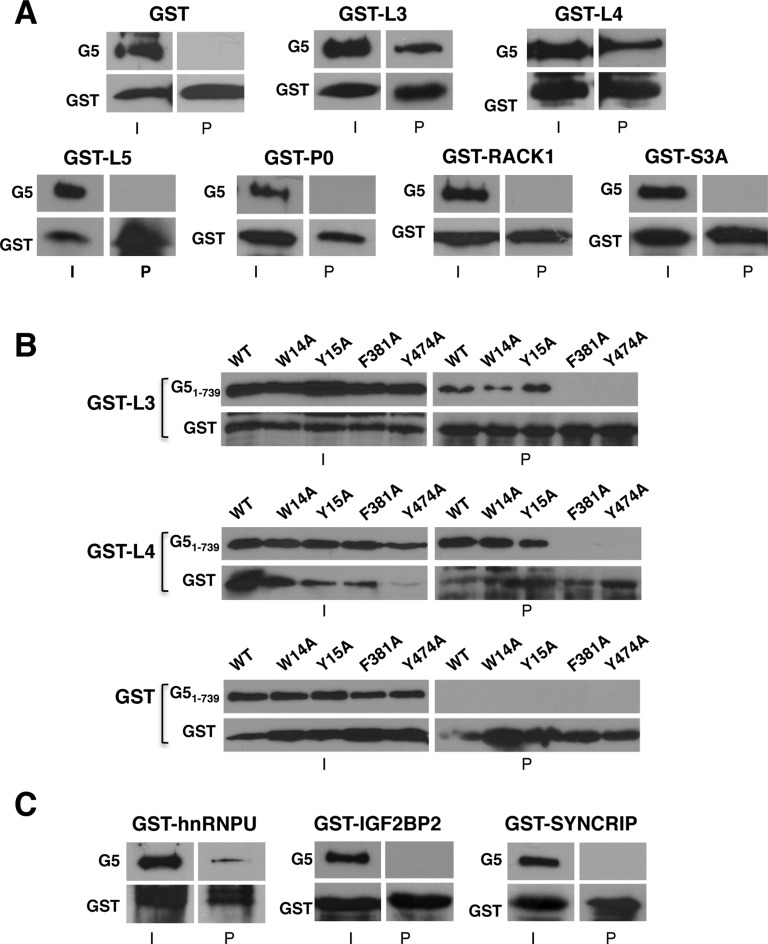
Based on the observation that the N-terminal domain of Gemin5 could interact with the 80S ribosome through the 60S subunit in vitro (Figure 4), we focused our attention for further analysis to ribosomal proteins L3 and L4 that we found associated with G5-Nter following RNase treatment (Supplementary Figure S5) to analyze protein–protein interaction. In addition, we selected two other proteins of the large ribosomal subunit (L5 and P0), and another two of the small ribosomal subunit (RACK1 and S3A), as controls. To this end, GST-fusions were purified from bacteria and used in a pull down assay with myc-tagged human Gemin5 (myc-G5). GST alone, used as negative control of the pull-down, did not interact with Gemin5 (Figure 5A). It should be noted that the GST fusions of the ribosomal proteins L3 and L4 showed a strong interaction with the full length Gemin5 protein (Figure 5A). In contrast, the other ribosomal proteins analyzed yielded undetectable binding (Figure 5A). Thus, these results reinforce the conclusion that there is a direct link between Gemin5 and specific components of the ribosome, namely L3 and L4.
Figure 5.
Pull-down assays reveal direct interaction of Gemin5 with the ribosomal proteins L3 and L4. (A) GST-pull-down assay of ribosomal proteins L3 and L4, resistant to RNase treatment during the TAP procedure, with myc-Gemin5. GST protein alone was used as negative control. Gemin5 was immunodetected by WB using anti-G5; the GST-fusion proteins were detected using anti-GST. I depicts input; P, pull-down. Proteins L5, P0, RACK1 and S3A were used as negative controls. (B) GST-pull-down assay of ribosomal proteins L3 and L4 with the indicated mutants of His-G51–739. His-G51–739 mutants were immunodetected with anti-His, and the GST-fusion proteins were detected using anti-GST. GST protein alone was used as negative control. (C) GST-pull-down assay of RBPs, resistant to RNase treatment during the TAP procedure, with myc-Gemin5. Independent pull-down assays were conducted at least three times.
To further reinforce these results, we used the mutant proteins His-G51–739, for which we showed a defect in binding to the 80S ribosome (see Figure 4E). As shown in Figure 5B, binding of F381A and Y474A to GST-L3 and GST-L4 was abolished, while W14A and Y15A retained the capacity to interact with both GST-L3 and GST-L4. None of these proteins showed binding to GST alone (Figure 5B). These results allowed us to conclude that there is a direct correlation between the interaction of Gemin5 with the ribosome and the binding of Gemin5 with L3 and L4, strongly suggesting that interaction of Gemin5 with the 80S (and the 60S subunit as well) is mediated by L3 or L4.
We also selected other RNA-binding proteins resistant to RNase treatment, identified with high score (hnRNP U, IGF2BP2, SYNCRIP). The GST-hnRNP U pull-down showed a positive binding with myc-G5 protein (Figure 5C), in agreement with previous data (48). Next, pull-down assays carried out with GST-IGF2BP2 and GST-SYNCRIP indicated that these proteins fail to interact directly with Gemin5 (Figure 5C). The lack of binding of these RBPs, which were selected according to their resistance to RNase treatment and reported capacity to modulate IRES activity, suggest that association of IGF2BP2 and SYNCRIP to Gemin5 was presumably due to indirect protein interactions within a large RNP complex.
Depletion and overexpression of Gemin5 alters the polysome/80S ratio and slightly, the global protein synthesis
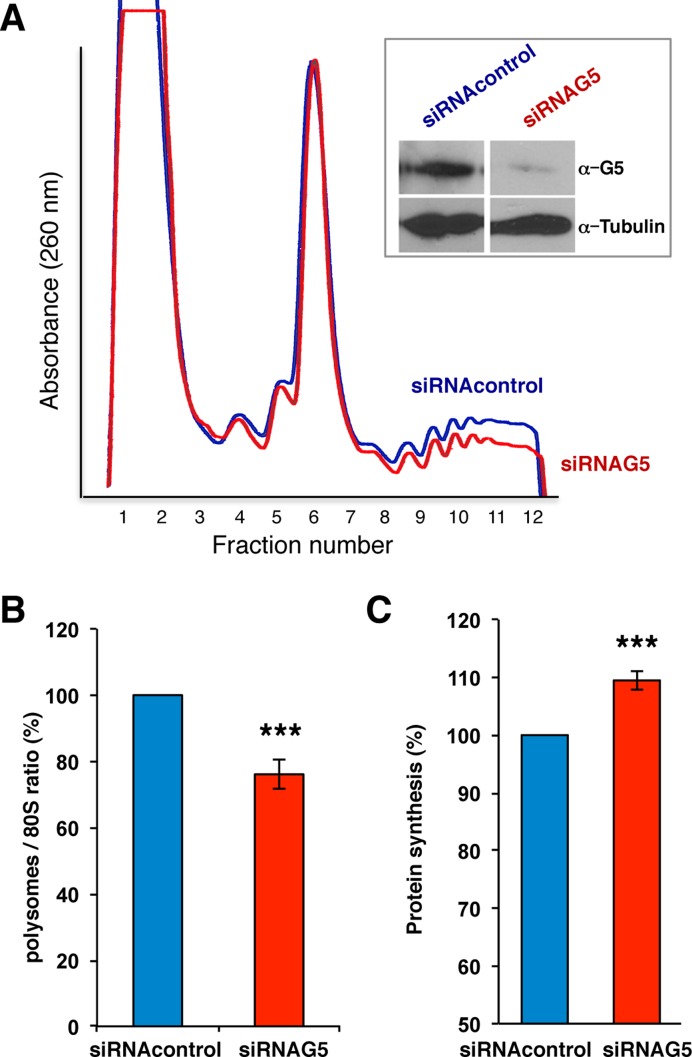
We next asked whether altering the levels of Gemin5 protein within cells influence the pattern of the polysome profile. To answer this question, lysates prepared from siRNA-Gemin5 depleted HEK293 cells (Figure 6A, inset), were fractionated on sucrose gradients in parallel to control siRNA extracts. Depletion of Gemin5 moderately altered the global polysome profile relative to the control siRNA (Figure 6A). The polysome/80S ratio was calculated by comparing the area under the 80S peak and the combined area under the polysome peaks. Accordingly, the ratio of polysome/80S showed a 24% decrease in three independent assays relative to the control siRNA (Figure 6B).
Figure 6.
Gemin5 siRNA depletion decreased the polysome/80S ratio, and enhanced global protein synthesis. (A) Polysome profiles (10–50% sucrose gradient) from cytoplasmic lysates of siRNA Gemin5 depleted cells (red line) and siRNA control cells (blue line). Gemin5 level in the lysates (siRNAG5) was determined by WB with anti-Gemin5 by comparison to HEK293 cells (siRNAcontrol). Tubulin was used as loading control. (B) Representation of the polysome/80S ratio (mean ± SD) obtained from three independent polysome profiles of Gemin5-depleted or control cells. (C) Representation of the intensity values of [35S]-met labeled proteins during a 3 h pulse in Gemin5 depleted and control cell extracts; values represent the mean ± SD obtained in three independent assays. Asterisks denote P-values (* P < 0.1; ** P < 0.05, *** P < 0.01).
This observation prompted us to determine the effect of Gemin5 depletion on global translation, monitoring the incorporation of [35S]-methionine in a 3 h pulse carried out at the end of the silencing treatment (e.g. 45–48 h post-siRNA transfection). Compared to the results obtained in the control cells, we observed an increase in the total protein synthesis (110%, see Figure 6C), in agreement with data previously reported using shRNAs targeting Gemin5 (26).
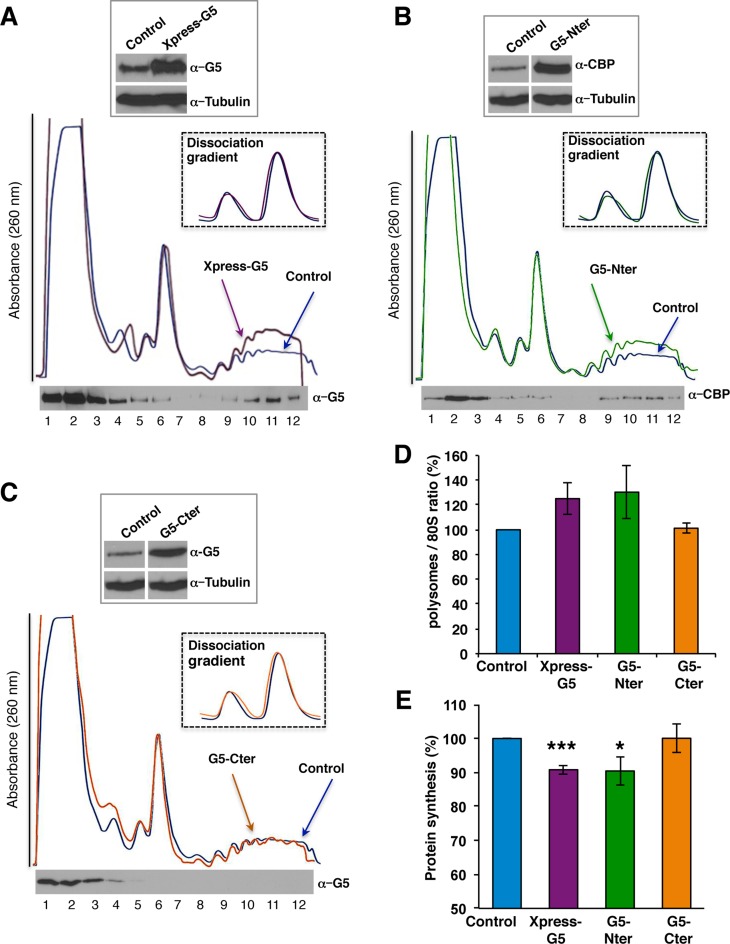
To further confirm these data, we conducted the reciprocal experiment. Overexpression of the Xpress-tagged full-length protein in HEK293 cells showed high levels of the tagged protein 24 h post-transfection (Figure 7A, top panel). Subsequent fractionation of soluble extracts on sucrose gradients in parallel to lysates derived from mock-transfected cells indicated a moderate alteration of the polysome profile, mainly affecting the heavy polysomes (Figure 7A). Notably, dissociation gradients conducted to monitor the relative amounts of 40S and 60S subunits (Figure 7A, inset) indicated that high expression levels of Gemin5 protein did not affect the amount of each of these ribosomal subunits. This result argues that the protein is unlikely involved in the biogenesis of the ribosomal subunits. Of interest, using anti-G5 antibody in a WB, the Xpress-G5 protein was clearly detected on the heavy polysome fractions (Figure 7A, bottom panel) with higher intensity than the level of the endogenous Gemin5 protein detected with the same antibody under the same conditions (see Figure 3B). Therefore, we conclude that overexpression of Gemin5 induced an increase on the heavy polysomes coincident with a higher intensity of the protein in the polysome fractions.
Figure 7.
Gemin5 overexpression alters the polysome/80S ratio. (A) Polysome profiles (10–50% sucrose gradient) from cytoplasmic lysates of HEK293 cells expressing Xpress-Gemin5 (purple line) and HEK293 control cells (blue line). (Top) Equal amounts of total protein were separated in SDS-PAGE to determine the expression of Xpress-G5 and Gemin5 by immunoblotting using anti-Gemin5 antibody. Tubulin was used as loading control. (Bottom) The distribution of Xpress-Gemin5 in the fractions of the gradient (15 μl) was immunoblotted using anti-Gemin5 antibody. The inset shows 40S and 60S peaks observed in a dissociation sucrose gradient profile (10–30%) in HEK293 control cells (blue line) compared to Xpress-G5 expression (purple line). Expression of (B) G5-Nter (green line) but not of (C) G5-Cter (orange line) alters the polysome profile, compared to the control cells (blue line). In both cases, the expression of the proteins, and the distribution of G5-Nter and G5-Cter on the gradient fractions (15 μl), was analyzed by WB using anti-CBP or anti-Gemin5 antibodies, respectively. Dissociation sucrose gradients profile showing the 40S and 60S peaks in HEK293 control cells (blue line) compared to G5-Nter (green line) and G5-Cter (orange line) transfected cells. (D) Representation of the mean polysome/80S ratio obtained from three independent polysome profiles determined in cells transfected with Xpress-G5, G5-Nter or G5-Cter compared to control cells. (E) Effect of Xpress-G5, G5-Nter and G5-Cter overexpression on global protein synthesis. The intensity of [35S]-methionine labeled proteins during a 3 h pulse was compared to the control cells; values represent the mean ± SD obtained in three independent assays. Asterisks denote P-values (*P < 0.1; **P < 0.05, ***P < 0.01).
To validate these results, we assayed the two moieties of the protein, G5-Nter and G5-Cter, of which only G5-Nter was shown to interact with ribosomes. The results indicate that overexpression of the G5-Nter protein altered the global polysome profile to the same extent than the full length Xpress-G5 (Figure 7B). In contrast, expression of G5-Cter protein failed to modify the polysome profile (Figure 7C). In addition, the G5-Nter protein but not the G5-Cter protein, was detected on the heavy polysome fractions, reinforcing the correlation between the change of the polysome profile and the presence of the protein in the heavy polysome fractions.
Independent overexpression experiments carried out with Xpress-G5 and G5-Nter showed an increase (30%) of the polysome/80S ratio relative to the control cells (Figure 7D), whereas the expression of G5-Cter was similar. However, the change in the ratio of polysome/80S correlated negatively with the effect on global synthesis of proteins, measured by incorporation of [35S]-methionine in a 3 h pulse (Figure 7E).
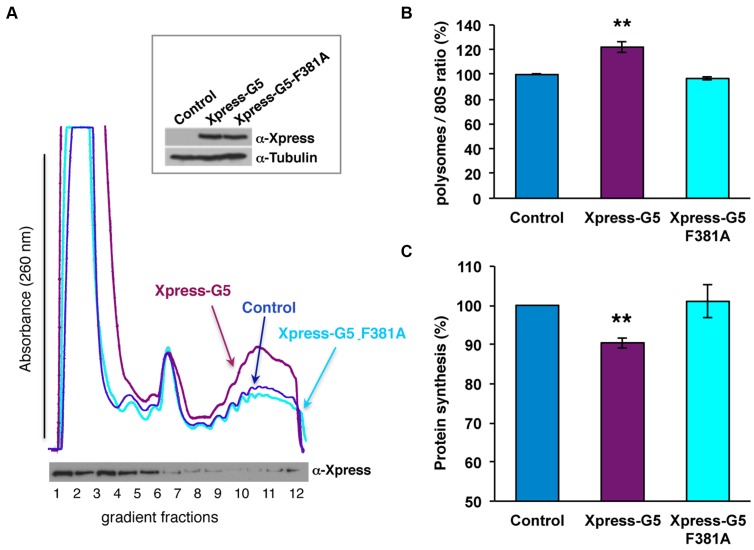
To further assess the implications of the residues involved in the interaction of Gemin5 with the ribosome we generated the mutant F381A in the context of the full length Gemin5 protein. Overexpression of Xpress-G5-F381A in HEK293 cells readily indicated that this mutation reverted the effect of the wild-type construct (Figure 8A). Indeed, the ratio of polysome/80S observed in cells expressing the mutant Xpress-G5-F381A yielded the same levels than those observed in control cells (Figure 8B), while the ratio was increased by 20% in the case of the wild-type Xpress-G5. In addition, the efficiency of global protein synthesis in Xpress-G5-F381A transfected cells was reverted to the levels of the control cells (Figure 8C).
Figure 8.
Gemin5 F381A overexpression reverts the effect on the polysome/80S ratio. (A) Polysome profiles (10–50% sucrose gradient) from cytoplasmic lysates of HEK293 cells expressing Xpress-Gemin5 (purple line), Xpress-Gemin5-F381A (pale blue line) and HEK293 control cells (blue line). (Top) Equal amounts of total protein were separated in SDS-PAGE to determine the expression of Xpress-G5 and Xpress-G5-F381A by immunoblotting using anti-Xpress antibody. Tubulin was used as loading control. (B) Representation of the mean polysome/80S ratio obtained from two independent polysome profiles determined in cells transfected with Xpress-G5 or Xpress-G5-F381A compared to control cells. (C) Effect of Xpress-G5 or Xpress-G5-F381A overexpression on global protein synthesis. The intensity of [35S]-methionine labeled proteins during a 3 h pulse was compared to the control cells; values represent the mean ± SD obtained in at least three independent assays. Asterisks denote P-values (*P < 0.1; **P < 0.05, ***P < 0.01).
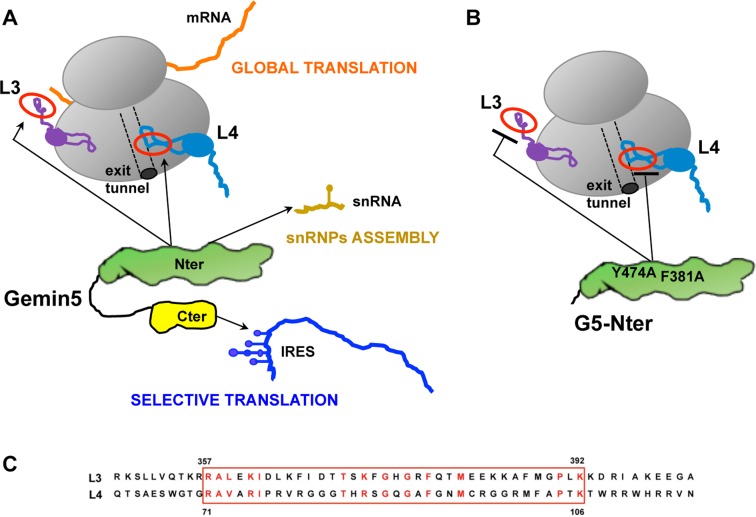
Thus, we conclude that there is a division of functions on the two moieties of the Gemin5 protein concerning translation control (Figure 9A). The ability of the N-terminal (G5-Nter) domain to interact with the ribosome particles through L3 or L4 proteins modulates global translation, while the ability of the C-terminal domain (G5-Cter) to bind directly with RNA motifs of IRES elements affects selective translation (33,45).
Figure 9.
(A) Model for the role of Gemin5 in RNA translation control. The functional domains G5-Nter and G5-Cter are shadowed in green and yellow, respectively. The Nter domain provides the capacity to interact with 80S ribosomes (grey) via the 60S subunit, and with the ribosomal proteins L3 (violet) and L4 (pale blue) as well, impacting on global mRNA translation (orange). The 5th WD motif is responsible for the delivery of snRNAs (pale brown) to the SMN complex (32), governing snRNPs assembly. Additionally, a non-canonical RNA-binding site at Cter provides the recognition of viral IRES elements (dark blue), thus impacting on selective translation (45). The association of Gemin5 to L3 and L4 could be mediated by a solvent accessible conserved motif (red circle). (B) Gemin5 mutants F381A and Y474A, located on the 7th and 9th WD motifs, impaired the binding to L3 and L4 and also abolished the interaction with the ribosome. (C) Amino acid sequence of the conserved motif present in the ribosomal proteins L3 and L4.
DISCUSSION
Gemin5 is a predominant cytoplasmic protein that has two distinct functional domains. At the N-terminus, a 13 WD40 repeat domain is responsible for the delivery of the SMN complex to snRNAs (32,53). In contrast, the C-terminal domain harbors a non-canonical RNA-binding site that mediates the interaction with a viral IRES element (45). This division of functions suggests the existence of multiple targets recognized by Gemin5, presumably assembled in distinct functional complexes. Our study shows a comprehensive overview of the protein network associated to Gemin5. An unbiased proteomic approach allowed us to identify the factors associated to the N-terminal and the C-terminal domains of Gemin5, and to emphasize the diversity of partners differentially associated to each domain. Importantly, we demonstrate the power of this global approach to identify distinct components of the proteome upon the disruption of RNA bridges that coordinate protein–protein interactions (Figure 2). More specifically, we show that L3 and L4 ribosomal proteins were associated to the N-terminal domain of Gemin5 even upon RNase treatment. These results highlight the dynamic nature of the Gemin5-riboproteome within the cell.
Notably, the number of proteins preferentially associated with the N-terminal domain generally exceeds that of the proteins associated to the C-terminal region. This result is in agreement with the fact that the N-terminal region contains a 13 WD40 repeat domain (51), which is considered a platform for protein–protein interactions (54,55). Hence, the association of a large number of partners with the N-terminal region of Gemin5 strongly suggests that this protein may participate in multiple pathways. In addition, we have observed that the proteins identified with high score cluster into subgroups of functional RNP complexes, as illustrated by the SMN complex (Figure 2B). This result indicates that, under the conditions used here, binding to one component may allow to capture RNP complexes. Our approach, however, showed that the number of peptides identified in the members of the SMN complex does not correlate with its proximity to Gemin5 in the model of the SMN complex (30,56). Notably, after RNase treatment mainly the endogenous Gemin5 protein copurified with the TAP-tagged G5-Nter. This result suggests that this protein has the capacity to oligomerize, and that the proteins of the SMN complex are bound through RNA bridges. Besides the SMN complex, proteins identified as Gemin5-associated factors include RBP, IRES-transacting factors and protein synthesis, among others. Although the vast majority of the identified proteins were found associated to the N-terminal domain of Gemin5 (Figure 2A), others (RAVER1, PCBP2, eIF3F or eIF3G) were differentially associated to the C-terminal domain (see Supplementary Figures S2, S3 and S4). The links of Gemin5 with RBPs support the view that defects in Gemin5 partners would compromise RNA metabolism processes.
The capacity to interact directly with ribosomes via its N-terminal domain reveals a novel feature of Gemin5
Accumulating evidences suggest the involvement of Gemin5 in cellular processes related to translation control (26,57,58). In addition to these properties, Gemin5 was described as a scaffolding protein with the capacity to interact with eIF4E (59) and m7GTP-resin (52), two features that also link Gemin5 with translation events. However, the mechanistic basis of this effect remains to be elucidated. Here, we have found for the first time that Gemin5 sediments with the total ribosome (R) fraction that contains ribosomal particles and translation factors among other proteins, but it is absent in the RSW fraction (Figure 3A), demonstrating that it is not an integral part of the ribosome. In agreement with the results obtained in the proteomic approach, Gemin5 was immunodetected in the light fractions of a polysome profile entering up to the 80S peak, as it also does eIF3b. These data are also consistent with the observation that Gemin5 binds to m7GTP-resin (59). Notably, Gemin5 was also detected in the heavy polysome fractions. Moreover, overexpression of both the Xpress-G5 and G5-Nter proteins in HEK293 cells showed a marked association to the heavy polysomes, increasing the polysome/80S ratio (Figure 7D). This effect correlated negatively with a decrease in global protein synthesis monitored by [35S]-methionine incorporation in a 3 h pulse at the end of the expression time (Figure 7E). Further confirming these observations, the reverse experiment revealed an increase in the global protein synthesis in siRNA-transfected cells concomitant with a decrease in the ratio of the polysome/80S (Figure 6B and C). These results allow us to hypothesize that the protein may have a stalling effect on translation elongation, consistent with its capacity to modulate negatively global protein synthesis (26). The inverse correlation between an increase of the ratio of polysome/80S and a decrease in translation efficiency has been documented (60). Mitotic cells contain the same amount of heavy polysomes than cycling cells, but they are less active, suggesting that stalling translating ribosomes during mitosis might allow rapid resumption of translation upon entry into the G1 phase. Furthermore, yeast mutants affected in rpL3 and Hpm1 displayed higher polysome/80S ratio compared to wild-type cells, but they have similar growth rates (61). Given that the selective translation of monosomes 80S versus polysomes has been reported (62), it is tempting to speculate that the increase in the polysome/80S ratio observed after overexpression of Gemin5 may be indicative of the preferential translation of selective mRNAs concomitant to the decrease in the elongation rate of the majority of mRNAs.
The direct interaction of Gemin5 with L3 and L4 ribosomal proteins is consistent with its effect on translation control
We have shown here for the first time that purified Gemin5 has the capacity to bind to 80S ribosome particles in vitro. More specifically, the full-length protein interacts with the 60S subunit, but not with the 40S ribosomal subunit. This feature of Gemin5 maps to the N-terminal domain of the protein within the region encompassing amino acids 1–739 (Figure 4C and D), specifically involving the residues F381 and Y474 (Figure 4E), which are located on the 7th and the 9th WD repeat motifs of the protein, respectively. In the absence of the Gemin5 crystal structure, we propose that these amino acid substitutions reorganize the tertiary structure of the N-terminal domain of Gemin5, negatively affecting its interaction with the ribosome. Further supporting the relevance of these residues in the role of Gemin5 in translation control, overexpression of the mutant Xpress-G5-F381A reverts the effect on the ratio of polysome/80S of the wild-type Xpress-G5, and also the effect on global protein synthesis. However, it is noteworthy that a moderate amount of this mutant protein was associated to the heavy polysome fractions (Figure 8A). This observation opens the possibility that the Gemin5 region flanked by amino acids 739 and 1287 (see Figure 1A) could also contribute to recruit the ribosomal machinery. This possibility, which is based on the large size of this protein, its potential posttranslational modifications within the cell and the large number of partners identified in the current study, remains to be elucidated in future studies.
It is well established that WD repeat motifs are platforms for many types of protein–protein interactions (54,55). RNAs bind to proteins to build RNPs complexes, which are subsequently guided to their site of action. We show here that upon exhaustive treatment with RNase A, the number of proteins copurifying with each functional domain of Gemin5 decreased in most groups, with the exception of the ribosome. Consistent with the capacity of the N-terminal region to bind 80S ribosomes, the number of identified peptides and the identity of the ribosomal proteins associated to the N-terminal domain generally remained constant after RNase treatment.
The ribosomal proteins L3 and L4, which were identified with the highest number of peptides in the proteomic analysis of G5-Nter after RNase digestion, interacted directly with Gemin5 in a GST-pull down assay (Figure 5A), suggesting that Gemin5 can form a binary complex with these ribosomal proteins independent of RNA bridges. Other ribosomal proteins (S3A, L5, P0 and RACK1, used as controls) were negative in the pull-down assay. The concomitant lack of binding of the Gemin5 mutants F381A and Y474A to GST-L3 and GST-L4 suggest that the binding of Gemin5 with L3 or L4 can be a plausible mechanism to explain its interaction with the ribosome.
L3 and L4 are located on different regions of the ribosome particle (63). Both, L3 and L4 are conserved proteins with terminal extensions located on the solvent side of the 80S ribosome (64–67). Since we have observed that the interactions between L3 or L4 with the F381A and Y474A mutants of Gemin5 are impaired, we hypothesized that the interaction can take place through a similar motif in each of these proteins. Amino acid sequence alignment of L3 and L4 evidenced a solvent accessible conserved motif (Figure 9C). For L3, this motif protrudes from the ribosome (64). L3 coordinates the binding of elongation factors and aminoacylated tRNAs (66,67). Thus, interaction of Gemin5 with L3 may interfere with its role in translation control. In L4 this motif forms part of the peptide exit tunnel wall (68). Hence, the interaction of Gemin5 with L4 within the peptide tunnel may explain the observed negative effect on global translation.
In summary, we envision that this large multifunctional protein has a dual role on translation control (Figure 9A), beyond its capacity to deliver the snRNAs into the SMN complex (31). These features rely on the capacity of Gemin5 to recognize multiple partners (both, RNAs and proteins) through different functional domains. The targets of Gemin5 include several RNA structural motifs, such as those of snRNAs recognized by the WD motifs at the N-terminal end (32), and those of IRES elements recognized by the non-canonical RNA-binding site at the C-terminal end (45). We showed here that the N-terminal domain of the protein mediates direct interactions with specific proteins, besides a network of proteins via RNA bridges. Thus, regarding the involvement of Gemin5 in translation control, the ribosome binding capacity of the N-terminal moiety enables Gemin5 to control global protein synthesis (Figure 9A), while the non-canonical RNA-binding motif located at the C-terminal end is responsible for the repressor effect of IRES-dependent translation (45). Hence, this protein could contribute to translation control in a very different manner using two domains, Nter and Cter, which have the capacity to interact with distinct partners within the cell.
Importantly, our study revealed a direct crosstalk between Gemin5 and the ribosomal proteins L3 and L4, and provided support for the view that Gemin5 may control translation elongation. The N-terminal Gemin51–739 mutants F381A and Y474A, which impaired the binding to L3 and L4, also abolished the interaction with the ribosome (Figure 9B). Silencing of Gemin5 induced an increase in global protein synthesis. In contrast, both the wild-type protein and the G5-Nter polypeptide, but not the G5-Cter, induced a reduction in global protein synthesis and remained associated to the heavy polysome fractions of a sucrose gradient. We propose that the association of Gemin5 to L3 or L4 could be mediated by a conserved solvent accessible motif (Figure 9A). In the case of L4, this motif is located near the tunnel exit (68), in agreement with the association of Gemin5 to heavy polysomes, and with the suggestion that Gemin5-L4 interaction can affect translation elongation. In the case of L3 this motif protrudes from the ribosome. However, L3 functions as an allosteric switch in coordinating binding of the elongation factors and aminocylated tRNAs (66,67). Hence, interaction of Gemin5 with L3 may interfere with the recruitment of factors needed for protein synthesis. Future studies aimed to understand further implications of these interactions in protein synthesis could provide hints about new pathways of gene regulation.
Supplementary Material
Acknowledgments
Work in the laboratory of EMS was supported by MINECO. The authors thank A-C Gingras and J.J. Berlanga for providing the vectors pcDNA3-CTAP and pcDNA3-NTAP, C. Xu and J. Min for the kind gift of the wild-type protein Gemin51-739 and the amino acid mutants W14A, Y15A, F381A and Y474A, J. Maillot, and M. Yusupov for providing information regarding L4 accessibility, M.A. Rodriguez-Gabriel for sharing with us the ISCO density gradient fractionator, A. Marina and the CBMSO Proteomics Unit for help with the MS/MS analysis and J.P.G. Ballesta, C. Gutierrez and G. Lozano for valuable comments on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
MINECO [BFU2011-25437, BFU2014-54564]; Institutional grant from Fundación Ramón Areces. Funding for open access charge: MINECO [BFU2014-54564].
Conflict of interest statement. None declared.
REFERENCES
- 1.Muller-McNicoll M., Neugebauer K.M. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat. Rev. Genet. 2013;14:275–287. doi: 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- 2.Maslon M.M., Heras S.R., Bellora N., Eyras E., Caceres J.F. The translational landscape of the splicing factor SRSF1 and its role in mitosis. Elife. 2014:e02028. doi: 10.7554/eLife.02028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ozgur S., Buchwald G., Falk S., Chakrabarti S., Prabu J.R., Conti E. The conformational plasticity of eukaryotic RNA-dependent ATPases. FEBS J. 2015;282:850–863. doi: 10.1111/febs.13198. [DOI] [PubMed] [Google Scholar]
- 4.Jonas S., Izaurralde E. The role of disordered protein regions in the assembly of decapping complexes and RNP granules. Genes Dev. 2013;27:2628–2641. doi: 10.1101/gad.227843.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckmann B.M., Horos R., Fischer B., Castello A., Eichelbaum K., Alleaume A.M., Schwarzl T., Curk T., Foehr S., Huber W., et al. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun. 2015;6:10127. doi: 10.1038/ncomms10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M., et al. Insights into RNA Biology from an Atlas of Mammalian mRNA-Binding Proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 7.Soto-Rifo R., Rubilar P.S., Limousin T., de Breyne S., Decimo D., Ohlmann T. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 2012;31:3745–3756. doi: 10.1038/emboj.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durie D., Lewis S.M., Liwak U., Kisilewicz M., Gorospe M., Holcik M. RNA-binding protein HuR mediates cytoprotection through stimulation of XIAP translation. Oncogene. 2011;30:1460–1469. doi: 10.1038/onc.2010.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee K.H., Woo K.C., Kim D.Y., Kim T.D., Shin J., Park S.M., Jang S.K., Kim K.T. Rhythmic interaction between Period1 mRNA and hnRNP Q leads to circadian time-dependent translation. Mol. Cell. Biol. 2012;32:717–728. doi: 10.1128/MCB.06177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goulet I., Boisvenue S., Mokas S., Mazroui R., Cote J. TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules. Hum. Mol. Genet. 2008;17:3055–3074. doi: 10.1093/hmg/ddn203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez G., Dury A.Y., Murray L.M., Biondi O., Tadesse H., El Fatimy R., Kothary R., Charbonnier F., Khandjian E.W., Cote J. A novel function for the survival motoneuron protein as a translational regulator. Hum. Mol. Genet. 2013;22:668–684. doi: 10.1093/hmg/dds474. [DOI] [PubMed] [Google Scholar]
- 12.Khandjian E.W., Corbin F., Woerly S., Rousseau F. The fragile X mental retardation protein is associated with ribosomes. Nat. Genet. 1996;12:91–93. doi: 10.1038/ng0196-91. [DOI] [PubMed] [Google Scholar]
- 13.Takagi M., Absalon M.J., McLure K.G., Kastan M.B. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 14.Chen E., Sharma M.R., Shi X., Agrawal R.K., Joseph S. Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol. Cell. 2014;54:407–417. doi: 10.1016/j.molcel.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Majzoub K., Hafirassou M.L., Meignin C., Goto A., Marzi S., Fedorova A., Verdier Y., Vinh J., Hoffmann J.A., Martin F., et al. RACK1 controls IRES-mediated translation of viruses. Cell. 2014;159:1086–1095. doi: 10.1016/j.cell.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hafren A., Eskelin K., Makinen K. Ribosomal protein P0 promotes Potato virus A infection and functions in viral translation together with VPg and eIF(iso)4E. J. Virol. 2013;87:4302–4312. doi: 10.1128/JVI.03198-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazumder B., Poddar D., Basu A., Kour R., Verbovetskaya V., Barik S. Extraribosomal l13a is a specific innate immune factor for antiviral defense. J. Virol. 2014;88:9100–9110. doi: 10.1128/JVI.01129-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinnebusch A.G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 19.Walsh D., Mohr I. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 2011;9:860–875. doi: 10.1038/nrmicro2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lozano G., Martinez-Salas E. Structural insights into viral IRES-dependent translation mechanisms. Curr. Opin. Virol. 2015;12:113–120. doi: 10.1016/j.coviro.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Lloyd R.E. Nuclear proteins hijacked by mammalian cytoplasmic plus strand RNA viruses. Virology. 2015;479-480:457–474. doi: 10.1016/j.virol.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez-Salas E., Francisco-Velilla R., Fernandez-Chamorro J., Lozano G., Diaz-Toledano R. Picornavirus IRES elements: RNA structure and host protein interactions. Virus Res. 2015;206:62–73. doi: 10.1016/j.virusres.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 23.Sagan S.M., Chahal J., Sarnow P. cis-Acting RNA elements in the hepatitis C virus RNA genome. Virus Res. 2015;206:90–98. doi: 10.1016/j.virusres.2014.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Au H.H., Jan E. Novel viral translation strategies. Wiley Interdiscip. Rev. RNA. 2014;5:779–801. doi: 10.1002/wrna.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson J.E., Pestova T.V., Hellen C.U., Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 26.Pacheco A., Lopez de Quinto S., Ramajo J., Fernandez N., Martinez-Salas E. A novel role for Gemin5 in mRNA translation. Nucleic Acids Res. 2009;37:582–590. doi: 10.1093/nar/gkn979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim M.S., Pinto S.M., Getnet D., Nirujogi R.S., Manda S.S., Chaerkady R., Madugundu A.K., Kelkar D.S., Isserlin R., Jain S., et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 29.Gates J., Lam G., Ortiz J.A., Losson R., Thummel C.S. rigor mortis encodes a novel nuclear receptor interacting protein required for ecdysone signaling during Drosophila larval development. Development. 2004;131:25–36. doi: 10.1242/dev.00920. [DOI] [PubMed] [Google Scholar]
- 30.Otter S., Grimmler M., Neuenkirchen N., Chari A., Sickmann A., Fischer U. A comprehensive interaction map of the human survival of motor neuron (SMN) complex. J. Biol. Chem. 2007;282:5825–5833. doi: 10.1074/jbc.M608528200. [DOI] [PubMed] [Google Scholar]
- 31.Yong J., Kasim M., Bachorik J.L., Wan L., Dreyfuss G. Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol. Cell. 2010;38:551–562. doi: 10.1016/j.molcel.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Battle D.J., Lau C.K., Wan L., Deng H., Lotti F., Dreyfuss G. The Gemin5 protein of the SMN complex identifies snRNAs. Mol. Cell. 2006;23:273–279. doi: 10.1016/j.molcel.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 33.Pineiro D., Fernandez N., Ramajo J., Martinez-Salas E. Gemin5 promotes IRES interaction and translation control through its C-terminal region. Nucleic Acids Res. 2013;41:1017–1028. doi: 10.1093/nar/gks1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Battle D.J., Kasim M., Wang J., Dreyfuss G. SMN-independent subunits of the SMN complex. Identification of a small nuclear ribonucleoprotein assembly intermediate. J. Biol. Chem. 2007;282:27953–27959. doi: 10.1074/jbc.M702317200. [DOI] [PubMed] [Google Scholar]
- 35.Chen G.I., Gingras A.C. Affinity-purification mass spectrometry (AP-MS) of serine/threonine phosphatases. Methods. 2007;42:298–305. doi: 10.1016/j.ymeth.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 36.Baltz A.G., Munschauer M., Schwanhausser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M., et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 37.Kessler S.M., Pokorny J., Zimmer V., Laggai S., Lammert F., Bohle R.M., Kiemer A.K. IGF2 mRNA binding protein p62/IMP2-2 in hepatocellular carcinoma: antiapoptotic action is independent of IGF2/PI3K signaling. Am. J. Physiol. Gastrointest. Liver Physiol. 2013;304:G328–G336. doi: 10.1152/ajpgi.00005.2012. [DOI] [PubMed] [Google Scholar]
- 38.Cox E.A., Bennin D., Doan A.T., O'Toole T., Huttenlocher A. RACK1 regulates integrin-mediated adhesion, protrusion, and chemotactic cell migration via its Src-binding site. Mol. Biol. Cell. 2003;14:658–669. doi: 10.1091/mbc.E02-03-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez-Gabriel M.A., Remacha M., Ballesta J.P. The RNA interacting domain but not the protein interacting domain is highly conserved in ribosomal protein P0. J. Biol. Chem. 2000;275:2130–2136. doi: 10.1074/jbc.275.3.2130. [DOI] [PubMed] [Google Scholar]
- 40.Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 41.Francisco-Velilla R., Fernandez-Chamorro J., Lozano G., Diaz-Toledano R., Martinez-Salas E. RNA-protein interaction methods to study viral IRES elements. Methods. 2015;91:3–12. doi: 10.1016/j.ymeth.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 42.Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 43.Roepstorff P., Fohlman J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed. Mass Spectrom. 1984;11:601. doi: 10.1002/bms.1200111109. [DOI] [PubMed] [Google Scholar]
- 44.Martinez-Azorin F., Remacha M., Ballesta J.P. Functional characterization of ribosomal P1/P2 proteins in human cells. Biochem. J. 2008;413:527–534. doi: 10.1042/BJ20080049. [DOI] [PubMed] [Google Scholar]
- 45.Fernandez-Chamorro J., Pineiro D., Gordon J.M., Ramajo J., Francisco-Velilla R., Macias M.J., Martinez-Salas E. Identification of novel non-canonical RNA-binding sites in Gemin5 involved in internal initiation of translation. Nucleic Acids Res. 2014;42:5742–5754. doi: 10.1093/nar/gku177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vilella M.D., Remacha M., Ortiz B.L., Mendez E., Ballesta J.P. Characterization of the yeast acidic ribosomal phosphoproteins using monoclonal antibodies. Proteins L44/L45 and L44′ have different functional roles. Eur. J. Biochem. 1991;196:407–414. doi: 10.1111/j.1432-1033.1991.tb15831.x. [DOI] [PubMed] [Google Scholar]
- 47.Pellizzoni L., Baccon J., Rappsilber J., Mann M., Dreyfuss G. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J. Biol. Chem. 2002;277:7540–7545. doi: 10.1074/jbc.M110141200. [DOI] [PubMed] [Google Scholar]
- 48.Yong J., Wan L., Dreyfuss G. Why do cells need an assembly machine for RNA-protein complexes. Trends Cell Biol. 2004;14:226–232. doi: 10.1016/j.tcb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 49.Meister G., Fischer U. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J. 2002;21:5853–5863. doi: 10.1093/emboj/cdf585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamazaki T., Chen S., Yu Y., Yan B., Haertlein T.C., Carrasco M.A., Tapia J.C., Zhai B., Das R., Lalancette-Hebert M., et al. FUS-SMN protein interactions link the motor neuron diseases ALS and SMA. Cell Rep. 2012;2:799–806. doi: 10.1016/j.celrep.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gubitz A.K., Mourelatos Z., Abel L., Rappsilber J., Mann M., Dreyfuss G. Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J. Biol. Chem. 2002;277:5631–5636. doi: 10.1074/jbc.M109448200. [DOI] [PubMed] [Google Scholar]
- 52.Bradrick S.S., Gromeier M. Identification of gemin5 as a novel 7-methylguanosine cap-binding protein. PLoS One. 2009;4:e7030. doi: 10.1371/journal.pone.0007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lau C.K., Bachorik J.L., Dreyfuss G. Gemin5-snRNA interaction reveals an RNA binding function for WD repeat domains. Nat. Struct. Mol. Biol. 2009;16:486–491. doi: 10.1038/nsmb.1584. [DOI] [PubMed] [Google Scholar]
- 54.Smith T.F., Gaitatzes C., Saxena K., Neer E.J. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- 55.Li D., Roberts R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol. Life Sci. 2001;58:2085–2097. doi: 10.1007/PL00000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neuenkirchen N., Englbrecht C., Ohmer J., Ziegenhals T., Chari A., Fischer U. Reconstitution of the human U snRNP assembly machinery reveals stepwise Sm protein organization. EMBO J. 2015;34:1925–1941. doi: 10.15252/embj.201490350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pineiro D., Fernandez-Chamorro J., Francisco-Velilla R., Martinez-Salas E. Gemin5: A Multitasking RNA-Binding Protein Involved in Translation Control. Biomolecules. 2015;5:528–544. doi: 10.3390/biom5020528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Workman E., Kalda C., Patel A., Battle D.J. Gemin5 Binds to the Survival Motor Neuron mRNA to Regulate SMN Expression. J. Biol. Chem. 2015;290:15662–15669. doi: 10.1074/jbc.M115.646257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fierro-Monti I., Mohammed S., Matthiesen R., Santoro R., Burns J.S., Williams D.J., Proud C.G., Kassem M., Jensen O.N., Roepstorff P. Quantitative proteomics identifies Gemin5, a scaffolding protein involved in ribonucleoprotein assembly, as a novel partner for eukaryotic initiation factor 4E. J. Proteome Res. 2006;5:1367–1378. doi: 10.1021/pr0504539. [DOI] [PubMed] [Google Scholar]
- 60.Sivan G., Kedersha N., Elroy-Stein O. Ribosomal slowdown mediates translational arrest during cellular division. Mol. Cell. Biol. 2007;27:6639–6646. doi: 10.1128/MCB.00798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Al-Hadid Q., Roy K., Chanfreau G., Clarke S.G. Methylation of yeast ribosomal protein Rpl3 promotes translational elongation fidelity. RNA. 2016;22:489–498. doi: 10.1261/rna.054569.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heyer E.E., Moore M.J. Redefining the translational status of 80S monosomes. Cell. 2016;164:757–769. doi: 10.1016/j.cell.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 63.Yusupova G., Yusupov M. High-resolution structure of the eukaryotic 80S ribosome. Annu. Rev. Biochem. 2014;83:467–486. doi: 10.1146/annurev-biochem-060713-035445. [DOI] [PubMed] [Google Scholar]
- 64.Anger A.M., Armache J.P., Berninghausen O., Habeck M., Subklewe M., Wilson D.N., Beckmann R. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- 65.Khatter H., Myasnikov A.G., Natchiar S.K., Klaholz B.P. Structure of the human 80S ribosome. Nature. 2015;520:640–645. doi: 10.1038/nature14427. [DOI] [PubMed] [Google Scholar]
- 66.Meskauskas A., Dinman J.D. Ribosomal protein L3: gatekeeper to the A site. Mol. Cell. 2007;25:877–888. doi: 10.1016/j.molcel.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mailliot J., Garreau de Loubresse N., Yusupova G., Meskauskas A., Dinman J.D., Yusupov M. Crystal structures of the uL3 mutant ribosome: Illustration of the importance of ribosomal proteins for translation efficiency. J. Mol. Biol. 2016;428:2195–2202. doi: 10.1016/j.jmb.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stelter P., Huber F.M., Kunze R., Flemming D., Hoelz A., Hurt E. Coordinated ribosomal L4 Protein assembly into the pre-ribosome is regulated by its eukaryote-specific extension. Mol. Cell. 2015;58:854–862. doi: 10.1016/j.molcel.2015.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.