Abstract
Synthesis of ribosomal subunits in eukaryotes is a complex and tightly regulated process that has been mostly characterized in yeast. The discovery of a growing number of diseases linked to defects in ribosome biogenesis calls for a deeper understanding of these mechanisms and of the specificities of human ribosome maturation. We present the 19 Å resolution cryo-EM reconstruction of a cytoplasmic precursor to the human small ribosomal subunit, purified by using the tagged ribosome biogenesis factor LTV1 as bait. Compared to yeast pre-40S particles, this first three-dimensional structure of a human 40S subunit precursor shows noticeable differences with respect to the position of ribosome biogenesis factors and uncovers the early deposition of the ribosomal protein RACK1 during subunit maturation. Consistently, RACK1 is required for efficient processing of the 18S rRNA 3′-end, which might be related to its role in translation initiation. This first structural analysis of a human pre-ribosomal particle sets the grounds for high-resolution studies of conformational transitions accompanying ribosomal subunit maturation.
INTRODUCTION
Protein synthesis is universally ensured by ribosomes, which are complex molecular machines made of ribosomal RNAs (rRNAs) and several tens of ribosomal proteins (RPs). While the overall ribosome structure is remarkably conserved across the three kingdoms of life, eukaryotic ribosomes are bigger and more complex than their prokaryotic counterparts. Compared to prokaryotes, human ribosomes contain ∼2650 additional nucleotides forming expansion segments of rRNA, and 26 supplementary RPs (1). Ribosome synthesis in eukaryotes has evolved into a sophisticated RNA processing and protein assembly process that requires several hundreds of trans-acting factors, hereafter called ribosome biogenesis factors (RBFs) (for review, (2,3)). The production of the two eukaryotic ribosomal subunits starts with the synthesis of a large ribosomal RNA precursor in the nucleolus that associates co-transcriptionally with most RPs and RBFs. This common precursor is then cleaved to generate the pre-40S and pre-60S particles, which then follow independent maturation pathways. The early nuclear pre-40S particles are rapidly exported to the cytoplasm (4), carrying a limited list of RBFs that are gradually released upon action of protein kinases (5–10). Several cytoplasmic maturation steps have been distinguished in yeast and in mammalian cells, until the final processing of the 18S rRNA 3′ end (7,9–13).
Defects in ribosome synthesis are now considered pathogenic in a growing family of genetic diseases or acquired syndromes, termed ribosomopathies (14,15). Ribosome biogenesis defects have been associated with human genetic diseases, such as Diamond-Blackfan anemia (DBA), Shwachman–Diamond syndrome or the Treacher–Collins syndrome. The cellular stress resulting from defective ribosome biogenesis triggers response pathways, including p53 activation and cell cycle arrest, which appear to be fatal to some physiological processes. Next generation sequencing techniques have recently revealed the presence of mutations in ribosomal proteins in several types of cancer, e.g. T-cell acute lymphoblastic leukemia and gastric cancer (16,17). Mutations in ribosomal protein genes may confer a familial predisposition to cancer, as shown with the link between RPS20 mutation and colon cancer (18). Understanding these inherited and sporadic pathologies requires an exhaustive characterization of ribosome biogenesis mechanisms in human cells.
Our knowledge of ribosome biogenesis in mammalian cells is only limited in comparison with the wealth of studies performed in baker's yeast. Advances in gene expression technologies in mammalian cells have recently unraveled substantial differences in the mechanisms of mammalian pre-ribosomal RNA (pre-rRNA) processing (for review (19)). While many ribosome biogenesis factors appear to have conserved functions, new ones have only been described in human cells (20–22), and the dynamics of their association with pre-ribosomes sometimes differs compared to yeast (5,6,23,24), suggesting changes in the spatial coordination of pre-ribosomal assembly. These significant mechanistic differences are likely linked, at least in part, to the emergence of new regulatory pathways in metazoans during evolution.
The full understanding of the molecular interplay between the actors of ribosome biogenesis is currently limited by a poor knowledge of the structure of pre-ribosomal particles at various maturation stages. Here, we have used cryo-electron microscopy (cryo-EM) and single particle analysis to determine the 3D structure of a human cytoplasmic pre-40S particle purified by tagging the ribosome biogenesis factor LTV1. We were able to position 5 out of the 6 stably bound RBFs onto this pre-40S particle. Although globally similar to its yeast counterpart, this structure highlights some differences in the position of the ribosome biogenesis factors. In addition, it shows the presence of RACK1, while this protein was not previously detected at this maturation stage in yeast (25). We subsequently show that RACK1 is required for the efficient processing of the 18S rRNA 3′-end. Given the role of RACK1 in translation initiation, we discuss the hypothesis that this last maturation step is kinetically coupled to translation initiation.
MATERIALS AND METHODS
TAP purification of human pre-40S particles
HEK293 FlpIn TRex cell lines expressing HASt-GFP or HASt-LTV1 have been described previously (26). The HEK293 FlpIn TRex cell line expressing RPS2-StHA was generated as described (26). The protocol described by Wyler et al. was adapted to purify pre-ribosomal particles from HASt-LTV1 HEK293 FlpIn TRex cells. Expression of N-terminally HASt-tagged or C-terminally StHA-tagged bait proteins in stable HEK293 cells was induced with tetracycline 24 h prior to harvest. Cells were detached with PBS containing 0.5 mM EDTA and lysed in lysis buffer (10 mM Tris–HCl, pH 7.5, 100 mM KCl, 2 mM MgCl2, 1 mM DTT, 0.5% NP-40, supplemented with protease and phosphatase inhibitors) using a dounce homogenizer. Lysed cells were centrifuged (4500 g, 12 min) and the lysate was incubated with StrepTactin beads (IBA) for 30 min in an overhead shaker. For electron microscopy studies, beads were washed six times with TAP buffer and eluted by incubation in TAP buffer supplemented with 2.5 mM d-desthiobiotin (Sigma). For subsequent analyses by silver staining and western blotting, beads were washed three times with TAP buffer (10 mM Tris–HCl, pH 7.5, 100 mM KCl, 2 mM MgCl2, 1 mM DTT, protease and phosphatase inhibitors) and bound material was eluted with SDS sample buffer in Mobicol spin columns (MoBiTec). Antibodies against RPL23a, NOB1, RPS3 and TSR1 have been described previously (5,9). Anti-RPS10 (ab151550) and anti-RPS26 (ab104050) antibodies were purchased from Abcam.
Cryo-electron microscopy
For cryo-EM, 3.5 μl of sample (at a final RNA concentration of ∼20 ng/μl as estimated by Nanodrop measurements), were deposited onto freshly glow-discharged holey carbon grids (Quantifoil R2/1 or R2/2) and loaded into the thermostatic chamber of a Leica EM-GP automatic plunge Freezer, set at 20°C and 95% humidity. Excess solution was blotted for 1.8–2.1 s with a Whatman filter paper no. 1, and the grid was immediately flash frozen in liquid ethane, cooled at -183°C. Grids were prepared and systematically checked at METI, the Toulouse cryo-EM facility. Cryo-EM image acquisition was performed at the Netherlands Center for Electron Nanoscopy (NeCEN), using a FEI Titan Krios Microscope operating at 300 kV. Using the FEI EPU software, images were automatically recorded on a Falcon II direct electron detector, with an electron dose of ∼30 e−/Å2, at a nominal magnification of 59 000× corresponding to a calibrated pixel size of 1.136 Å/pixel. Nominal defoci ranged from −1.6 to −2.5 μm.
Single particle analysis
Micrographs not suitable for further processing (i.e. zones containing no ice, or massive contamination) were discarded by visual checking. Contrast transfer function (CTF) parameters of the remaining micrographs were estimated using CTFFIND3 (27). Based on these parameters, micrographs showing signs of drift or astigmatism were removed, resulting in a dataset of 3812 micrographs. 103 247 particles were selected with e2boxer, the semi-automated particle picking command from EMAN2 (28), extracted in boxes of 384 × 384 pixels, which were subsequently binned by a factor 2 for faster computations. 2D and 3D classifications, as well as refinements were then performed using RELION 1.3 (29) (see Supplementary Material and Supplementary Figure S1). The consensus 3D structure here described was obtained from 54,436 particles, at a resolution of 22.9 Å for a FSC = 0.143, according to the gold-standard FSC procedure (30,31). This 3D structure was then masked with an ad hoc binarized soft mask (with a 10 pixels fall-off), by using the post-process command of RELION 1.3, which gave rise to a 19.4 Å resolution structure at FSC = 0.143 (see Supplementary Figure S2). Local resolution estimation was performed on the unsharpened final map with the ResMap software package (32), using a 2 Å step search between 5 and 30 Å resolution.
All 3D maps were displayed using Chimera (33), with a density threshold of 2σ, unless otherwise stated. For the rigid body docking of the mature 40S subunit, an atomic model of the human mature 40S subunit devoid of RPS26 and RPS10 was derived from the 3D structure published by Anger et al. (PDB code: 4V6X (1)). This structure was fitted into the HASt-LTV1 pre-40S particle density map, using the ‘Fit’ command of Chimera. For difference map computations, and the subsequent fitting of atomic models of various RBFs, the modified mature 40S subunit model was low-pass filtered to 21 Å resolution with the ‘Molmap’ command of Chimera. Difference maps between the 3D structures of the HASt-LTV1 pre-40S particle and the modified mature 40S subunit were calculated using either the ‘vop subtract’ option in Chimera, or the Diffmap program by N. Grigorieff. Both options yielded similar results. The difference map was superimposed to the modified mature 40S structure, and available X-ray or NMR structures of maturation factors were manually positioned in the density zones of difference maps according to their localization in yeast, as assessed by CRAC and cryo-EM analyses (8,25,34). This crude fitting was then refined with the ‘fit in map’ option of Chimera.
Knockdown of gene expression with small interfering RNAs
HeLa cells were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum and 1 mM sodium pyruvate (Sigma). The following 21-mer siRNAs (Eurogentec), whose efficiency was verified by qPCR, were used to knock down expression of the following human mRNAs in HeLa cells: RACK1 (GenBank accession number: NM_006098): 5′-CUCUGGAUCUCGAGAUAAAdTdT-3′ (siRNA rack1-1), 5′-CUGGGUGUGUGCAAAUACAdTdT-3′ (siRNA rack1-2), 5′-CCCACUUUGUUAGUGAUGUdTdT-3′ (siRNA rack1-3); RPS10 (GenBank accession number: NM_001203245): 5′-GAACCGGAUUGCCAUUUAUdTdT-3′ (siRNA rps10); RPS15 (GenBank accession number: NM_001308226): 5′-UCACCUACAAGCCCGUAAAdTdT-’3 (siRNA rps15); RPS26 (GenBank accession number: NM_001029): 5′-GGACAAGGCCAUUAAGAAAdTdT-3′ (siRNA rps26) (35). The siRNA solution (500 nM final concentration) was added to the cell suspension incubated on ice (10 × 106 cells in 200 μl of Na phosphate buffer, pH 7.25, 250 mM sucrose, 1 mM MgCl2). Electro-transformation was performed with square pulses at 240 V with a Gene Pulser (Bio-Rad) as described (36). A scramble siRNA (siRNA-negative control duplex; Eurogentec) was used as a control. Cells were then plated and grown in DMEM at 37°C for 48 h.
Analysis of ribosomes by sucrose density gradient centrifugation
The cells were treated for 10 min with 100 μg/ml cycloheximide (Sigma) 48 h post-transfection. After washing with PBS containing cycloheximide, they were mechanically disrupted with a Dounce homogenizer in buffer A (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT) containing cycloheximide. The cytoplasmic fraction was recovered by centrifugation for 10 min at 1000 g and at 4°C, and then further clarified by two successive centrifugations at 10 000 g. A volume corresponding to 1 mg total proteins was loaded on 10–50% (w/w) sucrose gradients, prepared with a Gradient Master former (BioComp Instruments). After centrifugation at 4°C and at 36 000 rpm for 105 min in a SW41 rotor (Optima L100XP ultracentrifuge; Beckman Coulter), the fractions were collected at OD254 nm with a Foxy Jr. gradient collector (Teledyne Isco).
RNA extraction and northern blot analysis of rRNA precursors
Total RNAs were extracted with Trizol from cell pellets containing 20–30 × 106 cells. For Northern blot analyses, RNAs were dissolved in formamide, denatured for 10 min at 70°C and separated on a 1.2% agarose gel containing 1.2% formaldehyde and Tri/Tri buffer (30 mM triethanolamine, 30 mM tricine, pH 7.9) (3 μg RNAs/lane). They were transferred to a Hybond N+ nylon membrane (GE Healthcare) by passive transfer and UV cross-linked. After pre-hybridization at 45°C in 6× SSC, 5× Denhardt's solution, 0.5% SDS, 0.9 μg/ml tRNA, the membrane was incubated overnight with the 5′-radiolabeled oligonucleotide probe. The following probes were used: 5′-ITS1 (5′-CCTCGCCCTCCGGGCTCCGTTAATGATC-3′); ITS2 (a mixture of ITS2b: 5′-CTGCGAGGGAACCCCCAGCCGCGCA-3′ and ITS2d/e: 5′-GCGCGACGGCGGACGACACCGCGGCGTC-3′); 18S (5′-TTTACTTCCTCTAGATAGTCAAGTTCGACC-3′); 3′ 18S (5′-TAATGATCCTTCCGCAGGTTCACCTACG-3′); 28S (5′-CCCGTTCCCTTGGCTGTGGTTTCGCTAGATA-3′). After washing for 10 min in 2× SSC, 0.1% SDS and in 1× SSC, 0.1% SDS, the membrane was exposed. Signals were acquired with a Typhoon Trio PhosphorImager (GE Healthcare) and quantified using the MultiGauge software.
RNase H cleavage assay and 3′ RACE analysis
For RNase H cleavage assays, 4 μg total RNAs were denatured at 95°C for 3 min with a reverse probe (5′-TTTACTTCCTCTAGATAGTCAAGTTCGACC-3′; 1 μl at 100 μM) hybridizing in the 3′ end of 18S rRNA (37). After annealing by cooling down to room temperature for 10 min, the reaction mixture was diluted to 30 μl with a reaction mix containing 1× RNase H reaction buffer, 65 μM DTT, 0.5 U/μl RNasin (Promega), and 50 U RNase H (New England Biolabs), and incubated at 37°C for 30 min. The reaction was then blocked by addition of 0.3 M sodium acetate, pH 5.2, and 0.2 mM EDTA. After phenol extraction and RNA precipitation, the samples were analyzed on a 12% polyacrylamide gel containing urea and TBE buffer. The samples were then electro-transferred to a nylon membrane (Hybond N+; Amersham) and revealed with radiolabeled probes.
3′ RACE analysis was adapted from Kiss and Filipowicz (38). Primers ITS1-Hs-RACE (5′- CGCGAATTCGATCATTAACGGAGCCCGGAG-3′) and ITS1+2-Hs-RACE (5′-CGCGAATTCACCTGCGGAAGGATCATTAAC-3′), which spans the 18S-ITS1 junction up to nucleotide 13 and to nucleotide 2 in the ITS1 respectively, were used for PCR amplification. The amplified fragments were sub-cloned, automatically sequenced and aligned with Jalview software (39).
Fluorescence in situ hybridization
Cells grown on glass cover slips were fixed with 4% paraformaldehyde in PBS for 15 min. After washing in PBS, the cells were permeabilized in 70% ethanol overnight at 4°C. They were then re-hydrated twice for 5 min in 2× SSC containing 10% formamide. The precursors to the 18S rRNA were localized by fluorescence in situ hybridization (FISH) with a 5′ ITS1 probe coupled to Cy3 (35). The cover slips were washed in 2× SSC containing 10% formamide, and DNA was counter stained with Hoechst 33342 (Invitrogen), before mounting in Mowiol 4.88 (PolyScience, Inc.). The slides were observed with an inverted microscope (IX-81; Olympus), equipped with a ×100, oil immersion objective (Plan Apochromat, 1.4 NA; Olympus) and an Orca Flash 4.0 camera with a pixel size of 6.5 μm (Hamamatsu), driven by MetaMorph (MDS Analytical Technologies).
RESULTS
Cryo-EM 3D structure of a human cytoplasmic pre-40S particle
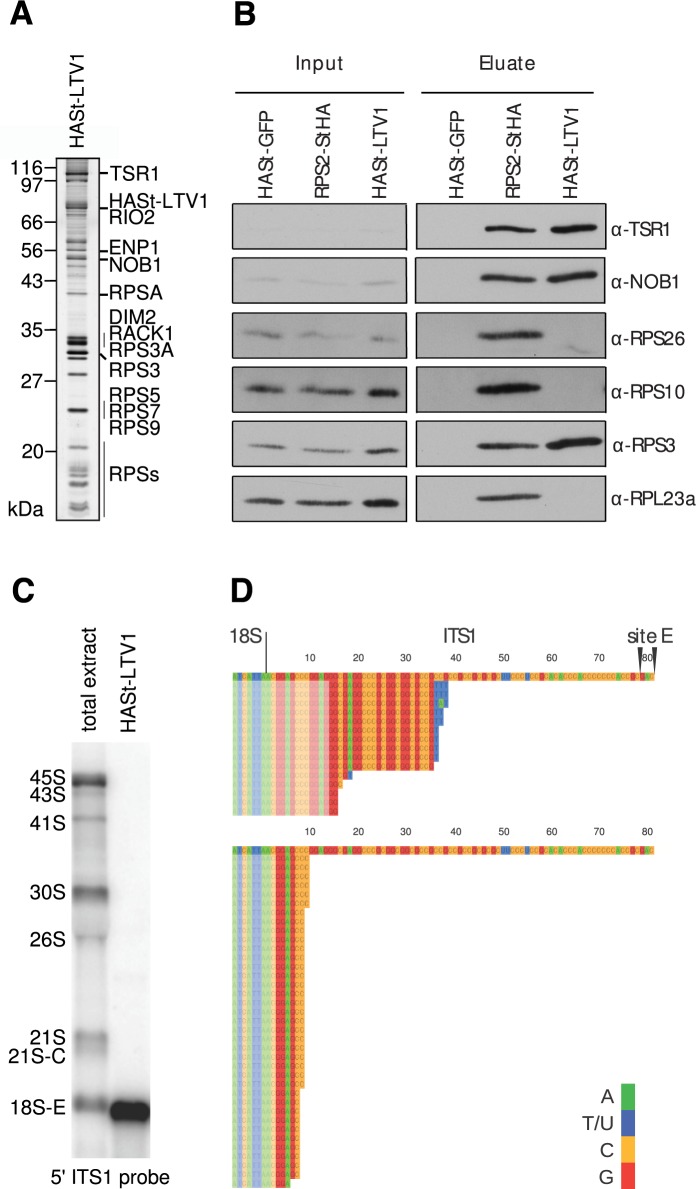
In order to decipher the cytoplasmic steps of the small ribosomal subunit assembly, human pre-40S particles were purified by tandem affinity chromatography using the ribosome biogenesis factor LTV1 as bait (26) (Figure 1). Previous proteomic and Western blot analyses have shown that HASt-LTV1 pre-40S particles contain at least six stably bound RBFs, namely HASt-LTV1, ENP1/Bystin, TSR1, RIO2, NOB1 and DIM2 (26). A migration pattern similar to that previously published was observed after SDS-PAGE and silver staining of the HASt-LTV1 purification (Figure 1A). Furthermore, Western blot analyses revealed that pre-40S particles purified by HASt-LTV1 are devoid of RPS10 and RPS26 (Figure 1B). This is similar to yeast pre-40S particles purified using tagged Rio2 as bait (25). Although LTV1 associates to pre-40S particles in the nucleus, this protein is mostly cytoplasmic (5,20). This suggests that the purified HASt-LTV1 pre-40S particles are predominantly associated to 18S-E pre-rRNA, the last precursor to 18S rRNA, which is released by cleavage at site E. Just after the cleavage, this precursor bears an ITS1 extension of 78–81 nucleotides in length, which is rapidly trimmed by a 3′-5′ exonuclease and exported to the cytoplasm (4,39). Production of 18S-E pre-rRNA is a prerequisite for the export of pre-40S particles, as attested by the absence of earlier 18S precursors in the cytoplasm. Consistently, both Northern blot and 3′-RACE experiments showed that pre-40S particles isolated by LTV1 affinity purification contain 18S-E pre-rRNAs bearing short 3′ ITS1 extensions (Figure 1C and D), as expected for cytoplasmic particles (39). A 3′ RACE primer annealing to 13 nucleotides in the ITS1 (Figure 1D, upper panel) revealed that the 3′ extensions were 15–35 nucleotides long, and most of them bore 1–3 non-templated uridines, which are cytoplasmic marks for the 18S-E pre-rRNAs (39). A 3′ RACE primer spanning only 2 nucleotides in the ITS1 highlighted 5–9 nt extensions, devoid of non-templated uridines (Figure 1D, lower panel). These data demonstrate that the cytoplasmic pre-40S particles used in this study contained heterogeneous 18S-E pre-rRNAs with very short ITS1 extensions.
Figure 1.
Biochemical characterization of human HASt-LTV1 pre-40S particles. (A), HASt-LTV1 purification product analyzed by silver staining. The bands from the SDS-PAGE gel were annotated according to (26). (B), Western blot analysis of RBFs and RPs in HASt-LTV1 pre-40S particles. Proteins copurifying with HASt tagged RPS2 or GFP are shown as controls. Total cell extracts (Input) and the corresponding purification products (Eluate) are displayed on the left and right panels, respectively. (C), Northern blot analysis of the RNA content of the HASt-LTV1 pre-40S particles revealed with the 5′ ITS1 probe (0.2μg/lane). Total RNAs were loaded for comparison (1.5 μg/lane). (D), 3′ RACE analyses of the RNAs extracted from HASt-LTV1 pre-40S particles. Sequences were aligned relative to the 3′ end of 18S rRNA followed by the first 81 nt of the ITS1, which are left by endonucleolytic cleavage at site E (top sequences). PCR was performed with two different forward primers spanning 13 nt (top panel) or 2 nt (bottom panel) into the ITS1, as indicated by semi-transparent blocks.
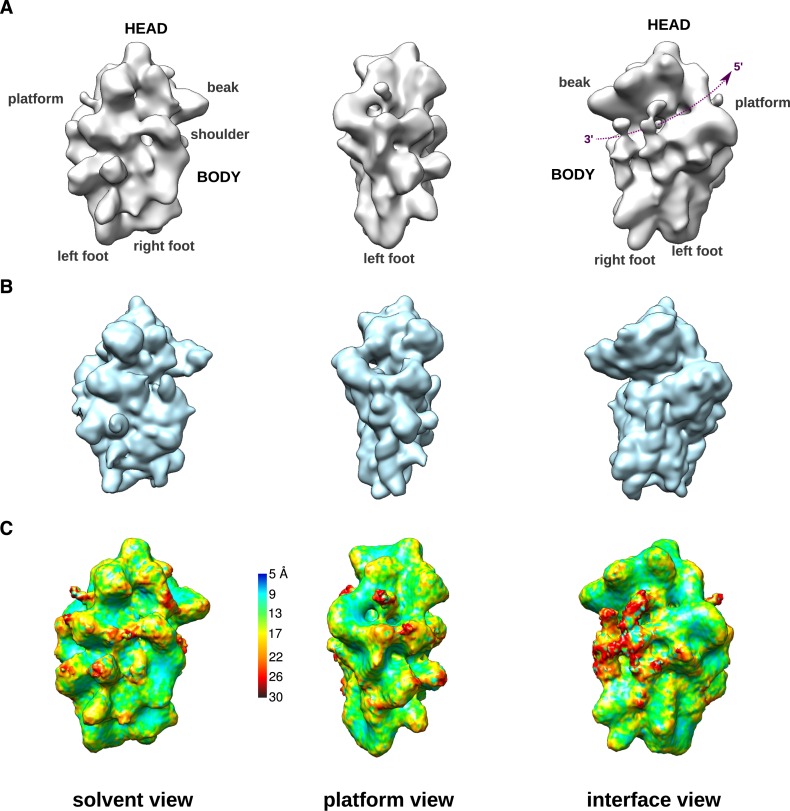
The 3D structure of purified HASt-LTV1 particles was determined using cryo-EM and single particle analysis (Figure 2A), yielding a 19 Å resolution according to the gold-standard FSC criterion (30,31), (Supplementary Figure S2A). This consensus 3D structure was obtained from ∼58 000 particles selected from an initial dataset of ∼103 000 particles. Further attempts to distinguish a finite number of structural conformers to further improve resolution were unsuccessful, which we assign to the strong conformational heterogeneity of these particles (see Supplementary Material and Supplementary Figure S1).
Figure 2.
Cryo-EM 3D structure of human HASt-LTV1 pre-40S particles. (A), Surface views of the cryo-EM 3D structure of HASt-LTV1 pre-40S particles, solved at 19 Å resolution. On the interface view, the mRNA groove located between the head and the body has been outlined by a purple dotted line arrow. (B), Surface views, displayed at identical angles, of the 3D structure of the human mature 40S subunit (PDB code: 4V6X) devoid of RPS10 and RPS26. This model was scaled to the same pixel size as in (A) and low-pass filtered at 21-Å resolution. (C), Surface views of the unfiltered final map of HASt-LTV1 pre-40S particles colored according to local resolution calculated with ResMap (32).
This structure resembles the mature small ribosomal subunit in its overall morphological features and dimensions, with some apparent additional densities (Figure 2A, to be compared to the mature 40S subunit structure filtered at an equivalent resolution on Figure 2B). Based on local resolution assays, the median resolution of the structure is around 15 Å, with seemingly more resolved regions forming small, scattered areas in the center of the structure (blue to green regions on Figure 2C and Supplementary Figure S2B). On the contrary, other parts of the structure harbor a lower level of detail, with resolution down to 26 Å (in red). Most of these ‘less-ordered’ regions correspond to extra-densities that are absent from the mature 40S subunit and that have been attributed to ribosome biogenesis factors (see below). This suggests that these proteins are loosely tethered to the pre-40S and may move around their anchoring point, and/or that their presence is substoichiometric among the observed pre-40S particles.
Structures of the human and yeast cytoplasmic pre-40S particles are globally conserved
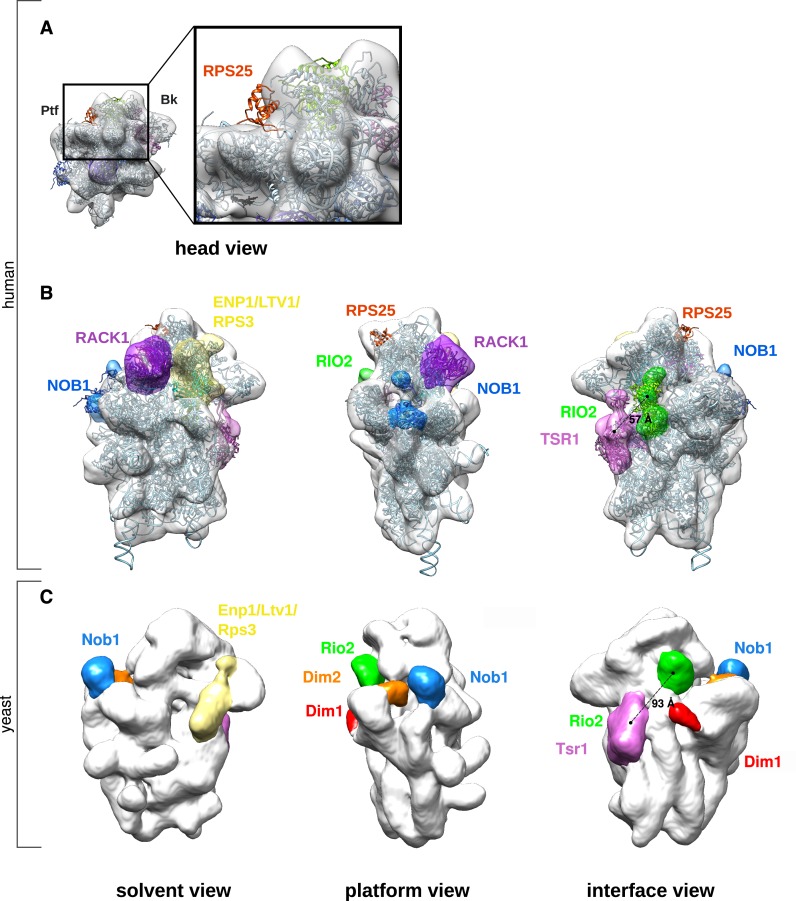
To analyze the structural model in more details, we performed rigid body docking of a 3D structure of the human 40S subunit structure (PDB code: 4V6X) into the pre-40S electron density map (Figures 3 and 4). RPS10 and RPS26, which are absent from these pre-40S particles (Figure 1B), were removed from the docked 40S subunit structure. The pre-40S particle electron density map accommodates all the components of the mature 40S subunit, with the exception of RPS25, whose structured residues are clearly out of this density map (Figure 3A, red ribbon). Nevertheless, previous mass spectrometry analyses have shown that RPS25 is associated with HASt-LTV1 (26). This suggests that, at this maturation step, RPS25 is not in its final position.
Figure 3.
Interpretation of the cryo-EM structure of human HASt-LTV1 pre-40S particles. (A), The electron density map of HASt-LTV1 pre-40S particles (white density) was fitted with the quasi-atomic 3D structure of the mature human 40S subunit devoid of RPS10 and RPS26 (ribbon style, pale blue unless otherwise stated). The view from the head (left panel) and the close-up of the region framed by a black rectangle (right panel) show that RPS25 in its mature conformation (red ribbon) cannot be fitted in the pre-40S density map. (B), Ribosome biogenesis factors associated to HASt-LTV1 pre-40S particles were positioned on the human pre-40S 3D structure according to CRAC data (8,34) and cryo-EM structures (10,25) obtained in yeast. Segmentation of the HASt-LTV1 pre-40S map was performed with the ‘Segger’ command (75) from Chimera (33). Available atomic models of ribosome biogenesis factors were fitted into their corresponding densities zones by rigid body docking. (C), As a comparison, the electron density map of yeast Rio2-TAP pre-40S particle (EMDB accession code: 1927) was aligned to the human HASt-LTV1 pre-40S particle, by using the ‘Fit’ command of Chimera. The density zones attributed to pre-ribosomal factors are displayed in similar colors in (B) and (C).
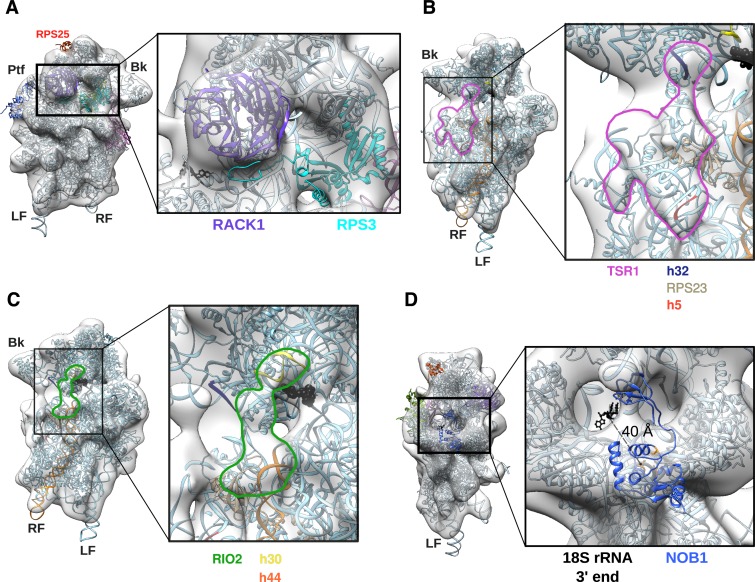
Figure 4.
Details of the 3D model of human HASt-LTV1 pre-40S particle. (A), Solvent view of the 3D model (left panel) and close-up of the framed head region (right panel). (B-C), Interface views (left panel) and close-ups (right panel) on the electron density map domain of the HASt-LTV1 pre-40S particle attributed to TSR1 (B, pink contour) and RIO2 (C, green contour). Both domains have been left empty to see the contacts with the components of the mature 40S subunit, i.e. for TSR1 (B): rRNA segments of h32 (dark blue), and h5 (red) and RPS23 (light grey). (C), Interactions between RIO2 and rRNA: h30 (yellow), and upper part of h44 (orange). Cross-linking site of yeast Rio2 on h31 as found by CRAC analyses (34) is represented by black beads. (D), Platform view of the HASt-LTV1 pre-40S 3D model (left panel) and close-up (right panel). Atoms of the 3′ end of 18S rRNA are represented in black. The NMR structure of P. horikoshii Nob1 (PDB code: 2LCQ) is in medium blue, while atoms of its catalytic site are displayed in orange.
This docking experiment also highlighted several zones of extra-densities on the pre-40S structure compared to the mature one (Figure 3B). One is located between the beak and the shoulder region and is terminated by a horn-like structure on the head of the pre-40S particle (yellow). Another partly fills the center of the platform domain (blue). In addition, two elongated extra-density regions are located on the interface domain, near the A and P tRNA interaction sites (pink and green). These extra-densities are also clearly visible when computing a difference map between the pre-40S and the docked mature human 40S subunit (Supplementary Figure S2C). They roughly correspond to the locations assigned to RBFs in 3D EM structures of yeast pre-40S particles purified at an equivalent maturation step (10,25) (Figure 3C), but display significant morphological differences as detailed below.
The ENP1:LTV1:RPS3 module
The horn on the head of the pre-40S particle (Figure 3B, pale yellow) likely corresponds to the domain formed by ENP1, LTV1 and RPS3 (40). This hypothesis is supported by the known position of RPS3 in the mature 40S subunit structure, CRAC analyses of the yeast pre-40S ribosomal RBFs (8,34), as well as the localization of these factors on the yeast pre-40S subunit (Figure 3C). Rigid body docking indicates that the majority of the RPS3 structure, as found in the mature ribosome, fits into the beak region of the human pre-40S particle (Figure 4A). This is a notable difference with the yeast cryo-EM structure of Rio2-TAP pre-40S particles, which does not accommodate RPS3 in its mature position, although RPS3 was detected by mass spectrometry (25). Indeed, volume-to-volume fitting assays show that the human pre-40S structure displays more density in the beak region than its yeast counterpart (Supplementary Figure S2D, grey arrowhead on surface view). Ltv1, Enp1 and Rps3 are known to form a very stable protein complex, which can be untethered from the beak of pre-40S particles by treatment with high concentrations of salt (40). This module may be more rigidly anchored to human pre-40S particles compared to its yeast counterpart, thus representing more electron density in the beak region of the human pre-40S structure.
The 60S interface module: TSR1 and RIO2
By analogy with the EM data obtained in yeast (25), we attributed the two extra-densities domains located on the 60S interface to TSR1 for the density closest to the beak and the A tRNA binding site (Figure 3B, pink), and to RIO2 for the density at the center of the pre-40S subunit (Figure 3B, green), respectively. Both proteins seem to bridge the head and the body of the pre-40S particle, and occlude the mRNA decoding groove (Figures 2A and 3B (interface views) and Figure 4B). In the human model, RIO2 and TSR1 appear significantly closer (57 Å) than in the yeast model (93 Å) (compare interface views in Figure 3B and C). The density attributed to TSR1 connects rRNA helices h32/34, located on the head, to RPS23 and rRNA helix h5, situated on the body of the small subunit precursor (Figure 4B). This density slightly bulges outwards and is closer to the beak than to the center of the body. In this position, TSR1 is expected to sterically hamper the binding of translation initiation factors such as eIF1A, DHX29 and eIF5b on the small ribosomal subunit (41–43).
The shape and position of the extra-density corresponding to kinase RIO2 are not strictly similar to yeast (compare Figure 3B and C, interface views, green; see also Supplementary Figure S2D, white and dark purple arrowheads on interface view). In yeast, Rio2 has been positioned perpendicularly to helix h44, nested into the mRNA groove over the platform domain and close to tRNA binding site E (25,44). Here, human RIO2 is parallel to helix h44, covering its upper part and the tRNA binding site P, and linking h44 to h30 on the beak of the particle (Figure 4C). Rigid body docking assays of the X-ray structure of Rio2 from fungus Chaetomium thermophilum (PDB code: 4GYI (44)) into the extra-density zone attributed to RIO2 yielded no satisfactory result (Supplementary Figure S3A, green). This suggests either a substoichiometric binding of this maturation factor on the pre-40S particle, or that RIO2 wobbles around an anchoring position.
The platform module: NOB1 and DIM2
The general aspect of the platform on the human pre-40S structure is similar to its yeast counterpart, with NOB1 forming a knob obstructing the platform ‘valley’ (Figure 3B and C). Nob1 is the enzyme ensuring the endonucleolytic cleavage of the last precursor to 18S rRNA in yeast (45–47) and its homolog is likely to play a similar role in human (39,48). The NMR structure of Nob1 from archaea P. horikoshii (PDB code: 2LCQ; Supplementary Figure S3B, blue) revealed two structurally independent domains, separated by a flexible linker (49). Its catalytic activity is carried by aspartates 12 and 82, and several other amino acids lining the Mn2+ pocket of the PIN domain (45,47,49). The PIN domain represents a large N-terminal region, whereas the smaller, globular C-terminal part of the protein is formed by the zinc ribbon domain. This archaeal Nob1 is composed of only 161 amino acids, which are highly conserved in its human counterpart, made of 412 amino acids. Using PSIPRED (50) and BioSERF (51), we performed secondary structure predictions and built a homology 3D model. The result suggests that the 3D structure of human NOB1 is also composed of these two conserved structural domains, interspersed by two long, unstructured stretches of more than 150 amino acids: a PIN domain-intrinsic loop and a C-terminal extension after the Zn-ribbon domain (Supplementary Figure S3B, yellow). Although NOB1 is ∼2.5 times heavier than its archaeal counterpart, both 3D structures at a resolution of ∼19 Å should give rise to similar electron density maps showing only the structured domains. On the basis of this similarity, we docked the archaeal Nob1 NMR solution structure into the human pre-40S electron density map (Figure 4D). The fitting experiments indicated that the zinc ribbon domain could form the small outgrowth poking out of the head. This position is in good agreement with recent findings showing that the zinc ribbon domain is sufficient to interact with helix 40 of the rRNA (34,49). The N-terminal PIN domain of NOB1 would then be nested on the platform ‘valley’. In this model, the 3′ end of 18S rRNA and the catalytic site of NOB1 are separated by at least 40 Å. This large distance indicates that at this maturation step, NOB1 cannot perform its endonucleolytic activity on the 18S-E pre-rRNA. Thus, we postulate that, as proposed for yeast (11,46), NOB1 must change position or undergo a conformational change of an unknown nature to reach and cleave the 3′ end of the 18S rRNA.
DIM2, also known as PNO1 (Partner of Nob1), is associated with NOB1 until a very late maturation step in yeast and human pre-40S particles (6,7,9,13). Though human DIM2 is associated with HASt-LTV1 particles, as shown by mass spectrometry and Western blot analyses (26), we did not distinguish any extra density zone large enough to position DIM2 with certainty. In yeast, Dim2 was assigned to a density in the vicinity of the platform/back of the head interface region, both by 3D EM structure (Figure 3C, orange) (25) and later on by CRAC analyses (8). Thus, because the platform domain of the human pre-40S particle appears globally bigger than its mature counterpart, we assume that DIM2 is present somewhere in this region of the particle (see Discussion).
RACK1 is stably bound to various precursors of the human small ribosomal subunit
The so-called ‘ear region’ of the human pre-40S structure presents a density of ∼67 nm3 that can almost entirely accommodate RACK1 (Figures 2A, 3B and 4A). This represents a distinct feature from yeast, since Asc1p/Rack1 was not found in the structure of yeast Rio2-TAP pre-40S particles and was proposed to associate very late in the maturation pathway (12,25). Nevertheless, RACK1 has been previously identified by mass spectrometry as part of human cytoplasmic pre-40S particles isolated by HASt-LTV1, as well as in nucle(ol)ar particles purified by using NOC4 (26) or C21ORF70 (data not shown) as baits. Moreover, RNAi screening for proteins involved in ribosome maturation revealed that RACK1 knockdown impairs the release of ENP1 from cytoplasmic pre-40S particles (20).
To confirm the robustness of the protruding density provisionally assigned to RACK1, we processed the same dataset of imaged particles to calculate a second 3D reconstruction. The 3D structure of the human 40S subunit (PDB code: 4V6X), devoid of RACK1, and low-pass filtered to 60 Å resolution, was used as initial reference. As expected from the presence of RACK1 on HASt-LTV1 particles, a protruding density in the ear region was restored in this 3D reconstruction (Supplementary Figure S3C, domain framed by a black ellipse; compare to the structure of the mature 40S subunit devoid of RACK1 and filtered to 21 Å resolution in Supplementary Figure S3D). The volume of this density was smaller (∼35 nm3), than in the reconstruction obtained by using the 3D structure of the human 40S subunit including RACK1 as initial reference. This suggests that the binding of RACK1 to the HASt-LTV1 pre-40S particles is either substoichiometric or that its position on the particle is fluctuating around a tethering area.
RACK1 depletion affects the ultimate steps of 18S-E pre-rRNA processing
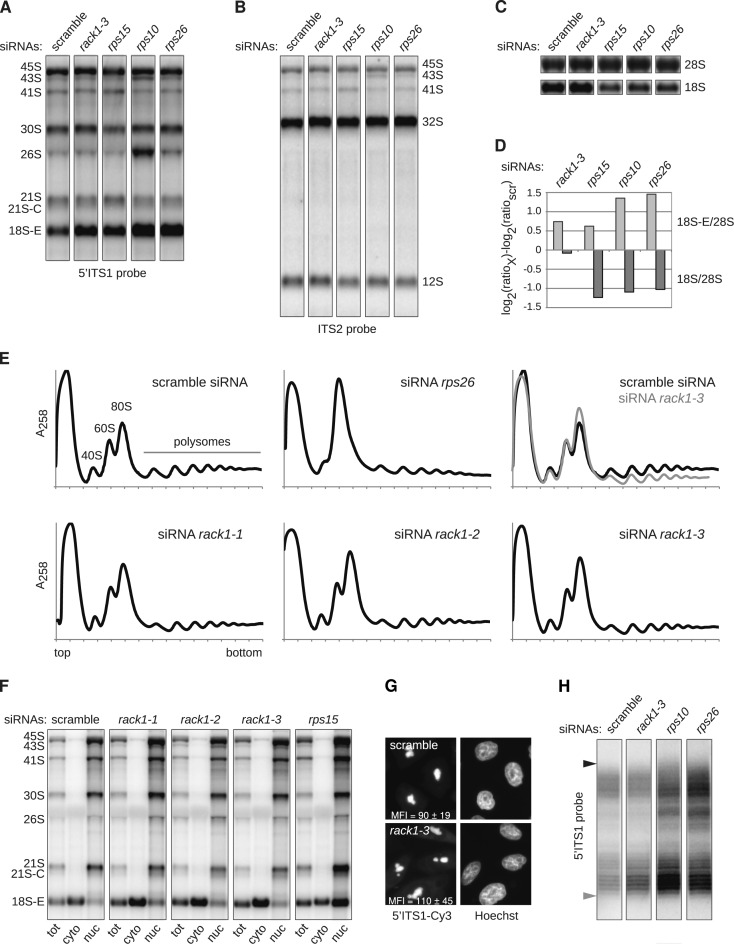
The presence of RACK1 in human pre-40S particles prompted us to investigate its role in 18S pre-rRNA processing. Depletion experiments were conducted with three different siRNAs targeting RACK1 (85–95% reduction of RACK1 mRNA levels, Supplementary Figure S4) and compared to the action of siRNAs directed against late-acting RPSs. After depletion for 48 hours, Northern blot analyses showed an accumulation of 18S-E pre-rRNA in all these samples compared to control cells treated with a scramble siRNA (Figure 5A and F). In contrast, the levels of both the precursors to the large subunit (12S and 32S pre-rRNAs) and the mature 28S rRNA were unchanged compared to control cells (Figure 5, B and C). After RACK1 knockdown, the 18S-E/28S ratio was comparable to that obtained after depletion of RPS15 (4,52), which is required for nuclear export of pre-40S particles, but not as high as that of cells lacking RPS10 or RPS26, two proteins needed for the cytoplasmic steps of 18S-E pre-rRNA processing (Figure 5A and D). However, this accumulation of 18S-E pre-rRNA after RACK1 knockdown did not lead to a strong imbalance of the 18S/28S rRNA ratio, which is observed upon knockdown of most RPSs, as illustrated with RPS10, RPS15 or RPS26 depletion (Figure 5C and D). These results indicate that RACK1 affects 18S-E pre-rRNA processing, but does not lead to a strong defect in 18S rRNA production. A similar phenotype was previously described for RPS25, while depletion of the 31 other RPSs strongly impairs 18S rRNA synthesis and 40S subunit accumulation (35). Consistently, loss of RACK1 only led to a moderate decrease of the free 40S subunit peak observed by polysome analysis, in contrast with the visible diminution of free 40S subunits yielded by RPS26 depletion (Figure 5E). However, RACK1-depleted cells showed a larger monosome peak (Figure 5E, overlay), together with a marked decrease of the polysome levels, compared to control cells. This phenotype suggests that knockdown of RACK1 has a general effect on translation.
Figure 5.
Analysis of the role of RACK1 in ribosome biogenesis. (A–C) Northern blot analysis of total RNAs extracted from HeLa cells treated for 48 h with the indicated siRNAs. Precursors to the 18S and 28S rRNAs were successively revealed with the 5′ ITS1 (A) and ITS2 probes (B), before visualizing the 18S and 28S rRNAs (C). (D) After quantification, the levels of 18S-E pre-rRNA and 18S rRNA relative to 28S rRNAs were assessed for each sample (76) and expressed relatively to the control (scr). (E), Cytoplasmic fractions prepared from siRNA-treated cells were separated on 10–50% sucrose gradients in order to evaluate the impact of the corresponding knockdown upon ribosomal subunit synthesis and assembly. (F), After treatment with different siRNAs for 48 h, HeLa cells were subjected to sub-cellular fractionation, and the 18S-E pre-rRNA precursors contained in each fraction were evidenced by northern blot. (G) The intracellular localization of the precursors to the 18S rRNA was also assessed by FISH experiments conducted with a 5′ ITS1 probe. The mean fluorescence intensity (MFI) in the cytoplasm was measured in 316 pixel (∼1.3 μm2) regions distributed over 18–20 cells taken with the same exposure time. The MFI values displayed in the picture correspond to the mean of 48 measurements (Student's t-test: P = 0.0005). (H) RNA samples displayed in (A) were hybridized to a primer complementary to the 3′ end of the 18S rRNA, and subjected to RNase H treatment prior to separation on a denaturing 12% polyacrylamide gel. After transfer to a nylon membrane, the 3′ extremities of the various 18S-E species were revealed with a 5′ ITS1 probe. The position of the longest 18S-E precursors, corresponding to an 81-nt ITS1 extension, is indicated by a black arrowhead. The shortest 18S-E pre-rRNAs revealed by the 5′ ITS1 probe corresponded to ITS1 extensions around 20 nt (gray arrowhead).
The maturation of the 18S-E pre-rRNA is a multi-step process starting in the nucleus and ending in the cytoplasm, which involves 3′-5′ exoRNase(s) and endonuclease NOB1 (21,39,48). In order to determine more accurately the step impacted by RACK1 depletion, we performed cell fractionation analyses. The loss of RACK1 induced accumulation of 18S-E pre-rRNA in the cytoplasm, whereas depletion of RPS15, a RPS required for nuclear export competency, resulted in nuclear accumulation of this species (Figure 5F). FISH experiments conducted with the 5′ ITS1 probe confirmed this increase of 18S-E pre-rRNA in the cytoplasm upon depletion of RACK1 compared to control cells (Figure 5G). Therefore, the loss of RACK1 did not noticeably affect nuclear export of 18S-E pre-rRNA, but delayed its final processing at site 3 in the cytoplasm. To further address this point, RNase H digestion assays were performed on total RNA samples in order to compare the patterns of the 18S-E pre-rRNA 3′ extremities (Figure 5H). In the control, the 5′ ITS1 probe revealed a ladder of bands, corresponding to ITS1 extensions spanning from 81 nt (i.e. position of site E; black arrow) to around 20 nt (grey arrow), as expected from 3′-5′ exonucleolytic processing. Depletion of RPS10 or RPS26 led to a strong accumulation of long ITS1 extensions, together with shorter 18S-E pre-rRNA species, meaning that the trimming of the ITS1 extension was impeded in the absence of these RPSs. Depletion of RACK1 led to a pattern in which only 18S-E pre-rRNAs bearing short ITS1 extensions were enriched. This result suggests that the absence of RACK1 does not affect the exonucleolytic processing of the ITS1, but slows down the kinetics of the endonucleolytic cleavage at site 3 that produces 18S rRNA.
DISCUSSION
Human HASt-LTV1 pre-40S particles display an overall flexible structure
By using cryo-EM and single particle analysis methods, we have solved the 3D structure of an early cytoplasmic intermediate of the human small ribosomal subunit at 19 Å resolution. This resolution is in the same range as the two structures of yeast cytoplasmic pre-40S particles currently available (10,25), despite the considerable challenge of purifying these particles in much lower amount from human cells. A major obstacle to reach high resolution is the well-known intrinsic flexibility of the small ribosomal subunit (53,54). Indeed, our many attempts to perform 3D classification to increase resolution did not give rise to distinct conformers (Supplementary Material and Supplementary Figure S1). This leads us to conclude that HASt-LTV1 pre-40S particles display continuous structural heterogeneity (55), and that high flexibility of Ltv1-TAP pre-40S particles prevails over its compositional heterogeneity. Thus, describing the full heterogeneity and determining high resolution 3D structure(s) of such pre-40S particles will require a much larger number of particles. Moreover, the use of light chemical cross-linking might help to reduce their flexibility (56). Nevertheless, the 19 Å resolution 3D structure presented here provides sufficient information to propose the positioning of five ribosomal biogenesis factors and identify RACK1 as an unexpected component of these particles.
The interplay between RPS3, LTV1 and ENP1
A few regions of the pre-40S particle structure showed extra-densities compared to the mature 40S subunit. These likely correspond to the presence of ribosome biogenesis factors stably bound to the HASt-LTV1 pre-40S particle (26). The protein composition of these human pre-40S particles is globally similar to that of yeast pre-40S particles purified at the same maturation step (25); only the methyltransferase DIM1 (DIMT1L) is absent from human HASt-LTV1 pre-40S particles, while present on yeast Rio2-TAP pre-40S particles. This is in good agreement with the observation that base modification by DIM1/Dim1 is already performed in the nucleus in human cells, whereas it takes place in the cytoplasm in yeast (24,57). In accordance with this compositional similarity, extra-densities present in the human HASt-LTV1 pre-40S particles have been positioned in the same areas as in the yeast Rio2-TAP pre-40S particles. Nevertheless, they show marked morphological differences.
Due to the known position of RPS3, we attributed the density located on the beak region of the human pre-40S structure to the module formed by ENP1/Bystin, LTV1 and RPS3. The C-terminal part of the RPS3 structure does not seem to be in its final position (Figure 4A), which would suggest a subsequent structural rearrangement upon maturation. Recent structures of an ‘open’ conformation of RPS3, bound to its chaperone Yar1 have led to a model of RPS3 integration into the subunit, in which RPS3 would first interact with the pre-40S particle via its C-terminal part (58), (59). Though this low resolution 3D structure prevents us to confirm this model, our data reinforce the general idea that the release of LTV1 and ENP1 from the pre-40S subunit after phosphorylation by CK1ϵ and δ (9,10) triggers a conformational change of RPS3, which finalizes its accommodation on the 40S subunit (25,40,58,59).
What prevents the pre-40S particles to initiate translation or associate with 60S subunits?
Human TSR1 is an 804 amino acid protein which structurally mimics the translation elongation factor SelB (60,61), and the translation initiation factor IF2/eIF5B (62). Though it belongs to the same family as the GTPase Bms1 and adopts a fold similar to translation factor GTPases such as domain II of elongation factor Tu (23,61), yeast Tsr1 does neither bear GTP-binding nor GTPase activity in vitro (11). Rigid body docking of the recently solved structure of yeast Tsr1 (pdb code 5IW7 (61) on the human pre-40S electron density map (Supplementary Figure S3A) shows that positioning of TSR1 would be highly similar to what was proposed in yeast (61), or for SelB association to the mature ribosome (62). As for SelB, however, the position of TSR1 on the human pre-40S structure does not seem to represent a steric hindrance per se for association of the pre-40S particle with a 60S subunit, unlike what was proposed for the yeast pre-40S particles (25). Nevertheless, TSR1 clearly impedes the binding of several translation factors, including initiation factors.
Close to TSR1 and covering the tRNA binding site P, we attributed another extra-density to RIO2. This position in the human model is, like in yeast, in good agreement with CRAC data, which show that Rio2 binds preferentially to rRNA residues of helix 31 (34). Rigid-body docking of the X-ray structure of Rio2 from fungus Chaetomium thermophilum (PDB code: 4GYI (44)) into this domain yielded no satisfactory result (Supplementary Figure S3A). This suggests either a substoichiometric binding of this maturation factor on the pre-40S particle, or that RIO2 wobbles around an anchoring position that corresponds to the extra-density visible on the structure. This very central location of RIO2 prevents tRNAs to be loaded on the pre-40S particle, therefore impeding translation initiation to occur. We hypothesize that RIO2 is a key player, if not the only one, in blocking the association of human pre-40S particles with 60S subunits through steric hindrance.
A structural rearrangement of the platform?
Though DIM2 is found associated with pre-40S particles isolated with HASt-LTV1 based on mass spectrometry and Western blot analyses, we could not identify any extra-density domain of the human pre-40S particle to be large enough to be attributed to DIM2 with certainty. This could be explained by the possibility that DIM2, although located at the surface of the pre-ribosomal particle, does not adopt a conformation stable enough to be visualized on a pre-40S particle at this maturation step. Alternatively, we cannot formally exclude that association of DIM2 is sub-stoichiometric. Considering the larger volume of the platform of the pre-ribosomal particle compared to the mature subunit, a third hypothesis would be that DIM2 is buried within the platform, occupying the space allotted to RPS26 in the mature small ribosomal subunit and somehow sequestering the 18S-E pre-rRNA 3′ end. This hypothesis would be in good agreement with CRAC analyses performed in yeast (8), which have shown that Dim2 displays strong interactions with rRNA helices h24, h28 and h45 in the platform domain. Moreover, this would reinforce the emerging idea that the platform undergoes significant remodeling events during cytoplasmic maturation steps (13,63).
RACK1 is associated to early, cytoplasmic pre-40S particles and facilitates the last maturation steps
RACK1 has been first described as a scaffolding protein involved in multiple cellular signaling pathways and cellular processes, and its function as a ribosomal protein has been only recently acknowledged (for review, (64,65)). RACK1 was found in the structure of mature yeast and mammalian 40S subunits, but its presence in yeast pre-40S particles has remained unclear. Asc1p, the yeast homolog of RACK1, was detected as a sub-stoichiometric component of 90S and pre-40S particles (66), but did not appear in the cryo-EM structure of pre-40S particles isolated by Rio2-TAP (25). It was co-purified with the ribosomal biogenesis factor Fap7p and proposed to incorporate into pre-40S particles at a late cytoplasmic maturation stage (12). Nevertheless, it was not found to co-precipitate with 20S pre-rRNA, the RNA component of pre-40S particles (67). Our results show the unmistakable presence of RACK1 in human HASt-LTV1 pre-40S particles, as predicted by mass spectrometry data (26). 3D reconstruction experiments obtained by using an initial model devoid of RACK1 saw only a partial density growing back in the RACK1 region, suggesting that this protein is either not in its mature position, or that it is sub-stoichiometrically associated to these pre-40S particles. The various structures generated by 3D classifications assays always harbored a density in the ear region (Supplementary Figure S1A and SD), even when using the model devoid of RACK1 as initial reference (not shown). This favors the first hypothesis, but will need additional techniques such as quantitative mass spectrometry to be confirmed.
Consistent with previous RNAi screening results showing the involvement of RACK1 in human pre-40S particle maturation (20), we demonstrate here that RACK1 is necessary for the efficient conversion of the 18S-E pre-rRNA to 18S rRNA, but that it is not strictly required for the formation of 40S ribosomal subunits. A similar phenotype was observed for RPS25, while all other RPSs are strictly necessary for 18S rRNA production (35). Both RACK1 and RPS25 are specific to eukaryotic ribosomes. They are positioned at the back of the head of the small subunit, above the platform, and were recently implicated in the regulation of translation initiation for specific classes of viral IRESs (68,69). RACK1 has long been proposed to play a role in the regulation of general translation by acting as a scaffolding protein for recruiting the active form of the PKC ß II kinase to the ribosome (70–72). It is tempting to speculate that accumulation of 18S-E pre-rRNA in RACK1-depleted cells could reflect a coupling between the final processing of the 3′ end of 18S rRNA and translation initiation. According to a recent structure of a mammalian 43S complex, the PCI/MPN core of the translation initiation factor eIF3 is positioned in the vicinity of the 18S rRNA 3′ end, with the subunit eIF3d contacting RACK1 (43). The carboxyl terminus of eIF3c was also found to interact with RACK1 by two-hybrid screening in yeast (73). Therefore, although not essential in yeast or in cultured mammalian cells, RACK1 might be an accessory factor for the recruitment of eIF3, which in turn could, for example, stimulate NOB1 activity by favoring the structural rearrangement of the platform mentioned above. Such interplay between eIF3 and processing of the 18S rRNA 3′ end was already suggested in S. cerevisiae, where the absence of Hcr1, the ortholog of eIF3j, resulted in an accumulation of 20S pre-rRNA (74). Furthermore, a recent genome-wide RNAi screening has indicated that components of the eIF3 complex were required for 40S subunit synthesis in human cells (22). Additional work is now necessary to explore this putative role of RACK1 in the coordination between the final steps of the 18S rRNA formation and translation initiation. Our findings showing that RACK1 is present in early, cytoplasmic pre-40S particles of human cells reinforces the idea that the dynamics of pre-40S particle maturation differs between yeast and mammals (23), which could be related to additional quality control or regulatory mechanisms.
COORDINATES
The electron density map of the human HASt-LTV1 pre-40S particle was deposited in the Electron Microscopy Data Bank (European Molecular Biology Laboratory-European Bioinformatics Institute, Cambridge, UK) with the accession code EMD-3300.
Supplementary Material
Acknowledgments
The Authors would like to thank S. Balor and V. Soldan (METI platform, Toulouse), and Dr R. Matadeen (NeCEN, Leiden) for technical support in cryo-EM grids preparation and data acquisition; Drs A. Kamgoué at LBME, N. Renon and all the CALMIP support team for help in parallel computing; Dr F. Weis for help with RELION; L. Devaud for participation in RACK1 characterization; Dr T. Wild for generation of the RPS2-StHA cell line; Dr A. Cook for her precious help with the SelB/Tsr1 fitting; Dr. F. Delavoie for critically reading the manuscript; and Drs B. Pertschy, G. Stahl, M. Faubladier, as well as N. Montel-Lehry, D. Rinaldi and R. Shayan for fruitful scientific discussions. MFOD is very grateful to Dr M.P. Rols (IPBS, Toulouse) for electro-transformation protocols of eukaryotic cells. NL was supported by a Ph.D. salary from the University of Toulouse-Paul Sabatier.
Authors contributions: N.L., M.F.O.D., C.M., U.K., P.E.G. and C.P.C. designed the experiments, interpreted the data and wrote the manuscript. C.M. purified pre-40S particles for cryo-EM and performed their molecular characterization. MFOD analyzed the RNA content of pre-40S particles, and supervised and performed the analysis of RACK1 function. N.L. and C.P.C. performed the cryo-electron microscopy experiments, data processing and molecular modeling. P.E.G. and C.P.C. coordinated the project.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation and the NCCR ‘RNA&Disease’ (to U.K. and C.M.); Agence Nationale de la Recherche [RiboPre40S];Association pour la Recherche sur le Cancer [PJA20131200432 to M.F.O.D.]; University of Toulouse-Paul Sabatier, and the CNRS. This work was granted access to the HPC resources of CALMIP supercomputing center under the allocation 2015-P1406. Funding for open access charge: Research group internal funding.
Conflict of interest statement. The authors declare no competing commercial interests in relation to the work presented here.
REFERENCES
- 1.Anger A.M., Armache J.-P., Berninghausen O., Habeck M., Subklewe M., Wilson D.N., Beckmann R. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- 2.Zemp I., Kutay U. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 2007;581:2783–2793. doi: 10.1016/j.febslet.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Woolford J.L., Baserga S.J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195:643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rouquette J., Choesmel V., Gleizes P.-E. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 2005;24:2862–2872. doi: 10.1038/sj.emboj.7600752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zemp I., Wild T., O'Donohue M.-F., Wandrey F., Widmann B., Gleizes P.-E., Kutay U. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J. Cell Biol. 2009;185:1167–1180. doi: 10.1083/jcb.200904048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Widmann B., Wandrey F., Badertscher L., Wyler E., Pfannstiel J., Zemp I., Kutay U. The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40S subunits. Mol. Biol. Cell. 2012;23:22–35. doi: 10.1091/mbc.E11-07-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira-Cerca S., Kiburu I., Thomson E., LaRonde N., Hurt E. Dominant Rio1 kinase/ATPase catalytic mutant induces trapping of late pre-40S biogenesis factors in 80S-like ribosomes. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku542. doi:10.1093/nar/gku542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turowski T.W., Lebaron S., Zhang E., Peil L., Dudnakova T., Petfalski E., Granneman S., Rappsilber J., Tollervey D. Rio1 mediates ATP-dependent final maturation of 40S ribosomal subunits. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku878. doi:10.1093/nar/gku878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zemp I., Wandrey F., Rao S., Ashiono C., Wyler E., Montellese C., Kutay U. CK1 and CK1 are components of human 40S subunit precursors required for cytoplasmic 40S maturation. J. Cell Sci. 2014;127:1242–1253. doi: 10.1242/jcs.138719. [DOI] [PubMed] [Google Scholar]
- 10.Ghalei H., Schaub F.X., Doherty J.R., Noguchi Y., Roush W.R., Cleveland J.L., Stroupe M.E., Karbstein K. Hrr25/CK1 -directed release of Ltv1 from pre-40S ribosomes is necessary for ribosome assembly and cell growth. J. Cell Biol. 2015;208:745–759. doi: 10.1083/jcb.201409056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lebaron S., Schneider C., van Nues R.W., Swiatkowska A., Walsh D., Böttcher B., Granneman S., Watkins N.J., Tollervey D. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat. Struct. Mol. Biol. 2012;19:744–753. doi: 10.1038/nsmb.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strunk B.S., Novak M.N., Young C.L., Karbstein K. A translation-like cycle Is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012;150:111–121. doi: 10.1016/j.cell.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hector R.D., Burlacu E., Aitken S., Le Bihan T., Tuijtel M., Zaplatina A., Cook A.G., Granneman S. Snapshots of pre-rRNA structural flexibility reveal eukaryotic 40S assembly dynamics at nucleotide resolution. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku815. doi:10.1093/nar/gku815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis S.R., Gleizes P.-E. Diamond Blackfan anemia: ribosomal proteins going rogue. Semin. Hematol. 2011;48:89–96. doi: 10.1053/j.seminhematol.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Danilova N., Gazda H.T. Ribosomopathies: how a common root can cause a tree of pathologies. Dis. Models Mech. 2015;8:1013–1026. doi: 10.1242/dmm.020529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Keersmaecker K., Atak Z.K., Li N., Vicente C., Patchett S., Girardi T., Gianfelici V., Geerdens E., Clappier E., Porcu M., et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat. Genet. 2013;45:186–190. doi: 10.1038/ng.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagarajan N., Bertrand D., Hillmer A.M., Zang Z.J., Yao F., Jacques P.-É., Teo A.S.M., Cutcutache I., Zhang Z., Lee W.H., et al. Whole-genome reconstruction and mutational signatures in gastric cancer. Genome Biol. 2012;13:R115. doi: 10.1186/gb-2012-13-12-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nieminen T.T., O'Donohue M.-F., Wu Y., Lohi H., Scherer S.W., Paterson A.D., Ellonen P., Abdel-Rahman W.M., Valo S., Mecklin J.-P., et al. Germline mutation of RPS20, encoding a ribosomal protein, causes predisposition to hereditary nonpolyposis colorectal carcinoma without DNA mismatch repair deficiency. Gastroenterology. 2014;147:595–598. doi: 10.1053/j.gastro.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henras A.K., Plisson-Chastang C., O'Donohue M.-F., Chakraborty A., Gleizes P.-E. An overview of pre-ribosomal RNA processing in eukaryotes: Pre-ribosomal RNA processing in eukaryotes. Wiley Interdisc. Rev.: RNA. 2014 doi: 10.1002/wrna.1269. doi:10.1002/wrna.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wild T., Horvath P., Wyler E., Widmann B., Badertscher L., Zemp I., Kozak K., Csucs G., Lund E., Kutay U. A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol. 2010;8:e1000522. doi: 10.1371/journal.pbio.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tafforeau L., Zorbas C., Langhendries J.-L., Mullineux S.-T., Stamatopoulou V., Mullier R., Wacheul L., Lafontaine D.L.J. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol. Cell. 2013;51:539–551. doi: 10.1016/j.molcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 22.Badertscher L., Wild T., Montellese C., Alexander L.T., Bammert L., Sarazova M., Stebler M., Csucs G., Mayer T.U., Zamboni N., et al. Genome-wide RNAi screening identifies protein modules required for 40S subunit synthesis in human cells. Cell Rep. 2015;13:2879–2891. doi: 10.1016/j.celrep.2015.11.061. [DOI] [PubMed] [Google Scholar]
- 23.Carron C., O'Donohue M.-F., Choesmel V., Faubladier M., Gleizes P.-E. Analysis of two human pre-ribosomal factors, bystin and hTsr1, highlights differences in evolution of ribosome biogenesis between yeast and mammals. Nucleic Acids Res. 2011;39:280–291. doi: 10.1093/nar/gkq734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zorbas C., Nicolas E., Wacheul L., Huvelle E., Heurgué-Hamard V., Lafontaine D.L.J. The human 18S rRNA base methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA modification are required for ribosome biogenesis. Mol. Biol. Cell. 2015;26:2080–2095. doi: 10.1091/mbc.E15-02-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strunk B.S., Loucks C.R., Su M., Vashisth H., Cheng S., Schilling J., Brooks C.L., Karbstein K., Skiniotis G. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011;333:1449–1453. doi: 10.1126/science.1208245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wyler E., Zimmermann M., Widmann B., Gstaiger M., Pfannstiel J., Kutay U., Zemp I. Tandem affinity purification combined with inducible shRNA expression as a tool to study the maturation of macromolecular assemblies. RNA. 2011;17:189–200. doi: 10.1261/rna.2325911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mindell J.A., Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 28.Tang G., Peng L., Baldwin P.R., Mann D.S., Jiang W., Rees I., Ludtke S.J. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 29.Scheres S.H.W. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheres S.H.W., Chen S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen S., McMullan G., Faruqi A.R., Murshudov G.N., Short J.M., Scheres S.H.W., Henderson R. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kucukelbir A., Sigworth F.J., Tagare H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2013;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Granneman S., Petfalski E., Swiatkowska A., Tollervey D. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA–protein cross-linking. EMBO J. 2010;29:2026–2036. doi: 10.1038/emboj.2010.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Donohue M.-F., Choesmel V., Faubladier M., Fichant G., Gleizes P.-E. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J. Cell Biol. 2010;190:853–866. doi: 10.1083/jcb.201005117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paganin-Gioanni A., Bellard E., Escoffre J.M., Rols M.P., Teissie J., Golzio M. Direct visualization at the single-cell level of siRNA electrotransfer into cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10443–10447. doi: 10.1073/pnas.1103519108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haag S., Kretschmer J., Bohnsack M.T. WBSCR22/Merm1 is required for late nuclear pre-ribosomal RNA processing and mediates N7-methylation of G1639 in human 18S rRNA. RNA. 2015;21:180–187. doi: 10.1261/rna.047910.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiss T., Filipowicz W. Small nucleolar RNAs encoded by introns of the human cell cycle regulatory gene RCC1. EMBO J. 1993;12:2913–2920. doi: 10.1002/j.1460-2075.1993.tb05953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Preti M., O'Donohue M.-F., Montel-Lehry N., Bortolin-Cavaillé M.-L., Choesmel V., Gleizes P.-E. Gradual processing of the ITS1 from the nucleolus to the cytoplasm during synthesis of the human 18S rRNA. Nucleic Acids Res. 2013;41:4709–4723. doi: 10.1093/nar/gkt160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schäfer T., Maco B., Petfalski E., Tollervey D., Böttcher B., Aebi U., Hurt E. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 2006;441:651–655. doi: 10.1038/nature04840. [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto H., Unbehaun A., Loerke J., Behrmann E., Collier M., Bürger J., Mielke T., Spahn C.M.T. Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV-IRES RNA. Nat. Struct. Mol. Biol. 2014;21:721–727. doi: 10.1038/nsmb.2859. [DOI] [PubMed] [Google Scholar]
- 42.Hashem Y., des Georges A., Dhote V., Langlois R., Liao H.Y., Grassucci R.A., Hellen C.U.T., Pestova T.V., Frank J. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell. 2013;153:1108–1119. doi: 10.1016/j.cell.2013.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.des Georges A., Dhote V., Kuhn L., Hellen C.U.T., Pestova T.V., Frank J., Hashem Y. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature. 2015 doi: 10.1038/nature14891. doi:10.1038/nature14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferreira-Cerca S., Sagar V., Schäfer T., Diop M., Wesseling A.-M., Lu H., Chai E., Hurt E., LaRonde-LeBlanc N. ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit. Nat. Struct. Mol. Biol. 2012;19:1316–1323. doi: 10.1038/nsmb.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pertschy B., Schneider C., Gnädig M., Schäfer T., Tollervey D., Hurt E. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J. Biol. Chem. 2009;284:35079–35091. doi: 10.1074/jbc.M109.040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamanna A.C., Karbstein K. An RNA conformational switch regulates pre-18S rRNA cleavage. J. Mol. Biol. 2011;405:3–17. doi: 10.1016/j.jmb.2010.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fatica A., Tollervey D., Dlakić M. PIN domain of Nob1p is required for D-site cleavage in 20S pre-rRNA. RNA. 2004;10:1698–1701. doi: 10.1261/rna.7123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sloan K.E., Mattijssen S., Lebaron S., Tollervey D., Pruijn G.J.M., Watkins N.J. Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing. J. Cell Biol. 2013;200:577–588. doi: 10.1083/jcb.201207131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Veith T., Martin R., Wurm J.P., Weis B.L., Duchardt-Ferner E., Safferthal C., Hennig R., Mirus O., Bohnsack M.T., Wohnert J., et al. Structural and functional analysis of the archaeal endonuclease Nob1. Nucleic Acids Res. 2012;40:3259–3274. doi: 10.1093/nar/gkr1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGuffin L.J., Bryson K., Jones D.T. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 51.Buchan D.W.A., Ward S.M., Lobley A.E., Nugent T.C.O., Bryson K., Jones D.T. Protein annotation and modelling servers at University College London. Nucleic Acids Res. 2010;38:W563–W568. doi: 10.1093/nar/gkq427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Léger-Silvestre I., Milkereit P., Ferreira-Cerca S., Saveanu C., Rousselle J.-C., Choesmel V., Guinefoleau C., Gas N., Gleizes P.-E. The ribosomal protein Rps15p is required for nuclear exit of the 40S subunit precursors in yeast. EMBO J. 2004;23:2336–2347. doi: 10.1038/sj.emboj.7600252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khatter H., Myasnikov A.G., Natchiar S.K., Klaholz B.P. Structure of the human 80S ribosome. Nature. 2015;520:640–645. doi: 10.1038/nature14427. [DOI] [PubMed] [Google Scholar]
- 54.Abeyrathne P.D., San Koh C., Grant T., Grigorieff N., Korostelev A.A. Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome. eLife. 2016;5:e14874. doi: 10.7554/eLife.14874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheres S.H.W., Gao H., Valle M., Herman G.T., Eggermont P.P.B., Frank J., Carazo J.-M. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat. Methods. 2007;4:27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- 56.Weis F., Giudice E., Churcher M., Jin L., Hilcenko C., Wong C.C., Traynor D., Kay R.R., Warren A.J. Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat. Struct. Mol. Biol. 2015;22:914–919. doi: 10.1038/nsmb.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lafontaine D., Vandenhaute J., Tollervey D. The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev. 1995;9:2470–2481. doi: 10.1101/gad.9.20.2470. [DOI] [PubMed] [Google Scholar]
- 58.Holzer S., Ban N., Klinge S. Crystal structure of the yeast ribosomal protein rpS3 in complex with its chaperone yar1. J. Mol. Biol. 2013;425:4154–4160. doi: 10.1016/j.jmb.2013.08.022. [DOI] [PubMed] [Google Scholar]
- 59.Mitterer V., Murat G., Réty S., Blaud M., Delbos L., Stanborough T., Bergler H., Leulliot N., Kressler D., Pertschy B. Sequential domain assembly of ribosomal protein S3 drives 40S subunit maturation. Nat. Commun. 2016;7:10336. doi: 10.1038/ncomms10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gelperin D., Horton L., Beckman J., Hensold J., Lemmon S.K. Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA. 2001;7:1268–1283. doi: 10.1017/s1355838201013073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCaughan U.M., Jayachandran U., Shchepachev V., Chen Z.A., Rappsilber J., Tollervey D., Cook A.G. Pre-40S ribosome biogenesis factor Tsr1 is an inactive structural mimic of translational GTPases. Nat. Commun. 2016;7:11789. doi: 10.1038/ncomms11789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leibundgut M., Frick C., Thanbichler M., Böck A., Ban N. Selenocysteine tRNA-specific elongation factor SelB is a structural chimaera of elongation and initiation factors. EMBO J. 2005;24:11–22. doi: 10.1038/sj.emboj.7600505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loc'h J., Blaud M., Réty S., Lebaron S., Deschamps P., Bareille J., Jombart J., Robert-Paganin J., Delbos L., Chardon F., et al. RNA Mimicry by the Fap7 Adenylate Kinase in Ribosome Biogenesis. PLoS Biol. 2014;12:e1001860. doi: 10.1371/journal.pbio.1001860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adams D.R., Ron D., Kiely P.A. RACK1, A multifaceted scaffolding protein: Structure and function. Cell Commun. Signal. 2011;9:22. doi: 10.1186/1478-811X-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gandin V., Senft D., Topisirovic I., Ronai Z.A. RACK1 Function in Cell Motility and Protein Synthesis. Genes Cancer. 2013;4:369–377. doi: 10.1177/1947601913486348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schäfer T., Strauss D., Petfalski E., Tollervey D., Hurt E. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 2003;22:1370–1380. doi: 10.1093/emboj/cdg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schütz S., Fischer U., Altvater M., Nerkurkar P., Peña C., Gerber M., Chang Y., Caesar S., Schubert O.T., Schlenstedt G., et al. A RanGTP-independent mechanism allows ribosomal protein nuclear import for ribosome assembly. eLife. 2014;3:e03473. doi: 10.7554/eLife.03473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Landry D.M., Hertz M.I., Thompson S.R. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 2009;23:2753–2764. doi: 10.1101/gad.1832209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Majzoub K., Hafirassou M.L., Meignin C., Goto A., Marzi S., Fedorova A., Verdier Y., Vinh J., Hoffmann J.A., Martin F., et al. RACK1 controls IRES-mediated translation of viruses. Cell. 2014;159:1086–1095. doi: 10.1016/j.cell.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ceci M., Gaviraghi C., Gorrini C., Sala L.A., Offenhäuser N., Marchisio P.C., Biffo S. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature. 2003;426:579–584. doi: 10.1038/nature02160. [DOI] [PubMed] [Google Scholar]
- 71.Grosso S., Volta V., Sala L.A., Vietri M., Marchisio P.C., Ron D., Biffo S. PKCβII modulates translation independently from mTOR and through RACK1. Biochem. J. 2008;415:77. doi: 10.1042/BJ20080463. [DOI] [PubMed] [Google Scholar]
- 72.Ruan Y., Sun L., Hao Y., Wang L., Xu J., Zhang W., Xie J., Guo L., Zhou L., Yun X., et al. Ribosomal RACK1 promotes chemoresistance and growth in human hepatocellular carcinoma. J. Clin. Invest. 2012;122:2554–2566. doi: 10.1172/JCI58488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kouba T., Rutkai E., Karaskova M., Valasek L.S. The eIF3c/NIP1 PCI domain interacts with RNA and RACK1/ASC1 and promotes assembly of translation preinitiation complexes. Nucleic Acids Res. 2012;40:2683–2699. doi: 10.1093/nar/gkr1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valasek L. Dual Function of eIF3j/Hcr1p in Processing 20 S Pre-rRNA and Translation Initiation. J. Biol. Chem. 2001;276:43351–43360. doi: 10.1074/jbc.M106887200. [DOI] [PubMed] [Google Scholar]
- 75.Pintilie G., Chiu W. Comparison of Segger and other methods for segmentation and rigid-body docking of molecular components in cryo-EM density maps. Biopolymers. 2012;97:742–760. doi: 10.1002/bip.22074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang M., Anikin L., Pestov D.G. Two orthogonal cleavages separate subunit RNAs in mouse ribosome biogenesis. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku787. doi:10.1093/nar/gku787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.