Abstract
RNA aptamers are synthetic oligonucleotide-based affinity molecules that utilize unique three-dimensional structures for their affinity and specificity to a target such as a protein. They hold the promise of numerous advantages over biologically produced antibodies; however, the binding affinity and specificity of RNA aptamers are often insufficient for successful implementation in diagnostic assays or as therapeutic agents. Strong binding affinity is important to improve the downstream applications. We report here the use of the phosphorodithioate (PS2) substitution on a single nucleotide of RNA aptamers to dramatically improve target binding affinity by ∼1000-fold (from nanomolar to picomolar). An X-ray co-crystal structure of the α-thrombin:PS2-aptamer complex reveals a localized induced-fit rearrangement of the PS2-containing nucleotide which leads to enhanced target interaction. High-level quantum mechanical calculations for model systems that mimic the PS2 moiety and phenylalanine demonstrate that an edge-on interaction between sulfur and the aromatic ring is quite favorable, and also confirm that the sulfur analogs are much more polarizable than the corresponding phosphates. This favorable interaction involving the sulfur atom is likely even more significant in the full aptamer-protein complexes than in the model systems.
INTRODUCTION
Molecular recognition plays a central role in all chemical and biological systems (1). In addition, molecular recognition between proteins and small molecules (2) or nucleic acids (3) could play a key role in the design of therapeutic agents, diagnostics and analytical tools (4–6). One in vitro method that is widely used to identify RNA-based molecular recognition molecules (RNA aptamers) that selectively bind to a protein through non-covalent interactions is known as the systematic evolution of ligands by exponential enrichment (SELEX) (7,8). RNA aptamers identified with SELEX often need to be characterized more fully by post-SELEX experiments in order to meet the desired application. For example, pegaptanib sodium (Macugen), a SELEX-derived RNA aptamer approved by the FDA for clinical use to treat age-related macular degeneration (9), was modified post selection using several different moieties to achieve high affinity binding (KD = 49 pM (10)) to human vascular endothelial growth factor-165 (VEGF165). Incorporation of a variety of chemical modifications potentially complicates and limits the downstream processes. However, routine application of SELEX to obtain molecules that are useful as therapeutics as well as diagnostics remains an elusive goal, as candidate molecules have to combine both affinity and specificity (11–13). Efforts using chemical modification to optimize RNA aptamers have been mainly directed at the ribose moiety (10,14). Although modification confers increased affinity in some cases, many modifications primarily address in vivo stability issues. Thus, achieving excellent binding affinity along with high specificity continues to be a challenge in the aptamer field.
The phosphate backbone of RNA plays a crucial role in mediating RNA–protein interactions (15,16). Phosphate groups are typically well-exposed on the surface of RNA and thus readily available for contacts to binding partners. This is certainly the case for interactions between RNA aptamers and protein targets (7,8) including natural nucleic acid motifs engaged in intimate contacts to protein partners such as the RNA stem-loop motif interacting with bacteriophage MS2 coat protein (17,18) as well as in vitro selected RNAs bound to protein targets such as thrombin (19). Numerous crystal structures determined to date show that the two non-bridging oxygen atoms of many phosphate groups in the RNA backbone commonly interact with amino acid side chains of the protein via H-bonds and/or salt bridges (Arg, Lys, His), thus contributing to both binding affinity and specificity (20–23). Conversely, formation of hydrophobic contacts between the phosphates of an RNA and the amino acids of proteins in complexes is impossible.
Replacement of two non-bridging phosphate oxygen atoms in nucleic acids by sulfur gives rise to a phosphorodithioate (PS2) linkage (24–26) that has been the subject of numerous studies in the context of PS2-modified DNAs (PS2-DNAs) (27–34). PS2-DNAs are capable of binding proteins with higher affinities than their native phosphate counterparts as observed by several research labs (35–38). In the case of RNA, the improved gene silencing activities in vitro and in vivo (39,40) as a result of introducing two PS2 linkages at the 3′-end of sense strand siRNAs were suggested to be a consequence of the higher affinity of PS2-RNA for Ago2 protein, caused by a hydrophobic effect (41). Although invoking a hydrophobic effect that underlies the higher binding affinity between a protein and a modified RNA is quite reasonable, given the more hydrophobic nature of the PS2 moiety compared to phosphate (41), support in the form of atomic-level structural evidence for this hypothesis has been missing thus far.
It is not clear that increased hydrophobicity due to the sulfur substitution can explain fully the dramatic increase in binding affinity observed for some PS2-modified aptamers. Sulfur is significantly more polarizable than oxygen, and enhanced, favorable polarization interactions may explain in part why PS2-aptamers may bind more tightly.
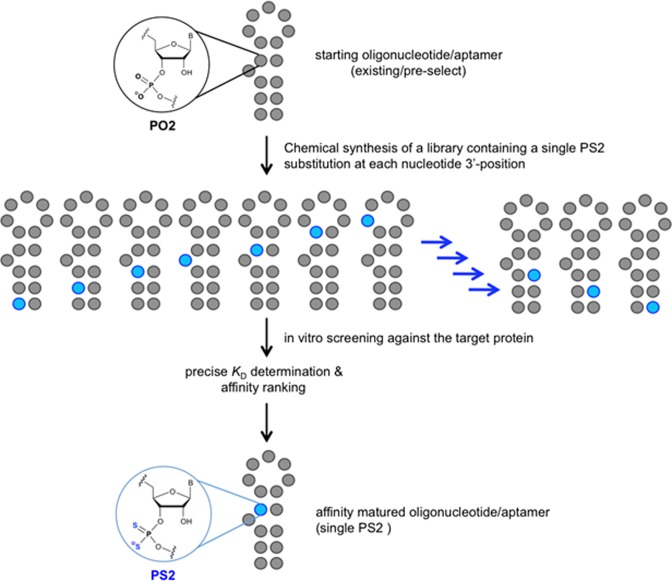
To investigate whether the hydrophobic effect and/or enhanced polarizability of the PS2 moiety could be used to modulate the affinity of an RNA aptamer toward its target protein, we tested the effect of individual PS2 substitutions on the binding constant (KD) between a target protein and an RNA aptamer by chemically synthesizing PS2-modified variants of the aptamer, each with a single PS2 modification in a different sequence location (PS2 walk) (Figure 1). The KD of each variant containing a single PS2 modification was compared to the native aptamer to measure the relative effect (relative KD). This approach identified modification hotspots along the phosphate backbone of the aptamer that brought remarkable enhancements in affinity toward the target protein (nanomolar to picomolar). The PS2 aptamer variants displayed little to no perturbations in global molecular structure and appeared to retain high specificity for their target. An X-ray co-crystal structure study suggests that the high gain in affinity involves an induced fit that stems from an altered, yet much preferred interaction between the PS2 moiety and neighboring protein side chains. A series of high-level quantum mechanical calculations for small model systems that serve as a proxy for the phosphate (phosphine oxide) and PS2 analog (phosphine sulfide) interactions with phenylalanine 232 (benzene) reveal that the sulfur substitution does indeed lead to enhanced binding affinity for this specific contact, and suggest that the effect will likely be more dramatic when we take into account the full protein environment near the PS2-phenylalanine interaction site.
Figure 1.
Schematic of the PS2-walk for mapping of RNA–protein interactions using an aptamer library of sequence variants each containing a single PS2 modification.
MATERIALS AND METHODS
Materials
5′-DMT-2′-O-TBDMS nucleoside (ABz, CAc, GiBu and U) phosphoramidite monomers, 5′-DMT-2′-O-methyl nucleoside (ABz, CAc, GiBu and U) phosphoramidite monomers and/or 5′-DMT-2′-F-nucleoside (ABz, CAc, GiBu and U) phosphoramidite monomers were purchased from Hongene Biotechnology USA, Inc. The 5′-biotin or 5′-fluorescein labeled aptamers were obtained by chemical coupling of corresponding phosphoramidite monomers purchased from Glen Research. The proteins used in this investigation were, human VEGF165 (Peprotech), human VEGF121 (Peprotech), human α-thrombin (Haematologic Technologies), bovine serum albumin (BSA) (Sigma Aldrich), IgE (Scripps Laboratories) and MAPK1 (Abnova). These proteins were handled according to manufacturers’ recommendations and aliquots were stored at −80 °C. All other chemicals and buffer components were obtained from Sigma-Aldrich. All solutions for in vitro assays and purifications were made using deionized/diethylpyrocarbonate (DEPC) treated water filtered through a 0.22 μm filter (Millipore). The aptamers were stored at −20 °C.
Aptamers and oligonucleotides
Modified and unmodified RNAs were synthesized on a 1μmole scale on an Expedite 8909 DNA/RNA Synthesizer using commercially available 5′-DMT-2′-O-TBDMS nucleoside (ABz, CAc, GiBu and U) phosphoramidite monomers, 5′-DMT-2′-O-methyl nucleoside (ABz, CAc, GiBu and U) phosphoramidite monomers and/or 5′-DMT-2′-F nucleoside (ABz, CAc, GiBu and U) phosphoramidite monomers as well as in house produced 5′-DMT-2′-O-TBDMS nucleoside (ABz, CAc, GiBu and U) thiophosphoramidite monomers or 5′-DMT-2′-O-methyl nucleoside (ABz, CAc, GiBu and U) thiophosphoramidite monomers (39). The 5′-DMT-2′-F nucleoside (ABz, CAc, GiBu and U) thiophosphoramidite monomers were synthesized by using the ‘Caruthers’ protecting group’ (42). All oligonucleotides were synthesized in DMT-off mode. For binding assays leading to affinity ranking, the RNAs were biotinylated at their 5′-end using Biotin-TEG (Glen Research), allowing immobilization onto a streptavidin (SA) coated sensor surface (this step enables kinetic characterization of crude RNA binding to proteins on a forteBIO Octet Red96 instrument). After completion of the synthesis, the solid support was suspended in ammonium hydroxide/methylamine solution (prepared by mixing one volume of ammonium hydroxide (28%) with one volume of 40% aqueous methylamine) and heated at 65 °C for 15 min to release the product from the support and to complete the removal of all protecting groups except the TBDMS group at the 2′-position. The solid support was filtered, and the filtrate was concentrated to dryness. For 2′-O-TBDMS RNA, the obtained residue was re-suspended in 115 μl of anhydrous dimethylformamide (DMF) and then heated for 5 min at 65 °C to dissolve the crude product. Triethylamine (TEA, 60 μl) was added to each solution, and the solutions were mixed gently. TEA·3HF (75 μl) was added to each solution, and the tubes were then sealed tightly and incubated at 65 °C for 2.5 h. The reaction was quenched with 1.75 ml of DEPC-treated water. Following deprotection, the oligonucleotides were desalted/buffer exchanged into ddH2O (using 3000 MWCO Amicon filters) and lyophilized to dryness. Purification was performed on an Amersham Biosciences P920 FPLC (Fast protein liquid chromatography) instrument fitted with a Mono Q 10/100 GL column. The PS2-RNAs were desalted using reverse-phase high performance liquid chromatography (HPLC) to yield the PS2-RNA final products. Mass of the modified RNAs were confirmed by ESI-MS. Folding of RNAs was achieved in ammonium acetate buffer (pH 7.4) or in reported binding buffer by heating the RNA oligonucleotides at 90 °C for 2 min, followed by slow cooling to room temperature (over 2 h).
BLI binding analyses, LOD and aptamer specificity
The binding affinities of select aptamers and their library variants were determined by biolayer interferometry (BLI) on a fortéBIO Octet Red96 instrument (Pall fortéBIO) at 30 °C. All aptamers were chemically labeled with biotin-TEG at the 5′-end allowing immobilization on to an SA coated sensor surface enabling a kinetic analysis of their binding events in a suitable running buffer. Each biotinylated aptamer was diluted to 500 nM (or 25 nM when applied in pure form) in a suitable buffer, heat denatured at 95 °C, slowly cooled down to room temperature just prior to the experiment. Samples were agitated at 1000 rpm. Tips were saturated with 500 nM (or 25 nM, if a purified form is used) biotinylated aptamers for 1 min, which typically resulted in capture levels of 0.4 ± 0.05 nm within a row of eight tips. Analytes (VEGF165 or thrombin) were prepared in the appropriate buffer as a dilution series (typically in the range from 0.5 to 6 nM in duplicate) along with the buffer blanks. Association was monitored for 300 s and dissociation was followed for 300 s into buffer alone. If slower off rates were observed, as in the case of tight binding, the dissociation was monitored for at least 500 s. The data were fit to a 1:1 binding model using fortéBIO Octet data analysis software. Kinetic constants were determined by integration of the experimental data using the differential rate equation dR/dt = ka·C·(Rmax-R)-kd·R to obtain ka and kd values simultaneously (R = observed response, Rmax = maximum response upon saturation, C = analyte concentration, ka = association rate constant, kd = dissociation rate constant). Then, the ratio between kd and ka gives the reported dissociation constants (kd/ka= KD). The goodness of the fit was judged by the reduced chi-square (χ2) values and by the R2 values approaching 1.
The relative KD values of the aptamers corresponding to a library are also expressed as the ratio of KD values (KDunsubstituted/KDsubstituted) and plotted against positioning of each nucleotide residue with a corresponding PS2 substitution. Relative KD of >1 (blue = 1000-fold) and <1 (red = 0.001-fold) denote an increase or decrease in binding affinity, respectively.
For VEGF165 biomarker limit of detection (LOD) experiments, the biomarker dilutions were made in PBST buffer (10 mM sodium phosphate pH 7.4, 150 mM NaCl, 0.04% tween 20) containing 0.01 mg/ml BSA. The dilutions ranging 3.8 fg/ml to 380 ng/ml of VEGF165 were tested against AF83-7 and AF83-1 at a loading concentration of 100 nM. Parallel referencing on the Octet was achieved by running a similar assay with a scrambled aptamer sequence as negative control. For each aptamer, the association phase was monitored for 1200 s using the high sensitivity kinetics (2 Hz, averaging by 50) mode and the observed Response on the Octet was plotted against biomarker concentration to obtain qualitative assessment of LODs.
For determining the selectivity of each aptamer, 10 nM each of proteins, human VEGF165, human VEGF121, human α-thrombin, BSA, IgE and MAPK1, were used.
SPRi binding analyses
A Dextran-based surface chemistry was used (amine coupling) to immobilize first extravidin protein and then the biotinylated RNA aptamers. The aptamers were immobilized with a surface plasmon resonance imaging (SPRi)-CFM microfluidic printer, which uses a flow to print biomolecules on the SPRi-Biochip surface. The interaction monitoring was carried out using a XelPleX SPRi system (Horiba Scientific) equipped with a continuous flow pump and an integrated auto sampler fully controlled by the EzSuite software. Distinguishable off-rates between the ‘nanomolar’ binder and the ‘picomolar’ binder were observed at the immobilization concentration of 1.25 μM for aptamers. Therefore, kinetic analysis was performed at 1.25 μM immobilization concentration. VEGF165 protein was injected at increasing concentrations using a 2-fold dilution series (from 0.35 to 22.6 nM) under injection parameters of 480 μl for the injection volume and of 240 μl/min for the flow rate (highest flow rate tested). A regeneration step using a 1.0 M NaCl solution was performed between each concentration injected (200 μl at 50 μl/min). The kinetic curves were analyzed using the ScrubberGen software. The SPRi signal obtained on reference spots (i.e. no binding control AF83B RNA aptamers spots) were used for referencing. Then, the data was fitted locally [i.e. Rmax (maximum of reflectivity) different for each curve] using a 1:1 interaction model (see Supplementary Figures SF-3a and SF-3b; orange curves correspond to the 1:1 model fits).
Melting temperature (Tm) and CD measurements
Aptamers were annealed before using, according to ‘Handling protocol for aptamers’ (http://www.amsbio.com/datasheets/2369FC-DY647.pdf from Amsbio). All absorption measurements were accomplished in a 1.0-cm path length cell with a Cintra 4040 spectrophotometer equipped with a Peltier Thermocell (GBC, Dandenong, Australia), with detector set at 260 nm. Tm measurements for VEGF165 aptamers (AF83-1 and AF83-7) were performed in phosphate buffered saline (PBS) buffer (10 mM sodium phosphate pH 7.4, 150 mM NaCl) and for thrombin aptamers (AF113-1 and AF113-18) in HSC buffer (20 mM HEPES-KOH pH 7.4, 125 mM NaCl, 2 mM CaCl2) at 2.0 μM final concentration of oligonucleotides. The first step of analysis was annealing from 90 to 15 °C, with a temperature gradient of 1 °C/min. Melting profiles were measured from 15 to 90 °C, with the temperature gradient of 0.5 °C/min. Each reported Tm is an average of values from at least three independent experiments.
Melting temperatures were assessed using two methods: (i) as the first derivative of the function describing the melting curve, using the melting software of Cintra 4040 and (ii) by fitting the obtained melting curves using MeltWin v.3.5 software. Thermodynamic analysis of data for VEGF165 aptamers (AF83-1 and AF83-7) and α-thrombin aptamers (AF113-1 and AF113-18) was performed using the MeltWin v.3.5 melt curve-processing program (results are average from three independent experiments).
Circular dichroism spectroscopy (CD) spectra were recorded on a CD6 dichrograph (Jabin-Yvon, Longjumeau, France) using cells with 0.5 cm path length, 2 nm bandwidth and 1–2 s integration time. Each spectrum was smoothed with a 24-point algorithm (included in the manufacturer′s software, version 2.2.1) after averaging of ten scans. The spectra from 200 to 350 nm were recorded at 25 °C in PBS buffer (10 mM sodium phosphate pH 7.4, 150 mM NaCl) for VEGF165 aptamers (AF83-1 and AF83-7) or in HSC buffer (20 mM HEPES-KOH pH 7.4, 125 mM NaCl, 2 mM CaCl2) for α-thrombin aptamers (AF113-1 and AF113-18). The concentration of the RNA aptamers was 2 μM. Measurements were performed as 10 scans taken for each sample.
Cellular binding and fluorescence imaging
The HT-29 colorectal cancer and MRC-5 human fetal lung fibroblast cells were plated at a seeding density of 105 cells/ml in RPMI-1640 (Lonza, Basel, Switzerland)/10% FCS (Seradigm, Randor,PA) and were allowed to attach for 48 h in a humidified hypoxic chamber (StemCell Technologies, Vancouver, Canada) containing 5% CO2, 1% O2 and 94% N2 (hypoxia) at 37 °C. The cells were dislodged by Accutase (Biolegend, San Diego, CA) and incubated with FITC labeled VEGF165 aptamers at different concentrations for 2 h at 37 °C in culture medium. The cells were then centrifuged for 5 min at 300xg and re-suspended in PBS for flow cytometry analysis immediately. Analysis was performed on a BD Accuri C flow cytometer. At least 10 000 events were collected for each sample. All the experiments for the binding assay were repeated at least three times.
The HT-29 cells and MRC-5 cells were seeded in a 8-well slide (EMD Millipore, Billerica, MA, USA) in RPMI-1640 (Lonza, Basel, Switzerland)/10% FCS (Seradigm, Randor, PA, USA). Cells were plated at a seeding density of 104 cells/ml and were allowed to attach for 48 h in a humidified hypoxic chamber (Stem Cell Technologies, Vancouver, Canada) containing 5% CO2, 1% O2 and 94% N2 (hypoxia) at 37 °C. Subsequently, the cells were incubated with FITC labeled VEGF165 aptamers at 37 °C for 2 h and washed with PBS buffer three times to remove unbound aptamers. Sequence specificity of the aptamers was determined using a scrambled sequence as control. Images of aptamer binding to cells were acquired using an Olympus FV1000 confocal microscope.
Limits of detection (LOD) experiments
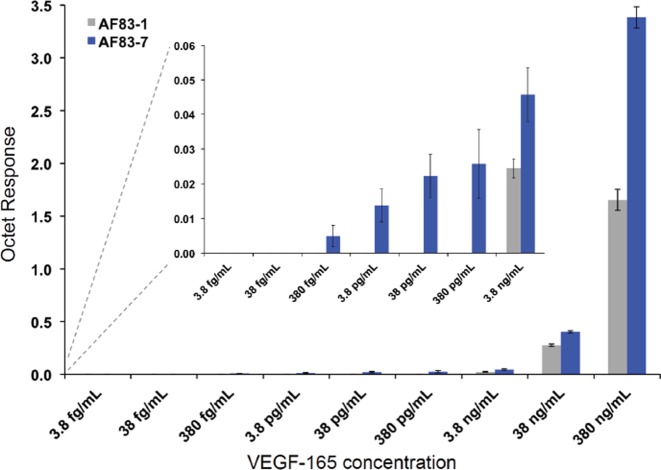
The LOD of the PS2-modified aptamers and their analogs containing no PS2 were investigated by BLI on a fortéBIO Octet Red96 instrument (Pall fortéBIO) at 30 °C using the high sensitivity data acquisition/quantitation mode (2 Hz, averaging by 50). All aptamers were chemically labeled with biotin-TEG at the 5′-end allowing immobilization onto a SA coated sensor surface enabling the detection of the analyte association in a suitable running buffer containing 0.01 mg/ml BSA. Each biotinylated aptamer was diluted to 100 nM in a suitable reconstitution buffer, heat renatured at 95 °C for 1.5 min, slowly cooled down to room temperature just prior to the experiment. Samples were agitated at 1000 rpm. Tips were saturated with 100 nM biotinylated aptamers for 5 min, which typically resulted in capture levels of 1.4 ± 0.05 nm within a row of eight tips. Aptamer loaded tips were subsequently equilibrated in the running buffer containing 0.01 mg/ml BSA for 5 min or until a steady baseline was observed. Analytes were prepared in the running buffer containing 0.01 mg/ml BSA as a dilution series (typically in the range from 3.8 fg/ml to 380 ng/ml in duplicates). Association was monitored for 1200 s and the maximum response obtained at the end of the association phase was recorded for analyte concentration. A row of eight sensors was saturated with 100 nM biotinylated, scrambled-aptamer sequence (negative control) and used for parallel referencing. The response-data were processed using ForteBio Octet data analysis software. The reference-subtracted final response data were plotted against analyte concentration to obtain the graphs shown in Figure 4. An average of six individual assays was performed for each aptamer in six different days or runs to be able to capture the true variability of the analytical method.
Figure 4.
The limit of detection (LOD) assay for AF83-1 and AF83-7 as observed by BLI. Results are expressed as mean ±SEM, n = 6.
Serum stability assay
Aptamer (1 μg) was incubated with 10% human serum in PBS buffer at 37 °C. Aliquots were drawn at various time points from 0 to 168 h (7 days) and immediately mixed with 2× gel loading denaturing buffer (Bio-Rad), frozen at −70 °C until analysis. Samples were analyzed by polyacrylamide gel electrophoresis using a 12% polyacrylamide/urea gel, and were stained with SYBR Gold (Invitrogen) nucleic acid gel stain. Stained bands were imaged with a FluorChem FC2 fluorescent image analyzer (Alpha Innotech) and quantified using the AlphaView software package. The percentage of intact aptamer was calculated as the percent ratio of band intensity = (band intensity at time t ÷ band intensity at 0 h) × 100%.
Crystallization of AF113-18:thrombin complex
We used PPACK (D-Phe-Pro-Arg chloromethylketone)-inhibited human thrombin to limit proteolysis during crystallization (Haemtech, VT, USA). The 25-nucleotide modified RNA aptamer was chemically synthesized as described above with all pyrimidine nucleotides carrying 2′-F substituents, two further nucleotides containing 2′-SeMe substituents and the 3′-phosphate group of U17 replaced by a PS2 moiety (5′-GGGAACAAAGCUSeGAAGUFS2ACUUSeACCC-3′). The protein:RNA complex was formed prior to crystallization as previously described (19). Typically, this was achieved by incubating frozen protein at 3 mg/ml with the aptamer in 150 mM sodium chloride, 2 mM calcium chloride and 20 mM HEPES, pH 7.3, for 10 min at 37 °C and then cooling on ice. The molar ratio for protein:RNA used was 1:1.5. Crystals were grown at 18 °C from sitting drops containing 1 μl of protein:RNA complex mixed with 1 μl of reservoir solution (10% [w/v] PEG-6000, 5% v/v 2-methyl-2,4-pentanediol (MPD), 100 mM HEPES, pH 7.5) and equilibrated against the reservoir solution using vapor diffusion (43). Crystals appeared within a week to 10 days. The crystals were cryo-protected using a solution containing the reservoir components and 20% glycerol and were then flash-frozen in liquid nitrogen.
Structure determination of AF113-18:thrombin complex
Diffraction data for the thrombin–aptamer complex were collected on the 21-ID-G beam line of the Life Sciences Collaborative Access Team (LS-CAT) at the Advanced Photon Source (APS), located at Argonne National Laboratory (Argonne, IL, USA), using a MARCCD 300 detector. The wavelength was 0.9785 Å and crystals were kept at 100 K during data collection. Diffraction data were integrated, scaled and merged using HKL2000 (44). Selected crystal data and refinement statistics are listed in Supplementary Table S5 (ST-5). The structure (SF-10) was determined by the molecular replacement method with the program Molrep (45) in the CCP4 suite of crystallographic software (46) using thrombin as the search model (PDB ID: 3DD2 (19); http://www.rcsb.org). Following initial positional and isotropic temperature factor refinement cycles with the program Refmac (47), the entire RNA aptamer molecule could be placed into the electron density map. At this stage, refinement was continued and the values for R-work and R-free (48) dropped to ∼35%. After six cycles of refinement in Refmac5, the PS2 moiety was built into Fourier (2Fo-Fc) sum and (Fo-Fc) difference electron density maps that were visualized with Coot (49) and the refinement continued after adaptation of the dictionary files. Further refinement was carried out after addition of further water molecules and ions. Final refinement parameters are summarized in ST-5. All crystallographic figures were generated using the program Chimera (50).
Model system calculations
All geometry optimizations for phosphine oxide, phosphine sulfide, benzene and the complexes, as well as potential energy surface scans for the complexes were performed at the MP2 level of theory with the aug-cc-pVDZ basis set. Final energies and static polarizability for all geometry-optimized molecules and complexes were computed at the CCSD(T) level of theory with the aug-cc-pVTZ basis set. Local minima on the potential energy surfaces were confirmed via frequency calculations, and static polarizability was calculated as the mean of the trace of the polarizability tensor. All calculations were performed with the Gaussian 09 package (51) and GaussView 5 (52) was used for analysis and visualization of results.
RESULTS
Identification of modification hotspots and affinity maturation of selected aptamers
The PS2-walk is useful for identifying modification hotspots and for producing aptamer variants with significantly enhanced binding affinity to a target protein, while retaining desired specificity and potentially improving biological function. The RNA aptamers to VEGF165 and α-thrombin used as reference starting points in this study were previously identified using the SELEX methodology. The VEGF165 RNA aptamer [AF83-1 in Supplementary Table S1 (ST-1)] has a reported KD of 2.1 nM (14). The RNA aptamer (AF113-1 in ST-2) used as the second reference in this study binds to the exosite-2 of α-thrombin with a reported KD of 2.8 nM (19,53).
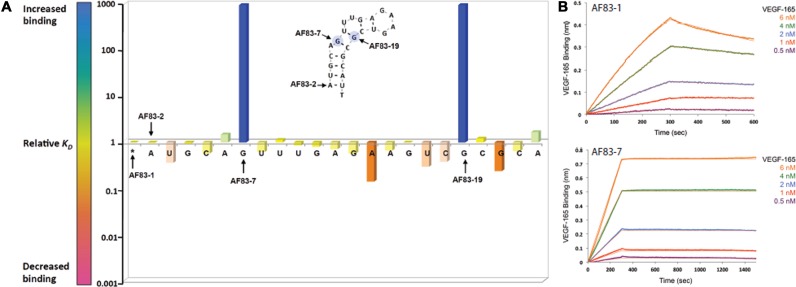
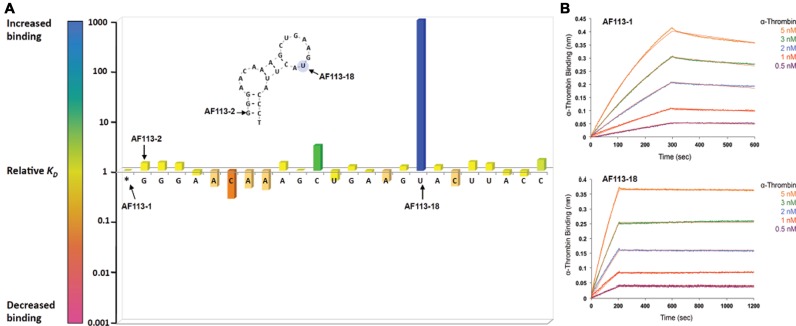
The aptamer libraries containing 5′-biotinylated variants of AF83-1 or AF113-1 (ST-1 and ST-2, respectively) were chemically synthesized using standard phosphoramidite chemistry with a PS2 substitution made for every PO2 moiety along the phosphate backbone, one occurrence per sequence. Kinetic characterization of the aptamer variant library by BLI allowed affinity ranking of PS2-modified variants with respect to their native counterpart. Evaluation consisted of using a serial dilution of the VEGF165 or α-thrombin protein screened against corresponding PS2 aptamer variants immobilized onto SA coated BLI sensors. The binding of AF83-1 or AF113-1 was also determined in parallel to the library variant candidates to determine whether sequences containing the PS2-modification are capable of enhancing the affinity compared to their native aptamer. Supplementary Figure (SF)-1 and ST-3 show the titration curves and characterization of binding performed using BLI for AF83-1 variants, respectively. Similarly, SF-2 and ST-4 show the characterization of AF113-1 variants. The ratio of KD values (KDunsubstituted/KDsubstituted) defines the relative KD of a variant to its native aptamer. Figures 2A and 3A plot the relative KD of each VEGF165 or α-thrombin variant, respectively against the position of each PS2 substitution. Relative values of KD of >1 (blue = 1000-fold) and <1 (red = 0.001-fold) denote an increase or decrease in binding affinity, respectively (see color scale bar in Figures 2A and 3A). A single PS2 substitution at most positions had only a relatively small effect on protein binding. However, a single PS2 substitution resulted in about thousand-fold enhancement in the variant's affinity over the native aptamer in at least one position for each reference aptamer. Variant affinities of AF83-7 for VEGF165 and AF113-18 for α-thrombin were chosen for further investigation.
Figure 2.
Identification of modification hotspots and characterization of high-affinity PS2 variants (A) the outcome of the PS2-walk corresponding to a series of PS2 variants of a reference aptamer of AF83-1. The RNA sequences generated for VEGF165 protein are listed in ST-1, and the affinity ranking kinetic data for each RNA sequence is reported in ST-3. (B) Binding of AF83-1 and AF83-7 to VEGF165 as measured by BLI.
Figure 3.
Identification of modification hotspots and characterization of high-affinity PS2 variants (A) the outcome of the PS2-walk corresponding to a series of PS2 variants of a reference aptamer of AF113-1. The RNA sequences generated for α-thrombin protein are listed in ST-2, and the affinity ranking kinetic data for each RNA sequence is reported in ST-4. (B) Binding of AF113-1 and AF113-18 to α-thrombin as measured by BLI.
The PS2 variants with the strongest binding affinity (AF83-7 and AF113-18) and the corresponding native aptamers (AF83-1 and AF113-1, respectively) were purified using a two-step procedure that involves MonoQ ion-exchange FPLC (54,55) and a PRP-1 reverse-phase HPLC. Polyacrylamide gel electrophoresis and mass spectrometry confirmed that the purified aptamers were of the proper length and identity. Pure native aptamers and variants were analyzed using BLI technology (Figure 2b (AF83-1 and AF83-7) and Figure 3b (AF113-1 and AF113-18)) to determine the KD (Table 1). The anti-VEGF165 variant aptamer AF83-7 revealed much higher affinity to VEGF165 than the native aptamer with a KD = 1.0 ± 0.1 pM according to BLI estimates, which is a ca. 1000-fold affinity enhancement compared to the native aptamer using just a single PS2 modification in the correct sequence location. SPRi verified the BLI-derived kinetic parameters for AF83-7 and AF113-18 as shown in SF-3a and SF-3b, respectively. Based on the SPRi estimates, the KD for AF83-7 is 8.1 ± 0.2 pM (mean ± SEM, n = 4). With SPRi technology, the signal observed at the end of the association step for the anti-VEGF165 sequences is proportional to the relative amounts of VEGF165 retained by the immobilized native aptamer or PS2 variant. AF83-7 generated a higher SPRi response signal upon binding to VEGF165 compared to the native aptamer (AF83-1). Further investigations into this observation using BLI allowed determination of the LOD of AF83-1 and AF83-7 for VEGF165 (Figure 4). The median LOD values for AF83-1 and AF83-7 are ≤ 3800.0 pg/ml (or ≤ 0.1 nM) and ≤ 0.38 pg/ml (or ≤ 10.0 fM), respectively (Table 2, LOD ≤ median, n = 6). This result demonstrates the ability of PS2-modified AF83-7 to detect VEGF165 at a much lower concentration than the native AF83-1, suggesting a superior capability of AF83-7 for use in a diagnostic capacity.
Table 1. Dissociation constants (KD) of selected PS2-modified aptamers binding to their targetTs.
| Target | Aptamer or PS2 aptamer | Aptamer ID | KD (pM) |
|---|---|---|---|
| VEGF165 | Aptamers with a single PS2 substitution | AF83-7 | 1.0 ± 0.1 |
| AF83-19 | 1.0 ± 0.1 | ||
| Reference aptamer | AF83-1 | 961.0 ± 25.0 | |
| Thrombin | Aptamers with a single PS2 substitution | AF113-18 | 1.8 ± 0.2 |
| Reference aptamer | AF113-1 | 1871.0 ± 36.0 |
Table 2. The limits of detection (LOD) of anti-VEGF165 aptamers (AF83-7 versus AF83-1).
| Aptamers for VEGF-165 | Aptamer ID | KD (pM) | LOD (M)a | LOD (pg/ml)a |
|---|---|---|---|---|
| Aptamer modified with a single PS2 | AF83-7 | 1 ± 0.1 | ≤10 × 10−15 | ≤0.38 |
| Native aptamer | AF83-1 | 961 ± 25 | ≤0.1 × 10−9 | ≤3800 |
aLOD values are expressed as median, n = 6.
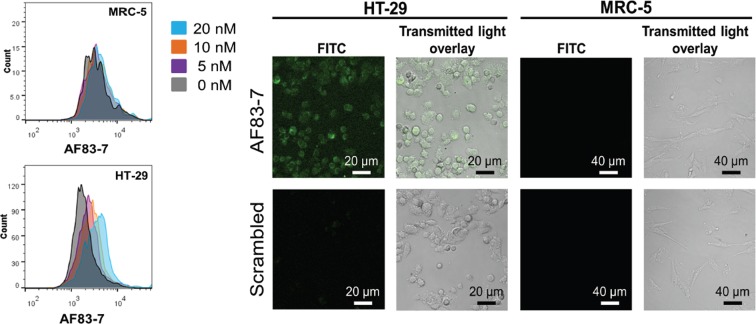
We also investigated binding and specificity of the PS2 variants at the cellular level, using a chemically synthesized AF83-7 labeled with fluorescein (FITC-AF83-7). HT-29 colorectal cancer cells and normal MRC-5 fibroblast cells (negative control) under hypoxic condition were incubated with a serial dilution of FITC-AF83-7 and analyzed using flow cytometry. Compared to techniques such as capillary electrophoresis or nitrocellulose filter binding, cell-based flow cytometry allows identification of aptamers that have the ability to bind to their target with high specificity in a more physiologically relevant environment (56,57). Figure 5 show concentration-dependent binding of FITC-AF83-7 to only HT-29 colorectal cancer cells. Flow cytometry analysis of MRC-5 fibroblast cells exposed to FITC-AF83-7 resulted in no change in the fluorescent signal upon increasing the PS2 variant concentration. The results demonstrate specific binding of AF83-7 under equilibrium conditions to cellular VEGF165 present in HT-29. Therefore, in addition to the enhanced affinity against VEGF165, cellular imaging experiments and flow cytometry data clearly suggest specific binding of the AF83-7 PS2 variant at the cellular level. The remarkably high affinity and specificity of AF83-7 for VEGF165 demonstrates the PS2 variants’ potential for use as a diagnostic agent in aptamer-mediated target cell biotherapy (5,58).
Figure 5.
Left panel: representative flow cytometry profiles of AF83-7 aptamer at different concentrations in HT-29 and MRC-5 cells. Profiles are generated based on the data from six independent experiments. Right panel: binding of fluorescein-labeled AF83-7 and a scrambled negative control aptamer to HT-29 and MRC-5 cells under hypoxic conditions. Images are representative of three independent experiments.
BLI studies comparing the native aptamer and the PS2 variants targeting human α-thrombin also revealed an enhanced affinity of the AF113-18 variant with an estimated KD = 1.0 ± 0.1 pM (ST-4). Further verification of BLI kinetic parameters using SPRi technology revealed a KD of 4.5 ± 0.2 pM (mean ± SEM, n = 4) for AF113-18 (SF-3b).
Secondary structure, specificity and serum stability of PS2-modified aptamers
Characterizing the high-affinity PS2 aptamer variants also included evaluating the effect of the PS2 modification on helicity, stability, and specificity to their target protein. CD spectra (SF-4) suggest no global influence on the structures of PS2-containing variants AF83-7 or AF113-18 relative to their native aptamer counterparts AF83-1 or AF113-1, respectively. In addition, thermodynamic parameters suggest unperturbed secondary structures (Table 3).
Table 3. Thermodynamic parameters of PS2-modified aptamers and reference aptamers.
| Target | Aptamer ID | ΔG°37 (kcal/mol) | ΔH° (kcal/mol) | ΔS° (cal/Kmol) | Tm (°C) |
|---|---|---|---|---|---|
| VEGF165 | AF83-7 | −9.1 ± 0.1 | −42.9 ± 2.6 | −108.9 ± 8.4 | 46.1 ± 0.1 |
| AF83-1 | −8.9 ± 0.2 | −34.2 ± 3.1 | −81.5 ± 9.3 | 45.1 ± 1.3 | |
| Thrombin | AF113-18 | −8.5 ± 0.2 | −30.3 ± 4.8 | −70.4 ± 15.3 | 44.6 ±1.4 |
| AF113-1 | −8.5 ± 0.1 | −30.5 ± 2.1 | −70.8 ± 6.6 | 44.9 ±1.1 |
In vitro specificity measurements using BLI included qualitative screening of aptamer variants AF83-7 and AF113-18 as well as the native aptamers for interaction with various proteins. Murine VEGF164, which has a high sequence homology to human VEGF165, bound AF83-7 as well as AF83-1, whereas human VEGF121, BSA, human α-thrombin, human IgE and MAPK-1 did not show any binding (SF-5) to either the variant or the native aptamer. This further validates the cellular binding data for AF83-7 (Figure 5). Similarly, AF113-18 variant and the native aptamer bound only human α-thrombin, whereas, human VEGF165, Murine VEGF164, BSA, human IgE and MAPK-1 did not show any binding (SF-5).
The anti-VEGF165 native aptamer AF83-1 and PS2 variant AF83-7 were incubated in vitro with normal human serum at 37 °C to measure their serum stability and to evaluate their potential usefulness as in vivo agents. The motivation for this experiment was to replicate an in vivo pharmacokinetic evaluation and provide a comparison of relative stabilities of the aptamers against endonucleases present in the plasma. AF83-1 was degraded at a relatively faster rate compared to the PS2-containing variant AF83-7 (SF-6), suggesting enhanced nuclease resistance of the variant aptamer containing just a single PS2 modification.
Structural insights into PS2-mediated affinity enhancements
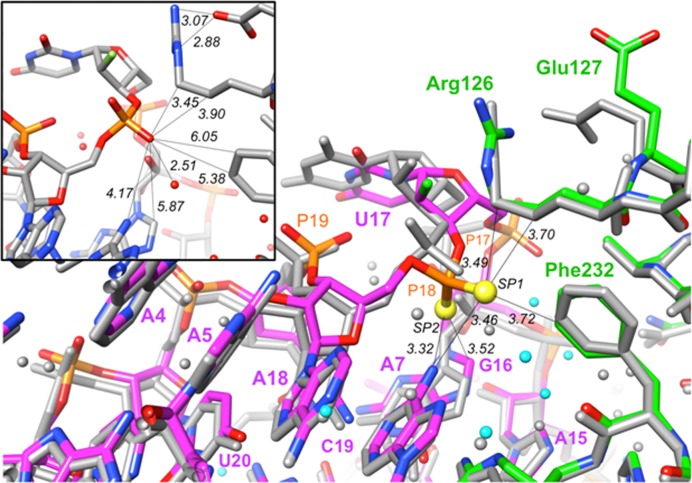
In order to identify potentially altered interactions in the PS2 aptamer variant that underlie the significantly enhanced binding affinity, we determined the crystal structure of AF113-18 bound to α-thrombin at 1.90 Å resolution. The structure was phased by molecular replacement, using the thrombin coordinates from the native complex (19) as the search model. Inspection of the complex shows no fundamental changes in terms of the site (exosite-2) on the thrombin surface occupied by the AF113-18 variant. Similarly, the conformations of the protein portion in the complexes with native AF113-1 aptamer and AF113-18 are very similar (Figure 6). The overall conformations of AF113-1 and AF113-18 in the two structures are also preserved. However, the base and sugar moieties of U17 in AF113-18 have moved by ca. 2.0 Å toward the protein and, both, the 5′-phosphate and 3′-PS2 moiety are rotated by ca. 90° relative to the corresponding phosphates in the native aptamer (Figure 6). In addition, the 5′-phosphate (P17) has undergone a shift of 2.8 Å and the shift of the 3′-PS2 (P18) amounts to 1.5 Å. These conformational changes are accompanied by multiple new interactions within the PS2 variant and between PS2 variant and thrombin. Regarding the former, the rotation of PS2 results in H-bonds between SP1 and SP2 and the N6 amino group of A7 (distances of 3.46 and 3.32 Å, respectively). These distances are longer than a typical H-bond between a phosphate-oxygen and an amino group (e.g. 2.8 Å), but it is important to keep in mind that the P-S bond is considerably longer than the P-O bond (1.95 versus 1.50 Å, respectively). Importantly, an additional hydrophobic contact is established between SP2 and C8 (H) of G16 (3.52 Å; the van der Waals radii for sulfur and oxygen are 1.80 and 1.50 Å, respectively). The attractive nature of this interaction is indicated by a shift of the guanine nucleobase toward the rotated PS2 moiety; unlike G16, the A15 and A18 nucleosides can be overlaid almost perfectly with the respective residues in the structure of the native complex. Newly established interactions between AF113-18 and thrombin are all hydrophobic in nature and include SP1…Cϵ2 (Phe232) (3.72 Å), SP1…Cγ (Arg126) (3.70 Å) and SP1…Cδ (Arg126) (3.49 Å). The guanidino moiety of Arg126 hovers above the ribose sugar of U17, and lies at some distance from the PS2 group (distance P…Cζ = 6.0 Å). The observed differences relative to the native complex (Figure 6, inset) are consistent with an induced fit mechanism underlying the tighter binding between AF113-18 and thrombin, whereby only the PS2-containing U17 undergoes a conformational change.
Figure 6.
Structural features underlying the tighter binding between AF113-18 PS2-modified RNA aptamer and α-thrombin. Superimposition of the crystal structures of the AF113-18:thrombin and native RNA:thrombin complexes. The color scheme is as follows: atoms in the AF113-18 RNA:thrombin complex are colored by atom, with RNA carbons in magenta and protein carbons in green, sulfur atoms are drawn as yellow spheres, 2′-fluorine atoms are light green and water molecules are cyan spheres of smaller radius; all RNA, protein and water oxygen atoms in the native complex are colored in gray. Selected residues and moieties are labeled, and selected contacts are marked with thin solid lines with distances in Å. Inset: The P18 environment in the structure of the native aptamer:thrombin complex. The drawing was generated with the program UCSF Chimera.
It is unlikely that PS2 modification at a site that is conformationally rigid and interacting with a region of the protein partner that also lacks the ability to flex will lead to substantial improvements in binding. Characteristics of the RNA backbone and thrombin binding site around P18 include some flexibility in the aptamer (U17 and P18), neighboring moieties (A7, G16) that can accommodate and stabilize the changes in conformation and stereoelectronics as a result of PS2 modification and amino acids on the thrombin surface that provide a hydrophobic environment situated in a shallow depression (SF-7). Rather than interacting with the guanidino moiety of Arg126, the PS2 moiety prefers methylene groups of the arginine side chain (Figure 6). Moreover, PS2 modification appears to result in the loss of the salt bridge between Arg126 and Glu127 seen in the native structure (19) (Figure 6, inset). Finally, water molecules are displaced because of PS2 modification and the more intimate interaction between the PS2 variant and thrombin. Thus, more water molecules are present in the vicinity of P18 in the structure of the native complex as compared to the AF113-18 complex structure despite similar resolutions. Although the crystal structure of AF83-7 or AF83-19 with VEGF165 is not available, we speculate that similarities exist between these PS2-modified aptamers (AF83-7 and AF83-9) and the AF113-8 aptamer and its interactions with thrombin.
Model system calculations
Optimized geometries for both phosphine oxide and phosphine sulfide are in excellent agreement with recently reported calculations (59) and crystal structures for related molecules (60). Full geometry optimizations for both the phosphine oxide/benzene and the phosphine sulfide/benzene complexes yielded structures with a face-centered geometry, i.e. the P = O and P = S bonds were approximately parallel to and centered over the face of the benzene ring. Detailed potential energy surface scans revealed additional, albeit less favorable, low-energy structures where the P = X bond was roughly coplanar with the benzene ring, forming an edge-on interaction. These results are conceptually similar to earlier computational studies for hydrogen sulfide and methanethiol interactions with benzene (61).
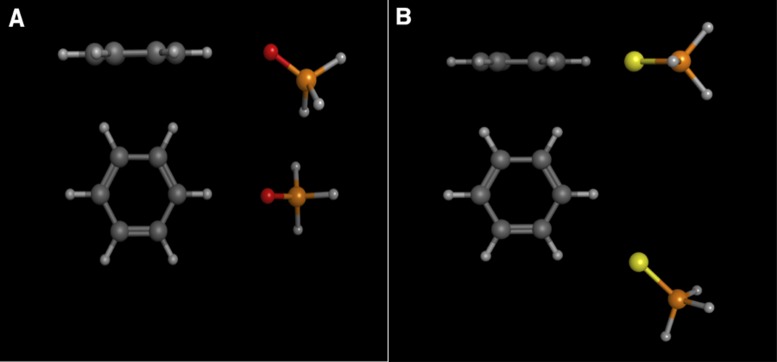
Figure 7A displays a local minimum structure for the phosphine oxide-benzene complex in an edge-on geometry. This structure is characteristic of a weak hydrogen bonding interaction, with a carbon–oxygen distance (i.e. donor–acceptor) of ∼3.35 Å, and a C—O—P angle of ∼145°. Figure 7B displays a low-energy structure for the phosphine sulfide-benzene complex in an edge-on geometry. This structure is not a proper local minimum, nor is it a true transition state (based on the eigenvalues computed in the frequency calculations). The potential energy surface scan reveals a flat, broad region on the surface with nearly degenerate energy values. This region of the potential energy surface defines a pathway that connects the global minimum geometry on the ‘top’ face of the benzene ring with the symmetry-related, degenerate energy minimum structure on the ‘bottom’ face of the benzene ring. During the potential energy surface scans, we varied the benzene ring—sulfur distance in 0.1 Å increments, and the angle between the benzene ring and the P = S bond vector in 0.5° increments. It is possible that a potential energy surface scan on a higher resolution grid might locate a proper transition state structure along this pathway. The structure displayed in Figure 7B, and numerous related, near energy-degenerate structures exhibit a ring carbon-sulfur distance range of 3.63–3.67 Å, and the ring plane—S—P angle ranges from ∼170-180°. These values correspond quite closely to those observed for the sulfur-phenylalanine interaction in the PS2-aptamer/protein crystal structure. The phosphine sulfide-benzene complexes are 0.4-0.5 kcal/mol more favorable than the edge-on local minimum energy structure identified for the phosphine oxide–benzene complex.
Figure 7.
(A) Side and top views of the local minimum energy structure for the phosphine oxide–benzene complex in an edge-on geometry. (B) Side and top views for a representative low-energy structure for the phosphine sulphide–benzene complex in an edge-on geometry. Carbon atoms are dark gray, phosphorous atoms are orange, oxygen atoms red, sulfur atoms yellow and hydrogen atoms are light gray.
While the low-energy, edge-on structures for the phosphine sulfide complex are in good agreement with the sulfur-phenylalanine interaction observed in the PS2-aptamer crystal structure, the edge-on local minimum structure for the phosphine oxide does not correspond well to the native aptamer complex. In the wild-type aptamer–protein crystal structure, the ribophosphate oxygen–phenylalanine distance is slightly longer than 5.3 Å, so we performed an additional geometry optimization for the phosphine oxide-benzene complex with the C—O distance constrained to this value. The interaction energy for this constrained complex is ∼2.5 kcal/mol less favorable than the phosphine sulfide-benzene complexes, suggesting that the sulfur-phenylalanine interaction observed in the crystal structure contributes substantially to the enhanced binding affinity observed for the PS2-aptamer relative to the wild-type aptamer.
We also calculated the static polarizability for both model compounds, yielding values of 7.3 Å3 for phosphine sulfide and 4.4 Å3 for phosphine oxide. As expected, the sulfur-containing molecule is significantly more polarizable and this should make a major contribution to the more favorable aptamer-protein interaction energy. The SP1 sulfur atom of the PS2-aptamer sits in a relatively nonpolar pocket formed by Phe-232 and the nonpolar side chain fragment of Arg-126, as shown in Figure 6 and SF-7. However, there are a number of charged residues in close proximity to this pocket, including Arg-233, Lys-235 and Lys-236. The proximity of these charged residues will likely contribute to a substantial favorable polarization contribution to the binding free energy for the PS2-aptamer as compared to the wild-type aptamer. This complex binding site environment is much too large to investigate with the quantum mechanical protocol we have used here, but merits more detailed study in future calculations.
DISCUSSION
Aptamers selected from combinatorial nucleic acid libraries have generally had normal phosphate backbones, and so would generally be unsuitable as drugs or diagnostics agents that are exposed to serum or cell supernatants because the nucleases degrade the phosphate backbone of nucleic acids. There are reports that phosphorothioate (PSO) modifications in the nucleic acid aptamers (either included in the selection library or added post selection) can provide modest improvements in the interaction with targets (27,29,62,63). However, the effect of PSO substitution cannot be predicted since the PSO substitution can lead to decreased (or increased) binding to a specific protein. In addition, the phosphorus of the PSO backbone modification is chiral and the stereocontrolled synthesis of a pure diastereoisomer of a PSO aptamer is not trivial (64,65). On the other hand, the chemical synthesis of PSO using phosphoramidite methodology typically results in a mixture of diastereoisomers (66) with a fairly limited influence on the affinity improvement (62). Indeed, substituting a mixture of diastereoisomers of PSO for PS2 in AF83-7 resulted in a 14-fold decrease in affinity (KDPSO = 13 100 ± 119 pM) relative to the native AF83-1 aptamer (KDPO2 = 961 ± 25 pM) (KD = mean ± SEM, n = 3) (SF-8); while, substituting a mixture of diastereoisomers of PSO for PS2 in AF113-18 revealed only a slight increase in affinity (KDPSO = 828 ± 13 pM) compared to the native AF113-1 (KDPO2 = 1871 ± 36 pM) (KD = mean ± SEM, n = 3) (SF-9). Our research has focused on the development of achiral, closely related mimics of natural nucleic acids. A particularly promising derivative has a PS2 internucleotide bridge (28,33). Strategically introduced PS2 modifications have significantly improved siRNA enhanced gene-silencing activity which was attributed to the enhanced loading into Ago2 as a result of the hydrophobic contributions of PS2 (39,41).
In this study, we describe a simple approach to assessing the effects of PS2 modified RNAs on two specific protein bindings. Using a modern BLI assay, we have shown that a single PS2 linkage can lead to nearly 1000-fold enhancement in target binding affinity while maintaining excellent target specificity. The atomic-resolution structural analysis establishes the basis for improved aptamer function, and the model system calculations reveal that the sulfur substitution does indeed contribute directly to a more favorable binding free energy, most likely via favorable polarization induced by charged side chains in close proximity to the PS2 binding pocket. In addition, a single PS2 modification also imparts improved nuclease resistance to the aptamer.
In many applications, RNA aptamers have distinct advantages over DNA aptamers and antibodies (4). RNA flexibility allows for a unique tertiary structure that can lead to tighter and more specific binding than for DNA aptamers. The structural adaptation of RNA aptamers also makes them smaller and somewhat easier to transfect into cells than DNA aptamers of the same nucleotide length (4). Unfortunately, unmodified RNA degrades more quickly than DNA in vivo and like the vast majority of their DNA counterparts, consequently, unmodified RNA aptamers typically do not perform well in downstream applications such as in diagnostic tests or as therapeutics. Numerous chemical modification strategies improve the in vivo stability of RNA aptamers against nucleases but few improve binding affinity to any significant degree (67). Beyond the PS2-walk for VEGF165 and α-thrombin RNA aptamer screening, we have reported on PS2/2′-OMe-PS2 modified siRNAs with pM binding affinity for human Ago2 protein (41). Literature reports demonstrate that the introduction of certain functional groups, especially hydrophobic moieties on the nucleobase that mimic amino acid side chains slows the off-rate of DNA aptamers (13,68). We have also initiated tests of the PS2-walk strategy for searching high affinity DNA aptamers such as thrombin DNA aptamer (69) and IgE DNA aptamer (70). In both cases, we have identified phosphates that result in significantly improved affinity for these targets upon PS2 replacement. This is in contrast to the modest change in affinity observed by Zandarashvili et al. for the simple case of the antennapedia homeodomain operator:receptor interaction and stresses the importance of local conformational flexibility in the DNA aptamer structure, as observed in the case of the best PS2-RNA binder for thrombin.
The findings presented in this article provide an opportunity to generate RNA aptamers of substantially improved affinity with a single PS2-moiety and without negatively affecting specificity. Our findings also provide crucial insights that could significantly accelerate the development of PS2-based RNA aptamers for diagnostics, therapeutics and applications beyond.
Supplementary Material
Acknowledgments
The authors thank Dr. David Gorenstein for his useful comments on the manuscript, and Mark Shumbera and Jeremy Pronchik for technical assistance. Dedicated to Prof. Fritz Eckstein on the occasion of the 50th anniversary of his first synthesis of a nucleoside phosphorothioate (71).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM086937 to X.Y.]; Brazilian National Council for Scientific and Technological Development (to R.Q.M); State of Rio de Janeiro Research Foundation (to R.Q.M); Centre of Molecular and Macromolecular Studies Statutory funds; Polish Academy of Sciences (to B.N.). Funding for open access charge: NIH [GM086937].
Conflict of interest statement. X.Y. is an employee of AM Biotechnologies LLC. The remaining authors declare no competing financial interests.
REFERENCES
- 1.Persch E., Dumele O., Diederich F. Molecular recognition in chemical and biological systems. Angew. Chem. Int. Ed. Engl. 2015;54:3290–3327. doi: 10.1002/anie.201408487. [DOI] [PubMed] [Google Scholar]
- 2.Breiten B., Lockett M.R., Sherman W., Fujita S., Al-Sayah M., Lange H., Bowers C.M., Heroux A., Krilov G., Whitesides G.M. Water networks contribute to enthalpy/entropy compensation in protein-ligand binding. J. Am. Chem. Soc. 2013;135:15579–15584. doi: 10.1021/ja4075776. [DOI] [PubMed] [Google Scholar]
- 3.Ellis J.J., Broom M., Jones S. Protein-RNA interactions: structural analysis and functional classes. Proteins. 2007;66:903–911. doi: 10.1002/prot.21211. [DOI] [PubMed] [Google Scholar]
- 4.Germer K., Leonard M., Zhang X. RNA aptamers and their therapeutic and diagnostic applications. Int. J. Biochem. Mol. Biol. 2013;4:27–40. [PMC free article] [PubMed] [Google Scholar]
- 5.Sun H., Zhu X., Lu P.Y., Rosato R.R., Tan W., Zu Y. Oligonucleotide aptamers: new tools for targeted cancer therapy. Mol. Ther. Nucleic Acids. 2014;3:e182. doi: 10.1038/mtna.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iqbal S.S., Mayo M.W., Bruno J.G., Bronk B.V., Batt C.A., Chambers J.P. A review of molecular recognition technologies for detection of biological threat agents. Biosens. Bioelectron. 2000;15:549–578. doi: 10.1016/s0956-5663(00)00108-1. [DOI] [PubMed] [Google Scholar]
- 7.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 8.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 9.Ng E.W., Shima D.T., Calias P., Cunningham E.T., Jr, Guyer D.R., Adamis A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug. Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 10.Ruckman J., Green L.S., Beeson J., Waugh S., Gillette W.L., Henninger D.D., Claesson-Welsh L., Janjic N. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998;273:20556–20567. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 11.Fukaya T., Abe K., Savory N., Tsukakoshi K., Yoshida W., Ferri S., Sode K., Ikebukuro K. Improvement of the VEGF binding ability of DNA aptamers through in silico maturation and multimerization strategy. J. Biotechnol. 2015;212:99–105. doi: 10.1016/j.jbiotec.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Gold L., Ayers D., Bertino J., Bock C., Bock A., Brody E.N., Carter J., Dalby A.B., Eaton B.E., Fitzwater T., et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimoto M., Yamashige R., Matsunaga K., Yokoyama S., Hirao I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat. Biotechnol. 2013;31:453–457. doi: 10.1038/nbt.2556. [DOI] [PubMed] [Google Scholar]
- 14.Burmeister P.E., Lewis S.D., Silva R.F., Preiss J.R., Horwitz L.R., Pendergrast P.S., McCauley T.G., Kurz J.C., Epstein D.M., Wilson C., et al. Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem. Biol. 2005;12:25–33. doi: 10.1016/j.chembiol.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 15.Draper D.E. Themes in RNA-protein recognition. J. Mol. Biol. 1999;293:255–270. doi: 10.1006/jmbi.1999.2991. [DOI] [PubMed] [Google Scholar]
- 16.Varani G. RNA-protein intermolecular recognition. Acc. Chem. Res. 1997;30:189–195. [Google Scholar]
- 17.Rowsell S., Stonehouse N.J., Convery M.A., Adams C.J., Ellington A.D., Hirao I., Peabody D.S., Stockley P.G., Phillips S.E. Crystal structures of a series of RNA aptamers complexed to the same protein target. Nat. Struct. Biol. 1998;5:970–975. doi: 10.1038/2946. [DOI] [PubMed] [Google Scholar]
- 18.Convery M.A., Rowsell S., Stonehouse N.J., Ellington A.D., Hirao I., Murray J.B., Peabody D.S., Phillips S.E., Stockley P.G. Crystal structure of an RNA aptamer-protein complex at 2.8 Å resolution. Nat. Struct. Biol. 1998;5:133–139. doi: 10.1038/nsb0298-133. [DOI] [PubMed] [Google Scholar]
- 19.Long S.B., Long M.B., White R.R., Sullenger B.A. Crystal structure of an RNA aptamer bound to thrombin. RNA. 2008;14:2504–2512. doi: 10.1261/rna.1239308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagai K. Recent advances in RNA-protein interaction studies. Mol. Biol. Rep. 1993;18:105–112. doi: 10.1007/BF00986764. [DOI] [PubMed] [Google Scholar]
- 21.Draper D.E., Reynaldo L.P. RNA binding strategies of ribosomal proteins. Nucleic Acids Res. 1999;27:381–388. doi: 10.1093/nar/27.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valegard K., Murray J.B., Stonehouse N.J., van den Worm S., Stockley P.G., Liljas L. The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein-RNA interactions. J. Mol. Biol. 1997;270:724–738. doi: 10.1006/jmbi.1997.1144. [DOI] [PubMed] [Google Scholar]
- 23.Valegard K., Murray J.B., Stockley P.G., Stonehouse N.J., Liljas L. Crystal structure of an RNA bacteriophage coat protein-operator complex. Nature. 1994;371:623–626. doi: 10.1038/371623a0. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen J., Brill W.K.D., Caruthers M.H. Synthesis and characterization of dinucleoside phosphorodithioates. Tetrahedron Lett. 1988;29:2911–2914. [Google Scholar]
- 25.Farschtschi N., Gorenstein D.G. Preparation of a deoxynucleoside thiophosphoramidite intermediate in the synthesis of nucleoside phosphorodithioates. Tetrahedron Lett. 1988;29:6843–6846. [Google Scholar]
- 26.Eldrup A.B., Bjergarde K., Felding J., Kehler J., Dahl O. Preparation of oligodeoxyribonucleoside phosphorodithioates by a triester method. Nucleic Acids Res. 1994;22:1797–1804. doi: 10.1093/nar/22.10.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X., Li N., Gorenstein D.G. Strategies for the discovery of therapeutic Aptamers. Exp. Op. Drug Disc. 2011;6:75–87. doi: 10.1517/17460441.2011.537321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X., Mierzejewski E. Synthesis of nucleoside and oligonucleoside dithiophosphates. New J. Chem. 2010;34:805–819. [Google Scholar]
- 29.Yang X., Gorenstein D.G. Progress in thioaptamer development. Curr. Drug Targets. 2004;5:705–715. doi: 10.2174/1389450043345074. [DOI] [PubMed] [Google Scholar]
- 30.Piotte M.E., Granger J.N., Cho Y., Gorenstein D.G. 1H 2D Nuclear magnetic resonance spectra of oligonucleotide phosphorodithioate, d(CGCTps2Tps2AAGCG). An unusual aairpin loop structure. J. Am. Chem. Soc. 1990;112:8632–8634. [Google Scholar]
- 31.Okruszek A., Sierzchala A., Feraron K.L., Stec W.J. Synthesis of oligo(deoxyribonucleoside phosphorodithioate)s by the dithiaphospholane approach. J. Org. Chem. 1995;60:6998–7005. [Google Scholar]
- 32.Cummins L., Graff D., Beaton G., Marshall W.S., Caruthers M.H. Biochemical and physicochemical properties of phosphorodithioate DNA. Biochemistry. 1996;35:8734–8741. doi: 10.1021/bi960318x. [DOI] [PubMed] [Google Scholar]
- 33.Marshall W.S., Caruthers M.H. Phosphorodithioate DNA as a potential therapeutic drug. Science. 1993;259:1564–1570. doi: 10.1126/science.7681216. [DOI] [PubMed] [Google Scholar]
- 34.Caruthers M.H., Beaton G., Wu J.V., Wiesler W. Chemical synthesis of deoxyoligonucleotides and deoxyoligonucleotide analogs. Meth. Enzymol. 1992;211:3–20. doi: 10.1016/0076-6879(92)11003-2. [DOI] [PubMed] [Google Scholar]
- 35.Tonkinson J.L., Guvakova M., Khaled Z., Lee J., Yakubov L., Marshall W.S., Caruthers M.H., Stein C.A. Cellular pharmacology and protein binding of phosphoromonothioate and phosphorodithioate oligodeoxynucleotides: a comparative study. Antisense Res. Dev. 1994;4:269–278. doi: 10.1089/ard.1994.4.269. [DOI] [PubMed] [Google Scholar]
- 36.Yang X., Fennewald S., Luxon B.A., Aronson J., Herzog N.K., Gorenstein D.G. Aptamers containing thymidine 3′-O-phosphorodithioates: synthesis and binding to nuclear factor-kappaB. Bioorg. Med. Chem. Lett. 1999;9:3357–3362. doi: 10.1016/s0960-894x(99)00600-9. [DOI] [PubMed] [Google Scholar]
- 37.Zandarashvili L., Nguyen D., Anderson K.M., White M.A., Gorenstein D.G., Iwahara J. Entropic enhancement of protein-DNA affinity by oxygen-to-sulfur substitution in DNA phosphate. Biophys. J. 2015;109:1026–1037. doi: 10.1016/j.bpj.2015.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Volk D.E., Yang X., Fennewald S.M., King D.J., Bassett S.E., Venkitachalam S., Herzog N., Luxon B.A., Gorenstein D.G. Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB. Bioorg. Chem. 2002;30:396–419. doi: 10.1016/s0045-2068(02)00510-2. [DOI] [PubMed] [Google Scholar]
- 39.Wu S.Y., Yang X., Gharpure K.M., Hatakeyama H., Egli M., McGuire M.H., Nagaraja A.S., Miyake T.M., Rupaimoole R., Pecot C.V., et al. 2′-OMe-phosphorodithioate-modified siRNAs show increased loading into the RISC complex and enhanced anti-tumour activity. Nat. Commun. 2014;5:3459–3462. doi: 10.1038/ncomms4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang X., Sierant M., Janicka M., Peczek L., Martinez C., Hassell T., Li N., Li X., Wang T., Nawrot B. Gene silencing activity of siRNA molecules containing phosphorodithioate substitutions. ACS Chem. Biol. 2012;7:1214–1220. doi: 10.1021/cb300078e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pallan P.S., Yang X., Sierant M., Abeydeera D.D., Hassell T., Martinez C., Janicka M., Nawrot B., Egli M. Crystal structure, stability and Ago2 affinity of phosphorodithioate-modified RNAs. RSC Adv. 2014;4:64901–64904. [Google Scholar]
- 42.Wiesler W.T., Caruthers M.H. Synthesis of phosphorodithioate DNA via sulfur-linked, base-labile protecting groups. J. Org. Chem. 1996;61:4272–4281. doi: 10.1021/jo960274y. [DOI] [PubMed] [Google Scholar]
- 43.Ke A., Doudna J.A. Crystallization of RNA and RNA-protein complexes. Methods. 2004;34:408–414. doi: 10.1016/j.ymeth.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 44.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 45.Vagin A., Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 46.Winn M.D., Isupov M.N., Murshudov G. N. Overview of the CCP4 suite and current developments. Acta Cryst. D. 2001;57:122–133. [Google Scholar]
- 47.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 48.Brunger A.T. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 50.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera – a visualization system for exploratory research and analysis. J. Comp. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 51.Frisch M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Scalmani G., Barone V., Mennucci B., Petersson G. A., et al. Gaussian 09, Revision D.01. Wallingford: Gaussian, Inc; 2009. [Google Scholar]
- 52.Dennington R., Keith T., Millam J. GaussView, Version 5. Shawnee Mission: Semichem, Inc; 2009. [Google Scholar]
- 53.White R., Rusconi C., Scardino E., Wolberg A., Lawson J., Hoffman M., Sullenger B. Generation of species cross-reactive aptamers using "toggle" SELEX. Mol. Ther. 2001;4:567–573. doi: 10.1006/mthe.2001.0495. [DOI] [PubMed] [Google Scholar]
- 54.Yang X. Solid-phase synthesis of oligodeoxynucleotide analogs containing phosphorodithioate linkages. Curr. Protoc. Nucleic Acid Chem. 2016;66:4.71.1–4.71.14. doi: 10.1002/cpnc.13. [DOI] [PubMed] [Google Scholar]
- 55.Yang X., Hodge R.P., Luxon B.A., Shope R., Gorenstein D.G. Separation of synthetic oligonucleotide dithioates from monothiophosphate impurities by anion-exchange chromatography on a Mono-Q column. Anal. Biochem. 2002;306:92–99. doi: 10.1006/abio.2001.5694. [DOI] [PubMed] [Google Scholar]
- 56.Davis K.A., Lin Y., Abrams B., Jayasena S.D. Staining of cell surface human CD4 with 2′-F-pyrimidine-containing RNA aptamers for flow cytometry. Nucleic Acids Res. 1998;26:3915–3924. doi: 10.1093/nar/26.17.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaur H., Yung L.Y. Probing high affinity sequences of DNA aptamer against VEGF165. PLoS One. 2012;7:e31196. doi: 10.1371/journal.pone.0031196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sullenger B.A., Gilboa E. Emerging clinical applications of RNA. Nature. 2002;418:252–258. doi: 10.1038/418252a. [DOI] [PubMed] [Google Scholar]
- 59.Alkorta I., Sánchez-Sanz G., Elguero J., Del Bene J.E. Pnicogen bonds between X=PH3 (X = O, S, NH, CH2) and phosphorus and nitrogen bases. J. Phys. Chem. A. 2014;118:1527–1537. doi: 10.1021/jp411623h. [DOI] [PubMed] [Google Scholar]
- 60.Preisenberger M., Pyykkö P., Shier A., Schmidbaur H. Isomerism of aurated phosphine sulfides, thiophosphinates, thiophosphonates, and thiophosphates: Structural and quantum chemical studies. Inorgan. Chem. 1999;38:5870–5875. [Google Scholar]
- 61.Ringer A.L., Senenko A., Sherrill C.D. Models of S/π interactions in protein structures: comparison of the H2S-benzene complex with PDB data. Protein Sci. 2007;16:2216–2223. doi: 10.1110/ps.073002307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dertinger D., Behlen L.S., Uhlenbeck O.C. Using phosphorothioate-substituted RNA to investigate the thermodynamic role of phosphates in a sequence specific RNA-protein complex. Biochemistry. 2000;39:55–63. doi: 10.1021/bi991769v. [DOI] [PubMed] [Google Scholar]
- 63.Jhaveri S., Olwin B., Ellington A.D. In vitro selection of phosphorothiolated aptamers. Bioorg. Med. Chem. Lett. 1998;8:2285–2290. doi: 10.1016/s0960-894x(98)00414-4. [DOI] [PubMed] [Google Scholar]
- 64.Yang X., Misiura K., Sochacki M., Stec W. J. Deoxyxylothymidine 3′-O-phosphorothioates: synthesis, stereochemistry and stereocontrolled incorporation into oligothymidylates. Bioorg. Med. Chem. Lett. 1997;7:2651–2656. [Google Scholar]
- 65.Sierzchala A., Okruszek A., Stec W.J. Oxathiaphospholane method of stereocontrolled synthesis of diribonucleoside 3′,5′-phosphorothioates. J. Org. Chem. 1996;61:6713–6716. doi: 10.1021/jo960811e. [DOI] [PubMed] [Google Scholar]
- 66.Burgers P.M., Eckstein F. Absolute configuration of the diastereomers of adenosine 5′-O-(1-thiotriphosphate): consequences for the stereochemistry of polymerization by DNA-dependent RNA polymerase from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1978;75:4798–4800. doi: 10.1073/pnas.75.10.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adler A., Forster N., Homann M., Goringer H.U. Post-SELEX chemical optimization of a trypanosome-specific RNA aptamer. Comb. Chem. High Throughput Screen. 2008;11:16–23. doi: 10.2174/138620708783398331. [DOI] [PubMed] [Google Scholar]
- 68.Davies D.R., Gelinas A.D., Zhang C., Rohloff J.C., Carter J.D., O'Connell D., Waugh S.M., Wolk S.K., Mayfield W.S., Burgin A.B., et al. Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets. Proc. Natl. Acad. Sci. U.S.A. 2012;109:19971–19976. doi: 10.1073/pnas.1213933109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Macaya R.F., Schultze P., Smith F.W., Roe J.A., Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. U.S.A. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Katilius E., Flores C., Woodbury N.W. Exploring the sequence space of a DNA aptamer using microarrays. Nucleic Acids Res. 2007;35:7626–7635. doi: 10.1093/nar/gkm922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eckstein F. Nucleoside Phosphorothioates. J. Amer. Chem. Soc. 1966;88:4292–4294. doi: 10.1021/ja00718a039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.