Abstract
Apoptosis, a form of programmed cell death, is a highly regulated process, the deregulation of which has been associated with the tumor initiation, progression, and metastasis in various cancers including breast cancer. Induction of apoptosis is a popular target of various therapies currently being tested or used for breast cancer treatment. Thus, identifying apoptotic mediators and regulators is imperative for molecular biologists and clinicians for benefit of patients. The regulation of apoptosis is complex and involves a tight equilibrium between the pro- and anti-apoptotic factors. Recent studies have highlighted the role of miRNAs in the control of apoptosis and their interplay with p53, the master guardian of apoptosis. Here, we summarize and integrate the data on the role of miRNAs in apoptosis in breast cancer and the clinical advantage it may offer for the prognosis or treatment of breast cancer patients.
Keywords: breast cancer, apoptosis, microRNA, apoptomiR, biomarker
Introduction
Breast cancer is the most common malignancy in women in western nations with a scary current estimate of one in eight US women likely to develop invasive breast cancer once in her lifetime (American Cancer Society, 2015; Ferlay et al., 2015). The lowest incidences of breast cancer are seen in Asia and Africa but the cases are steadily rising and poor survival rates remain a cause of grave concern (American Cancer Society, 2015; Ferlay et al., 2015). The various risk factors are age, density of the breast, family and personal health history, germline mutations in the tumor suppressor genes-BRCA1, BRCA2, and other breast cancer susceptibility genes, alcohol intake, endogenous estrogen levels, use of hormone replacement therapy, radiation exposure to the breast, obesity, and race/ethnicity (American Cancer Society, 2015; Ferlay et al., 2015). Breast cancer is a heterogeneous disease characterized by up to 21 distinct histological subtypes and at least four different molecular subtypes each with distinct presentations, response to treatment, and clinical outcomes (American Cancer Society, 2015; Ferlay et al., 2015).
There has been marked improvement in the breast cancer survival rates and decline in deaths owing to early detection, better understanding of the disease, and personalized treatment (Ferlay et al., 2015). Current breast cancer treatment modality involves surgery (both curative and preventive), radiation therapy, systemic chemotherapy, endocrine therapy, and targeted therapy that interfere with the cellular functions thereby inducing cell cycle arrest and eventually apoptosis, a form of programmed cell death (Gonzalez-Angulo et al., 2007; Florea and Busselberg, 2013). However, breast cancer still remains the leading cause of death from cancer among women. The most significant reasons for therapy failure are tumor metastases to distant sites and development of treatment resistance (Gonzalez-Angulo et al., 2007; Florea and Busselberg, 2013). It is often seen that metastatic and treatment resistant cells lose control over apoptosis and thus fail to die in response to various therapies. Identification of the mediators and regulators of the apoptotic pathway may help to tailor treatments for maximization of treatment efficacy leading to complete tumor remission and prevention of metastases (Kasibhatla and Tseng, 2003).
Recently, microRNAs (miRNAs) have emerged as critical regulators of apoptosis. miRNAs are small non-coding RNAs (~18–25 bases) regulating gene expression at the post-transcriptional and/or translational levels. Traditionally, they are known to bind to the 3′ untranslated region of the target transcript to bring about transcript degradation or inhibition of translation depending on the extent of complementarity (Bartel, 2004). However, recent research suggests that miRNA may bind anywhere in the transcript to regulate its levels and can also bring about increase in the transcript levels by various mechanisms (Antonio et al., 2006). It is increasingly becoming clear that miRNAs generate a complex network with mRNAs, snoRNAs, long non-coding RNAs, and RNA binding proteins to establish cellular transcriptome (Mattick and Makunin, 2006).
The generation of miRNA profiles of normal or patient samples using microarray, qPCR, or deep sequencing technology and its analyses has helped in identifying useful clinical correlations of miRNAs with potential applications in cancer diagnosis, prognosis or therapy (Hammond, 2006). Several human miRNAs have been reported to be significantly deregulated in at least one cancer type and have been shown to function as oncomiRs or tumor suppressor miRs. Such deregulation of miRNA expressions across various tumor types suggests that these miRNAs may be involved in crucial cellular pathways that are commonly deregulated in cancer development. In this review, we will focus on the recent progress of research on miRNA-mediated regulation of apoptosis in breast cancer, discuss pro- and anti-apoptotic miRNAs (ApoptomiRs), miRNA targets, and their interaction with the p53 pathway. Finally, we will discuss current/future cancer prognostics and treatment targets based on these apoptotic regulators.
Breast cancer and apoptosis
A critical balance between cell proliferation and apoptosis is essential for normal breast development. Accumulation of genetic or epigenetic mutations by various factors brings about disturbance in this balance leading to the development of breast tumors. Breast tumors are heterogeneous in their genetic composition and are histologically diverse based on receptor status, proliferation and differentiation markers that correlate to disease prognosis (Vakkala et al., 1999). The histological analyses guide clinicians in selecting adjuvant therapies for breast cancer treatment. Several studies have made attempts to correlate apoptosis and its associated molecular markers with known histologic and prognostic factors in breast cancer (Lipponen et al., 1994). A high apoptotic index has been correlated with increased tumor grade, aneuploidy, high mitotic index, negative status for estrogen receptor (ER), and progesterone receptor (PR), tumor necrosis and increased lymphocyte infiltration (Lipponen, 1999). The tumor suppressor gene, p53, and the BCL2 family have been widely studied in breast cancer. The mutant p53 correlates with negative ER status, high mitotic index, and the tumor grade, while BCL2 expression correlates to ER positive status, wild type p53, low mitotic index and low grade (Megha et al., 2002). Strikingly, expression of BCL2 is seen in ~80% of breast cancer patients. Contrary to expectations, high apoptotic index is significantly associated to poor prognosis while high BCL2 expression correlates to good prognosis of breast cancer patients (Silvestrini et al., 1996). Possible explanations may be that neither apoptotic index nor mutant p53 or BCL2 is independent predictor of survival in breast cancer patients. High apoptosis index also correlates to high proliferative index and negative ER status, which are poor prognostic factors. High apoptotic index may also put selective pressure on tumor cells leading to development of apoptosis resistant cells leading to tumor progression and decreased survival in patients (Frenzel et al., 2009). Similarly, BCL2, an estrogen regulated gene correlates with positive ER status, a good prognostic feature. Nevertheless, in vitro experiments with BCL2 overexpression or silencing in breast cancer cells confirm its pro-survival effects accounting for current pre-clinical and clinical trials involving BCL2 silencing for breast cancer treatment (Honma et al., 2015). Breast cancer cells show activation of the upstream apoptotic signaling such as an increase in Fas-L expression and presence of active caspases in cancer cells (Hengartner, 2001; Fulda and Debatin, 2006). However, upregulation of Inhibitors of apoptosis proteins (IAPs), X-linked inhibitor of apoptosis protein (XIAP), and Survivin inhibits activity of the caspases and blocks apoptotic signaling (Deveraux and Reed, 1999). Overall, the balance between the concentrations of pro- and anti-apoptotic proteins is an important regulatory factor for apoptotic regulation (Quail and Joyce, 2013).
Pro- and anti-apoptotic miRNAs in breast cancer
Deregulation of apoptosis is an important step in cancer as it allows the genetically unstable cells to survive and accumulate further mutations that eventually lead to tumorigenesis (Elmore, 2007). One of the mechanism by which miRNAs influence tumor development is by regulation of proteins involved in the apoptotic process. miRNAs that function to promote or inhibit apoptotis are called pro- and anti-apoptotic miRNAs, respectively. Table 1 enlists various miRNAs that have been reported to function as pro- or anti-apoptotic miRNAs in breast cancer.
Table 1.
List of apoptosis-associated miRNAs in breast cancer.
| miRNA | Target gene | Function of miRNA | Breast cancer cell lines/in vivo models | Citation | miRNA Cancer/normal | |
|---|---|---|---|---|---|---|
| Fold Change | p-value | |||||
| PRO-APOPTOTIC miRs | ||||||
| miR-7-5p | PSME3 | Inhibits cell proliferation and induces apoptosis in vitro and in vivo | MDA-MB-231, MCF7, and nude mice | Shi et al., 2015 | 2.08 | S |
| miR-15/16 | RPS6KB1 | Inhibits cell proliferation and promotes cell cycle arrest and caspase-3 dependent apoptosis | MDA-MB-231, MCF7 | Janaki Ramaiah et al., 2014 | 1.71, 1.44 | S, S |
| miR-15a-5p | SNCG, CCNE1 | Mediates cell cycle arrest and promotes cell apoptosis | MDA-MB-231, MDA-MB-231 | Luo et al., 2013 | 1.71 | S |
| miR-15a-3p | BCL2L1 | Inhibits the expression of BCL2L1 and activates caspase-3/7 activity to promote apoptosis | MDA-MB-231 | Druz et al., 2013 | ND | ND |
| miR-16-5p | CCND1, BCL2, METTL13 | Decreases cell growth and proliferation and induces apoptosis. Repression of METTL13 by miR-16 promotes apoptosis of cancer cells | MCF7 | Rivas et al., 2012 | 1.44 | S |
| mir-24-2-5p | BCL-2 | mir-24-2-5p induces apoptosis by targeting BCL-2 | MCF7 | Srivastava et al., 2011 | ND | ND |
| miR-26a | MTDH, EZH2, MCL1 | Induces cell apoptosis and suppresses tumorigenesis in vivo | MCF7, MDA-MB-231, and nude mice | Zhang et al., 2011; Gao et al., 2013 | 0.72 | S |
| Repression of MCL1 by miR-26a suppresses cell growth and proliferation | ||||||
| miR-26b | SLC7A11 | Impairs viability and induces cell apoptosis | MCF7 | Liu X. X. et al., 2011 | 0.83 | S |
| miR-31 | PRKCE (PKCϵ) | Enhances cell apoptosis, inhibits oncogenic NF-κB pathway and brings about indirect downregulation of BCL2 | MCF10A, MDA-MB-231. | Körner et al., 2013 | 1.07 | NS |
| miR-34a | BCL2, SIRT1, FRA1, LMTK3, AXL, NOTCH1, LDHA | Suppresses cells proliferation, glycolysis, migration, invasion, and induces apoptosis. Reduces cancer stem cell properties and increases sensitivity to doxorubicin treatment. | MCF7, MDA-MB-231, BT549, Hs578T, and nude mice | Mackiewicz et al., 2011; Li et al., 2013; Yang et al., 2013; Zhao et al., 2013; Park et al., 2014; Xiao et al., 2016 | 1.09 | NS |
| miR-124 | Ets-1 | Induces cell apoptosis, reduces cell proliferation, and colony formation. | MCF7, MDA-MB-231. | Li W. et al., 2014 | 1.1 | S |
| miR-125a | ELAVL1 | Promotes apoptosis and inhibits cell growth and proliferation | MCF7 | Guo et al., 2009 | 0.68 | S |
| miR-125b | MUC1, EPO, EPOR, ENPEP, CK2-α, CCNJ, MEGF9, ERBB2, ARID3B | miR-125b promotes DNA damage-induced apoptosis and regulates cell motility | ZR-75-1, BT549, MCF7, MDA-MB-231 | Rajabi et al., 2010; Akhavantabasi et al., 2012; Feliciano et al., 2013; Ferracin et al., 2013 | 0.27 | S |
| miR-145 | RTKN, c-Myc | Induces cell apoptosis and inhibits cancer cell growth | MCF7 | Sachdeva et al., 2009; Wang et al., 2009 | 0.17 | S |
| miR-146a/b | IRAK1, TRAF6 | Mediates tumor suppression and triggers apoptosis | MCF7 | Liu et al., 2015 | 1.11, 1.54 | NS, S |
| miR-290-3p | ARID4B | miR-290 enhances ER signaling and increases apoptosis thereby suppressing breast cancer progression | 6DT1, MVT-1 | Goldberger et al., 2013 | ND | ND |
| miR-486-5p | PIM-1 | Suppresses cell proliferation in vitro and in vivo, induces G0/G1 arrest, and promotes apoptosis | MDA-MB-231, T47D and nude mice | Zhang et al., 2014 | 0.07 | S |
| miR-497 | Bcl-w (Bcl2L2) | Inhibits cellular growth and enhances apoptosis, promotes G0/G1 cell phase arrest | MCF7 | Shen et al., 2012 | 0.38 | S |
| miR-502-5p | TRAF2 | Enhances early apoptosis and inhibits proliferation of breast cancer cells | MCF7, MDA-MB-231, and MCF-10A | Sun et al., 2014 | ND | ND |
| miR-769-3p | NDRG1 | Inhibits cell proliferation and enhances apoptosis | MCF7 | Luo et al., 2014 | ND | ND |
| miR-874 | CDK9 | Induces cell apoptosis and inhibits cell proliferation and brings about cell cycle arrest | MCF7, MDA-MB-231 | Wang L. et al., 2014 | 0.77 | S |
| miR-99a | mTOR | Induces cell apoptosis and suppresses cell viability | MCF7, MDA-MB-231 | Hu et al., 2014 | 0.22 | S |
| ANTI-APOPTOTIC miRs | ||||||
| miR-21 | BCL2, TIMP3, PDCD4, PTEN, TPM1, MASPIN, ANKRD46 | miR-21 inhibition suppress both cell growth in vitro and tumor growth in vivo miR-21 promotes invasion in breast cancer cells | MCF7, MDA-MB-231, MDA-MB-435, and nude mice | Zhu et al., 2007; Frankel et al., 2008; Yang et al., 2009; Song et al., 2010; Wang et al., 2011; Yan et al., 2011 | 4.81 | S |
| miR-24-3p | p27Kip1 | Promotes cell proliferation and inhibits cell apoptosis | MDA-MB-435, MDA-MB-468 | Lu et al., 2015 | 0.94 | NS |
| miR-100 | MTMR3 | Antagonism of miR-100 led to G2/M cell-cycle arrest and induce apoptosis | SK-BR-3 | He et al., 2015 | 0.23 | S |
| miR-155 | FOXO3a, SOCS1, RHOA | Induces cell survival and plays an important role in chemoresistance in breast cancer. | MCF7, MDA-MB-231 | Kong et al., 2008, 2010; Jiang et al., 2010 | 2.24 | S |
| miR-155 | TP53INP1 | miR-155 mediates cell proliferation and inhibits cell apoptosis | MCF7 | Zhang et al., 2013 | 2.24 | S |
| miR-96/182 | PFN1, FOXO1 | miR-182 promotes proliferation and invasion and inhibits apoptosis of breast cancer cells | MDA-MB-231, MCF7 | Guttilla and White, 2009; Liu et al., 2013 | miR-182 (5.31) | S |
| miR-96 (7.35) | S | |||||
| miR-196a | ANXA1 | Enhances cell proliferation, colony formation and suppresses apoptosis | T47D, MDA-MB-453, MDA-MB-231 | Luthra et al., 2008 | 5.12 | S |
| miR-221/222 | PTEN, PUMA, CASP3, p27Kip1 | Enforced expression of miR-221/222 promotes breast cancer cell proliferation, migration and invasion and inhibits apoptosis by targeting and blocking caspase-3 expression | MCF7, SKBR3, HCC1500, MDA-MB- 231, | Miller et al., 2008; Zhang et al., 2010; Ergun and Oztuzcu, 2014; Li et al., 2016 | miR-221 (0.81), miR-222 (0.79) | S, S |
| miR-210 | RAD52, GPD1L | miR-210 overexpression inhibits apoptosis | MCF7 | Crosby et al., 2009; Fasanaro et al., 2009 | 5.46 | S |
| miR-504 | P53 | miR-504 overexpression reduces p53 mediated apoptosis and cell cycle arrest in response to stress | MCF7 and nude mice | Hu et al., 2010 | 0.73 | S |
Pro-apoptomiRs
A total of 22 miRNAs have been reported so far to be involved in the induction of apoptosis suggesting them to be tumor suppressors. Tumor suppressor miRNAs prevent tumor progression by negatively regulating the expression of genes that promote cell proliferation, differentiation, migration, or apoptosis. We evaluated the expression of pro-apoptomiRs in breast cancer patients using StarBase software that analyzes TCGA data (Li J. H. et al., 2014). The data set included 683 breast cancer patients and 87 normal samples. Interestingly, we found that miR-26a, -26b, -125a, -125b, -145, -486-5p, -497, -874, and -99a that are known to function as proapoptomiRs were significantly downregulated in breast cancer patients. In the following section, we have discussed some pro-apoptotic miRNAs.
miR-15/16 cluster
miR-15/16 was shown to inhibit cell proliferation and promote cell cycle arrest and apoptosis in various breast cancer cell lines (Rivas et al., 2012; Druz et al., 2013; Luo et al., 2013; Janaki Ramaiah et al., 2014). The members of miR-15/16 cluster negatively regulate the anti-apoptotic protein BCL2, cell cycle regulators CCND1, CCNE1 and other proteins RPS6KB1, SNCG and METTL13 in breast cancer (refer Table 1).
miR-26a/b
miR-26a is downregulated in breast cancer specimens and cell lines and its transient transfection initiates apoptosis of breast cancer cell line, MCF7. Oncogenes, MTDH, and EZH2 (a chromatin regulator), were identified as direct targets of miR-26a. MCF7 xenografts with exogenous miR-26a show that a decrease in expression of both MTDH and EZH2 is accompanied by an increase in apoptosis (Zhang et al., 2011). MCL1, an anti-apoptotic member of the BCL2 family and SLC7A11, an amino acid transporter are other targets of miR-26a (Gao et al., 2013). Silencing of SLC7A11 mimics miR-26b aroused viability impairment and apoptosis in MCF7 cells.
miR-34a
Several groups have independently reported pro-apoptotic role of miR-34a in breast cancer (Mackiewicz et al., 2011; Li et al., 2013; Yang et al., 2013; Zhao et al., 2013; Park et al., 2014; Xiao et al., 2016). miR-34 overexpression suppresses cell proliferation, glycolysis, metastasis, invasion, stem cell properties, and promotes apoptosis and sensitivity to doxorubicin treatment in breast cancer (Mackiewicz et al., 2011; Li et al., 2013; Yang et al., 2013; Zhao et al., 2013; Park et al., 2014; Xiao et al., 2016). miR-34a is transcriptionally regulated by p53 and targets SIRT1 (a modulator of p53 activity) thus making a mir-34-p53 positive feedback loop (Li et al., 2013). Other targets of miR-34a are BCL2, FRA1, LMTK3, AXL, NOTCH1 and LDHA. miR-34a/AXL interaction reduces phospho-AKT expression, and impairs the motility of triple-negative breast cancer cells (Mackiewicz et al., 2011). miR-34a suppresses MCF7 cell proliferation in vitro and in vivo by targeting LMTK3 in an estrogen dependent manner (Zhao et al., 2013). Overall, miR-34a functions as a tumor suppressor in breast cancer as also reported for other cancers.
miR-125a/b
miR-125a/b promotes apoptosis and inhibits cell growth and proliferation. miR-125a functions as a tumor suppressor for breast cancer, with HuR as a direct and functional target (Guo et al., 2009). miR-125b perform its tumor suppressor function via the direct targeting of the 3′-UTRs of ENPEP, CK2-α, CCNJ, MUC1, and MEGF9 mRNAs. miR-125b regulates cell motility by targeting ERBB2 and ARID3B (Rajabi et al., 2010; Akhavantabasi et al., 2012; Feliciano et al., 2013; Ferracin et al., 2013). Overexpression of miR-125b markedly inhibits Taxol-induced cell cytotoxicity in MDA-MB-435, MDA-MB-231, and BT474 breast cancer cells by targeting 3′UTR of BAK1 mRNA, which is an effector of taxol induced apoptosis (Zhou et al., 2010).
Anti-apoptomiRs
A total of 11 miRNAs have been shown to inhibit apoptosis suggesting them to be oncomiRs in breast cancer. Among these miR-21, -155, -96, -182, -196a, and -210 were found to be highly upregulated in breast cancer patients (Table 1).
miR-21
miRNA 21 is overexpressed in many cancers and has been shown to have oncogenic activity. Si et al., 2007 showed that miR-21 knock down inhibits MCF7 cell growth in vitro and suppresses MCF7 cell derived breast tumor growth in a murine xenograft model (Si et al., 2007). This was associated with increased apoptosis and decreased cell proliferation by downregulation of anti-apoptotic BCL2. miR-21 was also shown to promote breast cancer cell invasion. Overall, miR-21 functions as an oncomiR and modulates tumorigenesis through regulation of genes such as ANKRD46, BCL2, TIMP3, PDCD4, PTEN, TPM1, and MASPIN in breast cancer (Zhu et al., 2007; Frankel et al., 2008; Yang et al., 2009; Song et al., 2010; Wang et al., 2011; Yan et al., 2011).
miR-155
miR-155 exerts its anti-apoptotic activity by downregulating FOXO3a for cell survival (Kong et al., 2010). Active form of FOXO3a resides in the nucleus and induces cell death by up-regulation of apoptotic proteins, such as BIM, p27, BNIP3, and 24p3 and repression of anti-apoptotic proteins, FLIP, and BCL-xL (Kong et al., 2010). miR-155 also targets TP531NP1, SOCS1, and RHOA and inhibits apoptosis and chemosensitivity in MCF7 cells (Kong et al., 2008, 2010; Jiang et al., 2010; Zhang et al., 2013).
miR-210
miR-210 is a hypoxia inducible miRNA that functions as an oncomiR in breast cancer. High miR-210 expression is a poor prognostic factor in breast cancer patients. miR-210 inhibition promotes apoptosis in breast cancer. It is known to target DNA repair protein RAD52 and a protein involved in mitochondrial function regulation, GPD1L (Crosby et al., 2009; Fasanaro et al., 2009).
miR-221/222
miR-221/222 are frequently up-regulated in human epithelial cancers. Enforced expression of miR-221/222 promotes breast cancer cell proliferation, migration, and invasion via targeting PTEN/AKT pathway. In breast cancer miR-221/222 directly targets pro-apoptotic proteins PUMA and CASP3 and cell cycle inhibitor, p27Kip1 to show its oncogenic effect (Zhang et al., 2010). miR-221/222 inhibition results in an increase in mitochondrial membrane potential and caspase-3/7 activity thereby leading to apoptosis in the A549 and MCF7 cell lines (Zhang et al., 2010).
Targets of apoptomiRs
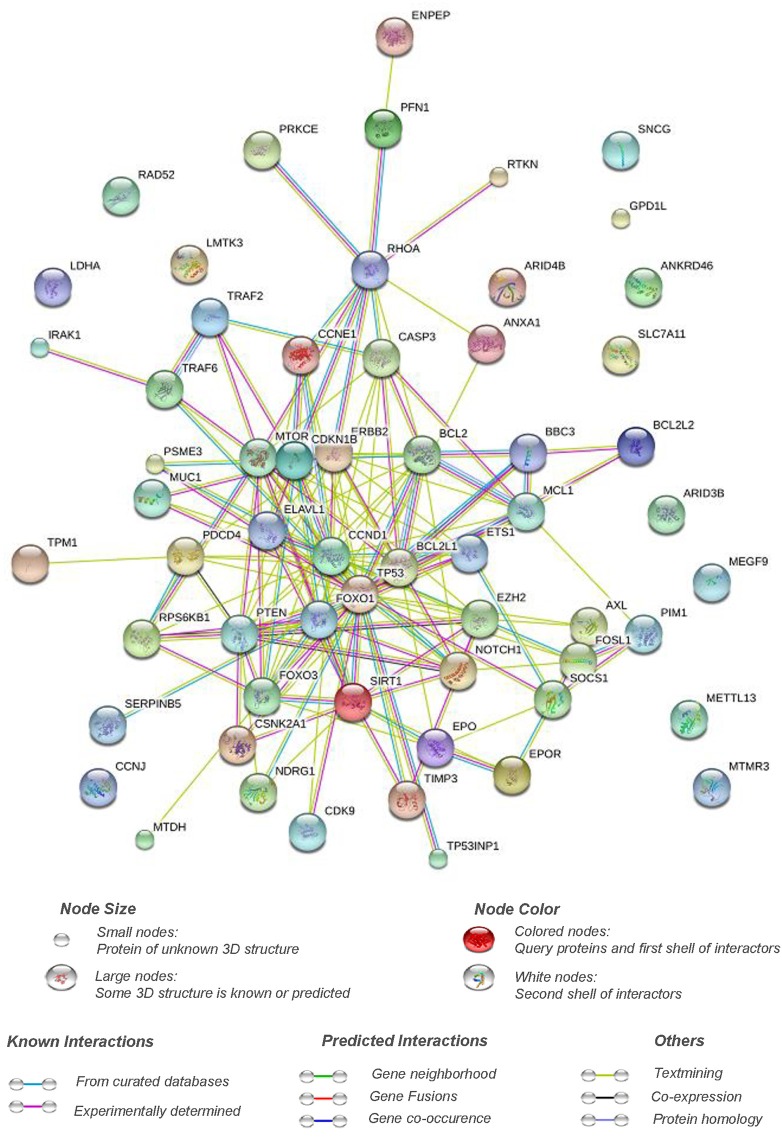
A total of 61 genes have been validated as direct targets of apoptomiRs. Interestingly, ~36% of these targets were found to be altered at the transcript (BCL2, CCNE1, CSKN2A1, ELAVL1, ERBB2, ETS1, EZH2, FOXO1, IRAK1, MUC1, MYC, NDRG1, PDCD4, SERPINB5, RPS6B1, SNCG, TIMP3, TP53, and TP53INP1) or the protein (BCL2, CASP3, CCNE1, CDKN1B, ERBB2, MUC1, MYC, PTEN, SERPINB5, TP53) levels in breast cancer based on analyses using genes to system breast cancer (G2SBC) database (Mosca et al., 2010). Whether the effects of the miRNAs on apoptosis are mediated through these targets has not been fully elucidated. Pathway analyses of the target genes of apoptomiRs using Reactome pathway browser identified transcriptional regulation by TP53 (CCNE1, CDKN1B, mTOR, CDK9, CSKN2A1, NDRG1, PTEN, PUMA, TP53, TP53INP1); signaling by nerve growth factor, NGF (IRAK1, ERBB2, CASP3, CDKN1B, RHOA, mTOR, FOXO1, PSME3, FOXO3, PRKCE, TRAF6, PTEN); and PI3K/AKT activation (CDKN1B, ERBB2, RHOA, mTOR, FOXO1, FOXO3, PTEN) as the top three overrepresented pathways (Joshi-Tope et al., 2005). NGF is known to be overexpressed in ~80% of breast tumor biopsies and has been implicated in enhanced survival, proliferation, migration, and inhibition of apoptosis in breast cancer (Molloy et al., 2011). Similarly, the pro-proliferative and anti-apoptotic effects of PI3K/AKT pathway are well documented. A protein interaction network analyses using STRING database (Von Mering et al., 2005) reveals that a strong interaction network with a high confidence (0.7) exists between the validated targets of the apoptomiRs suggesting that this set of the genes are biologically related (Figure 1).
Figure 1.
Pathway interaction network of targets of apoptomiRs in breast cancer. STRING interaction database (Von Mering et al., 2005) was used at setting of high confidence (0.7) to generate the interaction network of the list of the targets of apoptomiRs in breast cancer (Zhu et al., 2007; Frankel et al., 2008; Kong et al., 2008; Luthra et al., 2008; Miller et al., 2008; Crosby et al., 2009; Fasanaro et al., 2009; Guo et al., 2009; Guttilla and White, 2009; Sachdeva et al., 2009; Wang et al., 2009; Yang et al., 2009; Hu et al., 2010; Jiang et al., 2010; Kong et al., 2010; Rajabi et al., 2010; Song et al., 2010; Zhang et al., 2010; Liu X. X. et al., 2011; Mackiewicz et al., 2011; Srivastava et al., 2011; Wang et al., 2011; Yan et al., 2011; Zhang et al., 2011; Akhavantabasi et al., 2012; Rivas et al., 2012; Shen et al., 2012; Druz et al., 2013; Feliciano et al., 2013; Ferracin et al., 2013; Gao et al., 2013; Goldberger et al., 2013; Körner et al., 2013; Li et al., 2013; Liu et al., 2013; Luo et al., 2013; Yang et al., 2013; Zhang et al., 2013; Zhao et al., 2013; Ergun and Oztuzcu, 2014; Hu et al., 2014; Janaki Ramaiah et al., 2014; Luo et al., 2014; Li W. et al., 2014; Park et al., 2014; Sun et al., 2014; Wang L. et al., 2014; Zhang et al., 2014; He et al., 2015; Liu et al., 2015; Lu et al., 2015; Shi et al., 2015; Li et al., 2016; Xiao et al., 2016). The implication of the various connective lines based on different data sources are shown below the figure.
Interestingly, around 40% of the apoptomiR targets (BCL2, BCL2L1, BCl2L2, FOSL1, NOTCH1, TIMP3, TRAF2, TRAF6, ANXA1, CASP3, CDKN1B, FOXO1, FOXO3, IRAK1, MCL1, PTEN, PIM1, PSME3, PRKCE, PUMA, RHOA, SIRT1, TP53, TP53INP1, ERBB2, and ETS1) were found to be associated with regulation of apoptosis based on functional annotation clustering using DAVID microarray software (Huang et al., 2009a,b). Several of these belong to the p53 network or BCL2 family of proteins involved in control of apoptosis.
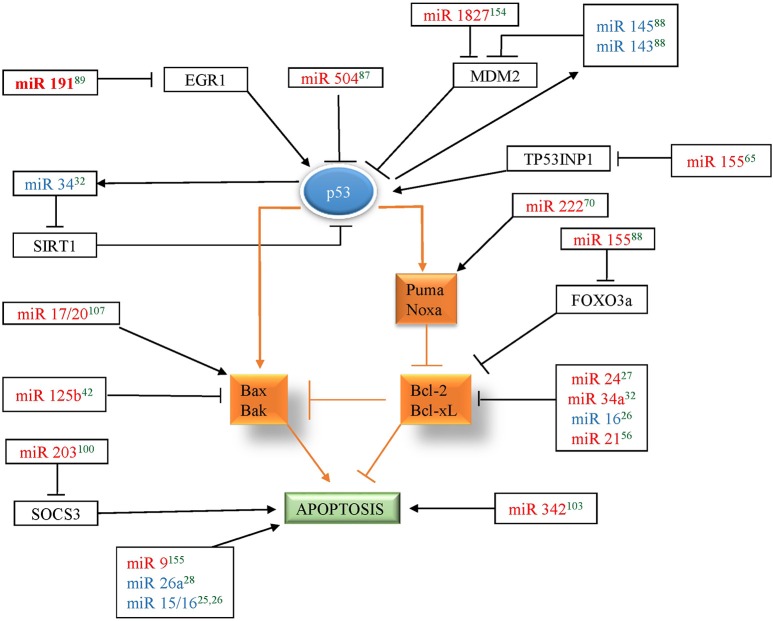
p53, is a tumor suppressor protein that mediates cellular responses to various stress signals like DNA damage, hypoxia, or aberrant oncogene expression by promoting cell-cycle checkpoints, DNA repair, cellular senescence, and apoptosis. In breast cancer, TP53 mutations have been associated with poor response to various therapies and thus poor prognosis. Several miRNAs were found to be associated with the p53 network by targeting p53 directly or indirectly (by targeting its upstream regulators or downstream mediators) to influence apoptosis in breast cancer. miR-504 was shown to directly target p53 and thus, inhibit p53 dependent apoptosis in breast cancer (Hu et al., 2010). MDM2 is an E3 ubiquitin-protein ligase that mediates ubiquitination of p53, leading to its proteasomal degradation and nuclear export. miR-143/145, and miR-1827 were found to directly target MDM2 in breast cancer and thus enhance p53-mediated stress responses, including apoptosis and senescence (Okamura et al., 2001). TP53INP1, SIRT1, and EGR1, regulators of p53 transcriptional activity, have also been shown to be direct targets of apoptomiRs- miR-155, miR-34, and miR-191, respectively, in breast cancer (Li et al., 2013; Nagpal et al., 2013; Zhang et al., 2013). Similarly, several miRNAs (miR-9, miR-17/20, 21, 125b, 155, 203, 222, and 342) affect the levels of p53 downstream genes to establish their effect on apoptosis. Additionally, p53 has been shown to induce the levels of several miRNAs involved in regulation of apoptosis in breast cancer. A total of six tumor suppressor miRNAs (miR-143, miR-145, miR-15, miR-16, miR-26a, and miR-34) have been shown to be upregulated by p53 in breast cancer (Sachdeva et al., 2009; Gao et al., 2013; Li et al., 2013; Janaki Ramaiah et al., 2014). Since miR-143/145 regulate MDM2 (regulator and transcriptional target of p53) and are also regulated by p53, it indicates existence of miR-143/145-MDM2-p53 feedback loop in breast cancer that controls cell proliferation and apoptosis (Okamura et al., 2001). Similarly, miR-34 is induced by p53 and targets SIRT1 (a modulator of p53 activity) thus making a mir-34-p53 positive feedback loop (Yamakuchi et al., 2008). Overall, apoptomiRs emerge as important regulators of the p53 network at multiple levels to regulate apoptosis in breast cancer (Figure 2).
Figure 2.
miRNAs in red color correspond to the miRNAs that regulate p53 in breast cancer and miRNAs in blue color corresponds to the miRNAs that are regulated by p53 in breast cancer. Arrow indicates activation and bar indicates inhibition. The references for each miRNA are marked as superscript in green color. 25(Druz et al., 2013), 26(Rivas et al., 2012), 27(Srivastava et al., 2011), 28(Zhang et al., 2011), 32(Li et al., 2013), 42(Feliciano et al., 2013), 56(Frankel et al., 2008), 65(Zhang et al., 2013), 70(Zhang et al., 2010), 87(Hu et al., 2010), 88(Okamura et al., 2001), 89(Nagpal et al., 2013), 100(Ru et al., 2011), 103(He et al., 2013), 107(Yu et al., 2014), 154(Zhang et al., 2016), 155(Zhou et al., 2015).
A number of BCL2 family members were found to be targeted by apoptomiRs by various groups. BCL2 family consists of both pro and anti-apoptotic proteins. BCL2, an antiapototic protein, was found to be targeted by four miRNAs (miR-21, -24, -34a, and -16) indicating that its levels are kept under tight control in breast tissue. Overexpression of miR-24, 34a, and 16 reduces BCL2 expression that leads to apoptosis through disruption of the mitochondrial membrane potential and the release of cytochrome C (Srivastava et al., 2011; Rivas et al., 2012; Li et al., 2013). Other anti-apoptotic members of the BCL2 family are also directly targeted by apoptomiRs- (BCL2L1- mir-15a-3p, BCL2L2- miR-497, MCL1- miR-26a; Shen et al., 2012; Druz et al., 2013; Gao et al., 2013). The level of BAK1, a pro-apoptotic BCL2 family protein, was found to be a direct target of oncomiR, miR-125b. PUMA, belonging to the BH3-only subgroup of BCL2 family protein was also found to be targeted by miR-221/222 (Zhang et al., 2010). miR-17/20 transduction of MCF7 cells has been shown to induce p53 and apoptosis characterized by activation of BAX, release of cytochrome C, and induction of BCL-X(S) in response to DNA damaging agents, like doxorubicin and tamoxifen. Overall, a multipronged attack by miRNAs seems to directly or indirectly target BCL2 family proteins or downstream targets in breast cancer.
CASP3 is a mediator of apoptosis that gets activated in apoptotic cell by both death ligand (extrinsic) and mitochondrial (intrinsic) pathways. It was found to be a direct target of miR-221/222 in breast cancer (Miller et al., 2008). Several cell cycle related genes (CCND1, CCNE1, CCNJ, CDK9, CDKN1B, and PSME3) also feature in the list suggesting important roles these apoptomiRs may play in maintaining balance between apoptosis and cell cycle. Finally, apoptomiRs target crucial genes involved mainly in the apoptotic or cell cycle related pathways in breast cancer. Fishing out more targets using advanced technologies will likely widen the range of the pathways affected by these targets.
miRNAs and drug resistance in breast cancer
The commonly used drugs for breast cancer treatment are Cyclophosphamide, Docetaxel, Doxorubicin, Epirubicin, Methotrexate, Paclitaxel, Cisplatin, Carboplatin, 5 Fluorouracil, Gemcitabine, etc. The patients are also given treatment based on the receptor status. The patients are tested for ER, Her2, and PR status. ER+ve patients receive tamoxifen (antagonist of ER signaling) and Her2+ve patients receive trastuzumab (herceptin), a monoclonal antibody against Her2, included in the chemotherapy regimen (Osborne, 1998; Ariazi et al., 2006). While the response of breast cancer patients to radiation, drug, or hormonal treatment has considerably improved the patient disease free survival and overall survival, chances of recurrence still remain. Since apoptosis mediated cancer cell death remains central to all the treatment strategies, the recurrence has been attributed to the development of resistance to apoptotic death and maintenance of cell viability by undergoing viable cellular responses such as cellular senescence (generates secretomes which can directly enhance the malignant phenotype) or autophagy (a state of reversible dormancy making it possible for cancer cells to survive and regrow at a later stage; Degenhardt et al., 2006).
Several mechanisms of chemoresistance in breast cancer have been reported in breast cancer such as upregulation of the ATP-binding cassette (ABC) transporters, drug inactivation or detoxification, alterations in genes related to cell cycle, apoptosis or DNA repair enzymes, epigenetic modifications, and activation of signaling pathways related to the progression of cancer (Baker et al., 2005; Pogribny et al., 2010). Recently, alterations of miRNA levels too have been linked to the development of drug resistance in breast cancer (Kovalchuk et al., 2008; Baylin, 2011). Very few miRNAs (miR-17/20, -21, -34a, -125b, -181a, -203, -218, -221, -222, -342) have been reported till date to affect the sensitivity to drugs (paclitaxel, cisplatin, doxorubicin, and taxol) or hormonal (tamoxifen and trastuzumab) treatment in various breast cancer cell lines (Table 2). These miRNAs affect the drug sensitivity by interfering in apoptotic death by affecting the levels of various apoptotic regulators (TP53, BCL2, BAX, BAK1, SURVIVIN, NOTCH1), members or regulators of AKT signaling pathway (AKT1, PTEN), ID4, NOTCH1 and SOCS3. Considering that the levels of several miRNAs change in response to drug treatment or are differentially regulated between sensitive versus resistant breast cancer cell lines, it is very likely that the list of miRNAs associated with drug resistance will expand. Some of these miRNAs may also function as biomarkers in predicting and monitoring drug response in breast cancer patients (Nana-Sinkam and Croce, 2013). These miRNAs are also being evaluated for their use in combination therapies to counteract drug resistance (Nana-Sinkam and Croce, 2013).
Table 2.
miRNAs associated with drug resistance in breast cancer.
| miRNA | Drug | Targets | Cell lines/in vivo models | Citation |
|---|---|---|---|---|
| miR-125b | Paclitaxel (Taxol) | Bcl-2 antagonist killer 1 (Bak1) is a direct target of miR-125b. miR-125b confers Taxol resistance through suppression of Bak1 expression | MDA-MB-435, MDA-MB-231, MDA-MB-436, MCF7, SKBR3, and BT474 | Zhou et al., 2010 |
| miR-203 | Cisplatin | Knockdown of miR-203 sensitized human breast cancer MCF7 cells to cisplatin-mediated apoptotic cell death | MCF7, ZR-75, and MDA-MB-231 | Ru et al., 2011 |
| SOCS3 is a novel target of miR-203 and plays an important role in cisplatin sensitivity of breast cancer cells | ||||
| miR-21 | Doxorubicin(ADR) | Dysregulation of miR-21 plays critical roles in the ADR (doxorubicin) resistance of breast cancer via targeting PTEN | MCF7 | Wang et al., 2011 |
| MIR-34a | Doxorubicin(ADR) | Ectopic miR-34a expression reduces cancer stem cell properties and increases sensitivity to doxorubicin treatment by directly targeting NOTCH1 | MCF7 | Li et al., 2012 |
| miR-342 | Tamoxifen | Overexpression of miR-342 upregulates the expression of ER-α mRNA and sensitizes the MCF7 cells to tamoxifen. miR-342 is down-regulated in breast tumors resistant to Tamoxifen. Reintroduction of miR-342 sensitizes refractory breast tumor cells to tamoxifen therapy. ID4 was identified as a putative target of miR-342 | MCF7, BT20, MDA-MB-231,T47D, HCC1937 | He et al., 2013; Crippa et al., 2014 |
| miR-221/222 | Trastuzumab | miR-221 mediated downregulation of PTEN confers trastuzumab resistance of HER2-positive breast cancers | SKBR3 and nude mice | Ye et al., 2014 |
| miR-181a | Adriamycin | miR-181a regulates the chemosensitivity to Adriamycin by targeting BCL2 in MCF7 and MCF7/ADR cells | MCF7 | Zhu et al., 2013 |
| miR-17/20 | Doxorubicin and Tamoxifen | miR-17/20 sensitized breast cancer cells to chemotherapy-induced apoptosis requires AKT1 | MCF7 | Yu et al., 2014 |
| miR-218 | Doxorubicin and Taxol | miR-218 is involved in the development of multi drug resistance in breast cancer cells via targeting SURVIVIN leading to evasion of apoptosis | MCF7 and nude mice | Hu et al., 2015 |
ApoptomiRs as biomarkers and therapeutic targets in breast cancer
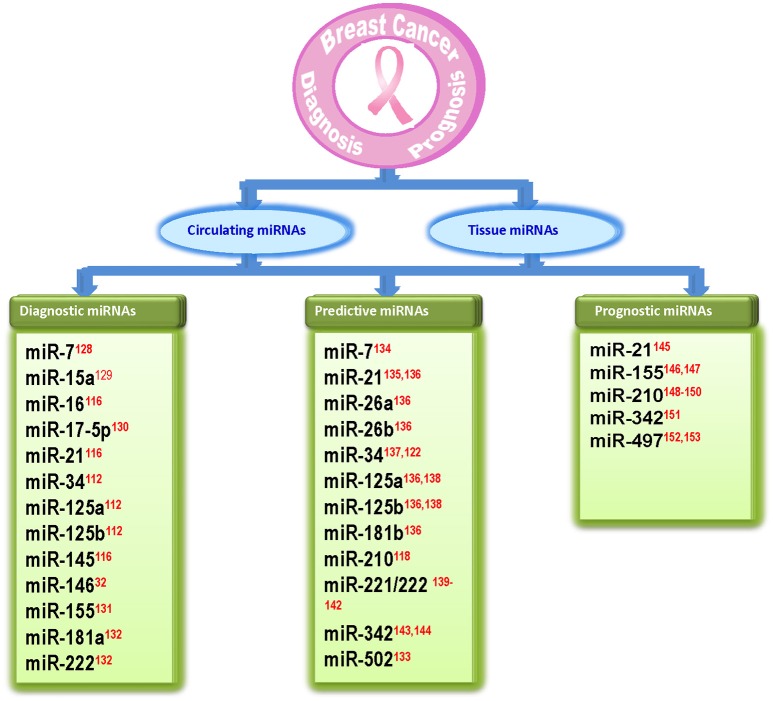
Early detection of breast cancer is important to reduce the risk of disease. Current diagnostic methods for early detection of breast cancer are still limited to some procedures such as tissue biopsies and histological examination using mRNA or protein biomarkers. However, there is still need for sensitive and specific markers. As miRNAs have small size, high specificity, and greater stability, they can be used as biomarkers with diagnostic (based on their ability to discriminate between normal and breast cancer patients), predictive [which are involved in response (sensitivity/resistance) to conventional breast cancer therapeutic strategies] or prognostic (which predict patient overall or disease-free survival) potential (Krutovskikh and Herceg, 2010; Abba et al., 2012; Schooneveld et al., 2015). Several studies have shown the role of miRNAs in diagnosis and prognosis of breast cancer. Iorio et al. (2005) identified a 13 miRNA signature that could differentiate breast cancer from normal breast tissue with 100% accuracy (Iorio et al., 2005). Blenkiron et al. (2007) identified 133 miRNAs that displayed aberrant expression levels in breast tumor tissues compared with normal breast tissues (Blenkiron et al., 2007). Recently several reports suggested that circulating miRNAs are highly stable in serum/plasma and levels of some miRNAs are specifically elevated in patients with breast cancer (Heneghan et al., 2010). In a recent study, nine circulating miRNAs were shown to be capable of discriminating between early-stage breast cancer and healthy controls (Kodahl et al., 2014). Another study reported three significantly overexpressed serum miRNAs in breast cancer patients as compared to breast cancer free controls (Ng et al., 2013). Recently, Bertoli et al. (2015) and Schooneveld et al. (2015) reported circulating and non-circulating miRNAs as breast cancer biomarkers. Several of the apoptomiRs have also been shown to serve as biomarkers in breast cancer (Abba et al., 2012; Bertoli et al., 2015; Figure 3). While some apoptomiRs have only diagnostic (miR-15a/16, -17, -181a, -145, -146) or predictive (miR-26a/b, -181b, -502) or prognostic (miR-497) potential, some have been proposed to serve as biomarkers for any two (miR-7, -34, -125a/b, -155, -222, -342) or all the three purposes (miR-21). Based on their presence and high stability in patient serum, capacity to reflect tumor status, and high clinical outcome, some apoptomiRs (miR-155, -21, -16, -222, and -210) are present as circulating miRNAs and can be used as potential biomarkers for breast cancer. miR-210 is a hypoxia regulated anti-apoptotic miRNA in breast cancer and high miR-210 baseline expression has been associated with poor prognosis and resistance to trastuzumab-included chemotherapy (Jung et al., 2012) High miR-155 levels too have been reported in tumor tissues sections and the serum of breast cancer patients and have been correlated to poor prognosis (Kong et al., 2014). Similarly, miR-21 upregulation is associated with taxol resistance and poor prognosis in breast cancer cells (Mei et al., 2010). miR-34 has been proposed to serve as a predictive marker for response to radiotherapy (Stankevicins et al., 2013). Kato et al. reported that low levels of miR-34a rendered breast cancer cells more resistant to radiotherapy (Kato et al., 2009).
Figure 3.
Diagram showing apoptosis related miRNAs that are identified as biomarkers in breast cancer. The references for each miRNA are marked as superscript in red color. 32(Li et al., 2013), 112(Iorio et al., 2005), 116(Ng et al., 2013), 118(Jung et al., 2012), 122(Kato et al., 2009), 128(Foekens et al., 2008), 129(Kodahl et al., 2014), 130(Volinia et al., 2006), 131(Wang F. et al., 2014), 132(Godfrey et al., 2013), 133(Lyng et al., 2012), 134(Chen and Bourguignon, 2014), 135(Mei et al., 2010), 136(Maillot et al., 2009), 137(Stankevicins et al., 2013), 138(Valabrega et al., 2007), 139(Zhao et al., 2008), 140(Wei et al., 2014), 141(Gan et al., 2014), 142(Rao et al., 2011), 143(He et al., 2013), 144(Cittelly et al., 2010), 145(Lee et al., 2011), 146(Song et al., 2012), 147(Kong et al., 2014), 148(Volinia et al., 2012), 149 (Li M. et al., 2014), 150(Wang J. et al., 2014), 151(Leivonen et al., 2014), 152(Shen et al., 2012), 153(Wang et al., 2013).
While successful miRNA delivery still remains a challenge, the prospects of using miRNAs as means and targets for breast cancer therapy remain attractive. There are two lines of action for therapeutic use of miRNAs: miRNA antagonists for miRNA inhibition and miRNA mimics for miRNA replacement therapy (Bader, 2012; Thorsen et al., 2012). Based on in vitro studies several miRNAs have reached preclinical trials and two miRNAs, have entered clinical trials. A locked nucleic acid-based antisense oligonucleotide against miR-122 (Miravirsen), successfully completed phase I clinical study and is undergoing phase II clinical trials for treatment of chronic hepatitis C virus infection (Janssen et al., 2013). In preclinical studies, Liu et al. reported that the injection of miR-34a mimic extended the survival of tumor-bearing mice (Liu C. et al., 2011). Another study demonstrated that systemic administration of miR-34 in a pancreatic xenograft cancer model significantly inhibited tumor growth and induced cancer cell apoptosis (Hu et al., 2013). A clinical trial using miR-34 mimic (MRX34) has already entered phase I clinical trial in patients with unresectable primary liver cancer or metastatic cancer with liver involvement (ClinicalTrials.gov Identifier: NCT01829971). Considering that miR-34 plays a major role in breast tumorigenesis and shows promising results in preclinical trials, miR-34 may soon enter clinical trials for treatment of breast cancer patients.
Conclusions
Mechanisms of cell death and its regulation during initiation, progression, and treatment of breast cancer are complex and still remain partially understood. The role of miRNAs as critical mediators of apoptotic cell death has been a revelation in the past decade and has given us means to manipulate cell death for breast cancer treatment or for disease monitoring. The current impediments for successful treatment are miRNA delivery issues, concerns over off-target effects and our incomplete knowledge of the functions of these miRNAs. Many of these miRNAs may have uninvestigated roles in other forms of cell death that may change the output based on the balance between the desired (apoptosis) and undesired (autophagy and necrosis) forms of cell death for breast cancer treatment. Nevertheless, miRNA delivery issues are slowly getting resolved, miRNAs are also being considered as means to target multiple aberrant pathways and certainly preclinical and clinical trials of some of these miRNAs look promising. The pool of apoptomiRs in breast cancer is likely to get bigger with advanced technologies such as large scale miRNA functional screens using 3D tumor models, thus giving us more probable candidates as novel targets for breast cancer therapy.
Author contributions
RK conceptualized the theme of the review and has written the review. SS compiled all the data on pro- and anti-apoptotic miRNAs and their correlation to drug resistance and has contributed in writing the review. PP and SA compiled the data of miRNA and p53 regulatory loop and contributed toward writing this section.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer KT and handling editor declared their shared affiliation, and the handling editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
This work was supported by the grant SB/S0/BB-0088/2013 from Science and Engineering Research Board, Department of Science & Technology, Government of India to RK. SS thanks Centre for Scientific and Industrial Research for Senior Research Fellowship.
References
- Abba M., Mudduluru G., Allgayer H. (2012). Micrornas in cancer: small molecules, big chances. Anticancer Agents Med. Chem. 12, 733–743. 10.2174/187152012802650273 [DOI] [PubMed] [Google Scholar]
- Akhavantabasi S., Sapmaz A., Tuna S., Erson-Bensan A. E. (2012). miR-125b targets ARID3B in breast cancer cells. Cell Struct. Funct. 37, 27–38. 10.1247/csf.11025 [DOI] [PubMed] [Google Scholar]
- American Cancer Society (2015). Breast Cancer Facts & Figures 2015-2016. Atlanta: American Cancer Society. [Google Scholar]
- Antonio M., Sanchez V., Liu J., Hannon G. J., Parker R. (2006). Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20, 515–524. 10.1101/gad.1399806 [DOI] [PubMed] [Google Scholar]
- Ariazi E. A., Ariazi J. L., Cordera F., Jordan V. C. (2006). Estrogen receptors as therapeutic targets in breast cancer. Curr. Top. Med. Chem. 6, 181–202. 10.2174/156802606776173483 [DOI] [PubMed] [Google Scholar]
- Bader A. G. (2012). miR-34 - a microRNA replacement therapy is headed to the clinic. Front. Genet. 3:120. 10.3389/fgene.2012.00120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker E. K., Johnstone R. W., Zalcberg J. R., El-Osta A. (2005). Epigenetic changes to the MDR1 locus in response to chemotherapeutic drugs. Oncogene 24, 8061–8075. 10.1038/sj.onc.1208955 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Baylin S. B. (2011). Resistance, epigenetics and the cancer ecosystem. Nat. Med. 17, 288–289. 10.1038/nm0311-288 [DOI] [PubMed] [Google Scholar]
- Bertoli G., Cava C., Castiglioni I. (2015). MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics 5, 1122–1143. 10.7150/thno.11543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenkiron C., Goldstein L. D., Thorne N. P., Spiteri I., Chin S. F., Dunning M. J., et al. (2007). MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 8:R214. 10.1186/gb-2007-8-10-r214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Bourguignon L. Y. (2014). Hyaluronan-CD44 interaction promotes c-Jun signaling and miRNA21 expression leading to Bcl-2 expression and chemoresistance in breast cancer cells. Mol. Cancer 13:52. 10.1186/1476-4598-13-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cittelly D. M., Das P. M., Spoelstra N. S., Edgerton S. M., Richer J. K., et al. (2010). Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol. Cancer 9:317. 10.1186/1476-4598-9-317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crippa E., Lusa L., De Cecco L., Marchesi E., Calin G. A., Radice P., et al. (2014). miR-342 regulates BRCA1 expression through modulation of ID4 in breast cancer. PLoS ONE 9:e87039. 10.1371/journal.pone.0087039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby M. E., Kulshreshtha R., Ivan M., Glazer P. M. (2009). MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 69, 1221–1229. 10.1158/0008-5472.CAN-08-2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K., Mathew R., Beaudoin B., Bray K., Anderson D., et al. (2006). Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10, 51–64. 10.1016/j.ccr.2006.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux Q. L., Reed J. C. (1999). IAP family proteins—suppressors of apoptosis. Genes Dev. 13, 239–252. 10.1101/gad.13.3.239 [DOI] [PubMed] [Google Scholar]
- Druz A., Chen Y. C., Guha R., Betenbaugh M., Martin S. E., Shiloach J. (2013). Large-scale screening identifies a novel microRNA, miR-15a-3p, which induces apoptosis in human cancer cell lines. RNA Biol. 10, 287–300. 10.4161/rna.23339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore S. (2007). Apoptosis: a review of programmed cell death. Toxicol. Pathol. 35, 495–516. 10.1080/01926230701320337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergun S., Oztuzcu S. (2014). miR-221: a critical player in apoptosis as a target of Caspase-3. Cancer Cell Microenviron. 1:e313 10.14800/ccm.313 [DOI] [Google Scholar]
- Fasanaro P., Greco S., Lorenzi M., Pescatori M., Brioschi M., Kulshreshtha R., et al. (2009). An integrated approach for experimental target identification of hypoxia-induced miR-210. J. Biol. Chem. 284, 35134–35143. 10.1074/jbc.m109.052779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feliciano A., Castellvi J., Artero-Castro A., Leal J. A., Romagosa C., Hernández-Losa J., et al. (2013). miR-125b acts as a tumor suppressor in breast tumorigenesis via its novel direct targets ENPEP, CK2-α, CCNJ, and MEGF9. PLoS ONE 8:e76247. 10.1371/journal.pone.0076247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., et al. (2015). Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, 359–386. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- Ferracin M., Bassi C., Pedriali M., Pagotto S., D'Abundo L., Zagatti B., et al. (2013). miR-125b targets erythropoietin and its receptor and their expression correlates with metastatic potential and ERBB2/HER2 expression. Mol. Cancer 12:130. 10.1186/1476-4598-12-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florea A.-M., Busselberg D. (2013). Breast cancer and possible mechanisms of therapy resistance. J. Local Glob. Health Sci. 2:9 10.5339/jlghs.2013.2 [DOI] [Google Scholar]
- Foekens J. A., Sieuwerts A. M., Smid M., et al. (2008). Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc. Natl. Acad. Sci. U.S.A. 105, 13021–13026. 10.1073/pnas.0803304105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel L. B., Christoffersen N. R., Jacobsen A., Lindow M., Krogh A., Lund A. H. (2008). Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 283, 1026–1033. 10.1074/jbc.M707224200 [DOI] [PubMed] [Google Scholar]
- Frenzel A., Grespi F., Chmelewskij W., Villunger A. (2009). Bcl2 family proteins in carcinogenesis and the treatment of cancer. Apoptosis 14, 584–596. 10.1007/s10495-008-0300-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda S., Debatin K.-M. (2006). Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene 25, 4798–4811. 10.1038/sj.onc.1209608 [DOI] [PubMed] [Google Scholar]
- Gan R., Yang Y., Yang X., Zhao L., Lu J., Meng Q. H. (2014). Downregulation of miR-221/222 enhances sensitivity of breast cancer cells to tamoxifen through upregulation of TIMP3. Cancer Gene Ther. 21, 290–296. 10.1038/cgt.2014.29 [DOI] [PubMed] [Google Scholar]
- Gao J., Li L., Wu M., Liu M., Xie X., Guo J., et al. (2013). MiR-26a inhibits proliferation and migration of breast cancer through repression of MCL-1. PLoS ONE 8:e65138. 10.1371/journal.pone.0065138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey A. C., Xu Z., Weinberg C. R., Getts R. C., Wade P. A., DeRoo L. A., et al. (2013). Serum microRNA expression as an early marker for breast cancer risk in prospectively collected samples from the Sister Study cohort. Breast Cancer Res. 15, R42. 10.1186/bcr3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberger N., Walker R. C., Kim C. H., Winter S., Hunter K. W. (2013). Inherited variation in miR-290 expression suppresses breast cancer progression by targeting the metastasis susceptibility gene Arid4b. Cancer Res. 73, 2671–2681. 10.1158/0008-5472.CAN-12-3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Angulo A. M., Morales-Vasquez F., Hortobagyi G. N. (2007). Overview of resistance to systemic therapy in patients with breast cancer. Adv. Exp. Med. Biol. 608, 1–22. 10.1007/978-0-387-74039-3_1 [DOI] [PubMed] [Google Scholar]
- Guo X., Wu Y., Hartley R. S. (2009). MicroRNA-125a represses cell growth by targeting HuR in breast cancer. RNA Biol. 6, 575–583. 10.4161/rna.6.5.10079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttilla I. K., White B. A. (2009). Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J. Biol. Chem. 284, 23204–23216. 10.1074/jbc.M109.031427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S. M. (2006). microRNA detection comes of age. Nat. Methods 3, 12–13. 10.1038/nmeth0106-12 [DOI] [PubMed] [Google Scholar]
- He T. Y. G., Yang L., Yang G., Chen Y., Zhang X. (2015). The role of miR-100 in regulating apoptosis of breast cancer cells. Sci. Rep. 5:11650. 10.1038/srep11650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. J., Wu J. Z., Ji M. H., Ma T., Qiao E. Q., Ma R., et al. (2013). miR-342 is associated with estrogen receptor-α expression and response to tamoxifen in breast cancer. Exp. Ther. Med. 5, 813–818. 10.3892/etm.2013.915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneghan H. M., Miller N., Lowery A. J., Sweeney K. J., Newell J., et al. (2010). Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann. Surg. 251, 499–505. 10.1097/SLA.0b013e3181cc939f [DOI] [PubMed] [Google Scholar]
- Hengartner M. O. (2001). Apoptosis: corralling the corpses. Cell 104, 325–328. 10.1016/S0092-8674(01)00219-7 [DOI] [PubMed] [Google Scholar]
- Honma N., Horii R., Ito Y., Saji S., Younes M., et al. (2015). Differences in clinical importance of Bcl-2 in breast cancer according to hormone receptors status or adjuvant endocrine therapy. BMC Cancer. 15:698. 10.1186/s12885-015-1686-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q. L., Jiang Q. Y., Jin X., Shen J., Wang K., Li Y. B., et al. (2013). Cationic microRNA-delivering nanovectors with bifunctional peptides for efficient treatment of PANC-1 xenograft model. Biomaterials 34, 2265–2276. 10.1016/j.biomaterials.2012.12.016 [DOI] [PubMed] [Google Scholar]
- Hu W., Chan C. S., Wu R., Zhang C., Sun Y., Song J. S., et al. (2010). Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol. Cell 38, 689–699. 10.1016/j.molcel.2010.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Xu K., Yagüe E. (2015). miR-218 targets survivin and regulates resistance to chemotherapeutics in breast cancer. Breast Cancer Res. Treat. 151, 269–280. 10.1007/s10549-015-3372-9 [DOI] [PubMed] [Google Scholar]
- Hu Y., Zhu Q., Tang L. (2014). MiR-99a antitumor activity in human breast cancer cells through targeting of mTOR expression. PLoS ONE 9:e92099. 10.1371/journal.pone.0092099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A. (2009a). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protoc. 4, 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A. (2009b). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio M. V., Ferracin M., Liu C. G., et al. (2005). MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 65, 7065–7070. 10.1158/0008-5472.CAN-05-1783 [DOI] [PubMed] [Google Scholar]
- Janaki Ramaiah M., Lavanya A., Honarpisheh M., Zarea M., Bhadra U., Bhadra M. P. (2014). MiR-15/16 complex targets p70S6 kinase 1 and controls cell proliferation in MDA-MB-231 breast cancer cells. Gene 552, 255–264. 10.1016/j.gene.2014.09.052 [DOI] [PubMed] [Google Scholar]
- Janssen H. L., Reesink H. W., Lawitz E. J., Zeuzem S., Rodriguez-Torres M., Patel K., et al. (2013). Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 368, 1685–1694. 10.1056/NEJMoa1209026 [DOI] [PubMed] [Google Scholar]
- Jiang S., Zhang H. W., Lu M. H., He X. H., et al. (2010). MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 70, 3119–3127. 10.1158/0008-5472.CAN-09-4250 [DOI] [PubMed] [Google Scholar]
- Joshi-Tope G., Gillespie M., Vastrik I., D'Eustachio P., Schmidt E., et al. (2005). Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 33, D428–D432. 10.1093/nar/gki072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung E. J., Santarpia L., Kim J., Esteva F. J., Moretti E., et al. (2012). Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer 118, 2603–2614. 10.1002/cncr.26565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasibhatla S., Tseng B. (2003). Why target apoptosis in cancer treatment? Mol. Cancer Ther. 2, 573–580. [PubMed] [Google Scholar]
- Kato M., Paranjape T., Müller R. U., Nallur S., Gillespie E., Keane K., et al. (2009). The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene 28, 2419–2424. 10.1038/onc.2009.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodahl A. R., Lyng M. B., Binder H., Cold S., Gravgaard K., Knoop A. S., et al. (2014). Novel circulating microRNA signature as a potential non-invasive multi-marker test in ER-positive early-stage breast cancer: a case control study. Mol. Oncol. 8, 874–883. 10.1016/j.molonc.2014.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W., He L., Coppola M., Guo J., Esposito N. N., Coppola D., et al. (2010). MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J. Biol. Chem. 285, 17869–17879. 10.1074/jbc.M110.101055 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kong W., He L., Richards E. J., Challa S., Xu C. X., Permuth-Wey J., et al. (2014). Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene 33, 679–689. 10.1038/onc.2012.636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W., Yang H., He L., Zhao J. J., Coppola D., et al. (2008). MicroRNA-155 is regulated by the transforming growth factor β/smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol. Cell Biol. 28, 6773–6784. 10.1128/MCB.00941-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körner C., Keklikoglou I., Bender C., Wörner A., Münstermann E., Wiemann S. (2013). MicroRNA-31 sensitizes human breast cells to apoptosis by direct targeting of protein kinase C epsilon (PKCepsilon). J. Biol. Chem. 288, 8750–8761. 10.1074/jbc.M112.414128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk O., Filkowski J., Meservy J., Ilnytskyy Y., et al. (2008). Involvement of microRNA-451 in resistance of the MCF7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol. Cancer Ther. 7, 2152–2159. 10.1158/1535-7163.MCT-08-0021 [DOI] [PubMed] [Google Scholar]
- Krutovskikh V. A., Herceg Z. (2010). Oncogenic microRNAs (OncomiRs) as a new class of cancer biomarkers. Bioessays 32, 894–904. 10.1002/bies.201000040 [DOI] [PubMed] [Google Scholar]
- Lee J. A., Lee H. Y., Lee E. S., Kim I., Bae J. W. (2011). Prognostic implications of microRNA-21 overexpression in invasive ductal carcinomas of the breast. J. Breast Cancer 14:269. 10.4048/jbc.2011.14.4.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivonen S. K., Sahlberg K. K., Mäkelä R., Due E. U., et al. (2014). High-throughput screens identify microRNAs essential for HER2 positive breast cancer cell growth. Mol. Oncol. 8, 93–104. 10.1016/j.molonc.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Lu Y., Wang H., Han X., Mao J., Li J., et al. (2016). miR-221/222 enhance the tumorigenicity of human breast cancer stem cells via modulation of PTEN/Akt pathway. Biomed. Pharmacother. 79, 93–101. 10.1016/j.biopha.2016.01.045 [DOI] [PubMed] [Google Scholar]
- Li J. H., Liu S., Zhou H., Qu L. H., Yang J. H. (2014). starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 42, D92–D97. 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Yuan L., Luo J., Gao J., Guo J., Xie X. (2013). MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin. Exp. Med. 13, 109–117. 10.1007/s10238-012-0186-5 [DOI] [PubMed] [Google Scholar]
- Li M., Ma X., Li M., Zhang B., et al. (2014). Prognostic role of microRNA-210 in various carcinomas: a syst ematic review and meta-analysis. Dis. Markers. 2014:106197. 10.1155/2014/106197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zang W., Liu P., Wang Y., Du Y., Chen X., et al. (2014). MicroRNA-124 inhibits cellular proliferation and invasion by targeting Ets-1 in breast cancer. Tumour Biol. 35, 10897–10904. 10.1007/s13277-014-2402-2 [DOI] [PubMed] [Google Scholar]
- Li X. J., Ji M. H., Zhong S. L., Zha Q. B., et al. (2012). MicroRNA-34a modulates chemosensitivity of breast cancer cells to adriamycin by targeting Notch1. Arch. Med. Res. 43, 514–521. 10.1016/j.arcmed.2012.09.007 [DOI] [PubMed] [Google Scholar]
- Lipponen P. (1999). Apoptosis in breast cancer: relationship with other pathological parameters. Endocr. Relat. Cancer 6, 13–16. 10.1677/erc.0.0060013 [DOI] [PubMed] [Google Scholar]
- Lipponen P., Aaltomaa S., Kosma V.-M., Syrjänen K. (1994). Apoptosis in breast cancer as related to histopathological characteristics and prognosis. Eur. J. Cancer 30A, 2068–2073. 10.1016/0959-8049(94)00342-3 [DOI] [PubMed] [Google Scholar]
- Liu C., Kelnar K., Liu B., Chen X., Calhoun-Davis T., Li H., et al. (2011). The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 17, 211–215. 10.1038/nm.2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wang Y., Li X., Zhang Y. J., Li J., Zheng Y. Q., et al. (2013). Expression and regulatory function of miRNA-182 in triple-negative breast cancer cells through its targeting of profilin 1. Tumour Biol. 34, 1713–1722. 10.1007/s13277-013-0708-0 [DOI] [PubMed] [Google Scholar]
- Liu R., Liu C., Chen D., Yang W. H., Liu X., Liu C. G., et al. (2015). FOXP3 controls an miR-146/NF-κB negative feedback loop that inhibits apoptosis in breast cancer cells. Cancer Res. 75, 1703–1713. 10.1158/0008-5472.CAN-14-2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. X., Li X. J., Zhang B., Liang Y. J., Zhou C. X., Cao D. X., et al. (2011). MicroRNA-26b is underexpressed in human breast cancer and induces cell apoptosis by targeting SLC7A11. FEBS Lett. 585, 1363–1367. 10.1016/j.febslet.2011.04.018 [DOI] [PubMed] [Google Scholar]
- Lu K., Wang J., Song Y., Zhao S., Liu H., et al. (2015). miRNA-24-3p promotes cell proliferation and inhibits apoptosis in human breast cancer by targeting p27Kip1. Oncol. Rep. 34, 995–1002. 10.3892/or.2015.4025 [DOI] [PubMed] [Google Scholar]
- Luo E. C., Chang Y. C., Sher Y. P., Huang W. Y., Chuang L. L., et al. (2014). MicroRNA-769-3p down-regulates NDRG1 and enhances apoptosis in MCF7 cells during reoxygenation. Sci. Rep. 4:e5908. 10.1038/srep05908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q., Li X., Li J., Kong X., Zhang J., Chen L., et al. (2013). MiR-15a is underexpressed and inhibits the cell cycle by targeting CCNE1 in breast cancer. Int. J. Oncol. 43, 1212–1218. 10.3892/ijo.2013.2034 [DOI] [PubMed] [Google Scholar]
- Luthra R., Singh R. R., Luthra M. G., Li Y. X., Hannah C., Romans A. M., et al. (2008). MicroRNA-196a targets annexin A1: a microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene 27, 6667–6678. 10.1038/onc.2008.256 [DOI] [PubMed] [Google Scholar]
- Lyng M. B., Laenkholm A. V., Sokilde R., et al. (2012). Global microRNA expression profiling of high-risk ER+ breast cancers from patients receiving adjuvant tamoxifen mono-therapy: a DBCG study. PLoS ONE 7:e36170. 10.1371/journal.pone.0036170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz M., Huppi K., Pitt J. J., Dorsey T. H., Ambs S., Caplen N. J. (2011). Identification of the receptor tyrosine kinase AXL in breast cancer as a target for the human miR-34a microRNA. Breast Cancer Res. Treat. 130, 663–679. 10.1007/s10549-011-1690-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillot G., Lacroix-Triki M., Pierredon S., et al. (2009). Widespread estrogen-dependent repression of micrornas involved in breast tumor cell growth. Cancer Res. 69, 8332–8340. 10.1158/0008-5472.CAN-09-2206 [DOI] [PubMed] [Google Scholar]
- Mattick J. S., Makunin I. V. (2006). Non-coding RNA. Hum. Mol. Genet. 15, R17–R29. 10.1093/hmg/ddl046 [DOI] [PubMed] [Google Scholar]
- Megha T., Ferrari F., Benvenuto A., Bellan C., Lalinga A. V., Lazzi S., et al. (2002). p53 mutation in breast cancer. Correlation with cell kinetics and cell of origin. J. Clin. Pathol. 55, 461–466. 10.1136/jcp.55.6.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei M., Ren Y., Zhou X., Yuan X. B., Han L., Wang G. X., et al. (2010). Downregulation of miR-21 enhances chemotherapeutic effect of taxol in breast carcinoma cells. Technol. Cancer Res. Treat. 9, 77–86. 10.1177/153303461000900109 [DOI] [PubMed] [Google Scholar]
- Miller T. E., Ghoshal K., Ramaswamy B., Roy S., Datta J., et al. (2008). MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J. Biol. Chem. 283, 29897–29903. 10.1074/jbc.M804612200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy N. H., Read D. E., Gorman A. M. (2011). Nerve growth factor in cancer cell death and survival. Cancers 3, 510–530. 10.3390/cancers3010510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca E., Alfieri R., Merelli I., Viti F., Calabria A., Milanesi L. (2010). A multilevel data integration resource for breast cancer study. BMC Syst. Biol. 4:76. 10.1186/1752-0509-4-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal N., Ahmad H. M., Molparia B., Kulshreshtha R. (2013). MicroRNA- 191, an estrogen-responsive microRNA, functions as an oncogenic regulator in human breast cancer. Carcinogenesis 34, 1889–1899. 10.1093/carcin/bgt107 [DOI] [PubMed] [Google Scholar]
- Nana-Sinkam S. P., Croce C. M. (2013). Clinical applications for microRNAs in cancer. Clin. Pharmacol. Ther. 93, 98–104. 10.1038/clpt.2012.192 [DOI] [PubMed] [Google Scholar]
- Ng E. K., Li R., Shin V. Y., Jin H. C., Leung C. P., Ma E. S., et al. (2013). Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS ONE 8:e53141. 10.1371/journal.pone.0053141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura S., Arakawa H., Tanaka T., Nakanishi H., et al. (2001). p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol. Cell 8, 85–94. 10.1016/S1097-2765(01)00284-2 [DOI] [PubMed] [Google Scholar]
- Osborne C. K. (1998). Tamoxifen in the treatment of breast cancer. N. Engl. J. Med. 339, 1609–1618. 10.1056/NEJM199811263392207 [DOI] [PubMed] [Google Scholar]
- Park E. Y., Chang E., Lee E. J., Lee H. W., Kang H. G., Chun K. H., et al. (2014). Targeting of miR34a-NOTCH1 axis reduced breast cancer stemness and chemoresistance. Cancer Res. 74, 7573–7582. 10.1158/0008-5472.CAN-14-1140 [DOI] [PubMed] [Google Scholar]
- Pogribny I. P., Filkowski J. N., Tryndyak V. P., Golubov A., Shpyleva S. I., Kovalchuk O. (2010). Alterations of microRNAs and their targets are associated with acquired resistance of MCF7 breast cancer cells to cisplatin. Int. J. Cancer 127, 1785–1794. 10.1002/ijc.25191 [DOI] [PubMed] [Google Scholar]
- Quail D. F., Joyce J. A. (2013). Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 11, 1423–1437. 10.1038/nm.3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajabi H., Jin C., Ahmad R., McClary A. C., Joshi M. D., Kufe D. (2010). Mucin 1 oncoprotein expression is suppressed by the miR-125b oncomir. Genes Cancer 1, 62–68. 10.1177/1947601909357933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao X., Di Leva G., Li M., Fang F., Devlin C., et al. (2011). MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene 30, 1082–1097. 10.1038/onc.2010.487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas M. A., Venturutti L., Huang Y. W., Schillaci R., Huang T. H., Elizalde P. V. (2012). Downregulation of the tumor-suppressor miR-16 via progestin-mediated oncogenic signaling contributes to breast cancer development. Breast Cancer Res. 14, R77. 10.1186/bcr3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru P., Steele R., Hsueh E. C., Ray R. B. (2011). Anti-miR-203 upregulates SOCS3 expression in breast cancer cells and enhances cisplatin chemosensitivity. Genes Cancer 2, 720–727. 10.1177/1947601911425832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M., Zhu S., Wu F., Wu H., Walia V., Kumar S., et al. (2009). p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc. Natl. Acad. Sci. U.S.A. 106, 3207–3212. 10.1073/pnas.0808042106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooneveld E. V., Wildiers H., Vergote I., Vermeulen P. B., Dirix L. Y., Van Laere S. J. (2015). Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. Breast Cancer Res. 17, 21. 10.1186/s13058-015-0526-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Li J., Xu L., Ma J., Li H., Xiao X., et al. (2012). miR-497 induces apoptosis of breast cancer cells by targeting Bcl-w. Exp. Ther. Med. 3, 475–480. 10.3892/etm.2011.428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Luo X., Li P., Tan J., Wang X., et al. (2015). miR-7-5p suppresses cell proliferation and induces apoptosis of breast cancer cells mainly by targeting REGγ. Cancer Lett. 358, 27–36. 10.1016/j.canlet.2014.12.014 [DOI] [PubMed] [Google Scholar]
- Si M.-L., Zhu S., Wu H., Lu Z., Wu F., Mo Y.-Y. (2007). miR-21-mediated tumor growth. Oncogene 26, 2799–2803. 10.1038/sj.onc.1210083 [DOI] [PubMed] [Google Scholar]
- Silvestrini R., Benini E., Veneroni S., Daidone M. G., Tomasic G., et al. (1996). p53 and bcl-2 expression correlates with clinical outcome in a series of node-positive breast cancer patients. J. Clin. Oncol. 14, 1604–1610. [DOI] [PubMed] [Google Scholar]
- Song B., Wang C., Liu J., Wang X., Lv L., Wei L., et al. (2010). MicroRNA-21 regulates breast cancer invasion partly by targeting tissue inhibitor of metalloproteinase 3 expression. J. Exp. Clin. Cancer Res. 29:29. 10.1186/1756-9966-29-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C. G., Wu X. Y., Wang C., Fu F. M., Shao Z. M. (2012). Correlation of miR-155 on formalin-fixed paraffin embedded tissues with invasiveness and prognosis of breast cancer. Zhonghua Wai Ke Za Zhi 50, 1011–1014. 10.3760/cma.j.issn.0529-5815.2012.11.012 [DOI] [PubMed] [Google Scholar]
- Srivastava N., Manvati S., Srivastava A., Pal R., Kalaiarasan P., Chattopadhyay S., et al. (2011). miR-24-2 controls H2AFX expression regardless of gene copy number alteration and induces apoptosis by targeting antiapoptotic gene BCL-2: a potential for therapeutic intervention. Breast Cancer Res. 13, R39. 10.1186/bcr2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankevicins L., Almeida da Silva A. P., Ventura Dos Passos F., Dos Santos Ferreira E., Menks Ribeiro M. C. G., David M., et al. (2013). MiR-34a is up-regulated in response to low dose, low energy X-ray induced DNA damage in breast cells. Radiat. Oncol. 8:231. 10.1186/1748-717X-8-231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L. L., Wang J., Zhao Z. J., Liu N., Wang A. L., et al. (2014). Suppressive role of miR-502-5p in breast cancer via downregulation of TRAF2. Oncol. Rep. 31, 2085–2092. 10.3892/or.2014.3105 [DOI] [PubMed] [Google Scholar]
- Thorsen S. B., Obad S., Jensen N. F., Stenvang J., Kauppinen S. (2012). The therapeutic potential of microRNAs in cancer. Cancer J. 18, 275–284. 10.1097/PPO.0b013e318258b5d6 [DOI] [PubMed] [Google Scholar]
- Vakkala M., Lahteenmaki K., Raunio H., Paakko P., Soini Y. (1999). Apoptosis during breast carcinoma progression. Clin. Cancer Res. 5, 319–324. [PubMed] [Google Scholar]
- Valabrega G., Montemurro F., Aglietta M. (2007). Trastuzumab: mechanism of action, resistance and future perspectives in HER2-overexpressing breast cancer. Ann. Oncol. 18, 977–984. 10.1093/annonc/mdl475 [DOI] [PubMed] [Google Scholar]
- Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., et al. (2006). A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261. 10.1073/pnas.0510565103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S., Galasso M., Sana M. E., Wise T. F., et al. (2012). Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc. Natl. Acad. Sci. U.S.A. 109, 1–6. 10.1073/pnas.1200010109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Mering C., Jensen L. J., Snel B., Hooper S. D., Krupp M., et al. (2005). STRING: known and predicted protein–protein associations integrated and transferred across organisms. Nucleic Acids Res. 33, D433–D437. 10.1093/nar/gki005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Hou J., Jin W., Li J., Yue Y., Jin H., et al. (2014). Increased circulating microRNA-155 as a potential biomarker for breast cancer screening: a meta-analysis. Molecules 19, 6282–6293. 10.3390/molecules19056282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhao J., Shi M., Ding Y., et al. (2014). Elevated expression of miR-210 predicts poor survival of cancer patients: a systematic review and meta-analysis. PLoS ONE 9:e89223. 10.1371/journal.pone.0089223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Gao W., Hu F., Xu Z., Wang F. (2014). MicroRNA-874 inhibits cell proliferation and induces apoptosis in human breast cancer by targeting CDK9. FEBS Lett. 588, 4527–4535. 10.1016/j.febslet.2014.09.035 [DOI] [PubMed] [Google Scholar]
- Wang S., Bian C., Yang Z., Bo Y., Li J., Zeng L., et al. (2009). miR-145 inhibits breast cancer cell growth through RTKN. Int. J. Oncol. 34, 1461–1466. 10.3892/ijo_00000275 [DOI] [PubMed] [Google Scholar]
- Wang S., Li H., Wang J., Wang D. (2013). Expression of microRNA-497 and its prognostic significance in human breast cancer. Diagn Pathol. 8:172. 10.1186/1746-1596-8-172 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang Z. X., Lu B. B., Wang H., Cheng Z. X., Yin Y. M. (2011). MicroRNA-21 modulates chemosensitivity of breast cancer cells to doxorubicin by targeting PTEN. Arch. Med. Res. 42, 281–290. 10.1016/j.arcmed.2011.06.008 [DOI] [PubMed] [Google Scholar]
- Wei Y., Lai X., Yu S., Chen S., Ma Y., Zhang Y., et al. (2014). Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast Cancer Res. Treat. 147, 423–431. 10.1007/s10549-014-3037-0 [DOI] [PubMed] [Google Scholar]
- Xiao X., Huang X., Ye F., Chen B., Song C., Wen J., et al. (2016). The miR-34a-LDHA axis regulates glucose metabolism and tumor growth in breast cancer. Sci. Rep. 6:21735. 10.1038/srep21735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M., Ferlito M., Lowenstein C. J. (2008). miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. U.S.A. 105, 13421–13426. 10.1073/pnas.0801613105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L. X., Wu Q. N., Zhang Y., Li Y. Y., Liao D. Z., et al. (2011). Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth. Breast Cancer Res. 13:e2803. 10.1186/bcr2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Li Y., Gao J., Zhang T., Li S., Luo A., et al. (2013). MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene 32, 4294–4303. 10.1038/onc.2012.432 [DOI] [PubMed] [Google Scholar]
- Yang Y., Chaerkady R., Beer M. A., Mendell J. T., Pandey A. (2009). Identification of miR-21 targets in breast cancer cells using a quantitative proteomic approach. Proteomics 9, 1374–1384. 10.1002/pmic.200800551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Bai W., Zhu H., Zhang X., Chen Y., Wang L., et al. (2014). MiR-221 promotes trastuzumab-resistance and metastasis in HER2-positive breast cancers by targeting PTEN. BMB Rep. 47, 268–273. 10.5483/BMBRep.2014.47.5.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Xu Z., Disante G., Wright J., et al. (2014). miR-17/20 sensitization of breast cancer cells to chemotherapy-induced apoptosis requires Akt1. Oncotarget 5, 1083–1090. 10.18632/oncotarget.1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Liu X. X., He J. R., Zhou C. X., Guo M., He M., et al. (2011). Pathologically decreased miR-26a antagonizes apoptosis and facilitates carcinogenesis by targeting MTDH and EZH2 in breast cancer. Carcinogenesis 32, 2–9. 10.1093/carcin/bgq209 [DOI] [PubMed] [Google Scholar]
- Zhang C., Liu J., Tan C., Yue X., Zhao Y., Peng J., et al. (2016). microRNA-1827 represses MDM2 to to positively regulate tumor suppressor p53 and suppress tumorigenesis. Oncotarget. 7, 8783–8796. 10.18632/oncotarget.7088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Zhang J., Zhang A., Wang Y., Han L., et al. (2010). PUMA is a novel target of miR-221/222 in human epithelial cancers. Int. J. Oncol. 37, 1621–1626. 10.3892/ijo_00000816 [DOI] [PubMed] [Google Scholar]
- Zhang C. M., Zhao J., Deng H. (2013). MiR-155 promotes proliferation of human breast cancer MCF7 cells through targeting tumor protein 53-induced nuclear protein 1. J. Biomed. Sci. 20:79. 10.1186/1423-0127-20-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Liu Z., Cui G., Wang X., Yang Z. (2014). MicroRNA-486-5p targeting PIM-1 suppresses cell proliferation in breast cancer cells. Tumour Biol. 35, 11137–11145. 10.1007/s13277-014-2412-0 [DOI] [PubMed] [Google Scholar]
- Zhao G., Guo J., Li D., Jia C., Yin W., Sun R., et al. (2013). MicroRNA-34a suppresses cell proliferation by targeting LMTK3 in human breast cancer MCF7 cell line. DNA Cell Biol. 32, 699–707. 10.1089/dna.2013.2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. J., Lin J., Yang H., Kong W., He L., Ma X., et al. (2008). MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J. Biol. Chem. 283, 31079–31086. 10.1074/jbc.M806041200 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhou E., Hui N., Shu M., Wu B., Zhou J. (2015). Systematic analysis of the p53-related microRNAs in breast cancer revealing their essential roles in the cell cycle. Oncol Lett. 10, 3488–3494. 10.3892/ol.2015.3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Liu Z., Zhao Y., Ding Y., Liu H., et al. (2010). MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J. Biol. Chem. 285, 21496–21507. 10.1074/jbc.M109.083337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Si M. L., Wu H., Mo Y. Y. (2007). MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J. Biol. Chem. 282, 14328–14336. 10.1074/jbc.M611393200 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Wu J., Li S., Ma R., et al. (2013). The function role of miR-181a in chemosensitivity to adriamycin by targeting Bcl-2 in low-invasive breast cancer cells. Cell Physiol. Biochem. 32, 1225–1237. 10.1159/000354521 [DOI] [PubMed] [Google Scholar]