Abstract
Packaging of DNA into chromatin affects all processes on DNA. Nucleosomes present a strong barrier to transcription, raising important questions about the nature and the mechanisms of overcoming the barrier. Recently it was shown that DNA sequence, DNA–histone interactions and backtracking by RNA polymerase II (Pol II) all contribute to formation of the barrier. After partial uncoiling of nucleosomal DNA from histone octamer by Pol II and backtracking of the enzyme, nucleosomal DNA recoils on the octamer, locking Pol II in the arrested state. Histone chaperones and transcription factors TFIIS, TFIIF and FACT facilitate transcription through chromatin using different molecular mechanisms.
Keywords: chromatin, nucleosome, RNA polymerase II, pausing, arrest, transcription, elongation, mechanism, structure, intermediates
Transcribing RNA polymerase II (Pol II) induces extensive chromatin remodeling facilitated by histone chaperones and elongation factors and accompanied by limited histone exchange (see [1] for review). At the same time, histones are fully evicted only from highly transcribed genes [1]; thus Pol II typically encounters nucleosomes during transcription of every ~200 bp DNA region. Nucleosomes remaining on transcribed genes form two types of barriers for transcribing Pol II [2,3]. In yeast and Drosophila each nucleosome presents a barrier where Pol II is paused after transcribing ~15 and ~50 bp from the nucleosome boundary [2,3]; these barriers are also universally observed in vitro [4]. A much higher barrier of the second type is formed when the active center of the enzyme is positioned ~10 bp upstream of the first (+1) transcribed nucleosome in Drosophila [3]. However, the relative contribution to this pause from the +1 nucleosome and negative elongation factors is not clear, particularly for highly expressed genes [3].
When Pol II encounters a barrier during transcript elongation, either DNA-bound proteins or DNA sequences that disfavor addition of the next NTP, polymerase backtracks by sliding the transcription bubble and RNA-DNA hybrid upstream along the template. This displaces the RNA 3′ end from the Pol II active site, resulting in transcriptional arrest. Rapid relief of arrest requires protein factor TFIIS, which acts along with the Pol II active center to drive cleavage of the transcript. This restores alignment of the 3′ end with the active center and releases the downstream RNA segment [5]. Arrest sites are rare within DNA but backtracking and arrest are general properties of Pol II complexes halted just downstream (~+17 to +32) of transcription start [6]. This is potentially important for the interaction of newly-initiated Pol II complexes with the +1 nucleosome.
A single nucleosome typically forms a high, asymmetrical barrier of the first type for Pol II transcription in vitro [4,7]; however, the putative regulatory −10 barrier of the second type observed in vivo has not been recapitulated in vitro. The strong +15 and +50 nucleosomal barriers are nucleosome-specific, Pol II-specific, and were described for all analyzed organisms, from yeast to human [4,8,9]. On any given DNA sequence wrapped onto a nucleosome, the barrier forms in distinct positions within the +(10–20) and +(40–50) regions [4]. Thus, these nucleosomal barriers are “universal signatures” of transcription through chromatin by Pol II both in vitro and in vivo [2–4].
Both DNA-histone interactions and Pol II pausing and backtracking contribute to formation of the barrier. Mapping of intranucleosomal DNA-histone interactions in single nucleosomes [10,11] identified two regions of strong interactions [+(25–35) and +(70–80)] that significantly affect the rate and efficiency of Pol II progression through a nucleosome, contributing to formation of the +15 and +50 nucleosomal barriers, respectively [4]. An additional DNA region +(89–102), the polar barrier sequence, determines overall affinity of DNA-histone interactions in a sequence-specific way, contributes to the +50 pausing and dictates overall height of the nucleosomal barrier to transcription [12,13]. The height of the barrier can also be strongly affected by a single nick in nucleosomal DNA [14].
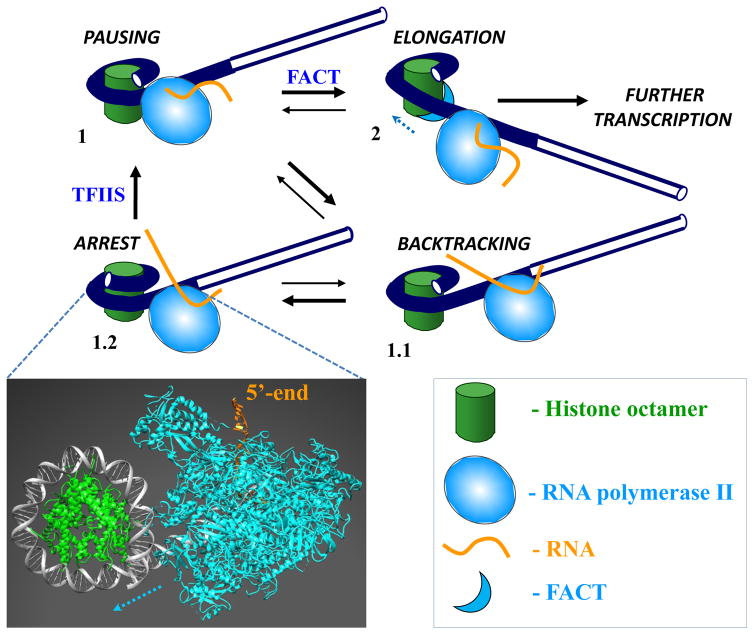
The nucleosomal barrier is largely relieved after Pol II advances beyond position +49. Initially a small, Pol II-containing intranucleosomal DNA loop (Ø-loop) forms on the surface of histone octamer at position +49 [12,15]. The Ø-loop is stabilized by Pol II-histone interactions that transiently and locally replace DNA-histone interactions [16]; the high efficiency of Ø-loop formation is characteristic for the Pol II-specific mechanism of transcription through chromatin [17]. Formation of the Ø-loop induces uncoiling of the ~100-bp DNA region in front of the enzyme allowing further transcription through the nucleosome and efficient survival of nearly all histones (with exception of one H2A/H2B dimer that is displaced by Pol II) during this process [12,15]. The high efficiency of histone survival during transcription is explained in part by allosterically stabilized intranucleosomal histone-histone interactions [18]. Recent structural analysis indicates that after Pol II encounters the strong +50 barrier, the enzyme backtracks and nucleosomal DNA re-coils on the octamer, locking Pol II in the arrested state (Fig. 1, [18]).
Figure 1. Structures of intermediates and the minimal kinetic scheme of Pol II transcription through the nucleosomal barrier.
As Pol II enters a nucleosome (intermediate 1), it pauses and can backtrack along DNA (1.1). Backtracked state is strongly stabilized by recoiling of DNA on the open histone octamer surface (1.2). Bulk of Pol II collides with nucleosomal DNA, forming a very tight complex that strongly inhibits recovery of Pol II from the backtracked state. Pol II can recover from arrest with help of TFIIS that facilitates Pol II-induced RNA cleavage, providing Pol II with another chance to proceed further in the nucleosome, accompanied by uncoiling of nucleosomal DNA and facilitated by transient, stepwise binding of histone chaperone FACT to H2A/H2B dimers (2). Insert: A model of yeast RNA polymerase II elongation complex (PDB 1Y1W) with the active center at the position +42 bp in the nucleosome (PDB 1KX5). The model was built using the published structure of the +42 complex with E. coli RNA polymerase [18] where the bacterial enzyme was replaced by Pol II. Dotted line indicates direction of transcription by Pol II.
Two general mechanisms should facilitate nucleosome traversal: holding Pol II in its active state, including facilitating recovery from arrest, and disrupting critical histone-DNA interactions. As noted, TFIIS mediates transcript cleavage to restart arrested polymerases and facilitates transcription through chromatin in vitro [8,19,20] (Fig. 1). In metazoans, TFIIF maintains Pol II’s catalytic readiness and thus substantially increases overall elongation rates. Both TFIIF and TFIIS are associated with the body of active genes [21]. In vitro studies showed that these two factors together modestly facilitate elongation through a single nucleosome. However, with a nucleosome containing a Sin mutant histone, which weakens the critical octamer-DNA interactions near the nucleosome dyad, elongation in the presence of TFIIF and TFIIS nearly matched the efficiency and rate of elongation on histone-free DNA [22,23]. These in vitro studies with a minimal transcription machinery demonstrate that efficient and rapid nucleosome traversal is clearly possible when Pol II is optimized and DNA unwrapping from the octamer is facilitated [24]. Histone chaperone FACT is an example of a factor that facilitates DNA unwrapping from H2A/H2B dimers to relieve the nucleosomal barrier and facilitate nucleosome traversal by Pol II [25,26] (Fig. 1). Histone acetylation [27] and/or multiple molecules of Pol II [28,29] also help to overcome the barrier, affecting different steps during transcription through chromatin in vitro. While wrapping of DNA on the central core of the histone octamer provides the primary block to transcript elongation, in vitro studies using histones lacking the N-terminal tails showed that the tails also contribute to the nucleosomal barrier [27,30].
Future studies in this area should address more fully the mechanisms through which Pol II overcomes the two classes of nucleosome-induced pauses described above. It has been suggested that the nearly universal pause by metazoan Pol II at ~50 nt downstream of transcription start is directly linked to the barrier imposed by the +1 nucleosome [3], consistent with the general tendency of Pol II to backtrack early in elongation [6]. While pausing ~10 bp upstream of a promoter-proximal nucleosome has not be observed in vitro, earlier studies did not incorporate known negative elongation factors, including NELF and DSIF (reviewed in [31]). Once Pol II has overcome promoter-proximal pausing, the polymerase will encounter barriers at ~15 and 50 bp within each downstream nucleosome [3]. Entry into productive elongation in vivo requires at least the activity of P-TEFb, but the full set of factors essential for pause relief and rapid long-range transcription has not been identified [31]. While proof of principle experiments with TFIIF and TFIIS have shown that the nucleosome is not an insurmountable barrier to elongation by Pol II [22], a major long term challenge will be to evaluate the roles of the much larger set of elongation-associate factors [31] in studies which require Pol II to rapidly and effectively traverse long arrays of nucleosomes in vitro.
Acknowledgments
This work was supported by NIH grant GM58650 to V.M.S., NSF grant MCB1121210 to D.S.L., and by the Russian Science Foundation (RSF grant No 14-24-00031). We apologize to colleagues whose work could not be cited due to space limitations.
References
- 1.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015;16:178–89. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 2.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–73. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weber CM, Ramachandran S, Henikoff S. Nucleosomes Are Context-Specific, H2A.Z-Modulated Barriers to RNA Polymerase. Mol Cell. 2014;53:819–30. doi: 10.1016/j.molcel.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 4.Bondarenko VA, Steele LM, Ujvari A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell. 2006;24:469–79. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–53. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 6.Luse DS. Promoter clearance by RNA polymerase II. Biochim Biophys Acta. 2013;1829:63–8. doi: 10.1016/j.bbagrm.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Izban MG, Luse DS. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J Biol Chem. 1992;267:13647–55. [PubMed] [Google Scholar]
- 8.Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell. 2005;18:97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 9.Izban MG, Luse DS. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991;5:683–96. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- 10.Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, Wang MD. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat Struct Mol Biol. 2009;16:124–9. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol Cell. 2002;9:541–52. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- 12.Kulaeva OI, Gaykalova DA, Pestov NA, Golovastov VV, Vassylyev DG, Artsimovitch I, Studitsky VM. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat Struct Mol Biol. 2009;16:1272–8. doi: 10.1038/nsmb.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaykalova DA, Nagarajavel V, Bondarenko VA, Bartholomew B, Clark DJ, Studitsky VM. A polar barrier to transcription can be circumvented by remodeler-induced nucleosome translocation. Nucleic Acids Res. 2011;39:3520–8. doi: 10.1093/nar/gkq1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pestov NA, Gerasimova NS, Kulaeva OI, Studitsky VM. Structure of Transcribed Chromatin is a Sensor of DNA Damage. Sci Adv. 2015;1:e1500021. doi: 10.1126/sciadv.1500021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulaeva OI, Hsieh FK, Chang HW, Luse DS, Studitsky VM. Mechanism of transcription through a nucleosome by RNA polymerase II. Biochim Biophys Acta. 2013;1829:76–83. doi: 10.1016/j.bbagrm.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang HW, Kulaeva OI, Shaytan AK, Kibanov M, Kuznedelov K, Severinov KV, Kirpichnikov MP, Clark DJ, Studitsky VM. Analysis of the mechanism of nucleosome survival during transcription. Nucleic Acids Res. 2014;42:1619–27. doi: 10.1093/nar/gkt1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang HW, Shaytan AK, Hsieh FK, Kulaeva OI, Kirpichnikov MP, Studitsky VM. Structural Analysis of the Key Intermediate Formed during Transcription through a Nucleosome. Trends Cell Mol Biol. 2013;8:13–23. [PMC free article] [PubMed] [Google Scholar]
- 18.Gaykalova DA, Kulaeva OI, Volokh O, Shaytan AK, Hsieh FK, Kirpichnikov MP, Sokolova OS, Studitsky VM. Structural analysis of nucleosomal barrier to transcription. Proc Natl Acad Sci USA. 2015;112:E5787–95. doi: 10.1073/pnas.1508371112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010;140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guermah M, Palhan VB, Tackett AJ, Chait BT, Roeder RG. Synergistic functions of SII and p300 in productive activator-dependent transcription of chromatin templates. Cell. 2006;125:275–86. doi: 10.1016/j.cell.2006.01.055. [DOI] [PubMed] [Google Scholar]
- 21.Cojocaru M, Jeronimo C, Forget D, Bouchard A, Bergeron D, Cote P, Poirier GG, Greenblatt J, Coulombe B. Genomic location of the human RNA polymerase II general machinery: evidence for a role of TFIIF and Rpb7 at both early and late stages of transcription. Biochem J. 2008;409:139–47. doi: 10.1042/BJ20070751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luse DS, Spangler LC, Ujvari A. Efficient and rapid nucleosome traversal by RNA polymerase II depends on a combination of transcript elongation factors. J Biol Chem. 2011;286:6040–8. doi: 10.1074/jbc.M110.174722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsieh FK, Fisher M, Ujvari A, Studitsky VM, Luse DS. Histone Sin mutations promote nucleosome traversal and histone displacement by RNA polymerase II. EMBO Rep. 2010;11:705–10. doi: 10.1038/embor.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325:626–8. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–3. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 26.Hsieh FK, Kulaeva OI, Patel SS, Dyer PN, Luger K, Reinberg D, Studitsky VM. Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc Natl Acad Sci USA. 2013;110:7654–9. doi: 10.1073/pnas.1222198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bintu L, Ishibashi T, Dangkulwanich M, Wu YY, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal elements that control the topography of the barrier to transcription. Cell. 2012;151:738–49. doi: 10.1016/j.cell.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulaeva OI, Hsieh FK, Studitsky VM. RNA polymerase complexes cooperate to relieve the nucleosomal barrier and evict histones. Proc Natl Acad Sci USA. 2010;107:11325–30. doi: 10.1073/pnas.1001148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin J, Bai L, Johnson DS, Fulbright RM, Kireeva ML, Kashlev M, Wang MD. Synergistic action of RNA polymerases in overcoming the nucleosomal barrier. Nat Struct Mol Biol. 2010;17:745–52. doi: 10.1038/nsmb.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ujvari A, Hsieh FK, Luse SW, Studitsky VM, Luse DS. Histone N-terminal tails interfere with nucleosome traversal by RNA polymerase II. J Biol Chem. 2008;283:32236–43. doi: 10.1074/jbc.M806636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–43. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]