Abstract
Kidney epithelial cells are common targets for human and rhesus cytomegalovirus (HCMV and RhCMV) in vivo, and represent an important reservoir for long-term CMV shedding in urine. To better understand the role of kidney epithelial cells in primate CMV natural history, primary cultures of rhesus macaque kidney epithelial cells (MKE) were established and tested for infectivity by five RhCMV strains, including two wild-type strains (UCD52 and UCD59) and three strains containing different coding contents in UL/b′. The latter strains included 180.92 [containing an intact RhUL128-RhUL130-R hUL131 (RhUL128L) locus but deleted for the UL/b′ RhUL148–rh167-loci], 68-1 (RhUL128L-defective and fibroblast-tropic) and BRh68-1.2 (the RhUL128L-repaired version of 68-1). As demonstrated by RhCMV cytopathic effect, plaque formation, growth kinetics and early virus entry, we showed that MKE were differentially susceptible to RhCMV infection, related to UL/b′ coding contents of the different strains. UCD52 and UCD59 replicated vigorously in MKE, 68-1 replicated poorly, and 180.92 grew with intermediate kinetics. Reconstitution of RhUL128L in 68-1 (BRh68-1.2) restored its replication efficiency in MKE as compared to UCD52 and UCD59, consistent with the essential role of UL128L for HCMV epithelial tropism. Further analysis revealed that the UL/b′ UL148-rh167-loci deletion in 180.92 impaired RhUL132 (rh160) expression. Given that 180.92 retains an intact RhUL128L, but genetically or functionally lacks genes from RhUL132 (rh160) to rh167 in UL/b′, its attenuated infection efficiency indicated that, along with RhUL128L, an additional protein(s) encoded within the UL/b′ RhUL132 (rh160)-rh167 region (potentially, RhUL132 and/or RhUL148) is indispensable for efficient replication in MKE.
Introduction
Human cytomegalovirus (HCMV) infection of epithelial cells (EP) is central to HCMV transmission, persistence and pathogenesis. Apart from the 0.6–0.7% rate of congenital infection, more than 99% of primary HCMV infections occur by horizontal transfer of bodily fluids containing infectious HCMV (e.g. breast milk, saliva, urine and genital fluids) from an infected individual to the mucosal EP of an uninfected host. Following haematogenous spread, progeny virions ultimately seed the EP of the salivary glands, mammary glands and genitourinary organs from which virus is excreted into bodily fluids. Within immunocompromised hosts, such as iatrogenically immunosuppressed transplant recipients, immunologically immature foetuses and AIDS patients, extensive HCMV-induced pathology can be observed in different types of EP, including retinal pigment EP (RPE), and alveolar, bronchial and intestinal EP, which contribute to organ dysfunction either directly by HCMV cytolysis or indirectly by HCMV-induced immunopathology (Landolfo et al., 2003). Accepting that EP constitute a central role in HCMV natural history, a full understanding of HCMV epithelial tropism is required for developing new intervention strategies that can interrupt HCMV horizontal transmission and limit HCMV pathology.
It is now evident that HCMV clinical isolates rapidly undergo mutations (rearrangements, point mutations, deletions) after limited passages in fibroblasts, particularly affecting RL13, UL36 and ORFs within UL/b′ (UL128–UL150). Notably, the epithelial/endothelial determinant locus (UL128-UL130-UL131A; hereafter UL128L) is especially susceptible to genetic alterations after only a few passages in cultured fibroblasts (Akter et al., 2003; Bradley et al., 2009; Cha et al., 1996; Cunningham et al., 2010; Dargan et al., 2010; Hahn et al., 2004; Prichard et al., 2001; Ryckman et al., 2006, 2008; Sinzger et al., 2008; Stanton et al., 2010). Proteins encoded by UL128L, together with glycoprotein H (gH) and L (gL), form a gH-anchored pentamer complex that mediates HCMV entry of EP and endothelial cells via receptor-dependent endocytosis. The genetic integrity of UL128L determines the level of virus entry, which, in turn, regulates the infectivity of HCMV in these cells (Bodaghi et al., 1999; Ryckman et al., 2006; Wang & Shenk, 2005; Wang et al., 2007). However, it has been noted that the anatomical and tissue origin of both EP and endothelial cells affects HCMV infection efficiency and pathogenesis (Esclatine et al., 2000; Fish et al., 1998; Jarvis & Nelson, 2007; Jarvis et al., 1999; Millard et al., 2010; Tugizov et al., 1996).
Both EP and endothelial cells are heterogeneous, exhibiting phenotypic differences and tissue-specific functions, while at the same time sharing common features like apico-basal polarity and tight junctions (TJ) (Elkouby-Naor & Ben-Yosef, 2010; Gonzalez-Mariscal et al., 2009). While endothelial cells with distinct anatomical and tissue origin displayed different susceptibility, productivity and cytopathic effects to HCMV infection (Fish et al., 1998; Jarvis & Nelson, 2007; Millard et al., 2010), the influence of EP origins has been similarly documented in a broad spectrum of viruses, including HCMV. It has been shown that HCMV entry in polarized RPE is mainly via the apical membrane, whereas in Caco-2 polarized intestinal epithelial cells, entry is via the basal surface (Esclatine et al., 2000; Jarvis et al., 1999; Tugizov et al., 1996). Accordingly, results related to HCMV infection in EP derived from one tissue might not be applicable to HCMV infection in EP derived from a different tissue. Put another way, biologically relevant EP should be used to address particular aspects of HCMV virology in the host, such as transmission, persistence and/or pathogenesis. Most HCMV epithelial tropism studies have been performed in RPE, which are critical in HCMV pathogenesis in immunocompromised individuals (Landolfo et al., 2003). However, it is not clear what extent infection of RPE plays in the course of HCMV infection in a healthy immunocompetent host. As such, conclusions drawn from infection studies of cultured RPE may not necessarily be applicable or relevant to other EP derived from other tissues without confirmation.
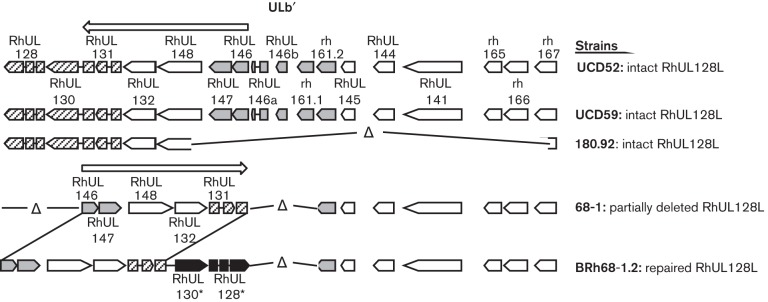
Akin to HCMV, the UL/b′ (RhUL128–rh167) region of the rhesus CMV (RhCMV) genome also undergoes rearrangements and mutations after passage on fibroblasts. Characterized genetic alterations in RhCMV include: (1) the loss of RhUL128 and RUL130 and three alpha-chemokine-like ORFs in laboratory-adapted strain 68-1, and (2) in the 180.92 strain, retention of the RhUL128 and RhUL130 genes but deletion of all ORFs from RhUL148 (rh159) to rh167 [ULb′ RhUL148 (rh159)–rh167] (Hansen et al., 2003; Oxford et al., 2008; Rivailler et al., 2006) (Fig. 1). Moreover, genetic repair of the RhUL128 and RhUL130 genes within 68-1 (the repaired variant, BRh68-1.2) has been shown to greatly improve its replication efficiency in rhesus RPE (RRPE) and even human RPE, as compared with both its parental 68-1 virus and the 180.92 strain (Lilja & Shenk, 2008). While these results clearly demonstrate the importance of RhUL128 and RhUL130, and presumably the intact RhUL128L, to RRPE tropism in vitro, additional studies have not yet been reported for other types of rhesus EP, particularly those that serve as sites of excretion during long-term RhCMV infection.
Fig. 1.
Protein-coding contents of ULb′ region of RhCMV UCD52, UCD59, 180.92, 68-1 and BRh68-1.2. Open triangles indicate the deleted and truncated genes; asterisks indicate the genes that have been reconstituted in the viral genome; and arrows show the transcription direction of RhUL128L, RhUL132 (rh160) and RhUL148 (rh159) in different RhCMV strains.
Prolonged viruria is a hallmark of HCMV-infected humans and RhCMV-infected macaques, and kidney epithelial cells have been documented as common targets of HCMV and RhCMV in vivo. Infected kidney EP likely represent an important source of virus excreted in urine (Britt & Alford, 1996; Ho, 1991; Yue & Barry, 2008). Moreover, recent in vivo studies on two wild-type (wt) strains UCD52 and UCD59 (both contain a full-length UL/b′), a RhUL128L-intact but UL/b′ RhUL148 (rh159)-rh167-deficient strain 180.92, and a RhUL128L-defective strain 68-1 have found that RhCMV dissemination and shedding frequency in body fluids, including urine, are dependent on the integrity of the UL/b′ region (Assaf et al., 2014; Oxford et al., 2008). Herein, to further understand the role of kidney EP in RhCMV natural history, we established a primary culture for macaque kidney EP (MKE) and investigated the infectivity of UCD52, UCD59, 180.92 and 68-1 in MKE in vitro. In addition, the RhUL128-repaired 68-1 variant, BRh68-1.2 (Fig. 1), was also included to test the function of RhUL128L in RhCMV infection of MKE. Our results showed that MKE are fully permissive to RhCMV infection and exhibit a differential susceptibility to different RhCMV strains. Moreover, the differential infectivity of BRh68-1.2, 180.92, UCD52, UCD59 and 68-1 in MKE indicated the essential role of RhUL128L in efficient infection of MKE by RhCMV, but also highlights the role of additional ORF in UL/b′ for conferring fully efficient EP tropism.
Results
Characterization of MKE
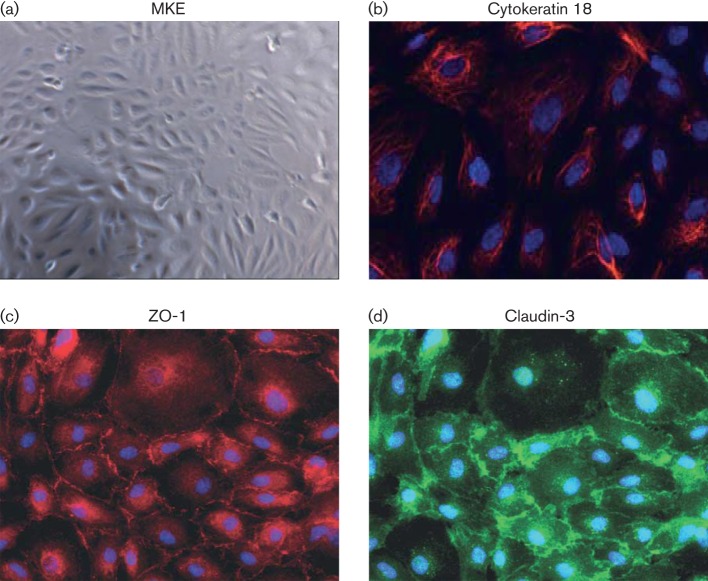
To establish MKE in culture, a kidney of a neonatal rhesus macaque was removed at necropsy, and single cell suspensions were prepared by mechanical and enzymatic dissociation and then placed in culture. After the initial plated cells reached confluence, dissociated kidney cell cultures were propagated in growth medium containing cis-4-hydroxy-l-proline (4-HP), a proline analogue that selectively impairs replication of fibroblasts within the cultured cells (Kao & Prockop, 1977). Following four serial subdivisions in 4-HP selective media, primary cultured MKE exhibited a uniform cobblestone-liked morphology, phenotypically consistent with epithelial cells (Fig. 2a). Cultured MKE were then evaluated for the expression of two TJ proteins, zonula occludens (ZO)-1 and claudin-3, and an EP-specific marker, cytokeratin 18 (Baer et al., 2006; Saitou et al., 1997). As shown in Fig. 2(b–d), MKE cell cultures uniformly expressed the cytokeratin 18, ZO-1 and claudin-3 proteins, consistent with an epithelial-derived origin.
Fig. 2.
Characterization of MKE. (a) Light microscopy image of MKE (×5). (b) Immunofluorescence staining of MKE with an epithelial-specific marker, cytokeratin 18 (red), ×40. (c, d) Immunofluorescence staining of MKE with tight junction markers ZO-1 (red) and claudin-3 (green), respectively; ×40. The nuclei of MKE were counterstained with DAPI (blue).
Interstrain differences in RhCMV CPE development and spread in MKE
The susceptibility of MKE to individual RhCMV strains was initially confirmed by different levels of pp65-2 and immediate early (IE)1-expressing cells and characteristic RhCMV cytopathic effect (CPE) at 96 h post-infection (p.i.) (data not shown). In addition, during virus propagation in MKE, UCD52-, UCD59- and BRh68-1.2-infected MKE (m.o.i. <0.01 p.f.u. per cell) displayed focal expansion of individual plaques, a pattern indicative of cell-to-cell spread (Adler et al., 2006; Scrivano et al., 2011), and progressed to ~100 % CPE, whereas 180.92 infection of MKE required an inordinately long time (~30 days p.i.) for the entire culture to exhibit RhCMV CPE. In contrast, as indicated by 68-1-expressed EGFP, 68-1-infected MKE were noted for scattered rounded-up cells and occasionally small foci (<10 cells), which never progressed to ~100 % CPE despite prolonged culture (data not shown). These observations suggested a differential replication and cell-to-cell spread competency of UCD52, UCD59, 180.92, BRh68-1.2 and 68-1 in MKE.
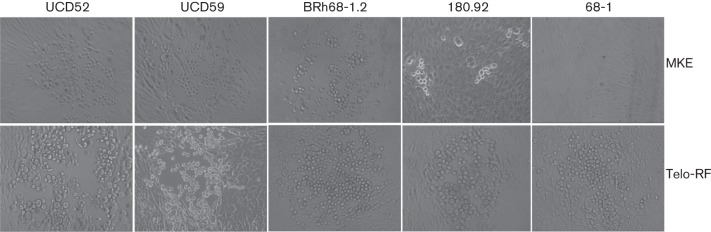
Subsequently, plaque formation assays were performed in MKE and telomerase-immortalized rhesus fibroblasts (Telo-RF) to further define the cell-to-cell spread capacity of each RhCMV strain (Chang et al., 2002). While all the infected Telo-RF cultures exhibited typical RhCMV plaques with all RhCMV strains, marked inter-strain differences were noted in MKE cultures (Fig. 3). UCD52, UCD59 and BRh68-1.2 cultures developed progressive discrete plaques, whereas 68-1-infected MKE were essentially normal in appearance, and only a few rounded cells were observed. MKE cultures infected with 180.92 exhibited an intermediate phenotype characterized by a discrete number of small foci of rounded cells and plaques that were far smaller in size than those observed with UCD52, UCD59 and BRh68-1.2 infection. These results supported that RhCMV strains display the differential spread capacities in MKE.
Fig. 3.
Virus cell-to-cell spread in MKE and Telo-RF was characterized using plaque formation assay. MKE and Telo-RF were infected with UCD52, UCD59, 180.92, BRh68-1.2 and 68-1, respectively, at an m.o.i. of 0.001, with the exception that infection of MKE with RhCMV 68-1 and 180.92 was carried out at an m.o.i. of 0.02. Plaque formation and size were evaluated by light microscopy at 10 days p.i. Representative plaques of individual RhCMV strains are shown (×10 magnification).
Differential RhCMV replication kinetics in MKE
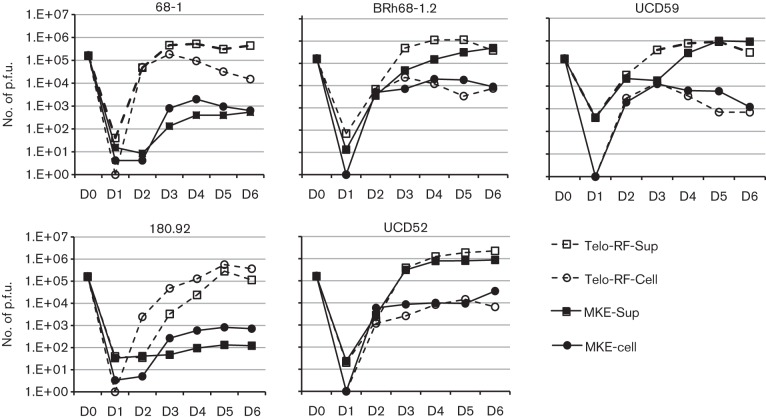
The replication competency of RhCMV strains was determined by performing one-step growth curves following infection of MKE and Telo-RF (0.5 m.o.i.). Differential growth kinetics were observed in MKE as compared to Telo-RF. UCD52, UCD59 and BRh68-1 exhibited comparable replication titres and kinetics in both MKE and Telo-RF, and cell-free virus titres were >1.5 logs higher than cell-associated virus titres in either cell type (Fig. 4), consistent with a lytic replication pattern. In contrast, although the patterns of replication for 68-1 and 180.92 in Telo-RF were comparable to the other tested strains regarding the peak titres and kinetics, their titres in MKE were markedly lower. Compared to UCD52, UCD59 and BRh68-1.2, the peak production of 68-1 in the supernatant of MKE was 900–1830-fold lower, demonstrating a severely restricted growth in MKE. Unexpectedly, the peak yield of cell-free 180.92 in the supernatant of MKE was even lower than that observed for 68-1. Moreover, a difference was also observed in that higher levels of cell-associated 180.92 progeny virions were detected after infection of Telo-RF and MKE cells. The distinction of greater cell-associated 180.92 versus cell-free 180.92, particularly in MKE, suggested a defect of virus release.
Fig. 4.
Short-term growth kinetics of RhCMV strains in MKE and Telo-RF. MKE and Telo-RF were infected in parallel with individual RhCMV strain at an m.o.i. of 0.5. Both supernatant fluids and cells were collected daily until 6 days p.i. The yielding of progeny viruses in these two compartments was measured by plaque assay in Telo-RF. D, Day. -Sup and -cell represent virus yielding in the supernatant fluids and cells, respectively.
However, as Lilja and Shenk have reported that 180.92 infection of RRPE yields more cell-free virus than RhCMV 68-1 at 8 and 12 days p.i. but not at 4 days p.i. (Lilja & Shenk, 2008), a prolonged growth study was performed with 180.92 and 68-1 (0.5 m.o.i.) following similar protocols (Dargan et al., 2010; Lilja et al., 2008). Beginning at 14 days p.i., the yielding of cell-free 180.92 exceeded those of 68-1, and remained ~50 % higher than that of 68-1 at 22 days p.i. (Fig. 5). The cell-free 180.92 virus was approximately equivalent to cell-associated virus at 22 daysp.i., and peak titres were coincident with the progressive development to ~100 % CPE in 180.92-infected cells (data not shown). In addition, compared to cell-associated 68-1, cell-free 68-1 was lower from 3–5 days p.i. in MKE before reaching equivalence at 6 days p.i. (Fig. 4) and much higher (~52-fold) at 22 days p.i. (Fig. 5). These data further demonstrated the difference between 68-1 and 180.92 replication in MKE.
Fig. 5.
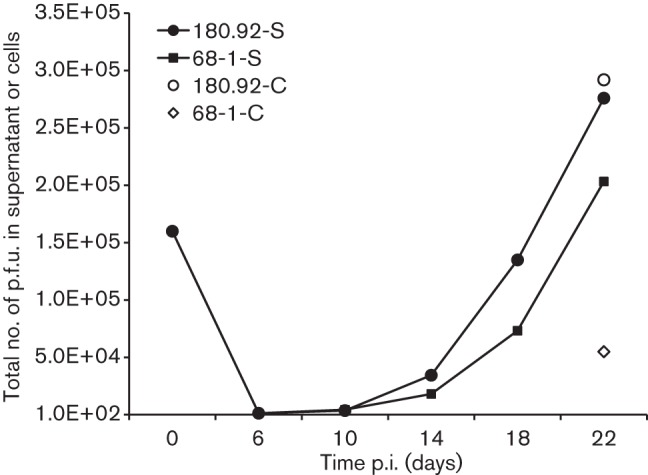
Long-term growth kinetics study of RhCMV 180.92 and 68-1 in MKE. MKE were infected with RhCMV 180.92 or 68-1 at an m.o.i. of 0.5. Supernatant fluids of infected MKE were collected every 4 days starting 6 days p.i. and the infected cells were harvested at 22 days p.i. The production of progeny viruses in these two compartments was measured by plaque assay in Telo-RF. S, Cell-free virus in the supernatant fluids; C, cell-associated virus.
Initial events are critical for efficient RhCMV replication MKE
As the block of infection of EP and endothelial cells with UL128L-defective HCMV strains has been observed at the early stage of infection, indicated by deficient expression of IE genes (Ryckman et al., 2006; Sinzger et al., 2000), it was important to characterize expression of IE1 during the initial stage of RhCMV infection in MKE. Telo-RF and MKE were infected with each strain (0.5 m.o.i.), and the fractions of IE1-expressing cells were quantified at 17 h p.i. to determine whether the differential replication efficiency of RhCMV strains in MKE was related to viral entry (Fig. 6). Infection of Telo-RF with RhCMV UCD52, UCD59 and BRh68-1.2 resulted in 85–89 % IE1-positive cells, whereas infection of MKE with these same three strains generated 47–61 % IE1-positive cells. The ratio of IE1-positive cells in Telo-RF : MKE (T : M) ranged from 1.5 to 1.9, consistent with comparably efficient viral entry into both cell types for UCD52, UCD59 and BRh68-1.2. As expected, 68-1 infection of MKE was highly inefficient (1.3 % IE1-positive), although IE1 expression in Telo-RF (83%; T : M = 66) was equivalent to UCD52, UCD59 and BRh68-1.2. IE1 expression following 180.92 infection of MKE was similarly inefficient (1.7 %), which was surprising given the presence of an sequence-identical RhUL128L to BRh68-1.2 (Lilja & Shenk, 2008). Unexpectedly, 180.92 also exhibited a reduced frequency of IE1-positive cells in Telo-RF (58 %; T : M = 33.9) compared to the other four RhCMV strains. Together, the results indicated that 180.92 and 68-1 infection of MKE exhibited a defect in a stage prior to IE1 expression, such as viral entry and/or post-entry nuclear translocation of viral DNA (Ryckman et al., 2006; Sinzger et al., 2000), and this defect was especially prominent in epithelial cells.
Fig. 6.
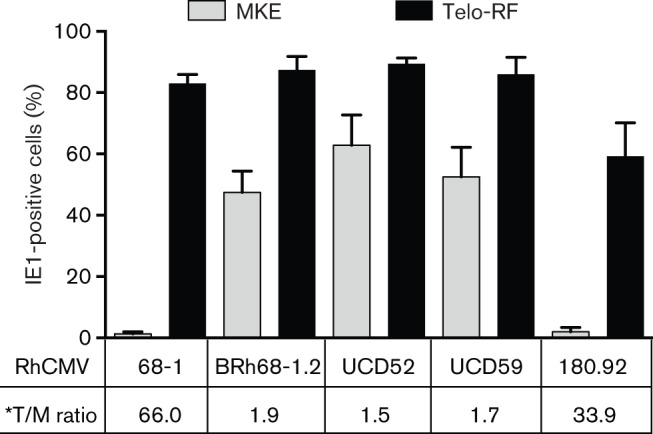
IE1 expression in Telo-RF and MKE at 17 h p.i. with different RhCMV strains (0.5 m.o.i.). Each bar represents the mean percentage of IE1-positive cells in five randomly chosen fields. *T : M ratio shows the ratios of IE1-positive Telo-RF (%, black bar) to IE1-positive MKE (%, grey bar).
Characterization of the UL/b′ region of MKE-propagated 180.92
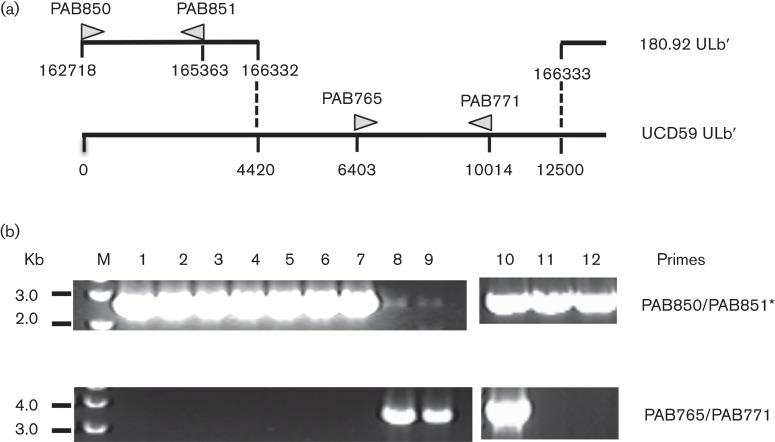
It has been reported that the original Telo-RF-propagated 180.92 stock comprised a mixed population of viral variants: a predominant population consisting of a UL/b′ truncated variant (Fig. 1), from which the annotated sequence was derived (Rivailler et al., 2006), and a minority population of a full-length UL/b′ variant (<15 %) (Assaf et al., 2014). Thus, it was important to determine the constituents of 180.92 variants after serial propagation in MKE. To this end, a variant-specific PCR (primers PAB765/PAB771) was performed to amplify an ~3.6 kb amplicon that corresponded to part of the deleted UL/b′ region within the truncated 180.92 variant (Fig. 7a). Out of 30 plaques picked from infected Telo-RF, only two plaques yielded the 3.6 kb fragment, consistent with the reported full-length variant (Fig. 7b) (Assaf et al., 2014), while in contrast, amplification of DNA from two stocks of 180.92 prepared after three passages in MKE, and five independent plaques prepared after four passages in MKE, did not detect any full-length UL/b′-specific amplicon under the same conditions, supporting the conclusion that serial passage of 180.92 in MKE resulted in a population of the truncated genetic variant (Fig. 7b). Meanwhile, RhUL128L was identified in all the prepared DNA samples by a PCR (primers PAB850/PAB851) that amplified a 2.6 kb RhUL128L amplicon (Fig. 7a, b). Moreover, sequence analysis of the RhUL128L amplicon confirmed that the coding regions of RhUL128, RhUL130 and RhUL131 of MKE-propagated 180.92 were identical to the annotated sequence of 180.92 (Rivailler et al., 2006) (data not shown). The reasons for losing the full-length variant after serial passages in MKE are unclear. However, infection with multiple genetically distinct strains of a virus is common in nature and can lead to positive (complementary) or negative (competitive) within-host virus–virus interactions (DaPalma et al., 2010; Read & Taylor, 2001). Given the low frequency (~6.7 %) of the full-length variant in the original Telo-RF-propagated stock, coupled with propagation of the 180.92 stock in MKE at exceedingly low m.o.i. (<0.01), it is possible that the loss of the full-length 180.92 variant in MKE virus stock may result from negative selection induced by the predominant truncated variant or frequency-dependent strain selection (Pepin & Hanley, 2008; Pepin et al., 2008; Perefarres et al., 2014; Renzette et al., 2011).
Fig. 7.
PCR analysis of MKE- and Telo-RF-propagated RhCMV 180.92. (a) Diagram represents UL/b′ of 180.92 (GenBank accession no. DQ120516) and its alignment to RhCMV-WT strain UCD59 ULb′ (GenBank accession #no. EU130540). PCR primer pair names, positions and sizes of their respective amplicons are indicated (∆). (b) PCR amplicons generated with primers PAB850/PAB851 and PAB765/PAB771. Lanes: M, DNA marker; 1–5, five individual plaques picked from 180.92 infected MKE; 6, MKE-propagated 180.92 at passage 3; 7, MKE-propagated 180.92 at passage 4; 8, MKE-propagated UCD52; 9, MKE-propagated UCD59; 10–12, representatives of 30 individual plaques picked from 180.92 infected Telo-RF. Asterisk indicates primers PAB850 and PAB851 contained mismatched nucleotides to UCD52 and UCD59, respectively (see Methods).
RhUL132 (rh160) transcription in infected MKE cells
RhCMV encodes an orthologue of HCMV UL132, named RhUL132 (rh160). RhUL132 is 30 % identical and 53 % similar to UL132 at the protein level, and is highly conserved in all the tested RhCMV strains with an amino acid identity of 96 %. As UL132 is transcribed at multiple initiation sites, including upstream of the co-transcribed UL146, UL147 and UL148 genes (He et al., 2012; Lurain et al., 2006), the transcription pattern of RhUL132 was characterized to determine the effect of the upstream RhUL148–rh167 deletion within 180.92 UL/b′. Two major bands (~2.3 kb and ~0.5 kb) were detected in MKE infected with 68-1, BRh68-1.2, UCD52 and UCD59, whereas only one band (~1.3 kb) was detected in 180.92-infected MKE (Fig. 8a). These results showed that genetic deletion in 180.92 UL/b′ altered the transcription of RhUL132. Sequence analysis of the amplicons from BRh68-1.2 and 180.92 showed that the start site of the larger transcript of BRh68-1.2 RhUL132 mapped to the predicted translation start codon of RhUL147 (rh158), and the start site of the smaller transcript mapped to an internal site within RhUL132, 177 nt downstream of the predicted start AUG of RhUL132. In contrast, the start site of the single 180.92 transcript mapped to a site 86 nt downstream of the predicted start codon resulting from the ectopic fusion of partial RhUL148 and rh167 ORF (Fig. 1) (Rivailler et al., 2006). The altered transcription start site of 180.92 RhUL132 suggested that 180.92 might be functionally devoid of RhUL132 protein (pRhUL132), although the coding region was extant.
Fig. 8.
RhUL132 (rh160) transcription and expression. (a) RhUL132 transcripts in RhCMV-infected MKE. Total RNA was extracted from RhCMV-infected MKE. RhUL132 transcripts were identified by 5′ RACE RT-PCR (see Methods). (b) pRhUL132 expression in extracellular RhCMV virions. Extracellular virions were purified from the supernatant fluids of infected Telo-RF and used for Western blot analysis. (c) pRhUL132 expression in RhCMV-infected MKE. RhCMV-infected MKE lysates were prepared after 90–100% of the cells showed CPE and used for Western blot analysis. Uninfected MKE lysates were used for cell control. pp65-2 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were used as internal controls. Asterisk indicates 68-1 was purified extracellular virion.
RhUL132 (rh160) protein expression in extracellular virions and in infected cells
To test this theory, the expression of pRhUL132 was further analysed in both infected-cell lysates and purified extracellular virions. In HCMV, Western blot analysis showed that there were two diffuse forms of UL132 protein – a larger form between 45 and 60 kDa and a smaller form between 22 and 28 kDa detected in infected cells and virions. Moreover, considerable variations were observed between different HCMV strains with respect to the diffuse migration of the large form and the ratios between the large and small forms of UL132 protein (Spaderna et al., 2005). Similar to the expression of gpUL132, two distinctly sized proteins were detected in the virions of BRh68-1.2 and 68-1, a larger diffuse 45–80 kDa band and a minor ~23 kDa band, whereas only the larger form was detected in 180.92, UCD52 and UCD59 virions (Fig. 8b). While the amounts of pRhUL132 were relatively less in UCD52 and UCD59 than observed in BRh68-1.2 and 68-1, after normalizing to the relative levels of pp65-2 protein, the expression of pRhUL132 was barely detectable in 180.92 virions. To exclude the possibility that the minimal amount of pRhUL132 in 180.92 virions resulted from a defect in packaging the protein into mature virions, infected cell lysates were also analysed to determine whether there was detectable pRhUL132 in 180.92-infected cells. While there were abundant, various sized forms of pRhUL132 in MKE infected with UCD52, UCD59 and BRh68-1.2, there was a complete absence of detectable pRhUL132 in MKE infected with 180.92 (Fig. 8c). On the other hand, same as the expression of pRhUL132 in RhCMV virions, extremely low levels of pRhUL132 were detected in 180.92-infected primary rhesus dermal fibroblasts (data not shown). These results are consistent with the interpretation that pRhUL132 was a virion structural protein, and its expression in 180.92-infected MKE cells was impaired.
Discussion
Since the overwhelming preponderance of HCMV dissemination from infected to uninfected individuals occurs by horizontal transmission of bodily fluids containing infectious virus across mucosal surfaces (Yue & Barry, 2008), it is important to define the mechanisms of EP infection to design intervention strategies that can impair HCMV epithelial tropism. For both HCMV and RhCMV, infection of kidney EP represents a salient viral reservoir for long-term virus shedding in urine of infected humans and monkeys. Herein, we defined conditions for MKE isolation and primary culture, and demonstrated their epithelial phenotype by epithelial morphology and immunostaining of TJ-specific and EP-specific markers (Fig. 2). As demonstrated by characteristic CPE, viral protein expression, plaque formation and growth kinetics, we have shown that, in accordance with the genetic composition of ORF within UL/b′, primary cultured MKE are differentially susceptible to productive RhCMV infection.
In HCMV, clinical strains contain a UL/b′ region (UL128–UL150) that encodes multiple ORFs involved in cell-type dependent replication, intra-host dissemination, latency and immune evasion (Bradley et al., 2009; Cheung et al., 2005; Goodrum et al., 2007; Hahn et al., 2004; Penfold et al., 1999; Prod'homme et al., 2010; Ryckman et al., 2006, 2008; Sinzger et al., 2008; Stanton et al., 2010; Tomasec et al., 2005; Umashankar et al., 2011). As viral determinants for HCMV entry of EP, UL128L-encoded proteins, form a pentamer with gH and gL mediating HCMV entry into EP via an endocytotic pathway (Bodaghi et al., 1999; Ryckman et al., 2008; Wang & Shenk, 2005; Wang et al., 2007). The UL128L is conserved in RhCMV, and its products are postulated to form a pentamer complex with RhCMV gH and gL (Wussow et al., 2013). In this study, we showed that, although the replication kinetics and magnitude of all the studied strains were generally comparable in Telo-RF, notable interstrain differences were observed in the infection of MKE with regard to the early stage of RhCMV infection, growth kinetics and cell-to-cell spreading. Two wt strains (UCD52 and UCD59) with an intact UL/b′ region (including RhUL128L) showed comparable growth efficiency in MKE as in Telo-RF, whereas the laboratory strain 68-1 with a defective RhUL128L and a deletion of RhUL146a, RhUL146b, rh161.1 and rh161.2 ORFs (Fig. 1) infected MKE poorly (Oxford et al., 2008). In contrast, BRh68-1.2, a 68-1 variant engineered to contain an intact RhUL128L (Lilja & Shenk, 2008), exhibited similar replication kinetics to UCD52 and UD59 in MKE, supporting the critical importance of RhUL128L and the minimal role of RhUL146a, RhUL146b, rh161.1 and rh161.2 ORFs in RhCMV infection of MKE in vitro. However, given the finding that RhCMV 180.92 contains an identical RhUL128L to BRh68-1.2 (Lilja & Shenk, 2008) and, compared to other tested strains, only exhibited intermediate growth efficiency in MKE, we argue that the presence of an intact RhUL128L does not in itself ensure efficient replication in MKE.
Sequence comparison of 180.92 and BRh68-1.2 reveals that the prominent genetic difference between 180.92 and BRh68-1.2 is that 180.92 lacks more ORFs within UL/b′ (Fig. 1 ) (Hansen et al., 2003; Lilja & Shenk, 2008; Rivailler et al., 2006). Moreover, our studies on the transcription and expression of RhUL132 have found that RhUL132 encodes a virion structural protein and is co-transcribed with two upstream adjacent genes from RhUL148 to RhUL147. As a result, despite the coding content of RhUL132 being extant in 180.92, the upstream genetic deletion of UL/b′ RhUL148–rh167 deleted the normal transcription initiation sites of RhUL132 and resulted in aberrant transcription and deficient expression of RhUL132 (Fig. 8). Together, the attenuated growth efficiency of 180.92 in MKE may result from genetic or functional absence of genes from RhUL132 to rh167. Interestingly, a previous study of a library of 68-1 mutants has shown that, despite the low replication efficiency of 68-1 in RRPE, mutagenesis in either RhUL132 or RhUL148 markedly suppress 68-1 replication in RRPE, whereas there is no aberrant phenotype associated with loss of RhUL147, RhUL144 or RhUL141 (Lilja et al., 2008). However, since the function of a protein in cell tropism can be affected by the genomic background of HCMV strains (Li et al., 2015), the role of these proteins in RhCMV infection of epithelial cells should be re-evaluated with an UL128L-intact RhCMV strain, such as BRh68-1.2. Future studies into the individual genes encoded by UL/b′ RhUL132–rh167, especially, RhUL132 and RhUL148, may provide new insights into virus–host interaction.
It should be noted that the reduced growth efficiency of RhCMV 180.92 in MKE observed in our study is in contrast to its vigorous growth in RRPE as reported previously (Lilja & Shenk, 2008). Although low levels of viral entry were detected in both MKE and RRPE after infection, large differences were observed in cell-free virus titres in both cell types. There are several factors that might explain the different infection patterns after infection of RRPE and MKE with 180.92. (1) As it has been noted, Telo-RF-propagated 180.92 contains two populations of viral variants – a predominant population consisting of a variant with a truncated UL/b′ region (Fig. 1) from which the annotated sequence was derived (Rivailler et al., 2006) (GenBank accession no. DQ120516), and a minor population (<15 %) of a full-length variant containing a complete UL/b′ (Assaf et al., 2014). In contrast, MKE-propagated 180.92 only contains the truncated variant (Fig. 7). (2) The in vitro infection phenotypes of HCMV progeny virions (i.e. cell-to-cell spread and cell tropism) are determined by the cell types in which the virus are propagated (Fish et al., 1998; Scrivano et al., 2011), and (3) the susceptibility of endothelial cells to HCMV infection is influenced by the anatomical origins of endothelial cells (Fish et al., 1998; Millard et al., 2010). Accordingly, the observed differences between 180.92 infection in MKE and RRPE could be related to the differences in preparation of 180.92 stocks (Telo-RF-propagated vs MKE-propagated) and tissue origins of EP (retina vs kidney). Nevertheless, it should be mentioned that the growth efficiency of RhCMV strains in MKE in vitro is commensurate to their in vivo shedding profiles (magnitude and frequency) after infection of macaques (Assaf et al., 2014; Oxford et al., 2011), suggesting the relevance of MKE in RhCMV natural transmission.
Overall, to the best of our knowledge, this is the first report to show that primary cultured MKE are fully permissive to RhCMV infection in vitro, and the replication efficiency of RhCMV strains in MKE is greatly impacted by the coding capacity of the UL/b′ region. In addition, our data suggested that an intact RhUL128L is essential, but insufficient, for RhCMV efficient replication in MKE.
Methods
Rhesus macaque kidney epithelial cell isolation and culture.
A kidney was collected at necropsy from a healthy neonatal rhesus macaque and placed in 5 ml cold Dulbecco’s modified Eagle's medium (DMEM) supplemented with 200 U ml-1 penicillin, 200 µg ml-1 streptomycin, 584 µg ml-1 l-glutamine, and 12.5 µg ml-1 of Fungizone (GIBCO; Invitrogen) before processing. Soon after the tissue was delivered to the laboratory, the tissue was transferred to a 100-mm dish containing 3 ml DMEM. Half of the outer cortex was excised from the rest of the kidney tissues and minced into a slurry with a scalpel. The minced tissue was then dissociated by incubation in 30–40 ml DMEM consisting of 0.2 U ml-1 Liberase Blendzyme #3 (Roche Diagnostics) and 0.1 % trypsin (Gibco; Invitrogen) for 2–3 h at 37 °C, with vigorous vortexing every 20–30 min. The cell suspension was centrifuged at 1500 r.p.m. for 5 min, and the supernatant was discarded. The cell pellet was re-suspended with DMEM supplemented with 10 % FCS/Super Calf serum (Gemini), 100 U ml-1 penicillin, 100 µg ml-1 streptomycin, 292 µg ml-1 l-glutamine and 2.5 µg ml-1 Fungizone. After filtration through a 70-µm cell strainer, the cell suspension was centrifuged again. The cell pellet was re-suspended in serum-free growth medium and dispensed into T-75 flasks. The cells were maintained at 37 °C in 5 % CO2 incubator until 90–100 % confluence was attained. Subsequently, the cells were passaged after trypsinization and cultured for four subdivisions in a selection medium containing 100 µg ml-1 4-HP (Sigma) to eliminate contaminating fibroblasts (Kao & Prockop, 1977). Thereafter, the cells were passaged in a 1 : 1 mixture of DMEM and Ham’s F-12 (D : F12) growth medium, containing epithelial cell growth supplement (EpiCGS; ScienceCell, Research Laboratories), 2 % FCS/Super Calf serum (Gemini), 1 mM sodium pyruvate, 25 mM HEPES, 100 U ml-1 penicillin, 100 µg ml-1 streptomycin and 292 µg ml-1 l-glutamine. Cells generally reached confluence within 3 to 4 days at which time cultures were trypsinized and subdivided at a 1 : 2 or 1 : 3 split ratio. These primary cultured MKE were frozen at passage 10 and were characterized for epithelial phenotype at passage 13. MKE from passage 13 to 20 were used for all the experiments reported here. Growth surfaces of culture glasswares were coated with a thin layer of bovine type-I collagen prior to cell plating following the manufacturer’s instructions (BD Biosciences).
Cells and viruses.
Telo-RF were routinely cultured in 10 % FCS/DMEM containing 100 µg ml-1 G418 (Invitrogen) (Chang et al., 2002; Kirchoff et al., 2002) and maintained in 2.5 % FCS/DMEM after RhCMV infection. Primary rhesus dermal fibroblast cells (RhDF) were cultured in 20 % FCS/DMEM and maintained in 10 % FCS/DMEM after infection with RhCMV.
Five RhCMV strains, UCD52, UCD59, 180.92, BRh68-1.2 and 68-1 were used for this study (Fig. 1). UCD52 and UCD59, containing full-length UL/b′ regions (GenBank accession no. GU552456 and EU130540, respectively), were isolates from the urine of SIV-infected monkeys and were passaged unknown times in MRC-5 and WI-38 and then passaged three times in RhDF (Oxford et al., 2011). RhCMV 180.92 was obtained from Dr Amitinder Kaur (New England Primate Research Center) after 13 passages in MRC-5 and primary rhesus macaque fibroblasts (Rivailler et al., 2006), and subsequently passaged twice in Telo-RF in our laboratory. Recently, it has been determined that in addition to the dominant variant containing a large deletion within UL/b′ (GenBank accession no. DQ120516), the RhCMV 180.92 virus stock also contained a minor variant with full-length UL/b′ (Assaf et al., 2014). BRh68-1.2 (a gift from Dr Thomas Shenk at Princeton University) was constructed by repairing RhUL128, RhUL130 and RhUL36 in the genome of 68-1, and the original virus stock was derived at the first passage after reconstitution of BAC-derived virus in Telo-RF (Lilja & Shenk, 2008; McCormick et al., 2010). RhCMV 68-1 (containing full-length RhUL36 gene) was obtained from the American Type Culture Collection (ATCC, VR-677) and has been passaged seven times in human diploid lung fibroblasts and four times in RhDF.
RhCMV strains UCD52, UCD59, 180.92 and BRh68-1.2 were all serially passed four times in MKE, and virus stocks were prepared as follows. Monolayers of MKE at 90–100 % confluence were co-cultured with individual RhCMV strains. The MKE were then either maintained in MKE maintenance medium (EpiCGS-free MKE growth medium), or subjected to a growth medium shock by alternatively exchanging part of the MKE supernatant with DMEM/2.5 % FCS and MKE maintenance medium (Ando et al., 1997). When the cells exhibited 90–100 % CPE, both cell-free and cell-associated virus were collected and filtered through a 0.45 µm filter to make virus stocks. The filtrate was stored in aliquots at −80 °C. An extracellular virus stock of 68-1 was prepared after first passage in Telo-RF and used for all the assays in this report. In addition, a recombinant 68-1 expressing EGFP-68-1 was used for testing the development and progress of CPE in infected MKE (Chang & Barry, 2003). RhCMV strains UCD52, UCD59 and 68-1 were not plaque purified. All the strains were titrated by plaque assay in Telo-RF.
To prepare purified RhCMV virions, RhDF were infected with individual RhCMV strains. When the cells displayed 90–100 % CPE, the supernatants from infected RhDF were collected and clarified by centrifugation at 5000 r.p.m. (6000 g) for 15 min in a Beckman JLA-16.250 rotor. The clarified supernatants were then pelleted by centrifugation at 13 000 r.p.m. (26 000 g) for 2 h. Each pellet was resuspended in cold PBS and layered over a 20 % d-sorbitol cushion and then centrifuged at 21 000 rpm (72 000 g) for 1 h at 4 °C in a Beckman SW41 rotor (Kudchodkar et al., 2004). The pelleted virions were resuspended in cold PBS and stored at −80 °C for Western blot analysis.
Immunofluorescence assays.
Immunofluorescence assays were performed to characterize cultured MKE and RhCMV infectivity in MKE following previously described protocols (Yue et al., 2006). The features of MKE were characterized by immunostaining of two TJ proteins, ZO-1 and claudin-3, and an epithelial-specific marker, cytokeratin-18 (Baer et al., 2006; Saitou et al., 1997). Infectivity of RhCMV strains in MKE and Telo-RF was examined by immunostaining of phosphoprotein 65-2 (pp65-2) and/or immediate early protein (IE)-1 at 17 h and 96 h p.i. with individual RhCMV strains (Yue et al., 2007, 2006). The antigen-specific primary antibodies include mouse anti-ZO-1 and rabbit anti-claudin-3 (Invitrogen Life Sciences), monoclonal mouse anti-human cytokeratin 18 (Dako), rabbit anti-IE1-exon 4 and mouse anti-pp65-2 (Lockridge et al., 1999; Yue et al., 2006). Alexa Fluor 568-conjugated and Alexa Fluor 488-conjugated secondary antibodies (Invitrogen Life Sciences) were used for fluorescent staining and SlowFade gold anti-fade reagent with DAPI (Invitrogen Life Sciences) was added to counterstain cell nuclei. Controls of auto-fluorescence or non-specific fluorescence were performed without the secondary antibodies or with normal rabbit serum and mouse serum.
The slides were viewed with a Zeiss Axioplan 2 microscope (Carl Zeiss). Digital images were captured and analysed by a Zeiss Axiocam System and Openlab software (In provision). RhCMV early entry was calculated by the mean percentage of IE1-positive cells to DAPI-positive cells collected from five randomly chosen fields.
Plaque assay.
Cells (6.0×104) of Telo-RF or MKE were seeded into each well of 24-well plates 1 day prior to the plaque assay. On the next day, triplicate wells were incubated with 10-fold serial dilutions of virus. After adsorption for 2 h at 37 °C, unattached virions were removed by rinsing the cells twice with plain DMEM or D : F12 medium. The cells in each well were then overlaid with 1 ml of 0.5 % SeaPlaque agarose (Cambrex Bio Science Rockland) in DMEM or D : F12 maintenance medium. The viral plaques on Telo-RF were counted at 7 days p.i. whereas the plaques on MKE were counted at 10 days p.i. under a light microscope. In addition, 30 individual plaques from RhCMV 180.92-infected Telo-RF and five from MKE were picked and expanded once for DNA extraction and PCR.
Growth kinetics of RhCMV strains.
Growth kinetics of RhCMV strains was performed by infection of Telo-RF and MKE at 0.5 m.o.i.. The short-term growth kinetics was done by daily collection of supernatants and cells for 6 days (UCD52, UCD59 and BRh68-1.2 showed ~100 %). The long-term growth kinetics of RhCMV 180.92 and 68-1 in MKE were done following a protocol modified from previous reports (Dargan et al., 2010; Lilja & Shenk, 2008). Briefly, 1 ml supernatant was collected every 4 days starting from 6 days p.i. Detached cells in the supernatant were removed by centrifugation and returned to the infected culture by resuspending in 1 ml fresh medium. All cells and supernatants were harvested at 22 days p.i. when maximum3 CPE (~ 95 %) were observed in RhCMV 180.92-infected MKE. Virus yielding at each time-point was titrated in triplicate by plaque assay in Telo-RF and the mean plaque numbers were reported in growth curves.
RhUL128L sequencing and PCR analysis of UL/b′ of RhCMV 180.92.
To confirm the sequence of RhUL128L in the RhCMV 180.92 strain, viral DNA was extracted from infected cells. RhUL128L (nt 162718–165363) was amplified with the primers PAB850 (5′-CTT CTC GTC GAC cac ttt att cag ccc ttg tat ggt ccg ca-3′) and PAB851 (5′-ATT CTC CCG CGG cat ggg tgc atg tcg tgt gtt tat cac tgt-3′) (lower case letters refer to 180.92-specific nucleotides whereas the italic letters indicate the mismatched nucleotides for both UCD52 and UCD59, and the bold and italic letter indicates the mismatched nucleotides for UCD52) (Fig. 7a) and cloned into TOPO-TA vector for sequencing. To test whether MKE serial passaged RhCMV 180.92 stock holds a minority population of the full-length ULb′ genetic variant (Assaf et al., 2014), a variant specific primer pair PAB765 (5′-gtg cga tgt aca ctc gca gga agt ct-3′) and PAB771 (5′-cac cat cgt aat ggt gag cct gac-3′) (Fig. 7a) were used to amplify a ~3.6 kb fragment corresponding to the deleted region in the annotated 180.92 genome (Rivailler et al., 2006). Since this region is also present in the UL/b′ of UCD52 (nt 6376–9984), and UCD59 (nt 6404–10014) genomes (Oxford et al., 2008), viral DNA of UCD52 and UCD59 were extracted from infected MKE and used as positive controls.
RhUL132 antibodies.
Rabbit polyclonal antibodies to RhUL132 peptide (NH4-RRSSPSSKKNRPNK-COOH) were a customized product made by GenScript USA (Accession no. ABV45243; GenScript). Briefly, RhUL132 peptide was synthesized and conjugated with keyhole limpet haemocyanin (RhUL132 peptide-KLH conjugate), which was used to immunize rabbits. The production of anti-RhUL132 antibodies was confirmed by peptide-specific ELISA and RhUL132 protein-specific Western blot analysis.
RhUL132 transcription and expression.
Total RNA was extracted from uninfected and infected MKE at 48 h p.i. (for UCD52, UCD59 and BRh68-1.2) or 72 h p.i. (for RhCMV 68-1, 180.92 and uninfected cell control) using the RNeasy Plus kit (Qiagen). Reverse transcription was performed with the SMARTer RACE cDNA amplification kit (Clontech), and the cDNA was amplified with universal primer A mix provided by the kit as the forward primer and a RhUL132-specific primer PAB 908 (5′-cat ctt agg cat ttc cca cga ttc gga tgg-3′ ) as the reverse primer (positions 166028–165999 of 68-1; GeneBank accession no. AY186194). The expression of RhUL132 in purified RhCMV virions, and infected MKE and Telo-RF lysates was detected by Western blotting using rabbit anti-RhUL132 peptide serum following a previously published protocol (Vogel et al., 1994). The expression of pp65-2 and GAPDH were used as internal controls. In addition, to eliminate the background of non-specific binding, rabbit anti-RhUL132 peptide serum was pre-incubated with a blot transferred with uninfected cell antigens prior to detecting RhUL132 expression in infected cells.
Acknowledgements
We would like to thank Dr Zhongmin Ma for capturing the images and Dr Thomas Shenk for providing BRh68-1.2 virus. This work was supported by funding from the National Institutes of Health to P. A. B. (AI063356, AI49342) and from the California National Primate Research Center (OD011107).
References
- Adler B., Scrivano L., Ruzcics Z., Rupp B., Sinzger C., Koszinowski U. (2006). Role of human cytomegalovirus UL131A in cell type-specific virus entry and release. J Gen Virol 87 2451–2460. 10.1099/vir.0.81921-0 [DOI] [PubMed] [Google Scholar]
- Akter P., Cunningham C., McSharry B. P., Dolan A., Addison C., Dargan D. J., Hassan-Walker A. F., Emery V. C., Griffiths P. D., Wilkinson G. W., Davison A. J. (2003). Two novel spliced genes in human cytomegalovirus. J Gen Virol 84 1117–1122. 10.1099/vir.0.18952-0 [DOI] [PubMed] [Google Scholar]
- Ando Y., Iwasaki T., Sata T., Soushi S., Kurata T., Arao Y. (1997). Enhanced cytopathic effect of human cytomegalovirus on a retinal pigment epithelium cell line, K-1034, by serum-free medium. Arch Virol 142 1645–1658. [DOI] [PubMed] [Google Scholar]
- Assaf B. T., Mansfield K. G., Strelow L., Westmoreland S. V., Barry P. A., Kaur A. (2014). Limited dissemination and shedding of the UL128 complex-intact, ul/b'-defective rhesus cytomegalovirus strain 180.92. J Virol 88 9310–9320. 10.1128/JVI.00162-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer P. C., Bereiter-Hahn J., Schubert R., Geiger H. (2006). Differentiation status of human renal proximal and distal tubular epithelial cells in vitro: Differential expression of characteristic markers. Cells Tissues Organs 184 16–22. 10.1159/000096947 [DOI] [PubMed] [Google Scholar]
- Bodaghi B., Slobbe-van Drunen M. E., Topilko A., Perret E., Vossen R. C., van Dam-Mieras M. C., Zipeto D., Virelizier J. L., LeHoang P., Bruggeman C. A., Michelson S. (1999). Entry of human cytomegalovirus into retinal pigment epithelial and endothelial cells by endocytosis. InvestOphthalmol Vis Sci 40 2598–2607. [PubMed] [Google Scholar]
- Bradley A. J., Lurain N. S., Ghazal P., Trivedi U., Cunningham C., Baluchova K., Gatherer D., Wilkinson G. W., Dargan D. J., Davison A. J. (2009). High-throughput sequence analysis of variants of human cytomegalovirus strains towne and AD169. J Gen Virol 90 2375–2380. 10.1099/vir.0.013250-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt W. J., Alford C. A. (1996). Cytomegalovirus. Fields Virology, 3rd edn, 2493–2523. Edited by Fields B. N., Kniepe D. N., Howley P. M. Philadelphia, PA: Lippincott-Raven. [Google Scholar]
- Cha T. A., Tom E., Kemble G. W., Duke G. M., Mocarski E. S., Spaete R. R. (1996). Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol 70 78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W. L., Kirchoff V., Pari G. S., Barry P. A. (2002). Replication of rhesus cytomegalovirus in life-expanded rhesus fibroblasts expressing human telomerase. J Virol Methods 104 135–146. [DOI] [PubMed] [Google Scholar]
- Chang W. L., Barry P. A. (2003). Cloning of the full-length rhesus cytomegalovirus genome as an infectious and self-excisable bacterial artificial chromosome for analysis of viral pathogenesis. J Virol 77 5073–5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung T. C., Humphreys I. R., Potter K. G., Norris P. S., Shumway H. M., Tran B. R., Patterson G., Jean-Jacques R., Yoon M., Spear P. G., Murphy K. M., Lurain N. S., Benedict C. A., Ware C. F. (2005). Evolutionarily divergent herpesviruses modulate T cell activation by targeting the herpesvirus entry mediator cosignaling pathway. Proc Natl Acad Sci U S A 102 13218–13223. 10.1073/pnas.0506172102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham C., Gatherer D., Hilfrich B., Baluchova K., Dargan D. J., Thomson M., Griffiths P. D., Wilkinson G. W., Schulz T. F., Davison A. J. (2010). Sequences of complete human cytomegalovirus genomes from infected cell cultures and clinical specimens. J Gen Virol 91 605–615. 10.1099/vir.0.015891-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaPalma T., Doonan B. P., Trager N. M., Kasman L. M. (2010). A systematic approach to virus–virus interactions. Virus Res 149 1–9. 10.1016/j.virusres.2010.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dargan D. J., Douglas E., Cunningham C., Jamieson F., Stanton R. J., Baluchova K., McSharry B. P., Tomasec P., Emery V. C., Percivalle E., Sarasini A., Gerna G., Wilkinson G. W., Davison A. J. (2010). Sequential mutations associated with adaptation of human cytomegalovirus to growth in cell culture. J Gen Virol 91 1535–1546. 10.1099/vir.0.018994-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkouby-Naor L., Ben-Yosef T. (2010). Functions of claudin tight junction proteins and their complex interactions in various physiological systems. Int Rev Cell Mol Biol 279 1–32. 10.1016/S1937-6448(10)79001-8 [DOI] [PubMed] [Google Scholar]
- Esclatine A., Lemullois M., Servin A. L., Quero A. M., Geniteau-Legendre M. (2000). Human cytomegalovirus infects caco-2 intestinal epithelial cells basolaterally regardless of the differentiation state. J Virol 74 513–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish K. N., Soderberg-Naucler C., Mills L. K., Stenglein S., Nelson J. A. (1998). Human cytomegalovirus persistently infects aortic endothelial cells. J Virol 72 5661–5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Mariscal L., Garay E., Lechuga S. (2009). Virus interaction with the apical junctional complex. Front Biosci 14 731–768. [DOI] [PubMed] [Google Scholar]
- Goodrum F., Reeves M., Sinclair J., High K., Shenk T. (2007). Human cytomegalovirus sequences expressed in latently infected individuals promote a latent infection in vitro. Blood 110 937–945. 10.1182/blood-2007-01-070078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn G., Revello M. G., Patrone M., Percivalle E., Campanini G., Sarasini A., Wagner M., Gallina A., Milanesi G., Koszinowski U., Baldanti F., Gerna G. (2004). Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J Virol 78 10023–10033. 10.1128/JVI.78.18.10023-10033.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S. G., Strelow L. I., Franchi D. C., Anders D. G., Wong S. W. (2003). Complete sequence and genomic analysis of rhesus cytomegalovirus. J Virol 77 6620–6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Ma Y., Qi Y., Jiang S., Wang N., Li M., Ji Y., Sun Z., Ruan Q. (2012). Characterization of human cytomegalovirus UL146 transcripts. Virus Res 163 223–228. 10.1016/j.virusres.2011.09.034 [DOI] [PubMed] [Google Scholar]
- Ho M. (1991). Cytomegalovirus: Biology and Infection. New York: Plenum Medical Book Co. [Google Scholar]
- Jarvis M. A., Wang C. E., Meyers H. L., Smith P. P., Corless C. L., Henderson G. J., Vieira J., Britt W. J., Nelson J. A. (1999). Human cytomegalovirus infection of caco-2 cells occurs at the basolateral membrane and is differentiation state dependent. J Virol 73 4552–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis M. A., Nelson J. A. (2007). Human cytomegalovirus tropism for endothelial cells: Not all endothelial cells are created equal. J Virol 81 2095–2101. 10.1128/JVI.01422-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao W. W., Prockop D. J. (1977). Proline analogue removes fibroblasts from cultured mixed cell populations. Nat New Biol 266 63–64. [DOI] [PubMed] [Google Scholar]
- Kirchoff V., Wong S., St J. S., Pari G. S. (2002). Generation of a life-expanded rhesus monkey fibroblast cell line for the growth of rhesus rhadinovirus (RRV). Arch Virol 147 321–333. [DOI] [PubMed] [Google Scholar]
- Kudchodkar S. B., Yu Y., Maguire T. G., Alwine J. C. (2004). Human cytomegalovirus infection induces Imycin-insensitive phosphorylation of downstream effectors of mTOR kinase. J Virol 78 11030–11039. 10.1128/JVI.78.20.11030-11039.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landolfo S., Gariglio M., Gribaudo G., Lembo D. (2003). The human cytomegalovirus. Pharmacol Ther 98 269–297. 10.1016/S0163-7258(03)00034-2 [DOI] [PubMed] [Google Scholar]
- Li G., Nguyen C. C., Ryckman B. J., Britt W. J., Kamil J. P. (2015). A viral regulator of glycoprotein complexes contributes to human cytomegalovirus cell tropism. Proc Natl Acad Sci U S A 112 4471–4476. 10.1073/pnas.1419875112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilja A. E., Chang W. L., Barry P. A., Becerra S. P., Shenk T. E. (2008). Functional genetic analysis of rhesus cytomegalovirus: Rh01 is an epithelial cell tropism factor. J Virol 82 2170–2181. 10.1128/JVI.02316-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilja A. E., Shenk T. (2008). Efficient replication of rhesus cytomegalovirus variants in multiple rhesus and human cell types. Proc Natl Acad Sci U S A 105 19950–19955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockridge K. M., Sequar G., Zhou S. S., Yue Y., Mandell C. P., Barry P. A. (1999). Pathogenesis of experimental rhesus cytomegalovirus infection. J Virol 73 9576–9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurain N. S., Fox A. M., Lichy H. M., Bhorade S. M., Ware C. F., Huang D. D., Kwan S. P., Garrity E. R., Chou S. (2006). Analysis of the human cytomegalovirus genomic region from UL146 through UL147A reveals sequence hypervariability, genotypic stability, and overlapping transcripts. Virol J 3, 4. 10.1186/1743-422X-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick A. L., Roback L., Livingston-Rosanoff D., St Clair C. (2010). The human cytomegalovirus UL36 gene controls caspase-dependent and -independent cell death programs activated by infection of monocytes differentiating to macrophages. J Virol 84 5108–5123. 10.1128/JVI.01345-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard A. L., Häberli L., Sinzger C., Ghielmetti M., Schneider M. K., Bossart W., Seebach J. D., Mueller N. J., Haberli L. (2010). Efficiency of porcine endothelial cell infection with human cytomegalovirus depends on both virus tropism and endothelial cell vascular origin. Xenotransplantation 17 274–287. 10.1111/j.1399-3089.2010.00594.x [DOI] [PubMed] [Google Scholar]
- Oxford K. L., Eberhardt M. K., Yang K. W., Strelow L., Kelly S., Zhou S. S., Barry P. A. (2008). Protein coding content of the ULb′ region of wild-type rhesus cytomegalovirus. Virology (Auckl) 373 181–188. 10.1016/j.virol.2007.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxford K. L., Strelow L., Yue Y., Chang W. L., Schmidt K. A., Diamond D. J., Barry P. A. (2011). Open reading frames carried on ULb′ are implicated in shedding and horizontal transmission of rhesus cytomegalovirus in rhesus monkeys. J Virol 85 5105–5114. 10.1128/JVI.02631-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfold M. E. T., Dairaghi D. J., Duke G. M., Saederup N., Mocarski E. S., Kemble G. W., Schall T. J. (1999). Cytomegalovirus encodes a potent a chemokine. Proc Natl Acad Sci U S A 96 9839–9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepin K. M., Hanley K. A. (2008). Density-dependent competitive suppression of sylvatic dengue virus by endemic dengue virus in cultured mosquito cells. Vector Borne Zoonotic Dis 8 821–828. 10.1089/vbz.2008.0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepin K. M., Lambeth K., Hanley K. A. (2008). Asymmetric competitive suppression between strains of dengue virus. BMC Microbiol 8. 10.1186/1471-2180-8-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perefarres F., Thebaud G., Lefeuvre P., Chiroleu F., Rimbaud L., Hoareau M., Reynaud B., Lett J. M. (2014). Frequency-dependent assistance as a way out of competitive exclusion between two strains of an emerging virus. Proc Biol Sci 281 2013–3374. 10.1098/rspb.2013.3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard M. N., Penfold M. E., Duke G. M., Spaete R. R., Kemble G. W. (2001). A review of genetic differences between limited and extensively passaged human cytomegalovirus strains. Rev Med Virol 11 191–200. [DOI] [PubMed] [Google Scholar]
- Prod'homme V., Sugrue D. M., Stanton R. J., Nomoto A., Davies J., Rickards C. R., Cochrane D., Moore M., Wilkinson G. W., Tomasec P. (2010). Human cytomegalovirus UL141 promotes efficient downregulation of the natural killer cell activating ligand CD112. J Gen Virol 91 2034–2039. 10.1099/vir.0.021931-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read A. F., Taylor L. H. (2001). The ecology of genetically diverse infections. Science 292 1099–1102. [DOI] [PubMed] [Google Scholar]
- Renzette N., Bhattacharjee B., Jensen J. D., Gibson L., Kowalik T. F. (2011). Extensive genome-wide variability of human cytomegalovirus in congenitally infected infants. PLoS Pathog 7 . 10.1371/journal.ppat.1001344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivailler P., Kaur A., Johnson R. P., Wang F. (2006). Genomic sequence of rhesus cytomegalovirus 180.92: insights into the coding potential of rhesus cytomegalovirus. J Virol 80 4179–4182. 10.1128/JVI.80.8.4179-4182.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckman B. J., Jarvis M. A., Drummond D. D., Nelson J. A., Johnson D. C. (2006). Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J Virol 80 710–722. 10.1128/JVI.80.2.710-722.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckman B. J., Rainish B. L., Chase M. C., Borton J. A., Nelson J. A., Jarvis M. A., Johnson D. C. (2008). Characterization of the human cytomegalovirus gH/gL/UL128-131 complex that mediates entry into epithelial and endothelial cells. J Virol 82 60–70. 10.1128/JVI.01910-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M., Ando-Akatsuka Y., Itoh M., Furuse M., Inazawa J., Fujimoto K., Tsukita S. (1997). Mammalian occludin in epithelial cells: its expression and subcellular distribution. Eur J Cell Biol 73 222–231. [PubMed] [Google Scholar]
- Scrivano L., Sinzger C., Nitschko H., Koszinowski U. H., Adler B. (2011). HCMV spread and cell tropism are determined by distinct virus populations. PLoS Pathog 7 . 10.1371/journal.ppat.1001256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinzger C., Kahl M., Laib K., Klingel K., Rieger P., Plachter B., Jahn G. (2000). Tropism of human cytomegalovirus for endothelial cells is determined by a post-entry step dependent on efficient translocation to the nucleus. J Gen Virol 81 3021–3035. 10.1099/0022-1317-81-12-3021 [DOI] [PubMed] [Google Scholar]
- Sinzger C., Digel M., Jahn G. (2008). Cytomegalovirus cell tropism. Curr Top Microbiol Immunol 325 63–83. [DOI] [PubMed] [Google Scholar]
- Spaderna S., Kropff B., Ködel Y., Shen S., Coley S., Lu S., Britt W., Mach M., Kodel Y. (2005). Deletion of gpUL132, a structural component of human cytomegalovirus, results in impaired virus replication in fibroblasts. J Virol 79 11837–11847. 10.1128/JVI.79.18.11837-11847.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton R. J., Baluchova K., Dargan D. J., Cunningham C., Sheehy O., Seirafian S., McSharry B. P., Neale M. L., Davies J. A., Tomasec P., Davison A. J., Wilkinson G. W. (2010). Reconstruction of the complete human cytomegalovirus genome in a BAC reveals RL13 to be a potent inhibitor of replication. J Clin Invest 120. 10.1172/JCI42955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasec P., Wang E. C., Davison A. J., Vojtesek B., Armstrong M., Griffin C., McSharry B. P., Morris R. J., Llewellyn-Lacey S., Rickards C., Nomoto A., Sinzger C., Wilkinson G. W. (2005). Downregulation of natural killer cell-activating ligand CD155 by human cytomegalovirus UL141. Nat Immunol 6 181–188. 10.1038/ni1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugizov S., Maidji E., Pereira L. (1996). Role of apical and basolateral membranes in replication of human cytomegalovirus in polarized retinal pigment epithelial cells. J Gen Virol 77 (Pt 1), 61–74. 10.1099/0022-1317-77-1-61 [DOI] [PubMed] [Google Scholar]
- Umashankar M., Petrucelli A., Cicchini L., Caposio P., Kreklywich C. N., Rak M., Bughio F., Goldman D. C., Hamlin K. L., Nelson J. A., Fleming W. H., Streblow D. N., Goodrum F. (2011). A novel human cytomegalovirus locus modulates cell type-specific outcomes of infection. PLoS Pathog 7 . 10.1371/journal.ppat.1002444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel P., Weigler B. J., Kerr H., Hendrickx A. G., Barry P. A. (1994). Seroepidemiologic studies of cytomegalovirus infection in a breeding population of rhesus macaques. Lab Anim Sci 44 25–30. [PubMed] [Google Scholar]
- Wang D., Shenk T. (2005). Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc Natl Acad Sci U S A 102 18153–18158. 10.1073/pnas.0509201102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Yu Q. C., Schröer J., Murphy E., Shenk T. (2007). Human cytomegalovirus uses two distinct pathways to enter retinal pigmented epithelial cells. Proc Natl Acad Sci U S A 104 20037–20042. 10.1073/pnas.0709704104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wussow F., Yue Y., Martinez J., Deere J. D., Longmate J., Herrmann A., Barry P. A., Diamond D. J. (2013). A vaccine based on the rhesus cytomegalovirus UL128 complex induces broadly neutralizing antibodies in rhesus macaques. J Virol 87 1322–1332. 10.1128/JVI.01669-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y., Kaur A., Zhou S. S., Barry P. A. (2006). Characterization and immunological analysis of the rhesus cytomegalovirus homologue (Rh112) of the human cytomegalovirus UL83 lower matrix phosphoprotein (pp65). J Gen Virol 87 777–787. 10.1099/vir.0.81516-0 [DOI] [PubMed] [Google Scholar]
- Yue Y., Kaur A., Eberhardt M. K., Kassis N., Zhou S. S., Tarantal A. F., Barry P. A. (2007). Immunogenicity and protective efficacy of DNA vaccines expressing rhesus cytomegalovirus glycoprotein B, phosphoprotein 65-2, and viral interleukin-10 in rhesus macaques. J Virol 81 1095–1109. 10.1128/JVI.01708-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y., Barry P. A. (2008). Rhesus cytomegalovirus a nonhuman primate model for the study of human cytomegalovirus. Adv Virus Res 72 207–226. 10.1016/S0065-3527(08)00405-3 [DOI] [PubMed] [Google Scholar]