Abstract
The results from molecular assays can be affected significantly by the preanalytic condition of cytologic samples. The authors review current knowledge on the use of cytologic samples for epidermal growth factor receptor (EGFR) mutation testing in non–small cell lung cancer with a focus on preanalytic parameters. A systematic electronic search of the MEDLINE database was performed to identify original articles that reported the use of cytologic samples for EGFR molecular analysis and included a minimum of 100 samples. The information collected included author(s), journal, and year of publication; number of patients and samples; sampling method; type of preparation; type of fixative; staining techniques; mutation analysis techniques; tumor cellularity; the percentage of tumor cells; data on DNA quantity, quality, and concentration; failed assays; and the mutation rate. EGFR mutation analysis was conducted on 4999 cytologic samples from 22 studies that fulfilled the inclusion criteria. Fine‐needle aspirates and pleural effusions were the most common types of specimens used. DNA was mainly extracted from cell blocks and smears, and the most commonly reported fixatives included formalin, ethanol, and CytoLyt. Cellularity assessments and DNA yields were available from 5 studies each. The average success rate for the assays that used cytologic specimens was 95.87% (range, 85.2%‐100%). The mutation rate ranged from 6% to 50.46%, and a higher mutation detection rate and lower numbers of insufficient cases were reported for pleural effusions and lymph node samples from endobronchial ultrasound‐guided transbronchial needle aspiration compared with histologic specimens. Low cellularity and a low percentage of tumor cells were associated with higher test failure rates. Future guidelines should consider the current data for specific recommendations regarding cytologic samples. Cancer (Cancer Cytopathol) 2015;123:633–643. © 2015 American Cancer Society.
Keywords: cell blocks, cytology, epidermal growth factor receptor, fine‐needle aspiration, molecular cytopathology, mutation analysis, non–small lung cancer, preanalytic
Short abstract
Preanalytic parameters for epidermal growth factor receptor mutation testing are reviewed in non–small cell lung cancer using 4999 cytologic samples from 22 studies. A higher mutation detection rate and lower numbers of insufficient cases are observed for pleural effusions and lymph node samples obtained using endobronchial ultrasound‐guided transbronchial needle aspiration compared with histologic specimens, and low cellularity and a lower percentage of tumor cells are associated with higher test failure rates. Future guidelines should consider the current data for specific recommendations regarding cytologic samples.
INTRODUCTION
Currently, patients who have non–small cell lung carcinoma (NSCLC) with an adenocarcinoma component should be routinely tested for 2 major genomic alterations—epidermal growth factor receptor (EGFR) mutations and anaplastic lymphoma kinase (ALK) rearrangements—to identify those who are eligible for treatment with the targeted drugs available, ie, EGFR tyrosine kinase inhibitors (TKIs) and ALK inhibitors, respectively. Testing for those specific alterations has become a global standard of care; and, as a consequence, the number of laboratory molecular analyses has exponentially increased.
Limited efficacy has been attained with targeted therapy because of primary or acquired resistance during the course of treatment with the EGFR TKIs gefitinib and erlotinib and with the ALK inhibitor crizotinib. This has triggered the development of novel inhibitors to circumvent resistance. In contrast to first‐generation drugs, second‐generation EGFR TKIs are irreversibly bound, which means that the drugs form a direct chemical covalent bond with EGFR. Because of the disappointing response rates produced by all 3 of the second‐generation TKIs tested (neratininib, afatinib and dacomitinib), a new class of third‐generation EGFR TKIs has emerged, and preliminary data have produced response rates of 60% in patients with a biopsy‐proven EGFR resistance mutation (a threonine to methionine substitution at codon 790 [T790M]). Different from previous generation drugs, although third‐generation TKIs produce potent inhibition of both activating EGFR mutations and the T790M mutation, wild‐type inhibition is close to zero, allowing the escalation of doses to concentrations that can effectively overcome acquired resistance.1
The list of therapeutically relevant genome alterations as potential markers to be added to our practice is likely to increase. Furthermore, increasing numbers of patients who are amenable to minimally invasive procedures also are expected and will add to the collection of serial samples over time, either for research or to guide clinical decisions, to document recurrence and/or resistance, and to check for alterations in the genomic profile.2 The samples obtained with minimally invasive procedures are limited, and they are challenged by the need to provide multiple data for various ancillary studies currently in use in clinical practice and also by the potential addition of future tests. The inclusion of biomarker testing adds to the complexity of lung cancer diagnostic algorithms and can affect the timeliness of treatment decisions, particularly when biopsies yield insufficient tissue for analysis.3 Delays may be avoided by incorporating reflex biomarker testing into diagnostic algorithms for NSCLC at the level of the pathologist and by further education of the specialists involved in obtaining diagnostic cancer specimens to ensure that such specimens are sufficient for molecular testing.3
Substantial challenges arise in aliquoting limited samples like cytologic specimens for the evaluation of multiple molecular markers, because different techniques are currently used for their detection. Thus, in addition to the immunohistochemistry studies frequently performed for subtyping and/or establishing the lung as the primary site, adequate and sufficient quality cellular material should be available to conduct EGFR mutation analyses and detect ALK rearrangements using immunohistochemistry and/or fluorescent in situ hybridization. Another recurrent issue has been the strategies used to balance the need to obtain an adequate amount and quality of tissue/cells for multiple ancillary tests, including molecular analyses, and the fixative procedures required to process specimens for routine diagnostic workup and storage. The versatility of cytologic samples in terms of different types of sample collection, preparations, and fixatives can be viewed as an advantage, but it also presents several problems, including infinite numbers of repeated test validations for molecular studies. The advantage is that, because the fixation and processing of cytologic samples are almost always performed immediately after sample collection, usually without delays (even when using formalin as fixative), cytologic samples (including cell blocks) are expected to have better preserved material and, consequently, nucleic acids for obtaining consistently reliable molecular results.4
Rapid advances in genomic sequencing technologies with novel, high‐throughput sequencing platforms have emerged and have enabled comprehensive molecular profiling, leading to the discovery of genomic alterations in various types of cancer with the potential to elucidate several mechanisms involved in cancer development and progression, including drug resistance, thus improving clinical care and patient outcomes. These techniques, using minimal material coupled with robust techniques for DNA amplification, have overcome some limitations regarding the amount of sample required for multiple assays and have generated opportunities for the use of cytologic specimens. Furthermore, they also allow for the detection of multiple different genomic alterations at the same time, which may save precious tissue‐derived materials.
Current guidelines and expert recommendations for molecular testing are predominantly focused on formalin‐fixed, paraffin‐embedded (FFPE) surgical specimens and small‐needle biopsies, with minimal attention to cytologic samples.5, 6 Multiple studies have reported the use of different cytologic preparations for EGFR mutation analysis in lung cancer.7 Like other biologic materials, cytologic samples are subject to various preanalytic conditions, including different collection, processing, and storage factors that can significantly alter their molecular composition and consistency.8 The objective of this review was to provide an update of the current knowledge on the use of cytologic specimens for the evaluation of EGFR mutation status in patients with NSCLC with a focus on preanalytic variables.
MATERIALS AND METHODS
Search Strategy and Study Eligibility
We performed a systematic computerized search in the MEDLINE database for articles dealing directly with the use of cytologic samples for EGFR molecular analysis. The last search was conducted on March 20, 2015. The following terms were used, in this exact format: (“FNA” or “fine‐needle aspiration” or “needle aspiration” or “cytology” or “cytopathology” or “cytologic material” or “cytological specimens” or “needle biopsy” or “small needle biopsy”) and (“EGFR” or “epidermal growth factor receptor”) and “lung.” The results were filtered for articles originally published in English; and, in total, 181 articles were retrieved. The articles were then selected and were included in the structured review based on the following criteria: 1) only original reports with information on the use of cytologic samples for EGFR molecular analysis (abstracts, letters to the editor, tutorials, editorials, reviews, case reports, and conference proceedings were excluded), 2) a minimal sample size of 100 cases/samples, 3) articles obtained with the search strategy, and 4) articles that were included in the reference lists of the articles identified with the search strategy that fulfilled criteria 1 and 2. Additional original articles of interest or relevant to the current review were selected only for the sake of introduction and discussion of results but were excluded from the data extraction.
Data Extraction
The following information was collected from each article: the first author, journal, and year of publication; the number of patients enrolled in the study and the number of samples; the number of primary tumors and metastases; the sampling method; the specific site of collection; the type of cytologic preparation; the type of fixative; the method(s) of staining; the amount of tumor cells/cellularity assessment; the percentage of tumor cells; the number/percentage of failed assays; and the percentage of mutated samples. If the reasons for failure were provided, then they also were retrieved from the articles. Because different analytic sensitivities of the molecular assays have an impact on the required tumor cell content of the specimens, analytic parameters, such as the techniques used for mutation analysis, data on DNA quantity (final DNA yield)/quality, and DNA concentration, also were extracted from the articles.
RESULTS
From 181 articles that were identified using the search strategy, 22 original studies (20 from the search and 2 from the references) fulfilled the inclusion criteria, and the complete published articles were used for data extraction (Fig. 1). In total, 4999 samples were analyzed for EGFR mutation from all articles (Table 1), and information on the number of enrolled patients was available for 19 studies.9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27 In 1 of these studies, information was available for all enrolled patients, including those who had histologic samples.18 In 3 studies,28, 29, 30 information on the number of patients was not available. Eleven studies included only cytologic samples, and 11 also reported the results from surgical/core‐needle biopsy specimens.10, 11, 14, 17, 18, 19, 21, 22, 25, 26, 27 The oldest study with more than 100 specimens was published in 2007,25 and the most recent series retrieved were published online in 2014.18, 19, 21, 22, 26, 29 The 2 largest series described more than 500 cytologic samples each.10, 22 According to the country of the institution or laboratory where EGFR testing was performed, the 22 articles were subdivided as follows: there were 5 articles from Italy,14, 19, 23, 28, 29 three from Japan,15, 25, 27 three from the United Kingdom (with 1 study that gathered samples from multiples institutions, mainly southeast Asian),16, 17, 20 two from Australia,11, 18 two from the United States,9, 30 and 1 each from Canada,22 the Netherlands,24 Slovakia,10 Hong‐Kong,13 Spain,12 and China.26 In the other study, although samples reportedly originated from various east Asian countries, no information was available on the nationality of the laboratory where EGFR testing was performed.21 The largest series were from Slovakia (n = 675 specimens) and Canada (n = 513 specimens). Data from those series encompassed consecutive specimens over 24 months (from 2010 to 2012).10, 22 Another large series, although it reported complete sequencing of 1717 cytologic and histologic specimens, had information on the specific biopsy type available for only 929 specimens, and 29.4% of these were cytologic samples.18
Figure 1.
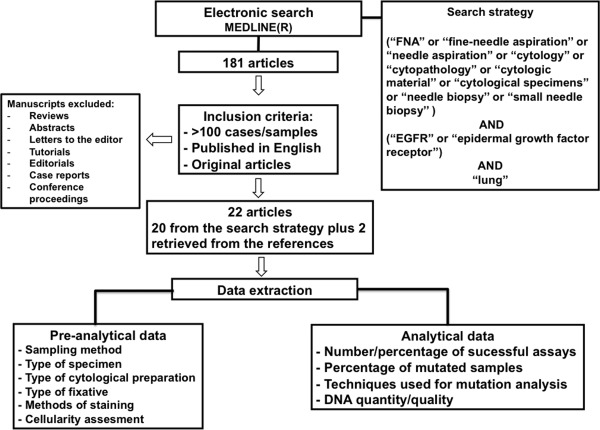
This is a flow diagram of the included and excluded articles and the data extracted.
Table 1.
Generic Data Retrieved From the Studies
| Reference | Country Where EGFR Testing Was Performed | No. of Cytologic Samples | No. of Histologic Samples | No. of Patients With Cytologic Samples |
|---|---|---|---|---|
| Allegrini 201228 | Italy | 108 | None | NA |
| Bellevicine 201429 | Italy | 362 | None | NA |
| Billah 20119 | United States | 209 | None | 209 |
| Hlinkova 201310 | Slovakia | 679 | 156 | 679 |
| Leslie 201411 | Australia | 168 | 142 | 168 |
| Lozano 201112 | Spain | 150 | None | 150 |
| Ma 201213 | Hong‐Kong | 269 | None | 269 |
| Mallapelle 201314 | Italy | 305 | 294 | 305 |
| Nakajima 201115 | Japan | 156 | None | 156 |
| Navani 201216 | United Kingdom | 119 | None | 119 |
| Pang 201217 | United Kingdoma | 165 | 505 | 165 |
| Peters 201418 | Australia | 274b | 655b | 2012c |
| Proietti 201419 | Italy | 161 | 265 | 161 |
| Rekhtman 201130 | United States | 128 | None | NA |
| Santis 201120 | United Kingdom | 131 | None | 131 |
| Shi 201421 | NAd | 169 | 1313 | 169 |
| Shiau 201422 | Canada | 513 | 1780 | 513 |
| Stella 201323 | Italy | 134 | None | 134 |
| Stigt 201324 | Netherlands | 126 | None | 126 |
| Takano 200725 | Japan | 117 | 130 | 117 |
| Wu 201426 | China | 434 | 101 | 434 |
| Yamada 201227 | Japan | 122 | 22 | 122 |
Abbreviations: EGFR, epidermal growth factor receptor; NA, not available.
Samples were from various East Asian countries.
This number was based on 929 samples that had a specified biopsy type; there were 901 additional samples for which this information was not provided.
The total number is indicated, including histologic samples.
Information on EGFR testing performed in individual countries was not available, but samples originated from various countries, including China (and Hong‐Kong), India, the Philippines, Taiwan, Thailand, and Vietnam.
Sampling Methods and Types of Specimens, Fixatives, and Cytologic Preparations
Several sampling methods and types of cytologic specimens have been reported. The 2 most frequent specimen types were fine‐needle aspirations (FNAs) (17 of 22 studies)9, 11, 12, 13, 14, 17, 18, 19, 21, 22, 23, 24, 25, 26, 27, 28, 30 and pleural fluids (11 of 22 studies).9, 11, 12, 13, 14, 18, 19, 22, 25, 26, 28, 30 In the vast majority of studies, FNAs of the lung were performed using computerized tomography guidance, although other sites, such as liver, bone, and lymph nodes, also have been described. Ultrasonography has been used for guiding FNA from different extrapulmonary sites. Three series reported only cytologic samples obtained by endobronchial ultrasound‐guided transbronchial needle aspiration (EBUS‐TBNA) from mediastinal lymph nodes.15, 16, 20, 31 Other methods/types of specimens included bronchial brushing, bronchial washing, peritoneal and pericardial fluids, bronchoalveolar lavage, and sputum. Only 1 study did not include information about the sampling method used29 (Table 2).
Table 2.
Types of Samples and Fixatives Used and Molecular Data
| Reference | Sampling Method | Type of Fixative | DNA Yield, ng/μL | Successful Assays, % | Mutation Rate, % |
|---|---|---|---|---|---|
| Allegrini 201228 | FNA, BB, BW, PF, PeF | CytoLyt‐PreservCyt (Cytyc Corp, Boxborough, Mass), ethanol, Dubosq‐Brasil, formalin | NA | 85.2 | 24 |
| Bellevicine 201429 | NA | CytoLyta | 60.94 (SM), 23.07 (LBC) | 96.6 | 12.2 |
| Billah 20119 | FNA, BAL, BW, PF, PeF | Carnoy, alcohol (SM); formalin and ethanol (CB) | NA | 93.8 | 19.4 |
| Hlinkova 201310 | BB, PF, BW | NA | 2.3–5.6b | 95c | 8 |
| Leslie 201311 | FNA, PF, PCF, PeF, BW, BB | Formalin | NA | 100 | 14.9 |
| Lozano 201112 | FNA, EBUS‐TBNA, fluids | Alcohold | 24.6 | 100 | 17.3 |
| Ma 201213 | FNA, PF, BB, BW, SP, others | NA | NA | 99.3 | 39.4 |
| Mallapelle 201314 | FNA, fluids, BB, BW, SP | NA | NA | 100 | 8.8 |
| Nakajima 201115 | EBUS‐TBNA | Allprotect Tissue Reagent (Qiagen, Venlo, Netherlands) | NA | 98.7 (CP), 100 (FC) | 26.9 |
| Navani 201216 | EBUS‐TBNA | Liquid fixative, formalin | NA | 90 | 6 |
| Pang 201217 | FNA, effusions, BB | Formaline | At least 20 | 87.7 | 4.2c |
| Peters 201418 | FNA, PF, PCF | NA | NA | 64f | 25.6 (FNA), 15.9 (fluids) |
| Proietti 201419 | FNA, PF, BB | Cytofix (BD Biosciences, Franklin Lakes, NJ)d | NA | 88.8 | 14.9 |
| Rekhtman 201130 | FNA, BB, BW, PF | CytoLyt | NA | 98 | 25 |
| Santis 201120 | EBUS‐TBNA | NA | NA | 95.5 | 10.3 |
| Shi 201421 | FNA, BW | NA | NA | 95.3 | 51.4g |
| Shiau 201422 | BW, BB, FNA, PF | Alcohol, formalin | NA | 95.5 | 26.1 |
| Stella 201323 | FNA | Formalin | NA | 100 | 9.7 |
| Stigt 201324 | FNA | Polyethylene glycol 3350 2% (Carbowax; Dow Chemical Company, Midland, Mich), formalin | NA | 96.8 | 12 |
| Takano 200725 | FNA, BB, BW, PF, PCF, SP | NA | NA | 97.7g | 41g |
| Wu 201426 | FNA, BB, BAL, PF, SP, EBUS‐TBNA | CytoLyt | 10‐935.3 | 100 | 50.4 |
| Yamada 201227 | FNA, BB | None | NA | 99 | 33.6 |
Abbreviations: BAL, bronchoalveolar lavage; BB, bronchial brushing; BW, bronchial washing; CB, cell block; CP, cell pellets; EBUS‐TBNA, tissue biopsy endobronchial ultrasound guided transbronchial needle aspiration; FC, fresh cells; FNA, fine‐needle aspiration; LBC, liquid‐based cytology; NA, not available; PCF, pericardial fluid; PeF, peritoneal fluid; PF, pleural fluid; SM, smears; SP, sputum.
This was used for LBC, no information was available for smears.
Information was for failed cases.
These included histologic samples.
Information was available only for smears.
Information was available only for cell blocks.
Information was available only for FNA.
Information included histologic samples.
The most commonly reported fixatives were formalin, ethanol, and CytoLyt (Cytyc Corp, Boxborough, Mass).9, 11, 12, 16, 17, 22, 23, 24, 26, 28, 29, 30 In 7 articles (7 of 22 studies), information about the fixative used was not available.10, 13, 14, 18, 20, 21, 25 The other fixatives described were Duboscq‐Brasil,28 Cytofix (BD Biosciences, Franklin Lakes, NJ),19 Carbowax 2% (Dow Chemical Company, Midland, Mich),24 and Allprotect Tissue Reagent (Qiagen, Venlo, Netherlands)15 (Table 2).
Cell blocks were the most commonly used type of cytologic preparation (15 of 22 studies)9, 11, 12, 13, 14, 16, 17, 18, 19, 20, 22, 23, 24, 28, 30 followed by direct smears (9 of 22 studies).9, 11, 12, 14, 17, 19, 25, 28, 29 Four articles described the use of fresh cells,12, 15, 23, 27 and 4 used liquid‐based cytology (LBC) preparations12, 26, 28, 29 (Fig. 2). Comparisons of failure rates for cell blocks and smears were available in only 3 studies. Two studies had concordant results with similar failure rates for smears (6.1%) and cell blocks (6.4%) in 1 study9; and, for the other study, although there were differences in the failure rates, they were not statistically significant (0% for FNA smears vs 4.7% for cell blocks).17 In addition, for the latter study, the rejection rate was lower for FNA smears (3.2%) than for FNA cell blocks (15%) and effusion cell blocks (8.2%). However, for the third study, the results were contradictory, because the rates for unamplified samples were higher for smears (27.3%) than for cell blocks (17.6%) and cell suspensions (0%).28
Figure 2.
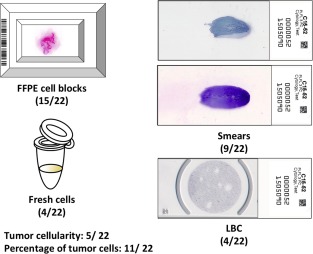
The types of cytologic preparations used for epidermal growth factor receptor mutation analysis are illustrated along with tumor cellularity and the percentage of tumor cells. Numbers in parentheses indicate the number of articles that reported such information out of 22 articles that were included in the current review. FFPE indicates formalin‐fixed, paraffin‐embedded; LBC, liquid‐based cytology.
Amount of Tumor Cells/Cellularity Assessment and Percentage of Tumor Cells
Only 5 studies reported cellularity assessments using different categories.9, 10, 22, 28, 29 Poor cellularity and a low tumor cell percentage (range, 1%‐20%) were the most common causes of test failure.10 Overall cell content was correlated significantly with the test success rate, and samples with minimal cell content had significantly lower test success rates than samples with small clusters to abundant cell content in a different study.22 One study reported tumor cellularity only for mutated and unamplified samples, with tumor cell numbers ranging from 16 to >1000 cells in the mutated samples.28 Cellularity had a good correlation with the amount of extracted DNA. Higher tumor cellularity was detected for smears compared with LBC samples (54% vs 31.4%). The test success rate for DNA amplification also was higher in smears.29
Eleven studies reported the percentage of tumor cells9, 10, 12, 13, 17, 22, 23, 24, 25, 28, 30 (Fig. 2). The majority of those studies defined a minimum for the percentage of tumor cells ranging from at least 1% to at least 50%.
Data on DNA Quantity/Final DNA Yield, DNA Quality, and DNA Concentration
Only 5 studies (22.7%) included information on DNA quantity/final DNA yield. In 2 of those studies, information was provided as the average DNA yield from the samples: One study used cell blocks, fresh cells, smears, and LBC samples and reported an average DNA yield of 24.6 ng/μL.12 In the other study, information was provided on the average DNA yield for each type of preparation used (smears, 60.94 ng/μL; LBC samples, 23.07 ng/μL).29 One study that used LBC samples reported that the DNA concentration ranged from 10.0 to 935.3 ng/μL.26 The other article that contained information on DNA quantity did not provide a range or a median but stated that the samples used for molecular analysis with polymerase chain reaction (PCR) followed by direct sequencing had at least 20 ng/μL of DNA.17 Finally, the fifth study only reported the DNA yield for failed tests (range, 2.3‐5.6 ng/μL).10
Number/Percentage of Successful Assays and Percentage of Mutated Samples
Information on the percentage of successfully amplified samples was available for all 22 articles. In 2 studies, however, information was reported for all types of samples, including histologic specimens.10, 25 In 1 study,18 the percentage of successfully amplified samples was reported for FNAs only (64%), although that study also used fluid cell blocks. However, the report indicates that the overall success rate for all samples was 85% and that FNAs were significantly more likely to result in failed assays than tissue biopsies or fluid cell blocks. For the other 19 articles listed in Table 1, the success rate for molecular assays using cytologic specimens ranged from 85.2% to 100%, and the total average success rate was 95.87% (Table 2).
The percentage of samples harboring EGFR mutations also was available for all articles listed in Table 2. In 3 of those studies, percentages were reported for all samples, including histologic specimens,17, 21, 25 and ranged from 4.2% to 51.4%. Another study provided information separately according to the type of specimen (FNAs, 25.6%; fluids, 15.9%).18 When we analyzed the remaining 18 articles for which we had information exclusively for cytologic specimens, the percentage of cytologic samples that contained EGFR mutations ranged from 6% to 50.46%, with an average of 20% (Table 2). The highest rejection rates in a large study were reported for sputum (50%) and bronchial washings/brushings (24.13%) followed by pleural effusions (20%), EBUS‐TBNA samples (19.3%), and FNAs (14.2%).14
Techniques Used for Mutation Analysis
Various PCR‐based techniques were used for EGFR mutation analysis, and this information was available for all studies. PCR followed by direct sequencing was the method of choice in 7 studies (31.8%).9, 11, 12, 13, 17, 18, 19 Real‐time PCR was used as the main method in 2 studies (9.1%): 1 study26 used real‐time PCR combining amplification‐refractory mutation system (ARMS) and a bi‐loop probe protocol, with some mutated samples confirmed by direct sequencing; and the other study also used real‐time PCR and ARMS, this time combined with scorpion chemistry.28 Fragment‐length assays and restriction fragment analyses were used in 4 studies (18.2%) to identify mutations in exon 21 and deletions in exon 1914, 22, 29, 30; and, in 1 of those studies, aberrant results were further processed and confirmed by direct sequencing.14
Other high‐sensitivity, PCR‐based methods, such as high‐resolution melting analysis (HRMA), ARMS, peptide nucleic acid‐locked nucleic acid, and coamplification at lower denaturation temperature (COLD)‐PCR, were the main methods used in 4 studies,10, 23, 24, 25 two studies,16, 21 two studies,15, 27 and 1 study,20 respectively. Three of the studies that used HRMA had confirmation of mutated or abnormal samples performed by Sanger sequencing,23, 24, 25 and the other study combined HRMA with mutant‐enriched PCR and sequencing.10 One study that used ARMS had some samples from an institution involved in the study alternatively investigated by MassARRAY spectrometry,16 and the study that used COLD‐PCR also had the samples evaluated by conventional PCR and demonstrated higher final sensitivity for COLD‐PCR when the 2 methods were compared (5%‐10% vs 30%).20
DISCUSSION
In this review, the data retrieved from 22 published articles have confirmed that different types of cytologic samples and different cytologic preparations have been used for EGFR mutation analysis with high rates of success. Although current guidelines recommend that priority should be given to histologic samples,5 the current evidence indicates that, overall, cytologic samples provide similar/comparable results.9, 14, 22 In fact, for some specific specimen types, the detection rate achieved in cytologic specimens is superior to that achieved in histologic samples.
Regarding sampling methods, 1 of the largest series demonstrated that, for mediastinal lymph nodes, cytologic samples obtained by EBUS‐TBNA produced test success rates similar to those of histologic samples obtained by mediastinoscopy but had a considerably higher mutation detection rate (34.4% vs 13.4%).22 Differences in mutation detection between cytologic and histologic specimens also were observed for distant lymph nodes (36% vs 21.3%, respectively) and for pleural fluids and pleural core‐needle biopsies (31.1% vs 21.2%, respectively).22 Those findings are supported by various other series demonstrating that the lowest insufficient rates were observed in body fluids and EBUS samples9, 14 and that the highest detection rates among cytologic samples were observed in pleural effusions and other body fluids.13, 28 The worst results have been reported from bronchial washings/brushings and sputum.14, 28 In another study, a greater proportion of cytologic samples were from metastatic lesions (60.1% cytologic samples compared with 29.6% noncytologic samples), reflecting the advantage of cytology sampling modalities in targeting lesions, including deep‐seated tumors under image guidance and with minimally invasive techniques.11 A high concordance in EGFR mutation status between multiple lesions (primary and metastatic) from the same patient has been reported in studies using cytologic and histologic samples.32, 33, 34 No differences were observed in response to TKI inhibitor treatment among patients who had EGFR mutations when the types of specimens used for molecular analysis (high‐cellularity histologic specimens or low‐cellularity cytologic specimens) were compared in a large clinical trial.35
Material from a tumor specimen sufficient to perform mutation testing was obtained in a high percentage of specimens (range, 90%‐99.3%) in series that dealt only with EBUS‐TBNA and endoscopic ultrasound samples.15, 16, 20, 24 EBUS‐TBNA samples also had failure rates similar to those reported in surgical biopsies.36 Cytologic specimens obtained by brushing using EBUS with a guide sheath under direct vision and ultrathin bronchoscopy have also been used successfully for analysis.27 It has been speculated that FNA may preferentially obtain less cohesive tumor cells or remove PCR inhibitors in the cellular milieu and that DNA fragmentation caused by longer formalin fixation for histologic specimens may contribute to the lower mutation rate.22 In addition, it has been hypothesized that the lack of formalin fixation may account for the complete absence of unsatisfactory test outcomes reported in FNAs or effusion direct smears that were tested for EGFR mutations.17 Another study that compared the use of cytologic specimens, bronchial biopsies, and surgical specimens demonstrated a significantly higher EGFR mutation rate when cytologic or surgical specimens were used compared with bronchial biopsies.37
It is noteworthy that, in contrast to the data described above, other series that have examined cytologic and histologic samples have produced different results. In a large series with samples from several different referring centers that used direct sequencing, which always requires substantial numbers and percentages of tumor cells because of low analytic sensitivity, FFPE cell blocks from fine‐needle biopsies were more likely to result in incomplete sequencing than either tissue biopsies or fluid cell blocks. In that study, the estimated failure rate for FNA was 36%. Among 24 FNA samples with a stated reason for test failure, 3 had no tissue in the sample, 11 had no tumor tissue, 7 had an amplification failure, and 3 could only be partially sequenced.18 For that specific study, no minimum cellularity or percentage of tumor cells was reported. A small study that compared lung core‐needle biopsy with FNA concluded that core‐needle biopsy provided more specimens that were sufficient for molecular testing (EGFR, Kirsten rat sarcoma viral oncogene homolog [KRAS], and ALK) than FNA.38 Similarly, in a different study, the presence of approximately <20% tumor cells in the sample occurred more frequently in cytologic samples (n = 14; 8.3%) compared with noncytologic samples (n = 3; 2.1%).9
Different fixatives have been used, and some studies have reported failure rates. No significant difference was reported in the test success rate between alcohol‐fixed versus formalin‐fixed cell blocks.22 EGFR mutation rates were consistent among different specimens collected in CytoLyt, and a comparison using paired LBC specimens (n =101) with corresponding histologic specimens demonstrated that FNA exhibited 100% concordance, whereas EGFR mutation status was discordant in 8 specimens, including 3 pleural effusions, 4 EBUS‐TBNA samples, and 1 bronchial brushing sample.26 One large series that used histologic and cytologic specimens demonstrated no difference in the rate of EGFR mutation detection between surgical and nonsurgical specimens, and none of the fixation methods used impaired the analysis.34 In contrast, formalin‐fixed samples and the only fresh sample described in 1 study were all amplified fully, whereas EGFR mutation analysis was not possible in 33.3% of the Duboscq‐Brasil−fixed samples or in 12.7% of the ethanol‐fixed samples. All LBC samples were suitable for analysis, and manual scraping from spray‐fixed specimens on glass slides allowed the collection of sufficient numbers of cells in 8 of 11 samples (72.7%).28 A comparison study concluded that, although reliable clinical genotyping could be performed using a variety of fixatives, spray and ethanol fixation resulted in both higher yield and higher DNA quality compared with air drying. In LBC methods, CytoLyt produced a 5‐fold higher yield than CytoRich Red, and Papanicolaou stain produced twice the yield of hematoxylin and eosin stain in all wet‐fixed material.39 Needle rinse fixed in a formalin‐free fixative (FineFix; Leica Biosystems, Nussloch, Germany) has also been used successfully.40
Cell blocks are as good a source of tumor cells for mutation testing as histologic samples, regardless of the fixative used (alcohol or formalin), necrosis, or specimen type.24 Other smaller studies have presented equivalent data.17, 30 The 2 major cytology preparations, smears and cell blocks, are equally suitable for molecular testing9, 12, 17, 19, 25; and, although the data are limited, cell blocks and smears apparently perform similarly in terms of the test failure rate. Reported failure rates by preparation type were available for 4 of the 9 studies that used smears. Two studies described similar failure rates for smears and cell blocks,9, 17 and a third study reported a higher rate of unamplified samples for smears.28 The fourth, a comparison study, demonstrated that smears provided higher DNA yield and more frequently were cell‐rich than LBC slides.29 However, differences in adequacy and in the mutation rate between the 2 sample types did not reach statistical significance. Approximately 5% of failed PCR reactions in DNA extracted from fresh cells, compared with 25% of DNA extracted from FFPE sections, was observed in a study that used FNA samples.23 In a different study, 2 samples that could not be analyzed because of PCR failure were poorly preserved specimens that had been submitted by outside institutions.30 Furthermore, a series that included specimens from different countries that used various methods of acquisition, preparation, and processing of tumor material because of different routine clinical practices demonstrated that the quality of most cytologic samples was such that mutation testing was successful, and definite results were obtained.21 A high frequency of sequence alterations has been reported because of formalin fixation of archival specimens.31
Although the vast majority of studies did not provide cellularity assessments and/or tumor cell percentages, low cellularity and low tumor cell percentages were associated with higher test failure rates for the studies that reported those parameters.9, 10, 20, 28 Although it was modest, a significant correlation between the estimated tumor cellularity and the allele frequency has been demonstrated.24 One study demonstrated that samples with sparse cellularity (<300 tumor cells) had a higher PCR failure rate than samples with normal cellularity (>1000 tumor cells).9 The only sample that failed DNA amplification in an EBUS‐TBNA series contained 100 cells per section.20 To evaluate the minimal number of cells required for successful EGFR DNA sequence analysis, 1 study compared the quality of DNA chromatograms. Although the best result was obtained with 100 cells, the differences in evaluability between the different cell counts (30, 50, and 100 cancer cells) were not statistically significant.41 In another study, among 16 unamplified samples, 12 had <200 tumor cells, and 9 of those had <50% tumor cells.28 For a large series, a low tumor cell percentage (range, 1%‐20%) was the most common cause of failure in 24 of the 42 tests that failed (57%).10 In a study that used 20% as the minimum percentage of tumor cells, the following reasons were listed for the rejection of cytologic specimens for testing: insufficient material for macrodissection, insufficient tumor cellularity for analytic sensitivity, and no tumor cells observed.17 Low copy number DNA template input (<50 tumor cells) in PCR can generate false mutations, mainly guanine to adenine transitions, in a sequence‐dependent manner.42 The 2 largest cytology series have recommended that, because it has been documented that even suboptimal samples can produce positive mutation results, all samples should be eligible for mutation testing, regardless of tumor amount.10, 22 However, suboptimal samples with a negative EGFR mutation result should be considered for repeat testing with an alternate sample.22, 43
In a study that reported on DNA quality, expressed according to the length of base pairs (bp) in PCR products, the samples were classified into 2 subgroups, DNA with 100 to 300 bp and DNA with 300 to 400 bp, using mean sample ages of 784 days and 354 days, respectively. All testing failures had short DNA fragments (100‐200 bp).24 The age and storage conditions of tissue blocks have also been implicated in testing failure as a result of DNA oxidation and fragmentation, with higher rates reported in nonamplificable samples that were more than 5 years old.44 Unfortunately, DNA yield/quantity was only available for a few articles, and only 1 study reported the average yield for each sample type.29 Novel advanced techniques that require high‐quality DNA for reliable results, such as next‐generation sequencing, have been described as successful for use on lung FNA specimens.45, 46 Although cell block sections yielded significantly higher DNA quantity than air‐dried and ethanol‐fixed smears, no significant difference was observed in next‐generation sequencing results for the 3 types of preparations tested.46 In the other study, only cell blocks were used, and genomic profiles also were successfully generated from all pulmonary FNAs.45
Guidelines on reporting preanalytic conditions of biospecimens in a thorough, accurate, and standardized manner have been published to improve the quality of research that uses human tissue.8 Recently, a proposed checklist of 30 items that should serve as a useful guide to investigators preparing proposals for studies that involve the use of omics‐based tests had 4 criteria related to specimen issues. The specimen issues included the following criteria (all are preanalytic parameters): methods for specimen collection, processing and storage conditions, the minimum amount of specimen required, and the quantity and quality of isolated cells or analytes needed for successful assay performance.47 In addition, a cross‐validation study of EGFR and KRAS mutation detection among 15 molecular laboratories in France has concluded that the accuracy of results depends more on sample quality than on differences in molecular sequencing procedures, and the findings have emphasized the need for preanalytic quality‐control programs.44
This review documents an increased number of large series reporting EGFR mutation analysis of cytologic specimens in the last 3 years and confirms the feasibility of testing using a variety of specimen types and preparations, some of which are superior in quality and have a higher mutation‐detection rate compared with histologic samples. All cytologic preparations have produced similar results. On the basis of currently available data, we suggest avoiding prioritizing FFPE cell blocks and histologic specimens, at least for some specific locations, such as pleural space and mediastinal lymph nodes. Current guidelines and expert recommendations should be revised to incorporate new evidence generated from large published studies and should take into consideration the versatility and variety of cytologic specimens, focusing on their respective differences in handling and processing.
FUNDING SUPPORT
No specific funding was disclosed.
CONFLICT OF INTEREST DISCLOSURES
The authors made no disclosures.
REFERENCES
- 1. Morgensztern D, Campo MJ, Dahlberg SE, et al. Molecularly targeted therapies in non‐small‐cell lung cancer annual update 2014. J Thorac Oncol. 2015;10:S1–S63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Basik M, Aguilar‐Mahecha A, Rousseau C, et al. Biopsies: next‐generation biospecimens for tailoring therapy. Nat Rev Clin Oncol. 2013;10:437–450. [DOI] [PubMed] [Google Scholar]
- 3. Lim C, Tsao MS, Le LW, et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall‐cell lung cancer. Ann Oncol. 2015;26:1415–1421. [DOI] [PubMed] [Google Scholar]
- 4. Garady C, Saieg MA, Ko HM, Geddie WR, Boerner SL, da Cunha Santos G. Epstein‐Barr virus encoded RNA detected by in situ hybridization using cytological preparations. Cytopathology. 2014;25:101–107. [DOI] [PubMed] [Google Scholar]
- 5. Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Mol Diagn. 2013;15:415–453. [DOI] [PubMed] [Google Scholar]
- 6. Kerr KM, Bubendorf L, Edelman MJ, et al. Second ESMO consensus conference on lung cancer: pathology and molecular biomarkers for non‐small‐cell lung cancer. Ann Oncol. 2014;25:1681–1690. [DOI] [PubMed] [Google Scholar]
- 7. da Cunha Santos G, Saieg MA, Geddie W, Leighl N. EGFR gene status in cytological samples of nonsmall cell lung carcinoma: controversies and opportunities. Cancer Cytopathol. 2011;119:80–91. [DOI] [PubMed] [Google Scholar]
- 8. Moore HM, Kelly AB, Jewell SD, et al. Biospecimen reporting for improved study quality (BRISQ). Cancer Cytopathol. 2011;119:92–101. [DOI] [PubMed] [Google Scholar]
- 9. Billah S, Stewart J, Staerkel G, Chen S, Gong Y, Guo M. EGFR and KRAS mutations in lung carcinoma: molecular testing by using cytology specimens. Cancer Cytopathol. 2011;119:111–117. [DOI] [PubMed] [Google Scholar]
- 10. Hlinkova K, Babal P, Berzinec P, et al. Evaluation of 2‐year experience with EGFR mutation analysis of small diagnostic samples. Diagn Mol Pathol. 2013;22:70–75. [DOI] [PubMed] [Google Scholar]
- 11. Leslie C, Giardina T, Carrello A, Spagnolo DV, Amanuel B. Detection of EGFR mutational profile by direct dideoxy sequencing in cytology and non‐cytology biopsy samples. Pathology. 2014;46:283–288. [DOI] [PubMed] [Google Scholar]
- 12. Lozano MD, Zulueta JJ, Echeveste JI, et al. Assessment of epidermal growth factor receptor and K‐ras mutation status in cytological stained smears of non‐small cell lung cancer patients: correlation with clinical outcomes. Oncologist. 2011;16:877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma ES, Ng WK, Wong CL. EGFR gene mutation study in cytology specimens. Acta Cytol. 2012;56:661–668. [DOI] [PubMed] [Google Scholar]
- 14. Malapelle U, Bellevicine C, De Luca C, et al. EGFR mutations detected on cytology samples by a centralized laboratory reliably predict response to gefitinib in non‐small cell lung carcinoma patients. Cancer Cytopathol. 2013;121:552–560. [DOI] [PubMed] [Google Scholar]
- 15. Nakajima T, Yasufuku K, Nakagawara A, Kimura H, Yoshino I. Multigene mutation analysis of metastatic lymph nodes in non‐small cell lung cancer diagnosed by endobronchial ultrasound‐guided transbronchial needle aspiration. Chest. 2011;140:1319–1324. [DOI] [PubMed] [Google Scholar]
- 16. Navani N, Brown JM, Nankivell M, et al. Suitability of endobronchial ultrasound‐guided transbronchial needle aspiration specimens for subtyping and genotyping of non‐small cell lung cancer: a multicenter study of 774 patients. Am J Respir Crit Care Med. 2012;185:1316–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pang B, Dettmer M, Ong CW, et al. The positive impact of cytological specimens for EGFR mutation testing in non‐small cell lung cancer: a single South East Asian laboratory's analysis of 670 cases. Cytopathology. 2012;23:229–236. [DOI] [PubMed] [Google Scholar]
- 18. Peters MJ, Bowden JJ, Carpenter P, Lewis J, Solomon B. Outcomes of an Australian testing programme for epidermal growth factor receptor mutations in non‐small cell lung cancer. Intern Med J. 2014;44:575–580. [DOI] [PubMed] [Google Scholar]
- 19. Proietti A, Ali G, Pelliccioni S, et al. Anaplastic lymphoma kinase gene rearrangements in cytological samples of non‐small cell lung cancer: comparison with histological assessment. Cancer Cytopathol. 2014;122:445–453. [DOI] [PubMed] [Google Scholar]
- 20. Santis G, Angell R, Nickless G, et al. Screening for EGFR and KRAS mutations in endobronchial ultrasound derived transbronchial needle aspirates in non‐small cell lung cancer using COLD‐PCR. PLoS One. 2011;6:e25191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shi Y, Au JS, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non‐small‐cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol. 2014;9:154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shiau CJ, Babwah JP, da Cunha Santos G, et al. Sample features associated with success rates in population‐based EGFR mutation testing. J Thorac Oncol. 2014;9:947–956. [DOI] [PubMed] [Google Scholar]
- 23. Stella GM, Scabini R, Inghilleri S, et al. EGFR and KRAS mutational profiling in fresh non‐small cell lung cancer (NSCLC) cells. J Cancer Res Clin Oncol. 2013;139:1327–1335. [DOI] [PubMed] [Google Scholar]
- 24. Stigt JA, tHart NA, Knol AJ, Uil SM, Groen HJ. Pyrosequencing analysis of EGFR and KRAS mutations in EUS and EBUS‐derived cytologic samples of adenocarcinomas of the lung. J Thorac Oncol. 2013;8:1012–1018. [DOI] [PubMed] [Google Scholar]
- 25. Takano T, Ohe Y, Tsuta K, et al. Epidermal growth factor receptor mutation detection using high‐resolution melting analysis predicts outcomes in patients with advanced nonsmall cell lung cancer treated with gefitinib. Clin Cancer Res. 2007;13:5385–5390. [DOI] [PubMed] [Google Scholar]
- 26. Wu CY, Hou LK, Ren SX, Su B, Chen G. High feasibility of liquid‐based cytological samples for detection of EGFR mutations in Chinese patients with NSCLC. Asian Pac J Cancer Prev. 2014;15:7885–7889. [DOI] [PubMed] [Google Scholar]
- 27. Yamada N, Oizumi S, Asahina H, et al. The peptide nucleic acid‐locked nucleic acid polymerase chain reaction clamp‐based test for epidermal growth factor receptor mutations in bronchoscopic cytological specimens of non‐small cell lung cancer. Oncology. 2012;82:341–346. [DOI] [PubMed] [Google Scholar]
- 28. Allegrini S, Antona J, Mezzapelle R, et al. Epidermal growth factor receptor gene analysis with a highly sensitive molecular assay in routine cytologic specimens of lung adenocarcinoma. Am J Clin Pathol. 2012;138:377–381. [DOI] [PubMed] [Google Scholar]
- 29. Bellevicine C, Malapelle U, Vigliar E, de Luca C, Troncone G. Epidermal growth factor receptor test performed on liquid‐based cytology lung samples: experience of an academic referral center. Acta Cytol. 2014;58:589–594. [DOI] [PubMed] [Google Scholar]
- 30. Rekhtman N, Brandt SM, Sigel CS, et al. Suitability of thoracic cytology for new therapeutic paradigms in non‐small cell lung carcinoma: high accuracy of tumor subtyping and feasibility of EGFR and KRAS molecular testing. J Thorac Oncol. 2011;6:451–458. [DOI] [PubMed] [Google Scholar]
- 31. Williams C, Ponten F, Moberg C, et al. A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am J Pathol. 1999;155:1467–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khode R, Larsen DA, Culbreath BC, et al. Comparative study of epidermal growth factor receptor mutation analysis on cytology smears and surgical pathology specimens from primary and metastatic lung carcinomas. Cancer Cytopathol. 2013;121:361–369. [DOI] [PubMed] [Google Scholar]
- 33. Mansuet‐Lupo A, Zouiti F, Alifano M, et al. Intratumoral distribution of EGFR mutations and copy number in metastatic lung cancer, what impact on the initial molecular diagnosis? J Transl Med. 2014;12:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Righi L, Cuccurullo A, Vatrano S, et al. Detection and characterization of classical and “uncommon” exon 19 epidermal growth factor receptor mutations in lung cancer by pyrosequencing. BMC Cancer. 2013;13:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang JC, Wu YL, Chan V, et al. Epidermal growth factor receptor mutation analysis in previously unanalyzed histology samples and cytology samples from the phase III Iressa Pan‐ASia Study (IPASS). Lung Cancer. 2014;83:174–181. [DOI] [PubMed] [Google Scholar]
- 36. Folch E, Yamaguchi N, VanderLaan PA, et al. Adequacy of lymph node transbronchial needle aspirates using convex probe endobronchial ultrasound for multiple tumor genotyping techniques in non‐small‐cell lung cancer. J Thorac Oncol. 2013;8:1438–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yi S, Zhuang Y, Zhou J, et al. A comparison of epidermal growth factor receptor mutation testing methods in different tissue types in non‐small cell lung cancer. Int J Mol Med. 2014;34:464–474. [DOI] [PubMed] [Google Scholar]
- 38. Schneider F, Smith MA, Lane MC, Pantanowitz L, Dacic S, Ohori NP. Adequacy of core needle biopsy specimens and fine‐needle aspirates for molecular testing of lung adenocarcinomas. Am J Clin Pathol. 2015;143:193–200; quiz 306. [DOI] [PubMed] [Google Scholar]
- 39. Dejmek A, Zendehrokh N, Tomaszewska M, Edsjo A. Preparation of DNA from cytological material: effects of fixation, staining, and mounting medium on DNA yield and quality. Cancer Cytopathol. 2013;121:344–353. [DOI] [PubMed] [Google Scholar]
- 40. Fassina A, Gazziero A, Zardo D, Corradin M, Aldighieri E, Rossi GP. Detection of EGFR and KRAS mutations on trans‐thoracic needle aspiration of lung nodules by high resolution melting analysis. J Clin Pathol. 2009;62:1096–1102. [DOI] [PubMed] [Google Scholar]
- 41. Savic S, Tapia C, Grilli B, et al. Comprehensive epidermal growth factor receptor gene analysis from cytological specimens of non‐small‐cell lung cancers. Br J Cancer. 2008;98:154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Akbari M, Hansen MD, Halgunset J, Skorpen F, Krokan HE. Low copy number DNA template can render polymerase chain reaction error prone in a sequence‐dependent manner. J Mol Diagn. 2005;7:36–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rafael OC, Aziz M, Raftopoulos H, Vele OE, Xu W, Sugrue C. Molecular testing in lung cancer: fine‐needle aspiration specimen adequacy and test prioritization prior to the CAP/IASLC/AMP Molecular Testing Guideline publication. Cancer Cytopathol. 2014;122:454–458. [DOI] [PubMed] [Google Scholar]
- 44. Beau‐Faller M, Degeorges A, Rolland E, et al. Cross‐validation study for epidermal growth factor receptor and KRAS mutation detection in 74 blinded non‐small cell lung carcinoma samples: a total of 5550 exons sequenced by 15 molecular French laboratories (evaluation of the EGFR mutation status for the administration of EGFR‐TKIs in non‐small cell lung carcinoma [ERMETIC] project—part 1). J Thorac Oncol. 2011;6:1006–1015. [DOI] [PubMed] [Google Scholar]
- 45. Young G, Wang K, He J, et al. Clinical next‐generation sequencing successfully applied to fine‐needle aspirations of pulmonary and pancreatic neoplasms. Cancer Cytopathol. 2013;121:688–694. [DOI] [PubMed] [Google Scholar]
- 46. Karnes HE, Duncavage EJ, Bernadt CT. Targeted next‐generation sequencing using fine‐needle aspirates from adenocarcinomas of the lung. Cancer Cytopathol. 2014;122:104–113. [DOI] [PubMed] [Google Scholar]
- 47. McShane LM, Cavenagh MM, Lively TG, et al. Criteria for the use of omics‐based predictors in clinical trials. Nature. 2013;502:317–320. [DOI] [PMC free article] [PubMed] [Google Scholar]


