Abstract
We have previously shown that physio/pathological levels of hydrogen peroxide (H2O2) stimulate translation from the hepatitis C virus (HCV) internal ribosome entry site (IRES) element in tissue-cultured cells. Here, using in vitro translation, we further show that H2O2 upregulates HCV IRES-dependent mRNA translation and correlates with an increase in intracellular oxidant level. Using Western blotting, immunocytochemistry, microscopy and affinity pulldown, we show that H2O2 stimulates HCV IRES-dependent translation and correlates with nuclear–cytoplasmic shuttling of the La autoantigen, resulting in enhanced binding of cytoplasmic La to HCV IRES RNA. The role of the La protein in H2O2-stimulated IRES-dependent translation is further confirmed by the ability of an anti-La antibody to suppress H2O2-activated IRES-dependent translation in vitro. This is further supported by the ability of an ectopically expressed dominant, negative La mutant protein to suppress H2O2-inducible IRES-mediated translation in Huh7 cells, transiently transfected with a bicistronic reporter and in a sub-genomic replicon cell line resembling a persistent infection. On the other hand, translation from the encephalomyocarditis virus IRES is diminished in the presence of H2O2, suggesting that H2O2 translational responsiveness is a specific property of the HCV IRES and is not a general phenomenon for all viral IRESs. Altogether, these results suggest that HCV adapts to physio/pathological oxidative stress in the host cell by mediating La cytoplasmic shuttling to enhance its IRES-dependent translation.
Keywords: hepatitis C virus, IRES, translation, hydrogen peroxide, oxidative stress, La
Introduction
The hepatitis C virus (HCV) is a single-stranded, positive-sense RNA virus belonging to the genus Hepacivirus of the family Flaviviridae (Chan, 2014). Its 9.6 kb genome encodes a single polypeptide, which is cleaved by the host and viral proteases into structural proteins core, envelopes E1 and E2, and non-structural proteins p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B. HCV poses a major threat to human health, causing chronic hepatitis, which can then progress into cirrhosis and/or hepatocellular carcinoma. There is now accumulating evidence that oxidative stress may be responsible for the pathogenesis of viral hepatitis and other forms of liver diseases (Paracha et al., 2013). Oxidative stress is a prominent clinical feature associated with HCV infection. Hepatitis C patients frequently present elevated pro-oxidant and reduced anti-oxidant levels in the blood and liver, with iron overload, increased lipid peroxidation and oxidative DNA damage and decreased hepatic glutathione (Fujita et al., 2008; Konishi et al., 2006; Paradis et al., 1997; Venturini et al., 2010). Proteomic and microarray analysis of liver biopsies revealed increased oxidative stress in hepatitis C samples (Diamond et al., 2007; Yamashita et al., 2001). Little is known about how the virus can survive in such a highly oxidative environment. Our previous demonstration that HCV translation was upregulated by physio/pathological doses of hydrogen peroxide (H2O2) in hepatocytes suggests a translational advantage of the virus in an oxidative environment, enabling the virus to survive and establish chronicity (MacCallum et al., 2006).
Translation from the HCV genome is mediated by an internal ribosome entry site (IRES) element (Tsukiyama-Kohara et al., 1992). The IRES element mediates an alternative form of translation distinctive from that of cap-dependent translation of the vast majority of cellular genes, allowing selective translation of genes under conditions when global protein synthesis is compromised (Jackson, 2005). Cap-dependent translation is regulated by a canonical set of eukaryotic initiation factors (eIFs). In contrast, translation from the IRES element requires only a limited number of eIFs. In addition, IRES-dependent translation is regulated by unique sets of IRES trans-acting factors (ITAFs).
A number of putative ITAFs have been identified for the HCV IRES element, including the autoantigen La (Costa-Mattioli et al., 2004). La is pivotal in mediating translation from a number of IRESs (Costa-Mattioli et al., 2004; Holcik & Korneluk, 2000; Petz et al., 2012). La plays a critical role in HCV IRES-dependent translation initiation, by binding to and altering the conformation of the IRES element to orchestrate assembly of the ribosomal complex (Pudi et al., 2004).
ITAF modification is an important aspect in the regulation of IRES activity under stress conditions, using mechanisms such as nuclear–cytoplasmic shuttling, protein cleavage, phosphorylation and increased protein expression (Brenet et al., 2009; Lewis & Holcik, 2008; Sella et al., 1999; Shiroki et al., 1999). La modification has also been implicated in playing a regulating role in IRES-dependent translation initiation under various stress conditions (Petz et al., 2012; Shiroki et al., 1999; Zhang et al., 2012). In this study, we therefore investigated the role of La and the mechanism by which La upregulates HCV IRES-dependent translation under oxidative stress conditions.
Results
H2O2 induces host-factor change to stimulate IRES-dependent translation
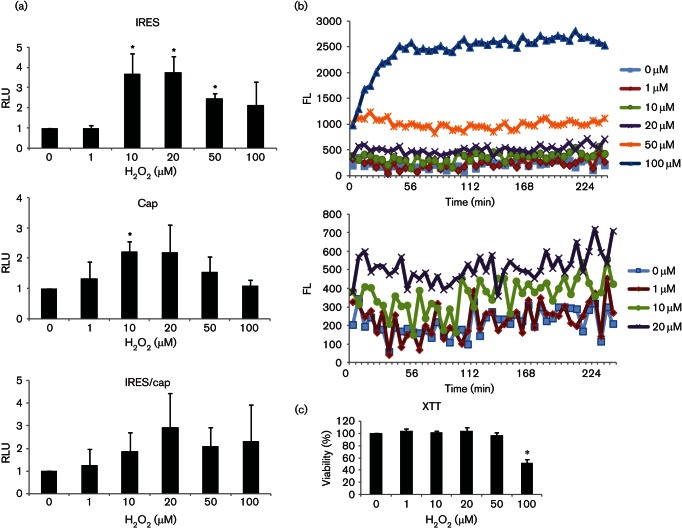
Using Huh7 cells transiently transfected with the bicistronic reporter pRL1b mRNA in which translation of the Renilla luciferase and firefly luciferase is driven by cap-dependent and HCV IRES-dependent translation, respectively, we have previously shown that H2O2 stimulates translation from the HCV IRES in tissue-cultured cells (MacCallum et al., 2006). This translational upregulation could be brought about by H2O2-induced change in the host factor and/or in the RNA template. In order to determine the role of host factor in H2O2-activated IRES-dependent translation, we performed an in vitro study using cytosolic extracts harvested from H2O2-treated Huh7 cells to prime in vitro translation programmed with the same bicistronic RNA template containing the HCV IRES. Huh7 cells were treated with 0 µM, 1 µM, 10 µM, 20 µM, 50 µM and 100 µM of H2O2 for 1 h. Cytoplasmic S10 fractions extracted from 10 µM, 20 µM, 50 µM and 100 µM H2O2-treated cells were able to prime and enhance in vitro translation from the HCV IRES over that of the untreated control, suggesting that host-factor change in response to H2O2 is responsible for H2O2-activated IRES-dependent translation (Fig. 1a). A low level of exogenous H2O2 (1 µM), which did not result in any increase in intracellular oxidants, also did not stimulate IRES-dependent translation, confirming that elevated intracellular oxidant level is essential to stimulate IRES-dependent translation (Fig. 1a, b). Furthermore, a slight elevation in intracellular oxidant level, as induced by 10 µM of H2O2, was sufficient to stimulate IRES-dependent translation. However, higher intracellular oxidant levels, as induced by 50 µM and 100 µM of H2O2, stimulated IRES-dependent translation to a lesser extent than lower intracellular oxidant levels of H2O2, as induced by 10 µM and 20 µM of H2O2, due to the cytotoxic effect of higher doses of H2O2 (Fig. 1a, c). Note that although the XTT viability test did not detect significant cell death at 50 µM of H2O2, a low degree of apoptosis was always visible at this concentration, similar to what we have observed before (MacCallum et al., 2006). A concentration of 100 µM of H2O2 caused significant cell death (41–58 %), resulting in significant variation in translational activity. This might explain the insignificant increase of IRES-dependent translation in Fig. 1, but significant increase of IRES-dependent translation in Fig. 6. Despite this variation, this concentration of H2O2 consistently enhanced IRES-dependent translation. In contrast, H2O2 also stimulated cap-dependent translation, but the results were not consistent and, hence, insignificant in most cases (Figs 1 and 6). This inconsistency in cap-dependent translation gave rise to insignificant increase in the IRES/cap ratio in Fig. 1, but significant increase in the IRES/cap ratio in Fig. 6. Importantly, the increase in the IRES/cap ratio was consistent in cells treated with 10–100 µM of H2O2, confirming differential upregulation of IRES-dependent translation by physio/pathological levels of H2O2 in vitro.
Fig. 1.
H2O2 stimulates translation in vitro. (a) Cytosolic S10 fractions extracted from Huh7 cells treated with doses of H2O2, as indicated, for 1 h, were used to prime in vitro translation programmed with the bicistronic pRL1b reporter transcript. The HCV IRES and cap-translational activities were measured by firefly and Renilla luciferase activities, respectively, and expressed relative to the untreated controls, which are set as 1. The IRES/cap ratio is represented by the ratio of firefly-to-Renilla luciferase activities and is expressed relative to the untreated control, which is set as 1. The values obtained represent the mean±sem of three independent experiments, performed in duplicates. RLU, Relative luciferase units. (b) A representation of three independent dichlorofluorescin fluorometric assays, performed in quadruplicates, showing the kinetics of reactive oxygen species (ROS) generation in Huh7 cells (19 000 per well/96-well plate) after treatment with doses of H2O2, as indicated. The bottom part of the graph is enlarged and depicted below to show ROS generation in the lower range of H2O2. FL, Fluorescence units. (c) XTT assay showing viability of Huh7 cells (19 000 per well/96-well plate) after treatment with doses of H2O2, as indicated, for 24 h. The values obtained represent the mean±sem of three independent experiments, performed in quadruplicates, and are expressed relative to the untreated control, which is set as 100 %. Significance of the difference *P<0.05.
Fig. 6.
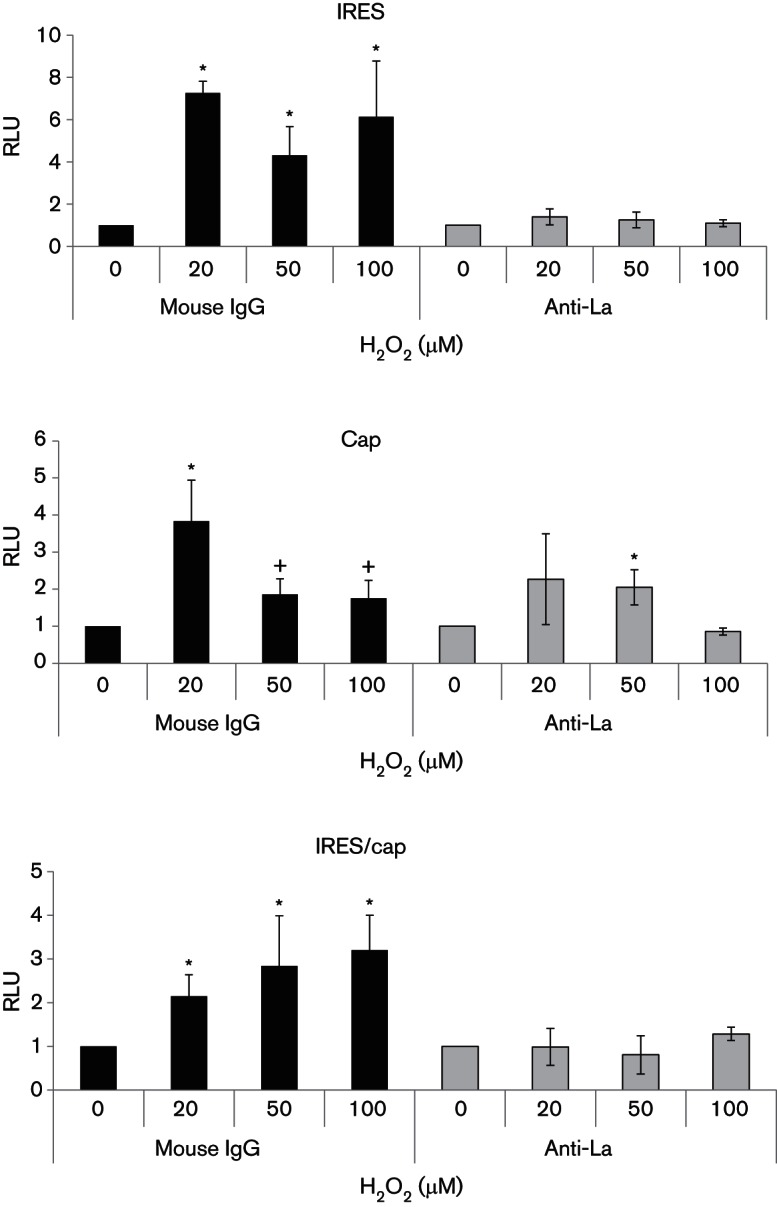
La antibody inhibits H2O2-activated translation in vitro. Cytosolic S10 fractions extracted from Huh7 cells treated with doses of H2O2, as indicated, for 1 h were incubated with 1 µl of the anti-La antibody SW5, or the control mouse IgG for 1 h on ice before being used to prime in vitro translation programmed with the bicistronic pRL1b reporter transcript. The HCV IRES and cap-translational activities were measured by firefly and Renilla luciferase activities and expressed relative to their respective 0 µM H2O2 controls, which are set as 1. The IRES/cap ratio is represented by the ratio of firefly-to-Renilla luciferase activities and is expressed relative to their respective 0 µM H2O2 control, which is set as 1. The values obtained represent the mean±sem of three independent experiments, performed in duplicates. Significance of the difference, *(P<0.05), +(P<0.1). RLU, Relative luciferase units.
H2O2 upregulates IRES-dependent translation and correlates with increased cytosolic La level
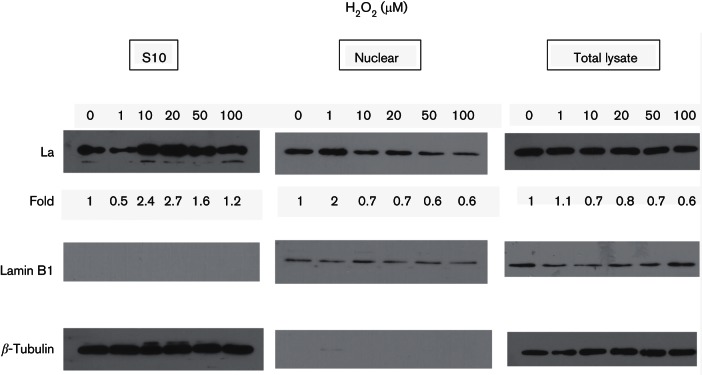
La is an important ITAF for many IRESs (Costa-Mattioli et al., 2004; Holcik & Korneluk, 2000; Petz et al., 2012). La undergoes post-translational changes in response to oxidative stress and is responsible for the oxidative stimulation of translation from the cellular IRES for Nrf2 (Zhang et al., 2012). As La is a key ITAF for HCV IRES-dependent translation (Costa-Mattioli et al., 2004), we therefore sought to examine whether H2O2-induced change in the La protein could account for the stimulation of HCV IRES-dependent translation. Cytoplasmic shuttling of ITAF is a rapid means of regulation of IRES-dependent translation under stress conditions (Lewis & Holcik, 2008); we therefore measured the amount of La protein in cytosolic extracts in cells exposed to H2O2 using Western blotting. Cytosolic extracts from 10 µM, 20 µM, 50 µM and 100 µM H2O2-treated cells displayed increases in the La protein level over that of the untreated control (Fig. 2). Importantly, the levels of cytoplasmic La correlated with the degrees of IRES-dependent translational stimulation by H2O2 (Figs 1a and 2). A very low level of H2O2 of 1 µM, which did not result in any increase in intracellular oxidants, also did not induce translational upregulation or led to an increase in cytoplasmic La level (Figs 1a, b and 2). Low levels of H2O2, at 10 µM and 20 µM, which resulted in the highest stimulation of IRES-dependent translational activity, also caused the accumulation of the highest level of La in the cytoplasm (Figs 1a and 2). High levels of H2O2, at 50 µM and 100 µM, which resulted in an intermediate level of stimulation of IRES-dependent translational activity, also caused the accumulation of an intermediate level of La in the cytoplasm. Examination of the nuclear fractions using lamin B1 as a marker, revealed a corresponding decrease in the La protein nuclear levels in cells treated with 10 µM, 20 µM, 50 µM and 100 µM H2O2, compared with that of the untreated cells (Fig. 2). We noticed a reduced cytoplasmic La level and an elevated nuclear La level in cells treated with 1 µM H2O2, although the significance of this is currently unknown and will need further investigation. The La protein levels from total lysates remained similar to that of the untreated control in all cases. Altogether, these results suggest that the increase in cytosolic La level could be a result of cytoplasmic shuttling. The absence of lamin B1 – a nuclear marker – in the S10 fractions, and β-tubulin – a cytoplasmic marker – in the nuclear fractions, confirms the purity of the fractionation process. The absence of the nuclear protein lamin B1 in the S10 fractions also indicates that the increase in cytoplasmic La levels is not a result of membrane leakage and is likely a result of H2O2-induced cytoplasmic shuttling.
Fig. 2.
H2O2 increases the cytoplasmic La level. Western blots showing the levels of the La protein and the internal loading control β-tubulin/lamin B1 in the cytoplasmic S10, nuclear extracts and total lysates, in Huh7 cells treated with doses of H2O2, as indicated, for 1 h. The following concentrations were used: anti-La antibody SW5 (1 : 5000); β-tubulin (1 : 5000; clone TUB2.1, Sigma); lamin B1 (1 : 1000; D9V6H, Cell Signaling); HRP-conjugated anti-mouse antibody (1 : 5000; Cell Signaling); HRP-conjugated anti-rabbit antibody (1 : 500; Cell Signaling). The levels of La were quantified using ImageJ, normalized against β-tubulin (for S10 fractions and total lysates)/lamin B1 (for nuclear fractions) and expressed as fold increase relative to the 0 µM H2O2 control.
H2O2 induces La cytoplasmic shuttling
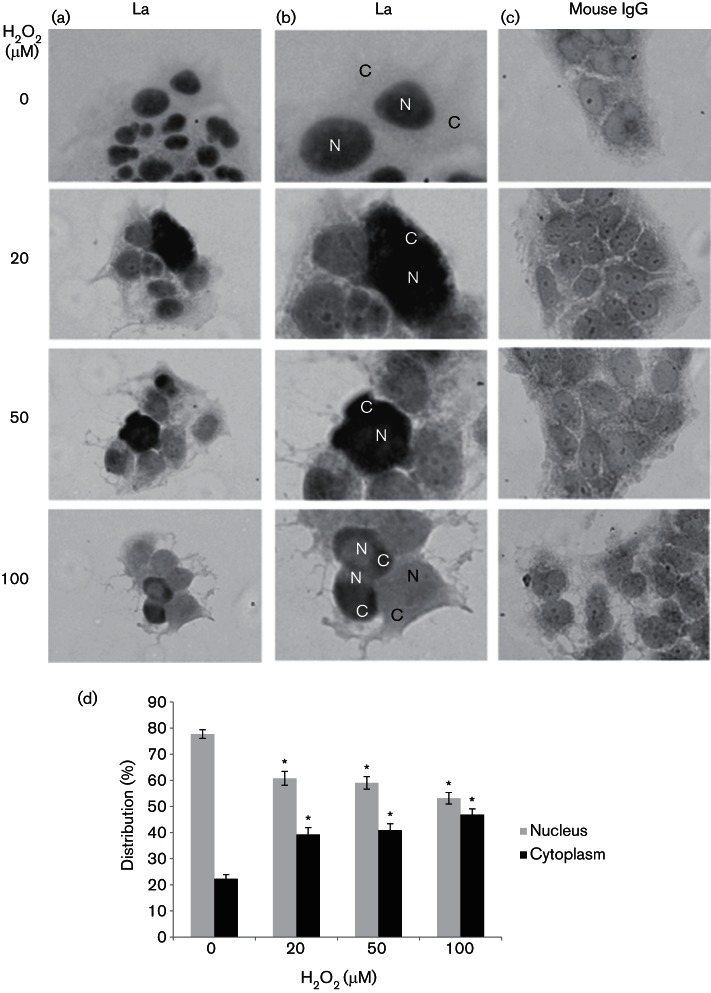
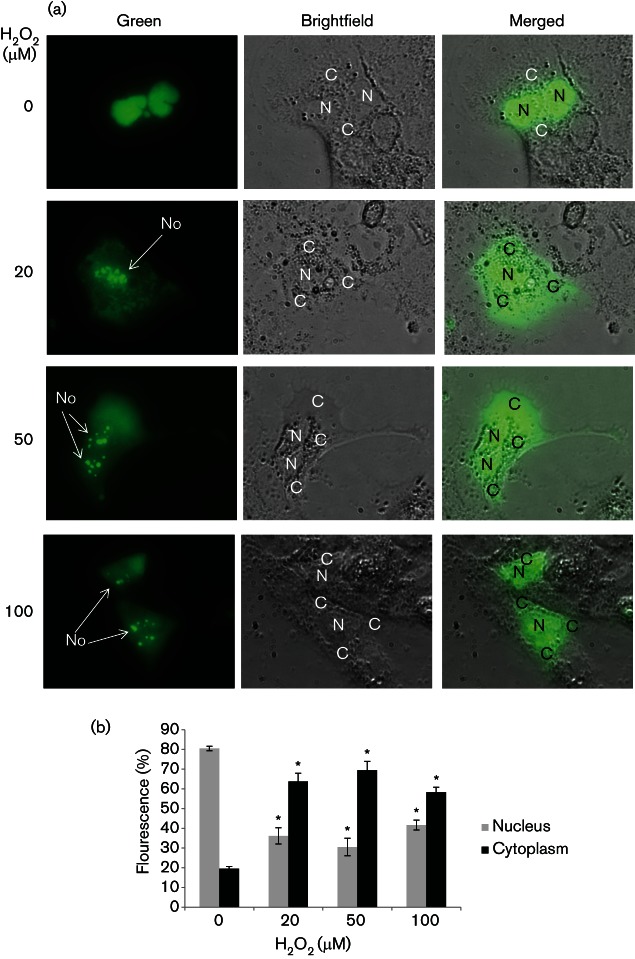
We then studied whether H2O2 was able to induce endogenous La re-localization to the cytoplasm, using immunocytochemistry. In untreated cells, the La protein displayed as dark nuclei staining, consistent with the fact that La is an abundant nuclear protein (Fig. 3) (Wolin & Cedervall, 2002). In Huh7 cells treated with 20 µM, 50 µM and 100 µM H2O2, the nuclear staining was significantly diminished to 61, 59 and 53 %, respectively; compared with that of 78 % in untreated cells. Staining became diffuse and was distributed in both the nucleus and the cytoplasm. Occasionally, dark cytoplasmic staining was observed. Significantly more cytoplasmic staining was detected in 20 µM, 50 µM and 100 µM H2O2-stressed cells (39 , 41 and 47 %); compared with that of 22 % in non-stressed cells. Altogether, these results suggest that the La protein has re-shuttled to the cytoplasm. Further confirmation was obtained using a GFP-tagged La protein transiently transfected into Huh7 cells (Fig. 4). The GFP–La fusion protein assumed a predominantly nuclear localization in non-stressed cells, resembling that of the endogenous La protein distribution (Fig. 4a). In cells treated with 20 µM, 50 µM and 100 µM of H2O2, a proportion of the GFP–La had migrated to the cytoplasm, hence, the fluorescence became diffuse. The nucleus was not clearly defined, however, the nucleoli (No) were visible. The nuclear/cytoplasmic (N/C) ratio of the GFP–La protein significantly changed from 80/20 % in non-stressed cells to 36/64 % , 31/69 % and 42/58 % in 20 µM, 50 µM and 100 µM H2O2-stressed cells, respectively, confirming cytoplasmic re-localization of the La protein upon exposure to H2O2 (Fig. 4b).
Fig. 3.
H2O2 induces La cytoplasmic shuttling. Immunocytochemistry showing the subcellular localizations of the La autoantigen in Huh7 cells treated with doses of H2O2, as indicated, and in untreated control for 1 h. (a) Representative views of clusters of cells as stained by the anti-La antibody, SW5. (b) Enlarged views of (a) showing details of the staining by the anti-La antibody, SW5. N, Nucleus; C, cytoplasm. (c) Clusters of cells stained with the isotypic control mouse IgG. (d) Percentage distribution of nuclear and cytoplasmic staining of the La protein. The values obtained represent the mean±sem of three independent experiments. Between 140 and 220 cells per treatment were analysed from each of the three repeats. Significance of the difference *P<0.05.
Fig. 4.
H2O2 induces GFP–La cytoplasmic re-localization. (a) Live cell imaging showing subcellular localizations of the GFP–La fusion protein in Huh7 cells treated with doses of H2O2, as indicated, and in untreated control for 30 min to 2 h. Huh7 cells were transfected with the plasmid expressing GFP–La for 24 h in 24-well plates, then split into 96-well glass-bottomed plate (Nunc) at a density of 19 000 cells per well for 24 h before subjected to treatment with H2O2. N, Nucleus; C, cytoplasm; No, nucleolus. (b) Percentage distribution of nuclear and cytoplasmic fluorescence of the GFP–La protein. The values obtained represent the mean±sem of three independent experiments. Twenty individual cells per treatment were analysed from each of the three repeats. Significance of the difference *P<0.05.
Increased cytosolic La corresponds to enhanced La-IRES binding
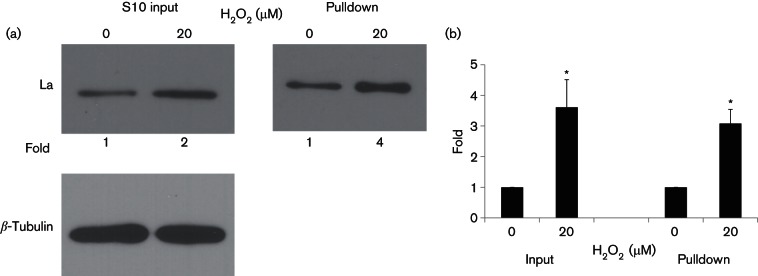
To examine whether the increase in cytoplasmic La level would enhance its binding to IRES RNA to activate HCV IRES-dependent translational activity, we performed an in vitro binding assay using the IRES fragment. Biotinylated IRES RNA pulled down three (2.5–4) times significantly more La protein from the cytosolic extract of 20 µM H2O2-treated cells, compared with that from the untreated control, confirming that the increase in cytoplasmic La level resulted in enhanced binding to the HCV IRES RNA to stimulate IRES-dependent translational activity (Fig. 5).
Fig. 5.
H2O2 increases binding of La to HCV IRES. Cytoplasmic S10 fractions from untreated and Huh7 cells treated with 20 µM of H2O2 for 1 h were pulled down by biotinylated HCV IRES RNA, separated on 12 % SDS-PAGE and probed for the La protein using the anti-La antibody 3B9 (1 : 5000) and HRP-conjugated anti-mouse secondary antibody (1 : 5000). The original S10 fractions were run as inputs. β-Tubulin (1 : 5000) is the internal loading control. The levels of La were quantified using ImageJ. In the S10 input, the levels of La were normalized against β-tubulin and expressed as fold increase relative to the 0 µM H2O2 control. (a) A representation of the Western blot. (b) The levels of the La protein pulled down by biotinylated HCV IRES RNA. The values obtained represent the mean±sem of three independent experiments. Significance of the difference *P<0.05.
La antibody blocks IRES H2O2 responsiveness in vitro
To study whether the increased binding of cytosolic La to HCV IRES RNA was responsible for the enhanced translational activity in H2O2-treated cells, we examined whether abolishing the effect of La would be able to block IRES responsiveness to H2O2. To inhibit La function, we added a La-specific antibody, SW5, to the in vitro translation reaction. Addition of the anti-La antibody, SW5, but not the isotypic control mouse IgG, to 20 µM, 50 µM and 100 µM of H2O2-treated cytosolic S10 fractions reduced the IRES-dependent translational activity to the same level as that primed by the 0 µM H2O2-treated S10 fraction in in vitro translation reactions, suggesting that anti-La antibody was able to abolish/reduce the stimulatory effect of H2O2 on IRES-dependent translation (Fig. 6). Note that the data have been plotted relative to their respective 0 µM controls for ease of comparison. Examination of the absolute values revealed generally lower readings in antibody-treated samples, in agreement with a role of the La protein in HCV IRES-mediated translation (Costa-Mattioli et al., 2004). In contrast, the anti-La antibody only exerted insignificant reduction of the stimulatory effect of H2O2 on the cap-dependent translational activity in some but not all concentrations of H2O2, compared with that of the control mouse IgG. Importantly, the addition of the anti-La antibody, but not that of the control mouse IgG, significantly reduced the IRES/cap ratios in the H2O2-treated samples to the same level as that of the 0 µM H2O2 sample.
La-DN mutant blocks IRES responsiveness to H2O2 ex vivo
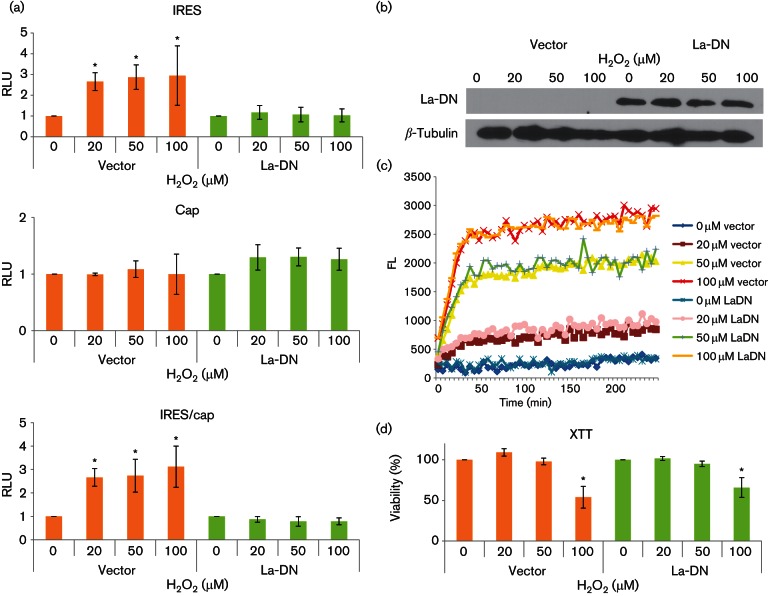
To confirm the role of La in HCV IRES H2O2 responsiveness ex vivo, we made use of a dominant-negative (DN) mutant to block the function of endogenous La. Oligomerization is required for La function and the La 226–348 mutant consisting of the dimerization domain functions as a DN mutant, as has been demonstrated before (Costa-Mattioli et al., 2004; Craig et al., 1997; Holcik & Korneluk, 2000). The La-DN mutant, but not that of the vector control, was able to suppress HCV IRES-dependent translational activation as well as the IRES/cap ratio as a result of H2O2 treatment (20 µM, 50 µM and 100 µM) in Huh7 cells to the same level as that of the 0 µM H2O2 sample (Fig. 7a). Note that the data have been plotted relative to their respective 0 µM controls for ease of comparison. Examination of the absolute values revealed generally lower readings in La-DN-transfected samples, in agreement with a role of the La protein in HCV IRES-mediated translation (Costa-Mattioli et al., 2004). There was no significant increase in cap-dependent translation in cells transfected with either La-DN mutant or the vector control, confirming the specific effect of H2O2 on IRES-mediated translation in an ex vivo model. Expression of the La-DN mutant was confirmed by the detection of the myc-tag (Fig. 7b). We also confirmed that expression of the La-DN mutant did not significantly alter cells’ responses to H2O2 challenge compared with that of the vector control, in terms of generation of intracellular oxidants or viability, excluding the possibility that expression of the La-DN might have an effect on cell stress response (Fig. 7c, d).
Fig. 7.
La-DN inhibits H2O2-activated translation ex vivo. (a) Relative translational activities in Huh7 cells transfected with either the dominant-negative La (La-DN) mutant or the control vector for 24 h, split into a 24-well plate at a density of 1.2×105 for 24 h before being transfected with the bicistronic pRL1b RNA transcript for 4 h followed by treatment with the indicated doses of H2O2 for 4 h. The HCV IRES and cap-translational activities were measured by firefly and Renilla luciferase activities, normalized with respect to total protein, and expressed relative to their respective 0 µM H2O2 controls, which are set as 1. The IRES/cap ratio is represented by the ratio of firefly to Renilla luciferase activities and is expressed relative to their respective 0 µM H2O2 control, which is set as 1. The values obtained represent the mean±sem of three independent experiments, performed in triplicates. RLU, Relative luciferase units. (b) Western blots showing expression of La-DN (detected by the myc-tag antibody clone A-14 Santa Cruz 1 : 1000; HRP-conjugated anti-rabbit antibody, 1 : 1000) at 48 h post-transfection and the internal loading control β-tubulin (1 : 5000; HRP-conjugated anti-mouse antibody 1 : 1000). (c) A representation of three independent dichlorofluorescin fluorometric assays, performed in triplicates, showing the kinetics of ROS generation in plasmid-transfected Huh7 cells (1.2×105 per well in a 24-well plate) after treatment with doses of H2O2, as indicated. FL, Fluorescence units. (d) XTT assay showing viability of plasmid-transfected Huh7 cells (1.2×105 per well in a 24-well plate) after treatment with doses of H2O2, as indicated, for 24 h. The values obtained represent the mean±sem of three independent experiments, performed in triplicates, and are expressed relative to the respective 0 µM H2O2 controls, which are set as 100 %. Significance of the difference *P<0.05.
La-DN mutant blocks IRES responsiveness to H2O2 in a sub-genomic replicon cell line
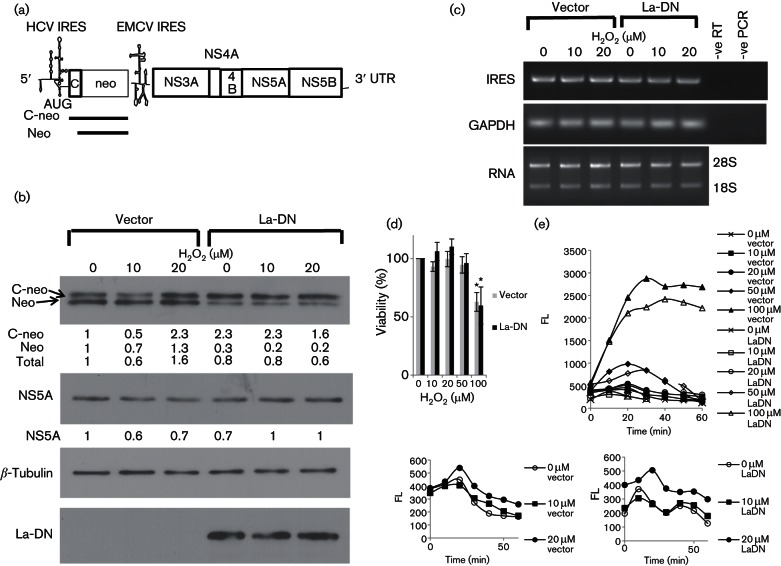
To examine the relevance of the responsiveness of IRES-mediated translation to H2O2 to the HCV life cycle, we made use of a cell line, Huh8, which harbours an HCV sub-genomic replicon resembling a persistent infection (Blight et al., 2000) (Fig. 8a). The sub-genomic replicon consists of a bicistronic transcript in which expression of the core (1–15 aa)-neomycin phosphotransferase (C-neo) fusion protein is under the translational control of the HCV IRES, whereas expression of the HCV NS3-NS5B is under the translational control of the encephalomyocarditis virus (EMCV) IRES. The effects of H2O2 on HCV IRES-mediated translation were compared in Huh8 cells transfected with either a plasmid expressing the La-DN protein or the empty vector using Western blotting on whole-cell lysates. We noticed a 26/28 kDa doublet of neo protein (Fig. 8b). The ~2 kDa difference in size would correspond to the estimated 1.6 kDa molecular weight of the N-terminal C peptide MSTNPKPQRKTKGRA. We reckoned that the upper band represents the C-neo fusion protein translated from the authentic start codon of the HCV IRES, whereas the lower band corresponds to neo protein translated from the internal neo start codon. This is possible because the start codon of the neo gene is retained in the C-neo fusion gene and is placed in a favourable Kozak context (Kozak, 1987) (Fig. 8a). Importantly, translation from the HCV IRES was increased by ~twofold in cells exposed to 20 µM H2O2 over that of the untreated cells in vector-transfected cells, confirming that translation from the HCV IRES is responsive to H2O2 in the context of viral replication resembling a persistent infection. Moreover, the effect of H2O2 was specific towards translation from the authentic start codon as it induced a 2.3-fold increase in the C-neo protein as compared with only a 1.3-fold increase in the neo protein (translated from the internal start codon). In contrast to Huh7 cells, which responded to 10 µM of H2O2 by increasing HCV IRES translational activity from a transiently transfected bicistronic reporter transcript (see Fig. 1a), treatment of vector-transfected Huh8 cells with 10 µM of H2O2 was not enough to increase the HCV IRES translational activity compared with that of the untreated control, as measured by the C-neo/neo levels (Fig. 8b). It is tempting to speculate that the two cell lines have different thresholds of H2O2 responsiveness. Indeed, 10 µM of exogenous H2O2 was sufficient to raise the intracellular oxidant level in Huh7 cells but not that in Huh8 cells (compare Figs 1b and 8e). To exclude the possibility that the increase in C-neo expression in 20 µM H2O2-treated cells was a result of increased replication of the sub-genomic replicon induced by H2O2, we analysed the sub-genomic replicon RNA levels using reverse transcription (RT)-PCR and primers specific for the HCV RNA. We showed similar levels of the sub-genomic replicon RNA in cells regardless of H2O2 treatments, confirming that H2O2 induced translation from the HCV IRES in the sub-genomic replicon cell line (Fig. 8c). In contrast, translation of the NS5A protein from the EMCV IRES was reduced upon H2O2 treatments, indicating that translation from the EMCV IRES is inhibited by H2O2 and responsiveness to H2O2 is specific to the HCV IRES element and is not a general phenomenon for all viral IRESs.
Fig. 8.
La-DN inhibits H2O2-activated translation in a sub-genomic replicon cell line. (a) Diagrammatic representation of the sub-genomic replicon used in this study. The sub-genomic replicon consists of a bicistronic transcript in which expression of the core (1–15 aa)-neomycin phosphotransferase (C-neo) fusion protein is under the translational control of the HCV IRES element, whereas expression of the HCV non-structural (NS) 3-NS5B polyprotein is under the translational control of the EMCV IRES element. (b) Western blots showing the levels of the C-neo/neo proteins, NS5A, myc-tag dominant-negative La (La-DN) mutant protein and the internal loading control β-tubulin in whole-cell lysates of Huh8 cells transfected with either the empty vector or a vector expressing the La-DN mutant protein and treated with doses of H2O2, as indicated, for 1 h. The following concentrations were used: (anti-neo antibody clone 4B4D1, Sigma, 1 : 1000; anti-mouse HRP 1 : 500); (NS5A clone 9E10 1 : 50; anti-mouse HRP 1 : 1000); (anti-myc clone A-14, Santa Cruz 1 : 1000; anti-rabbit HRP 1 : 1000); (anti-β-tubulin clone TUB2.1, Sigma 1 : 1000; anti-mouse HRP 1 : 1000). The levels of C-neo, neo and NS5A were quantified using ImageJ, normalized against β-tubulin and expressed as fold increase relative to the 0 µM H2O2 vector control. (c) Agarose gel electrophoresis showing the amplified fragments from RT-PCR using primers specific for the HCV IRES element and the internal control glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in Huh8 cells transfected with either the empty vector or a vector expressing the La-DN mutant protein and treated with doses of H2O2, as indicated, for 1 h. –ve RT, Negative control for the RT reaction; –ve PCR, negative control for the PCR. The bottom panel shows equal quantity of RNA had been used in the RT-PCR. The bands are the 28S and 18S ribosomal RNAs. (d) XTT assay showing viability of Huh8 cells (16 000 per well/96-well plate) after treatment with doses of H2O2, as indicated, for 24 h. The values obtained represent the mean ± sem of three independent experiments, performed in quadruplicates, and are expressed relative to the untreated control, which is set as 100 %. Significance of the difference *P<0.05. (e) A representation of three independent dichlorofluorescin fluorometric assays, performed in quadruplicates, showing the kinetics of ROS generation in Huh8 cells (16 000 per well/96-well plate) after treatment with doses of H2O2, as indicated. The bottom part of the graph is enlarged and depicted below to show ROS generation in the lower range of H2O2. FL, Fluorescence units.
We then investigated whether La-DN mutant could inhibit HCV IRES H2O2 responsiveness by using transient transfection of Huh8 with a plasmid expressing the La-DN mutant (Holcik & Korneluk, 2000). We do not know why there was a preferential translation from the internal neo start codon in the vector-transfected cells, but from the authentic C-neo start codon in the La-DN-transfected cells, however, comparison of total C-neo and neo levels in untreated cells showed that expression of La-DN did reduce the total neo level, confirming that the La-DN mutant did exhibit a DN effect on HCV IRES-mediated translation (Fig. 8b). The modest DN effect could be a result of cultivation of Huh8 cells in the presence of G418 (neomycin) and, hence, it was essential to maintain expression of the neomycin-resistant protein, i.e. C-neo/neo. Importantly, we showed that expression of La-DN abolished responsiveness of HCV IRES-mediated translation to 20 µM of H2O2, confirming that La plays a role in H2O2-induced translation from the HCV IRES in the sub-genomic replicon. The sub-genomic replicon RNA levels remained similar regardless of transfection or treatments, confirming that the effect of H2O2 is at the level of translation and not at the level of viral replication (Fig. 8c). We also confirmed that expression of the DN-La mutant did not significantly alter cells’ responses to H2O2 challenge compared with that of the vector control, in terms of generation of intracellular oxidants or viability, excluding the possibility that expression of the La-DN might have an effect on cell stress response (Fig. 8d, e).
Discussion
In this study, we showed that translation from the HCV IRES was upregulated by a range of physio/pathological levels of H2O2 that could be encountered by the virus during its prolonged phase of chronic infection, which has significance to HCV survival in an environment with continuously fluctuating levels of oxidants. This is similar to the human immunodeficiency virus (HIV), which also establishes a chronic infection and is associated with elevated oxidative stress in patients and enhanced IRES-dependent translation upon oxidative stress, suggesting that these viruses can adapt to and utilize oxidative stress to aid in their translation (Gendron et al., 2011; Jack & Chan, 2011; MacCallum et al., 2006; Stehbens, 2004). This could be a novel survival mechanism used by viruses to evade oxidative stress and establish chronicity.
The ability to respond to H2O2 is specific to certain viruses and is not a general characteristic of all viral IRESs, as we showed that translation from the EMCV IRES was inhibited by H2O2. The HCV IRES element is more closely related to the picornaviral IRESs than the HIV IRES. It has been speculated that HCV acquired an IRES element from picornavirus in the distant past by recombination (Belsham, 2009; Hellen & de Breyne, 2007). The HCV IRES has been grouped with Type IV picornaviral IRESs to form a class of HCV-like IRESs based on structural and functional similarity (Sweeney et al., 2012). The fact that both HCV and HIV (and some cellular IRESs – see below), but not EMCV IRES-mediated translation is responsive to H2O2, leads us to speculate that IRES responsiveness to oxidative stress is a convergent function evolved with persistent infection/stress response. However, the EMCV IRES is a type II picornaviral IRES, which is structurally and functionally distinct from the HCV-like IRES (Belsham, 2009). Interestingly, basal translation from the poliovirus IRES (a type I picornaviral IRES) is regulated by the La protein – the same ITAF that is responsible for H2O2-inducible translation from the HCV and the cellular nuclear factor erythroid-2-related factor 2 (Nrf2) IRESs (Belsham, 2009; Costa-Mattioli et al., 2004; Li et al., 2010). Therefore, further studies need to be carried out using picornaviral and HCV-like IRESs to see whether responsiveness to oxidative stress is a function preserved in all HCV-like IRESs and some picornaviral IRESs, regardless of whether they establish an acute or chronic infection or it is a function evolved with persistent infection.
Apart from the HCV IRES, other IRESs that have been investigated thus far also exhibit translational upregulation in response to oxidative stress (Daba et al., 2012; Gendron et al., 2011; Giudetti et al., 2013; Li et al., 2010; Yeh et al., 2011; Zhang et al., 2010, 2012). Still, exactly how oxidative stress stimulates IRES-dependent translation is far from clear. Despite being collectively known as IRES, each IRES is unique in terms of sequence, structure, use of eIF and ITAF, mechanism of translation and response to stress (Plank & Kieft, 2012). Thus, it is anticipated that the mechanisms used to respond to oxidative stress would be as diverse as the IRES itself. So far, a positive regulatory mechanism and a derepression mechanism have been proposed for the H2O2-responsive Nrf2 and HIV IRES-dependent translation, respectively (Gendron et al., 2011; Li et al., 2010). We, and others have shown that the La protein is responsible for translational upregulation from the HCV and Nrf2 IRESs during transient and prolonged oxidative stress (Zhang et al., 2012; and this study). The mechanistic similarity between HCV and Nrf2 IRESs suggests that IRES oxidative stress responsiveness could be an evolutionarily conserved homeostatic/adaptive response. Indeed, translation from the IRESs of Nrf2 and ferritin, both of which are important regulators in restoring redox balance, is stimulated by pro-oxidants (Daba et al., 2012; Li et al., 2010; Zhang et al., 2012). A protective response to oxidative stress was also mediated by Sp1 IRES-dependent translational upregulation in a pathological setting of ischaemic insults (Yeh et al., 2011).
Post-translational modification is commonly used to regulate ITAF function during stress (Brenet et al., 2009; Lewis & Holcik, 2008; Sella et al., 1999; Shiroki et al., 1999). La is an abundant nuclear protein, and nuclear–cytoplasm shuttling represents a rapid means of regulation of its function (Lewis & Holcik, 2008). Cytoplasmic export of La is accelerated during poliovirus infection, apoptosis and immune activation, by the cleavage of the C-terminal nuclear localization signal by viral serine protease, caspase and granzymes (Huang et al., 2007; Romero et al., 2009; Shiroki et al., 1999). For one of the doses of H2O2 we used (100 µM) to induce oxidative stress, which also caused apoptosis, however, our Western blots did not reveal any noticeable truncation of the La protein within the time frame of our study, suggesting that oxidative stress-induced apoptotic cleavage of the La protein is not responsible for La shuttling to stimulate HCV IRES-dependent translation in Huh7 cells in our case. Indeed, it has been found that cytoplasmic shuttling of cleaved La during apoptosis actually inhibits, rather than stimulates, HCV IRES-dependent translation (Romero et al., 2009). In another study, a 10 min transient H2O2 treatment of an apoptotic dose of H2O2 (100 µM) on HeLa cells was also sufficient to induce La shuttling without truncation (Zhang et al., 2012). This is further supported by the demonstration, in our study, that La shuttling also occurred in non-apoptotic doses of H2O2, 10 µM and 20 µM, in Huh7 cells (Figs 2, 3 and 4). Thus, a mechanism involving transportation of full-length La is highlighted. La is a phosphoprotein and its shuttling between nucleus and cytoplasm is controlled by nuclear localization signal and nuclear export signal (Bayfield et al., 2010). It is possible that de/phosphorylation shuttles La upon stress by altering the nuclear retention element structure to unmask the nuclear export signal (Intine et al., 2003). Dephosphorylation of La has been demonstrated in a number of cell types in response to diverse apoptotic signals (Rutjes et al., 1999). During tumour growth-factor-beta stimulation of epithelial cells and epithelial to mesenchymal transition, platelet-derived growth factor (PDGF) mediated La cytoplasmic shuttling via the MARK/ERK signalling to stimulate translation from the laminin B1 IRES, suggesting the involvement of a phosphorylation event (Petz et al., 2012). Indeed, it has been shown that PDGF-stimulated Akt-phosphorylation of La mediated its nuclear export to regulate a subset of mRNA translation (Brenet et al., 2009). However, a transient stress with 100 µM of H2O2 did not result in phosphorylation of La in HeLa cells, which might account for its cytoplasmic relocation to stimulate translation from the Nrf2 IRES (Zhang et al., 2012). Altogether, they imply diverse mechanisms used to export nuclear La upon different stress signals and/or in different cell types.
Intriguingly, we detected a neo protein translated from an internal AUG start codon in addition to the C-neo fusion protein translated from the authentic HCV IRES AUG start codon when we used a sub-genomic replicon cell line resembling a persistent infection to study the response of the HCV IRES-mediated translation to H2O2 (Fig. 8). It is well known that translation from the HCV IRES can utilize alternative ORFs and internal AUG and non-AUG start codons, reflecting translational flexibility (Boumlic et al., 2011).
On one hand, HCV replication is modulated by oxidative stress (Choi et al., 2004; McCartney et al., 2008); on the other hand, La also plays a role in viral replication (Kumar et al., 2013). However, we still do not yet know whether oxidative stress exerts its effect on viral replication via La. It will be interesting to study, in the future, how oxidative stress affects the dynamics of translation, replication and its role in translation–replication switch.
It is generally believed that H2O2 inhibits cap-dependent translation (Grant, 2011). Some studies on H2O2- responsive IRES-dependent translation also detected downregulation of cap-dependent translation (Gendron et al., 2011; Giudetti et al., 2013). In contrast, we, and others have previously detected simultaneous upregulation of cap- and IRES-dependent translation (with increased IRES/cap ratio), suggesting that cap-dependent translation could be upregulated by H2O2 (Li et al., 2010; MacCallum et al., 2006). In this study, we detected an inconsistent response of cap-dependent translation to H2O2. It is well known that susceptibility of cells to H2O2 fluctuates (Dickinson & Chang, 2011; Veal et al., 2007). A major determining factor is the balance between endogenous pro- and anti-oxidant levels/species and the redox buffering capacity, which is, in turn, very much affected by the passage number (age, metabolism, speed and ability to adapt) of the cells. Therefore, the response of cap-dependent translation to H2O2 could be more varied than previously thought (Davies, 1999; Grant, 2011).
In conclusion, HCV adapts to oxidative stress in the host cell by increasing translation from its IRES as a result of oxidative stress-induced cytoplasmic shuttling of its main ITAF, La. Further work is required to delineate the H2O2-responsive element in the HCV IRES and to decipher the mechanism by which full-length La re-shuttles to the cytoplasm during oxidative stress.
Methods
Cell culture.
Huh7 and Huh8 cells were maintained in Dulbecco’s modified Eagle’s medium, 10 % FCS, 100 µg penicillin ml−1, 100 µg streptomycin ml−1, 4 mM glutamate and 1× non-essential amino acids. Huh8 cultures were supplemented with 100 mg ml−1 G418 (Melford). To induce oxidative stress, H2O2 (30 % w/w solution, Sigma) was added exogenously.
XTT viability assay.
XTT assay was performed according to the manufacturer’s instruction (Cell Proliferation Kit II, Roche). Cells seeded in 24-well/96-well plates were treated with H2O2 for 24 h. XTT was added and readings were taken at 450 nm using a 650 nm reference filter (Bio-Tek Synergy HT).
Reactive oxygen species (ROS) measurement.
The generation of intracellular ROS was measured using the probe 2′,7′-dichlorofluorescin diacetate (DCFH-DA, Sigma) and a fluorimeter (Berthold Twinkle), as described previously (MacCallum et al., 2006).
Plasmids.
The bicistronic construct, pRL1b, encoding an upstream Renilla luciferase gene under the control of the T7 promoter, and a downstream firefly luciferase gene under the control of the HCV IRES has been previously described (Collier et al., 1998). To generate the sense IRES fragment used in affinity pulldown, the BamHI fragment containing the HCV IRES was excised from the plasmid pRL1b and subcloned into pcDNA3.1 (Invitrogen) to create the plasmid pIRES. The orientation of the insert has been confirmed by restriction digestion. The green fluorescent protein (gfp)-La plasmid was kindly provided by Junji Sagara (Ayukawa et al., 2000). The plasmid encoding a dominant negative mutant of the La protein (La-DN) and its empty vector were kindly provided by Martin Holcik (Holcik & Korneluk, 2000).
Transfection.
Transfection with DNA plasmids was performed using Fugene HD (Promega) according to the manufacturer’s instructions. Transfection with RNA transcripts was performed using Trans IT-mRNA transfection kit (Mirus), according to the manufacturer’s instructions.
Extraction of total lysates, cytoplasmic S10 and nuclear fractions.
Cells were trypsinized and washed with ice-cold PBS four times. A fraction of the pelleted cells was removed for total protein extraction in RIPA buffer (50 mM Tris pH 8, 150 mM NaCl, 1 % NP40, 0.5 % Na deoxycholate, 0.1 % SDS) or directly into SDS-PAGE loading buffer (62.5 mM Tris pH 6.8, 2 % SDS, 5 % β-mercaptoethanol, 10 % glycerol, 0.002 % bromophenol blue) for 10 min at 100 °C followed by clarification for 10 min at 14 000 rpm, 4 °C. To obtain S10, the remaining cell pellet was lysed in ice-cold hypotonic buffer (10 mM K-Hepes pH 7.5, 10 mM KOAc, 1.5 mM MgOAc, 2.5 mM DTT) for 10 min. The lysate was centrifuged at 1000 g for 5 min to pellet the nuclei and cell debris. The supernatant was then centrifuged at 10 000 g for 20 min to obtain the cytoplasmic S10 fraction. To obtain the nuclear fraction the pellet was washed twice with hypotonic buffer and then resuspended in RIPA buffer. Nuclear fraction was extracted by rotation for 30 min, 4 °C; followed by incubation at 100 °C for 10 min and then clarification for 20 min at 14 000 rpm, 4 °C. The protein concentrations of the S10 fractions were determined using the Bio-Rad Bradford protein assay according to the manufacturer’s instructions. The protein concentrations of the total lysates and nuclear fractions were determined using the BCA kit (Sigma) or the RC-DC kit (Bio-Rad) according to the manufacturers’ instructions.
In vitro transcription.
The bicistronic reporter RNA template used in in vitro translation reactions was generated by in vitro transcription from the T7 promoter of pRL1b using the Ambion’s mMESSAGE mMACHINE transcription kit according to the manufacturer’s instructions. Biotinylated RNA used in in vitro affinity pulldown reactions was synthesized from linearized pIRES plasmid DNA using the Ambion’s T7 Megascript transcription kit according to the manufacturer’s instructions, with the addition of 7.5 mM ATP, 7.5 mM CTP, 7.5 mM GTP, 5.6 mM UTP and 1.9 mM biotin-16-uridine-5′-triphosphate (Roche).
In vitro translation.
In vitro translation was carried out at 37 °C for 1 h using equal quantities of the S10 extracts and 0.4 µg of the RNA template in 10 µl of reaction mixture containing 6.4 mM creatine phosphate, 0.08 µg creatine phosphokinase, 1.6 mM DTT, 14.2 mM Hepes-KOH pH 7.2, 0.2 mM spermidine, 170 mM KOAc, 1.5 mM MgOAc, 1 mM ATP, 20 µM GTP, 20 µM complete amino acids mix (Promega) and 0.2 µl of RNase inhibitor (New England Biolabs). An aliquot of the reaction (5–10 µl) was used to measure the firefly and Renilla luciferase activities.
Dual luciferase assay.
The activities of firefly and Renilla luciferases were measured in relative light units over 10 s with a luminometer (Berthold Lumat LB9507), using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions.
RNA affinity pulldown.
Biotinylated RNA (10 µg) was mixed with 100 µg of S10 fraction in 500 µl binding buffer containing 10 mM Tris pH 7.4, 150 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 1 : 100 protease inhibitor cocktail (Sigma), 0.05 % NP40, 30 µg yeast tRNA (Sigma), 1 µl RNase inhibitor (New England Biolabs) for 30 min at room temperature followed by 2 h at 4 °C with slow rotation. To pulldown biotinylated RNA, 100 µl (1 : 1 slurry) of pre-washed streptavidin agarose beads (CL-4B, Sigma) were added to each reaction, and incubation continued at 4 °C for 2 h with slow rotation. The beads were then washed five times with binding buffer and the pellets resuspended in 20 µl of SDS-PAGE loading buffer for electrophoretic separation on 12 % SDS-PAGE gel and Western blotting.
Western blotting.
Western blotting was performed as previously described (Chan & Egan, 2005, 2009). Proteins separated on 12 % SDS-PAGE gel were incubated with primary antibodies, followed by HRP-conjugated secondary antibodies (Cell Signaling) in 5 % semi-skimmed milk (Marvel)/0.1 % Tween 20/TBS. Immunocomplexes were detected with the Uptilight HS HRP chemiluminescence system (Uptima).
Immunocytochemistry.
Cells grown on chamber slides (Falcon) were fixed in ice-cold methanol for 1 h. After blocking for 1 h in 1 % horse serum/2 % BSA/PBS, cells were incubated with anti-La antibody SW5 (1 : 10) or isotypic control, and then HRP-conjugated secondary antibody (1 : 50; Cell Signaling) for 1 h each in blocking buffer. Colour was developed using SIGMA FAST™ DAB (3,3′-diaminobenzidine tetrahydrochloride).
Microscopy.
Immunocytochemistry images were collected on a Zeiss Axioskop upright microscope using a 40×/0.95 Plan Apochromat objective and captured using an Axiocam colour CCD camera through Zeiss Axiovision software. GFP images were collected on an Zeiss Axiovert inverted microscope using a 60×/1.25 Plan Neofluar objective and captured using a Sony CCD video camera through Zeiss Axiovision software. Images were then processed and analysed using ImageJ (http://rsb.info.nih.gov/ij).
RNA extraction.
RNA was extracted from cultured cells using RNA-Bee (AMSBIO) according to the manufacturer’s instruction. RNA was quantified using a Nanodrop 1000.
Reverse transcription (RT)-PCR.
RNA (1 µg) was incubated with 1.5 µl of 20 µM antisense (AS) primer at 70 °C for 5 min and then added to a total volume of 20 µl of RT reaction consisting of 50 mM Tris pH 8.3, 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 0.24 mM dNTP, 3 µl DMSO, 0.1 µl RNase inhibitor (New England BioLabs) and 1 µl of Moloney murine leukemia virus reverse transcriptase (Promega) at 42 °C for 1 h. An aliquot (1 µl) of RT reaction was used in a total volume of 20 µl of PCR consisting of 20 mM Tris pH 8.8, 10 mM (NH4)2SO4, 10 mM KCl, 2 mM MgSO4, 0.1 % Triton-X-100, 0.2 mM dNTP, 1.25 µM each of sense and AS primers and 0.1 µl Taq polymerase (New England BioLabs). PCR was carried out with a 2 min denaturation and 25 cycles of (94 °C/25 s; 55 °C/35 s; 68 °C/1 min) followed by a 7 min extension at 68 °C. The sense and AS primers used to amplify the IRES fragment were 5′-caccagatctcactcccctgtgaggaacta-3′ and 5′-tgcccagtcatagccgaatag-3′. The sense and AS primers used to amplify the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) fragment were 5′-cctgttcgacagtcagccg-3′ and 5′-cgaccaaatccgttgactcc-3′.
Statistics.
Statistical analyses were performed using ANOVA. A P value <0.05 was considered significant.
Acknowledgements
I thank Richard Elliott (deceased) for providing the plasmid pRL1b; Martin Holcik for the La-DN construct; Junji Sagara for the gfp-La plasmid; Michael Bachmann for the La antibodies and Charlie Rice for the Huh7 and Huh8 cell lines and the NS5A antibody. Special thanks goes to Steven Marsden for his help with microscopy. Finally, I would like to express my sincere gratitude to Ian Roberts for his continuous support.
References
- Ayukawa K., Taniguchi S., Masumoto J., Hashimoto S., Sarvotham H., Hara A., Aoyama T., Sagara J.(2000). La autoantigen is cleaved in the COOH terminus and loses the nuclear localization signal during apoptosis. J Biol Chem 275 34465–34470. 10.1074/jbc.M003673200 [DOI] [PubMed] [Google Scholar]
- Bayfield M. A., Yang R., Maraia R. J.(2010). Conserved and divergent features of the structure and function of La and La-related proteins (LARPs). Biochim Biophys Acta 1799 365–378. 10.1016/j.bbagrm.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsham G. J.(2009). Divergent picornavirus IRES elements. Virus Res 139 183–192. 10.1016/j.virusres.2008.07.001 [DOI] [PubMed] [Google Scholar]
- Blight K. J., Kolykhalov A. A., Rice C. M.(2000). Efficient initiation of HCV RNA replication in cell culture. Science 290 1972–1974. 10.1126/science.290.5498.1972 [DOI] [PubMed] [Google Scholar]
- Boumlic A., Vassilaki N., Dalagiorgou G., Kochlios E., Kakkanas A., Georgopoulou U., Markoulatos P., Orfanoudakis G., Mavromara P.(2011). Internal translation initiation stimulates expression of the ARF/core+1 open reading frame of HCV genotype 1b. Virus Res 155 213–220. 10.1016/j.virusres.2010.10.007 [DOI] [PubMed] [Google Scholar]
- Brenet F., Socci N. D., Sonenberg N., Holland E. C.(2009). Akt phosphorylation of La regulates specific mRNA translation in glial progenitors. Oncogene 28 128–139. 10.1038/onc.2008.376 [DOI] [PubMed] [Google Scholar]
- Chan S. W.(2014). Establishment of chronic hepatitis C virus infection: translational evasion of oxidative defence. World J Gastroenterol 20 2785–2800. 10.3748/wjg.v20.i11.2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. W., Egan P. A.(2005). Hepatitis C virus envelope proteins regulate CHOP via induction of the unfolded protein response. FASEB J 19 1510–1512. 10.1096/fj.04-3455fje [DOI] [PubMed] [Google Scholar]
- Chan S.-W., Egan P. A.(2009). Effects of hepatitis C virus envelope glycoprotein unfolded protein response activation on translation and transcription. Arch Virol 154 1631–1640. 10.1007/s00705-009-0495-5 [DOI] [PubMed] [Google Scholar]
- Choi J. N., Lee K. J., Zheng Y. Y., Yamaga A. K., Lai M. M., Ou J. H.(2004). Reactive oxygen species suppress hepatitis C virus RNA replication in human hepatoma cells. Hepatology 39 81–89. 10.1002/hep.20001 [DOI] [PubMed] [Google Scholar]
- Collier A. J., Tang S. X., Elliott R. M.(1998). Translation efficiencies of the 5′ untranslated region from representatives of the six major genotypes of hepatitis C virus using a novel bicistronic reporter assay system. J Gen Virol 79 2359–2366. 10.1099/0022-1317-79-10-2359 [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M., Svitkin Y., Sonenberg N.(2004). La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol Cell Biol 24 6861–6870. 10.1128/MCB.24.15.6861-6870.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig A. W., Svitkin Y. V., Lee H. S., Belsham G. J., Sonenberg N.(1997). The La autoantigen contains a dimerization domain that is essential for enhancing translation. Mol Cell Biol 17 163–169. 10.1128/MCB.17.1.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daba A., Koromilas A. E., Pantopoulos K.(2012). Alternative ferritin mRNA translation via internal initiation. RNA 18 547–556. 10.1261/rna.029322.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies K. J. A.(1999). The broad spectrum of responses to oxidants in proliferating cells: a new paradigm for oxidative stress. IUBMB Life 48 41–47. 10.1080/713803463 [DOI] [PubMed] [Google Scholar]
- Diamond D. L., Jacobs J. M., Paeper B., Proll S. C., Gritsenko M. A., Carithers R. L., Larson A. M., Yeh M. M., Camp D. G., et al. (2007). Proteomic profiling of human liver biopsies: hepatitis C virus-induced fibrosis and mitochondrial dysfunction. Hepatology 46 649–657. 10.1002/hep.21751 [DOI] [PubMed] [Google Scholar]
- Dickinson B. C., Chang C. J.(2011). Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat Chem Biol 7 504–511. 10.1038/nchembio.607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Sugimoto R., Ma N., Tanaka H., Iwasa M., Kobayashi Y., Kawanishi S., Watanabe S., Kaito M., Takei Y.(2008). Comparison of hepatic oxidative DNA damage in patients with chronic hepatitis B and C. J Viral Hepat 15 498–507. 10.1111/j.1365-2893.2008.00972.x [DOI] [PubMed] [Google Scholar]
- Gendron K., Ferbeyre G., Heveker N., Brakier-Gingras L.(2011). The activity of the HIV-1 IRES is stimulated by oxidative stress and controlled by a negative regulatory element. Nucleic Acids Res 39 902–912. 10.1093/nar/gkq885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudetti A. M., Damiano F., Gnoni G. V., Siculella L.(2013). Low level of hydrogen peroxide induces lipid synthesis in BRL-3A cells through a CAP-independent SREBP-1a activation. Int J Biochem Cell Biol 45 1419–1426. 10.1016/j.biocel.2013.04.004 [DOI] [PubMed] [Google Scholar]
- Grant C. M.(2011). Regulation of translation by hydrogen peroxide. Antioxid Redox Signal 15 191–203. 10.1089/ars.2010.3699 [DOI] [PubMed] [Google Scholar]
- Hellen C. U. T., de Breyne S.(2007). A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: evidence for modular exchange of functional noncoding RNA elements by recombination. J Virol 81 5850–5863. 10.1128/JVI.02403-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M., Korneluk R. G.(2000). Functional characterization of the X-Linked Inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of la autoantigen in XIAP translation. Mol Cell Biol 20 4648–4657. 10.1128/MCB.20.13.4648-4657.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Ida H., Arima K., Nakamura H., Aramaki T., Fujikawa K., Tamai M., Kamachi M., Kawakami A., et al. (2007). La autoantigen translocates to cytoplasm after cleavage during granzyme B-mediated cytotoxicity. Life Sci 81 1461–1466. 10.1016/j.lfs.2007.09.017 [DOI] [PubMed] [Google Scholar]
- Intine R. V., Tenenbaum S. A., Sakulich A. L., Keene J. D., Maraia R. J.(2003). Differential phosphorylation and subcellular localization of la RNPs associated with precursor tRNAs and translation-related mRNAs. Mol Cell 12 1301–1307. 10.1016/S1097-2765(03)00429-5 [DOI] [PubMed] [Google Scholar]
- Jack S. C., Chan S.-W.(2011). The role of PERK and GCN2 in basal and hydrogen peroxide-regulated translation from the hepatitis C virus internal ribosome entry site. Virus Genes 43 208–214. 10.1007/s11262-011-0629-1 [DOI] [PubMed] [Google Scholar]
- Jackson R. J.(2005). Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem Soc Trans 33 1231–1241. 10.1042/BST0331231 [DOI] [PubMed] [Google Scholar]
- Konishi M., Iwasa M., Araki J., Kobayashi Y., Katsuki A., Sumida Y., Nakagawa N., Kojima Y., Watanabe S., et al. (2006). Increased lipid peroxidation in patients with non-alcoholic fatty liver disease and chronic hepatitis C as measured by the plasma level of 8-isoprostane. J Gastroenterol Hepatol 21 1821–1825. 10.1111/j.1440-1746.2006.04420.x [DOI] [PubMed] [Google Scholar]
- Kozak M.(1987). An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15 8125–8148. 10.1093/nar/15.20.8125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Ray U., Das S.(2013). Human la protein interaction with GCAC near the initiator AUG enhances hepatitis C virus RNA replication by promoting linkage between 5′ and 3′ untranslated regions. J Virol 87 6713–6726. 10.1128/JVI.00525-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S. M., Holcik M.(2008). For IRES trans-acting factors, it is all about location. Oncogene 27 1033–1035. 10.1038/sj.onc.1210777 [DOI] [PubMed] [Google Scholar]
- Li W., Thakor N., Xu E. Y., Huang Y., Chen C., Yu R., Holcik M., Kong A.-N.(2010). An internal ribosomal entry site mediates redox- sensitive translation of Nrf2. Nucleic Acids Res 38 778–788. 10.1093/nar/gkp1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum P. R., Jack S. C., Egan P. A., McDermott B. T., Elliott R. M., Chan S.-W.(2006). Cap-dependent and hepatitis C virus internal ribosome entry site-mediated translation are modulated by phosphorylation of eIF2 under oxidative stress. J Gen Virol 87 3251–3262. 10.1099/vir.0.82051-0 [DOI] [PubMed] [Google Scholar]
- McCartney E. M., Semendric L., Helbig K. J., Hinze S., Jones B., Weinman S. A., Beard M. R.(2008). Alcohol metabolism increases the replication of hepatitis C virus and attenuates the antiviral action of interferon. J Infect Dis 198 1766–1775. 10.1086/593216 [DOI] [PubMed] [Google Scholar]
- Paracha U. Z., Fatima K., Alqahtani M., Chaudhary A., Abuzenadah A., Damanhouri G., Qadri I.(2013). Oxidative stress and hepatitis C virus. Virol J 10 251. 10.1186/1743-422X-10-251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis V., Kollinger M., Fabre M., Holstege A., Poynard T., Bedossa P.(1997). In situ detection of lipid peroxidation by-products in chronic liver diseases. Hepatology 26 135–142. 10.1002/hep.510260118 [DOI] [PubMed] [Google Scholar]
- Petz M., Them N., Huber H., Beug H., Mikulits W.(2012). La enhances IRES-mediated translation of laminin B1 during malignant epithelial to mesenchymal transition. Nucleic Acids Res 40 290–302. 10.1093/nar/gkr717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank T.-D., Kieft J. S.(2012). The structures of nonprotein-coding RNAs that drive internal ribosome entry site function. Wiley Interdiscip Rev RNA 3 195–212. 10.1002/wrna.1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudi R., Srinivasan P., Das S.(2004). La protein binding at the GCAC site near the initiator AUG facilitates the ribosomal assembly on the hepatitis C virus RNA to influence internal ribosome entry site-mediated translation. J Biol Chem 279 29879–29888. 10.1074/jbc.M403417200 [DOI] [PubMed] [Google Scholar]
- Romero V., Fellows E., Jenne D. E., Andrade F.(2009). Cleavage of La protein by granzyme H induces cytoplasmic translocation and interferes with La-mediated HCV-IRES translational activity. Cell Death Differ 16 340–348. 10.1038/cdd.2008.165 [DOI] [PubMed] [Google Scholar]
- Rutjes S. A., Utz P. J., van der Heijden A., Broekhuis C., van Venrooij W. J., Pruijn G. J.(1999). The La (SS-B) autoantigen, a key protein in RNA biogenesis, is dephosphorylated and cleaved early during apoptosis. Cell Death Differ 6 976–986. 10.1038/sj.cdd.4400571 [DOI] [PubMed] [Google Scholar]
- Sella O., Gerlitz G., Le S. Y., Elroy-Stein O.(1999). Differentiation-induced internal translation of c-sis mRNA: analysis of the cis elements and their differentiation-linked binding to the hnRNP C protein. Mol Cell Biol 19 5429–5440. 10.1128/MCB.19.8.5429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroki K., Isoyama T., Kuge S., Ishii T., Ohmi S., Hata S., Suzuki K., Takasaki Y., Nomoto A.(1999). Intracellular redistribution of truncated La protein produced by poliovirus 3C(pro)-mediated cleavage. J Virol 73 2193–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehbens W. E.(2004). Oxidative stress in viral hepatitis and AIDS. Exp Mol Pathol 77 121–132. 10.1016/j.yexmp.2004.04.007 [DOI] [PubMed] [Google Scholar]
- Sweeney T. R., Dhote V., Yu Y., Hellen C. U.(2012). A distinct class of internal ribosomal entry site in members of the Kobuvirus and proposed Salivirus and Paraturdivirus genera of the Picornaviridae. J Virol 86 1468–1486. 10.1128/JVI.05862-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K., Iizuka N., Kohara M., Nomoto A.(1992). Internal ribosome entry site within hepatitis C virus RNA. J Virol 66 1476–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veal E. A., Day A. M., Morgan B. A.(2007). Hydrogen peroxide sensing and signaling. Mol Cell 26 1–14. 10.1016/j.molcel.2007.03.016 [DOI] [PubMed] [Google Scholar]
- Venturini D., Simão A. N., Barbosa D. S., Lavado E. L., Narciso V. E., Dichi I., Dichi J. B, Colado Simao A. N.(2010). Increased oxidative stress, decreased total antioxidant capacity, and iron overload in untreated patients with chronic hepatitis c. Dig Dis Sci 55 1120–1127. 10.1007/s10620-009-0833-1 [DOI] [PubMed] [Google Scholar]
- Wolin S. L., Cedervall T.(2002). The La protein. Annu Rev Biochem 71 375–403. 10.1146/annurev.biochem.71.090501.150003 [DOI] [PubMed] [Google Scholar]
- Yamashita T., Kaneko S., Hashimoto S., Sato T., Nagai S., Toyoda N., Suzuki T., Kobayashi K., Matsushima K.(2001). Serial analysis of gene expression in chronic hepatitis c and hepatocellular carcinoma. Biochem Biophys Res Commun 282 647–654. 10.1006/bbrc.2001.4610 [DOI] [PubMed] [Google Scholar]
- Yeh S. H., Yang W. B., Gean P. W., Hsu C. Y., Tseng J. T., Su T. P., Chang W. C., Hung J. J.(2011). Translational and transcriptional control of Sp1 against ischaemia through a hydrogen peroxide-activated internal ribosomal entry site pathway. Nucleic Acids Res 39 5412–5423. 10.1093/nar/gkr161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Wang X., Xiao Z., Liu W., Chen B., Dai J.(2010). Mapping of the minimal internal ribosome entry site element in the human embryonic stem cell gene OCT4B mRNA. Biochem Biophys Res Commun 394 750–754. 10.1016/j.bbrc.2010.03.064 [DOI] [PubMed] [Google Scholar]
- Zhang J., Thai Nho D., Kappeler K., Tsaprailis G., Chen Q. M.(2012). La autoantigen mediates oxidant induced de novo Nrf2 protein translation. Mol Cell Proteomics 11 015032. 10.1074/mcp.M111.015032 [DOI] [PMC free article] [PubMed] [Google Scholar]