Abstract
The accurate prediction of a ligand–protein complex structure is important for computer-assisted drug development. Although many docking methods have been developed over the last three decades, the success of binding structure prediction remains greatly limited. The purpose of this study was to demonstrate the usefulness of molecular dynamics (MD) simulation in assessing a docking pose predicted using a docking program. If the predicted pose is not unstable in an aqueous environment, MD simulation equilibrates the system and removes the ligand from the predicted position. Here we investigated two proteins that are important potential therapeutic targets: β2 adrenergic receptor (β2AR) and PR-Set7. While β2AR is rigid and its ligands are very similar to the template ligand (carazolol), PR-Set7 is very flexible and its ligands vary greatly from the template ligand (histone H4 tail peptide). On an empirical basis, we usually expect that the docking prediction is accurate when the protein is rigid and its ligands are similar to the template ligand. The MD analyses in this study clearly suggested such a tendency. Furthermore, we discuss the possibility that the MD simulation can predict the binding pose of a ligand.
Keywords: G-protein coupled receptor, histone methyltransferase, ligand–protein complex, drug design, molecular dynamics simulation
A central task in the early stage of drug discovery research is to identify new active compounds by evaluating interactions with a target protein. However, this undertaking continues to remain challenging [1,2] and many techniques have thus been developed to overcome such problems. For example, high-throughput screening (HTS) is an experimental technique to systematically evaluate the activity of compounds [3]. Although the HTS technique plays an essential role in the discovery of new hit compounds, associated costs tend to be very high. In this context, many computational methods have been developed as alternatives to experimental methods [4].
In most computational methods, potentially active compounds are selected from a chemical library by evaluating compound–protein interactions. A typical example is docking, which uses the three-dimensional structure of the compound–protein complex to evaluate any potential interaction [5,6]. Although docking has become a popular method because of advancements in X-ray crystallography technology and computational power, the prediction accuracy of the docking software is still rather limited.
The docking calculation consists of two steps. In the first step, the docking program searches for the binding pose of the ligand molecule in the pocket of the target protein, and then, in the second step, binding affinity is evaluated with a simple empirical scoring function. However, a significant problem associated with the second step is the limited accuracy of the empirical score [7]. The empirical score is calculated for a single predicted structure, although any dynamic effects can be essential [8]. Thus, it was proposed that molecular dynamics (MD) simulation could be used in the actual drug development procedure to improve the prediction of binding free energy to increase the efficiency of drug development [9]. MD simulation enables accurate prediction of free energy for several systems [10].
In this study, the main purpose was to investigate the first stage of docking by assessing the binding pose prediction by MD simulation and to demonstrate that MD simulation can provide useful information to complement the docking prediction. One advantage of MD simulation is that it takes into account the natural motion of the protein, whereas docking usually utilizes a single structure obtained by experiment. The MD simulation equilibrates the system to achieve a stable conformation. If the initial structure is energetically unstable, the system appropriately changes the conformation in subsequent MD simulations. Otherwise, the ligand-binding structure will only change slightly compared with the initial structure.
MD simulation has already been used to evaluate the docked complex structure [11–13]. Cavalli et al. [12] showed that MD simulation discriminated between the stable and unstable docked poses of a ligand (propidium) in acetylcholinesterase (HuAChE). Two binding complexes with the most stable MD trajectories agreed with the experimental structures, whereas the MD trajectories of the other complexes were unstable. In the case of inhibitors that selectively act on CDK4, but not on CDK2 [13], MD simulation showed that the docked complex of CDK4 was more stable than that of CDK2, probably because the inhibitors were exposed to bulk water much more in CDK2 than in CDK4. (Water molecules often play important roles in protein function [14].) These MD simulations were typically shorter than 5 ns and never considered the longer time-scale collective motions, probably because an inaccurate force field may result in an unrealistic long-time scale MD simulation. However, MD simulations exceeding 100 ns are sometimes required to equilibrate the protein structure.
Recently, it was suggested that sufficiently long MD simulations can predict the three-dimensional structures of small proteins [15]. In addition, a better protein force field FUJI has since been developed [16] and reproduced the experimental conformational distribution [9,17]. These results indicated that a long-time scale MD simulation with the all-atom model should be more reliable. In this study, we utilize MD simulation exceeding 100 ns with the reliable FUJI force field. To demonstrate the usefulness of long MD simulation from a broad viewpoint, we analyzed two characteristic proteins: β2 adrenergic receptor (β2AR) [18] and PR-Set7 [19]. These proteins essentially involve motions longer than 100 ns.
Although both of these proteins are potential therapeutic targets that have attracted considerable attention from both experimental and computational aspects, their features are totally different. β2AR is embedded in the cellular membrane and mainly consists of seven transmembrane helices. A small compound is bound in the pocket of this protein to regulate signaling activation. PR-Set7 is a soluble protein which binds the histone H4 tail and a small compound, which is usually a methyl-group donor. PR-Set7 catalyzes the methylation of the H4 tail in the cell. PR-Set7 contains a large cleavage for binding of the H4 tail. Several inhibitors have been designed to prevent PR-Set7 from binding to the H4 tail in the cleavage.
In the present study, we performed docking calculations for the two target proteins and several known ligands. Then, long MD trajectories were calculated starting from the complex structures predicted with the docking software. In the remaining part of this article, we assess the docking predictions with MD simulations and demonstrate that MD simulation can provide complementary information about the binding complex structure.
Materials and Methods
β2 adrenergic receptor
β2AR, an important therapeutic target in the treatment of asthma and cardiovascular disease [20], is one of the most extensively studied members of the superfamily of G protein-coupled receptors (GPCR) and plays important regulatory roles in a variety of cells and organ systems. A high-resolution crystal structure of β2AR has already been solved and many agonists and antagonists have also been identified [21–23].
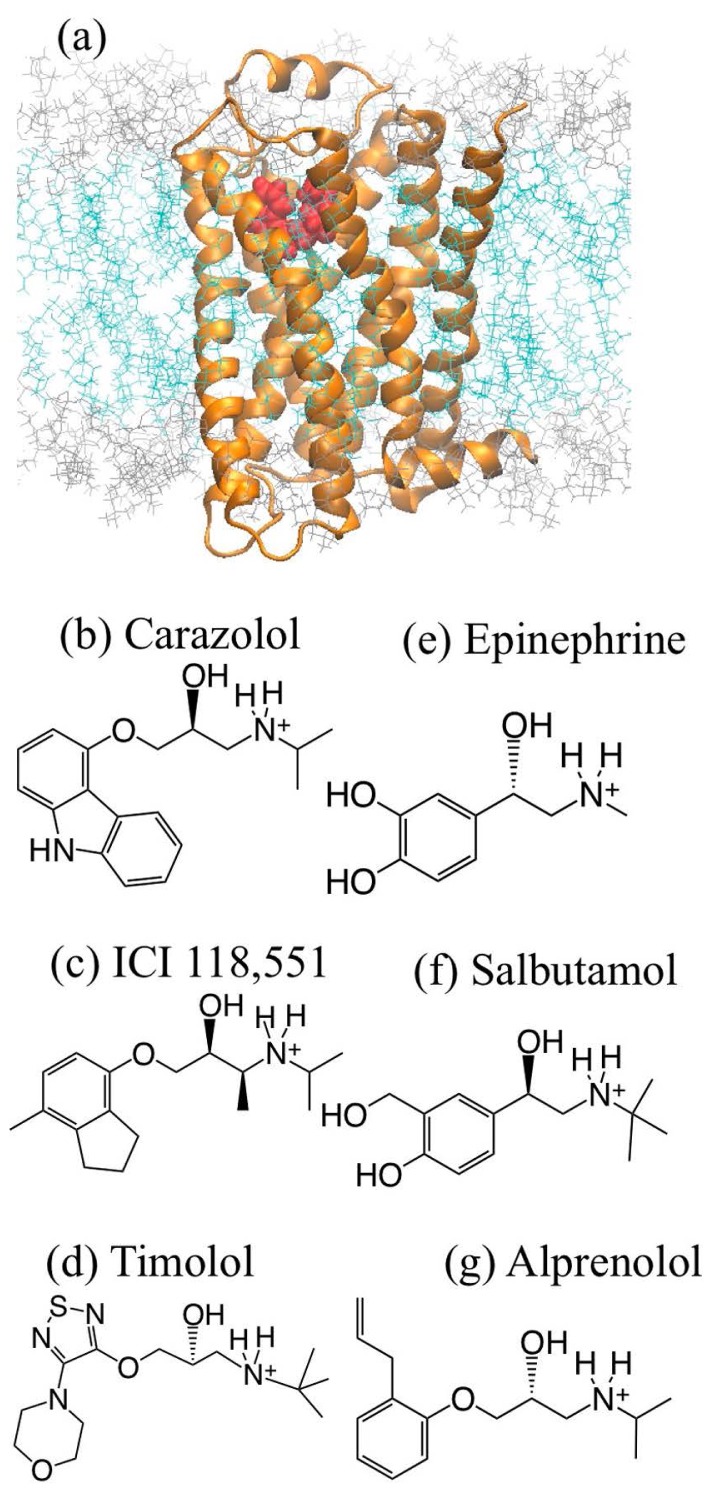
In this study, six compounds [carazolol, ICI 118,551, timolol, epinephrine (adrenaline), salbutamol, and alprenolol] were adopted as ligands of β2AR. While carazolol, ICI 118,551, and timolol are known as inverse agonists, epinephrine and salbutamol are agonists and alprenolol is a neutral antagonist. To predict the binding poses of these ligands, a high-resolution crystal structure of β2AR was used as a starting point of this research [protein data bank (PDB) entry: 2RH1] [18]. The fused T4-lysozyme was removed and the β2AR residues (K227-L230 and K263-F264) were remodeled with MODELLER [24] to connect the fusion terminals. Using this structural data, the six ligands were docked to the pocket of β2AR. Although carazolol was already included in the crystal structure, this ligand was redocked to the crystal structure with the docking program.
Although the ligands have different effects, they share several common structures (Fig. 1). With respect to the electrostatic features, a protonated amine is one of common structures of the ligands. The amine is linked to a large ring structure on one hand and to an aliphatic group on the other hand. If the examined ligand is quite similar to that included in the co-crystal (template ligand), the most reliable binding prediction method is to use the ligand pose of the co-crystal structure as a reference in addition to the protein structure. Thus, the POSIT program ver. 3.1.0.5 [25], which employs this strategy, was used in this step. To predict the binding pose, POSIT changes the ligand pose to maximize similarity with the reference ligand pose. Here Tanimoto Combo (TC) was used as a similarity score. The pose with the best score was selected as a predicted docking pose.
Figure 1.
Structures of β2AR and ligands: (a) Orange ribbon and red balls represent β2AR and carazolol, respectively, and cyan and gray lines are the hydrophobic and hydrophilic portions of the POPC bilayer; (b) carazolol; (c) ICI 118,551; (d) timolol; (e) epinephrine; (f) salbutamol; (g) alprenolol.
To assess the docking poses obtained in the above procedure, MD simulations were performed. First, the ligand-binding β2AR structures were embedded into the equilibrated 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayer using the InflateGRO2 software [26]. Then, water molecules and ions were added to the systems. Each system consisted of a ligand, β2AR protein, 225 POPC lipids, 12,293 TIP3P water molecules, 75 Na+ ions, and 77 Cl− ions. After energy minimization, 150 ns MD trajectories were conducted with positional restraints on heavy atoms of the protein and ligand under a constant NpT condition (T=298 K and p=1 atm) to equilibrate the systems. Next, all the restraints were eliminated and three MD trajectories were calculated for 1,000 ns. Here the Nose–Hoover thermostat, with a relaxation time of 1 ps, and the semi-isotropic Berendsen barostat, with a relaxation time of 2 ps, were used. The cutoff radius of the Lennard-Jones (LJ) and electrostatic interactions were set to 1.2 nm. The MD time step was set to 3 fs.
Here and throughout this article, the MD simulations were performed with GROMACS ver. 4.6.5 [27]. All chemical bond lengths were constrained using the LINCS algorithm. The temperature was set to 298 K to simulate in vitro systems as observed in many previous studies [12,13]. To describe long-range electrostatic interactions, the particle mesh Ewald (PME) method was used. While the TIP3P water model was employed, the FUJI force field parameters were consistently used both for proteins and ligands [16,28]. Atomic charges of the ligands were determined by multi-conformational restrained electrostatic potential (RESP) fitting with HF/6–31G(d) calculations [note that only the on-site charge model of LIG-1 was based on the HF/6–31+G(d) calculations]. The Gaussian09 program [29] was used to perform all ab initio calculations. The Lipid14 force field was used for the POPC bilayers [30].
PR-Set7
PR-Set7 is a monomethyltransferase that modifies a lysine located in the histone tail (H4K20) [31,32]. The loss of PR-Set7 decreases the H4K20me1 level and results in several cell-cycle defects, including improper DNA replication [33]. Deletion of PR-Set7 is catastrophic to the development of the mouse embryo. PR-Set7 is closely related to human cancer progression, indicating its potential as a target for cancer therapy. As PR-Set7 has been extensively studied, its three-dimensional structure is available in PDB and several inhibitors have already been developed [34].
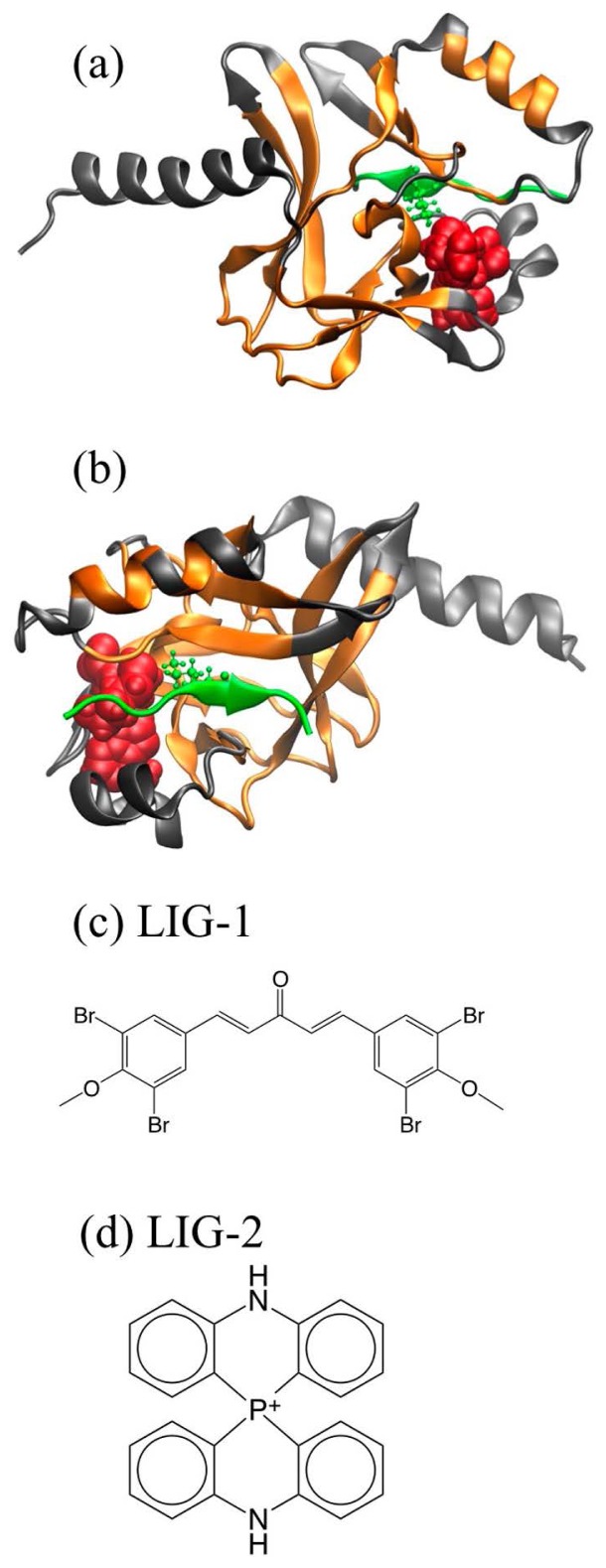
An X-ray crystal structure of PR-Set7 was already solved at a resolution of 0.145 nm (PDB entry: 1ZKK) [19], which was adopted in this study (Fig. 2). In this structure, PR-Set7 binds a histone H4 tail peptide and a second ligand, namely S-adenosylhomocysteine (SAH). In the cell, PR-Set7 binds to the methyl donor S-adenosylmethionine (SAM) and methylates K20H4 and SAM is converted to SAH. In the context of drug design, the structure of the inhibitor-binding PR-Set7 complex is very important. In this study, the binding structures were computationally predicted and examined for two inhibitors: One is a bromo-methoxy-phenyl derivative (compound 5 of Ref. [34]), while the other is a compound developed by Kodama et al. [formula (II) with X=P, R1i=H (i=1…7) in patent: WO2011-010715]. Hereafter, these inhibitors will be denoted as LIG-1 and LIG-2, respectively. Figure 2 shows the structures of the ligands as well as that of the protein. Because LIG-1 selectively binds to PR-Set7, the binding site is expected to be located around the K20-binding site. Also, the development process of LIG-2 indicated that the binding site of LIG-2 would be near the K20-binding site. Therefore, the search area of docking in this study was limited to the vicinity of K20.
Figure 2.
Structures of PR-Set7 and ligands: (a) yellow and gray ribbons are the skeletal core and flexible segments of the PR-Set7, respectively. Red balls represent SAH. Green ribbon is the H4 tail peptide, and the ball-and-stick is K20. (b) Same as in (a), but shown from the opposite direction. (c) LIG-1 and (d) LIG-2.
FRED ver. 3.0.1 [35], a conventional docking program that only refers to the receptor part of the crystal structure, was used to predict the binding structures of these inhibitors at the atomic level. In this case, the H4 tail peptide was significantly larger than the two inhibitors, and the similarities to the inhibitors were very low. Therefore, it becomes ineffective to refer to the binding pose in the co-crystal structure. It was also found that the FRED prediction was slightly better than the POSIT prediction for ligands with low similarity [25]. Here Chemgauss4 was used as an empirical scoring function, and the pose with the best score was selected as the predicted docking pose for each ligand.
To evaluate the docking poses, we performed the MD simulations starting from the docking poses. First, the ligand–protein complex structures were solvated with ca. 14,000 TIP3P water molecules, 42 Na+ ions, and 40 Cl− ions. After energy minimization, the systems were equilibrated by 0.1 ns MD simulations with position restraints of heavy protein and ligand atoms. Then, the restraints were removed, and MD simulations were conducted for 300 ns. The temperature and pressure were controlled at 298 K and 1 atm by the Nose–Hoover (τT=1.0 ps) and isotropic Berendsen (τp=2.0 ps) methods, respectively. The MD time step was set to be 2 fs. The LJ interaction was switched off in the region from 0.8 to 0.9 nm.
Skeletal core and flexible segment
To specify the ligand position with respect to the protein, it is useful to define the protein frame. A simple example is the ligand-root mean square deviation (L-RMSD), which is defined as RMSD calculated after the protein frame is fit to the reference frame. Thus, L-RMSD includes information about the translational and rotational motions of the ligand with respect to the protein and can clearly characterize the binding/dissociation processes. However, if a large part of the protein is flexible, the protein frame can be changed because of the large conformational change of the flexible part. Therefore, a large conformational change can affect L-RMSD analysis; although the ligand is stably bound in the pocket of the protein, the effect of the conformational change on the protein frame also influences L-RMSD value. In this study, because large parts of PR-Set7 are flexible, discussion of this artificial error is essential.
To eliminate this artificial error from the analysis, we introduced the concept of a “skeletal core” using root mean square fluctuation (RMSF) analysis. The set of PR-Set7 residues, for which backbone RMSF values are less than 0.15 nm, is defined as the skeletal core and the remaining part is considered as the flexible segment. Thus, the skeletal core of PR-Set7 consists of G217-D221, G229-I250, A255-K256, R258-E259, L261-Y262, C270-F275, Y282-R288, R292-D311, and P316-Y336. By definition, the skeletal core never undergoes large conformational changes, and the frame of the skeletal core (skeletal frame) is insensitive to motion of the flexible protein.
In case of PR-Set7, the skeletal core consists of 87 residues, while the remaining 74 residues constitute the flexible segment. Thus, 46% of the PR-Set7 residues are the flexible segment, which are capable of considerable movements that affect the protein frame. Therefore, it was worth using the skeletal frame for PR-Set7. Notably, this definition was based on the all-atom MD simulations of PR-Set7 without any ligands (apo form). L-RMSD was calculated after the skeletal core residues were fit to the reference.
In contrast, only 20 residues located in the loop regions and terminal regions were categorized as the flexible segment in the β2AR systems, while the remaining 262 residues formed the skeletal core. Because most of the β2AR residues (93%) were not capable of substantial movement, the protein frame was not greatly affected by conformational changes of the protein. L-RMSD of the protein frame is quite similar to that of the skeletal frame. Therefore, the standard protein frame was used for analysis of β2AR.
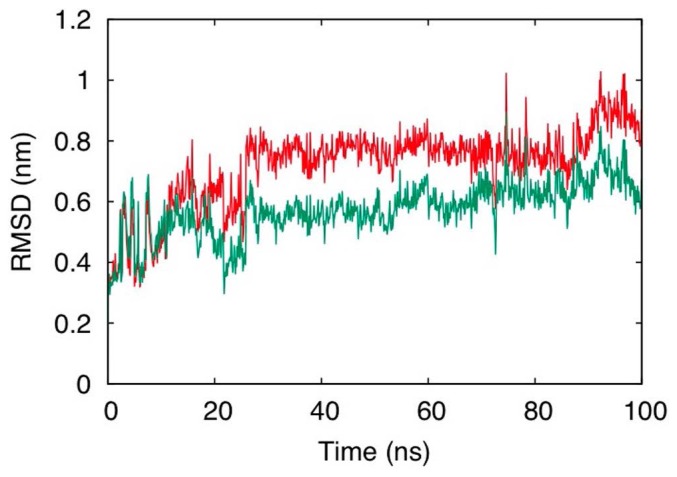
Figure 3 shows two L-RMSD curves calculated for an identical MD trajectory of a LIG-1-binding PR-Set7 system with two different frames. The backbone of the whole protein was used in one case (standard frame) and only the backbone of the skeletal core was used in the other (skeletal frame) to fit the protein structure. In the initial 10 ns, movement of the flexible segment is not substantial, and L-RMSD in the standard frame is almost identical to that in the skeletal frame. At approximately 20 ns, LIG-1 appears to move slightly back to the initial binding region, which decreases L-RMSD of the standard frame to 0.4 nm. In contrast, L-RMSD in the skeletal frame stayed at ca. 0.6 nm, indicating that the ligand does not approach the initial binding region. The motion of the flexible segment greatly changed the frame in the former and gave the illusion that the ligand moved toward the initial position in the standard frame. In this context, the concept of the skeletal core is useful to avoid such a misunderstanding.
Figure 3.
L-RMSDs in the standard frame (green line) and in the skeletal frame (red line); The L-RMSDs were calculated for the LIG-1-binding PR-Set7 systems.
Results and Discussion
Docking structures
β2AR
For β2AR, we investigated the following six ligands: carazolol, ICI 118,551, timolol, epinephrine (adrenaline), salbutamol, and alprenolol. The docking of carazolol can be regarded as a self-docking test because the reference crystal structure was the β2AR–carazolol complex. The docking of this ligand was very successful. L-RMSD between the docking prediction and reference crystal structure is 0.01 nm and the TC score is 1.97 (Table 1). In the energy-minimized structure (the initial structure of the MD simulations), the positively-charged amine group is bound by D113 and N312, for which the short-ranged Coulomb interactions are −199.4 and −67.9 kJ/mol, respectively (Table 2). The two residues have significantly larger electrostatic interactions than the others. Although the LJ interaction of V114 is dominant (−20.7 kJ/mol), F289, F290, and Y199 also have strong LJ interactions with carazolol. This energetic analysis is consistent with the structural features of the co-crystal.
Table 1.
Properties and docking results for β2AR ligands
| Carazolol | ICI 118,551 | Timolol | Salbutamol | Epinephline | Alprenolol | |
|---|---|---|---|---|---|---|
| Effect | Inverse agonist | Inverse agonist | Inverse agonist | Agonist | Agonist | Neutral antagonist |
| MW | 299.3 | 278.4 | 317.4 | 240.3 | 184.2 | 250.3 |
| TC | 1.97 | 1.53 | 1.51 | 0.98 | 0.81 | 1.46 |
Table 2.
Interactions between carazolol and β2AR (kJ/mol)
| Noc | Coulomb | LJ | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Initiala | Finalb | Initiala | Finalb | |||||
|
|
|
|
|
|||||
| Residue | Interaction | Residue | Interaction | Residue | Interaction | Residue | Interaction | |
| 1 | D113 | −199.4 | D113 | −164.3 | V114 | −20.7 | V114 | −22.3 |
| 2 | N312 | −67.9 | N312 | −65.6 | F289 | −14.1 | F289 | −15.6 |
| 3 | S203 | −9.6 | S203 | −19.7 | F290 | −13.5 | F193 | −15.5 |
| 4 | N293 | −10.0 | Y199 | −12.8 | N293 | −10.3 | ||
| 5 | F193 | −11.7 | V117 | −9.5 | ||||
| 6 | N293 | −11.4 | ||||||
| 7 | S203 | −10.1 | ||||||
| 8 | S204 | −9.2 | ||||||
| 9 | T110 | −8.3 | ||||||
Data were taken at t=0. The structure at t=0 is the structure predicted by docking.
Data were averaged over the three trajectories from t=800–1,000 ns.
Ranking residues whose interaction energy are lower than −8 kJ/mol.
The ligands ICI 118,551 and timolol are also inverse agonists with a similar molecular weight (MW) to that of carazolol (Table 1) and a positively-charged amine group (Fig. 1). The structural features of the predicted binding poses are consistent with those obtained by X-ray crystallography experiments [22,23]. Energetic analysis shows that this moiety is bound by D113 and N312 in both systems, as observed in the carazolol-binding system. In the ICI 118,551-binding system, V114, F289, and Y199 have strong LJ interactions, although the LJ interaction of F290 is much weaker than that of the carazolol-binding system (Table 3). In the timolol-binding system, V114, F290, and Y199 have strong LJ interactions as observed in the carazolol-binding system, although the interaction of F289 is slightly weaker (Table 4). The TC values for ICI 118,551 and timolol are therefore very large (1.53 and 1.51, respectively).
Table 3.
Interactions between ICI 118,551 and β2AR (kJ/mol)
| No | Coulomb | LJ | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Initial | Final | Initial | Final | |||||
|
|
|
|
|
|||||
| Residue | Interaction | Residue | Interaction | Residue | Interaction | Residue | Interaction | |
| 1 | D113 | −156.4 | D113 | −166.6 | V114 | −20.7 | V114 | −21.0 |
| 2 | N312 | −67.7 | N312 | −46.8 | F289 | −12.9 | F193 | −17.7 |
| 3 | W109 | −11.9 | F289 | −11.2 | ||||
| 4 | Y199 | −11.0 | F290 | −11.2 | ||||
| 5 | F193 | −11.0 | V117 | −8.8 | ||||
| 6 | T110 | −10.4 | ||||||
| 7 | Y316 | −9.1 | ||||||
| 8 | V117 | −8.0 | ||||||
Details are the same as in Table 2.
Ranking residues whose interaction energy is lower than −8 kJ/mol.
Table 4.
Interactions between timolol and β2AR (kJ/mol)
| No | Coulomb | LJ | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Initial | Final | Initial | Final | |||||
|
|
|
|
|
|||||
| Residue | Interaction | Residue | Interaction | Residue | Interaction | Residue | Interaction | |
| 1 | D113 | −180.0 | D113 | −176.6 | V114 | −22.4 | V114 | −25.7 |
| 2 | N312 | −68.0 | N312 | −68.4 | F290 | −14.9 | F193 | −16.7 |
| 3 | N293 | −12.9 | F289 | −15.3 | ||||
| 4 | S203 | −12.9 | F290 | −13.5 | ||||
| 5 | Y199 | −11.8 | V117 | −11.3 | ||||
| 6 | V117 | −11.5 | Y199 | −8.9 | ||||
| 7 | W109 | −11.5 | N293 | −8.7 | ||||
| 8 | F289 | −10.8 | ||||||
| 9 | S204 | −9.4 | ||||||
| 10 | F193 | −9.2 | ||||||
| T110 | −8.8 | |||||||
Details are the same as in Table 2.
Ranking residues whose interaction energy is lower than −8 kJ/mol.
Epinephrine and salbutamol are agonists; however, both have a conserved, positively-charged amine. Energetic analysis shows that the moiety is bound by D113 and N312, although S203 shows a strong electrostatic interaction with epinephrine (Table 5). One of the most characteristic differences between the inverse agonists and agonists was MW. MWs of agonists are much lower than those of the inverse agonists (Table 1). Therefore, the TC scores are much lower for the agonists than for the inverse agonists. The TC score strongly correlates with MW (R = 0.83 for the six ligands of β2AR). Overall LJ interactions became small compared with those of the inverse agonist systems. Although V114 remained in the dominant role for the epinephrine- and salbutamol-binding systems, many important LJ interactions were missing (Tables 5 and 6).
Table 5.
Interactions between epinephrine and β2AR (kJ/mol)
| No | Coulomb | LJ | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Initial | Final | Initial | Final | |||||
|
|
|
|
|
|||||
| Residue | Interaction | Residue | Interaction | Residue | Interaction | Residue | Interaction | |
| 1 | D113 | −179.0 | D113 | −179.2 | V114 | −9.8 | V114 | −12.9 |
| 2 | S203 | −60.2 | N312 | −32.5 | F193 | −9.3 | F193 | −9.6 |
| 3 | N312 | −58.9 | T110 | −8.5 | F289 | −9.5 | ||
Details are the same as in Table 2.
Ranking residues whose interaction energy is lower than −8 kJ/mol.
Table 6.
Interactions between salbutamol and β2AR (kJ/mol)
| No | Coulomb | LJ | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Initial | Final | Initial | Final | |||||
|
|
|
|
|
|||||
| Residue | Interaction | Residue | Interaction | Residue | Interaction | Residue | Interaction | |
| 1 | D113 | −179.3 | D113 | −174.8 | V114 | −16.4 | V114 | −17.5 |
| 2 | N312 | −61.5 | N312 | −60.2 | W109 | −11.4 | F193 | −16.3 |
| 3 | S207 | −16.8 | Y199 | −9.9 | T110 | −9.8 | F289 | −11.6 |
| 4 | F193 | −9.8 | F290 | −8.7 | ||||
| 5 | F290 | −9.2 | ||||||
| 6 | Y316 | −8.1 | ||||||
Details are the same as in Table 2.
Ranking residues whose interaction energy is lower than −8 kJ/mol.
Alprenolol is a neutral antagonist with a larger MW than those of the agonists but smaller than those of the inverse agonists. Accordingly, the TC score of the neutral antagonist is higher than that of the agonists and lower than that of the inverse agonists (Table 1). The predicted binding pose was similar to that obtained by X-ray crystallography experiments [22]. As observed with the other ligands, the conserved amine group is bound by D113 and N312 (Table 7). Although F193, Y199, and N293 have large LJ interactions, as with carazolol, the LJ interaction of V114 became weak and that of Y308 became more significant. In summary, the docking predictions maintain essential interactions of the carazolol-binding system, but energetic analyses shows that the details of the binding modes are dependent on the ligand species.
Table 7.
Interactions between alprenolol and β2AR (kJ/mol)
| No | Coulomb | LJ | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Initial | Final | Initial | Final | |||||
|
|
|
|
|
|||||
| Residue | Interaction | Residue | Interaction | Residue | Interaction | Residue | Interaction | |
| 1 | D113 | −191.1 | D113 | −145.6 | Y308 | −14.7 | F193 | −17.1 |
| 2 | N312 | −40.0 | N312 | −40.4 | F193 | −14.6 | V114 | −16.8 |
| 3 | Y199 | −13.9 | F290 | −10.3 | ||||
| 4 | N293 | −13.8 | V117 | −8.8 | ||||
| 5 | F289 | −10.8 | F289 | −8.1 | ||||
| 6 | T110 | −10.2 | ||||||
| 7 | F290 | −9.7 | ||||||
| 8 | V114 | −8.8 | ||||||
| 9 | N312 | −8.0 | ||||||
Details are the same as in Table 2.
Ranking residues whose interaction energy is lower than −8 kJ/mol.
PR-Set7
In the crystal structure, PR-Set7 binds to the H4 tail peptide (A15–D24) and SAH, and the ligands (LIG-1 and LIG-2) are designed to inhibit the binding of the H4 tail. However, because the ligands investigated here are significantly smaller than the H4 tail peptide, the energetic analysis is focused on the region from H18 to L22. Note that K20 of the H4 tail is methylated by PR-Set7 in the cell and probably plays an important role in recognizing the H4 tail by PR-Set7 (Table 8). E259 of PR-Set7 has the strongest Coulombic interaction with the partial peptide (H18-L22) because of the existence of a salt bridge between E259 and R21 of the H4 tail. Y274 and Y336 also have large Coulombic interactions with the partial H4 tail peptide and mainly interact with K20 of the H4 tail peptide. Among the LJ interactions, Y274, Y273, and Y336 of PR-Set7 have the three strongest interactions. In particular, Y273 and Y336 primarily interact with K20H4. In contrast, SAH is mainly bound by R228, G299, N298, K226, and Y271 through Coulombic interactions, whereas W349 and K226 are bound through LJ interactions (Supplementary Table S1).
Table 8.
Interactions between H4 tail (H18–L22) and PR-Set7 (kJ/mol)
| No | Coulomb | LJ | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| PR-Set7 residue | H4 tail (H18–L22) | K20H4 | PR-Set7 residue | H4 tail (H18–L22) | K20H4 | |
| 1 | E259 | −117.8 | 0.0 | Y274 | −33.7 | −6.0 |
| 2 | Y274 | −21.9 | −14.5 | Y273 | −31.5 | −17.4 |
| 3 | Y336 | −21.7 | −10.5 | Y336 | −31.4 | −21.5 |
| 4 | C270 | −20.7 | −7.3 | D338 | −21.3 | −2.5 |
| 5 | D338 | −17.1 | +0.1 | H347 | −12.7 | 0.0 |
| 6 | Y245 | −14.0 | −14.0 | F275 | −11.9 | −0.2 |
| 7 | M272 | −11.8 | −8.6 | |||
| 8 | G337 | −10.1 | −1.4 | |||
| 9 | S343 | −10.1 | 0.0 | |||
| 10 | C270 | −8.5 | −4.7 | |||
| 11 | L350 | −8.0 | −0.4 | |||
Ranking residues whose interaction energy is lower than −8 kJ/mol.
The present docking calculation suggests that LIG-1 should be mainly bound by Y273, Y274, and Y336 through the LJ interactions (Table 9). These binding interactions are also involved in binding of K20H4. However, there are no strong electrostatic interactions between LIG-1 and the PR-Set7 residues.
Table 9.
Interactions between LIG-1 and PR-Set7 (kJ/mol)
| Noc | Coulomb | LJ | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Initiala | Finalb | Initiala | Finalb | |||||
|
|
|
|
|
|||||
| Residue | Interaction | Residue | Interaction | Residue | Interaction | Residue | Interaction | |
| 1 | (No Strong Interaction) | (No Strong Interaction) | Y273 | −32.8 | Y274 | −22.2 | ||
| 2 | Y274 | −20.6 | Q276 | −17.3 | ||||
| 3 | Y336 | −17.7 | W349 | −12.8 | ||||
| 4 | M272 | −12.2 | F275 | −10.3 | ||||
| 5 | T307 | −9.0 | D338 | −9.6 | ||||
| 6 | D335 | −8.4 | ||||||
| 7 | C270 | −8.1 | ||||||
Data were taken for the structure at t=0. The structure at t=0 is the structure predicted by docking.
Averaged over three trajectories (250–300 ns).
Ranking residues whose interaction energy is lower than −8 kJ/mol.
It is predicted that LIG-2 should be bound near the K20-binding region. However, while C270, Y336, M272, and Y271 contribute to LIG-2 binding mainly through the LJ interactions, only Y336 shows a significant interaction with K20 of the H4 tail peptide (Table 10). Although C270 and M272 display a moderate interaction with K20H4 through LJ interactions, the interaction with Y271 is negligible. As observed with LIG-1, LIG-2 displays no strong electrostatic interactions in the predicted structure.
Table 10.
Interactions between LIG-2 and PR-Set7 (kJ/mol)
| No | Coulomb | LJ | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Initial | Final | Initial | Final | |||||
|
|
|
|
|
|||||
| Residue | Interaction | Residue | Interaction | Residue | Interaction | Residue | Interaction | |
| 1 | (No Strong Interaction) | D338 | −16.4 | C270 | −22.5 | Y336 | −32.1 | |
| 2 | Y336 | −21.7 | M272 | −19.9 | ||||
| 3 | M272 | −19.7 | Y271 | −12.8 | ||||
| 4 | Y271 | −14.0 | Y273 | −12.7 | ||||
| 5 | Y273 | −9.5 | H347 | −12.5 | ||||
| 6 | D338 | −9.2 | C270 | −12.1 | ||||
| 7 | Y274 | −9.0 | Y274 | −11.6 | ||||
| 8 | D338 | −10.0 | ||||||
Details are the same as in Table 9.
MD trajectories starting from the docking structures
When an MD trajectory starts from an unstable configuration, the system departs from the initial structure and usually migrates to a neighboring energy minimum. In the case of a ligand–protein complex, the ligand is finally located far from the initial position. In a MD simulation starting from a stable energy minimum, the system fluctuates around the initial conformation and the ligand continues to have initial binding modes.
β2AR
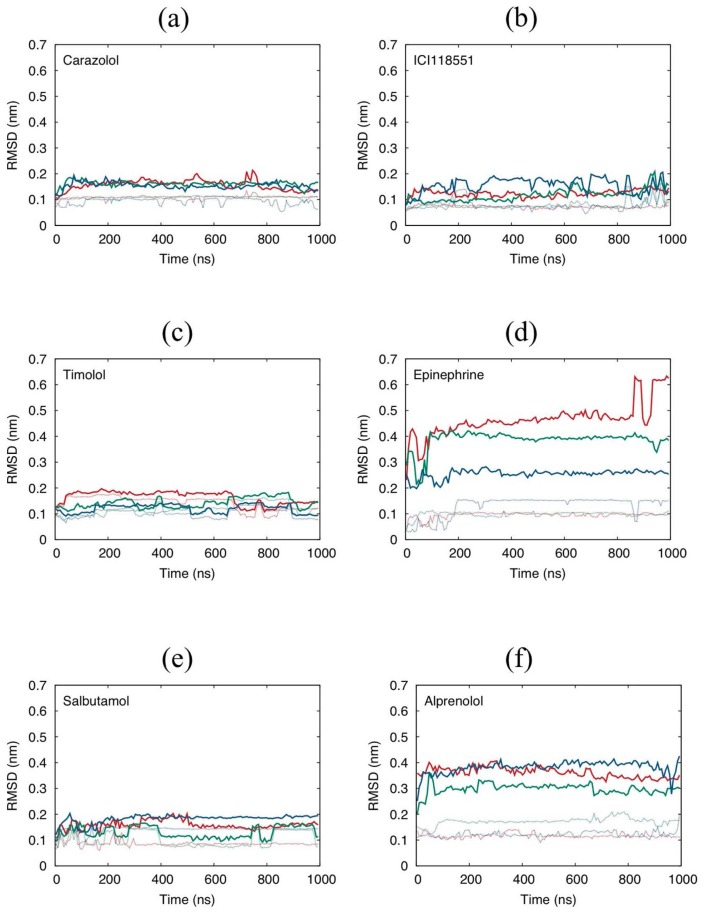
In the case of the inverse agonist-binding β2AR systems, the L-RMSD values are almost always less than 0.2 nm, which is largely attributed to the deviation of the ligand shapes (Fig. 4a–c). The ligands fluctuate around the predicted docking structures throughout the 1,000 ns period. Accordingly, although the ligands maintain strong interactions, some moderate interactions are lost or weakened during the MD simulations (Tables 2–4). These results indicate that the predicted binding poses should be reasonable, although they might be slightly different from the actual structures, mainly because of the thermal fluctuation in the aqueous environment.
Figure 4.
L-RMSDs of β2AR systems calculated for (a) carazolol, (b) ICI118551, (c) timolol, (d) epinephrine, (e) salbutamol, and (f) alprenolol. The dotted lines are the RMSDs of the ligand with fitted to the ligand frame. Red, green, and blue represent the trajectory samples 0, 1, and 2, respectively. The RMSD values are averaged over every 7.5 ns.
Although salbutamol is an agonist with a significantly lower TC score than the inverse agonists, the docking calculation predicted that salbutamol has similar intermolecular interactions (e.g., D113, N312, and V114). In MD simulations, salbutamol fluctuates around the initial structure (Fig. 4e) maintaining the dominant interactions (Table 6). Salbutamol interacts with S207 through electrostatic interactions in the initial conformation. However, this interaction is lost, and an interaction with Y199 is formed in the MD simulations.
Epinephrine is much smaller than the inverse agonists and has a low TC score in the docking calculation. L-RMSD analysis shows that this ligand moves much more freely than the inverse agonists, and the final structures of the 1,000 ns MD simulations differ from one another (Fig. 4d). Although the MD simulations might not have reached a sufficiently equilibrated state, epinephrine continues to display electrostatic interactions with D113 and N312 and LJ interactions with V114 and F193, as observed in the inverse agonist-binding and salbutamol-binding systems (Table 5). S203 initially bound epinephrine through a strong electrostatic interaction, although this interaction was lost releasing the benzene ring moiety.
In contrast to the inverse agonists, the agonists only have a few effective LJ interactions. The Coulombic interactions of the agonists are also much weaker than those of the inverse agonists. These features imply that the agonists bind to inactive β2AR more weakly than the inverse agonists. This finding is consistent with the fact that the inverse agonists stabilized the inactive form of β2AR, but the agonists helped activate β2AR. As β2AR transfers from the inactive to the active form, the interactions between the agonist and β2AR will be enhanced [21].
Alprenolol is a neutral antagonist with a greater MW than those of the agonists. As indicated by the TC score, the binding pose of alprenolol is more similar to that of carazolol than that of the agonists, but less than that of ICI 118,551 and timolol (Table 1). L-RMSD increased rapidly in the MD simulations (Fig. 4f). Accordingly, the LJ interactions are mainly affected, although D113 and N312 did maintain strong Coulombic interactions in the MD simulations. For example, the LJ interactions of Y308 and Y199 are lost, but that of V114 is greatly enhanced (Table 7). This observation was mainly attributed to the large conformational change of alprenolol. Although the vinyl group of alprenolol was initially oriented to the extracellular side (up-conformation), it rotated down to the intracellular side (down-conformation). Because this change was observed in all three MD simulations, we considered that the vinyl group prefers the down-conformation in the thermally fluctuating environment of the cell, although it took the down-conformation in the crystal environment (see also Supplementary Figs. S1–S2).
If the co-crystal structure is available and the investigated ligands are similar to the ligand of the co-crystal structure, the binding modes of the template ligand offer useful information to predict a docking pose. In fact, the correlation factor between the final L-RMSD value and TC was −0.46 for the six β2AR ligands. However, as observed in the alprenolol-binding β2AR system, a high similarity (high TC score) does not always indicate that the docking is entirely reliable.
PR-Set7
LIG-1 and LIG-2 of PR-Set7 are typical examples which display low similarity to the template ligand (the H4 tail peptide), which is much larger than LIG-1 and LIG-2. Thus, for these ligands, we employed the FRED docking program, which does not refer to the pose of the template ligand. As discussed above, the predicted binding interactions of LIG-1 and LIG-2 are also found to be important in the H4 tail peptide-binding system.
The H4 tail peptide is stably bound by PR-Set7 in the MD simulations. Thus, L-RMSD for the partial peptide (H18–L22) is always less than 0.2 nm. Also, L-RMSD of K20 is almost always approximately 0.15 nm (Supplementary Fig. S3). Accordingly, many of the dominant interactions listed in Table 8 are conserved in the MD simulations.
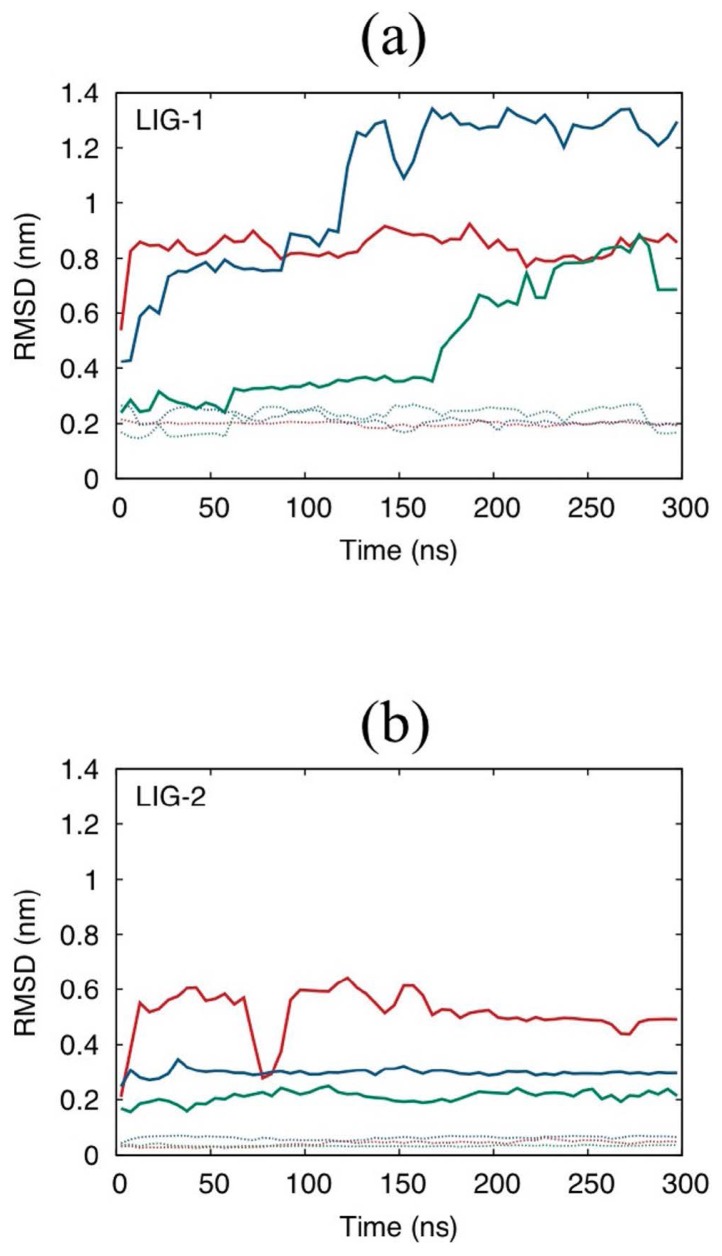
In contrast, LIG-1 moved considerably in the H4 tail-binding cleavage with enhancement of the interaction from Q276. Accordingly, L-RMSD exceeds 0.6 nm in all MD simulations (Fig. 5a). While L-RMSD began to gradually increase from t = 150 ns in one MD simulation, the ligand moved quickly after t=0 in the other MD simulations. Thus, almost all of the important binding interactions in the initial structure are lost in the last 50 ns. Therefore, the predicted binding pose is not sufficiently accurate to discuss the binding interactions at the atomic level. Only the LJ interaction of Y274 continues to be important in the MD simulations (Table 9). Because Y274 contributed to binding K20 in the H4 peptide-binding system, the final binding poses still indicate that LIG-1 can inhibit binding of the H4 tail.
Figure 5.
L-RMSDs of the PR-Set7 systems calculated for (a) LIG-1 and (b) LIG-2. The dotted lines are the RMSDs of the ligand with fitting the ligand frame. Red, green, and blue represent the trajectory samples 0, 1, and 2, respectively. The initial structures are the docking structures predicted for the crystal protein structure. The RMSD values are averaged over every 6 ns.
The L-RMSD values for LIG-2 greatly increases but not to the same extent compared with that of the LIG-1-binding system (Fig. 5b). Therefore, LIG-2 might bind near the predicted pose, although the pose prediction is not sufficiently accurate. In the MD simulations, PR-Set7 conserves some of the dominant interactions with Y336 and M272, which also play important roles in binding of the H4 tail, particularly around K20 (Table 10). Because D338 and H347 are located in the flexible segment, it is found that D338 and H347 approach LIG-2 and form strong interactions in two simulations, while the helix including D338 and H347 is drastically unfolded in the other (see also Supplementary Figs. S4 and S5a and b).
Influence of the target protein structure
Another difference between the PR-Set7 systems and β2AR was the flexibility of the protein structure, as discussed in the Materials and Methods section. If the reference structure of the target protein is changed, a different docking prediction will be obtained. The importance of the target protein structure is also suggested by the fact that the self-docking test is usually more accurate than the cross-docking test. The flexible segment occupied the large part of PR-Set7, and some residues of the flexible segment contributed to binding the H4 tail. D338, H347, G337, S343, and L350, which showed strong interactions with the partial H4 tail peptide, are in the flexible segment, although many of the important interactions are attributed to the skeletal core residues (Table 8).
To investigate the influence of the protein structure on docking prediction, we adopted a PR-Set7 structure equilibrated by a 102 ns MD simulation, where the system consists of a protein, solvated waters, and salts, but no ligands. In this study, the equilibrated structure is referred to as the “apo structure.” The time step was set to 3 fs, and the other conditions of the MD simulation were the same as those explained in the Materials and Methods section.
The docking calculation with FRED shows that the binding site should be located at the outer part of the H4 tail-binding cleavage because the cleavage of the apo structure is significantly narrower than that of the crystal structure (Supplementary Fig. S5). Thus, the energetic analysis suggests that although Y274 has strong LJ interactions with LIG-2, implying that this binding pose can inhibit binding of K20H4 to PR-Set7, many important binding interactions are in fact lost (Supplementary Table S2). Also, the total interaction energy of this docking pose is less than that of the previous subsection in which LIG-2 is docked to the crystal structure. Therefore, we considered that the docking structure for the crystal structure should be more likely than that for the apo structure. However, MD analysis suggests that the former docking structure is not sufficiently stable and LIG-2 moves significantly within the cleavage. It was possible that the MD simulations starting from the latter docking structure lead to a better docking pose. Thus, we conducted seven MD simulations starting from the docking structure for the apo PR-Set7.
Although we expected that LIG-2 should enter the deep region of the cleavage, only one trajectory (sample 3) indicated that LIG-2 should stay near the K20-binding site and maintain interaction with Y274. The L-RMSD analyses for all the other MD trajectories shows that LIG-2 should dissociate from PR-Set7 (Supplementary Fig. S6). In five of the remaining six MD simulations, LIG-2 is again bound near the SAH-binding site after dissociation of the H4 tail binding site (Supplementary Fig. S7). LIG-2 has large interactions with R228 or W349 in the last 50 ns, on average. In particular, the trajectory (sample 5) has strong interactions with R228, K226, Y271, W349, and N298, which are important for binding of SAH, and the total interaction energy was comparable to that of the MD simulation starting from the structure predicted for the crystal structure. This result indicates the possibility that LIG-2 might contribute to the inhibition of SAM-binding, at least to a minor extent, in addition to inhibiting H4 tail binding. In the remaining MD simulation (sample 6), LIG-2 is weakly bound by PR-Set7. Therefore, this binding structure may not be important in the ligand–protein interactions in this system (see also Supplementary Tables S3–S9).
Concluding Remarks
Here we investigated whether the long MD simulation was suitable to assess the binding pose predicted by standard docking programs through demonstrations using two protein systems: β2AR and PR-Set7. In both cases, although MD simulations longer than 100 ns were required to sufficiently equilibrate the systems, MD simulations less than 5 ns were often used in the typical MD assessment [12,13]. The former has a rigid protein structure and the ligands are similar to the template ligand, whereas the latter has large flexible segments and the sizes of the ligands differ from that of the template ligand (the H4 tail peptide).
In case of the inverse agonists of β2AR, molecular similarities to the template ligand (carazolol) were very high. Thus, the predicted poses were quite stable and the MD simulations showed that the ligand never underwent large displacements while maintaining the important interactions. The 3D structure of salbutamol was also similar to that of the template, and the predicted pose was stable in the MD simulations, although the total interaction energy was less than that of the inverse agonist. Compared with salbutamol, epinephrine has a lower similarity to the template and was the smallest β2AR ligand used in this study. In contrast to the inverse agonists, while a few important interactions were maintained, the ligand moved considerably in the binding pocket. As the inactive form was investigated in this study, the agonists may not interact with β2AR particularly well. When the protein is activated, the agonists will interact with β2AR more strongly. Although alprenolol had a high TC score (high similarity), the ligand considerably moved in the MD simulations. In particular, the vinyl moiety rotated downward toward the intracellular region, while it was toward the extracellular region in the docked structure.
The ligands of PR-Set7 are exceedingly different from the template H4 tail peptide. Therefore, accuracy of the pose prediction was decreased and the ligands greatly departed from the predicted docking pose in the MD simulations. However, because some of the key interactions were conserved in the MD simulations, the final complex structures can still contribute to inhibition of H4 tail binding. We may consider that the MD simulation can identify improved binding poses, although free energy calculations are required to conclude that the refined binding pose is correct and compounds with the same scaffold are promising for synthesis. The binding free energy calculated with one of the LIG-1-binding structures refined by the long MD simulations agreed with the experimental binding free energy very closely (unpublished data).
The target protein structure is critical for the success of docking. For this study, we created an apo structure using a MD simulation to investigate the influence of protein structure on the docking calculation of LIG-2 and found that the ligand–protein interactions for the binding structure predicted for the apo structure were much weaker than that for the crystal structure. Also, we observed that the ligand dissociated from the protein in many of the subsequent MD calculations. The results of these analyses suggest that the apo structure is less preferable for docking than the crystal structure. However, it is interesting that after the dissociation, the ligand bound the protein in the vicinity of the SAH-binding domain in five MD simulations. This implies that LIG-2 can affect SAM-binding.
Because protein flexibility, which is not taken into account in the standard docking method, plays an important role, many flexible docking methods [11,36,37] have been proposed, some of which involve MD calculations [11–13] to generate new protein conformations. Although flexible docking methods improve docking predictions to some extent, they were not as successful as expected, probably because of difficulty in assessing the large number of conformations of the protein [38]. Another approach when considering the protein flexibility is the two-step protocol involving MD assessment as the second step, which is the most practical and convenient [11]. In previous studies, MD simulations for checking docked structures, typically shorter than 5 ns, are not sufficiently long to investigate membrane proteins (e.g., β2AR) or highly-flexible proteins (e.g., PR-Set7). In this study, we demonstrated that MD simulations longer than 100 ns can yield useful information about the reliability of docking prediction and are also useful in assessing the predicted pose for these proteins. In addition, we developed a concept of the skeletal core, which provides a better protein frame to describe a ligand dislocation. In future, MD simulation will be more widely used to directly identify binding poses due to the rapid evolution of computational power [39].
Significance.
Computational prediction of the ligand-protein complex structure is a key to success in structure-based drug design. Although many computational (“docking”) methods have been proposed to predict ligand-protein complex structures, accuracy of the prediction is still limited. In this study, we demonstrate that molecular dynamics (MD) simulation can be used to assess binding structures predicted by docking and to provide complementary information. Also, we found that stability of the predicted ligand pose was moderately correlated to ligand similarity.
Supplementary Information
Acknowledgement
This work was supported by grants from HPCI strategic program field 1 (hp120297, hp130006, hp140228, and hp150230) and the FIRST Kodama project. T. Y. also thanks grant-in-aids for Scientific Research (JSPS KAKENHI (S)15H05752 and (C)15KT0103). The authors also wish to acknowledge Prof. T. Kodama for the valuable discussion on PR-Set7, Profs. Kozasa and Suzuki for the helpful discussion about GPCR, and Dr. Sato (OpenEye Scientific Software Inc.) for technical advice regarding the docking calculation.
Footnotes
Conflicts of Interest
T. S., M. I. M., T. Y., and H. F. declare that they have no conflict of interest.
Author Contributions
T. S. and H. F. performed the docking and MD calculations of the PR-Set7 systems. M. I. M. and T. Y. conducted the docking and MD simulations of the β2AR systems. T. Y. and T. S. also developed the concept of the skeletal core. T. Y., T. S., and M. I. M. wrote the manuscript.
References
- 1.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 2.Drews J. Drug discovery: a historical perspective. Science. 2000;287:1960–1964. doi: 10.1126/science.287.5460.1960. [DOI] [PubMed] [Google Scholar]
- 3.Macarron R, Banks MN, Bojanic D, Burns DJ, Cirovic DA, Garyantes T, et al. Impact of high-throughput screening in biomedical research. Nat Rev Drug Discov. 2011;10:188–195. doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 4.Taft CA, Da Silva VB, Tomich De Paula Da Silva CH. Current topics in computer-aided drug design. J Pharm Sci. 2008;97:1089–1098. doi: 10.1002/jps.21293. [DOI] [PubMed] [Google Scholar]
- 5.Cheng T, Li Q, Zhou Z, Wang Y, Bryant SH. Structure-based virtual screening for drug discovery: a problem-centric review. AAPS J. 2012;14:133–141. doi: 10.1208/s12248-012-9322-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyne PD. Structure-based virtual screening: an overview. Drug Discov Today. 2002;7:1047–1055. doi: 10.1016/s1359-6446(02)02483-2. [DOI] [PubMed] [Google Scholar]
- 7.Warren GL, Andrews CW, Capelli A-M, Clarke B, LaLonde J, Lambert MH, et al. A critical assessment of docking programs and scoring functions. J Med Chem. 2006;49:5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 8.Yamashita T, Ueda A, Mitsui T, Tomonaga A, Matsumoto S, Kodama T, et al. Molecular dynamics simulation-based evaluation of the binding free energies of computationally designed drug candidates: importance of the dynamical effects. Chem Pharm Bull. 2014;62:661–667. doi: 10.1248/cpb.c14-00132. [DOI] [PubMed] [Google Scholar]
- 9.Yamashita T, Ueda A, Mitsui T, Tomonaga A, Matsumoto S, Kodama T, et al. The feasibility of an efficient drug design method with high-performance computers. Chem Pharm Bull. 2015;63:147–155. doi: 10.1248/cpb.c14-00596. [DOI] [PubMed] [Google Scholar]
- 10.Fujitani H, Tanida Y, Ito M, Jayachandran G, Snow CD, Shirts MR, et al. Direct calculation of the binding free energies of FKBP ligands. J Chem Phys. 2005;123:084108. doi: 10.1063/1.1999637. [DOI] [PubMed] [Google Scholar]
- 11.Alonso H, Bliznyuk AA, Gready JE. Combining docking and molecular dynamic simulations in drug design. Med Res Rev. 2006;26:531–568. doi: 10.1002/med.20067. [DOI] [PubMed] [Google Scholar]
- 12.Cavalli A, Bottegoni G, Raco C, De Vivo M, Recanatini M. A computational study of the binding of propidium to the peripheral anionic site of human acetylcholinesterase. J Med Chem. 2004;47:3991–3999. doi: 10.1021/jm040787u. [DOI] [PubMed] [Google Scholar]
- 13.Park H, Yeom MS, Lee S. Loop flexibility and solvent dynamics as determinants for the selective inhibition of cyclin-dependent kinase 4: comparative molecular dynamics simulation studies of CDK2 and CDK4. ChemBioChem. 2004;5:1662–1672. doi: 10.1002/cbic.200400214. [DOI] [PubMed] [Google Scholar]
- 14.Yamashita T, Voth GA. Insights into the mechanism of proton yransport in cytochrome c oxidase. J Am Chem Soc. 2012;134:1147–1152. doi: 10.1021/ja209176e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindorff-Larsen K, Piana S, Dror RO, Shaw DE. How Fast-Folding Proteins Fold. Science. 2011;334:517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- 16.Fujitani H, Matsuura A, Sakai S, Sato H, Tanida Y. High-level ab initio calculations to improve protein backbone dihedral parameters. J Chem Theory Comput. 2009;5:1155–1165. doi: 10.1021/ct8005437. [DOI] [PubMed] [Google Scholar]
- 17.Yamashita T. Improvement in empirical potential functions for increasing the utility of molecular dynamics simulations. JPS Conf Proc. 2015;5:010003. [Google Scholar]
- 18.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Couture JF, Collazo E, Brunzelle JS, Trievel RC. Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase. Genes Dev. 2005;19:1455–1465. doi: 10.1101/gad.1318405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Talan MI, Ahmet I, Xiao R-P, Lakatta EG. β2 AR agonists in treatment of chronic heart failure: long path to translation. J Mol Cell Cardiol. 2011;51:529–533. doi: 10.1016/j.yjmcc.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasmussen SGF, Choi H-J, Fung JJ, Pardon E, Casarosa P, Chae PS, et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wacker D, Fenalti G, Brown MA, Katritch V, Abagyan R, Cherezov V, et al. Conserved binding mode of human β2 adrenergic receptor inverse agonists and antagonist revealed by x-ray crystallography. J Am Chem Soc. 2010;132:11443–11445. doi: 10.1021/ja105108q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola V-P, Chien EYT, et al. A specific cholesterol binding site is established by the 2.8 Å structure of the human β2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 25.Kelley BP, Brown SP, Warren GL, Muchmore SW. POSIT: flexible shape-guided docking for pose prediction. J Chem Inf Model. 2015;55:1771–1780. doi: 10.1021/acs.jcim.5b00142. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt TH, Kandt C. LAMBADA and InflateGRO2: efficient membrane alignment and insertion of membrane proteins for molecular dynamics simulations. J Chem Inf Model. 2012;52:2657–2669. doi: 10.1021/ci3000453. [DOI] [PubMed] [Google Scholar]
- 27.Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujitani H, Tanida Y, Matsuura A. Massively parallel computation of absolute binding free energy with well-equilibrated states. Phys Rev E. 2009;79:021914. doi: 10.1103/PhysRevE.79.021914. [DOI] [PubMed] [Google Scholar]
- 29.Gaussian 09. Gaussian, Inc; Wallingford, CT, USA: 2009. [Google Scholar]
- 30.Dickson CJ, Madej BD, Skjevik AA, Betz RM, Teigen K, Gould IR, et al. Lipid14: the amber lipid force field. J Chem Theory Comput. 2014;10:865–879. doi: 10.1021/ct4010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jorgensen S, Schotta G, Sorensen CS. Histone H4 Lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 2013;41:2797–2806. doi: 10.1093/nar/gkt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beck DB, Oda H, Shen SS, Reinberg D. PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev. 2012;26:325–337. doi: 10.1101/gad.177444.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brustel J, Tardat M, Kirsh O, Grimaud C, Julien E. Coupling mitosis to DNA replication: the emerging role of the histone H4-lysine 20 methyltransferase PR-Set7. Trends Cell Biol. 2011;21:452–460. doi: 10.1016/j.tcb.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 34.Valente S, Lepore I, Dell-Aversana C, Tardugno M, Castellano S, Sbardella G, et al. Identification of PR-SET7 and EZH2 selective inhibitors inducing cell death in human leukemia U937 cells. Biochimie. 2012;94:2308–2313. doi: 10.1016/j.biochi.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 35.McGann M. FRED pose prediction and virtual screening accuracy. J Chem Inf Model. 2011;51:578–596. doi: 10.1021/ci100436p. [DOI] [PubMed] [Google Scholar]
- 36.Davis IW, Baker D. ROSETTALIGAND docking with full ligand and receptor flexibility. J Mol Biol. 2009;385:381–392. doi: 10.1016/j.jmb.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 37.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comp Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamashita T, Fujitani H. On accurate calculation of the potential of mean force between antigen and antibody: a case of the HyHEL-10-hen egg white lysozyme system. Chem Phys Lett. 2014;609:50–53. [Google Scholar]
- 39.Shan Y, Kim ET, Eastwood MP, Dror RO, Seeliger MA, Shaw DE. How does a drug molecule find its target binding site? J Am Chem Soc. 2011;133:9181–9183. doi: 10.1021/ja202726y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.