Abstract
The Escherichia coli chaperonin GroEL is an essential molecular chaperone that mediates protein folding in association with its cofactor, GroES. It is widely accepted that GroEL alternates the GroES-sealed folding-active rings during the reaction cycle. In other words, an asymmetric GroEL–GroES complex is formed during the cycle, whereas a symmetric GroEL–(GroES)2 complex is not formed. However, this conventional view has been challenged by the recent reports indicating that such symmetric complexes can be formed in the GroEL–GroES reaction cycle. In this review, we discuss the studies of the symmetric GroEL–(GroES)2 complex, focusing on the molecular mechanism underlying its formation. We also suggest that GroEL can be involved in two types of reaction cycles (asymmetric or symmetric) and the type of cycle used depends on the concentration of non-native substrate proteins.
Keywords: molecular chaperone, protein folding, protein–protein interaction
Chaperonin GroEL
Protein folding is assisted by a number of molecular chaperones in vivo [1,2]. Chaperonins, a ubiquitous class of molecular chaperones, form double-ring complexes that mediate the folding of nascent and denatured proteins (non-native substrate proteins) in an ATP-dependent manner. There are two distinct groups of chaperonins; group I chaperonins, found in bacteria and within mitochondria and chloroplasts, originating from endosymbiotic bacteria and group II chaperonins, in archaea and the eukaryotic cytoplasm [3–5]. The best characterized chaperonin is the E. coli chaperonin, GroEL, which is associated with its cofactor, GroES. The GroEL–GroES system is the only chaperone in E. coli that is indispensable for growth at all temperatures [6]. Proteomic studies demonstrate that the system is essential for the productive folding of ~80 E. coli proteins [7,8] and assists in the folding of an even larger number of proteins [9,10].
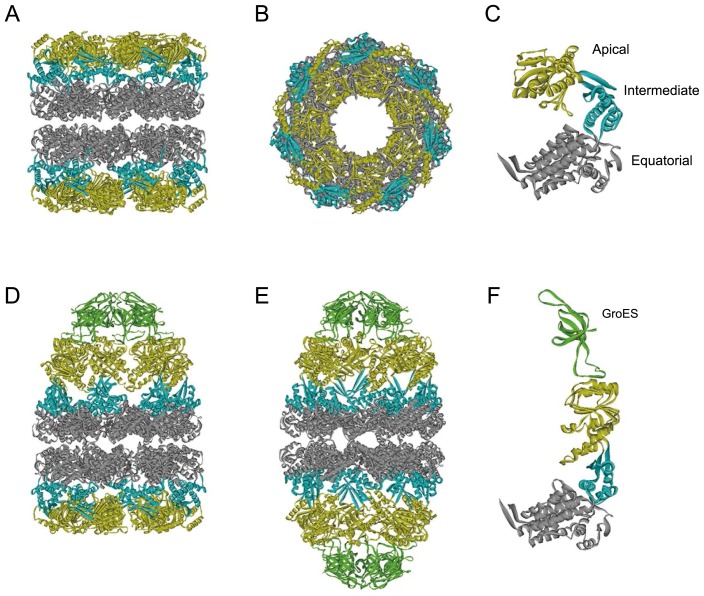
GroEL is a tetradecamer of 57 kDa subunits, arranged as two stacked, seven-member rings, each containing a large cavity (Fig. 1A and B). Each subunit is comprised of three domains: the apical, intermediate, and equatorial domain (Fig. 1C). The apical domain is involved in the binding to non-native substrate proteins and GroES (Fig. 1D and F). The equatorial domain contains the ATP-binding site and is involved in intra- and inter-ring interactions. The intermediate domain connects the equatorial and apical domains of each subunit and transfers the ATP-induced conformational changes from the equatorial to the apical domain [11,12]. GroES is arranged as a dome-shaped single heptameric ring composed of 10-kDa subunits. It caps one or both the ends of the GroEL cavities, forming chamber(s), in which non-native substrate proteins are encapsulated for folding (Fig. 1D–F). The chamber can accommodate proteins of up to 60 kDa [13].
Figure 1.
Crystal structures of GroEL and GroEL-GroES complexes.
(A–C) Side (A) and top (B) views of GroEL tetradecamer and its subunit structure (C) [Protein Data Bank (PDB) code: 1GRL] [63]. The apical, intermediate, and equatorial domains are in yellow, blue, and gray, respectively. (D, E) Side views of an asymmetric GroEL–GroES complex (PDB code: 1AON) [12] and a symmetric GroEL–(GroES)2 complex (PDB code: 3WVL) [50]. (F) Subunit structure of GroEL in the GroES-bound ring (PDB code: 1AON). The GroES subunit is shown in green.
Accepted model for the GroEL–GroES reaction cycle
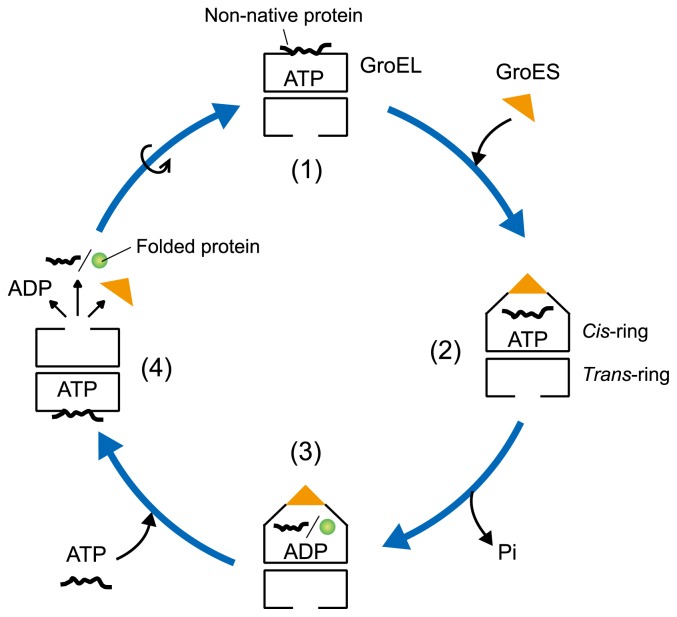
The widely accepted model for the GroEL–GroES reaction cycle is shown in Figure 2. First, one of the GroEL rings captures a non-native substrate protein via its hydrophobic sites, and GroES binds to the same ring (the cis-ring) in an ATP-dependent manner (Fig. 1D and F). GroES binding induces the displacement of the captured protein into the GroEL cavity, where the productive folding proceeds. Next, the hydrolysis of ATP in the cis-ring is followed by ATP binding to the opposite ring (the trans-ring). This results in the dissolution of the cis-ring, thereby releasing GroES and a (partially) folded protein. At the same time, the second GroES binds to the trans-ring to reorient a new cis-ring and starts the next ATPase cycle [14,15]. Because GroES binds alternatively to each ring of GroEL (two-stroke model), an asymmetric GroEL–GroES complex (also called the bullet-shaped complex; Fig. 1D), in which one GroES is bound to one end of GroEL, exists throughout the reaction cycle. In contrast, a symmetric GroEL–(GroES)2 complex (also known as the football-shaped complex; Fig. 1E), in which two GroES molecules simultaneously cap both ends of GroEL, is not formed. The origin of the GroEL–GroES interaction cycle has been explained by the conformational changes of GroEL. These changes are reflected in the binding and hydrolysis of ATP with positive intra-ring cooperativity and negative inter-ring cooperativity [16,17].
Figure 2.
Schematic model for the GroEL–GroES reaction cycle using an asymmetric complex.
In contrast, the symmetric complex has been identified using electron microscopic examination [18–26], chemical cross-linking [27,28], analytical ultracentrifugation [29], and fluorescence-based detection [30,31]. Taguchi et al. [32] found that the symmetric complex is formed when the GroEL ATPase cycle is stopped by beryllofluoride (BeFx), a structural analog of inorganic phosphate. However, the symmetric complex has been considered as a non-significant complex formed under non-physiological conditions or an unproductive dead-end complex. The view that the functional chaperonin complex is asymmetric has been widely accepted [14,15,33].
The symmetric complex can be formed during the GroEL–GroES reaction cycle
We previously examined the GroEL–GroES interaction in the reaction cycle and found that the cycle is significantly affected by the presence of non-native substrate proteins [34–36]. In the presence of these proteins, we then attempted to monitor the GroEL–GroES interaction using Förster resonance energy transfer, without stopping the reaction. As a result, we found that the symmetric and asymmetric complexes coexist in the presence of non-native substrate proteins and that the formation of the symmetric complex is promoted by increasing the concentration of these proteins [37,38]. On the other hand, in the absence of non-native substrate protein, the symmetric complex is not formed [38]. Our findings have been confirmed by other research groups in similar experimental systems [39–41]. The available results indicate that the symmetric complex appears as an intermediate in the presence of sufficient amounts of non-native substrate protein. It is likely that the symmetric complex has been overlooked because most of the previous studies have been performed either with or without small amounts of non-native substrate protein.
Based on a previous report [32], we assumed that the second GroES can associate with the trans-ring of the ATPbound asymmetric complex. We then performed similar experiments using an ATPase-defective mutant (GroELD398A). GroELD398A undergoes a conformational change, in the manner of wild-type GroEL, to bind GroES upon the binding of ATP, although the ATPase activity in this mutant is significantly reduced (~2% of the wild-type level) [14]. We found that GroELD398A forms the symmetric complex when both rings are occupied with ATP [37]. At the same time, Koike-Takeshita et al. [42] also demonstrated that GroELD398A forms a symmetric complex in the presence of ATP and GroES. These findings are surprising; the accepted model (Fig. 2) assumes that GroELD398A forms an asymmetric complex in the presence of ATP and GroES, and the ATP-bound complex cannot bind non-native substrate protein and GroES to the trans-ring [15]. Thus, the accepted model has been challenged [4,5,43].
The symmetric complex is a functional intermediate
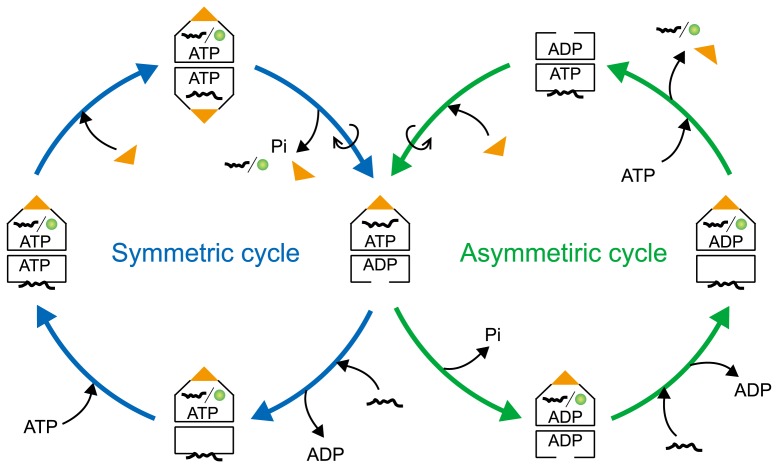
We then attempted to probe the GroEL–GroES interaction cycle via the symmetric complex using a single-molecule assay [44]. The assay allows the direct observation and characterization of the symmetric complexes during the reaction cycle. We found that the first GroES that interacts with GroEL does not always dissociate from the symmetric complex before the dissociation of the second GroES molecule, i.e. the dissociation of GroES molecules from this complex can occur in a random order. It is likely that GroES dissociates from the GroEL ring, in which ATP hydrolysis occurs. We also found that GroEL exited in three different states: as GroEL alone, as the asymmetric complex, and as the symmetric complex. This finding indicates the existence of two reaction cycles in the GroEL–GroES interaction: an asymmetric cycle (GroEL ↔ asymmetric complex) and a symmetric cycle (asymmetric complex ↔ symmetric complex) (Fig. 3; the details will be discussed below).
Figure 3.
Schematic model for the GroEL–GroES reaction cycle using asymmetric and symmetric complexes. GroEL mainly goes through the asymmetric cycle in the presence of a low concentration of non-native substrate protein. At high concentrations of non-native substrate protein, GroEL passes through the symmetric cycle.
Several studies have suggested that non-native substrate proteins can be encapsulated and folded in both rings of GroEL at the same time [24,25,32,42]. To understand how the protein folding proceeds in the symmetric complex, we previously visualized protein folding in this complex, employing a single-molecule assay [45]. We demonstrated that both rings in the symmetric complex actively assist in the refolding of GFP molecules. Furthermore, the kinetics of GFP refolding in each ring is in excellent agreement with that in the asymmetric complex. In other words, the same reactions occur in both rings of the symmetric complex. As the dissociation of GroES molecules from the symmetric complex can occur in a random order [44], the two rings might operate as parallel processing machines, indicating the lack of inter-ring communication in this complex.
Structure of the symmetric complex
The symmetric complex can be formed when both rings of GroEL are occupied with ATP [32,37,42]. Nojima and Yoshida [46] found that GroEL can adopt a conformation in which the two rings open in the presence of ATP. This finding indicates that the binding of ATP to both rings of GroEL results in the formation of the symmetric complex. Koike-Takeshita et al. [47] indicated that ATPγS, a non-hydrolyzable ATP analog, was relatively efficient, whereas AMP-PNP, another non-hydrolyzable ATP analog, was not effective in the formation of the symmetric complex. It is considered that ATP plays a unique role in the induction of structural rearrangements of GroEL, as has been suggested previously [14,48].
Recently, three groups determined the structure of the symmetric complex at atomic resolution [49–51]. As expected, the structure has an American football-like shape, showing that two GroES molecules bind to the two rings of GroEL (Fig. 1E). Fei et al. [49] determined the crystal structures of the symmetric complexes formed in the presence of ATP and BeFx. These complexes contain 14 ATP-analog (ADP-BeFx) molecules at the nucleotide-binding sites, with no significant negative cooperativity between the two rings. Koike-Takeshita et al. [50] determined the structure of the symmetric complex using an ATPase-deficient GroEL mutant (GroELD52A/D398A, with the activity <0.01% of the wild type). This mutant protein forms an extremely stable symmetric complex with a half-life of ~6 days [47]. In the crystal structure, the 14 nucleotide-binding sites are occupied by ATP. Importantly, in both symmetric complexes, the interactions between the two GroEL rings are reduced from those in the asymmetric complex. This reduction in the interactions can be attributed to the impaired inter-ring negative cooperativity in the symmetric complex. The importance of inter-ring interactions in the allosteric mechanism of GroEL has been confirmed using site-specific mutagenesis [52–56]. It has been also reported that a mutant GroEL (GroELE461K), with rearranged inter-ring electrostatic contacts and decreased negative cooperativity between the rings, forms the symmetric complex more easily than the wild-type molecule forms [56]. Nisemblat et al. [51] reported the crystal structure of the symmetric complex of human mitochondrial chaperonin, with a mutation (mHsp60E321K), stabilizing the complex [57]. As mHsp60E321K and cochaperonin (mHsp10) were mixed together with ATP and subjected to crystallization, mHsp60E321K hydrolyzed the ATP in the crystallization drop, resulting in the structure containing 14 ADP molecules. On the basis of these results, it is likely that group I chaperonins share the inherent “functional symmetry.”
Mechanism of the formation of the symmetric complex
What is the mechanism by which non-native substrate protein promotes the formation of the symmetric complex? The most probable explanation is that non-native substrate protein facilitates the dissociation of ADP from the transring of GroEL, leading to the association of ATP and the second GroES with this ring.
Kinetic studies have revealed that ADP remains in the GroEL ring even after GroES has been detached [58,59]. We have shown that ADP prevents the association of ATP with the trans-ring of GroEL and strongly inhibits the association of the second GroES [37,38]. These findings indicate that GroES cannot associate with the trans-ring of GroEL until ADP dissociates from this ring. Lorimer et al. demonstrated that the association of non-native substrate protein with the trans-ring promotes ADP/ATP exchange [39,59,60]. We also found that non-native substrate protein accelerates the association of the second GroES with the trans-ring of GroEL in the presence of ADP; this association is significantly reduced in the absence of non-native substrate protein [38]. On the basis of the available data, we propose a mechanism by which the non-native substrate protein promotes the symmetric complex formation (Fig. 3). ATP hydrolysis in one of the rings of GroEL results in the dissociation of GroES because the loss of the γ-phosphate decreases the affinity between GroEL and GroES [61] (Fig. 3, Symmetric cycle, right). Subsequently, the association of non-native substrate protein dissociates ADP from the trans-ring of GroEL (Fig. 3, Symmetric cycle, lower), leading to the association of ATP and the second GroES with this ring (Fig. 3, Symmetric cycle, left and upper). This series reaction can be enhanced by increasing the concentration of non-native substrate protein (the concentration depends on its affinity for GroEL) [38,40]. In the resultant GroEL–GroES complex (Fig. 3, Symmetric cycle, upper), the cis-ring and the trans-ring become indistinguishable. The previously published model of inter-ring negative cooperativity is correct but incomplete in that it does not apply to high concentrations of non-native substrate protein.
Model for the GroEL–GroES reaction cycle with the symmetric complex and the physiological significance of this complex
Based on our findings [37,38,44,45], we propose a schematic model of the GroEL–GroES reaction cycle, shown in Figure 3. The model consists of two cycles: the “asymmetric cycle” and the “symmetric cycle.” In the presence of non-native substrate protein at a low concentration, GroEL mainly goes through the asymmetric cycle due to the inhibitory effect of ADP in the trans-ring. However, a high concentration of non-native substrate protein causes a switch to the symmetric cycle as the protein weakens the inhibitory effect of ADP and facilitates the formation of the symmetric complex. Our model does not contradict the accepted model (Fig. 2), but rather shows that the GroEL–GroES system can work in a different mode in the presence of high concentrations of non-native substrate protein.
As indicated in previous reports [23,24,27], the symmetric complex is the advantageous form for protein folding because both cavities in the complex actively assist the process. Therefore, we expect the symmetric complex to be active when the amount of non-native substrate protein increases, and GroEL prevents the accumulation of these proteins in E. coli, e.g., under stress conditions. Interestingly, at elevated temperatures, the negative cooperativity between the two GroEL rings appears to be decreased [62]. This would also facilitate the formation of the symmetric complex. We also found that the ATPase activity of GroEL is higher when the levels of the symmetric complex increase [38]. GroEL does not have to employ the maximum ATPase activity in the presence of a small amount of non-native substrate protein. In other words, GroEL does not form symmetric complexes at low levels of non-native substrate protein, thus preventing unnecessary ATP consumption. These might be the reasons why GroEL functions as a double-ring structure. The next challenge is to find the direct evidence that the symmetric complexes are formed in E. coli, leading to the understanding their physiological significance.
Acknowledgements
The authors would like to thank all collaborators who contributed to the work described here. This study was partly supported by Grants-in-Aid for Scientific Research (B) (15H04358 to TF) and Scientific Research on Innovative Areas (15H01527 to TF) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. This study was also supported by the Center of Innovation Program from Japan Science and Technology Agency and by a grant from the Mitsubishi Foundation.
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest associated with this manuscript.
Ethics Standard
The authors declare that this manuscript is original and unpublished and is not currently being considered for publication elsewhere. This manuscript has been read and approved by the authors.
Author Contribution
R. I. and T. F. co-wrote the manuscript.
References
- 1.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 2.Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol. 2013;14:630–642. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horwich AL, Fenton WA, Chapman E, Farr GW. Two families of chaperonin: physiology and mechanism. Annu Rev Cell Dev Biol. 2007;23:115–145. doi: 10.1146/annurev.cellbio.23.090506.123555. [DOI] [PubMed] [Google Scholar]
- 4.Skjærven L, Cuellar J, Martinez A, Valpuesta JM. Dynamics, flexibility, and allostery in molecular chaperonins. FEBS Lett. 2015;589:2522–2532. doi: 10.1016/j.febslet.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 5.Hayer-Hartl M, Bracher A, Hartl FU. The GroEL-GroES chaperonin machine: a nano-cage for protein folding. Trends Biochem Sci. 2016;41:62–76. doi: 10.1016/j.tibs.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Fayet O, Ziegelhoffer T, Georgopoulos C. The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. J Bacteriol. 1989;171:1379–1385. doi: 10.1128/jb.171.3.1379-1385.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujiwara K, Ishihama Y, Nakahigashi K, Soga T, Taguchi H. A systematic survey of in vivo obligate chaperonin-dependent substrates. EMBO J. 2010;29:1552–1564. doi: 10.1038/emboj.2010.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niwa T, Fujiwara K, Taguchi H. Identification of novel in vivo obligate GroEL/ES substrates based on data from a cell-free proteomics approach. FEBS Lett. 2016;590:251–257. doi: 10.1002/1873-3468.12036. [DOI] [PubMed] [Google Scholar]
- 9.Houry WA, Frishman D, Eckerskorn C, Lottspeich F, Hartl FU. Identification of in vivo substrates of the chaperonin GroEL. Nature. 1999;402:147–154. doi: 10.1038/45977. [DOI] [PubMed] [Google Scholar]
- 10.Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang HC, Stines AP, et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 11.Braig K, Otwinowski Z, Hegde R, Boisvert DC, Joachimiak A, Horwich AL, et al. The crystal structure of the bacterial chaperonin GroEL at 2.8 Å. Nature. 1994;371:578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- 12.Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 13.Sakikawa C, Taguchi H, Makino Y, Yoshida M. On the maximum size of proteins to stay and fold in the cavity of GroEL underneath GroES. J Biol Chem. 1999;274:21251–21256. doi: 10.1074/jbc.274.30.21251. [DOI] [PubMed] [Google Scholar]
- 14.Rye HS, Burston SG, Fenton WA, Beechem JM, Xu Z, Sigler PB, et al. Distinct actions of cis and trans ATP within the double ring of the chaperonin GroEL. Nature. 1997;388:792–798. doi: 10.1038/42047. [DOI] [PubMed] [Google Scholar]
- 15.Rye HS, Roseman AM, Chen S, Furtak K, Fenton WA, Saibil HR, et al. GroEL-GroES cycling: ATP and nonnative polypeptide direct alternation of folding-active rings. Cell. 1999;97:325–338. doi: 10.1016/s0092-8674(00)80742-4. [DOI] [PubMed] [Google Scholar]
- 16.Horovitz A, Willison KR. Allosteric regulation of chaperonins. Curr Opin Struct Biol. 2005;15:646–651. doi: 10.1016/j.sbi.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Saibil HR, Fenton WA, Clare DK, Horwich AL. Structure and allostery of the chaperonin GroEL. J Mol Biol. 2013;425:1476–1487. doi: 10.1016/j.jmb.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 18.Llorca O, Marco S, Carrascosa JL, Valpuesta JM. The formation of symmetrical GroEL-GroES complexes in the presence of ATP. FEBS Lett. 1994;345:181–186. doi: 10.1016/0014-5793(94)00432-3. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt M, Rutkat K, Rachel R, Pfeifer G, Jaenicke R, Viitanen P, et al. Symmetric complexes of GroE chaperonins as part of the functional cycle. Science. 1994;265:656–659. doi: 10.1126/science.7913554. [DOI] [PubMed] [Google Scholar]
- 20.Engel A, Hayer-Hartl MK, Goldie KN, Pfeifer G, Hegerl R, Müller S, et al. Functional significance of symmetrical versus asymmetrical GroEL-GroES chaperonin complexes. Science. 1995;269:832–836. doi: 10.1126/science.7638600. [DOI] [PubMed] [Google Scholar]
- 21.Llorca O, Carrascosa JL, Valpuesta JM. Biochemical characterization of symmetric GroEL-GroES complexes: Evidence for a role in protein folding. J Biol Chem. 1996;271:68–76. doi: 10.1074/jbc.271.1.68. [DOI] [PubMed] [Google Scholar]
- 22.Llorca O, Marco S, Carrascosa JL, Valpuesta JM. Conformational changes in the GroEL oligomer during the functional cycle. J Struct Biol. 1997;118:31–42. doi: 10.1006/jsbi.1996.3832. [DOI] [PubMed] [Google Scholar]
- 23.Llorca O, Marco S, Carrascosa JL, Valpuesta JM. Symmetric GroEL-GroES complexes can contain substrate simultaneously in both GroEL rings. FEBS Lett. 1997;405:195–199. doi: 10.1016/s0014-5793(97)00186-5. [DOI] [PubMed] [Google Scholar]
- 24.Sparrer H, Rutkat K, Buchner J. Catalysis of protein folding by symmetric chaperone complexes. Proc Natl Acad Sci USA. 1997;94:1096–1100. doi: 10.1073/pnas.94.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beißinger M, Rutkat K, Buchner J. Catalysis, commitment and encapsulation during GroE-mediated folding. J Mol Biol. 1999;289:1075–1092. doi: 10.1006/jmbi.1999.2780. [DOI] [PubMed] [Google Scholar]
- 26.Ranson NA, Clare DK, Farr GW, Houldershaw D, Horwich AL, Saibil HR. Allosteric signalling of ATP hydrolysis in GroEL-GroES complexes. Nat Struct Mol Biol. 2006;13:147–152. doi: 10.1038/nsmb1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Azem A, Kessel M, Goloubinoff P. Characterization of a functional GroEL14(GroES7)2 chaperonin hetero-oligomer. Science. 1994;265:653–656. doi: 10.1126/science.7913553. [DOI] [PubMed] [Google Scholar]
- 28.Azem A, Diamant S, Kessel M, Weiss C, Goloubinoff P. The protein-folding activity of chaperonins correlates with the symmetric GroEL14(GroES7)2 heterooligomer. Proc Natl Acad Sci USA. 1995;92:12021–12025. doi: 10.1073/pnas.92.26.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Behlke J, Ristau O, Schönfeld HJ. Nucleotide-dependent complex formation between the Escherichia coli chaperonins GroEL and GroES studied under equilibrium conditions. Biochemistry. 1997;36:5149–5156. doi: 10.1021/bi962755h. [DOI] [PubMed] [Google Scholar]
- 30.Török Z, Vigh L, Goloubinoff P. Fluorescence detection of symmetric GroEL14(GroES7)2 heterooligomers involved in protein release during the chaperonin cycle. J Biol Chem. 1996;271:16180–16186. doi: 10.1074/jbc.271.27.16180. [DOI] [PubMed] [Google Scholar]
- 31.Gorovits BM, Ybarra J, Seale JW, Horowitz PM. Conditions for nucleotide-dependent GroES-GroEL interactions: GroEL14(GroES7)2 is favored by an asymmetric distribution of nucleotides. J Biol Chem. 1997;272:26999–27004. doi: 10.1074/jbc.272.43.26999. [DOI] [PubMed] [Google Scholar]
- 32.Taguchi H, Tsukuda K, Motojima F, Koike-Takeshita A, Yoshida M. BeFx stops the chaperonin cycle of GroEL-GroES and generates a complex with double folding chambers. J Biol Chem. 2004;279:45737–45743. doi: 10.1074/jbc.M406795200. [DOI] [PubMed] [Google Scholar]
- 33.Hayer-Hartl MK, Ewalt KL, Hartl FU. On the role of symmetrical and asymmetrical chaperonin complexes in assisted protein folding. Biol Chem. 1999;380:531–540. doi: 10.1515/BC.1999.068. [DOI] [PubMed] [Google Scholar]
- 34.Taguchi H, Ueno T, Tadakuma H, Yoshida M, Funatsu T. Single-molecule observation of protein–protein interactions in the chaperonin system. Nat Biotechnol. 2001;19:861–865. doi: 10.1038/nbt0901-861. [DOI] [PubMed] [Google Scholar]
- 35.Ueno T, Taguchi H, Tadakuma H, Yoshida M, Funatsu T. GroEL mediates protein folding with a two successive timer mechanism. Mol Cell. 2004;14:423–434. doi: 10.1016/s1097-2765(04)00261-8. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki M, Ueno T, Iizuka R, Miura T, Zako T, Akahori R, et al. Effect of the C-terminal truncation on the functional cycle of chaperonin GroEL: implication that the C-terminal region facilitates the transition from the folding-arrested to the folding-competent state. J Biol Chem. 2008;283:23931–23939. doi: 10.1074/jbc.M804090200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sameshima T, Ueno T, Iizuka R, Ishii N, Terada N, Okabe K, et al. Football-and bullet-shaped GroEL-GroES complexes coexist during the reaction cycle. J Biol Chem. 2008;283:23765–23773. doi: 10.1074/jbc.M802541200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sameshima T, Iizuka R, Ueno T, Funatsu T. Denatured proteins facilitate the formation of the football-shaped GroEL-(GroES)2 complex. Biochem J. 2010;427:247–254. doi: 10.1042/BJ20091845. [DOI] [PubMed] [Google Scholar]
- 39.Ye X, Lorimer GH. Substrate protein switches GroE chaperonins from asymmetric to symmetric cycling by catalyzing nucleotide exchange. Proc Natl Acad Sci USA. 2013;110:E4289–4297. doi: 10.1073/pnas.1317702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang D, Ye X, Lorimer GH. Symmetric GroEL:GroES2 complexes are the protein-folding functional form of the chaperonin nanomachine. Proc Natl Acad Sci USA. 2013;110:E4298–4305. doi: 10.1073/pnas.1318862110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haldar S, Gupta AJ, Yan X, Miličić G, Hartl FU, Hayer-Hartl M. Chaperonin-assisted protein folding: relative population of asymmetric and symmetric GroEL:GroES complexes. J Mol Biol. 2015;427:2244–2255. doi: 10.1016/j.jmb.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 42.Koike-Takeshita A, Yoshida M, Taguchi H. Revisiting the GroEL-GroES reaction cycle via the symmetric intermediate implied by novel aspects of the GroEL(D398A) mutant. J Biol Chem. 2008;283:23774–23781. doi: 10.1074/jbc.M802542200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taguchi H. Reaction cycle of chaperonin GroEL via symmetric “football” intermediate. J Mol Biol. 2015;427:2912–2918. doi: 10.1016/j.jmb.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 44.Sameshima T, Iizuka R, Ueno T, Wada J, Aoki M, Shimamoto N, et al. Single-molecule study on the decay process of the football-shaped GroEL-GroES complex using zeromode waveguides. J Biol Chem. 2010;285:23159–23164. doi: 10.1074/jbc.M110.122101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takei Y, Iizuka R, Ueno T, Funatsu T. Single-molecule observation of protein folding in symmetric GroEL-(GroES)2 complexes. J Biol Chem. 2012;287:41118–41125. doi: 10.1074/jbc.M112.398628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nojima T, Yoshida M. Probing open conformation of GroEL rings by cross-linking reveals single and double open ring structures of GroEL in ADP and ATP. J Biol Chem. 2009;284:22834–22839. doi: 10.1074/jbc.M109.020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koike-Takeshita A, Mitsuoka K, Taguchi H. Asp-52 in combination with Asp-398 plays a critical role in ATP hydrolysis of chaperonin GroEL. J Biol Chem. 2014;289:30005–30011. doi: 10.1074/jbc.M114.593822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Motojima F, Yoshida M. Discrimination of ATP, ADP, and AMPPNP by chaperonin GroEL: hexokinase treatment revealed the exclusive role of ATP. J Biol Chem. 2003;278:26648–26654. doi: 10.1074/jbc.M300806200. [DOI] [PubMed] [Google Scholar]
- 49.Fei X, Ye X, LaRonde NA, Lorimer GH. Formation and structures of GroEL:GroES2 chaperonin footballs, the protein-folding functional form. Proc Natl Acad Sci USA. 2014;111:12775–12780. doi: 10.1073/pnas.1412922111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koike-Takeshita A, Arakawa T, Taguchi H, Shimamura T. Crystal structure of a symmetric football-shaped GroEL: GroES2-ATP14 complex determined at 3.8 Å reveals rearrangement between two GroEL rings. J Mol Biol. 2014;426:3634–3641. doi: 10.1016/j.jmb.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 51.Nisemblat S, Yaniv O, Parnas A, Frolow F, Azem A. Crystal structure of the human mitochondrial chaperonin symmetrical football complex. Proc Natl Acad Sci USA. 2015;112:6044–6049. doi: 10.1073/pnas.1411718112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yifrach O, Horovitz A. Two lines of allosteric communication in the oligomeric chaperonin GroEL are revealed by the single mutation Arg196→Ala. J Mol Biol. 1994;243:397–401. doi: 10.1006/jmbi.1994.1667. [DOI] [PubMed] [Google Scholar]
- 53.Boisvert DC, Wang J, Otwinowski Z, Horwich AL, Sigler PB. The 2.4 Å crystal structure of the bacterial chaperonin GroEL complexed with ATPγS. Nat Struct Biol. 1996;3:170–177. doi: 10.1038/nsb0296-170. [DOI] [PubMed] [Google Scholar]
- 54.Aharoni A, Horovitz A. Inter-ring communication is disrupted in the GroEL mutant Arg13→Gly; Ala126→Val with known crystal structure. J Mol Biol. 1996;258:732–735. doi: 10.1006/jmbi.1996.0282. [DOI] [PubMed] [Google Scholar]
- 55.Sot B, Galán A, Valpuesta JM, Bertrand S, Muga A. Salt bridges at the inter-ring interface regulate the thermostat of GroEL. J Biol Chem. 2002;277:34024–34029. doi: 10.1074/jbc.M205733200. [DOI] [PubMed] [Google Scholar]
- 56.Sewell BT, Best RB, Chen S, Roseman AM, Farr GW, Horwich AL, et al. A mutant chaperonin with rearranged inter-ring electrostatic contacts and temperature-sensitive dissociation. Nat Struct Mol Biol. 2004;11:1128–1133. doi: 10.1038/nsmb844. [DOI] [PubMed] [Google Scholar]
- 57.Parnas A, Nisemblat S, Weiss C, Levy-Rimler G, Pri-Or A, Zor T, et al. Identification of elements that dictate the specificity of mitochondrial Hsp60 for its co-chaperonin. PLoS One. 2012;7:e50318. doi: 10.1371/journal.pone.0050318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Madan D, Lin Z, Rye HS. Triggering protein folding within the GroEL-GroES complex. J Biol Chem. 2008;283:32003–32013. doi: 10.1074/jbc.M802898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grason JP, Gresham JS, Widjaja L, Wehri SC, Lorimer GH. Setting the chaperonin timer: the effects of K+ and substrate protein on ATP hydrolysis. Proc Natl Acad Sci USA. 2008;105:17334–17338. doi: 10.1073/pnas.0807429105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grason JP, Gresham JS, Lorimer GH. Setting the chaperonin timer: a two-stroke, two-speed, protein machine. Proc Natl Acad Sci USA. 2008;105:17339–17344. doi: 10.1073/pnas.0807418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaudhry C, Farr GW, Todd MJ, Rye HS, Brunger AT, Adams PD, et al. Role of the γ-phosphate of ATP in triggering protein folding by GroEL-GroES: function, structure and energetics. EMBO J. 2003;22:4877–4887. doi: 10.1093/emboj/cdg477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Llorca O, Galán A, Carrascosa JL, Muga A, Valpuesta JM. GroEL under heat-shock. Switching from a folding to a storing function. J Biol Chem. 1998;273:32587–32594. doi: 10.1074/jbc.273.49.32587. [DOI] [PubMed] [Google Scholar]
- 63.Bartolucci C, Lamba D, Grazulis S, Manakova E, Heumann H. Crystal structure of wild-type chaperonin GroEL. J Mol Biol. 2005;354:940–951. doi: 10.1016/j.jmb.2005.09.096. [DOI] [PubMed] [Google Scholar]