Highlights
-
•
The AgNP was successfully synthesised from soil isolated bacterium Bacillus subtilis.
-
•
The synthesised AgNPs were confirmed by UV-Spec, FTIR, TEM and XRD.
-
•
The AgNPs were successfully treated microalga to disrupt its cell wall to release lipids and carbohydrates.
-
•
The cell wall damages evidenced by LDH assay and SEM.
Keywords: Chlorella vulgaris, Cell wall disruption, Bacillus subtilis, Silver nanoparticle, Carbohydrates, Lipids
Abstract
Microalgae are the fledging feedstocks yielding raw materials for the production of third generation biofuel. Assorted and conventional cell wall disruption techniques were helpful in extracting lipids and carbohydrates, nevertheless the disadvantages have led the biotechnologists to explore new process to lyse cell wall in a faster and an economical manner. Silver nanoparticles have the ability to break the cell wall of microalgae and release biomolecules effectively. Green synthesis of silver nanoparticles was performed using a novel bacterial isolate of Bacillus subtilis. Characterisation of nanosilver and its effect on cell wall lysis of microalgae were extensively analysed. Cell wall damage was confirmed by lactate dehydrogenase assay and visually by SEM analysis. This first piece of research work on direct use of nanoparticles for cell wall lysis would potentially be advantageous over its conventional approaches and a greener, cost effective and non laborious method for the production of biodiesel.
1. Introduction
Alternate transport fuel is an emerging research worldwide due to rapidly decreasing fossil fuel resources and raising environmental concern on global warming. Fossil fuels are not only dominant in energy sectors and global economy but also responsible for life threatening infection by the accumulation of toxic gases while burning mainly from transport division which contributes to 21% CO2 emissions [1], [2], [3], [4]. Hence, promising alternate to petrodiesel are considered to be biodiesel and bioethanol, termed as “green” fuels, as they are ecofriendly, non-toxic, renewable and biodegradable in nature [5], [6], [7].
Biofuel is generally produced from vegetable oil and agricultural crops [8]. However rise in food prices, consumption of more arable land and water for cultivation create serious problems of utilizing them [9]. Moreover, high lignin content in plant biomass makes saccharification difficult during bioethanol production [9], [10]. Nowadays microalgae have been recognised as the third generation biofuel feedstock by many scientists and advantageous to plant based feedstock due to their simple structure, fast growth rate, can be grown in non agricultural land and waste water, accumulate large amount of lipids and carbohydrates [11], [12]. However, biorefinery process of microalgae has several steps including species selection, mode of cultivation, harvesting, and cell disruption. Among all these steps cell disruption is the principal step because algal cell wall generally consist of multilayered rigid cell wall [13], [14], [15], which restrict the complete extraction of compound of interest from microalgae. Therefore, to extract lipid and carbohydrate from algal cells, the cell wall must be disrupted to smallest pieces using pretreatment process using organic solvents [16]. Methods of cell wall disruption and extracting solvents decide the efficiency of oil and carbohydrate extraction from microalgae [13].
Many pretreatment methods can be used for microalgal cell disruption such as physical treatment including ultrasonication [14], [17], thermal [13], [18], microwave [13], [14], [19], bead milling [14], [20] and cryogrinding [13], [21], chemical methods including acid [22], [23], alkaline [24], solvent soaking [21] and osmotic shock [13] and enzymatic treatments [10], [25], [26], [27], [28]. Nevertheless, these methods have some limitations; physical or mechanical methods are not effective and not suitable for large-scale operations [27], chemical methods cause pollution [29] and also destruct other cellular components [27], and enzymatic treatment is cost intensive and very slow process [17].
The disadvantages can be overcome by an efficient alternate method and it has to be developed for better microalgae cell disruption. One such potential alternate is the use of nanotechnology in biotechnological applications. Nanoparticles have carved their own niche in renewable energy sector having been implemented in various processes. Metal nanoparticles are recognized to be effective catalytic agents in chemical and biological sciences. Due to nanoparticles’ tiny structure, they penetrate and easily interact with the biomolecules acting upon them efficiently [30]. To the best of our knowledge and from extensive literature survey, no report on cell wall lysis of microalgae using silver nanoparticles for biofuel synthesis had been published. The main objectives of this study are the extracellular synthesis of silver nanoparticle (AgNP) from a novel bacterial isolate B.subtilis (KF681508) and evaluate its effects on cell wall rupture of fresh water microalga to extract lipid and carbohydrate from Chlorella vulgaris for biofuel production.
2. Materials and methods
2.1. Microalgal culture
Fresh water microalga Chlorella vulgaris was obtained from CAS in Botany, University of Madras (Guindy Campus), Chennai, Tamilnadu, India. It was grown in 14 L PBR using sterile Bold’s Basal Medium (BBM) consisting of (g/L) NaNO3 (0.25), K2HPO4 (0.075), KH2PO4 (0.175), NaCl (0.025), CaCl2.2H2O (0.025), MgSO4·7H2O (0.075), EDTA.2Na (0.05), KOH (0.031), FeSO4·7H2O (0.005), H3BO3 (0.008), ZnSO4·7H2O (0.0015), MnCl2.4H2O (0.0003), MoO3 (0.00025), CuSO4·5H2O (0.0003), Co(NO3)2·6H2O (0.0001) and mixing was provided by sparging air (0.3vvm) from the bottom of the PBR. The lighting was supplied by cool-white fluorescent light with an intensity of 5000 lux under 12:12 light/dark cycle.
2.2. Isolation and molecular identification of bacterium for AgNP synthesis
Silver nanoparticle producing bacteria was isolated from soil sample that was collected from the Annamalai University campus, Tamilnadu, India, using serial dilution technique and 0.1 ml sample was spreaded on Nutrient Agar plates (Hi Media Laboratories Ltd., Mumbai, India) and incubated at 37 °C for 24 h. After incubation, the plates were checked for white, swarmed mucoid colonies and then biochemical characterisation was performed to tentatively identify the isolate as Bacillus sp. The culture was finally subjected to strain identification and was confirmed using 16SrRNA primers: 27 F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492 R (5′- TAC GGT TAC CTT GTT ACG ACT T-3′). The gene sequence obtained from the organism was compared with other Bacillus strains for pairwise identification using NCBI-BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and multiple sequence alignments of the sequences were performed using Clustal Omega version of EBI (www.ebi.ac.uk/Tools/msa/clustalo). Finally Phylogenetic tree was constructed by Clustal Omega of EBI (www.ebi.ac.uk/Tools/phylogeny/clustalw2_phylogeny) using neighbor joining method.
2.3. Biosynthesis of silver nanoparticles
The bacterial isolate was cultivated in LB broth for 24 h at 37 °C and applied for silver nanoparticle synthesis. After incubation, the cell free supernatant was collected by centrifugation at 10,000 rpm for 10 min. In a 250 ml conical flask, 50 ml of 1 mM AgNO3 solution was added to 100 ml of culture supernatant. The control was the culture supernatant without AgNO3 solution. The flasks were then left for 48 h at room temperature in brightness. The colour change from pale yellow to brown was visually checked, which indicated the extracellular synthesis of AgNPs. The AgNPs were obtained by centrifugation at 12000 rpm for 10 min, washing the pellet with double distilled water and repeating the protocol thrice. The obtained AgNP pellet was then lyophilized for characterisation study.
2.4. Characterisation of AgNPs
Primary characterization of AgNP was performed by UV–vis Spectrophotometer (SL-159, Elico, India) with wavelengths ranging between 200 and 600 nm. FT-IR spectra of the nanoparticle using KBr pellet was obtained by FTIR Spectroscope (Bruker Optics, GmBH, Germany). Morphology of AgNP was observed through TEM (Jeol, Japan) and the XRD pattern was measured using diffractometer (Xpert-Pro, England) at room temperature with current of 30 mA, voltage of 40 kV and the peaks were recorded at 2θ.
2.5. Pretreatment of microalga using AgNPs
C.vulgaris culture was harvested when it reached stationary phase by centrifugation at 5000 rpm for 10 min. Then the microalgal paste was dried using Lyophilization and the dried biomass was subjected to pretreatment process. Various concentrations of AgNPs such as 50,75, 100, 125, 150 and 200 μg were added to the test tube contain 1 g of microalgal biomass with sterile distilled water. This mixture was then incubated at various time intervals of 10, 20, 30, 40 and 50 min in an orbital shaker at 100 rpm.
After the pretreatment process, the biomass slurry was subjected to drying at 60 °C for 8 h using hot air oven and the intracellular oil was extracted by Bligh and Dyer method [31] and oil extraction yield (%w/w) was calculated as,
The extracted oil from untreated microalgal biomass was considered as control and yield was compared with test sample.
The total carbohydrate content was determined, after extracting oil, using phenol sulfuric acid method [32]. A volume of 5 ml sample was used for the procedure and untreated sample was used as control.
2.6. Lactate dehydrogenase (LDH) assay for cell wall damage
LDH experiment was carried out according to Pakrashi et al., [33]. About 1 mL of the interacted cell suspensions was centrifuged at 8000 rpm for 10 min. A 100 μL of the supernatant was then collected and 100 μL of 30 Mm sodium pyruvate was added followed by 2.8 mL of 0.2 M Tris–HCl. Finally, a 100 μL of 6.6 mM NaDH was added just before measuring the decrease in absorbance at 340 nm as a function of 10 readings using UV–vis spectrophotometer (SL-159, ELICO Ltd, India).
2.7. Scanning Electron Microscope (SEM) Analysis
The effects of AgNP pretreated microalgal cells were subjected to morphological analysis for cell wall damage. After the pretreatment process, a small amount of sample was taken from the test tube, dried and observed with Scanning Electron Microscope (JSM 5610, Jeol, Japan).
3. Results and discussion
3.1. Molecular identification of AgNPs producing bacteria using 16S rRNA
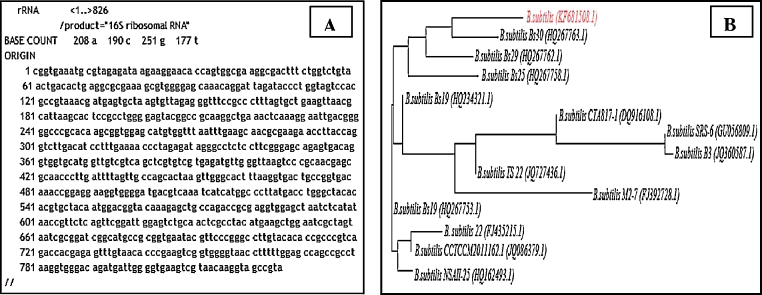
The AgNP producing bacteria was identified by molecular identification using 16 S rRNA and the amplified gene sequence was compared with BLAST database, acquiring sequences displaying maximum homology. The strain of the study exhibited maximum percentage of similarity, 100%, with the sequences of other B.subtilis strains with a high score. The target rRNA was aligned with all homologous sequences using Clustal W2 and a phylogenetic tree was eventually constructed (Fig. 1A and B). The phylogenetic analysis confirmed that the isolated strain was B.subtilis. The nucleotide sequence of the organism, referred to as has been deposited in the GenBank database under the accession number KF681508.1.
Fig. 1.
16S rRNA sequence (A) and phylogenetic tree of B.subtilis (B).
3.2. Extracellular synthesis of AgNPs
The formation of silver nanoparticles was started within few minutes at room temperature and it was confirmed by colour changes from pale yellow to dark brown at the end of incubation time (Fig. 2). There was no colour change in control which was maintained in the same environmental condition. It was corroborated with the results reported by Saravanan et al. (2011), Shivaji et al. (2011) and Gopinath and Velusamy [34], [35], [36]. The brown colour confirms that it was due to the reduction of Ag+ which indicates the formation of AgNPs.
Fig. 2.

change of color from yellow to brown indicating the formation of silver nanoparticle.
The mechanism of the synthesis of silver nanoparticles is due to the presence of enzyme “Nitrate reductase” [37] and secretion of cofactor NADH and NADH dependent enzymes which involved in reduction of Ag+ to Ag0 [30], [38], [39]. The reduction mediated by the presence of the enzyme in the organism has been found to be responsible for the synthesis. The process gets initiated with the electron transfer from NADH to NAD+ catalysed by NADH- dependent reductase as electron carrier, which reduces Ag+ ions to metallic silver [30], [40].
3.3. Characterisation of AgNPs
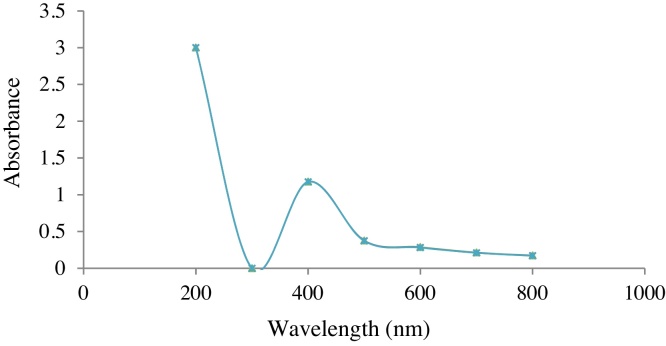
The visual observations were confirmed with the analysis by UV-Visible spectroscopy, which is a unique and simple protocol analysing samples with their optical properties (Fig. 3). An intense absorption peak was noted at 400 nm which confirmed the presence of AgNPs. The negatively charged polysaccharides in the supernatant bonded with nanosilver will confine free electrons of the nanoparticles in a smaller volume. This results in a high free electron density leading to high plasmon frequency. Hence a sharp peak was observed at a lower wavelength [41].
Fig. 3.
UV-Visible spectrum of AgNP.
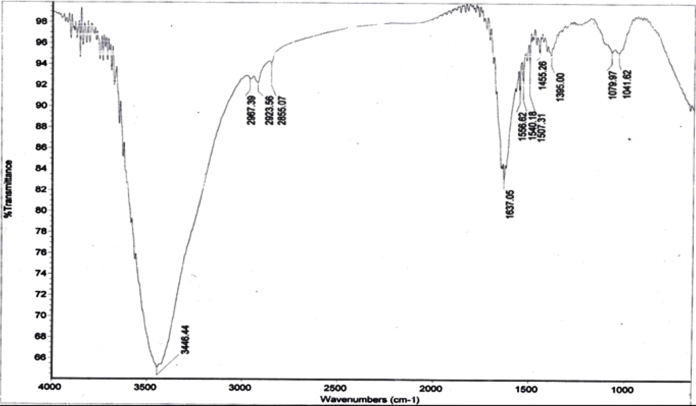
The spectrum of AgNP depicted the varied range of absorption from 3446–1022 cm−1 (Fig. 4). A broad absorption band was observed at 3446.44 cm−1 which indicated the hydroxyl stretch. Intense peaks at 2967.91, 2923.64 and 2855.07 cm−1 denoted the symmetric CH3 and CH2 stretches. Absorption peaks at 1653.05 indicated carbonyl groups of amide I bands and 1556.62 and 1540.18 cm−1 marked the presence of N-H stretching of amide II bands. An intense peak at 1263.51 cm−1 indicated C-O stretching. Absorption peaks at 1079.97 cm−1 were peaks for C-O-C stretching for all sugar moieties. Additional peak at 1455.26 cm−1 designated the presence of AgNPs. The protein (amide) denoting bands showed that the silver nanoparticles were capped and protected by the proteins present in the supernatant. Several results suggested that proteins were the major constituent capping and stabilizing AgNPs by electrostatic forces [42], [43], [44], [45], [46].
Fig. 4.
FTIR spectrum of microbially synthesised silver nanoparticle.
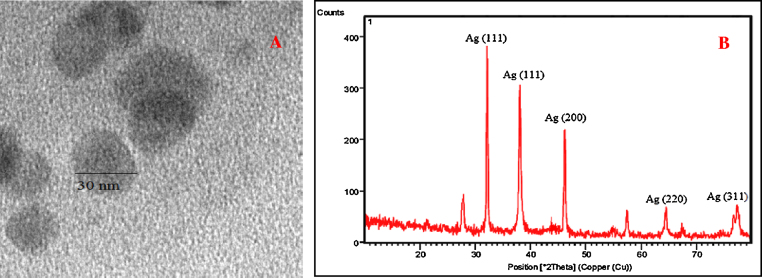
Morphology of the produced nanoparticle was studied using TEM analysis and the structures were found to be spherical (Fig. 5a). XRD was performed to study the crystallinity of the nanoparticle. The XRD pattern of the silver nanoparticle is shown in Fig. 5b. Five sharp peaks could be seen at 2θ = 32, 38, 46, 64.3 and 77.5 which correlated to the characteristic peaks of silver corresponding to the three d- spacing (111), (111), (200), (220) and (311) respectively. The intensity of the peaks denoted high degree of crystallinity of silver nanoparticle produced. Diffraction peaks have indicated that the size of AgNP would be very small and are crystalline in nature. The size was calculated using Debye-Scherrer’s equation represented as,
| d = Kα λ/β cos θ |
where, d is the average size, β is the full width half maxima, θ is the Bragg’s angle, λ is the wavelength of CuKα in radians. The size of the crystallite AgNP estimated from the equation was 31 nm.
Fig. 5.
(A) illustrating the structure of AgNP through TEM and (B) XRD pattern of AgNP produced from soil isolate B.subtilis VVS (KF681508).
3.4. Effect of AgNPs treatment on carbohydrate and oil yield
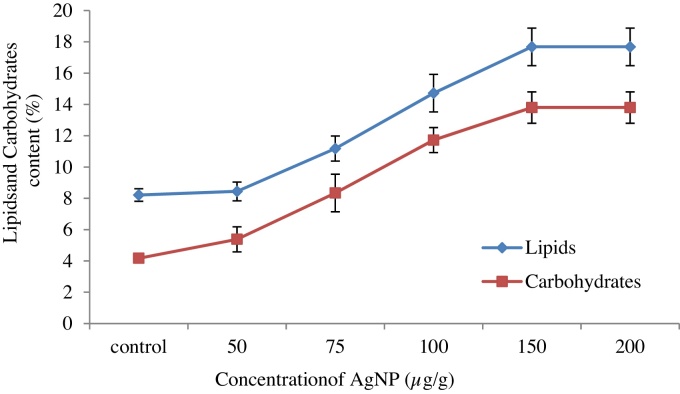
The amount of total carbohydrate and oil extracted from C.vulgaris was considered as an indication of the efficiency of cell disruption by the silver nanoparticle. The effects of various concentrations of AgNPs were analyzed in the ranges of 50–200 μg/g and their efficiency on extraction of product yield was shown in Fig. 6. A significant difference was found between the various studied concentrations and this study proved that nanoparticle pretreatment method was able to disrupt C.vulgaris to release intracellular oil and total carbohydrate. C.vulgaris has a maximum lipid content of 15-18% and a carbohydrate content of 16% under natural environmental conditions. The intracellular oil extraction was increased from 8.44% to 17.68% by increasing the concentration of AgNP from 50 μg/g to 150 μg/g. Similarly, maximum carbohydrate yield was found to be 13.8% with the same concentration of AgNP. However, the yield was constant when the concentration of AgNPs reached beyond the 150 μg/g. This is in agreement with the previous results reported by Hazani et al. [47]. They analysed effect of various concentrations of AgNPs such as 5,10, 25, 50,100, 200 and 400 mg/L on C.vulgaris and D. tertiolecta for toxicity study and found that 100 mg/L showed maximum cell lyses than the other concentrations with the short reaction time. Another study conducted by Palomares et al. [48] also reported that higher concentration of nanoparticle lysed the cell wall of cyanobacterium Anabaena with a shorter period of reaction time. This indicated that oil and carbohydrate yield was increased with increasing concentration of silver nanoparticles due to availability of more silver nanoparticles which highly rupture the microalgal cell membrane.
Fig. 6.
Effect of different concentration of AgNPs on oil and carbohydrate yield.
This mechanism can be explained as the small size and extremely large surface area of nanoparticles enables them to make strong contact with the cell wall of microorganisms [47], [49] and lyses molecules in the cell wall by forming “pits/holes”, cause release of intracellular molecules [50], [51]. Dash et al. further described that cell wall is mainly composed of cellulose and protein complex and is the primary site for the interaction of nanoparticles [52]. Thus, the nanoparticles are adhered on the surface and cover the microalgae to a greater extent than the bulk particles leads to cell wall rupture.
Currently many reports have been published on enzymatic treatment on microalgae cell disruption for extraction of lipid and carbohydrate due to its ecofriendly nature. However, different algae contain different cell wall composition and have different enzyme specificity hence all cell wall destructing enzymes cannot act on all the algal species. It was confirmed by our previous studies conducted on acid and enzymatic pretreatment processes to disrupt cell wall of two marine microalgae C.salina and N.oculata for extraction of intracellular oil [11], [13]. For C.salina, the cellulase pretreatment showed highest yield of lipid when compared to the acid pretreatment, whereas for N.oculata acid pretreatment showed maximum lipid yield. The difference was apparently due to variance of cell wall composition of these two different genera. Generally Nannochloropsis sp. contains algaenans, a trilaminar outer cell wall and cellulose. Algaenans are mainly composed of aliphatic biopolymer composed of long chain numbered ω 9-unsaturated ω-hydroxy fatty acids monomers varying chain length from 30 to 34 carbon atoms [15], [20]. Algaenan is easily soluble in acid using HCl or H2SO4 but it is resistant to cellulase hydrolysis [15]. In the other hand Chlorella contains trilaminar rigid cell wall matrix which is only made up of cellulose and easily degraded by enzyme and acid [15], [20], [53].
It was further confirmed by a study on investigation of the enzyme specificity on pretreatment process for extracting carbohydrate for bioethanol productions by Hong et al. (2014) [54]. They analysed the effect of four commercially produced enzymes Lactozym, Spirizyme, Viscozyme, and AMG (glucoamylase) on hydrolysis of red seaweed (Gelidium amansii), brown seaweed (Laminaria japonica), and green seaweed (Codium fragile). They resulted that Lactozym produced highest carbohydrate yield on red and green seaweeds, whereas Spirizyme produced highest carbohydrate yield on brown seaweed. Therefore, in this paper we report for the first time that silver nanoparticle can potentially be used to lyse cell wall of microalgae to release biofuel feedstocks, lipid and carbohydrate, in a dose and time dependent manner.
3.5. Effect of treatment time on carbohydrate and oil yield
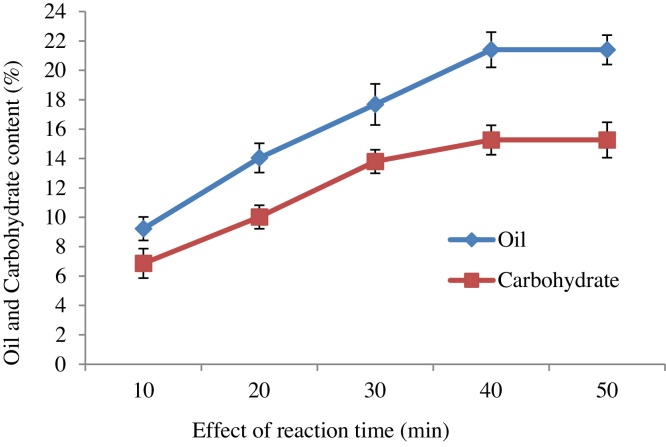
For the effect of pretreatment time on cell wall damages of C.vulgaris, the time period considered ranged between of 10 and 50 min. The oil yield was increased from 9.22% to 21.4% by increasing the reaction time of 10 min to 40 min. Similarly the total carbohydrate yield was increased from 6.86% to 15.26% by increasing the reaction time (Fig. 7). It was corroborated with the earlier report done by Lee et al. (2013) which evaluated the effect of different reaction time (1,5,10,15 and 20 min) on cell membrane damages of four marine microalgal species include Cochlodinium polykrikoides, Chattonella marina, Heterosigma akashiwo and Chattonella marina using aminoclay nanoparticles [54]. They found that all the microalgae cells were completely ruptured by increasing the reaction time to 20 min from 1 min. This result revealed that increasing the reaction time resulted in increasing in product yield by escalating contact of AgNP particles on surface of microalgae cells leading to immediate cell wall destruction of C.vulgaris.
Fig. 7.
Effect of different reaction time on oil and carbohydrate yield.
3.6. Analysis of cell membrane damage using lactate dehydrogenase (LDH) assay
The cell wall disruption of microalgae using AgNPs on C.vulgaris was further evidenced by LDH assay. Effect of different concentration of AgNPs and different reaction time on releasing LDH from microalgal cell wall was shown in Table 1. Different concentrations of AgNPs such as 50, 75, 100, 150 and 200 μg/g were exposed to analyse cell wall damage and found 0.9 ± 0.1, 2.8 ± 0.3, 9.46 ± 0.8, 11.2 ± 0.2 and 11.2 ± 0.4% of LDH release respectively. Simultaneously the effect of reaction time on LDH release was also analysed in the ranges of 10, 20, 30, 40 and 50 min and found the LDH content as 6.09 ± 0.6, 10.2 ± 0.8, 18.08 ± 0.3, 23.60 ± 0.6 and 23.62 ± 0.2% respectively. This result indicated that increasing concentration with increasing reaction time resulted in increased LDH content. Our current findings was highly corroborate with the report done by Pakrashi et al. (2013) that showed higher membrane damage with higher concentrations of aluminium oxide nanoparticles and higher reaction time on freshwater algal isolate Chlorella ellipsoides [33].
Table 1.
Different AgNPs concentration and reaction time on LDH content of C.vulgaris.
| Concentration (μg/g) | LDH (%) |
Time (min) | LDH (%) |
|---|---|---|---|
| 50 | 0.9 ± 0.1 | 10 | 6.09 ± 0.6 |
| 75 | 2.8 ± 0.3 | 20 | 10.2 ± 0.8 |
| 100 | 9.46 ± 0.8 | 30 | 18.08 ± 0.3 |
| 150 | 11.2 ± 0.2 | 40 | 23.60 ± 0.6 |
| 200 | 11.2 ± 0.4 | 50 | 23.62 ± 0.2 |
3.7. SEM analysis
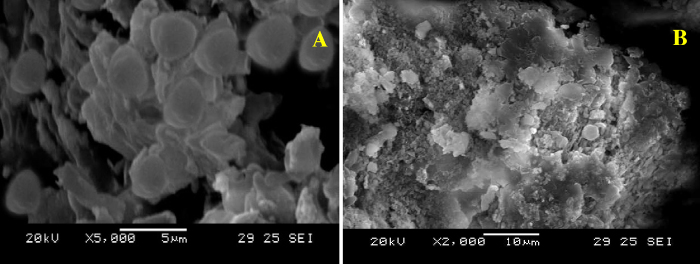
The Scanning Electron Microscope (SEM) study was used to analyze the direct evidence of cell wall destruction by silver nanoparticle treatment. Untreated cells of C.vulgaris appeared intact without any cell destruction, whereas nanoparticle treated cells were destroyed and observed as distorted structures with complete cell wall lysis (Fig. 8a and b). This is due to the disruption of the lipid bilayer and cellulose composition of cell wall of microalgae by surface interactions of sites of nanoparticles on microalgae [55].
Fig. 8.
SEM images C.vulgaris—intact cells (a); disrupted cells due to cell wall lysis (b).
4. Conclusion
The present article dealt with the involvement of silver nanoparticles in lysing cell wall of C.vulgaris. This study reports the green and simple synthesis of silver nanoparticles using silver nitrate as the substrate and culture supernatant as the producing agent. The supernatant, consisting of polysaccharides and proteins, served as the reducing and stabilising agent of nanosilver production. Characterisation study exhibited that the formed nanoparticles were crystalline and spherical. These AgNPs were utilised for breaking down the cell wall of the microalga. Maximum of 150 μg/g and 40 min of AgNPs and time respectively were required for the process. LDH assay and SEM analysis revealed that AgNPs bound and distorted the composition of cell wall thus rupturing them leading to the extrusion of lipids and carbohydrates. This research work could be helpful as an ecofriendly and essential protocol by overcoming the difficulties faced while using conventional techniques. Further studies could be carried out to learn its wider applications in other areas of renewable energy technology using microalgae.
Declaration of interest
The authors declare no conflict of interest.
Acknowledgments
The authors sincerely thanks Professor Dr. R. Rangasamy, Director of Centre for Advanced Study (CAS) in Botany, University of Madras (Guindy Campus), Chennai for providing the microalgae culture.
References
- 1.Mutanda T., Ramesh D., Karthikeyan S., Kumari S., Anandraj A., Bux F. Bioprospecting for hyper-lipid producing microalgal strains for sustainable biofuel production. Bioresour. Technol. 2011;102:57–70. doi: 10.1016/j.biortech.2010.06.077. [DOI] [PubMed] [Google Scholar]
- 2.Yen H.W., Hu I.C., Chen C.Y., Ho S.H., Lee D.J., Chang J.S. Microalgae-based biorefinery—from biofuels to natural products. Bioresour. Technol. 2013;135:166–174. doi: 10.1016/j.biortech.2012.10.099. [DOI] [PubMed] [Google Scholar]
- 3.Ullah K., Ahmad M., Sofia Kumar S.V., Lu P., Harvey A., Zafar M., Sultana S. Assessing the potential of algal biomass opportunities for bioenergy industry: a review. Fuel. 2015;143:414–423. [Google Scholar]
- 4.Bai F., Yan W., Zhang S., Yu D., Bai L. Immobilized lipase of reconstructed oil bodies and its potential application in biodiesel production. Fuel. 2014;128:340–346. [Google Scholar]
- 5.Demirbas A., Demirbas M.F. Importance of algae oil as a source of biodiesel. Energy Convers. Manag. 2011;52:163–170. [Google Scholar]
- 6.Zarei A., Amin N.A.S., Kiakalaieh A.T., Zain N.A.M. Immobilized lipase-catalyzed transesterification of Jatropha curcas oil: optimization and modeling. J. Taiwan Inst. Chem. E. 2014;45:444–451. [Google Scholar]
- 7.Jiang Y., Liu X., Chen Y., Zhou L., He Y., Ma L., Gao J. Pickering emulsion stabilized by lipase-containing periodic mesoporous organosilica particles: a robust biocatalyst system for biodiesel production. Bioresour. Technol. 2014;153:278–283. doi: 10.1016/j.biortech.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Surendhiran D., Vijay M., Sirajunnisa A. Biodiesel production from marine microalga Chlorella salina using whole cell yeast immobilized on sugarcane bagasse. J. Environ. Chem. Eng. 2014;2:1294–1300. [Google Scholar]
- 9.Ho S.H., Huang S.W., Chen C.Y., Hasunuma T., Kondo A., Chang J.S. Bioethanol production using carbohydrate-rich microalgae biomass as feedstock. Bioresour. Technol. 2013;135:191–198. doi: 10.1016/j.biortech.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 10.Kim K.H., Choi I.S., Kim H.M., Wi S.G., Bae H.J. Bioethanol production from the nutrient stress-induced microalga Chlorella vulgaris by enzymatic hydrolysis and immobilized yeast fermentation. Bioresour. Technol. 2014;153:47–54. doi: 10.1016/j.biortech.2013.11.059. [DOI] [PubMed] [Google Scholar]
- 11.Surendhiran D., Vijay M. Evaluation of various pretreatment process for extracting oil from Chlorella salina under nitrogen rich and depleted conditions. Int. J. Environ. Bioenergy. 2013;7:129–142. [Google Scholar]
- 12.Peralta-Ruiz Y., González-Delgado A.D., Kafarov V. Evaluation of alternatives for microalgae oil extraction based on energy analysis. Appl. Energy. 2013;101:226–236. [Google Scholar]
- 13.Surendhiran D., Vijay M. Effect of various pretreatment for extracting intracellular lipid from Nannochloropsis oculata under nitrogen replete and depleted conditions. ISRN Chem. Eng. 2014 [Google Scholar]
- 14.Zheng H., Yin J., Gao Z., Huang H., Ji X., Dou C. Disruption of Chlorella vulgaris cells for the release of biodiesel producing lipids: a comparison of grinding, ultrasonication, bead milling, enzymatic lysis, and microwaves. Appl. Biochem. Biotechnol. 2011;164:1215–1224. doi: 10.1007/s12010-011-9207-1. [DOI] [PubMed] [Google Scholar]
- 15.Rodrigues M.A., da Silva Bon E.P. Evaluation of Chlorella (Chlorophyta) as source of fermentable sugars via cell wall enzymatic hydrolysis. Enzyme Res. 2011 doi: 10.4061/2011/405603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho H.S., Oh Y.K., Park S.C., Lee J.K., Park J.Y. Effects of enzymatic hydrolysis on lipid extraction from Chlorella vulgaris. Renewable Energy. 2013;15:156–160. [Google Scholar]
- 17.Zhao G., Chen X., Wang L., Zhou S., Feng H., Chen W.N., Lau R. Ultrasound assisted extraction of carbohydrates from microalgae as feedstock for yeast fermentation. Bioresour. Technol. 2013;128:337–344. doi: 10.1016/j.biortech.2012.10.038. [DOI] [PubMed] [Google Scholar]
- 18.Kita K., Okada S., Sekino H., Imou K., Yokoyama S., Amano T. Thermal pre-treatment of wet microalgae harvest for efficient hydrocarbon recovery. Appl. Energy. 2010;87:2420–2423. [Google Scholar]
- 19.Balasubramanian S., Allen J.D., Kanitkar A., Boldor D. Oil extraction from Scenedesmus obliquus using a continuous microwave system—design, optimization, and quality characterization. Bioresour. Technol. 2011;102:3396–3403. doi: 10.1016/j.biortech.2010.09.119. [DOI] [PubMed] [Google Scholar]
- 20.Ryckebosch E., Muylaert K., Foubert I. Optimization of an analytical procedure for extraction of lipids from microalgae. J. Am. Chem. Soc. 2012;89:189–198. [Google Scholar]
- 21.Serive B., Kaas R., Bérard J.B., Pasquet V., Laurent Picot L., Cadoret J.P. Selection and optimisation of a method for efficient metabolites extraction from microalgae. Bioresour. Technol. 2012;124:311–320. doi: 10.1016/j.biortech.2012.07.105. [DOI] [PubMed] [Google Scholar]
- 22.Harun R., Danquah M.K., Forde G.M. Microalgal biomass as a fermentation feedstock for bioethanol production. J. Chem. Technol. Biotechnol. 2010;85:199–203. [Google Scholar]
- 23.Ho S.H., Huang S.W., Chen C.Y., Hasunuma T., Kondo A., Chang J.S. Bioethanol production using carbohydrate-rich microalgae biomass as feedstock. Bioresour. Technol. 2013;135:191–198. doi: 10.1016/j.biortech.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 24.Harun R., Jason W.S.Y., Cherrington T., Danquah M.K. Exploring alkaline pre-treatment of microalgal biomass for bioethanol production. Appl. Energy. 2011;88:3464–3467. [Google Scholar]
- 25.Choi S.P., Nguyen M.T., Sim S.J. Enzymatic pretreatment of Chlamydomonas reinhardtii biomass for ethanol production. Bioresour. Technol. 2010;101:5330–5336. doi: 10.1016/j.biortech.2010.02.026. [DOI] [PubMed] [Google Scholar]
- 26.Garcia J.M.R., Acien Fernandez F.G., Sevilla J.M.F. Development of a process for the production of l-amino-acids concentrates from microalgae by enzymatic hydrolysis. Bioresour. Technol. 2012;112:164–170. doi: 10.1016/j.biortech.2012.02.094. [DOI] [PubMed] [Google Scholar]
- 27.Jin G., Yang F., Hu C., Shen H., Zhao Z.K. Enzyme-assisted extraction of lipids directly from the culture of the oleaginous yeast Rhodosporidium toruloides. Bioresour. Technol. 2012;111:378–382. doi: 10.1016/j.biortech.2012.01.152. [DOI] [PubMed] [Google Scholar]
- 28.Li S., Zhang H., Han D., Row K.H. Optimization of enzymatic extraction of polysaccharides from some marine algae by response surface methodology. Korean J. Chem. Eng. 2012;29:650–656. [Google Scholar]
- 29.Surendhiran D., Sirajunnisa A.R., Vijay M. An alternative method for production of microalgal biodiesel using novel Bacillus lipase. 3 Biotech. 2015 doi: 10.1007/s13205-014-0271-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sirajunnisa A.R., Surendhiran D. Nanosilver fabrication mediated by exopolysaccharides from Pseudomonas fluorescens. Am. J. PharmTech Res. 2014;4:727–742. [Google Scholar]
- 31.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Phys. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 32.Sirajunnisa A.R., Vijayagopal V., Viruthagiri T. Medium optimization for the production of exopolysaccharide by Bacillus subtilis using synthetic sources and agro wastes. Turk. J. Biol. 2013;37:280–288. [Google Scholar]
- 33.Pakrashi S., Dalai S., Prathna T.C., Trivedi S., Myneni R., Raichur A.M., Chandrasekaran N., Mukherjee A. Cytotoxicity of aluminium oxide nanoparticles towards fresh water algal isolate at low exposure concentrations. Aquat. Toxicol. 2013;13:132–133. doi: 10.1016/j.aquatox.2013.01.018. 34–45. [DOI] [PubMed] [Google Scholar]
- 34.Saravanan M., Vemu A.K., Barik S.K. Rapid biosynthesis of silver nanoparticles from Bacillus megaterium (NCIM 2326) and their antibacterial activity on multi drug resistant clinical pathogens. Colloids Surf. B. 2011;88:325–331. doi: 10.1016/j.colsurfb.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 35.Shivaji S., Madhu S., Singh S. Extracellular synthesis of antibacterial silver nanoparticles using psychrophilic Bacteria. Process Biochem. 2011;46:1800–1807. [Google Scholar]
- 36.Gopinath V., Velusamy P. Extracellular biosynthesis of silver nanoparticles using Bacillus sp: GP-23 and evaluation of their antifungal activity towards Fusarium oxysporum. Spectrochim. Acta A. 2013;106:170–174. doi: 10.1016/j.saa.2012.12.087. [DOI] [PubMed] [Google Scholar]
- 37.Anthony K.J.P., Murugan M., Gurunathan S. Biosynthesis of silver nanoparticles from the culture supernatant of Bacillus marisflavi and their potential antibacterial activity. J. Ind. Eng. Chem. 2014;20:1505–1510. [Google Scholar]
- 38.Rajamanickam K., Sudha S.S., Francis M., Sowmya T., Rengaramanujam J., Sivalingam P., Prabakar K. Microalgae associated Brevundimonas sp. MSK 4 as the nano particle synthesizing unit to produce antimicrobial silver nanoparticles. Spectrochim. Acta A. 2013;113:10–14. doi: 10.1016/j.saa.2013.04.083. [DOI] [PubMed] [Google Scholar]
- 39.Ahmad A., Mukherjee P., Mandal D., Senapati S., Khan M.I., Kumar R., Sastry M. Enzyme mediated extracellular synthesis of CdS nanoparticles by the fungus, Fusarium oxysporum. J. Am. Chem. Soc. 2002;124:12108–12109. doi: 10.1021/ja027296o. [DOI] [PubMed] [Google Scholar]
- 40.Kathiresan K., Manivannan S., Nabeel M.A., Dhivya B. Studies on silver nanoparticles synthesized by a marine fungus, Penicillium fellutanum isolated from coastal mangrove sediment. Colloids Surf. B. 2009;71:133–137. doi: 10.1016/j.colsurfb.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 41.Dallas P., Sharma V.K., Zboril R. Silver polymeric nanocomposites as advanced antimicrobial agents: classification, synthetic paths, applications, and perspectives. Adv. Colloid Interface Sci. 2011;166:119–135. doi: 10.1016/j.cis.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 42.Sathyanarayanan G., Kiran G.S., Joseph S. Synthesis of silver nano particles by polysaccharide bioflocculant produced from marine Bacillus subtilis. Colloids Surf. B. 2013;102:13–20. doi: 10.1016/j.colsurfb.2012.07.032. [DOI] [PubMed] [Google Scholar]
- 43.Kwon C., Park B., Kim H., Jung S. Green synthesis of silver nanoparticles by sinorhizobial octasaccharide isolated from Sinorhizobium meliloti. Bull. Korean Chem. Soc. 2009;30:1651–1654. [Google Scholar]
- 44.Anghel L., Balasoiu M., Ishchenko L.A., Stolyar S.V., Kurkin T.S., Rogachev A.V., Kuklin A.I., Kovalev Y.S., Raikher Y.L., Iskhakov R.S., Duca G. Characterization of bio-synthesized nanoparticles produced by Klebsiella oxytoca. J. Phys. Conf. Ser. 2012;35:1–7. [Google Scholar]
- 45.Silambarasan S., Jayanthi A. Biosynthesis of silver nanoparticles using Pseudomonas fluorescens. Res. J. Biotechnol. 2013;8:71–75. [Google Scholar]
- 46.Huang H., Yang X. Synthesis of polysaccharide-stabilized gold and silver nanoparticles: a green method. Carbohydr. Res. 2004;339:2627–2631. doi: 10.1016/j.carres.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 47.Hazani A.A., Ibrahim M.M., Shehata A.I., El-Gaaly G.A., Daoud M., Fouad D., Rizwana H., Nadine M.S. Moubayed ecotoxicity of Ag-nanoparticles on two microalgae, Chlorella Vulgaris and Dunaliella tertiolecta. Arch. Biol. Sci. 2013;65:1447–1457. [Google Scholar]
- 48.Palomares I.R., Boltes K., Pinas F.F., Leganes F., Calvo E.G., Javier Santiago J., Rosal R. Physicochemical characterization and ecotoxicological assessment of CeO2 nanoparticles using two aquatic microorganisms. Toxicol. Sci. 2011;119:135–145. doi: 10.1093/toxsci/kfq311. [DOI] [PubMed] [Google Scholar]
- 49.Wong K.K.Y., Liu X. Silver nanoparticles—the real silver bullet in clinical medicine. MedChemComm. 2010;1:125–131. [Google Scholar]
- 50.Sondi I., Sondi B.S. Silver nanoparticles as antimicrobial agent: a case study on E. coli as a model for gram-negative bacteria. J. Colloid Interface Sci. 2004;275:177–182. doi: 10.1016/j.jcis.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 51.Li Q., Mahendra S., Lyon D.Y., Brunet L., Liga M.V., Li D., Alvarez P.J.J. Antimicrobial nanomaterials for water disinfection and microbial control: potential applications and implications. Water Res. 2008;42:4591–4602. doi: 10.1016/j.watres.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 52.Dash A., Singh A.P., Chaudhary B.R., Singh S.K., Dash D. Effect of silver nanoparticles on growth of eukaryotic green algae. Nano-Micro Lett. 2012;4:158–165. [Google Scholar]
- 53.Fu C.C., Hung T.C., Chen J.Y., Su C.H., Wu W.T. Hydrolysis of microalgae cell walls for production of reducing sugar and lipid extraction. Bioresour. Technol. 2010;101:8750–8754. doi: 10.1016/j.biortech.2010.06.100. [DOI] [PubMed] [Google Scholar]
- 54.Hong I.W., Jeon H., Lee S.B. Comparison of red, brown and green seaweeds on enzymatic saccharification process. J. Ind. Eng. Chem. 2014;20:2687–2691. [Google Scholar]
- 55.Lee Y.C., Jin E.S., Jung S.W., Kim Y.M., Chang K.S., Yang J.W., Kim S.W., Kim Y.O., Shin H.J. Utilizing the algicidal activity of aminoclay as a practical treatment for toxic red tides. Sci. Rep. 2013 doi: 10.1038/srep01292. [DOI] [PMC free article] [PubMed] [Google Scholar]