Graphical abstract
Keywords: Fluorescent reporter protein, Co-expression, Non-viral gene delivery, Mammalian cell, Poly(2-dimethylamino) ethyl methacrylate
Highlights
-
•
4 cell lines, 4 promoters, and 3 gene products were studied, i.e. 48 combinations.
-
•
Distinct cell line depended effects were observed.
-
•
Jurkat cell results tended to differ from that obtained with the other cells.
-
•
Co-transfection works well in CHO cells, but fails in up to 80% of Jurkat cells.
-
•
High transfection efficiency in CHO and HEK cells is maintained in spite of pDNA dilution.
Abstract
Non-viral transfection protocols are typically optimized using standard cells and reporter proteins, potentially underestimating cellular or transgene effects. Here such effects were studied for two human (Jurkat, HEK-293) and two rodent (CHO-K1, L929) cell lines and three fluorescent reporter proteins. Expression of the enhanced green fluorescent protein (EGFP) was studied under the control of the human elongation factor 1 alpha promoter and three viral promoters (SV40, SV40/enhancer, CMV), that of ZsYellow1 (yellow fluorescence) and mCherry (red fluorescence) for the CMV promoter. Results varied with the cell line, in particular for the Jurkat cells. Pair-wise co-transfection of the CMV controlled transgenes resulted in a significant fraction of monochromatic cells (EGFP for EGFP/YFP and EGFP/RFP co-transfections, YFP in case of YFP/RFP co-transfections). Only Jurkat cells were almost incapable of expressing YFP. Dilution of the plasmid DNA with a non-expressed plasmid showed cell line dependent effects on transfection efficiency and/or expression levels.
1. Introduction
Recombinant protein production in mammalian cells requires the introduction of the respective DNA sequences into the cells’ nuclei (‘transfection’). For biotechnological applications, non-viral transfection methods, involving, e.g., polycations such as PEI (poly(ethyleneimine)) or PDMAEMA (poly(2-dimethylamino) ethyl methacrylate), are preferred [1], [2]. According to current understanding, the role of the polycation is to compact and charge-compensate the plasmid DNA (formation of ‘polyplexes’), thereby facilitating uptake by the cells. Depending on its chemistry, the transfection agent may then also aid endosomal escape (e.g. via the ‘proton sponge’ effect described for PEI [3]), transport through the cytosol, and finally release in the perinuclear area. In spite of some undeniable progress over the last decade [4], non-viral transfection agents are still orders of magnitude less efficient than viral ones [5]. This has led to intensive research on the effect of chemistry and structure of non-viral transfection agents. In this context, it is generally assumed that the polycation determines the transfection efficiency, i.e. the percentage of cells that take up the DNA, but transfects the DNA indiscriminately of the encoded information. Differences in transgene expression strength, on the other hand, are determined by features of the transfected DNA such as the promoter. Promoters are known to show some variability in different cell lines. However, the compliance of their performance with some general trends, e.g. in regard to relative strength and species-dependency, is typically assumed [6], [7], [8], [9].
Since the PDMAEMA chemistry is particularly suited to the synthesis of well-defined homopolymers with varied topologies, much of our knowledge on the structure-function-relationship of polycationic transfection agents was derived from experiments involving PDMAEMAs [2]. From these studies it was inter alia deduced that non-linear polycations are significantly less cytotoxic than linear ones of the same size and chemistry [10]. Based on this observation, our group has recently demonstrated that PDMAEMA-based star-shaped nanoparticles have high potential for transfecting mammalian cells including primary human blood cells [11], [12]. Fine-tuning of the respective transfection protocols in terms of optimizing efficiency and biocompatibility typically involved the transfection buffer in terms of composition, ionic strength, and pH as well as the N/P ratio (amount of nitrogen in the polymer/amount of phosphate in pDNA) of the polyplexes. Pronounced heterogeneities in transgene expression were observed among the cells of a given transfection batch in these experiments, while the establishment of a truly generic transfection protocol has so far been elusive [13].
The basis for the investigation of transfection outcomes has been changed some years ago by the advent of fluorescent reporter proteins [14]. These transgenes allow a direct statistical evaluation of the distribution of the expression strength over the individuals of a (living) cell population by flow cytometry. In consequence it becomes possible to differentiate whether a given amount of transgene is produced by a small number of high producers within the population or by a large number expressing low levels of the protein. In contrast, only average values can be determined in the case of reporter proteins requiring enzymatic conversion of added substrates for detection, such as luciferase or ß-galactosidase [15], since these assays are by necessity performed in the respective cell lysates.
In view of the widespread use of recombinant reporter proteins as tools, surprisingly little can be found in the literature in terms of a systematic investigation of their transfection taking the various putative impact factors into account. An area where this could be of particular importance is the co-transfection of a fluorescent reporter with another (fluorescent) transgene, where interference or competition could bias the results. For instance, the combination of two or more fluorescent reporters is an important tool in cell and tissue analytics (imaging). Molecular biosensors are used to study cellular and molecular heterogeneity or the long-term biological effects of signaling in stem cell research [16]. Fluorescent proteins can also be paired for quantitative multiparameter imaging of live systems in vivo and in vitro or for fluorescence resonance energy transfer (FRET) studies. Recognized advantages of the “two-color”-approach include the possibility of photo-switching as well as of bimolecular fluorescence complementation (BiFC) [17], [18]. Since flow cytometry can be set up to quantify several fluorescent dyes in parallel, it is a suitable technique for studying such effects.
Here, a popular reporter transgene, namely enhanced green fluorescent protein (EGFP) under the control of one out of four different promoters was initially transfected into two human and two rodent cell lines to test for putative promoter effects. Subsequently, plasmids encoding for this or two other fluorescent proteins each under the control of the cytomegalovirus (CMV) immediate early promoter were transfected or (pair-wise) co-transfected into the cells. Using three different fluorescent transgenes allowed us to statistically quantify specific effects on transfection efficiency as well as on the distribution of transgene expression strength by flow cytometry. To our knowledge, this is the first time that the co-expression strength distribution of independently transfected reporter proteins was determined in parallel.
2. Materials and methods
2.1. Materials
If not otherwise indicated, we used PAA Laboratories (Cölbe, Germany) or Greiner bio-one (Frickenhausen, Germany) as supplier for cell culture materials and Sigma-Aldrich for chemicals. Fetal calf serum (FCS) was from Biochrom AG (Berlin, Germany). Dulbecco’s Phosphate-Buffered Saline without Ca2+ and Mg2+ (DPBS) was from Lonza (Visp, Switzerland). HBG buffer (20 mM Hepes, 5 wt% glucose, pH 5.5) was prepared in house and sterilized by filtration. Cell culture media R10 (RPMI 1640 without glutamine, add 10 vol% fetal calf serum, 2 mM l-glutamine, 100 IU/mL Penicillin/100 μg/mL Streptomycin), MEM10 (MEM Earle’s without l-glutamine/FCS, add 10 vol% FCS, 4 mM l-glutamine, 100 IU/mL Penicillin/100 μg/mL Streptomycin), and Opti-MEM were from Lonza (Cologne, Germany), Biochrom AG (Berlin, Germany), and Thermo Fisher Scientific (Dreieich, Germany), respectively. For pre-equilibration, media were incubated for 1–4 h in a standard mammalian cell culture incubator (37 °C, 5% CO2, 95% humidity).
2.2. Cryogenic transmission electron microscopy (cryo-TEM)
For cryo-TEM studies, a drop (∼2 μL) of the aqueous micellar solution (concentration ca. 0.5 g/L) was placed on a lacey carbon-coated copper TEM grid (200 mesh, Science Services, Munich, Germany), where most of the liquid was removed with filter paper, leaving a thin film. The specimens were shock vitrified by rapid immersion into liquid ethane in a temperature-controlled freezing unit (Zeiss Cryobox, Carl Zeiss NTS GmbH, Oberkochen, Germany) and cooled to approximately 90 K. The temperature was monitored and kept constant in the chamber during the entire preparation. After freezing the specimen were inserted into a cryo-transfer holder (CT3500, Gatan GmbH, Munich, Germany) and transferred to a Zeiss EM922 OMEGA EFTEM instrument (Carl Zeiss NTS GmbH). Measurements were carried out at approximately 90 K. The electron microscope was operated at an acceleration voltage of 200 kV. Zero-loss filtered images (ΔE = 0 eV) were taken under reduced dose conditions. All images were recorded digitally by a bottom mounted CCD camera system (Ultrascan 1000, Gatan GmbH) and processed with a digital imaging processing system (Gatan Digital Micrograph 3.9 for GMS 1.4, Gatan GmbH).
2.3. Transfection agent
The transfection agent was a poly(1,2-butadiene)-block-PDMAEMA (B290D240) block-copolymer as described in [19]. B290D240 consists of a hydrophobic polybutadiene block with an average length of 290 monomeric units and a polycationic PDMAEMA block with an average length of 240 monomeric units. The number average molecular weight, Mn, of the molecule is 54 kDa, the polydispersity (Mw/Mn) is <1.07. Stable star-shaped micelles of B290D240 were formed by dissolving the block copolymer in THF and dialyzing the solution against DPBS as described elsewhere [11]. B290D240 micelles were prepared as a 10.7 mg/mL stock solution in sterile DPBS.
2.4. Aggregation number calculation
Aggregation numbers (Nagg) of the micelles were calculated according to:
with mcore = mass of the core, mPBchain = mass of one polybutadiene chain, NA = Avogadro’s constant, ρPB = density of polybutadiene (= 1 g/cm3), Rcore = radius of the core (as determined by TEM data), and MPBchain = molecular weight of one polybutadiene chain (=15,700 g/mol).
2.5. Cell lines and maintenance
CHO-K1 (adherent, CCL-61, ATCC), Jurkat (human leukemia T cells, suspension, TIB-152, ATCC), and HEK-293 (adherent, CRL-1573, ATCC) cells were maintained in R10, L929 cells (adherent, murine fibroblast, CCL-1, ATCC) in MEM10, as suggested by the supplier. Cells were cultivated in the standard mammalian cell culture incubator.
2.6. Plasmids
Plasmids were pEGFP-N1 (4.7 kb) encoding for the enhanced green fluorescent protein (EGFP) driven by the cytomegalovirus (CMV) immediate early promoter, pmCherry-N1 (4.7 kb) encoding for the mutant fluorescent protein derived from the tetrameric Discosoma sp. red fluorescent protein (mCherry, RFP) driven by the immediate early CMV promoter, pZsYellow1-N1 (4.7 kb) encoding for the yellow fluorescent protein (ZsYellow1, YFP) driven by the immediate early CMV promoter, all from Clontech Laboratories, Inc. (Mountain View, CA) and pIVEX23UK (4.4 kb, Roche, Mannheim, Germany) encoding for a mouse urokinase under the control of the bacteriophage T7 promoter. The latter was used as control plasmid as due to the chosen promoter the gene product is not expressed in mammalian cells. In addition, plasmids pSV40-EGFP-N1 (4.1 kb) encoding for EGFP driven by the simian virus 40 early promoter (SV40) and pSV40/enhEGFP-N1 (4.3 kb) encoding for EGFP driven by the SV40 promoter/enhancer were constructed by excising the coding sequence of EGFP out of pEGFP-N1 and subcloning it into the pGL3-promoter and pGL3-control plasmids (both from Promega, Mannheim, Germany) after deletion of the luciferase cDNA from these plasmids. pEF1α-EGFP-N1 (7.0 kb) encoding for EGFP driven by the elongation factor 1 alpha promoter (EF-1α) was constructed by subcloning the coding sequence of EGFP into plasmid pEAK8 (Edge BioSystem, Gaithersburg, MD).
Plasmids were amplified in Escherichia coli (LB medium) using standard laboratory techniques. The EndoFree Plasmid Kit (Giga Prep/Maxi Prep) from QIAGEN (Hilden, Germany) was used for purification (quality control: >80% supercoiled topology (agarose gel; 1% TAE) and A260/A280 ≥ 1.8). Purified plasmids were solubilized in sterile PCR-water (Sigma-Aldrich).
2.7. N/P-ratio calculation
N/P-ratios were calculated according to:
with [N] = concentration of nitrogen residues in mM.
2.8. Transfection
For transfection of adherent cells (CHO-K1, L929, HEK-293), the cells were harvested by trypsinization upon reaching 80% confluency and seeded at a density of 2 × 105 cells in 2 mL growth medium per well in six-well plates 24 h prior to transfection. CHO-K1 and L929 cells were rinsed with DPBS one hour prior to transfection and supplemented with 1 mL Opti-MEM. Afterwards the plate was put back into the incubator. In the meantime, polyplexes of pDNA and B290D240 were prepared in a final volume of 200 μL by first diluting the desired amount of pDNA stock solution with HBG buffer to a final concentration of approximately 10 μg/mL in a 1.5 mL reaction tube followed by the addition of the necessary amount of B290D240 stock solution in a single drop (max. volume: 20 μL) to achieve the intended N/P ratio (7.5 for CHO-K1, 10 for L929 cells). For the co-transfection experiments, the indicated amounts of both pDNAs were mixed before the addition of the B290D240. Afterwards, the mixture was vortexed for 10 s and incubated for 20 min at room temperature. 1 mL Opti-MEM was added, followed by vortexing and 10 min incubation at room temperature. The polyplex mixture was then added drop-wise to the cells and distributed by gently rocking the plate. Cultures were then put back into the incubator. 4 h later the medium was aspirated and replaced by 2 mL of fresh pre-equilibrated growth medium.
HEK-293 cells (adherent) were transfected using a slightly modified protocol (cells adhere only weakly, washing would have led to considerable losses). Briefly, HEK-293 cells were harvested and reseeded as described above for CHO-K1 and L929 cells, but then maintained under regular growth conditions right up to the time of transfection. The supernatant was cautiously aspirated and immediately replaced by 1.2 mL of the polyplex mixture (N/P-ratio 7.5, drop-wise addition). Subsequently, HEK-293 cells were treated as described above for the CHO-K1 and L929 cells.
For the transfection of Jurkat cells (suspension), cells were harvested by centrifugation (200g, 5 min) from cultures in the exponential growth phase (viability >90%) two hours prior to transfection. Cells were washed twice with DPBS, and seeded at 2 × 105 cells per well in 1 mL Opti-MEM (6-well plate). The plate was put back into the incubator for one hour. Polyplex preparation and addition to the cells were as given for the adherent cells. N/P-ratios were 7.5 in these experiments. After polyplex addition, the plates were placed into the incubator for 4 h and subsequently centrifuged (200g, 5 min). The supernatant was carefully aspirated and replaced by 2 mL of fresh pre-equilibrated growth medium.
Transfection experiments used in the comparisons were always done in parallel using one polyplex preparation (N/P-ratio 7.5); an exception was made for the L929 cells, where a polyplex preparation with an N/P-ratio of 10 was prepared instead and used in parallel. N/P-ratios were chosen according to standard protocols previously established in house for the different cell lines. In general, transfection efficiencies increase with the N/P-ratio, while the culture viability decreases. In order to choose the N/P-ratio to be used for transfection, the ratio is step-wise increased from the value sufficient to charge-compensate the pDNA (N/P = 3 in case of B290D240, as verified by zeta potential measurements of the formed polyplexes, Zetasizer Nano ZS, Malvern, Herrenberg, Germany), until either no further improvement in transfection efficiency is observed or the culture viability drops below a value of 70%. The fact that polyplexes used for transfection had a positive net-charge was verified by zeta potential measurements.
2.9. Flow cytometry
For flow cytometry (Cytomics FC500, Beckman Coulter, Krefeld, Germany), adherent cells were harvested by trypsinization (5 min), resuspended in the original culture supernatant to include detached/dead cells, recovered by centrifugation (200g, 5 min) and resuspended in 500 μL DPBS containing 1 μg/mL propidium iodide (PI) to counterstain the dead cells (except cells transfected with pmCherry-N1, where the red fluorescence caused by PI would have been indistinguishable from that of mCherry). Jurkat cells were directly recovered by centrifugation and resuspended in the 500 μL DPBS containing the PI. Forward scatter (FSC), side scatter (SSC), green fluorescence (em 510 nm), red fluorescence (em 620 nm) and yellow fluorescence (em 550 nm) were recorded. For this, the flow cytometer filter block configuration was modified to allow the simultaneous optical separation of green and yellow fluorescence (EGFP/YFP), which cannot be separated with the standard filters configuration due an extensive spectral overlap. To assure comparability between the obtained data, care was taken to use the same instrument settings in all experiments involving a particular reporter protein. Cells were initially evaluated by scatter properties (FSC/SSC) in order to select a region representing single, non-apoptotic cells, while disregarding dead cells, debris and cellular aggregates. Cells ‘transfected’ at N/P = 0, i.e. in the absence of the transfection agent, were used to set the measurement parameters. Histogram plots of the respective fluorescence intensities (log scale) were used to estimate the percentage of transfected cells and the expression strength distribution according to: low producers (L): fluorescence intensity between 1 and 10 a.u.; middle producers (M): fluorescence intensity between 10 and 100 a.u.; high producers (H): fluorescence intensity >100 a.u.) in the non-apoptotic cell population; with a.u.: arbitrary units.
2.10. Statistical analysis
Group data are reported as mean ± SD. One-way ANOVA was used to determine whether data groups differed significantly from each other. Statistical significance was defined as p < 0.05.
3. Results and discussion
3.1. The transfection agent
To exclude any influence of the transfection agent, experiments were exclusively conducted with B290D240, a polycationic agent, which has previously been described for its ability to transfect siRNA into CHO cells [11]. In aqueous solution B290D240 forms stable star-shaped micelles with the polybutadiene block forming a hydrophobic inner core, as confirmed by cryogenic TEM. This micellar structure can therefore be considered a somewhat unusual yet functional representation of the general design principle of a star-shaped polycationic agent, which otherwise [11], [12] has been embodied in the form of polycationic arms covalently grown from a solid central core. B290D240 is by now routinely used for transfection in our group, as it gives superior results for a large number of cells, some of which are included in this publication.
Evaluation of approximately 100 micelles by image analysis of TEM data allowed calculating an average core diameter of Dcore = 18.2 ± 2.5 nm, i.e. a core radius Rcore of 9.1 nm, for the micelles. Under the assumption that the core of the micelles is spherical and consists only of polybutadiene, an average aggregation number Nagg of 120 was calculated for the micelles. Compared, e.g. to the gold standard among the polycationic transfection agents, namely PEI, B290D240 has a very low polydispersity (<1.1), while according to the TEM data, the corresponding micelles have a narrow size distribution. This should reduce the contribution of the transfection agent’s heterogeneity to the variability of the experiments.
3.2. Influence of the promoter
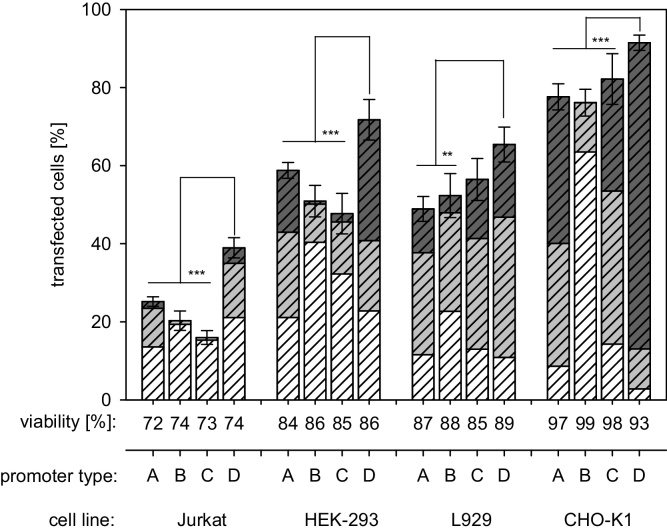
The influence of the promoter on the transfection outcome for a given cell line under otherwise identical experimental conditions was tested using the enhanced green fluorescent protein (EGFP) as reporter gene product. The transfection efficiency (percentage of cells expressing the transgene within the population) and the distribution of the expression strength (high, middle, and low producers) among the successfully transfected cells was analyzed 48 h post transfection. Results are given in Fig. 1 together with the culture viabilities.
Fig. 1.
Promoter effect on transfection efficiency and expression strength distribution. EGFP expression under the control of the cellular (A = pEF1α-EGFP-N1) and the three viral (B = pSV40-EGFP-N1, C = pSV40/enhEGFP-N1, D = pEGFP-N1, i.e. CMV) promoters was quantified 48 h post transfection. The height of the bar corresponds to the overall transfection efficiency, i.e. the percentage of transgene expressing cells within the population. The transfected cell population was further divided into: low producers (white, fluorescence signal between 1 and 10 a.u.), middle producers (grey, fluorescence signal between 10 and 100 a.u.), and high producers (dark grey, fluorescence signal >100 a.u.). Group data are reported as mean ± s.d. from three or more independent experiments. Statistical significance is indicated by * (p < 0.05).
Transfection efficiencies in Fig. 1 vary considerably in spite of the fact that the same transfection agent and conditions had been used. For a given transfection agent some dependency of the transfection efficiency on the on cell type has to be expected. The promoter, on the other hand, should have an effect on the strength of the transgene expression. This is, e.g., seen in case of the CHO-K1 cells for the three viral promoters. Expression was weakest under the control of the SV40 promoter (Fig. 1, bar B). Better expression was obtained in the presence of the SV40/enhancer (Fig. 1, bar C). The CMV promoter was by far the most effective in the CHO-K1 cells, leading to almost 80% of high producers (Fig. 1, bar D). Interestingly, the human cellular promoter EF-1α performed at least as well as the SV40/enhancer in the CHO-K1 cells (Fig. 1, bar A).
A promoter effect on the transfection efficiency, i.e. DNA uptake, was not expected, since for a given cell type/transfection agent, DNA uptake should be independent of the sequence details of the transfected DNA. If anything, the plasmid size has in the past been shown to affect DNA uptake. However, this was not the case here, where the percentage of transfected cells was similar for pEF1α-EGFP-N1 (7.0 kb) and pSV40-EGFP-N1 (4.1 kb) or pSV40/enhEGFP-N1 (4.3 kb). The significantly higher percentage of transfected cells observed in all experiments involving the CMV promoter thus has to be noted. It is possible that this is an artifact and that the apparent higher percentage of transgene expressing cells found for the CMV promoter is instead simply related to a stronger transgene expression. Only a few low producers were obtained with the CMV promoter. For the other promoters the low producer fraction was much larger. It is possible that in such cases some very low producers are lost during gating, thereby reducing the percentage of transfected cells. However, a direct effect of the viral CMV promoter on DNA uptake in addition to gene expression cannot be excluded at this point.
Trends observed in the second rodent cell line (L929) for the promoter effect on transfection efficiency and expression strength were similar, with the exception of the human cellular promoter EF-1α, for which the lowest transfection efficiency was observed in case of the L929 cells. This we interpret as a size effect. As mentioned above, plasmid size differences are known to cause differences in transfection efficiency.
In the two human cell lines, the CMV and the human cellular promoter EF-1α show a similar distribution of the expression strength over high, middle and low producers, but the transfection efficiency is again significantly lower in case of the larger pEF1α-EGFP-N1 plasmid. If one compares the performance of the three viral promoters in the human cell lines, expression driven by the SV40 promoter is again weaker than for the CMV promoter, while contrarily to the rodent cells, expression strength is not improved in the human cells by the presence of the SV40/enhancer. In addition to the – expected – influence on the expression strength, however, in the human cells as well we see a pronounced promoter effect in the total transfection efficiency between the CMV promoter and the three others, arguing once more for a promoter effect on non-viral DNA uptake. Incidentally we never observed any silencing of the CMV promoter as observed by Quin et al. [20] in our experiments.
3.3. Influence of the transgene
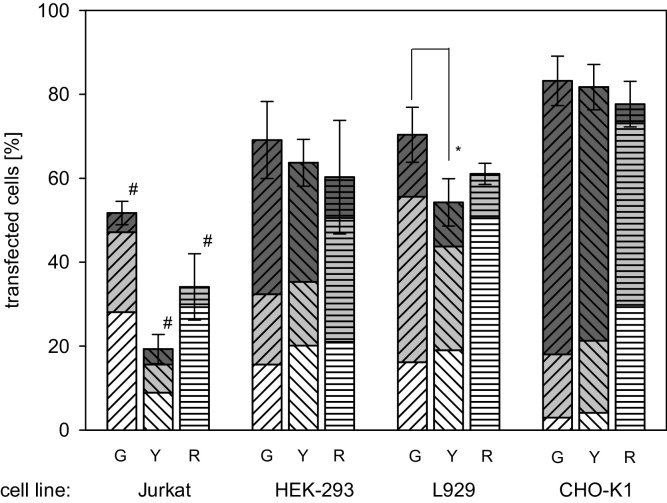
In order to investigate putative effects of the reporter transgene, two additional fluorescent proteins, namely YFP (yellow fluorescent protein) and RFP (red fluorescent protein) were expressed under the control of the immediate early CMV promoter in the investigated cell lines. Experiments were again performed in parallel for the four cell lines keeping experimental conditions as similar as possible. Results are summarized in Fig. 2. Since it is not possible to distinguish between the fluorescence of PI and RFP, cell viabilities could only be determined for cells transfected with EGFP and YFP. However, all determinable viability values were in the same range as those observed before and we assume this to be the case for RFP-transfected cells as well.
Fig. 2.
Transgene effect on transfection efficiency and expression strength distribution. Expression of EGFP (G, diagonal top right lines), YFP (Y, diagonal top left lines) and RFP (R, horizontal lines) all under the control of the intermediate early CMV promoter was quantified 48 h post transfection. The height of the bar corresponds to the overall transfection efficiency, i.e. the percentage of transgene expressing cells within the population. The transfected cells population was further divided into: low producers (white, fluorescence signal between 1 and 10 a.u.), middle producers (grey, fluorescence signal between 10 and 100 a.u.), and high producers (dark grey, fluorescence signal >100 a.u.). Group data are reported as mean ± s.d. from three or more independent experiments and the statistical significance is indicated by * (p < 0.05). Statistical significance within a cell line is indicated by # (p < 0.05).
Transfection efficiencies in these experiments showed no statistically relevant variation in HEK-293 and CHO-K1 cells, while the criterion for statistical significance, defined as p < 0.05, was just slightly surpassed in case of the L929 cells. This result is not surprising since the three vectors belong to the same plasmid family and therefore have a similar backbone/size.
However, in spite of the fact that the same strong (CMV) promoter was used to drive expression in all three vectors, differences in the expression strength were observed. Whereas EGFP and YFP showed a similar distribution over low, middle and high producers, RFP expression was less strong in all investigated cell lines (almost no high producers, mainly low producers). Codon usage in case of EGFP has been optimized for mammalian and in case of YFP even for human cells. This is not the case for RFP. However, it is unlikely that the lower expression observed for RFP is due to suboptimal codon usage, as was verified by analysis of the cDNA sequence [21]. Problems with incomplete maturation, which have been reported for the tetrameric parent protein DsRed [22], [23], should not occur for the monomeric mutant mCherry (RFP), which has been reported to have a halftime for maturation of 10 min (DsRed: 10 h). However, since we consistently observe ‘low RFP expression’ in all investigated cell lines, it is possible that this is simple due to difficulties in detecting the red fluorescence, i.e. a lower ‘brightness’ of the protein. According to Shaner et al., mCherry is also more sensitive to photobleaching than, e.g., EGFP [24].
While a consistently lower strength of transgene expression in the case of mCherry can therefore be attributed to the reporter protein itself, this is not the case for the transfection results obtained for the Jurkat cells. While the comparatively low transfection efficiencies found for EGFP (50%) and RFP (34%) were within the range seen above for EGFP expression under the CMV promoter, transfection of the Jurkat cells with YFP was almost impossible (transfection efficiencies <20%). It should be noted that in spite of the low overall transfection efficiency, the expression strength distribution of the few YFP-transfected Jurkat cells showed a significant fraction of high producers. In fact, in the Jurkat cells the relative size (percentage) of the high producer fraction was higher for YFP than for the other two investigated fluorescent proteins. The optimized codon usage might be in that case responsible for a more efficient translation of YFP in human cells. Thus with pZsYellow1-N1 we have an example for a plasmid which in spite of a similar backbone is much more difficult to transfect into Jurkat cells than pEGFP-N1 or pmCherry-N1, but which in the few successfully transfected cells is expressed with superior strength.
Moreover, in spite of their size B290D240 micelles were found to be very biocompatible. In particular, the direct correlation between transfection efficiency and cytotoxicity reported for most non-viral transfection agents was not seen in this case. Even Jurkat cells, which are known to be sensitive to polycationic transfection agents, showed viabilities >70% post transfection, whereas the values for the other investigated cell lines were routinely above 80% (>90% in case of the CHO cells).
3.4. Pair-wise co-transfection of the reporter genes
Experiments involving more than one reporter protein have significance beyond the investigation of promoter/transgene effects. However, as far as we could ascertain, the co-expression of two fluorescent proteins in mammalian cells has so far not been studied in a systematic manner. Zhu et al. [25] reported the simultaneous detection and quantification of EGFP, YFP, and CFP (cyan fluorescent protein) expressed from di- and tricistronic constructs after transfection with Lipofectamine in HEK 293 cells. However, the authors only gave the MFI (mean fluorescence intensity) for the cultures and did not discriminate between transfected and non-transfected cells or low, medium and high producers.
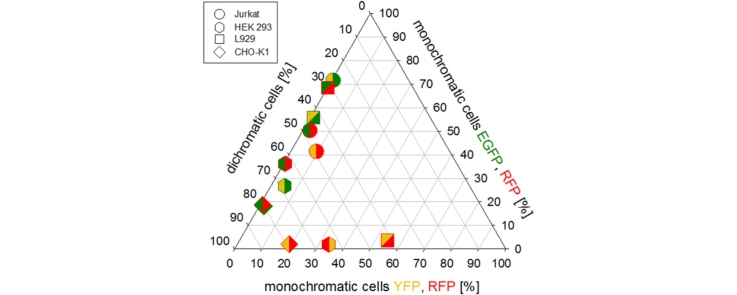
In order to investigate effects in co-transfection, the plasmids encoding for the three fluorescent proteins under the control of the CMV promoter were transfected pair-wise into the cells. Equal amounts (by weight) of plasmid were used, while keeping the total amount of plasmid DNA the same as in the previous experiments. The results are summarized in Fig. 3. In this figure the percentage of bichromatographic cells within the population as well as the percentages of cells expressing only one of the respective reporter proteins (“monochromatic” cells) are shown. Within each subpopulation, the expression strength is again indicated by shading. Please note that in this figure the overall transfection efficiency corresponds to the sum of all transfected subpopulations, i.e. the percentage of bichromatographic cell plus the percentage of monochromatic cells of subtype 1 and the monochromatic cells of subtype 2. Subpopulations with a strength ≤1% are not shown in this presentation.
Fig. 3.
Co-transfection experiments, involving pair-wise application of two encoding plasmid DNAs. Equal amounts (by weight) of pDNA encoding for the respective reporter proteins under the control of the intermediate early CMV promoter were used, with the total amount of pDNA equaling that used in the experiments involving only one reporter protein under otherwise identical experimental conditions. Expression of EGFP (diagonal top right lines), YFP (diagonal top left lines) and RFP (horizontal lines) under the control of the intermediate early CMV promoter was quantified 48 h post transfection. Indicated are bichromatic cells expressing both proteins and monochromatic cells expressing only one of the proteins. Transfected cells population was further divided into: low producers (white, fluorescence signal between 1 and 10 a.u.), middle producers (grey, fluorescence signal between 10 and 100 a.u.), and high producers (dark grey, fluorescence signal >100 a.u.).
For all investigated cells, except the Jurkat cells, total transfection efficiencies were highest for cells transfected with EGFP/YFP, where they were roughly in the same order of magnitude as in the experiments involving only EGFP, see Fig. 1 for comparison. In case of the EGFP/RFP transfection, overall transfection efficiencies were slightly lower, while the lowest values were obtained in the experiments involving YFP/RFP. Moreover, instead of obtaining only – or at least mainly – bichromatic cells, the co-expression experiments unexpectedly resulted in significant numbers of monochromatic cells. In general, the propensity for co-expression depended on both the cell type and the reporter protein(s). For instance, the CHO-K1 cells consistently showed the highest co-expression rates (≥80% of the transfected cells), whereas this fraction could be as low as 30% in case of the Jurkat or L929 cells. In the co-expression experiments involving EGFP, the majority of the monochromatic cells expressed EGFP and not the respective other protein. In the YFP/RFP experiments, all investigated cells, except for the Jurkat cells, expressed either both proteins or only YFP. The fraction of monochromatic RFP-expressing cells was less than 2% of the transfected cells.
Results from the Jurkat cells, on the other hand, point again towards a general difficulty with YFP. In the EGFP/YFP-co-transfections an unusual high fraction (>70%) of the transfected cells were monochromatic and expressed EGFP, while in the EGFP/RFP co-expression experiments the percentage of monochromatic (green) cells was only 50% of the transfected cells. Moreover, the Jurkat cells were the only ones, where a significant number of the transfected cells (>40%) expressed only the red fluorescent protein after co-transfection with YFP/RFP.
When cells expressing one or both fluorescence proteins were further classified into high, middle, and low producers, Fig. 3, it became clear that high producers were mainly found among the bichromatic cells, whereas the monochromatic cells from the same population tended to be middle and low producers. Thus cells that strongly expressed the fluorescent proteins often were able to co-express both proteins, while the inability to co-express the two fluorescent proteins coincides with a low overall expression level.
3.5. Effect of pDNA dilution
If one considers transfection efficiencies per individual transgene, for EGFP similar levels were reached in the co-transfection experiments in spite of the fact that only half as much pDNA had been used per individual reporter protein compared to the experiments involving only one type of pDNA. For example, the CHO-K1 cell population in Fig. 3 co-transfected with pEGFP-N1 and pZsYellow1-N1 contained a total of 83% EGFP positive cells (66.7% bichromatic cells expressing both proteins, 15.1% monochromatic ones expressing only EGFP). It is thus unlikely that the reduction in expression observed for the other two fluorescent proteins in the co-transfection experiments was due to the fact that less encoding pDNA was transfected in these experiments. Such a reduction in protein expression as a function of the amount of transfected plasmid has, e.g. been observed by Liu et al. [26]. Instead, since in our case all proteins were under the control of the same – strong – CMV promoter, a “saturation effect”, e.g. due to competition for some transcription factors, could not be excluded.
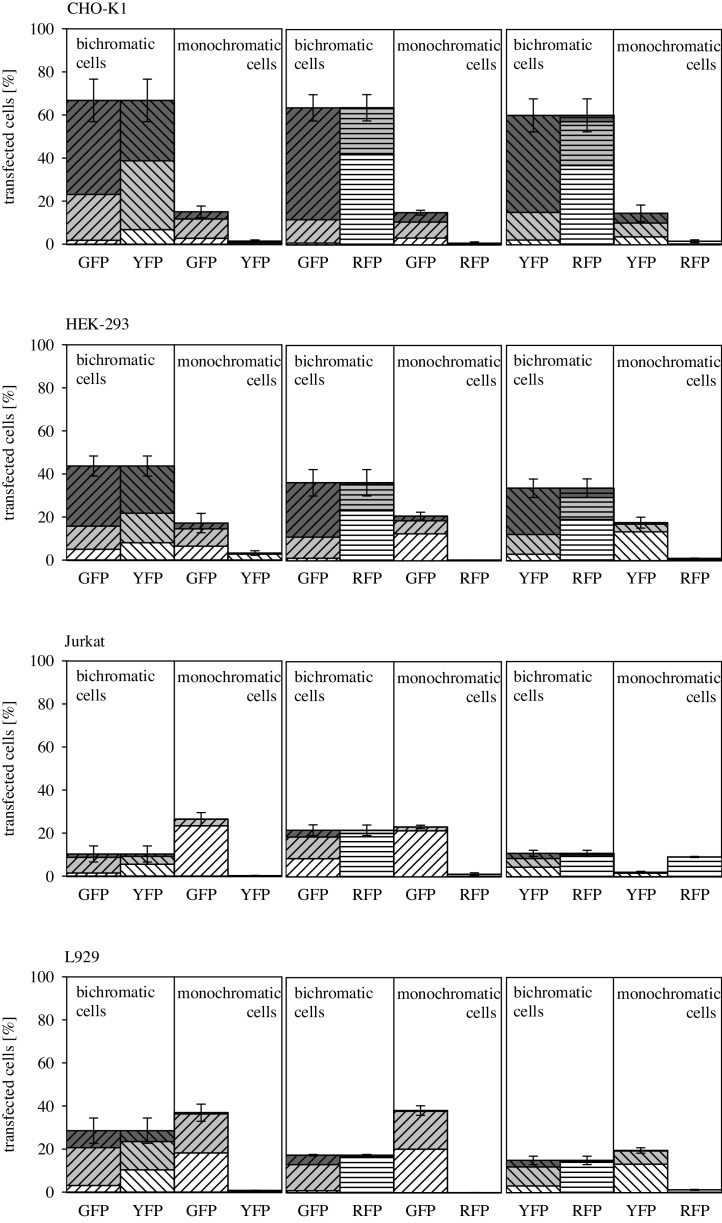
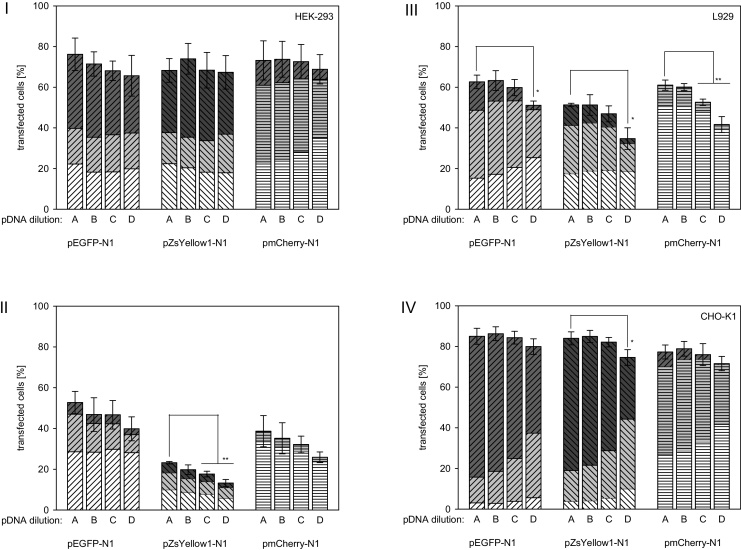
Subsequently, we therefore analyzed the effect of specific plasmid content in a transfection cocktail. A simple reduction of the pDNA amount was not suitable for this purpose, since this would have affected the N/P-ratio, which is know to affect both transfection efficiency and culture viability [27]. Adding varied amounts of polyplexes to the cells was also no option. Instead, a pDNA (pIVEX23UK) encoding for a mouse urokinase under the control of the T7 promoter, was used to dilute the pDNAs encoding for the various fluorescent proteins. T7 is a bacteriophagic promoter and requires the presence of the bacteriophage T7 RNA polymerase for activity. Unless mammalian cells are genetically modified to express this polymerase, which was not the case in our experiments, this promoter is not active [28], [29]. The presence of pIVEX23UK should therefore not interfere with the expression of EGFP, YFP or RFP by the cells. The results of the experiments are summarized in Fig. 4. Bars A–D represent experiments in which 0, 25, 50, or 75 wt% of the specific pDNA had been replaced by pIVEX23UK. In other words, bar A can be directly compared to the experiments summarized in Fig. 1, while the specific plasmid amounts in the experiments corresponding to bar C were identical to the ones used in the experiments summarized in Fig. 3.
Fig. 4.
Effect of pDNA dilution on transfection efficiency and expression strength distribution. (I) HEK-293, (II) Jurkat, (III) L929, (IV) CHO-K1. Expression of EGFP (diagonal top right lines), YFP (diagonal top left lines) and RFP (horizontal lines) under the control of the intermediate early CMV promoter was quantified 48 h post transfection with polyplexes containing decreasing amounts of encoding pDNA. For dilution of the specific pDNA at constant total pDNA in the transfection cocktail, polyplexes were prepared replacing 0, 25, 50, and 75 wt% (A–D) of the encoding DNA by a non-expressed control plasmid (pIVEX23UK). Transfected cells were further divided into: low producers (white, fluorescence signal between 1 and 10 a.u.), middle producers (grey, fluorescence signal between 10 and 100 a.u.), and high producers (dark grey, fluorescence signal >100 a.u.). Group data are reported as mean ± s.d. from three or more independent experiments. Statistical significance is indicated by * (p < 0.05).
Some differences are again observed as a function of the cell and transgene type. CHO-K1 cells, for example, showed constant transfection efficiencies independent of the dilution, with the possible exception of perhaps the very lowest plasmid content (25 wt% of the original, bar D), where a statistical difference in transfection efficiency can be observed at least for pZsYellow1-N1. The expression strength distribution, on the other hand, was increasingly shifting away from the high producers in the CHO cell experiments as the plasmid was diluted. Thus, the number of cells that take up the plasmid seems to be similar regardless of dilution, but high producers are more likely to occur when the specific plasmid is not diluted. This would argue against the hypothesis of a transcription factor shortage at higher plasmid concentrations, since more plasmid obviously still results in more protein. Hence such a shortage is also unlikely to be directly responsible for the decreased protein expression in some of the co-expression experiments discussed above.
In the HEK-293 cells, on the other hand, neither the transfection efficiency nor the expression strength is affected by the plasmid dilution, except perhaps in case of RFP, where the fraction of high and middle producers diminishes in case of the highest plasmid dilution (bar D). A similar behavior, i.e. comparable transfection efficiency and expression level distribution except for the highest dilution, is observed for the L929 cells, but in this case for all three fluorescent proteins, while Jurkat cells consistently show a statistically significant decrease in transfection efficiency, but not in the expression strength distribution with dilution. A mechanistic interpretation of these observations would require a detailed study of the intracellular events, which is far beyond the scope of this work. However, a clear conclusion can be drawn regarding the necessity to carefully evaluate the applicability of fluorescence reporter proteins (pairs) for any given cellular system.
4. Conclusions
The expression of a recombinant protein depends for a given cell line and transfection protocol on both the promoter and the transgene sequence. No general tendencies could be observed for this behavior in our experiments, not even among the two investigated human and rodent cell lines, respectively. ‘Standard’ conditions such as CMV promoter, EGFP as reporter gene product, and the chosen standard transfection protocol worked well for the CHO-K1 cells for which they had been originally developed, but were not always equally successful for the other investigated cells. Moreover, based on past experience in our group with the development of transfection protocols, the effect achievable with optimizing a given protocol is much smaller than the differences observed here as a function of the promoter/transgene type. We therefore propose that any development of a transfection protocol for mammalian cells should start with a chemometric multi-parameter investigation involving cell type, promoter type, transgene, and last but not least also the transfection agent. This aspect was not included in our investigation, but it is likely that not all transfection agents show similar performance for all cell types.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
This work was funded by the Upper Franconian Trust (Oberfrankenstiftung, Bayreuth, Germany) grant P-Nr.: 03847. Ullrich Stahlschmidt cloned the additional plasmids, while Andrea Schott and Philipp Neßbach supported this study by purifying some of the plasmids.
References
- 1.Boussif O., Lezoualc’h F., Zanta M.A., Mergny M.D., Scherman D., Demeneix B. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. U. S. A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agarwal S., Zhang Y., Maji S., Greiner A. PDMAEMA based gene delivery materials. Mater. Today. 2012;15:388–393. [Google Scholar]
- 3.Akinc A., Thomas M., Klibanov A.M., Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J. Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 4.Li S.-D., Huang L. Non-viral is superior to viral gene delivery. J. Control. Release. 2007;123:181–183. doi: 10.1016/j.jconrel.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 5.Pezzoli D., Chiesa R., De Nardo L., Candiani G. We still have a long way to go to effectively deliver genes! J. Appl. Biomater. Funct. Mater. 2012;10:82–91. doi: 10.5301/JABFM.2012.9707. [DOI] [PubMed] [Google Scholar]
- 6.Chan K.K.-K., Wu S.M., Nissom P.M., Oh S.K.W., Choo A.B.H. Generation of high-level stable transgene expressing human embryonic stem cell lines using Chinese hamster elongation factor-1 alpha promoter system. Stem Cells Dev. 2008;17:825–836. doi: 10.1089/scd.2007.0233. [DOI] [PubMed] [Google Scholar]
- 7.Tokushige K., Moradpour D., Wakita T., Geissler M., Hayashi N., Wands J.R. Comparison between cytomegalovirus promoter and elongation factor-1 alpha promoter-driven constructs in the establishment of cell lines expressing hepatitis C virus core protein. J. Virol. Methods. 1997;64:73–80. doi: 10.1016/s0166-0934(96)02143-x. [DOI] [PubMed] [Google Scholar]
- 8.Gopalkrishnan R.V., Christiansen K.A., Goldstein N.I., DePinho R.A., Fisher P.B. Use of the human EF-1alpha promoter for expression can significantly increase success in establishing stable cell lines with consistent expression: a study using the tetracycline-inducible system in human cancer cells. Nucleic Acids Res. 1999;27:4775–4782. doi: 10.1093/nar/27.24.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Addison C.L., Hitt M., Kunsken D., Graham F.L. Comparison of the human versus murine cytomegalovirus immediate early gene promoters for transgene expression by adenoviral vectors. J. Gen. Virol. 1997;78:1653–1661. doi: 10.1099/0022-1317-78-7-1653. [DOI] [PubMed] [Google Scholar]
- 10.Schallon A., Jérôme V., Walther A., Synatschke C.V., Müller A.H.E., Freitag R. Performance of three PDMAEMA-based polycation architectures as gene delivery agents in comparison to linear and branched PEI. React. Funct. Polym. 2010;70:1–10. [Google Scholar]
- 11.Schallon A., Synatschke C.V., Jérôme V., Müller A.H.E., Freitag R. Nanoparticulate nonviral agent for the effective delivery of pDNA and siRNA to differentiated cells and primary human T lymphocytes. Biomacromolecules. 2012;13:3463–3474. doi: 10.1021/bm3012055. [DOI] [PubMed] [Google Scholar]
- 12.Majewski A.P., Stahlschmidt U., Jérôme V., Freitag R., Müller A.H.E., Schmalz H. PDMAEMA-grafted core–shell–corona particles for nonviral gene delivery and magnetic cell separation. Biomacromolecules. 2013;14:3081–3090. doi: 10.1021/bm400703d. [DOI] [PubMed] [Google Scholar]
- 13.van Gaal E.V.B., van Eijk R., Oosting R.S., Kok R.J., Hennink W.E., Crommelin D.J.A. How to screen non-viral gene delivery systems in vitro? J. Control. Release. 2011;154:218–232. doi: 10.1016/j.jconrel.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Shaner N.C., Steinbach P.A., Tsien R.Y. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 15.Jiang T., Xing B., Rao J. Recent developments of biological reporter technology for detecting gene expression. Biotechnol. Genet. Eng. Rev. 2008;25:41–75. doi: 10.5661/bger-25-41. [DOI] [PubMed] [Google Scholar]
- 16.Endele M., Schroeder T. Molecular live cell bioimaging in stem cell research. Ann. N. Y. Acad. Sci. 2012;1266:18–27. doi: 10.1111/j.1749-6632.2012.06560.x. [DOI] [PubMed] [Google Scholar]
- 17.Müller-Taubenberger A., Anderson K.I. Recent advances using green and red fluorescent protein variants. Appl. Microbiol. Biotechnol. 2007;77:1–12. doi: 10.1007/s00253-007-1131-5. [DOI] [PubMed] [Google Scholar]
- 18.Ibraheem A., Campbell R.E. Designs and applications of fluorescent protein-based biosensors. Curr. Opin. Chem. Biol. 2010;14:30–36. doi: 10.1016/j.cbpa.2009.09.033. [DOI] [PubMed] [Google Scholar]
- 19.Schacher F., Müllner M., Schmalz H., Müller A.H.E. New block copolymers with poly(N,N-dimethylaminoethyl methacrylate) as a double stimuli-responsive block, macromol. Chem. Phys. 2009;210:256–262. [Google Scholar]
- 20.Qin J.Y., Zhang L., Clift K.L., Hulur I., Xiang A.P., Ren B.-Z. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS One. 2010;5:e10611. doi: 10.1371/journal.pone.0010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genscript, Rare Codon Analysis – Rare Codon Analysis Tool – Gene Analysis Tool, (2015). http://www.genscript.com/cgi-bin/tools/rare_codon_analysis (accessed 6.11.15).
- 22.Baird G.S., Zacharias D.A., Tsien R.Y. Biochemistry mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. U. S. A. 2000;97:11984–11989. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gross L.A., Baird G.S., Hoffman R.C., Baldridge K.K., Tsien R.Y. The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. U. S. A. 2000;97:11990–11995. doi: 10.1073/pnas.97.22.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N.G., Palmer A.E., Tsien R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 25.Zhu J., Musco M.L., Grace M.J. Three-color flow cytometry analysis of tricistronic expression of eBFP, eGFP, and eYFP using EMCV-IRES linkages. Cytometry. 1999;37:51–59. [PubMed] [Google Scholar]
- 26.Liu C., Dalby B., Chen W., Kilzer J.M., Chiou H.C. Transient transfection factors for high-level recombinant protein production in suspension cultured mammalian cells. Mol. Biotechnol. 2008;39:141–153. doi: 10.1007/s12033-008-9051-x. [DOI] [PubMed] [Google Scholar]
- 27.Synatschke C.V., Schallon A., Jérôme V., Freitag R., Müller A.H.E. Influence of polymer architecture and molecular weight of poly(2-(dimethylamino)ethyl methacrylate) polycations on transfection efficiency and cell viability in gene delivery. Biomacromolecules. 2011;12:4247–4255. doi: 10.1021/bm201111d. [DOI] [PubMed] [Google Scholar]
- 28.Elroy-Stein O., Moss B. Cytoplasmic expression system based on constitutive synthesis of bacteriophage T7 RNA polymerase in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 1990;87:6743–6747. doi: 10.1073/pnas.87.17.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghaderi M., Sabahi F., Sadeghi-Zadeh M., Khanlari Z., Jamaati A., Mousavi-Nasab D. Construction of an eGFP expression plasmid under control of T7 promoter and IRES sequence for assay of T7 RNA polymerase activity in mammalian cell lines. Iran. J. Cancer Prev. 2014;7:137–141. [PMC free article] [PubMed] [Google Scholar]