Highlights
-
•
Medium optimization for MPA production using P. brevicompactum by one-factor-at-a-time and CCD methods.
-
•
CCD afforded a 40% higher MPA titer than one-factor-at-a-time method.
-
•
The titer was nearly 6-fold higher compared to un-optimized medium.
Keywords: Mycophenolic acid, Immunosuppressant, Penicillium brevicompactum, Fermentation optimization, CCD
Abstract
Production of mycophenolic acid (MPA) by submerged fermentation using the microfungus Penicillium brevicompactum MTCC 8010 is reported here. Screening experiments were used to identify: the suitable media composition; the optimal initial pH; and the optimal incubation temperature to maximize the production of MPA in batch cultures. The initial concentrations of the selected sources of carbon (glucose), nitrogen (peptone) and the precursors (methionine, glycine) were then optimized by: (1) one-at-a-time variation of factors; and (2) a central composite design (CCD) of experiments, in a 12-day batch culture at an initial pH of 5.0, an incubation temperature of 25 °C, and an agitation speed of 200 rpm. The medium optimized using the one-at-a-time variation yielded a peak MPA titer of 1232 ± 90 mg/L. The medium optimized by the CCD method yielded a 40% higher MPA titer of 1737 ± 55 mg/L. The latter value was nearly 9-fold greater than the titer achieved prior to optimization.
1. Introduction
Mycophenolic acid (6-(4-hydroxy-6-methoxy-7-methyl-3oxophthalanyl)-4-methyl-4-hexenic acid, C17H20O6, MPA) is a fungal secondary metabolite that was first isolated in 1893 as an antibiotic against Bacillus anthracis [1], [2]. MPA is produced by many Penicillium species [3], [4], [5], [6], [7], [8] as well as other fungi [6], [9]. MPA and its derivatives are commercially used as frontline immunosuppressive agents to prevent rejection of transplant organs [10], [11], [12]. Commercial immunosuppressants based on MPA include CellCept (mycophenolate mofetil; Roche) and Myfortic (mycophenolate sodium; Novartis). In 2014, the sales of Myfortic were around US$ 543 million [13]. Similarly, in 2013 US$ 938 million worth of CellCept was sold [14]. Mycophenolate mofetil in combination with cyclosporine A (CyA) has been shown to reduce the incidence of graft rejection to 17% compared to a 60% rejection when CyA was used alone.
Here we report on the production of mycophenolic acid (MPA) by Penicillium brevicompactum MTCC 8010 (Institute of Microbial Technology, Chandigarh). The batch fermentation conditions are optimized to maximize the production of MPA. Two optimization approaches are compared: (1) the conventional one-at-a-time variation of factors; and (2) a simultaneous variation of the multiple factors based on a statistically robust central composite experiment design.
The relevant aspects of biosynthesis of MPA by P. brevicompactum have been discussed in the literature [1], [15], [16]. The chemistry, synthesis and modifications of MPA for improving the biological activity have been previously reviewed [17]. In submerged batch fermentations, low-intensity pulsed ultrasound has enhanced the production of MPA by P. brevicompactum [18]. Production in this fungus may also be influenced by the intensity and wavelength of the prevailing light [19], although most commercial submerged fermentations are inevitably conducted in the dark. Production of MPA by solid-state fermentation has been extensively reported in literature [7], [20], [21], but solid state fermentations are generally not used for the commercial production of medicinal products.
2. Materials and methods
2.1. Microorganism and inoculum preparation
The microfungus Penicillium brevicompactum MTCC 8010 used in this work was obtained from the Institute of Microbial Technology, Chandigarh, India. The culture was maintained aseptically on potato dextrose agar (PDA) slants. A spore suspension was used as inoculum. The spores scarped from the surface of a slant were transferred to PDA petri plates and incubated at 25 °C for 3-5 days. The spores from the plates were harvested aseptically with a sterile loop and dispersed in sterile distilled water to obtain a concentration of 107 spores per milliliter. A hemocytometer was used for the spore counts.
2.2. Media
The basal medium had the following composition (g/L): glucose 10, glycine 2, methionine 0.5, KH2PO4 2, MgSO4·7H2O 1, and 1 mL/L of a trace element solution. The latter contained the following (g/L): FeSO4·7H2O 2.2, CuSO4·5H2O 0.3, ZnSO4·7H2O 2.4, MnSO4·4H2O 0.16, and (NH4)2MoO4 0.2. The media components except glycine, methionine and trace element solution, were autoclaved separately at 121 °C for 15 min after the pH had been adjusted to 6.0 with 2 M HCl or 2 M NaOH solutions. Solutions of glycine, methionine and the trace elements were sterilized using a sterile 0.2 μm membrane filter (Millipore; www.emdmillipore.com).
2.3. Fermentations
Batch fermentations were carried out in 250 mL shake flasks each containing 50 mL of the culture medium. The medium always contained the following components (per L): MgSO4·7H2O 1 and the trace element solution (1 mL) specified earlier. This medium was supplemented with the required concentrations of the specified carbon source, the nitrogen source, the phosphate source and the potential precursors. The initial pH was adjusted to the required value. This medium (50 mL) was inoculated with 0.5 mL of the above specified spore suspension. Incubation occurred at the specified temperature. All fermentations were lasted for 12 days. The agitation speed of the platform shaker was 200 rpm. All fermentations were run in duplicate.
2.4. Analytical methods
2.4.1. Mycophenolic acid concentration
The MPA concentration was measured in the culture supernatant at the end of fermentation. The pH of the filtered supernatant was adjusted to 2.0 with 1 M HCl. This supernatant (50 mL) was extracted twice with ethyl acetate (100 mL for each extraction). The total duration of extraction was 30 min. The organic phase was recovered, pooled and evaporated at reduced pressure in a rotary evaporator (Rotavapor, R-200; www.buchi.com). The residue was dissolved in 1 mL of methanol, filtered through a 0.45 μm filter and measured via high performance liquid chromatography (HPLC) (Waters Alliance HPLC; www.waters.com). A C18 column (Symmetry® C18, 5 μm, 4.6 × 250 mm) was used at 40 °C. The mobile phase was water and acetonitrile (50: 50 by vol), pH 3.0, at a flow rate of 0.5 mL/min. The injection volume was 10 μL. A photodiode array detector was used at a wavelength of 220 nm [22].
A standard curve (data not shown) was prepared using HPLC-grade authentic MPA (Sigma-Aldrich, USA). A HPLC chromatogram of MPA is shown in Fig. S1. A fresh curve was made each time a batch of the samples was analyzed. The stock solution of MPA (1 mg/mL) was prepared in methanol and stored at −20 °C. When required, the solution was serially diluted with methanol to provide MPA concentrations in the range of 2.5–125 μg/mL. All measurements of MPA were in triplicate.
2.4.2. Glucose concentration
The glucose concentration was measured in the culture supernatant as reducing sugars by the dinitrosalicylic acid (DNS) method [23]. A suitably diluted sample (1 mL) was mixed with 1 mL of the DNS reagent, held at 90 °C for 10 min, and cooled. The absorbance was measured at 540 nm against a blank of deionized water treated the same way as the sample. The measured absorbance and the dilution factor were used to estimate the reducing sugar concentration by comparing with a calibration curve made using dilutions of a standard aqueous solution of glucose.
2.4.3. Cell mass concentration
The fungal biomass in the fermentation broth was quantified by the dry cell weight method. A 50 mL sample of the fermentation broth was filtered through Grade 50 cheese cloth (purchased from the local market). The biomass was washed with 50 mL of water and the resulting cake was dried at 60–65 °C for at least 48 h [7], [24]. The cake was cooled in a desiccator and weighed.
2.5. Optimization of fermentation medium using one-factor-at-a-time method
2.5.1. Carbon sources
The specified carbon sources (glucose, sucrose, fructose, maltose, lactose, glycerol, xylose, sorbitol, mannitol, starch and carboxymethyl cellulose) were studied at an initial concentration of 10 g/L. The carbon source that proved to be the best was examined further at various initial concentrations in the range 10–100 g/L. The other components of the production medium were held at the following initial concentrations (g/L): tryptone 5, glycine 2, methionine 0.5, KH2PO4 2, and MgSO4·7H2O 1. All media were supplemented with the earlier specified solution of trace elements (1 mL/L). The fermentation conditions were as follows: 27 °C; an initial pH of 6.0; an agitation speed of 200 rpm; and a fermentation duration of 12 days. All fermentations were carried out in duplicate.
2.5.2. Nitrogen sources
In the screening of the nitrogen sources, the carbon source was always glucose at an initial concentration of 60 g/L. The various organic and inorganic nitrogen sources (urea, ammonium nitrate, tryptone, peptone and asparagine) were screened individually as nitrogen sources. The initial concentration of a nitrogen source was always 10 g/L. Peptone was further examined at initial concentrations in the range of 10–40 g/L. The other components of the medium were held at the following initial concentrations (g/L): glycine 2, methionine 0.5, KH2PO4 2, and MgSO4·7H2O 1. All media were supplemented with the earlier specified solution of trace elements (1 mL/L). The fermentation conditions were as follows: 27 °C; an initial pH of 6.0; an agitation speed of 200 rpm; and a fermentation duration of 12 days. All fermentations were carried out in duplicate.
2.5.3. Phosphorous sources
Four different phosphate sources (KH2PO4, K2HPO4, NaH2PO4 and Na2HPO4) were screened individually at an initial concentration of 6 g/L. The other components of the medium were held at the following initial concentrations (g/L): glucose 60, peptone 20, glycine 2, methionine 0.5, and MgSO4·7H2O 1. All media were supplemented with the earlier specified solution of trace elements (1 mL/L). The other conditions were: 27 °C incubation temperature, 200 rpm agitation speed, and an initial pH of 6.0.
The phosphorous source that proved to be the best was examined further at various initial concentrations in the range 2–10 g/L. The other media components and the fermentation conditions were as specified above in this section. All fermentations lasted 12 days and were carried out in duplicate.
2.5.4. The precursors
Methionine is a known precursor of MPA [1], [3], [25], [26]. It is the main source of the methyl groups in MPA and directly enters the MPA biosynthesis pathway. Glycine was thought to be a potential precursor. The effects of the initial concentrations of these two amino acids on the production MPA and biomass were evaluated separately. In separate experiments the initial concentration of methionine was varied in the range of 0–3 g/L. Similarly, the initial concentration of glycine was varied in the range of 0–12 g/L. The initial pH value was 6.0 and the incubation temperature was 27 °C. The agitation speed was 200 rpm. All media contained the following (g/L): glucose 60, peptone 20, KH2PO4 6 and MgSO4·7H2O 1. The media were supplemented with glycine (2 g/L) in the experiments in which the initial concentration of methionine was varied. Otherwise, the media were supplemented with methionine (1.5 g/L) and the different specified concentrations of glycine. All media were supplemented with the earlier specified solution of trace elements (1 mL/L). All fermentations were run in duplicate for 12 days.
2.5.5. The initial pH
The initial medium composition was always as follows (g/L): glucose 60, peptone 20, glycine 6, methionine 1.5, KH2PO4 6, and MgSO4·7H2O 1. All media were supplemented with the earlier specified solution of trace elements (1 mL/L). The initial pH was adjusted to 4–9 in separate experiments. The other fermentation conditions were fixed as follows: 12 days at 27 °C and 200 rpm. All fermentations were carried out in duplicate.
2.5.6. Incubation temperature
The composition of the fermentation medium was fixed as specified above for the pH screening experiments. The initial pH was 5.0. The fermentations lasted 12 days and the agitation speed was 200 rpm. The incubation temperature in separate experiments was varied in the range of 20–35 °C. Fermentations were carried out in duplicate.
2.6. Optimization of the screened factors by a central composite design
The design and analysis of the experiments used the Design-Expert® software (Stat-Ease Inc, USA; trial version 9). In view of the results of the earlier screening experiments, the following conditions were fixed at the specified values: an incubation temperature of 25 °C, an incubation time of 12 days, and an initial pH of 5.0. The medium always contained the following: KH2PO4 6 g/L, MgSO4·7H2O 1 g/L, and the earlier specified trace element solution (1 mL/L). A spore suspension (0.5 mL, 107 spores/mL) was used to inoculate 50 mL of the culture medium in 250 mL shake flasks. The flasks were held at 200 rpm. For these conditions, the following four factors were examined through a central composite experiment design [7]: the concentrations of glucose, peptone, methionine and glycine. The concentration ranges of the various factors are shown in Table 1. The response was the concentration of mycophenolic acid. A full factorial central composite design with (24) 16 runs, 6 center points and 8 star points was used. This led to a total of 30 experiments (Table 2), all performed in duplicate.
Table 1.
Concentration ranges of the factors used in central composite design.
| Factor | Actual levels of coded factors |
||
|---|---|---|---|
| −1 | 0 | +1 | |
| Glucose (g/L) | 40 | 60 | 80 |
| Peptone (g/L) | 10 | 20 | 30 |
| Methionine (g/L) | 1 | 1.5 | 2 |
| Glycine (g/L) | 4 | 6 | 8 |
Table 2.
Experimental setup and results for the central composite design matrix.
| Run | Glucose (g/L) | Peptone (g/L) | Methionine (g/L) | Glycine (g/L) | Predicted MPA (mg/L) | Measured MPA (mg/L) |
|---|---|---|---|---|---|---|
| 1 | 60 | 20 | 1.5 | 8 | 1441.9 | 1388.4 |
| 2 | 80 | 30 | 1.0 | 8 | 1548.9 | 1603.1 |
| 3 | 80 | 30 | 2.0 | 4 | 1448.1 | 1493.0 |
| 4 | 60 | 20 | 1.5 | 6 | 1552.3 | 1550.6 |
| 5 | 60 | 20 | 1.5 | 6 | 1552.3 | 1537.1 |
| 6 | 80 | 30 | 1.0 | 4 | 1462.0 | 1456.0 |
| 7 | 80 | 30 | 2.0 | 8 | 1632.7 | 1560.1 |
| 8 | 40 | 20 | 1.5 | 6 | 1405.9 | 1432.7 |
| 9 | 60 | 30 | 1.5 | 6 | 1617.9 | 1618.9 |
| 10 | 60 | 20 | 1.5 | 6 | 1552.3 | 1585.3 |
| 11 | 80 | 10 | 1.0 | 8 | 1395.8 | 1398.3 |
| 12 | 80 | 10 | 1.0 | 4 | 1300.1 | 1300.6 |
| 13 | 40 | 30 | 2.0 | 4 | 1544.6 | 1547.1 |
| 14 | 60 | 20 | 1.5 | 6 | 1552.3 | 1580.2 |
| 15 | 80 | 10 | 2.0 | 8 | 1341.5 | 1405.2 |
| 16 | 40 | 10 | 2.0 | 4 | 1275.6 | 1224.1 |
| 17 | 80 | 10 | 2.0 | 4 | 1148.1 | 1118.5 |
| 18 | 40 | 10 | 2.0 | 8 | 1114.2 | 1125.3 |
| 19 | 40 | 10 | 1.0 | 8 | 1143.3 | 1101.0 |
| 20 | 60 | 20 | 1.5 | 6 | 1552.3 | 1575.0 |
| 21 | 60 | 20 | 1.5 | 6 | 1552.3 | 1578.5 |
| 22 | 40 | 30 | 2.0 | 8 | 1374.4 | 1376.6 |
| 23 | 60 | 20 | 1.5 | 4 | 1479.1 | 1501.6 |
| 24 | 60 | 20 | 2.0 | 6 | 1599.2 | 1628.5 |
| 25 | 60 | 10 | 1.5 | 6 | 1406.8 | 1375.0 |
| 26 | 60 | 20 | 1.0 | 6 | 1620.7 | 1560.5 |
| 27 | 40 | 30 | 1.0 | 4 | 1533.3 | 1472.3 |
| 28 | 80 | 20 | 1.5 | 6 | 1483.8 | 1426.2 |
| 29 | 40 | 10 | 1.0 | 4 | 1402.4 | 1480.0 |
| 30 | 40 | 30 | 1.0 | 8 | 1265.5 | 1300.2 |
2.7. Validation of the response surface model
The predicted optimal values of the factors were identified using the response surface model and the point prediction feature of the Design-Expert® software. A medium with the predicted optimal composition was formulated and validation experiments were run in triplicate. The measured response was compared to the predicted response to validate the optimal values.
3. Results and discussion
3.1. Screening experiments and one-at-a-time variation of factors
3.1.1. The carbon source
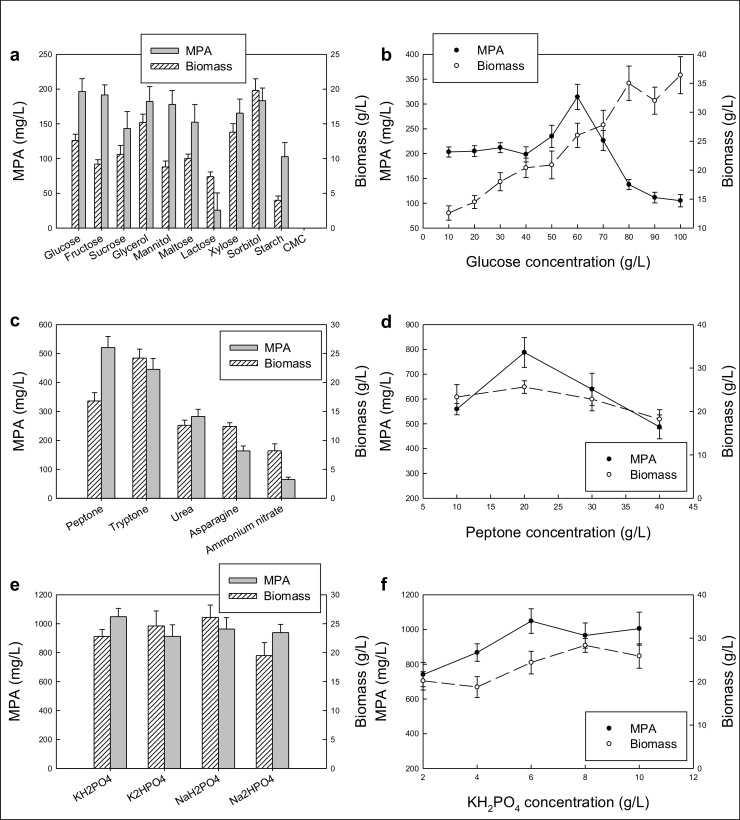
As shown in Fig. 1a, the values of the MPA titers were the highest with the carbon sources of glucose, fructose, glycerol, mannitol, xylose and sorbitol. All the carbon sources in Fig. 1a were used at an identical initial concentration of 10 g/L. The average values of the MPA titer was the highest with glucose, although statistically not significantly different relative to the other carbon sources noted earlier. There was no growth and MPA production on carboxymethyl cellulose (CMC) (Fig. 1a). Sorbitol yielded the highest biomass concentration relative to the other carbon sources (Fig. 1a). As the objective was the production of MPA, glucose was selected as the carbon source of choice. The equally high titers of MPA with several of the carbon sources (Fig. 1a) suggest that the carbon source per se did not directly contribute to the synthesis of MPA.
Fig. 1.
Effects of the following on the production of the biomass and MPA: (a) the type of the carbon source at an initial concentration of 10 g/L; (b) the initial concentration of glucose; (c) the type of the nitrogen source at an initial concentration of 10 g/L; (d) the initial concentration of peptone; (e) the type of the phosphorous source at an initial concentration of 6 g/L; (f) the initial concentration of KH2PO4.
The effects of the initial concentration of glucose on the production of MPA and biomass were tested. The results are shown in Fig. 1b. An increase in concentration of glucose over the range of 10–100 g/L resulted in a progressive increase in the final concentration of the biomass. This notwithstanding, a distinct peak in the concentration of MPA was seen at a glucose concentration of 60 g/L (Fig. 1b). Therefore this concentration was taken as the optimal value. The results in Fig. 1b suggest that the production of MPA is not growth associated, as expected for a secondary metabolite. At 60 g/L initial glucose concentration, the MPA titer was 314 ± 25 mg/L.
3.1.2. The nitrogen source
The influence of the different nitrogen sources on the production of biomass and MPA is shown in Fig. 1c. All the nitrogen sources were tested at an initial concentration of 10 g/L. The carbon source was glucose at an initial concentration of 60 g/L. The organic nitrogen sources proved to be distinctly better than the inorganic nitrogen source of ammonium nitrate (Fig. 1c) for the production of the biomass and the MPA. Tryptone yielded the highest final biomass concentration, but peptone yielded a higher average value of the MPA titer of 521 ± 38 mg/L. Although the MPA titers obtained with tryptone and peptone were not significantly different, the biomass specific MPA production was higher with peptone relative to tryptone. (The biomass concentration obtained with peptone was about 69% of the biomass concentration obtained with tryptone as the nitrogen source; Fig. 1c.) In view of this, peptone was taken to be the best source of nitrogen among the sources tested.
The effects of the initial concentration of peptone on the production of MPA and biomass were tested (Fig. 1d). In the peptone concentration range of 10–40 g/L, the final biomass concentration was not substantially affected by peptone (Fig. 1d). Nevertheless, a distinct peak occurred in the MPA titer at an initial peptone concentration of 20 g/L (Fig. 1d). Therefore, peptone at an initial concentration of 20 g/L was taken to be the optimal nitrogen sources.
3.1.3. The phosphorous source
The effect of the type of the phosphorous (P) source on the production of the biomass and MPA is shown in Fig. 1e. There were no particularly substantial differences in the performance of the four P-sources. All the P-sources provided phosphorous in the form of phosphate. As all the P-source were used at the same concentration of 6 g/L and the molar masses of the source salts were different, the concentration of phosphate in media formulated with the different salts were slightly different. (The molar concentrations of phosphate were in the range of 34.5–50.0 mM) These small differences in the concentration of phosphate did not cause clear distinctions in the final concentrations of the biomass and MPA (Fig. 1e).
In view of these results, potassium dihydrogen phosphate (KH2PO4) was arbitrarily selected as the preferred source of phosphorus. Further experiments were carried out with different initial concentrations of this P-source to identify its optimal level. The results are shown in Fig. 1f. The final biomass concentration generally increased with the increasing initial concentration of phosphate, but beyond a KH2PO4 concentration of 6 g/L there was barely any impact of the concentration on the titer of MPA (Fig. 1f). Therefore, a KH2PO4 concentration of 6 g/L was taken as optimal.
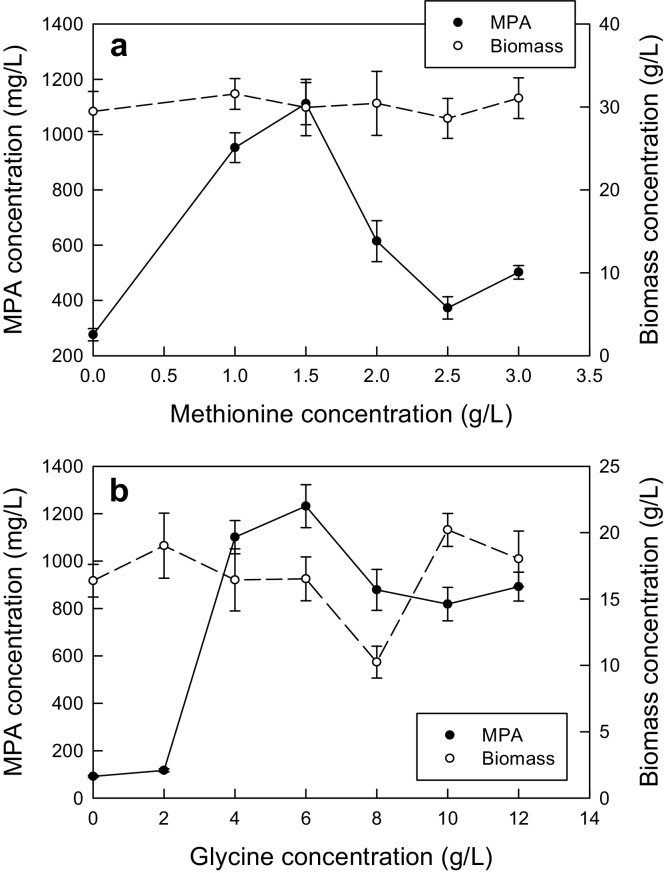
3.1.4. The precursors
In separate experiments, different initial concentration of methionine and glycine were evaluated for their possible impact on the production of the biomass and MPA. The results are shown in Fig. 2. Increasing concentration of methionine in the range of 0–3 g/L barely had any effect on the final biomass concentration (Fig. 2a), but strongly impacted the production of MPA. The MPA titer peaked at a methionine concentration of 1.5 g/L (Fig. 2a). In these experiments the glycine concentration was fixed at 2 g/L. Based on the data in Fig. 2a, a methionine concentration of 1.5 g/L was taken as optimal.
Fig. 2.
Effects of the initial concentrations of methionine (a) and glycine (b) on the production of the biomass and MPA.
An increase in the initial concentration of glycine from 0 to 6 g/L strongly enhanced the MPA titer, but not the biomass concentration (Fig. 2b). Higher concentrations (>6 g/L) of glycine actually caused some decline in the MPA titer (Fig. 2b). Therefore, a glycine concentration of 6 g/L was taken as optimal. In these experiments the methionine concentration was always 1.5 g/L.
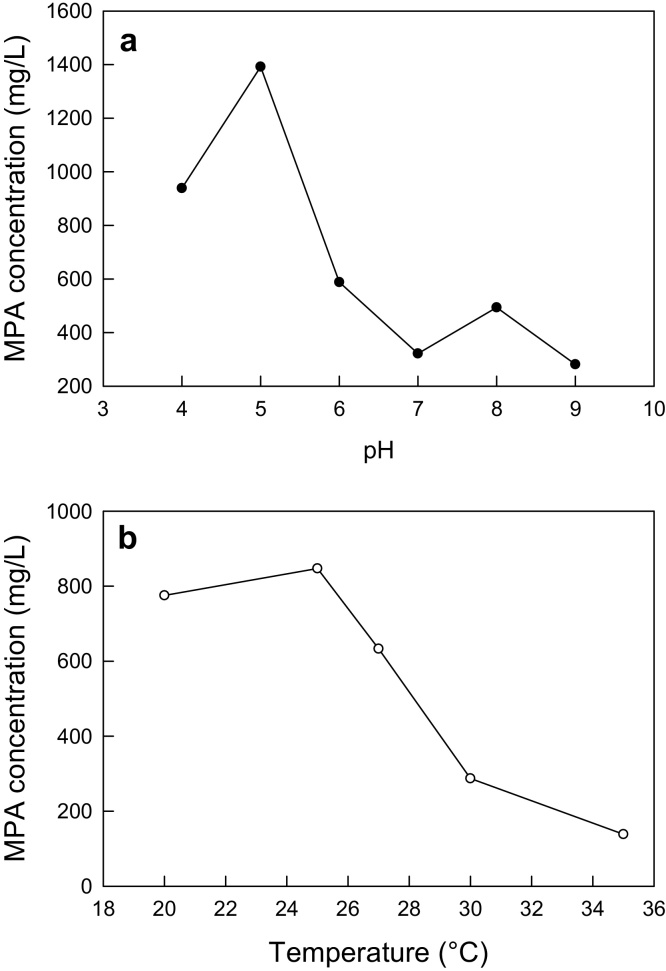
3.1.5. The initial pH and the incubation temperature
The effect of the initial pH on the production of MPA is shown in Fig. 3a. An initial pH of 5 clearly maximized the MPA titer relative to the other pH values. Therefore, a pH of 5 was taken as optimal. An incubation temperature of 25 °C proved to be best for maximizing the production of MPA (Fig. 3b) and was taken to be optimal.
Fig. 3.
Effects of pH (a) and incubation temperature (b) on the concentration of MPA.
Thus, based on one-at-a-time variation of the specified fermentation parameters, the optimal values for maximizing the titer of MPA (1232 mg/L) in a 12-day fermentation were as follows: an initial pH of 5.0; an incubation temperature of 25 °C; an agitation speed of 200 rpm; the MgSO4·7H2O concentration of 1 g/L; the initial concentrations of glucose, peptone and KH2PO4 at 60 g/L, 20 g/L and 6 g/L, respectively; and the initial concentrations of methionine and glycine at 1.5 g/L and 6 g/L, respectively.
3.2. Central composite design
Response surface methodology using central composite design was employed to determine the optimal levels of the four selected factors that affected MPA production. The relevant factors were the initial concentrations of glucose, peptone, methionine and glycine. Each factor was tested at three levels (Table 1). The center point levels were kept at the values found to be optimal via the one-at-a-time variation of factors. The other media components and the fermentation conditions were kept fixed at the following values: a temperature of 25 °C; a fermentation duration of 12 days; an initial pH of 5.0; an agitation speed of 200 rpm; the KH2PO4 concentration of 6 g/L; the MgSO4·7H2O concentration of 1 g/L; and the earlier specified trace element solution at 1 mL/L. A total of 30 runs (each in duplicate) were carried out with the values of the factors as noted in Table 2. The predicted and measured responses (MPA concentration) of the fermentations are shown in Table 2.
The response surface model was as follows:
| (1) |
Where A (g/L) is the concentration of glucose, B (g/L) is the concentration of peptone, C (g/L) is the concentration of methionine and D (g/L) is the concentration of glycine.
The maximum MPA concentration of 1629 mg/L was measured in run 24 (Table 2). The predicted MPA titer for this run was 1599 mg/L, or within ± 2% of the measured value (Table 2). The ANOVA of the response surface model (Eq. (1)) is shown in Table 3. The F-value for the model (i.e. Eq. (1)) was >14 (Table 3), suggesting that the model was significant and the probability of the F-value of the model being due to experimental noise was less than 0.01% (Table 3). The initial concentrations of glucose and peptone had a significant effect on the response, but not the initial concentrations of methionine and glycine (Table 3). However, glycine and glucose interacted to significantly affect the response (Table 3). Similarly, the interactive effect of peptone and methionine was significant (Table 3, P < 0.05). The determination coefficient (R2) of the model (Eq. (1)) was 0.9289, therefore the model could explain >92% of the variation in the predicted response of the MPA concentration.
Table 3.
Analysis of variance (ANOVA) for the response surface model (Eq. (1)).
| Source | SS | DF | MS | F-value | P-value (Prob > F) |
|---|---|---|---|---|---|
| Model | 1.137 × 105 | 14 | 43,833.28 | 14.01 | <0.0001 |
| Glucose (A) | 27,352.04 | 1 | 27,352.04 | 8.74 | 0.0098 |
| Peptone (B) | 2.004 × 105 | 1 | 2.004 × 105 | 64.06 | <0.0001 |
| Methionine (C) | 2082.97 | 1 | 2082.97 | 0.67 | 0.4273 |
| Glycine (D) | 6237.81 | 1 | 6237.81 | 1.99 | 0.1784 |
| AB | 958.14 | 1 | 958.14 | 0.31 | 0.5882 |
| AC | 632.84 | 1 | 632.84 | 0.20 | 0.6593 |
| AD | 1.258 × 105 | 1 | 1.258 × 105 | 40.22 | <0.0001 |
| BC | 19,054.45 | 1 | 19,054.45 | 6.09 | 0.0261 |
| BD | 77.09 | 1 | 77.09 | 0.025 | 0.8774 |
| CD | 9538.21 | 1 | 9538.21 | 3.05 | 0.1013 |
| A2 | 2992.48 | 1 | 2992.48 | 9.56 | 0.0074 |
| B2 | 4137.62 | 1 | 4137.62 | 1.32 | 0.2682 |
| C2 | 8598.97 | 1 | 8598.97 | 2.75 | 0.1181 |
| D2 | 21,863.37 | 1 | 21,863.37 | 6.99 | 0.0184 |
| Residual | 46,936.58 | 15 | 3129.11 | ||
| Lack of fit | 45,072.33 | 10 | 4507.23 | 12.09 | 0.0066 |
| Pure error | 1864.24 | 05 | 372.85 | ||
| Cor. Total | 6.606 × 105 | 29 |
SS—Sum of squares; DF—degrees of freedom; MS—mean sum of squares.
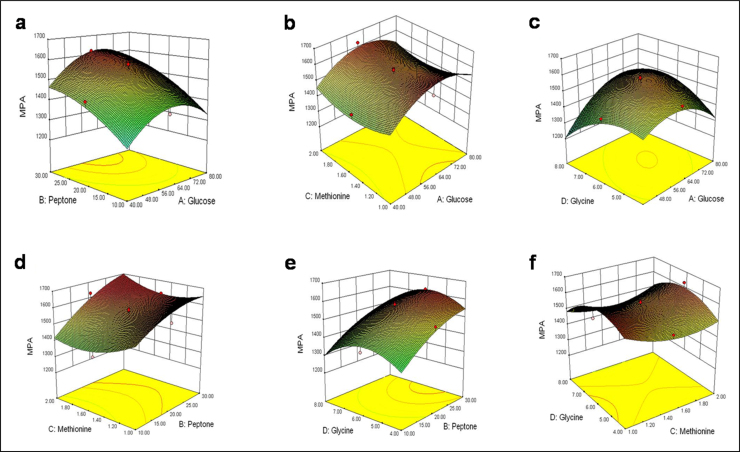
The response surface plots in Fig. 4 illustrate the effects of pair-wise combination of the four factors, on the response of MPA concentration. In generating these plots, the fixed factors were kept at their center point levels (Table 1). The plots in Fig. 4 provide a visual indication of how any two factors interactively affected the response. The center points of the contour plots (not shown) corresponding to Fig. 4 were used to identify the following values of the optimum concentrations (g/L): glucose 66, peptone 30, methionine 2, and glycine 6.3.
Fig. 4.
Response surface plots for the interactive effects of the following on the production of mycophenolic acid (MPA): glucose and peptone (a) at 1.5 g/L methionine and 6 g/L glycine; glucose and methionine (b) at 20 g/L peptone and 6 g/L glycine; glucose and glycine (c) at 20 g/L peptone and 1.5 g/L methionine; peptone and methionine (d) at 60 g/L glucose and 6 g/L glycine; peptone and glycine (e) at 60 g/L glucose and 1.5 g/L methionine; methionine and glycine (f) at 60 g/L glucose and 20 g/L peptone. MPA is in mg/L. All other concentrations are in g/L.
3.2.1. Validation of the response surface model
Validation runs were carried out with the values of the factors fixed at the above identified optimal levels. The measured response was compared to the model (Eq. (1)) predicted response. The measured MPA concentration (Fig. S2) under the optimized conditions was 1737.2 ± 54.5 mg/L. For the same conditions, the model predicted concentration was 1704.2 mg/L. Therefore, the model prediction agreed with measured data within ± 2% of the measured value.
3.3. A comparison of the optimization approaches
The optimal values of the factors obtained by the two methods of optimization were different in terms of some of the key constituents of the medium (i.e. glucose, peptone, methionine and glycine). The CCD-optimized medium was richer than the conventionally optimized medium in terms of the concentrations of these key components. Nevertheless, the CCD-optimized medium yielded an MPA titer of 1737 ± 55 mg/L, a value nearly 40% greater than the peak MPA titer of 1232 ± 90 mg/L attained using the conventionally optimized medium and nearly 9-fold greater than the titer (196 ± 18 mg/L) achieved in unoptimized medium (glucose 10 g/L, glycine 2 g/L, methionine 0.5 g/L, KH2PO4 2 g/L, MgSO4.7H2O 1 g/L, and 1 mL/L of a trace element solution). Further the product was confirmed as MPA by mass spectroscopy, m/z: 343.11 (Figs. S3, S4). These differences were attributed to the ability of the CCD-based optimization to account for the interactive effects of the experimental factors on the response (i.e. the MPA titer) [27], [28], [29]. Unlike the conventional method, the CCD-based method was statistically sound. By providing a response surface model (Eq. (1)), the CCD-based optimization enabled the prediction of the MPA titer for any combination of values of the factors within the experimental space.
3.4. A comparison with the literature
The MPA titers reported in the literature [7] for the batch and continuous submerged cultures of the various strains of P. brevicompactum are summarized in Table 4.
Table 4.
Mycophenolic acid production in various submerged fermentation processes using P. brevicompactum strains.a
| Process | Microorganism | Fermentation time (h) | MPA concentration (g/L) | Reference |
|---|---|---|---|---|
| Shake flask (250 mL) batch culture | P. brevicompactum ATCC 16024 | 336 | 1.7 | [31] |
| P. brevicompactum No. 5-1b | 336 | 5.3 | [31] | |
| P. brevicompactum ATCC 16024 | 240 | 5.91 | [30] | |
| P. brevicompactum MUCL 19011 | 280 | 1.38 | [7] | |
| P. brevicompactum MUCL 19011c | 280 | 3.63 | [7] | |
| P. brevicompactum MTCC 8010 | 280 | 1.73 | This work | |
| Bioreactor (5 L) batch culture | P. brevicompactum ATCC 16024 | 264 | 2.31 | [30] |
| Bioreactor (2 L) batch culture | P. brevicompactum MUCL 19011 | 240 | 1.37 | [7] |
| Bioreactor (2 L) continuous cultured | P. brevicompactum MUCL 19011 | 120 | 1.46 | [7] |
| Rotating fibrous bed bioreactor (5 L)e | P. brevicompactum ATCC 16024 | 338 | 5.70 | [30] |
The highest reported titer of 5.91 g/L was obtained in shake flasks in an optimized medium using the strain P. brevicompactum ATCC 16024 [30] (Table 4). For the same strain, grown in the same optimized medium, the titer was reduced to 2.31 g/L in a 5 L conventional stirred batch bioreactor (Table 4). The mycelial biomass accumulated at the bioreactor walls and the impeller and this made control of the pH and dissolved oxygen difficult [30]. Shear associated-damage to the mycelium in the stirred bioreactor possibly contributed to its poor performance relative to the shake flasks [30]. For the same strain and medium, the cultivation in a 5 L rotating fibrous bed bioreactor with the biomass immobilized in the bed improved control of the fermentation conditions and the MPA titer increased to 5.7 g/L (Table 4), or fairly close to the results of the shake flask culture [30]. Thus, the same fungal strain in the same optimized medium can give quite different MPA titers depending on the kind of bioreactor system used. In comparison with the shake flask data of Xu and Yang [30], the MPA titer obtained by Ozaki et al. [31] with the same strain (ATCC 16024) was only 1.7 g/L (Table 4), presumably because of the different media compositions used in these studies [31]. The titer in the present study was 1.73 g/L, or comparable to value reported by Ozaki et al. [31] for the wildtype strain ATCC 16024. The use of a mutant of the strain ATCC 16024 (the strain No. 5-1 in Table 4), increased the MPA titer by nearly 3-fold relative to the wildtype [31].
The differences in the attainable MPA titers are at least partly strain related. For example, using the strain MUCL 19011, Ardestani et al. [7] never exceeded a titer of 3.63 g/L in shake flasks and in the batch and continuous operational modes of a stirred tank bioreactor (Table 4). Although in the present study a titer of 1.73 g/L was comparable to the shake flask data of Ozaki et al. [31] for a wildtype strain, the MPA productivity in the present work was ∼20% higher at 0.006 g L−1 h−1 compared to a productivity of 0.005 g L−1 h−1 for shake flask cultures of Ozaki et al. [31]. This was because of the different times at which the reported titers were achieved (Table 4).
4. Conclusion
For a 12-day batch culture (an initial pH of 5.0, an incubation temperature of 25 °C, and an agitation speed of 200 rpm), the optimal medium composition identified via the conventional one-at-a-time variation of factors was different to the optimal composition identified through the central composite design of experiments. Using the conventional method, the composition was as follows (g/L): (1) glucose 60, peptone 20, KH2PO4 6, methionine 1.5, and glycine 6.0. Based on the CCD method, the optimal composition was as follows (g/L): glucose 66, peptone 30, KH2PO4 6, methionine 2, and glycine 6.3. In addition, both the optimized media contained equal levels of MgSO4·7H2O (1 g/L) and a trace element solution of a specified composition (1 mL/L). The CCD-optimized medium was richer than the conventionally optimized medium in concentrations of the key components of glucose, peptone, methionine and glycine. The CCD-optimized medium yielded an MPA titer of 1737 ± 55 mg/L, or 40% higher than the conventionally optimized medium. Therefore, the CCD-based optimization was concluded to be superior to the conventional method. The CCD-optimization had the other important advantage of being statistically robust [27], [32]. Unlike the conventional method, the CCD-based method provided a response surface model which could be used to predict the MPA titers for a wide range of values of the relevant factors.
Conflict of interest
All authors declare that he/she has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Acknowledgments
GP and MP gratefully acknowledge Department of Biotechnology (DBT), New Delhi, India for the award of Senior Research Fellowship. SS gratefully acknowledges Department of Science and Technology (DST), New Delhi, India for the award of INSPIRE fellowship.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.btre.2016.07.003.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Regueira T.B., Kildegaard K.R., Hansen B.G., Mortensen U.H., Hertweck C., Nielsen J. Molecular basis for mycophenolic acid biosynthesis in Penicillium brevicompactum. Appl. Environ. Microbiol. 2011;77:3035–3043. doi: 10.1128/AEM.03015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley R. Mycophenolic acid: a one hundred year odyssey from antibiotic to immunosuppressant. Chem. Rev. 2000;100:3801–3825. doi: 10.1021/cr990097b. [DOI] [PubMed] [Google Scholar]
- 3.Muth W.L., Nash C.H. Biosynthesis of mycophenolic acid: purification and characterization of S-adenosyl-L-methionine: demethylmycophenolic acid O-methyltransferase. Antimicrob. Agents Chemother. 1975;8:321–327. doi: 10.1128/aac.8.3.321. (Updated) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lafont P., Debeaupuis J.P., Gaillardin M., Payen J. Production of mycophenolic acid by Penicillium roqueforti strains. Appl. Environ. Microbiol. 1979;37:365–368. doi: 10.1128/aem.37.3.365-368.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vinokurova N.G., Ivanushkina N.E., Kochkina G.A., Arinbasarov M.U., Ozerskaya S.M. Production of mycophenolic acid by fungi of the genus Penicillium link. Appl. Biochem. Microbiol. 2005;41:83–86. [PubMed] [Google Scholar]
- 6.Puel O., Tadrist S., Galtier P., Oswald I.P., Delaforge M. Byssochlamys nivea as a source of mycophenolic acid. Appl. Environ. Microbiol. 2005;71:550–553. doi: 10.1128/AEM.71.1.550-553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ardestani F., Fatemi S.S.A., Yakhchali B., Hosseyni S.M., Najafpour G. Evaluation of mycophenolic acid production by Penicillium brevicompactum MUCL 19011 in batch and continuous submerged cultures. Biochem. Eng. J. 2010;50:99–103. http://dx.doi.org/10.1016/j.bej.2010.03.008 [Google Scholar]
- 8.Ismaiel A.A., Ahmed A.S., El-Sayed E.-S.R. Optimization of submerged fermentation conditions for immunosuppressant mycophenolic acid production by Penicillium roqueforti isolated from blue-molded cheeses: enhanced production by ultraviolet and gamma irradiation. World J. Microbiol. Biotechnol. 2014;30:2625–2638. doi: 10.1007/s11274-014-1685-1. [DOI] [PubMed] [Google Scholar]
- 9.Séguin V., Gente S., Heutte N., Vérité P., Kientz-Bouchart V., Sage L., Goux D., Garon D. First report of mycophenolic acid production by Eurotium repens isolated from agricultural and indoor environments. World Mycotoxin J. 2014;7:321–328. [Google Scholar]
- 10.Kitchin J.E.S., Pomeranz M.K., Pak G., Washenik K., Shupack J.L. Rediscovering mycophenolic acid: a review of its mechanism, side effects, and potential uses. J. Am. Acad. Dermatol. 1997;37:445–449. doi: 10.1016/s0190-9622(97)70147-6. [DOI] [PubMed] [Google Scholar]
- 11.Badrick A.C., Hanson G.R., Jones C.E. Chaperoning the immunosuppressant mycophenolic acid through the gastrointestinal tract: a role for copper. Appl. Magn. Reson. 2009;36:231–236. [Google Scholar]
- 12.Arns W., Cibrik D.M., Walker R.G., Moura d G., Budde K., Mueller E.A., Vincenti F. Therapeutic drug monitoring of mycophenolic acid in solid organ transplant patients treated with mycophenolate mofetil: review of the literature. Transplantation. 2006;82:1004–1012. doi: 10.1097/01.tp.0000232697.38021.9a. [DOI] [PubMed] [Google Scholar]
- 13.Novartis, Full year; product; sales 2014. (accessed 29.04.16) https://www.novartis.com/investors/financial-data/product-sales.
- 14.Roche Finance Report, 2013. F. Hoffmann-La Roche Ltd, 4070 Basel, Switzerland, (n.d.). (accessed 29.04.16) http://www.roche.com/inv-update-2014-01-30-e.pdf.
- 15.Hansen B.G., Mnich E., Nielsen K.F., Nielsen J.B., Nielsen M.T., Mortensen U.H., Larsen T.O., Patil K.R. Involvement of a natural fusion of a cytochrome P450 and a hydrolase in mycophenolic acid biosynthesis. Appl. Environ. Microbiol. 2012;78:4908–4913. doi: 10.1128/AEM.07955-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W., Cao S., Qiu L., Qi F., Li Z., Yang Y., Huang S., Bai F., Liu C., Wan X., Li S. Vol. 16. 2015. pp. 565–569. (Functional Characterization of MpaG’, the O-methyltransferase Involved in the Biosynthesis of Mycophenolic Acid). [DOI] [PubMed] [Google Scholar]
- 17.Cholewinski G., Malachowska-Ugarte M., Dzierzbicka K. The chemistry of mycophenolic acid–synthesis and modifications towards desired biological activity. Curr. Med. Chem. 2010;17:1926–1941. doi: 10.2174/092986710791163920. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y., Ang W.T., Xing J., Zhang J., Chen J. Applications of ultrasound to enhance mycophenolic acid production. Ultrasound Med. Biol. 2012;38:1582–1588. doi: 10.1016/j.ultrasmedbio.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 19.Shu C.H., Peng J.C., Tsai C.C. Effects of light intensity and light wavelength on the production of mycophenolic acid by Penicillium brevicompactum in batch cultures. Enzyme Microb. Technol. 2010;46:466–471. doi: 10.1016/j.enzmictec.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Sadhukhan K., Ramana Murthy A.K., Ajaya Kumar M.V., Mohan R., Vandana E.V.S., Bhar G., Venkateswara Rao C. Optimization of mycophenolic acid production in solid state fermentation using response surface methodology. J. Ind. Microbiol. Biotechnol. 1999;22:33–38. [Google Scholar]
- 21.Alani F., Grove J.A., Anderson W.A., Moo-Young M. Mycophenolic acid production in solid-state fermentation using a packed-bed bioreactor. Biochem. Eng. J. 2009;44:106–110. [Google Scholar]
- 22.Elbarbry F.A., Shoker A. Simple high performance liquid chromatographic assay for mycophenolic acid in renal transplant patients. J. Pharm. Biomed. Anal. 2007;43:788–792. doi: 10.1016/j.jpba.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 23.Miller G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959;31:426. [Google Scholar]
- 24.Ardestani F., Fatemi S.S., Yakhchali B., Hosseyni S.M. The effect of methionine and acetate concentrations on mycophenolic acid production by Penicillium bervicompactum MUCL 19011 in submerged culture. World Acad. Sci. Eng. Technol. 2009;56:808–811. [Google Scholar]
- 25.Bedford C.T., Fairlie J.C., Knittel P., Money T., Phillips G.T. Sequence studies in biosynthesis; mycophenolic acid. J. Chem. Soc. D: Chem. Commun. 1971;7:323–324. [Google Scholar]
- 26.Bedford C.T., Knittel P., Money T., Phillips G.T., Salisbury P. Biosynthesis of mycophenolic acid. Can. J. Chem. 1973;51:694–697. [Google Scholar]
- 27.Patil M.D., Shinde K.D., Patel G., Chisti Y., Banerjee U.C. Use of response surface method for maximizing the production of arginine deiminase by Pseudomonas putida. Biotechnol. Rep. 2016;10:29–37. doi: 10.1016/j.btre.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chuprom J., Bovornreungroj P., Ahmad M., Kantachote D., Dueramae S. Approach toward enhancement of halophilic protease production by Halobacterium sp. strain LBU50301 using statistical design response surface methodology. Biotechnol. Rep. 2016;10:17–28. doi: 10.1016/j.btre.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mouafi F.E., Abo Elsoud M.M., Moharam M.E. Optimization of biosurfactant production by Bacillus brevis using response surface methodology. Biotechnol. Rep. 2016;9:31–37. doi: 10.1016/j.btre.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Z.N., Yang S.T. Production of mycophenolic acid by Penicillium brevicompactum immobilized in a rotating fibrous-bed bioreactor. Enzyme Microb. Technol. 2007;40:623–628. [Google Scholar]
- 31.Ozaki H., Ishihara M., Kida T., Yamanaka S., Shibai H. Mycophenolic acid production by drug-resistant and methionine or glutamic-acid requiring mutants of Penicillium brevicompactum. Agric. Biol. Chem. 1987;51:2509–2514. [Google Scholar]
- 32.Sathishkumar R., Ananthan G., Iyappan K., Stalin C. A statistical approach for optimization of alkaline lipase production by ascidian associated—Halobacillus trueperi RSK CAS9. Biotechnol. Rep. 2015;8:64–71. doi: 10.1016/j.btre.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.