Graphical Abstract
This review summarizes research into the metal-binding properties of catalytic DNAzymes, towards the goal of understanding the structural properties leading to metal ion specificity. Progress made and insight gained from a range of biochemical and biophysical techniques are covered, and promising directions for future investigations are discussed.
Keywords: Metalloenzymes, deoxyribozymes, catalytic DNA, metal ion selectivity, metal-nucleic acid interactions, metal sensors
1. Introduction
DNAzymes, also known as deoxyribozymes, are oligomers of DNA whose sequence provides them with catalytic functionality. They are analogous to ribozymes, which are RNA molecules with enzymatic functions naturally found in all forms of life. In contrast, DNAzymes are not known to be naturally occurring; they were first identified in 1994 through in vitro selection from a large pool of random DNA sequences (Figure 1).1
Figure 1.
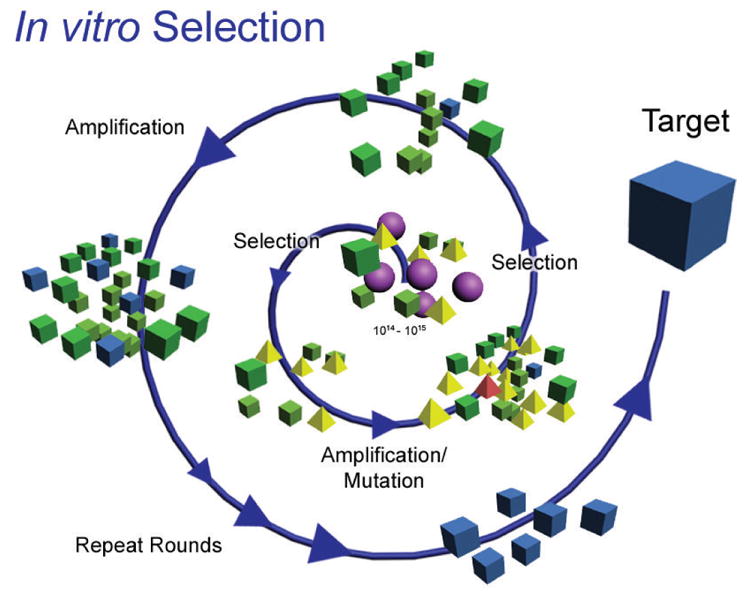
A representative scheme for in vitro selection of metal ion-dependent DNAzymes. A combinatorially large pool of random nucleotide sequences (center) with defined primer-binding regions is put through successive rounds of selection (activity towards the target metal ion of interest), amplification (using polymerase chain reaction, PCR), and mutation (using mutagenic PCR). Figure adapted with permission from ref 2
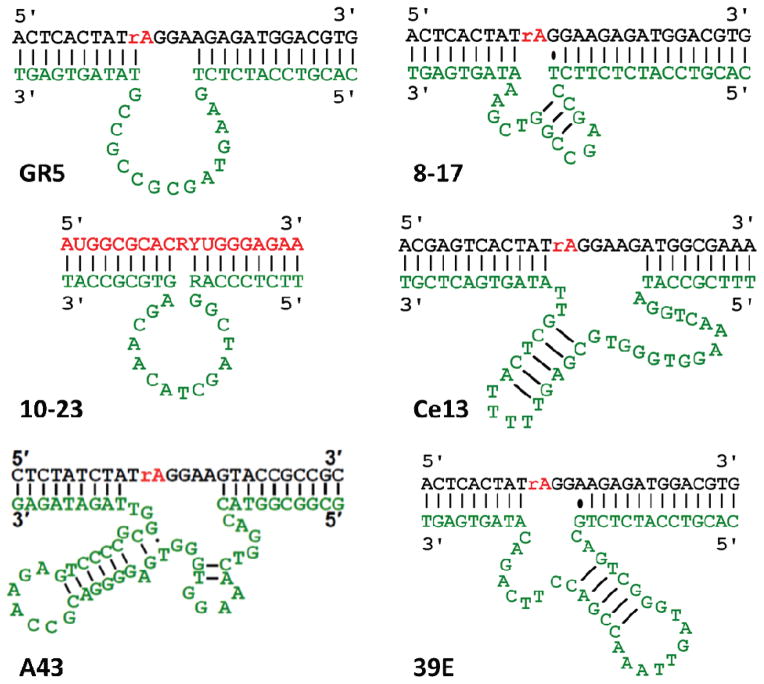
The specific type of catalytic functionality available to DNAzymes depends on the sequence of the DNA, and in many cases also on the presence of additional cofactors. The most common class of cofactors for DNAzyme catalysis is metal ions.3–7 A number of DNAzymes that utilize metal ions to carry out catalysis are shown in Table 1. The sequences and proposed secondary structures of some DNAzymes are shown in Figure 2.
Table 1.
Examples of different reactions catalyzed by DNAzymes in the presence of different metal ions
| Reaction type | Metal cofactor | Refs |
|---|---|---|
| RNA cleavage | Pb2+, Mg2+, Zn2+, Ca2+, Mn2+, Ni2+, Mg2+, UO2+, Co2+, Na+, trivalent lanthanides | 1,8–24 |
| RNA phosphorothioate cleavage | Cd2+ | 25 |
| DNA cleavage (oxidative) | Cu2+ | 26–28 |
| DNA cleavage (hydrolytic) | Zn2+, Mn2+, Ce3+/Eu3+/Yb3+ | 29–32 |
| Phosphoramidate bond cleavage | Mg2+ | 33 |
| DNA ligation | Cu2+, Zn2+, Mn2+ | 34,35 |
| RNA ligation (2′–5′) | Mg2+ | 36 |
| RNA ligation (3′–5′) | Zn2+, Mg2+ | 37,38 |
| RNA ligation (branch formation) | Mn2+, Mg2+ | 39–42 |
| RNA ligation (lariat formation) | Mn2+ | 39 |
| Nucleopeptide linkage formation | Mn2+, Mg2+ | 43–45 |
| DNA phosphorylation | Mn2+, Cu2+ | 46 |
| DNA capping | Mg2+, Cu2+ | 47,48 |
| DNA depurination | Ca2+ | 49 |
| Diels-Alder reaction | Ca2+ | 50 |
| Porphyrin metallation | Cu2+, Zn2+ | 51,52 |
| Amide hydrolysis | Zn2+, Mn2+, or Mg2+ | 53 |
| Peptide phosphatase activity | Zn2+ | 54 |
| Tyrosine kinase activity | Zn2+, Mn2+, Mg2+ | 55,56 |
| Friedel-Crafts reaction | Cu2+ | 57 |
Figure 2.
Selected metal ion-dependent DNAzymes, showing predicted secondary structure. Enzyme strands are highlighted in green, substrate strands in black (DNA) and red (RNA). R = purine (A, G), Y = pyrimidine (C, T). Dashes indicate Watson-Crick base pairs, dots indicate wobble pair interactions.
Hence, DNAzymes are an emerging class of metalloenzymes in biology that perform many different reactions with rates that can sometimes compete with other known types of enzymes – for instance, the 10-23 DNAyzme catalyzes RNA cleavage with a catalytic efficiency (kCat/kM = 109 M−1 min−1) rivaling protein ribonucleases.9 In contrast to the rich knowledge on interaction of metals with proteins and RNA, DNA-metal interactions are far less well understood. Thus, studying DNA-metal interactions is an important step in improving our understanding of this class of metalloenzymes.
While it may not be a surprise that DNAzymes can bind metal ions due to their highly negatively charged phosphodiester backbone, many studies in the past 15 years have shown that DNAzymes can be identified that bind a metal ion selectively.14,18,58 As a result, these DNAzymes have been converted into sensors that are able to report on the presence of the necessary metal ion cofactor in environmental samples, as well as within cells.18,59,60 In this approach, the catalytic activity of the DNAzyme is directly read out by coupling the catalyzed reaction (e.g. RNA cleavage via transesterification) to a reporting modality. In its simplest form this readout signal can be something as direct as fluorescence increase; however, many additional strategies have been reported, including fluorescence,14,17–19,21,24,25,58–61 luminescence,62–65 electrochemical signal,66–69 MRI relaxivity,70,71 and methods utilizing conjugated protein enzymes to produce metabolites such as glucose.72,73 A number of these strategies have been discussed in more depth in recent reviews and will not be covered in detail in this review.3,4,74,75
In this review we focus on characterization methods that have been utilized or which show promise for investigating details of metal-DNA interactions, including metal specificity of DNAzymes, metal-dependent DNA folding, and structural features of DNA-metal interactions. We have separated these techniques into three broad categories: a) biochemical assays to obtain information about sequence features that are responsible for catalysis and metal selectivity, b) structural characterization to obtain information about 3D structure of DNAzymes, and c) spectroscopic studies of the metal-binding sites. At each section, we discuss the results of recent studies using these methods. Finally, we conclude with a perspective on the future directions of the DNAzyme field.
2. Biochemical approaches towards understanding metal ion selectivity
Biochemical approaches are the most ubiquitous methods for studying DNAzymes, as they typically apply directly to DNA and are thus closest to the strategies employed during DNAzyme selection.76 As with the biochemical study of protein enzymes, many mutagenic approaches can be utilized to better understand critical regions or bases in a DNAzyme sequence. The impact of alterations to a DNAzyme sequence is most often assessed by direct measurement of the catalytic activity, i.e., the catalytic rate of a DNAzyme in the presence of a particular metal ion. Comparing the catalytic activity of a particular DNAzyme in the presence of different metal ions allows the determination of the selectivity of that DNAzyme for a particular metal ion, which is critical for sensing applications.2 By introducing deletions, mutations, or modified nucleotides, one can directly probe the locations, bases, and individual functional groups, respectively, that are critical for metal-dependent DNAzyme catalysis.
2.1 Biochemical studies of divalent metal ion-dependent DNAzymes
The first DNAzyme isolated by in vitro selection was a Pb2+-dependent DNAzyme, reported by Breaker and Joyce,1 later named the GR5 DNAzyme (Figure 2).58 Soon after this work, Santoro and Joyce carried out in vitro selection of DNAzymes capable of cleaving all-RNA substrates in the presence of Mg2+. This selection resulted in two structurally distinct scaffolds, termed the 8-17 and 10-23 DNAzymes (Figure 2).9 Both DNAzymes have relatively short sequences within their predicted catalytic core regions – 13 and 15 nucleotides, respectively. In order to better define the sequence requirements, a reselection protocol was carried out in which the originally identified catalytic core was partially randomized and in vitro selection was repeated on this pool. Initial analysis of the sequence diversity from this process indicated a predicted stem-loop secondary structure for 8-17 with a highly invariant AGC loop, and a 4–5 nt unpaired region with slight variability. In contrast, the 10-23 DNAzyme was found to have nearly no tolerated sequence variations.9
Subsequently, two other independent in vitro selections by the Faulhammer and Lu groups resulted in the identification of new DNAzymes sharing the same motif as the 8-17 DNAzyme.8,11 These DNAzymes were investigated by Peracchi group, who identified that the catalytic activity of both DNAzymes as well as the original 8-17 DNAzyme was faster with Ca2+ than with Mg2+.77 Mutations of a specific ACGA turn motif appeared to reduce the selectivity for Ca2+ over Mg2+.77
Although the 8-17 DNAzyme was obtained through in vitro selection in the presence of different metal ions such as Mg2+ and Zn2+, it was later shown to display much higher activity with Pb2+ than with other metal ions.78 This high activity with lead was utilized to create the first DNAzyme-based catalytic beacon sensor for lead ions.78 Later studies identified that the selectivity of GR5 for Pb2+ over other metal ions is considerably greater than that of the 8-17 DNAzyme.58 A comparative analysis of the GR5 and 8-17 DNAzymes has led to a proposal that the 8-17 DNAzyme is a particular variant of GR5 DNAzyme with additional peripheral motifs that allows other metals to also support catalytic activity.79 A major barrier towards practical applications of these DNAzymes for detection of metal ions is that the hybridization and dehybridization of the DNA binding arms are highly dependent on temperature. To overcome this limitation, mutagenesis has been made to make the 8-17 DNAzyme more resistant to temperature variations.80
To broaden the scope of metal ion cofactors used by DNAzymes to transition metal ions, Lu and coworkers selected DNAzymes for Co2+ using a protocol designed to select DNAzymes responsive to Co2+ but not Zn2+.15 In this protocol, the random pool of DNA sequences was subject to selection towards the target ion, Co2+, but sequences that were also active with Zn2+ were removed. This process, named negative selection or counter selection resulted in sequences with more activity for Co2+ over Zn2+ or Pb2+ improving the metal ion selectivity of the resulting DNAzyme.15 Later, in a more detailed truncation study of this sequence, Lu and coworkers found that the Loop II stem-loop structure in the periphery of the DNAzyme sequence was a major factor in relative activity of the DNAzyme towards Co2+ instead of Zn2+ or Pb2+. Due to the distance between cleavage site and peripheral domains, this result indicates the importance of a very significant level of structural complexity in determining metal ion selectivity.81
The 39E DNAzyme was identified in a selection attempt to obtain DNAzymes active towards the uranyl ion (UO22+).14,82 UO22+ is the most stable chemical form of uranium in water, and is of great interest for environmental monitoring and remediation applications.14 The 39E DNAzyme shows excellent (> 106-fold) selectivity for uranyl over other metal ions; this DNAzyme was converted into a catalytic beacon sensor whose detection limit for uranyl (45 pM) exceeded the limit of detection that can be achieved by inductively-coupled plasma mass spectrometry (ICP-MS).14 Analysis and mutation of the 39E sequence by Lu and coworkers yielded an optimized form, which consists of a bulged stem-loop structured.82 Interestingly, a G-A wobble pair adjacent to the cleavage site was found to be important for DNAzyme activity, which could indicate the formation of a uranyl-specific binding site at this noncanonical base pair.82
2.2 Biochemical studies of trivalent and monovalent metal ion-dependent DNAzymes
A great deal of recent study has investigated DNAzymes that are active in the presence of targets beyond divalent metal ions. Silverman and coworker selected DNA-hydrolyzing sequences in the presence of both Zn2+ and a trivalent lanthanide ion, such as Ce3+, Eu3+, or Yb3+. They identified several sequences that required both metal ions for activity. Interestingly, two of the identified sequences were found to be active even in the absence of Zn2+ and had significant selectivity for Yb3+ over the other lanthanides.32 Liu and coworkers selected a number of RNA-cleaving DNAzyme sequences responding to trivalent lanthanide ions. One such sequence, termed the Ce13 DNAzyme (Figure 2), was found to have very broad substrate scope, showing high activity with any of the trivalent lanthanide ions, as well as with Y3+.19 Further in vitro selections identified other sequences with different patterns of lanthanide reactivity20,21; a ratiometric approach combining multiple sequences with different response patterns to distinguish between light and heavy lanthanide ions has been demonstrated.21
Another class of targets that has recently undergone investigation is monovalent metal ions. Lu and coworkers identified a DNAzyme, called the A43 DNAzyme that catalyzed RNA cleavage specifically in the presence of Na+ (Figure 2). The A43 DNAzyme was found to have over 10000-fold selectivity for Na+ over even other monovalent metal ions, and even higher selectivity over divalent and trivalent metal ions. This DNAzyme was truncated and converted into a catalytic beacon sensor to measure intracellular sodium in HeLa cells.18
A comparison of the sequences of the A43 and Ce13 DNAzymes revealed significant similarity and further analysis by both Lu and Liu groups uncovered a simultaneous requirement for Na+ as well as lanthanide ions for the activity of Ce13 DNAzyme.83,84 Interestingly, this Na+-dependent activity is distinct from other reported lanthanide-responsive DNAzymes, which show no change in activity even in the absence of Na+.84 At the same time, although the A43 DNAzyme does not function without Na+,18,83 its activity was shown to be accelerated in the presence of high concentrations of Ce3+.84 An analysis of the kinetics of Ce13 DNAzyme showed that it is first-order with respect to both Na+ and Ce3+, suggesting the presence of independent binding sites for the two metal ions.83,84 Systematic screening of mutants partway between the sequences of Ce13 and A43 DNAzymes identified a single nucleotide in the A43 DNAzyme sequence, G23, which is absolutely critical for the Ce3+-independent activity of the A43 DNAzyme, as well as a Na+-binding loop containing 8 conserved guanines.84
2.3 Biochemical studies using modified nucleotides
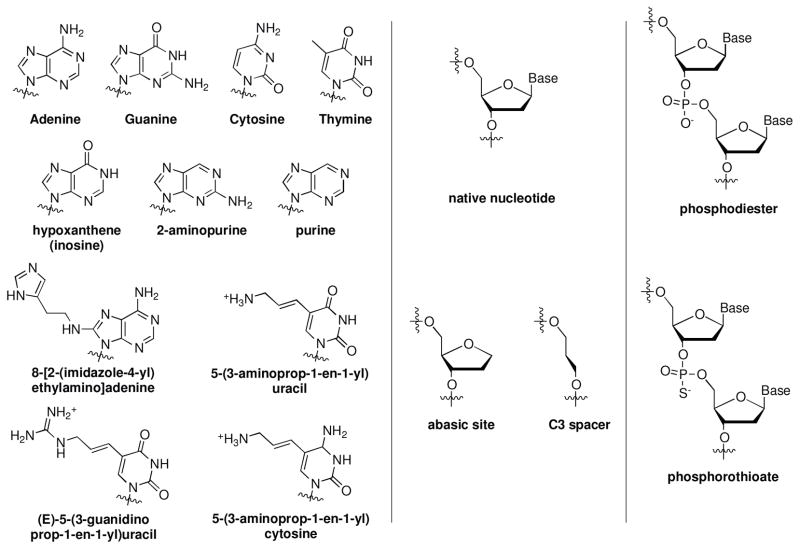
Modified or unnatural bases provide the researchers with an opportunity to tune individual functional groups of nucleotides with exceptional precision. Therefore, they can be used to elucidate the role of specific functional groups in enabling catalytic activity and in interacting with metal ion cofactors. Several modified bases have been used to study DNAzymes (see Figure 3).
Figure 3.
Selected structures of modified bases used in the study of DNAzymes.
Kurreck and coworker replaced bases in different regions of the 10-23 DNAzyme by hypoxanthine and 2-aminopurine and determined that the 2-amino and 6-oxo groups of G14 and the exocyclic group of A5 are essential for the activity.85 Clerici and coworkers carried out substitutions at the A6, G7, C13, and G14 positions in the 8-17 DNAzyme with a variety of unnatural nucleobases including hypoxanthine and 2-aminopurine (Figure 4).86 The study suggested a crucial role for the N7 site of A6, O6 site of G7, and the N2 site of G14. Xi and coworkers used abasic nucleotides or C3 spacers in different positions in the 10-23 and 8-17 DNAzymes and showed that, in addition to the known essential nucleotides, other positions within the DNAzyme can be of importance.87 Liu and coworkers replaced G23 in NaA43 DNAzyme to hypoxanthine and demonstrated the importance of the Nl position in G23 in the activity.84 They also showed that by incorporating a modified phosphorothioate in their DNA pool, they could enhance the affinity for Cd2+ a thiophilic target, resulting in a Cd2+-dependent DNAzyme with over 100,000-fold selectivity for Cd2+ over Zn2+. Obtaining such selectivity for Cd2+ has been shown to be difficult without the use of this modified ligand.25 Interestingly, Perrin and coworkers obtained a divalent metal-independent DNAzyme by incorporating a cationic amine, an imidazole and a guanidine functional group to their DNA pool.88
Figure 4.
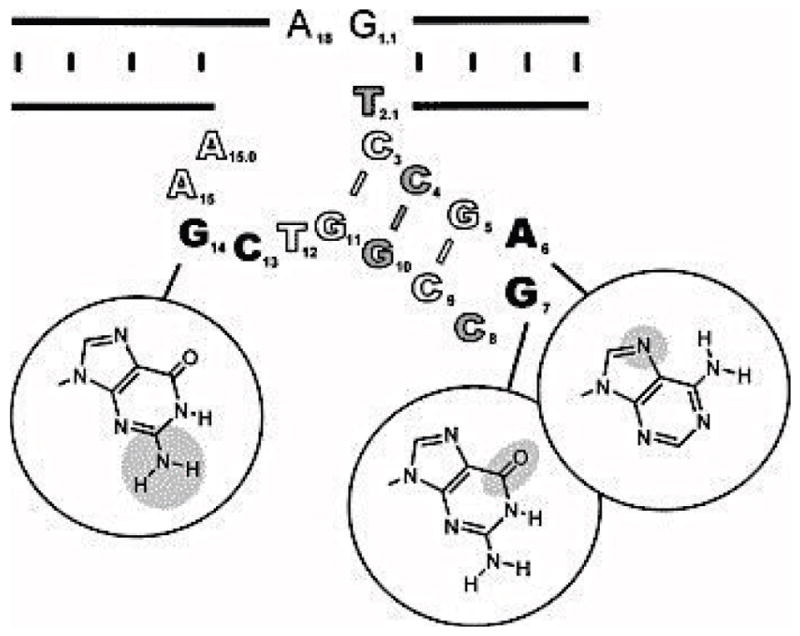
A scheme of conserved sequences of the 8-17 DNAzyme revealed by mutational analysis. Mutations of the bases shown in black to any other natural base resulted in >100-fold decrease in activity. Removal of the functional groups highlighted in gray circles caused >100-fold decrease in activity. Residues shown in gray are those whose replacement by modified bases caused >10-fold decrease in activity. Residues shown in white were more tolerant to mutagenesis. Figure adapted with permission from ref 86.
2.4 Insight from kinetic analysis into metal specificity
In addition to providing invaluable information about rates of reaction, activity assays can be used to investigate the substrate scope of a given DNAzyme and the specificity of the DNAzyme towards a particular metal cofactor, with possible hints about the mode of action. For example, if a DNAzyme is functional with both Cu2+, a redox active metal ion, and Zn2+, a redox inactive metal ion, with similar rates, it can be deduced that the role of the metal ion may not rely on metal redox-activity. Concentration-dependent curves of the rate can be used to find out the number of metal ions required for activity. It has been shown that in systems requiring more than one metal ion, the rate equation can be used as a guide to deduce the relationship between the different metal ions. Liu and coworkers showed that Ce13 DNAzyme has a linear rate dependence to both Ce3+ and Na+ (k = [Na+][Ce3+]), which suggests that the two metal binding sites are independent of each other.83,84
Much information can be deduced from analyzing the observed trends in the activity of a DNAzyme in the presence of different metal ions and its binding affinity to those metals. For example, preference for transition metal ions over alkaline earth metal ions can reveal information about the ligands to the metal ion. Alkaline earth metal ions are known to have lower affinities to nucleic acid ligands, as they tend to retain their hydration shell and bind via outer-sphere coordination.89,90 Transition metal ions in contrast can bind to O or N groups of bases through both outer- and inner-sphere interactions in addition to the non-bridging phosphate oxygen group.90–92 Comparing the trends of the binding affinity and activity of the DNAzymes in the presence of different metal ions can provide information about the metal-DNA interaction. Similarity between these two trends is an indication of binding being a rate-limiting step. Liu and coworkers showed that, in the lanthanide-responsive Ce13 DNAzyme, these two trends are not consistent and hence binding is not the rate-limiting step in RNA cleavage.93 Similarly, Lu and coworkers compared the result of binding affinity and activity of several metal ions to the 8-17 DNAzyme and identified a relationship between increased charge density and higher affinity, suggesting a role for electrostatic interactions in metal binding. Pb2+ was observed to be an exception to the observed trend, suggesting a possible alternative mechanism for Pb2+-dependent activity.94
2.5 Biochemical studies of hemin-dependent peroxidase DNAzymes
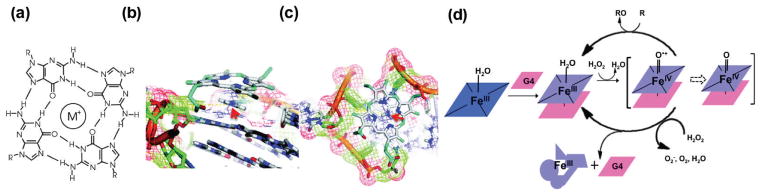
Activity assays can inform on DNA-metal interactions in the specific case of peroxidase-mimicking DNAzymes. In these DNAzymes, a hemin cofactor is coordinated to a G-quadruplex motif present in the DNAzymes (see Figure 5a–c). These DNAzymes can perform peroxidation (one-electron oxidation) and peroxygenation (two-electron oxidation) in an efficient manner, with activity comparable to natural peroxidases and P450 monooxygenases.95,96 This robust activity can be used to assess the effect of mutagenesis or changes in the enzyme structure and hence DNAzyme-hemin interaction in a fast and reliable manner. Sen and coworkers used isoguanine (iG) multi-stranded DNA and RNA to find out how the hemin is activated by the G-quadruplex. They found out that iG pentaplexes show peroxidase activity but that iG-quadruplexes do not. Based on a combination of computational and experimental evidence, they conclude that the planarity of the binding motif (G-quadruplex or iG-pentaplex) is an important factor in activating the hemin cofactor.97 In a detailed study, Shangguan and coworkers compared the kinetics of G-quadruplex hemin DNAzymes with horseradish peroxidase (HRP). They found out that the DNAzyme has a broader substrate specificity than HRP and a higher inactivation rate as well, mostly due to hemin degradation. They suggest that this higher degradation rate is because the hemin is more exposed in DNAzymes. They also proposed a possible catalytic mechanism and inactivation pathway for G-quadruplex/hemin-dependent DNAzymes (shown in Figure 5d).98
Figure 5.
(a) Schematic of a G-quadruplex, with a monovalent cation M+ (e.g., Na+ or K+) in the center; (b, c) A docked structural model of hemin cofactor stacked on top of a G-quadruplex (Bcl-2 DNA oligomer). Adapted with permission from ref 96; (d) A proposed mechanism of reaction and inactivation of G-quadruplex/hemin DNAzyme. Figure adapted with permission from ref 98.
2.6 Reaction product analysis
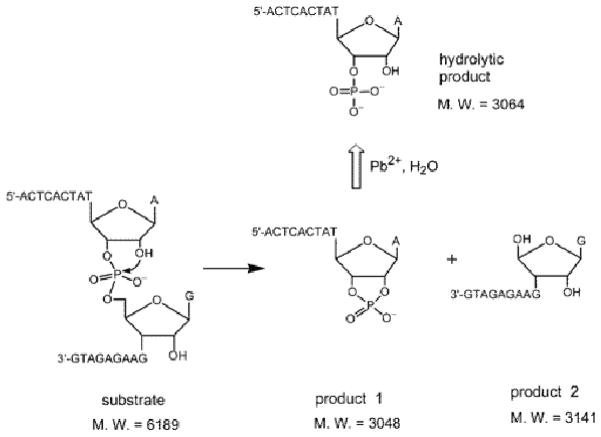
Mass spectroscopy can identify the reaction products of a DNAzyme and provide insight into how the metal ion affects or controls DNAzyme catalysis. Such information can be helpful in identifying different interaction modes between a DNAzyme and its metal cofactor. Lu and coworkers performed MALDI-TOF mass spectroscopy on the cleaved RNA products of the 8-17 DNAzyme with different added metal ions. In the presence of Zn2+ or Mg2+, the reaction goes through a 2′,3′-cyclic phosphate at the end of the 5′-product.8 However, this moiety is changed to a 3′ (or 2′) monophosphate in the case of Pb2+ (Figure 6).99
Figure 6.
Proposed mechanism of substrate cleavage by the 17E DNAzyme and the calculated mass of substrate and products. Figure adapted with permission from ref 99.
3. Structural characterization of DNAzymes
3.1 Structural characterization using NMR
Biochemical study of protein and ribozyme catalysis has been greatly enhanced by the ability to obtain high-resolution three-dimensional structures from methods including X-ray crystallography and nuclear magnetic resonance (NMR). In contrast, structural determination of DNAzymes is comparatively underexplored. Such paucity in structural information on DNAzymes is proposed to be due to the flexibility of the enzyme strand and the conformational changes that are associated with its activity.100 Reports on NMR structures of DNAzymes are rare. In fact, to our knowledge only a single structural assignment of a DNAzyme via NMR has been reported.101 Choi and coworkers performed 1H NMR on an RNA-cleaving DNAzyme with an uncleavable substrate under typical reaction conditions containing the Mg2+ cofactor. The authors were able to assign all the imino protons and then using sequential assignment assigned protons from the bases of one binding arm, but the catalytic core could not be assigned in this way.101
3.2 Structural characterization using X-ray crystallography
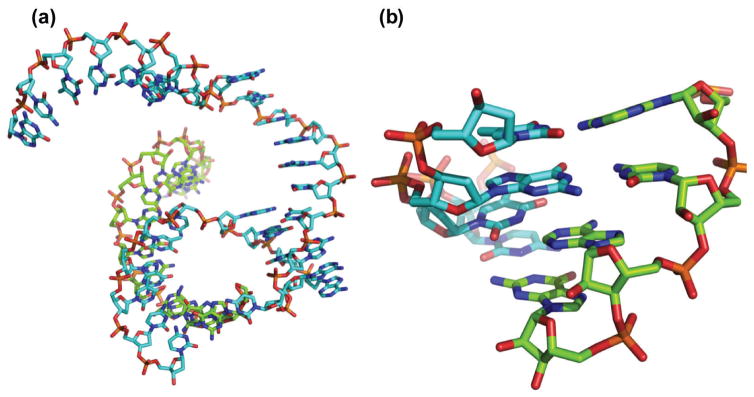
In 1999, Joyce and coworkers reported an X-ray crystal structure of the 10-23 DNAzyme at 3.0 Å resolution (Figure 7). This crystal revealed an interesting nucleic acid fold in which the helices formed two four-helix junctions stabilized by extensive base-stacking interactions and possibly by metal ions. However, this structure was shown to not be the active form of the DNAzyme, and therefore structural insight into the mechanism of action or the role of metal ions in the 10-23 DNAzyme was ultimately limited.102
Figure 7.
(a) Crystal structure of inactive 10-23 DNAzyme and (b) detailed view showing the extensive base-stacking network in the junction. (PDB ID: 1BR3) DNAzyme is shown in cyan and substrate RNA strand is shown in green.
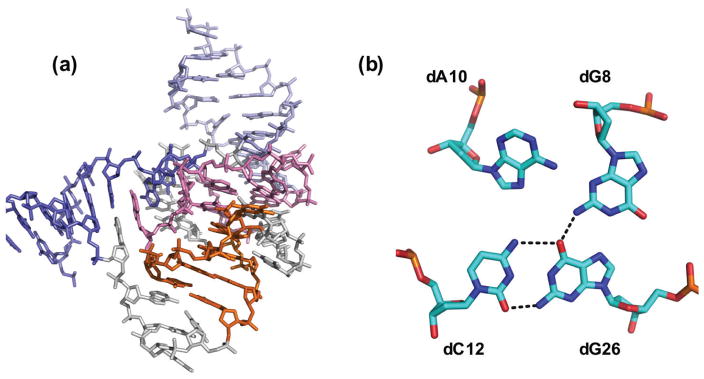
In 2016, Höbartner, Pena and coworkers reported the first crystal structure of an active RNA-ligating DNAzyme at 2.8 Å resolution (Figure 8).103 This study enabled them to propose, for the first time, a molecular mechanism of action for a DNAzyme. The work suggested that the lack of the 2′ hydroxyl group in DNAzymes, which is the most common reactive group in ribozymes, is compensated for the more flexible backbone of DNA, made possible by the lack of the same hydroxyl group.103
Figure 8.
The crystal structure of 9DB1 DNAzyme color-coded based on the helices in the structure. (PDB ID: 5CKK] An example of multiple stacking and hydrogen bonding interaction in the DNAzyme is shown on the right.
Although no electron density was observed that would clearly correspond to the metal ion cofactor in the active site, a number of interesting features suggest roles of particular functional groups in metal binding. The backbone phosphate group of nucleotide A13 was found to be located 3.1 Å from the ligation junction. They further investigated this nucleotide by installing phosphorothioate modifications at the backbone site. For both cofactor metal ions (Mg2+ and Mn2+), a major loss of activity was observed with only the Sp phosphorothioate diastereomer, but no loss of activity was observed with the Rp diastereomer. While no restoration of activity (thio effect) could be demonstrated with the more thiophilic Cd2+ ion,103 the observed lack of thio effect does not conclusively rule out metal coordination to the non-bridging pro-Sp phosphate oxygen.104 Further work will be required to address the specific role of metal ions in this and other DNAzymes, including obtaining more crystal structures of DNAzymes with metal-binding sites in the active sites.
3.3 Structural characterization using photocrosslinking and photocleavage
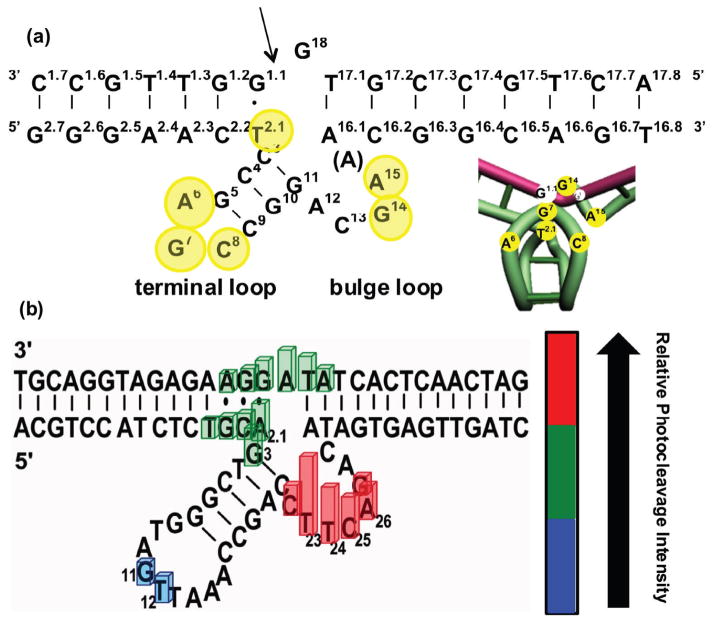
At the same time as the ongoing efforts to obtain crystal or NMR structures of DNAzymes in their active form, several studies used contact crosslinking experiments to obtain information of the global fold of DNAzymes, the required features for DNA-metal interaction and subsequent activity. In this method, the contact points between the substrate strand and the enzyme strand, and the enzymes strands with itself, are mapped. Changes in the folding result in changes of contact points. These studies, while not as direct as a crystal or NMR structure, shed light on several structural aspects of metal-mediated DNA activity. Liu and Sen used contact photo-crosslinking to study the 8-17 DNAzyme. They used a 32P-labeled enzyme strand and then mapped the formation of radioactive products after photo-crosslinking by irradiation at 365 nm.105,106 In one study, different nucleotides in the 8-17 DNAzyme were replaced with different thio- or halogen-substituted base analogues in the presence of Mg2+. The use of these modified bases allowed photochemical production of contact crosslinks. By analysis of the resulting crosslinking patterns, this technique was used to produce a model of the folded 8-17 DNAzyme (Figure 9a). In this model, the scissile base in substrate is sandwiched between the wobble thymine in the DNAzyme and the two loops around it.105 In order to separate the interactions between the enzyme and substrate strand and those within the enzyme strand, they also repeated the experiment with a labeled substrate strand and unlabeled enzyme strand. Interestingly they showed that no global folding of the DNAzyme is required for its lead-dependent cleavage activity.106 In another study, Sekhon and Sen showed that replacement of the scissile phosphodiester with phosphorothioate adds chirality and can thus provide insight into the arrangement of nucleobases in the active site of the DNAzyme.107 Using this method they provided evidence for the importance of the presence and position of the unpaired C13 base in the enzyme strand. They also demonstrated that the C3 base played a critical role in the DNAzyme’s activity, and may be involved in acid-base catalysis.107
Figure 9.
Photocleavage and photocrosslinking analysis of DNAzyme structures, (a) Predicted secondary structure and model of three-dimensional structure of 8-17 DNAyzme produced by photocrosslinking analysis. The enzyme strand is shown in green and the substrate strand is shown in pink. Yellow and white circles indicate specific nucleotides. (b) Summary of the results of uranyl photocleavage studies of 39E DNAzyme. Bar heights are proportional to relative distance from uranyl. Colors show intensity of photocleavage: red (high), green (intermediate), and blue (low). Figure (a) is adapted with permission from ref 105, and figure (b) is adapted with permission from ref 108.
In a related study, Lu and coworkers investigated the active site of the 39E and 8-17 DNAzymes using uranyl as a photochemical probe (Figure 9b).108 By irradiation of uranyl at 300–420 nm, nearby single-stranded DNA can be nicked; separation by PAGE then allows sites of cleavage to be readily visualized. Taking advantage of the specific binding of uranyl to the 39E DNAzyme, the metal-binding domains could be visualized by the pattern of cleavage. The observed corresponding patterns differed for the 8-17 DNAzyme, which lacks a specific binding site for UO22+. The locations of photocleavage correlated with regions known to be critical for activity, and deletions of these regions altered the pattern of photocleavage and reduced the uranyl binding affinity, demonstrating the importance of those loop regions for uranyl binding.108
4. Spectroscopic characterization of metal-binding site in DNAzymes
In addition to the biochemical and structural characterization approaches described previously, many spectroscopic techniques have begun to be employed to characterize DNAzymes. These methods are largely metal ion-focused techniques adapted from inorganic and bioinorganic chemistry that have been used previously in studies of inorganic complexes, metalloproteins and metallo-ribozymes, and which can provide a wealth of information on metal-binding sites in DNAzymes as well. It is important to note that some of these techniques are applicable only to certain types of metal ions. For instance, electron paramagnetic resonance (EPR) relies on the presence of unpaired electron spin, limiting its application to DNAzymes that utilize paramagnetic metal ions.
4.1 X-ray absorption spectroscopy (XAS)
Extended X-ray absorption fine structure (EXAFS) is an extensively used method to study metal binding centers in proteins, RNA, and DNA EXAFS is dependent on the ejection of a core electron upon X-ray irradiation in a metal ion and the scattered patterns of interaction of this photoelectron with the nearby electrons in non-excited atoms.109 EXAFS provides information about the geometry of the metal-binding sites, such as the type and distance of the ligands, the number of metal ions in the site, and some possible second shell interactions. Moreover, since the atoms of different elements have different energies, each will absorb photons with different energies, and thus EXAFS is element-specific. X-ray absorption near-edge spectroscopy (XANES) can provide additional information about the oxidation state of the metal ion.109
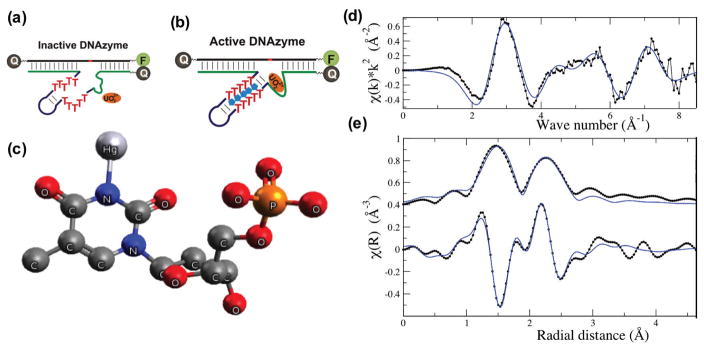
EXAFS has been used to study DNA-metal interactions. Shafer and coworkers have used EXAFS to study the interaction of Pb2+ with the thrombin aptamer.110 They demonstrated that the Pb2+ ion is coordinated to the O6 position of 6 guanines within the G-quadruplex core of the aptamer, in good agreement with crystallography data.110 Although this study was performed on an aptamer and not a DNAzyme, it aptly demonstrates the power of EXAFS in studying DNA-metal interactions. Furthermore, as described previously (Figure 5), the G-quadruplex motif is known to be present in the metal-dependent peroxidase-mimicking DNAzymes,111 and thus similar approaches can be used to study the DNA-metal interactions of those DNAzymes. Lu and coworkers have used EXAFS to study the interaction of DNA with mercury in a 10-bp double stranded DNA as a model of a catalytic mercury-dependent DNAzyme (Figure 10).112 This 10-bp DNA shared the same mercury-binding site to the larger catalytic DNA.113 Their results showed one tight binding affinity site for mercury, consistent with the observed one to one binding of the mercury ion to the DNAzyme.112,113
Figure 10.
(a) Secondary structure of inactive and Hg(II)-binding activated DNAzymes. The Hg(II) binding to the T-T repeats in the stem activate the DNAzyme., (b). (c) Model of thymidine paired with Hg(II) ion used to fit the experimentally obtained EXAFS curve (d) and its Fourier transforms (e). Figures a and b are adapted with permission from ref 112 and (c–e) are obtained with permission from ref 113.
4.2 Fluorescent resonance energy transfer (FRET)
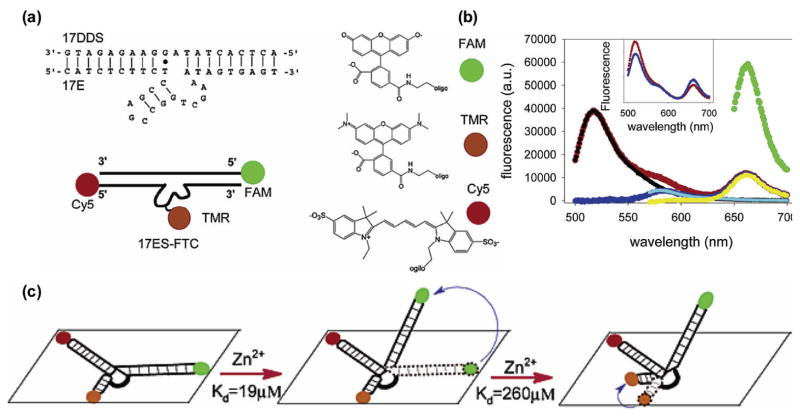
Fluorescent resonance energy transfer spectroscopy (FRET) has also been used to study DNA-metal interactions. Taking advantage of the ease of modification of DNA bases, one can label any two different sites in DNA with FRET pairs and monitor the FRET signal upon metal addition or reaction. Careful placement of the FRET pairs can provide significant insight about the interplay between the metal ion and enzymatic reactivity.114 Lu and coworkers used a trifluorophore-labeled 8-17 DNAzyme to monitor the conformational changes in the DNAzyme upon Zn2+ addition.115 This technique demonstrated that the folding of DNAzyme to its active form is a two-step process, each linked to association of one zinc ion (Figure 11).115 Later, single-molecule FRET (smFRET) was used to study the metal-dependent folding of the 8-17 DNAzyme, by immobilization of the enzyme strand to a surface. Surprisingly, it was observed that Pb2+-dependent cleavage of the substrate doesn’t result in the same global folding observed with Zn2+-dependent folding, indicating that DNAzymes may be able to carry out lock-and-key target recognition for metal ions, in addition to the more common induced fit mechanism.116
Figure 11.
(a) The 8-17 DNAzyme with uncleavable substrate strand (17DDS) and the terminal FRET pair modifications and their structures. (b) An example of spectral fitting with trifluorophore-modified DNAzyme. (c) Proposed two-step mechanism of DNAzyme folding in the presence of Zn2+ ions. Figures are adapted with permission from ref 115.
4.3 Circular dichroism (CD)
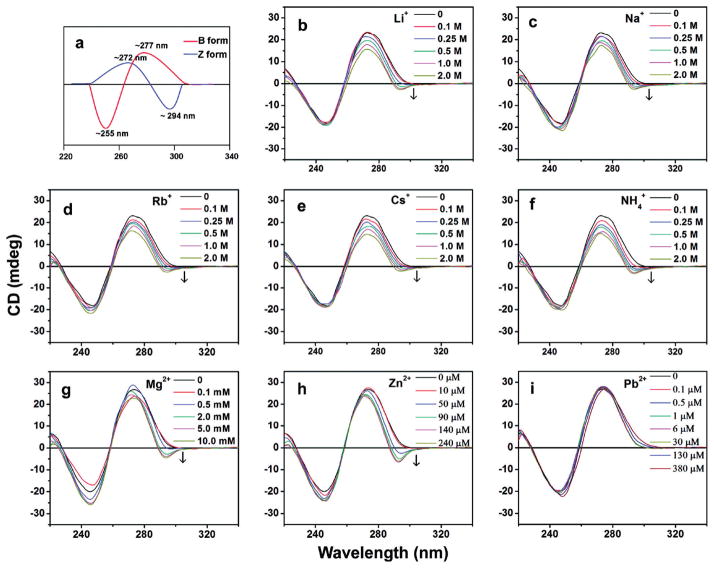
Another commonly used technique for obtaining structural information about DNA-metal interactions in DNAzymes is circular dichroism (CD). CD is a differential spectroscopy technique, which shows the difference between left- and right-handed circularly polarized light.117 Any chiral molecule will have a non-zero CD spectrum. Biological samples, including DNA, have many chiral centers and hence have robust CD spectra. CD is very sensitive to secondary structures of macromolecules and therefore can be used to monitor transitions in secondary structure during DNA-metal interactions.117 Lu and coworkers showed the formation of Z-DNA in the 8-17 DNAzyme in the presence of monovalent metal ions, and several divalent metal ions (Figure 12).94 Such structural changes were not observed in controls with no enzyme or strand or an inactive form of the DNAzyme with a single mutation. Taira and coworkers used CD to associate structural element formation with activity and they showed that in metal-dependent DNAzymes, higher activity corresponds well with increased occurrence of B-helix structure, as monitored by CD.118 Cieslak et al. used a combination of CD, FRET and fluorescence anisotropy to study the folding of the 10-23 DNAzyme in the presence of Mg2+.119 When the binding arms are correctly designed, the 10-23 DNAzyme is known to inhibit expression of protein transcripts both in vitro and in vivo, but the initial in vitro kinetic measurements were performed under Mg2+ concentrations much higher than physiological conditions.119 In this paper, the author showed that the formation of the active DNAzyme happens in multiple stages and in a Mg2+-dependent manner. They also demonstrated that under intracellular conditions, the DNAzyme is in a partially folded state that cannot effectively bind the RNA substrate well, and thus the observed activity in vitro might result simply from an antisense effect rather than by catalysis.119
Figure 12.
(a) Representative CD spectra of B- and Z-form of DNA. (b–i) CD spectral changes of 8-17 DNAzyme upon titration with several monovalent and divalent metal ions reveal structural alterations upon metal-binding. Figure adapted with permission from ref 94
4.4 Sensitized luminescence
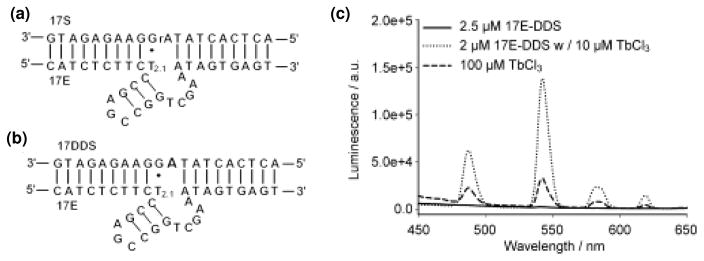
Analysis of the sensitized luminescence emission of lanthanides has been used to study nucleic acid-metal interactions in both ribozymes and DNAzymes. These studies have been possible due to the ability of Tb3+ to bind at many metal binding sites. Upon binding, the sensitized luminescence at 290 nm increases due to energy transfer from DNA/RNA.120,121 Thus Tb3+ luminescence is highly sensitive to binding and provides a tool to study the metal binding site in DNAzymes. Lu and coworkers showed that the luminescence of Tb3+ is enhanced only in the active 8-17 DNAzyme and showed that it binds to an unpaired region in the DNAzyme (Figure 13).121 Lifetime luminescence measurements in water and D2O showed that the Tb3+ ion is coordinated to about 6 water molecules and 2–3 ligands from DNA. Competition between divalent metal ions and Tb3+ suggested that Pb2+ might bind to a different site compared to other metal ions such as Zn2+.121 Liu and coworkers showed that guanines in the loop of the sodium-dependent A43 DNAzyme are critical for binding to Na.84 Liu and coworkers also used the same strategy to investigate the affinity of the lanthanide-dependent Ce13 DNAzyme for different lanthanides. They showed that the binding affinities are symmetric across the series with Nd3+ and Ho3+ having the highest affinity. Since the activity of the DNAzyme with different lanthanides was not consistent with the observed binding trend, the authors suggested that metal binding is not the rate limiting step of the reaction.93
Figure 13.
(a) Structure of active 8-17 DNAzyme. (b) 17E-DDS, the inactive form of 8-17 DNAzyme in which the substrate is not cleavable. (c) Increase in Tb3+ sensitized absorption upon binding of Tb3+ to DNAzyme. Figures are adapted with permission from ref 121.
5. Concluding remarks
In this review we have described the commonly used methods for characterization of metal ion-DNAzyme interactions and how such methods have been applied towards better understanding this fascinating type of metalloenzymes. The increasing use of DNAzymes as metal ion sensors both for environmental samples and more recently for cellular detection has in part led to increased interest in the properties underlying their function and selectivity.
Achieving a better understanding of the main contributors to metal ion selectivity in DNAzymes not only opens new avenues in bioinorganic chemistry, but also may allow the design and improvement of metal ion sensors based on these molecules. As shown in the previous sections, a number of novel characterization techniques have already been applied towards understanding this selectivity, and future work will likely build upon the established suite of techniques already in widespread use in studying metalloproteins.
In order to realize the full potential of metallo-DNAzymes, we believe the field will vastly benefit from focus on the following future directions.
Comparative analysis of DNAzvmes with similar metal ion specificity. As shown early in the history of DNAzymes, selections for disparate targets can sometimes serendipitously identify sequences that are also active for unintended targets. The clearest example of such event is the independent selection of the 8-17 DNAzyme motif by multiple research groups under different in vitro selection conditions.8,9,11,77 In the process, researchers identified a number of functionally similar but non-identical sequences, whose comparison enabled the identification of features critical for observed differences in metal ion selectivity.77,79 More recent work has been carried out to investigate the comparative selectivity of multiple Na+-responsive DNAzymes, which could in turn report on structural features critical for selective monovalent metal ion binding by DNA.83,84 Employment of next-generation sequencing technology may make it possible to track large-scale changes in the pool of sequences during the in vitro selection process. This method would allow observation of the gain or loss of particular domains as a property of selection stringency or selection round, and could provide a wealth of information for such comparative analysis.
Incorporation of modified nucleotides to study structural features responsible for metal selectivity and activity on the level of individual functional groups. Modified nucleotides have been incorporated into DNAzymes for a number of applications, e.g. as high-affinity metal-binding ligands during selection,122 or for improved stability for cellular sensing applications.59 The ease of synthesis of DNAzymes as well as the straightforward design of modified nucleotides allows a great deal of fine-tuning of the chemical properties beyond what is achievable with the four basic nucleotides, on the scale of individual functional groups. An excellent example is from the recent use of specific diastereomer phosphorothioates to investigate the three-dimensional structure of DNAzymes and improve catalytic activity.25,103 Especially when combined with higher-resolution structural information, these studies can greatly elucidate the nature of the binding pockets used by DNAzymes to selectively bind metal ions.
In vitro selection and study of DNAzvmes that are active in the presence of spectroscopically rich paramagnetic metal ions. Most of the DNAzymes that have been selected so far bind metal ions that often do not display rich spectroscopic features, such as alkali and alkaline earth metal ions. As a result, there has been an overall lack of spectroscopic investigation of metal-binding sites in DNAzymes. Obtaining new DNAzymes that can specifically bind spectroscopically rich paramagnetic metal ions such as iron, manganese and nickel will not only expand the scope of DNAzyme reactivity, but will also allow the use of the full range of spectroscopic techniques used with metalloproteins and ribozymes, such as electron paramagnetic resonance (EPR).123,124
In conclusion, much progress has been made in understanding the interactions of metal ions with DNAzymes. With the employment of better in vitro selection methods using modified nucleotides and more advanced biochemical, structural and spectroscopic techniques, we will gain much deeper understanding of metal-binding sites in DNAzymes. This understanding will pave the way for wider applications of this burgeoning family of metalloenzymes as a powerful new class of sensors for both environmental and cellular metal detection.
Highlights.
Reviews metal-dependent DNAzymes and identification of features underlying metal ion specificity
Biochemical analysis has identified conserved sequences and secondary structure necessary for metal ion discrimination
Many methods for structural characterization have been investigated
Spectroscopic studies allow probing of the metal binding sites in DNAzymes and catalytic mechanisms, including “lock-and-key” mode of catalysis
Acknowledgments
The Lu group research mentioned in this review has been supported by the U.S. National Institutes of Health (Grant ES016865), by the Office of Science (BER), the U.S. Department of Energy (DE-FG02-08ER64568), and National Science Foundation (CTS-0120978, CMMI 0749028 and DMR-0117792).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Breaker RR, Joyce GF. A DNA enzyme that cleaves RNA. Chem Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 2.Wernette DP, Liu J, Bohn PW, Lu Y. Functional-DNA-Based Nanoscale Materials and Devices for Sensing Trace Contaminants in Water. MRS Bull. 2008;33:34–41. [Google Scholar]
- 3.Lan T, Lu Y. In: Interplay between Metal Ions and Nucleic Acids; Sigel A, Sigel H, Sigel ORK, editors. Springer Netherlands; Dordrecht: 2012. pp. 217–248. [Google Scholar]
- 4.Xiang Y, Lu Y. DNA as sensors and imaging agents for metal ions. Inorg Chem. 2014;53:1925–1942. doi: 10.1021/ic4019103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao Z, Lu Y. Metal Complex-DNA Interactions; John Wiley & Sons, Ltd; 2009. pp. 395–414. [Google Scholar]
- 6.Lu Y. New transition-metal-dependent DNAzymes as efficient endonucleases and as selective metal biosensors. Chem Eur J. 2002;8:4588–4596. doi: 10.1002/1521-3765(20021018)8:20<4588::AID-CHEM4588>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 7.Lu Y. Metalloprotein and metallo-DNA/RNAzyme design: current approaches, success measures, and future challenges. Inorg Chem. 2006;45:9930–9940. doi: 10.1021/ic052007t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Zheng W, Kwon AH, Lu Y. In vitro selection and characterization of a highly efficient Zn(II)-dependent RNA-cleaving deoxyribozyme. Nucl Acids Res. 2000;28:481–488. doi: 10.1093/nar/28.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geyer CR, Sen D. Evidence for the metal-cofactor independence of an RNA phosphodiester-cleaving DNA enzyme. Chem Biol. 1997;4:579–593. doi: 10.1016/s1074-5521(97)90244-1. [DOI] [PubMed] [Google Scholar]
- 11.Faulhammer D, Famulok M. The Ca2+ Ion as a Cofactor for a Novel RNA-Cleaving Deoxyribozyme. Angew Chem Int Ed. 1996;35:2837–2841. [Google Scholar]
- 12.Feldman AR, Sen D. A new and efficient DNA enzyme for the sequence-specific cleavage of RNA. J Mol Biol. 2001;313:283–294. doi: 10.1006/jmbi.2001.5058. [DOI] [PubMed] [Google Scholar]
- 13.Cruz RP, Withers JB, Li Y. Dinucleotide junction cleavage versatility of 8-17 deoxyribozyme. Chem Biol. 2004;11:57–67. doi: 10.1016/j.chembiol.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Brown AK, Meng X, Cropek DM, Istok JD, Watson DB, Lu Y. A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity. Proc Natl Acad Sci USA. 2007;104:2056–2061. doi: 10.1073/pnas.0607875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruesehoff PJ, Li J, Augustine AJ, 3rd, Lu Y. Improving metal ion specificity during in vitro selection of catalytic DNA. Comb Chem High Throughput Screen. 2002;5:327–335. doi: 10.2174/1386207023330264. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Mei SH, Brennan JD, Li Y. Assemblage of signaling DNA enzymes with intriguing metal-ion specificities and pH dependences. J Am Chem Soc. 2003;125:7539–7545. doi: 10.1021/ja035208+. [DOI] [PubMed] [Google Scholar]
- 17.Mei SH, Liu Z, Brennan JD, Li Y. An efficient RNA-cleaving DNA enzyme that synchronizes catalysis with fluorescence signaling. J Am Chem Soc. 2003;125:412–420. doi: 10.1021/ja0281232. [DOI] [PubMed] [Google Scholar]
- 18.Torabi SF, Wu P, McGhee CE, Chen L, Hwang K, Zheng N, Cheng J, Lu Y. In vitro selection of a sodium-specific DNAzyme and its application in intracellular sensing. Proc Natl Acad Sci USA. 2015;112:5903–5908. doi: 10.1073/pnas.1420361112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang PJ, Lin J, Cao J, Vazin M, Liu J. Ultrasensitive DNAzyme beacon for lanthanides and metal speciation. Anal Chem. 2014;86:1816–1821. doi: 10.1021/ac403762s. [DOI] [PubMed] [Google Scholar]
- 20.Huang PJ, Vazin M, Matuszek Z, Liu J. A new heavy lanthanide-dependent DNAzyme displaying strong metal cooperativity and unrescuable phosphorothioate effect. Nucl Acids Res. 2015;43:461–469. doi: 10.1093/nar/gku1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang PJ, Vazin M, Liu J. In vitro selection of a new lanthanide-dependent DNAzyme for ratiometric sensing lanthanides. Anal Chem. 2014;86:9993–9999. doi: 10.1021/ac5029962. [DOI] [PubMed] [Google Scholar]
- 22.Saran R, Chen Q, Liu J. Searching for a DNAzyme Version of the Leadzyme. J Mol Evol. 2015;81:235–244. doi: 10.1007/s00239-015-9702-z. [DOI] [PubMed] [Google Scholar]
- 23.Lam JC, Kwan SO, Li Y. Characterization of non-8-17 sequences uncovers structurally diverse RNA-cleaving deoxyribozymes. Mol Biosyst. 2011;7:2139–2146. doi: 10.1039/c1mb05034f. [DOI] [PubMed] [Google Scholar]
- 24.Shen Y, Chiuman W, Brennan JD, Li Y. Catalysis and rational engineering of trans-acting pH6DZ1, an RNA-cleaving and fluorescence-signaling deoxyribozyme with a four-way junction structure. Chembiochem. 2006;7:1343–1348. doi: 10.1002/cbic.200600195. [DOI] [PubMed] [Google Scholar]
- 25.Huang PJ, Liu J. Rational evolution of Cd2+-specific DNAzymes with phosphorothioate modified cleavage junction and Cd2+ sensing. Nucl Acids Res. 2015;43:6125–6133. doi: 10.1093/nar/gkv519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carmi N, Balkhi SR, Breaker RR. Cleaving DNA with DNA. Proc Natl Acad Sci USA. 1998;95:2233–2237. doi: 10.1073/pnas.95.5.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carmi N, Breaker RR. Characterization of a DNA-cleaving deoxyribozyme. Bioorg Med Chem. 2001;9:2589–2600. doi: 10.1016/s0968-0896(01)00035-9. [DOI] [PubMed] [Google Scholar]
- 28.Carmi N, Shultz LA, Breaker RR. In vitro selection of self-cleaving DNAs. Chem Biol. 1996;3:1039–1046. doi: 10.1016/s1074-5521(96)90170-2. [DOI] [PubMed] [Google Scholar]
- 29.Chandra M, Sachdeva A, Silverman SK. DNA-catalyzed sequence-specific hydrolysis of DNA. Nat Chem Biol. 2009;5:718–720. doi: 10.1038/nchembio.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao Y, Chandra M, Silverman SK. Functional compromises among pH tolerance, site specificity, and sequence tolerance for a DNA-hydrolyzing deoxyribozyme. Biochemistry. 2010;49:9630–9637. doi: 10.1021/bi1013672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao Y, Allen EC, Silverman SK. Merely two mutations switch a DNA-hydrolyzing deoxyribozyme from heterobimetallic (Zn2+/Mn2+) to monometallic (Zn2+-only) behavior. Chem Commun. 2011;47:1749–1751. doi: 10.1039/c0cc04575f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dokukin V, Silverman SK. Lanthanide ions as required cofactors for DNA catalysts. Chem Sci. 2012;3:1707–1714. doi: 10.1039/C2SC01067D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burmeister J, von Kiedrowski G, Ellington AD. Cofactor-Assisted Self-Cleavage in DNA Libraries with a 3′–5′-Phosphoramidate Bond. Angew Chem Int Ed. 1997;36:1321–1324. [Google Scholar]
- 34.Cuenoud B, Szostak JW. A DNA metalloenzyme with DNA ligase activity. Nature. 1995;375:611–614. doi: 10.1038/375611a0. [DOI] [PubMed] [Google Scholar]
- 35.Sreedhara A, Li Y, Breaker RR. Ligating DNA with DNA. J Am Chem Soc. 2004;126:3454–3460. doi: 10.1021/ja039713i. [DOI] [PubMed] [Google Scholar]
- 36.Flynn-Charlebois A, Wang Y, Prior TK, Rashid I, Hoadley KA, Coppins RL, Wolf AC, Silverman SK. Deoxyribozymes with 2′–5′ RNA ligase activity. J Am Chem Soc. 2003;125:2444–2454. doi: 10.1021/ja028774y. [DOI] [PubMed] [Google Scholar]
- 37.Purtha WE, Coppins RL, Smalley MK, Silverman SK. General deoxyribozyme-catalyzed synthesis of native 3′–5′ RNA linkages. J Am Chem Soc. 2005;127:13124–13125. doi: 10.1021/ja0533702. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Silverman SK. Directing the outcome of deoxyribozyme selections to favor native 3′–5′ RNA ligation. Biochemistry. 2005;44:3017–3023. doi: 10.1021/bi0478291. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Silverman SK. Deoxyribozymes that synthesize branched and lariat RNA. J Am Chem Soc. 2003;125:6880–6881. doi: 10.1021/ja035150z. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Silverman SK. Characterization of deoxyribozymes that synthesize branched RNA. Biochemistry. 2003;42:15252–15263. doi: 10.1021/bi0355847. [DOI] [PubMed] [Google Scholar]
- 41.Coppins RL, Silverman SK. Rational modification of a selection strategy leads to deoxyribozymes that create native 3′–5′ RNA linkages. J Am Chem Soc. 2004;126:16426–16432. doi: 10.1021/ja045817x. [DOI] [PubMed] [Google Scholar]
- 42.Coppins RL, Silverman SK. A DNA enzyme that mimics the first step of RNA splicing. Nat Struct Mol Biol. 2004;11:270–274. doi: 10.1038/nsmb727. [DOI] [PubMed] [Google Scholar]
- 43.Pradeepkumar PI, Hobartner C, Baum DA, Silverman SK. DNA-catalyzed formation of nucleopeptide linkages. Angew Chem. 2008;47:1753–1757. doi: 10.1002/anie.200703676. [DOI] [PubMed] [Google Scholar]
- 44.Wong OY, Pradeepkumar PI, Silverman SK. DNA-catalyzed covalent modification of amino acid side chains in tethered and free peptide substrates. Biochemistry. 2011;50:4741–4749. doi: 10.1021/bi200585n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sachdeva A, Silverman SK. DNA-catalyzed serine side chain reactivity and selectivity. Chem Commun. 2010;46:2215–2217. doi: 10.1039/b927317d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang W, Billen LP, Li Y. Sequence diversity, metal specificity, and catalytic proficiency of metal-dependent phosphorylating DNA enzymes. Chem Biol. 2002;9:507–517. doi: 10.1016/s1074-5521(02)00127-8. [DOI] [PubMed] [Google Scholar]
- 47.Li Y, Liu Y, Breaker RR. In vitro selection of deoxyribozymes with DNA capping activity. Nucleic Acids Symp Ser. 1999:237–238. doi: 10.1093/nass/42.1.237. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, Liu Y, Breaker RR. Capping DNA with DNA. Biochemistry. 2000;39:3106–3114. doi: 10.1021/bi992710r. [DOI] [PubMed] [Google Scholar]
- 49.Sheppard TL, Ordoukhanian P, Joyce GF. A DNA enzyme with N-glycosylase activity. Proc Natl Acad Sci USA. 2000;97:7802–7807. doi: 10.1073/pnas.97.14.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chandra M, Silverman SK. DNA and RNA can be equally efficient catalysts for carbon-carbon bond formation. J Am Chem Soc. 2008;130:2936–2937. doi: 10.1021/ja7111965. [DOI] [PubMed] [Google Scholar]
- 51.Li Y, Sen D. The modus operandi of a DNA enzyme: enhancement of substrate basicity. Chem Biol. 1998;5:1–12. doi: 10.1016/s1074-5521(98)90082-5. [DOI] [PubMed] [Google Scholar]
- 52.Li Y, Sen D. Toward an efficient DNAzyme. Biochemistry. 1997;36:5589–5599. doi: 10.1021/bi962694n. [DOI] [PubMed] [Google Scholar]
- 53.Zhou C, Avins JL, Klauser PC, Brandsen BM, Lee Y, Silverman SK. DNA-Catalyzed Amide Hydrolysis. J Am Chem Soc. 2016 doi: 10.1021/jacs.5b12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chandrasekar J, Silverman SK. Catalytic DNA with phosphatase activity. Proc Natl Acad Sci USA. 2013;110:5315–5320. doi: 10.1073/pnas.1221946110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walsh SM, Sachdeva A, Silverman SK. DNA catalysts with tyrosine kinase activity. J Am Chem Soc. 2013;135:14928–14931. doi: 10.1021/ja407586u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dokukin V, Silverman SK. A modular tyrosine kinase deoxyribozyme with discrete aptamer and catalyst domains. Chem Commun. 2014;50:9317–9320. doi: 10.1039/c4cc04253k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mohan U, Burai R, McNaughton BR. In vitro evolution of a Friedel-Crafts deoxyribozyme. Org Biomol Chem. 2013;11:2241–2244. doi: 10.1039/c3ob40080h. [DOI] [PubMed] [Google Scholar]
- 58.Lan T, Furuya K, Lu Y. A highly selective lead sensor based on a classic lead DNAzyme. Chem Commun. 2010;46:3896–3898. doi: 10.1039/b926910j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hwang K, Wu P, Kim T, Lei L, Tian S, Wang Y, Lu Y. Photocaged DNAzymes as a general method for sensing metal ions in living cells. Angew Chem. 2014;53:13798–13802. doi: 10.1002/anie.201408333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu P, Hwang K, Lan T, Lu Y. A DNAzyme-gold nanoparticle probe for uranyl ion in living cells. J Am Chem Soc. 2013;135:5254–5257. doi: 10.1021/ja400150v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiang Y, Tong A, Lu Y. Abasic site-containing DNAzyme and aptamer for label-free fluorescent detection of Pb(2+) and adenosine with high sensitivity, selectivity, and tunable dynamic range. J Am Chem Soc. 2009;131:15352–15357. doi: 10.1021/ja905854a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mun H, Jo EJ, Li T, Joung HA, Hong DG, Shim WB, Jung C, Kim MG. Homogeneous assay of target molecules based on chemiluminescence resonance energy transfer (CRET) using DNAzyme-linked aptamers. Biosens Bioelectron. 2014;58:308–313. doi: 10.1016/j.bios.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 63.Hu L, Liu X, Cecconello A, Willner I. Dual switchable CRET-induced luminescence of CdSe/ZnS quantum dots (QDs) by the hemin/G-quadruplex-bridged aggregation and deaggregation of two-sized QDs. Nano Lett. 2014;14:6030–6035. doi: 10.1021/nl503299f. [DOI] [PubMed] [Google Scholar]
- 64.Liu X, Freeman R, Golub E, Willner I. Chemiluminescence and chemiluminescence resonance energy transfer (CRET) aptamer sensors using catalytic hemin/G-quadruplexes. ACS Nano. 2011;5:7648–7655. doi: 10.1021/nn202799d. [DOI] [PubMed] [Google Scholar]
- 65.Freeman R, Liu X, Willner I. Chemiluminescent and chemiluminescence resonance energy transfer (CRET) detection of DNA, metal ions, and aptamer-substrate complexes using hemin/G-quadruplexes and CdSe/ZnS quantum dots. J Am Chem Soc. 2011;133:11597–11604. doi: 10.1021/ja202639m. [DOI] [PubMed] [Google Scholar]
- 66.Shen L, Chen Z, Li Y, He S, Xie S, Xu X, Liang Z, Meng X, Li Q, Zhu Z, Li M, Le XC, Shao Y. Electrochemical DNAzyme sensor for lead based on amplification of DNA-Au bio-bar codes. Anal Chem. 2008;80:6323–6328. doi: 10.1021/ac800601y. [DOI] [PubMed] [Google Scholar]
- 67.Yang X, Xu J, Tang X, Liu H, Tian D. A novel electrochemical DNAzyme sensor for the amplified detection of Pb2+ ions. Chem Commun. 2010;46:3107–3109. doi: 10.1039/c002137g. [DOI] [PubMed] [Google Scholar]
- 68.Tang Q, Yuan Y, Xiao X, Guo P, Hu J, Ma D, Gao Y. DNAzyme based electrochemical sensors for trace uranium. Microchim Acta. 2013;180:1059–1064. [Google Scholar]
- 69.Guo Q, Bao Y, Yang X, Wang K, Wang Q, Tan Y. Amplified electrochemical DNA sensor using peroxidase-like DNAzyme. Talanta. 2010;83:500–504. doi: 10.1016/j.talanta.2010.09.054. [DOI] [PubMed] [Google Scholar]
- 70.Xu W, Xing H, Lu Y. A smart T1-weighted MRI contrast agent for uranyl cations based on a DNAzyme-gadolinium conjugate. Analyst. 2013;138:6266–6269. doi: 10.1039/c3an01182h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu L, Yin H, Ma W, Wang L, Kuang H, Xu C. MRI biosensor for lead detection based on the DNAzyme-induced catalytic reaction. J Phys Chem B. 2013;117:14367–14371. doi: 10.1021/jp4087656. [DOI] [PubMed] [Google Scholar]
- 72.Xiang Y, Lu Y. Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nat Chem. 2011;3:697–703. doi: 10.1038/nchem.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xiang Y, Lu Y. An invasive DNA approach toward a general method for portable quantification of metal ions using a personal glucose meter. Chem Commun. 2013;49:585–587. doi: 10.1039/c2cc37156a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang XB, Kong RM, Lu Y. Metal ion sensors based on DNAzymes and related DNA molecules. Annu Rev Anal Chem. 2011;4:105–128. doi: 10.1146/annurev.anchem.111808.073617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Torabi SF, Lu Y. Functional DNA nanomaterials for sensing and imaging in living cells. Curr Opin Biotechnol. 2014;28:88–95. doi: 10.1016/j.copbio.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ihms HE, Lu Y. In vitro selection of metal ion-selective DNAzymes. Methods Mol Biol. 2012;848:297–316. doi: 10.1007/978-1-61779-545-9_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peracchi A. Preferential activation of the 8–17 deoxyribozyme by Ca(2+) ions. Evidence for the identity of 8-17 with the catalytic domain of the Mg5 deoxyribozyme. J Biol Chem. 2000;275:11693–11697. doi: 10.1074/jbc.275.16.11693. [DOI] [PubMed] [Google Scholar]
- 78.Li J, Lu Y. A Highly Sensitive and Selective Catalytic DNA Biosensor for Lead Ions. J Am Chem Soc. 2000;122:10466–10467. [Google Scholar]
- 79.Saran R, Liu J. A comparison of two classic Pb2+-dependent RNA-cleaving DNAzymes. Inorg Chem Front. 2016 [Google Scholar]
- 80.Nagraj N, Liu J, Sterling S, Wu J, Lu Y. DNAzyme catalytic beacon sensors that resist temperature-dependent variations. Chem Commun. 2009:4103–4105. doi: 10.1039/b903059j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nelson KE, Ihms HE, Mazumdar D, Bruesehoff PJ, Lu Y. The importance of peripheral sequences in determining the metal selectivity of an in vitro-selected Co(2+) -dependent DNAzyme. Chembiochem. 2012;13:381–391. doi: 10.1002/cbic.201100724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brown AK, Liu J, He Y, Lu Y. Biochemical characterization of a uranyl ion-specific DNAzyme. Chembiochem. 2009;10:486–492. doi: 10.1002/cbic.200800632. [DOI] [PubMed] [Google Scholar]
- 83.Torabi SF, Lu Y. Identification of the Same Na(+)-Specific DNAzyme Motif from Two In Vitro Selections Under Different Conditions. J Mol Evol. 2015;81:225–234. doi: 10.1007/s00239-015-9715-7. [DOI] [PubMed] [Google Scholar]
- 84.Zhou W, Zhang Y, Huang PJ, Ding J, Liu J. A DNAzyme requiring two different metal ions at two distinct sites. Nucl Acids Res. 2016;44:354–363. doi: 10.1093/nar/gkv1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zaborowska Z, Furste JP, Erdmann VA, Kurreck J. Sequence requirements in the catalytic core of the “10-23” DNA enzyme. J Biol Chem. 2002;277:40617–40622. doi: 10.1074/jbc.M207094200. [DOI] [PubMed] [Google Scholar]
- 86.Peracchi A, Bonaccio M, Clerici M. A mutational analysis of the 8-17 deoxyribozyme core. J Mol Biol. 2005;352:783–794. doi: 10.1016/j.jmb.2005.07.059. [DOI] [PubMed] [Google Scholar]
- 87.Wang B, Cao L, Chiuman W, Li Y, Xi Z. Probing the function of nucleotides in the catalytic cores of the 8-17 and 10-23 DNAzymes by abasic nucleotide and C3 spacer substitutions. Biochemistry. 2010;49:7553–7562. doi: 10.1021/bi100304b. [DOI] [PubMed] [Google Scholar]
- 88.Hollenstein M, Hipolito CJ, Lam CH, Perrin DM. A self-cleaving DNA enzyme modified with amines, guanidines and imidazoles operates independently of divalent metal cations (M2+) Nucl Acids Res. 2009;37:1638–1649. doi: 10.1093/nar/gkn1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Holbrook SR, Sussman JL, Warrant RW, Church GM, Kim SH. RNA-ligant interactions. (I) Magnesium binding sites in yeast tRNAPhe. Nucl Acids Res. 1977;4:2811–2820. doi: 10.1093/nar/4.8.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jack A, Ladner JE, Rhodes D, Brown RS, Klug A. A crystallographic study of metal-binding to yeast phenylalanine transfer RNA. J Mol Biol. 1977;111:315–328. doi: 10.1016/s0022-2836(77)80054-5. [DOI] [PubMed] [Google Scholar]
- 91.Brown RS, Dewan JC, Klug A. Crystallographic and biochemical investigation of the lead(II)-catalyzed hydrolysis of yeast phenylalanine tRNA. Biochemistry. 1985;24:4785–4801. doi: 10.1021/bi00339a012. [DOI] [PubMed] [Google Scholar]
- 92.Rubin JR, Wang J, Sundaralingam M. X-ray diffraction study of the zinc(II) binding sites in yeast phenylalanine transfer RNA. Preferential binding of zinc to guanines in purine-purine sequences. Biochim Biophys Acta. 1983;756:111–118. doi: 10.1016/0304-4165(83)90030-2. [DOI] [PubMed] [Google Scholar]
- 93.Lin WT, Huang PJ, Pautler R, Liu J. The group trend of lanthanides binding to DNA and DNAzymes with a complex but symmetric pattern. Chem Commun. 2014;50:11859–11862. doi: 10.1039/c4cc05551a. [DOI] [PubMed] [Google Scholar]
- 94.Mazumdar D, Nagraj N, Kim HK, Meng X, Brown AK, Sun Q, Li W, Lu Y. Activity, folding and Z-DNA formation of the 8-17 DNAzyme in the presence of monovalent ions. J Am Chem Soc. 2009;131:5506–5515. doi: 10.1021/ja8082939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sen D, Poon LC. RNA and DNA complexes with hemin [Fe(III) heme] are efficient peroxidases and peroxygenases: how do they do it and what does it mean? Crit Rev Biochem Mol Biol. 2011;46:478–492. doi: 10.3109/10409238.2011.618220. [DOI] [PubMed] [Google Scholar]
- 96.Poon LC, Methot SP, Morabi-Pazooki W, Pio F, Bennet AJ, Sen D. Guanine-rich RNAs and DNAs that bind heme robustly catalyze oxygen transfer reactions. J Am Chem Soc. 2011;133:1877–1884. doi: 10.1021/ja108571a. [DOI] [PubMed] [Google Scholar]
- 97.Shumayrikh N, Huang YC, Sen D. Heme activation by DNA: isoguanine pentaplexes, but not quadruplexes, bind heme and enhance its oxidative activity. Nucl Acids Res. 2015;43:4191–4201. doi: 10.1093/nar/gkv266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang X, Fang C, Mei H, Chang T, Cao Z, Shangguan D. Characterization of G-quadruplex/hemin peroxidase: substrate specificity and inactivation kinetics. Chem Eur J. 2011;17:14475–14484. doi: 10.1002/chem.201101941. [DOI] [PubMed] [Google Scholar]
- 99.Brown AK, Li J, Pavot CM, Lu Y. A lead-dependent DNAzyme with a two-step mechanism. Biochemistry. 2003;42:7152–7161. doi: 10.1021/bi027332w. [DOI] [PubMed] [Google Scholar]
- 100.Borman S. After Two Decades of Trying, Scientists Report First Crystal Structure of a DNAzyme. C&EN. 2016;94:3. [Google Scholar]
- 101.Choi YJ, Han HJ, Lee JH, Suh SW, Choi B-S. Synthesis and NMR Studies of the RNA-Cleaving DNAzyme. Bull Korean Chem Soc. 2000;21:955–956. [Google Scholar]
- 102.Nowakowski J, Shim PJ, Prasad GS, Stout CD, Joyce GF. Crystal structure of an 82-nucleotide RNA-DNA complex formed by the 10-23 DNA enzyme. Nat Struct Biol. 1999;6:151–156. doi: 10.1038/5839. [DOI] [PubMed] [Google Scholar]
- 103.Ponce-Salvatierra A, Wawrzyniak-Turek K, Steuerwald U, Hobartner C, Pena V. Crystal structure of a DNA catalyst. Nature. 2016;529:231–234. doi: 10.1038/nature16471. [DOI] [PubMed] [Google Scholar]
- 104.Frederiksen JK, Piccirilli JA. Identification of catalytic metal ion ligands in ribozymes. Methods. 2009;49:148–166. doi: 10.1016/j.ymeth.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu Y, Sen D. A contact photo-cross-linking investigation of the active site of the 8-17 deoxyribozyme. J Mol Biol. 2008;381:845–859. doi: 10.1016/j.jmb.2008.06.036. [DOI] [PubMed] [Google Scholar]
- 106.Liu Y, Sen D. Local rather than global folding enables the lead-dependent activity of the 8-17 deoxyribozyme: evidence from contact photo-crosslinking. J Mol Biol. 2010;395:234–241. doi: 10.1016/j.jmb.2009.11.028. [DOI] [PubMed] [Google Scholar]
- 107.Sekhon GS, Sen D. A stereochemical glimpse of the active site of the 8-17 deoxyribozyme from iodine-mediated cross-links formed with the substrate’s scissile site. Biochemistry. 2010;49:9072–9077. doi: 10.1021/bi1013547. [DOI] [PubMed] [Google Scholar]
- 108.Cepeda-Plaza M, Null EL, Lu Y. Metal ion as both a cofactor and a probe of metal-binding sites in a uranyl-specific DNAzyme: a uranyl photocleavage study. Nucl Acids Res. 2013;41:9361–9370. doi: 10.1093/nar/gkt694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yano J, Yachandra VK. X-ray absorption spectroscopy. Photosyn Res. 2009;102:241–254. doi: 10.1007/s11120-009-9473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Smirnov IV, Kotch FW, Pickering IJ, Davis JT, Shafer RH. Pb EXAFS studies on DNA quadruplexes: identification of metal ion binding site. Biochemistry. 2002;41:12133–12139. doi: 10.1021/bi020310p. [DOI] [PubMed] [Google Scholar]
- 111.Travascio P, Li Y, Sen D. DNA-enhanced peroxidase activity of a DNA-aptamer-hemin complex. Chem Biol. 1998;5:505–517. doi: 10.1016/s1074-5521(98)90006-0. [DOI] [PubMed] [Google Scholar]
- 112.Liu J, Lu Y. Rational design of “turn-on” allosteric DNAzyme catalytic beacons for aqueous mercury ions with ultrahigh sensitivity and selectivity. Angew Chem. 2007;46:7587–7590. doi: 10.1002/anie.200702006. [DOI] [PubMed] [Google Scholar]
- 113.Ravel B, Slimmer SC, Meng X, Wong GCL, Lu Y. EXAFS studies of catalytic DNA sensors for mercury contamination of water. Radiat Phys Chem. 2009;78:S75–S79. [Google Scholar]
- 114.Kim HK, Liu J, Li J, Nagraj N, Li M, Pavot CM, Lu Y. Metal-dependent global folding and activity of the 8-17 DNAzyme studied by fluorescence resonance energy transfer. J Am Chem Soc. 2007;129:6896–6902. doi: 10.1021/ja0712625. [DOI] [PubMed] [Google Scholar]
- 115.Liu J, Lu Y. FRET study of a trifluorophore-labeled DNAzyme. J Am Chem Soc. 2002;124:15208–15216. doi: 10.1021/ja027647z. [DOI] [PubMed] [Google Scholar]
- 116.Kim HK, Rasnik I, Liu J, Ha T, Lu Y. Dissecting metal ion-dependent folding and catalysis of a single DNAzyme. Nat Chem Biol. 2007;3:763–768. doi: 10.1038/nchembio.2007.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kypr J, Kejnovska I, Renciuk D, Vorlickova M. Circular dichroism and conformational polymorphism of DNA. Nucl Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ota N, Warashina M, Hirano K, Hatanaka K, Taira K. Effects of helical structures formed by the binding arms of DNAzymes and their substrates on catalytic activity. Nucl Acids Res. 1998;26:3385–3391. doi: 10.1093/nar/26.14.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cieslak M, Szymanski J, Adamiak RW, Cierniewski CS. Structural rearrangements of the 10-23 DNAzyme to beta 3 integrin subunit mRNA induced by cations and their relations to the catalytic activity. J Biol Chem. 2003;278:47987–47996. doi: 10.1074/jbc.M300504200. [DOI] [PubMed] [Google Scholar]
- 120.Feig AL, Scott WG, Uhlenbeck OC. Inhibition of the hammerhead ribozyme cleavage reaction by site-specific binding of Tb. Science. 1998;279:81–84. doi: 10.1126/science.279.5347.81. [DOI] [PubMed] [Google Scholar]
- 121.Kim HK, Li J, Nagraj N, Lu Y. Probing metal binding in the 8-17 DNAzyme by TbIII luminescence spectroscopy. Chem Eur J. 2008;14:8696–8703. doi: 10.1002/chem.200701789. [DOI] [PubMed] [Google Scholar]
- 122.Santoro SW, Joyce GF, Sakthivel K, Gramatikova S, Barbas CF., 3rd RNA cleavage by a DNA enzyme with extended chemical functionality. J Am Chem Soc. 2000;122:2433–2439. doi: 10.1021/ja993688s. [DOI] [PubMed] [Google Scholar]
- 123.Edwards TE, Sigurdsson ST. EPR spectroscopic analysis of U7 hammerhead ribozyme dynamics during metal ion induced folding. Biochemistry. 2005;44:12870–12878. doi: 10.1021/bi050549g. [DOI] [PubMed] [Google Scholar]
- 124.Kisseleva N, Khvorova A, Westhof E, Schiemann O. Binding of manganese(II) to a tertiary stabilized hammerhead ribozyme as studied by electron paramagnetic resonance spectroscopy. RNA. 2005;11:1–6. doi: 10.1261/rna.7127105. [DOI] [PMC free article] [PubMed] [Google Scholar]