Abstract
Malectin-like domain-containing receptor-like kinases (MRLK) constitute a large and divergent family of proteins in plants; however, little is known about the role of MRLKs in fruit growth and development. In this study, we characterized MRLK family genes in diploid strawberry, Fragaria vesca. Based on an analysis of malectin-like domain and a search in the strawberry genome and NCBI database, we identified 62 FvMRLKs in the strawberry genome, and classified these genes into six subfamilies with distinct malectin domains in the extracellular regions of the encoded proteins. Gene expression analysis indicated that more than 80% of the FvMRLKs were expressed in various tissues, with higher levels in roots than in other organs. Thirty-three FvMRLKs were found to be expressed in fruits during the early stages of development, and over 60% of these exhibited dramatic decreases in expression during fruit growth and development. Moreover, the expression of some FvMRLKs was sensitive to both environmental and internal cues that play critical roles in regulating strawberry fruit development and ripening. Collectively, this study provides valuable insight into the FvMRLKs gene family and its role in regulating strawberry fruit development and ripening.
Introduction
Receptor-like kinases (RLKs) in plants comprise a large family of proteins with putative amino-terminal extracellular domains and carboxyl-terminal intracellular kinase domains [1, 2]. A phylogenetic analysis of the model plant Arabidopsis thaliana identified more than 610 proteins containing RLK domain sequences. RLKs can be further divided into more than 44 subfamilies based on their extracellular domain structures [3, 4]; plant RLKs were initially categorized into three major types, the S-domain type, the leucine-rich repeat type, and the epidermal growth factor (EGF)-like type [5, 6]. CrRLK1 was later identified in Catharanthus roseus and represents a unique subfamily of RLKs [5].
The Arabidopsis genome contains 17 CrRLK1-like RLK genes. The CrRLK1L subfamily can be further classified into six subclasses [7, 8].The functions of several members of the CrRLK1L subfamily have been determined in Arabidopsis [7]. Feronia (FER) was first identified for its role in fertilization [9]. Recent research demonstrated that FER directly interacts with guanine nucleotide exchange factors (ROPGEFs), which regulate RHO GTPase signaling and mediate the production of reactive oxygen species (ROS) at the entrance point of the female gametophyte, which induces pollen tube rupture and sperm release [10–12]. Besides its critical roles in fertilization, FER has been suggested to be a key regulator of the crosstalk among abscisic acid (ABA), brassinosteroids (BR), and ethylene signaling [13]. Moreover, FER was reported to be involved in a variety of important processes, such as root hair elongation[14,15], ethylene biosynthesis [16], starch accumulation[17], seed development [18], pathogen resistance[19, 20], and vegetative growth [21–23].Theseus 1 (THE1) was identified in a screen for suppressors that attenuated the short hypocotyl phenotype of dark-grown seedlings [24–26].THE1 functions as a cell wall integrity sensor that mediates the disruption of cellulose synthesis [24–26].Named after the male consort of Feronia, Anxur1 (ANX1) and Anxur2 (ANX2) are closely related to FER. Like FER, ANX1 and ANX2 are pollen-specific CrRLK1Ls that play important roles in the regulation of pollen tube rupture and sperm release [27]. HERCULES 1 (HERK1) is also a member of the CrRLK1L subfamily and functions redundantly with THE1 to regulate plant growth and development [21, 22].
Acting as RLKs, CrRLK1Ls signaling is based on a reaction with specific ligands in the extracellular region of their conserved domains; therefore, analysis of the conserved domains of CrRLK1Ls is particularly important for a deeper understanding of the mechanisms underlying CrRLK1L signaling. Sequence analysis of FER, THE1, ANXS, HERK1, and several other closely related proteins indicated that an important feature of their extracellular sequences is the existence of a conserved domain, named the malectin or malectin-like domain. Malectin is a membrane-anchored protein of the endoplasmic reticulum. It recognizes and binds Glc2-N-glycan, thereby regulating the production and secretion of N-glycosylated proteins [28–30]. The existence of the malectin domain provides an important clue for exploring the signaling mechanisms of RLKs. CrRLK1Ls are commonly referred to as malectin domain-containing RLKs [7, 25, 31]. Given the importance of CrRLK1Ls, genome-wide identification and analysis of the CrRLK1Lgene subfamily has been conducted in several plant species, including Oryza sativa (rice) [32] and Gossypium (cotton) [33]. As mentioned above, the Arabidopsis genome contains 17 CrRLK1-like RLK genes. However, a search for malectin and malectin-like domains in Arabidopsis revealed more than 30 genes, indicating that malectin and malectin-like domain containing RLKs exist outside of the CrRLK1Lgene family. Therefore, it is more suitable to categorize the malectin and malectin-like domain containing RLKs into a unique subfamily (referred as MRLK subfamily) and a genome-wide analysis of this gene subfamily would provide insight into their functions in plants.
Flesh fruit production accounts for a substantial fraction of the world’s agricultural output. Since regulating fruit development and ripening is a key challenge in fleshy fruit production, understanding the molecular basis for fruit development and ripening is of great interest. While signaling mechanisms have been extensively studied for the regulation of plant growth and development and responses to abiotic and biotic stresses, limited information is available in fleshy fruits, especially non-climacteric fruits. Fleshy fruits are physiologically classified as climacteric or non-climacteric. Strawberry is becoming a model plant for basic research of non-climacteric fruits (i.e., fruits that ripen without increased ethylene production).To explore the molecular basis of strawberry plant growth and development, with a focus on fruit growth and development, we conducted a genome-wide analysis of the MRLK subfamily in diploid strawberry, Fragaria vesca. We found that the F. vesca genome contains more than 60 MRLKs, about 50% of which are expressed in the fruit. Among the MRLKs expressed in fruit, more than 60% were expressed at relatively high levels only in the early stages of fruit development, with transcript levels declining dramatically as fruit development progressed. Furthermore, many of these genes were sensitive to both environmental and internal cues controlling fruit development and ripening. These findings provide valuable insight into the roles of FvMRLKs in the regulation of flesh fruit development and ripening.
Materials and Methods
Identification of MRLK proteins in Fragaria vesca
Annotated strawberry MRLK proteins were identified in the following three public databases: the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/), the Strawberry Genome Browser (https://strawberry.plantandfood.co.nz/index.html), and PLAZA (http://bioinformatics.psb.ugent.be/plaza/). Specifically, to identify the MRLK gene family, we first identified the HMM profile of the malectin domain (pfam11721) and malectin-like domain (pfam12819) from the protein families database (http://pfam.janelia.org); Based on the HMM profile of the malectin and malectin-like domains, we obtained the MRLK protein candidates by searching the database of strawberry genome (https://strawberry.plantandfood.co.nz/index.html) and the MRLK protein candidates were further identified and characterized in the NCBI database for a final confirmation of the MRLK proteins. The lengths of amino acid sequences, molecular weights, and isoelectric points of the putative proteins were calculated using tools provided at the ExPasy website (http://web.expasy.org/protparam/). The signal peptides of the strawberry MRLK proteins were predicted based on the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/) [34]. Information of the Arabidopsis MRLK gene family was obtained from The Arabidopsis Information Resource (http://www.arabidopsis.org/) and used for the comparative study.
Phylogenetic analysis and classification of the MRLK protein family in Fragaria vesca
All identified MRLK genes (FvMRLKs) in Fragaria vesca were classified into different groups based on the alignment of FvMRLK proteins using ClustalX 2.1with default settings [35]. The phylogenetic trees were constructed using the neighbor-joining method implemented in MEGA 5.0[36]. Bootstrap values were calculated for 1000 iterations.
Chromosomal location of FvMRLKs
All FvMRLK genes were mapped to strawberry chromosomes based on information available at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/). The map was drafted using Mapchart software (http://www.wageningenur.nl/en.htm) [37]. Tandem duplicated FvMRLK genes in the strawberry genome were identified by checking their physical locations on individual chromosomes and were defined as adjacent paralogous genes with no more than one intervening gene. Physical chromosomal locations were graphically represented to scale on the seven chromosomes.
Analysis and distribution of conserved motifs and exon-intron structures
The exon-intron organization of the FvMRLK genes was determined using PLAZA (http://bioinformatics.psb.ugent.be/plaza/). The Fancy Gene online tool (http://bio.ieo.eu/fancygene/) was used to illustrate the exon-intron structure of the FvMRLKs [38]. The Expectation Maximization for Motif Elicitation online program (http://meme-suite.org/) [39] for DNA and protein sequence analysis was used to identify conserved motifs in the 62 FvMRLK-deduced proteins. The optimized parameters of Expectation Maximization for Motif Elicitation were set as follows: the number of repetitions, any; the maximum number of motifs, 50; and the optimum width of each motif, between 6 and 300 residues.
Plant materials and treatments
The strawberry (Fragaria vesca‘Ruegen’) plants were cultivated at 22±1°C in a 13/11 h light/dark photoperiod in the Tissue Culture Center at Beijing University of Agriculture. To investigate FvMRLK expression in different strawberry tissues, the roots, stems, leaves, and fruits were independently sampled and analyzed using qRT-PCR. Root samples were taken from white, new roots, stem samples were taken from new creeping stems, leaf samples were the leaves 15 days old, fruit samples were taken from medium green fruits. To study the expression of FvMRLKs at different developmental stages, strawberry fruits were divided into five developmental stages, i.e., SG: small green stage; MG: medium green stage; LG: large green stage; TN: turn stage; and FR: full red stage, and were independently sampled. At each developmental stage, ten representative fruits were sampled, snap-frozen in liquid nitrogen, and kept at -80°C for subsequent analysis. The expression of the FvMRLK genes in the five fruit developmental stages was analyzed using qRT-PCR. MG fruits were selected to study some of the high-expression genes of fruit in response to various environmental cues and internal signals, including heat treatment (40°C,4 h), cold treatment (4°C,48 h), dehydration treatment (25°C,8 h), ABA treatment (25°C,100μM, 6 h), IAA treatment (25°C,500μM, 6 h),and sucrose treatment (25°C,100mM, 6 h).The fruits were cut in half with one-half used for the treatment and the other half as a control. The controls were maintained at 25°C and100% relative humidity. Three biological replicates were used in all experiments. The expression data in the heat-map were calculated as log2 fold change in treated vs. untreated samples.
RNA preparation and quantitative RT-PCR analysis
Total RNA was isolated using a Plant RNA Kit (Omega) according to the manufacturer’s instructions. Quantitative RT-PCR was conducted on a LightCycler 96 Real Time PCR System (Roche Diagnostics GmbH, Mannheim City, Germany) using SYBR-Green (Takara, Dalian, China).The oligonucleotide primers were designed based on the predicted coding region using Beacon designer software and are listed in S1 Table. Primers were checked using the BLAST tool of NCBI and the dissociation curve was analyzed after the PCR reaction for primer specificity. In addition, the specificity of PCR products was verified by cloning the relative amplicons in the pMD19-T vector (Takara), sequencing, and aligning them onto the reference genome. Each reaction was carried out in a volume of 20μL, which contained 10μL SYBR-Green master mix, 8.6μL ddH2O, 1μL diluted template (1μL of the generated first-strand cDNA was diluted in 9 μL ddH2O), and 0.2μL of each gene-specific primer. The following program conditions were used: 95°C for 600 s (pre-denaturation) followed by 40 cycles at 95°C for 20 s (denaturation), 53°C for 20 s (primer annealing), and 72°C for 20 s (extension and data acquisition). Three technical replicates were performed for each biological replicate. The actin-7 gene (LOC101313051), which maintained almost constant expression levels under all experimental conditions, was used as an internal control.
Results
Identification and annotation of MRLK genes in the strawberry genome
To identify FvMRLK subfamily genes, we first obtained the HMM profile of the malectin domain (pfam11721) and malectin-like domain (pfam12819) from the pfam protein families database. Using the malectin or malectin-like domain information, we searched three databases, the National Centre for Biotechnology Information, the Strawberry Genome Browser, and PLAZA. We identified a total of 100 candidate FvMRLK genes in the Fragaria vesca genome. The 100 FvMRLK candidates were further examined by searching the malectin or malectin-like domain in the NCBI database and 62 members were confirmed to be FvMRLKs, which were named FvMRLK1 to FvMRLK62 based on their chromosome location. The characteristics of the FvMRLK proteins are listed in Table 1 and include the deduced protein length, molecular weight, isoelectric point (PI), aliphatic index, and the grand average of hydropathicity. The deduced length of the FvMRLK proteins ranged from 132 (FvMRLK16) to 1,955 (FvMRLK 51) amino acid residues, whereas the PI ranged from 4.54 (FvMRLK33) to 8.88 (FvMRLK55). Thirty-one FvMRLK members contained a signal peptide (S2 Table).
Table 1. Strawberry MRLK genes identified in the Fragaria vesca.
| Proposed name | Gene | Locus name | Chr | ORF (aa) | MW(kDa) | pI | Ai | GRAVY | strand | Group |
|---|---|---|---|---|---|---|---|---|---|---|
| FvMRLK 1 | gene26561 | XP_004288865 | 1 | 1016 | 114893.9 | 6.72 | 75.53 | -0.624 | + | VI |
| FvMRLK 2 | gene11357 | XP_004287503 | 1 | 890 | 98998.4 | 5.62 | 89.15 | -0.188 | - | III |
| FvMRLK 3 | gene28050 | XP_011462047 | 1 | 639 | 70994.6 | 5.98 | 93.79 | -0.179 | - | IV |
| FvMRLK 4 | gene24016 | XP_011463697 | 1 | 858 | 95229.8 | 5.83 | 86.93 | -0.164 | + | II |
| FvMRLK 5 | gene24017 | XP_004289187 | 1 | 814 | 90517.6 | 6.19 | 88.5 | -0.153 | + | II |
| FvMRLK 6 | gene16334 | XP_011465668 | 1 | 1187 | 128931.7 | 7.77 | 79.94 | -0.2 | + | IV |
| FvMRLK 7 | gene05569 | XP_004288317 | 1 | 783 | 87567.3 | 4.76 | 99.03 | -0.122 | + | III |
| FvMRLK 8 | gene20704 | XP_011468416 | 1 | 1086 | 123288.5 | 6.43 | 80.22 | -0.723 | - | VI |
| FvMRLK 9 | gene21850 | XP_004289649 | 2 | 1461 | 161379 | 7.16 | 77.67 | -0.293 | + | II |
| FvMRLK10 | gene17333 | XP_011458098 | 2 | 844 | 92261.1 | 5.3 | 96.34 | 0.012 | + | II |
| FvMRLK11 | gene17626 | XP_011458187 | 2 | 868 | 96529.3 | 6.1 | 100.48 | -0.079 | - | III |
| FvMRLK12 | gene11624 | XP_011459850 | 2 | 848 | 93736.8 | 5.95 | 91.84 | -0.241 | + | VI |
| FvMRLK13 | gene19789 | XP_004293373 | 3 | 971 | 108344.2 | 8.4 | 89.17 | -0.366 | + | VI |
| FvMRLK14 | gene34059 | XP_011461991 | 3 | 135 | 15483.9 | 9.65 | 96.74 | -0.248 | + | IV |
| FvMRLK15 | gene24842 | XP_011461995 | 3 | 1789 | 197989.3 | 759 | 95.41 | -0.015 | + | IV |
| FvMRLK16 | gene32988 | XP_011460577 | 3 | 132 | 14904.1 | 7.82 | 97.42 | -0.071 | + | V |
| FvMRLK17 | gene29734 | XP_011460577 | 3 | 681 | 74929.4 | 6.57 | 92.35 | -0.072 | + | V |
| FvMRLK18 | gene28878 | XP_011460644 | 3 | 916 | 102254.3 | 6.04 | 79.6 | -0.343 | - | II |
| FvMRLK19 | gene30033 | XP_011462047 | 3 | 1007 | 110147.1 | 6.6 | 88.68 | -0.169 | + | IV |
| FvMRLK20 | gene27319 | XP_011460815 | 3 | 1014 | 111185.8 | 5.62 | 94.62 | -0.101 | - | IV |
| FvMRLK21 | gene27321 | XP_011460818 | 3 | 1011 | 110937.6 | 5.41 | 92.2 | -0.091 | + | IV |
| FvMRLK22 | gene27326 | XP_011460817 | 3 | 966 | 106406.5 | 6.2 | 91.15 | -0.162 | + | IV |
| FvMRLK23 | gene27328 | XP_004295664 | 3 | 1007 | 110243.5 | 5.65 | 94.14 | -0.062 | + | IV |
| FvMRLK24 | gene27329 | XP_011460821 | 3 | 931 | 102026.6 | 5.24 | 90.09 | -0.144 | + | IV |
| FvMRLK25 | gene27330 | XP_011460823 | 3 | 759 | 83739.9 | 5.97 | 87.76 | -0.236 | + | IV |
| FvMRLK26 | gene25018 | XP_011461160 | 3 | 1145 | 125496.7 | 6.12 | 91.15 | -0.092 | - | II |
| FvMRLK27 | gene25031 | XP_011463354 | 3 | 239 | 27018.6 | 4.66 | 84.9 | -0.091 | + | V |
| FvMRLK28 | gene14004 | XP_004294625 | 3 | 1169 | 125860.1 | 5.37 | 71.2 | -0.357 | - | V |
| FvMRLK29 | gene14005 | XP_011462143 | 3 | 577 | 62611.1 | 4.75 | 74.33 | -0.353 | - | V |
| FvMRLK30 | gene14010 | XP_004294627 | 3 | 832 | 91480.3 | 5.54 | 83.73 | -0.116 | + | II |
| FvMRLK31 | gene28048 | XP_011462168 | 3 | 493 | 54445.5 | 4.84 | 93.33 | -0.145 | - | IV |
| FvMRLK32 | gene28580 | XP_004296071 | 3 | 484 | 53260.3 | 5.25 | 85.76 | -0.217 | + | V |
| FvMRLK33 | gene05121 | XP_004296244 | 3 | 468 | 52082.6 | 4.54 | 73.12 | -0.453 | + | I |
| FvMRLK34 | gene05124 | XP_004296245 | 3 | 860 | 94587.5 | 6.28 | 81.28 | -0.239 | + | I |
| FvMRLK35 | gene22909 | XP_011463352 | 4 | 756 | 85062.9 | 5.26 | 86.89 | -0.321 | + | III |
| FvMRLK36 | gene28915 | XP_011460364 | 4 | 731 | 81731.3 | 6.06 | 100.01 | 0.001 | - | IV |
| FvMRLK37 | gene05892 | XP_004298162 | 4 | 576 | 62901.4 | 5.2 | 91.37 | -0.097 | - | V |
| FvMRLK38 | gene03872 | XP_004298306 | 4 | 602 | 66979.6 | 5.29 | 86.45 | -0.189 | - | V |
| FvMRLK39 | gene04553 | XP_004298352 | 4 | 791 | 88436.4 | 7.32 | 83.64 | -0.296 | + | I |
| FvMRLK40 | gene23070 | XP_004298555 | 4 | 375 | 42757 | 5.02 | 84.19 | -0.36 | - | III |
| FvMRLK41 | gene22902 | XP_004297625 | 4 | 835 | 92486 | 5.95 | 86.66 | -0.147 | - | III |
| FvMRLK42 | gene22907 | XP_011463354 | 4 | 1609 | 181729 | 5.63 | 86.52 | -0.329 | + | III |
| FvMRLK43 | gene22908 | XP_011463354 | 4 | 791 | 89658.1 | 5.97 | 89.67 | -0.273 | + | III |
| FvMRLK44 | gene26060 | XP_004301135 | 5 | 1434 | 161363.1 | 6.56 | 82.37 | -0.346 | + | II |
| FvMRLK45 | gene09312 | XP_011465700 | 5 | 489 | 53813.9 | 5.87 | 86.97 | -0.064 | + | V |
| FvMRLK46 | gene16583 | XP_004304753 | 6 | 878 | 96629.5 | 5.74 | 76.99 | -0.369 | + | I |
| FvMRLK47 | gene13568 | XP_004302537 | 6 | 883 | 96718.3 | 6.03 | 75.54 | -0.236 | - | I |
| FvMRLK48 | gene18098 | XP_011466646 | 6 | 969 | 107689.9 | 5.7 | 88.14 | -0.31 | + | III |
| FvMRLK49 | gene25622 | XP_004305253 | 6 | 762 | 85854.7 | 5.17 | 83.39 | -0.358 | + | III |
| FvMRLK50 | gene04244 | XP_004306057 | 6 | 885 | 98830.2 | 6.5 | 78.73 | -0.287 | + | I |
| FvMRLK51 | gene04245 | XP_004306058 | 6 | 1955 | 216885.6 | 7.02 | 79.46 | -0.254 | + | I |
| FvMRLK52 | gene04246 | XP_004306059 | 6 | 841 | 93694 | 6.31 | 82.87 | -0.239 | + | I |
| FvMRLK53 | gene04247 | XP_011467877 | 6 | 833 | 92932.8 | 6.65 | 81.46 | -0.26 | + | I |
| FvMRLK54 | gene04250 | XP_004306062 | 6 | 827 | 92049.3 | 5.76 | 85.83 | -0.198 | - | I |
| FvMRLK55 | gene19246 | XP_011468989 | 7 | 458 | 50587.8 | 8.88 | 92.93 | 0.017 | + | II |
| FvMRLK56 | gene23363 | XP_004307450 | 7 | 814 | 90488.8 | 7.22 | 92.16 | -0.044 | - | II |
| FvMRLK57 | gene23366 | XP_011470602 | 7 | 568 | 63963.8 | 7.52 | 80.44 | -0.278 | - | V |
| FvMRLK58 | gene26704 | XP_004308888 | 7 | 848 | 94034.1 | 6.38 | 91.8 | -0.081 | - | II |
| FvMRLK59 | gene12571 | XP_004307857 | 7 | 592 | 64822 | 4.57 | 87.75 | -0.091 | - | V |
| FvMRLK60 | gene13200 | XP_011470689 | 7 | 829 | 91709.2 | 6.63 | 88.85 | -0.153 | - | II |
| FvMRLK61 | gene06940 | XP_011457436 | Un | 931 | 104255.7 | 5.87 | 89.52 | -0.229 | - | III |
| FvMRLK62 | gene23790 | XP_004309619 | Un | 1664 | 185177.7 | 6.25 | 95.77 | -0.12 | - | IV |
Abbreviations: Ai, aliphatic index; Chr, chromosome numbers; GRAVY, grand average of hydropathicity; MW, molecular weight; ORF, open reading frame; pI, isoelectric point; UN, unknown chromosome.
Chromosomal distribution and exon-intron organization of the FvMRLK genes
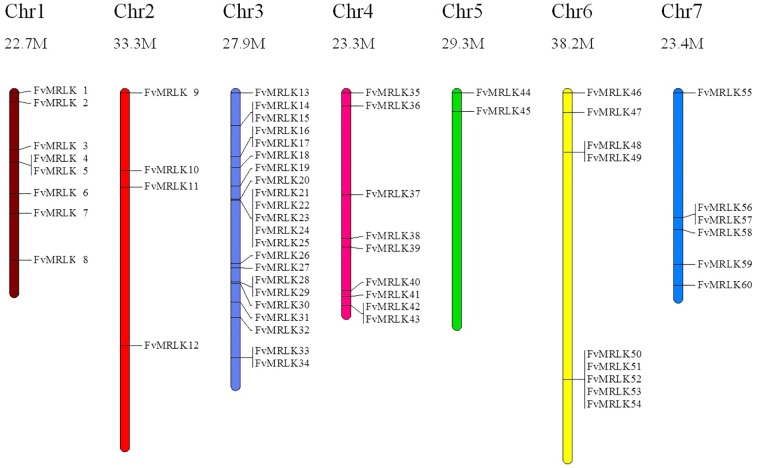
The FvMRLK genes were mapped across the chromosomes of the strawberry genome. As shown in Fig 1, 60 of the 62 FvMRLK genes were distributed on chromosomes 1 through 7. Chromosome 3 (22 genes) contained the largest number of FvMRLK genes and chromosome 5 (2 genes) contained the smallest number of FvMRLK genes. We were unable to map two FvMRLKs (FvMRLK61 and FvMRLK62) to any of the chromosomes. Based on the definition of a gene cluster by Holub [40], the FvMRLKs were placed into ten clusters. Among the ten clusters, five were found on chromosome 3 and two on chromosomes 6, whereas only one cluster was located on chromosomes 1, 4, and 7. Tandem and segmental duplications have been suggested as two main causes for gene family expansion in plants [41]. Five clusters of FvMRLKs with tandem duplications were found to be located on chromosome 3. To characterize the evolution of the FvMRLK family genes in strawberry, we analyzed the exon-intron structures of all FvMRLKs. We found that the intron numbers varied greatly (from 0 to 47) (Fig 2) amongst members of this gene family. For example, while FvMRLK5 had no introns, FvMRLK62 had 47. Interestingly, it appeared that the FvMRLKs within the same group generally had a similar exon-intron structure.
Fig 1. Chromosomal distribution of the FvMRLK genes.
Chromosome numbers are provided at the top of each chromosome together with the approximate size. The FvMRLKs were named FvMRLK01 to FvMRLK62 based on their order on the chromosomes.
Fig 2. Structures and exon–intron composition of the FvMRLK genes.

Names of genes are indicated on the left. Exons, represented by gray boxes, are drawn to scale. Dashed lines connecting two exons represent an intron. Malectin and malectin-like domains in the FvMRLK proteins are marked in blue and red, respectively.
Phylogenetic analysis, classification, and motifs of the strawberry MRLK gene family
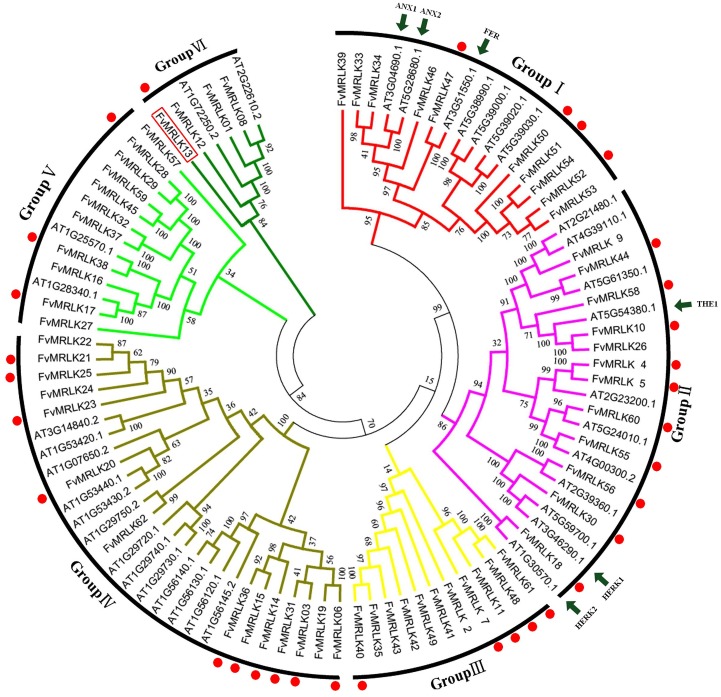
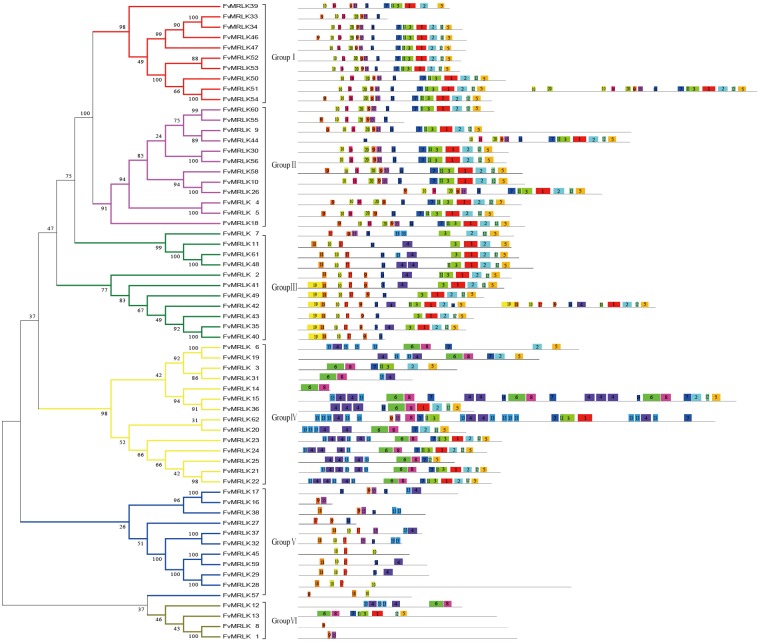
To determine the phylogenetic relationship between strawberry and Arabidopsis MRLKs, we constructed an unrooted phylogenetic tree based on alignments of the full-length protein sequences. The phylogenetic analyses showed that FvMRLKs could be classified into six large groups. As shown in Fig 3, groupI, II, III, IV, V, and VI contained 10, 12, 7, 14, 7, and 4 members, respectively. A major difference in the protein structure among the different groups was the number of malectin and malectin-like domains. For example, while most of the members of group I contained two malectin domains and one malectin-like domain, most of the members of group II contained one malectin domain and one malectin-like domain. Furthermore, while most of the members of group III and V contained one malectin-like domain, most of the members of group IV and VI contained only one malectin domain. Using the Multiple Expectation Maximization for Motif Elicitation online tool (http://meme-suite.org/), we identified 20 conserved motifs in the FvMRLK subfamily (Fig 4 and Table 2). Motif 6, 8, 9, 10, 14, 15, 16, 17, 18, 19, and 20 represented pivotal motifs of the malectin domains in strawberry and these conserved motifs were distributed widely amongst all FvMRLK groups. As shown in Fig 4, the FvMRLKs within the same group were found to share a similar motif composition. For example, whereas motif 19 was only found in group III, motif 10,15, and 20 were found mainly in groupI and II.
Fig 3. Phylogenetic tree of the MRLKs based on an alignment of strawberry and Arabidopsis protein sequences.
The un-rooted phylogenetic tree was constructed with ClustalX 2.1 using the neighbour-joining method implemented in MEGA 5.0. Reliability of the predicted tree was tested using bootstrapping with 1000 replicates. Branch lines with different colors represent different MRLK groups. The functionally-elucidated homologs from Arabidopsis are labeled by arrows and the FvMRLKs expressed in fruits are labeled by red-spots.
Fig 4. Phylogenetic tree of deduced FvMRLK proteins associated with the motif compositions in the amino-acid sequences.
The phylogenetic trees were constructed using the neighbour-joining method implemented in MEGA 5.0. Reliability of the predicted tree was tested using bootstrapping with 1000 replicates. Numbers at the nodes indicate how often the group to the right appeared among bootstrap replicates. The motif composition related to each FvMRLK protein is displayed on the right-hand side. The motifs, numbered 1–20, are displayed in different colored boxes. The sequence information for each motif is provided in Table 2.
Table 2. Analysis and distribution of conserved motifs in FvMRLKs.
| Motif | Sites | E-value | Width | Best possible match |
|---|---|---|---|---|
| 1 | 38 | 5.1E-1440 | 55 | L[ST]W[KE]QRL[EQ]I[CA][IV][GD]AA[RK]GL[HE]YLHTG[AC][KA]PTI[IV]HRD[VI]K[ST]TNILLDE[NK][LFW]V[AP]K[VI][SA]DFGLSK |
| 2 | 42 | 4.8E-1120 | 41 | TAV[KA]G[ST]FGYL[DA]PEY[YFA][RM]R[QG]QLT[ED]KSDVYSFG[VI]VL[LF]E[VI][LVI][CS][GA]RP |
| 3 | 39 | 7.9E-947 | 41 | HEFQ[TN]EIE[ML]LS[KR][LI][RH]HR[HN]LVSLIGYCD[ED][GK][GN][EQ][ML]ILVY[ED]YM[AE]NG |
| 4 | 50 | 2.1E-677 | 41 | [NS][LI][EQ]T[LI]DLS[NS]NN[LF][TS]GS[LI]PE[SF]L[GA][NK][LI][TP]TLKxL[ND][LI]SFNS[LF][ST]Gx[ILV]P |
| 5 | 44 | 1.3E-560 | 34 | KF[IGV]E[VIT]A[LE][KL]C[LTV]x[ED]S[GAP][VA][DE]RP[ST]M[SG]DV[LV][WS][NME]LE[GY]ALQ[LI][QE] |
| 6 | 17 | 1.9E-560 | 70 | [ILS]SL[TR]YY[GA][FL][CG]L[AEG]NGNYTV[KN]LHFAEI[MV][FIL][RLT][DN][ND][RT][TS]Y[KSY][SG][LV] GRR[IV]FD[VI]YIQ[GD]K[LR]VLKDF[ND]I[AR]KEAGGx[GD]K[EA][VI][IV]KEF |
| 7 | 37 | 8.0E-572 | 29 | F[ST][LF]A[EQ][IL][KQ]AATNNFD[EP]A[NL][KV][IL]G[EV]GGFG[PNK]VYK |
| 8 | 18 | 1.0E-345 | 41 | V[TSN][ENV][NS]TLEI[RH][FL][FY]WAGKGT[TC][CGN][IV]P[AR]RGTYG[PS]LISAIS[VA]T[PS]DF[KE]P |
| 9 | 50 | 2.60E-211 | 15 | [PS]N[FG]xYL[VI]RLHF[CA][ED][IF][EV] |
| 10 | 44 | 1.30E-188 | 16 | K[SD][GSA][YFV][AP]F[IV][NS][AG][IL]E[VL][VR][SP][LM]P |
| 11 | 33 | 2.40E-165 | 15 | TK[VI]A[VI]KR[LG][NS]PxS[EK]QG |
| 12 | 38 | 4.60E-130 | 15 | [CL][QH][EK]KGx[LI][ED][QE][IL][VI]DPRL |
| 13 | 50 | 3.80E-186 | 21 | TxLQx[LI]DLTGNYL[ST]G[PT]IPSE[IL] |
| 14 | 47 | 5.70E-138 | 15 | L[DP][AP][IL]LN[GA]LEI[LYF]K[IL][SN][ND] |
| 15 | 29 | 1.10E-103 | 21 | L[GN]Q[RL]VF[DN][VI][YF][IV]N[NG]QT[AV]ESD[AL]D[VLI] |
| 16 | 23 | 3.80E-102 | 15 | AL[EQ][TM][MV]YR[LVI]N[VM]GG[PS]L[IV] |
| 17 | 19 | 1.70E-97 | 15 | [IY]RYPDD[PV][YFH]DR[IF]W[DY][PS][FY] |
| 18 | 18 | 1.40E-96 | 20 | [KS]GT[KR]YL[IV]RA[ST]F[YV]YGNYDG[KQ][NGS] |
| 19 | 7 | 7.30E-144 | 57 | SYTEQTTGINYISD[AT]GF[IV]DTGESKLVL[PR][EY][YV][RK]D[NT]YQQPYW[SG][LV]RSFPEG[TI]RNCY[KR]INVT |
| 20 | 23 | 9.50E-120 | 23 | [YE][ISV]APK[EW]VYKT[AG]RSMGDN[KN]TINK[NS] |
FvMRLK expression profiles in strawberry
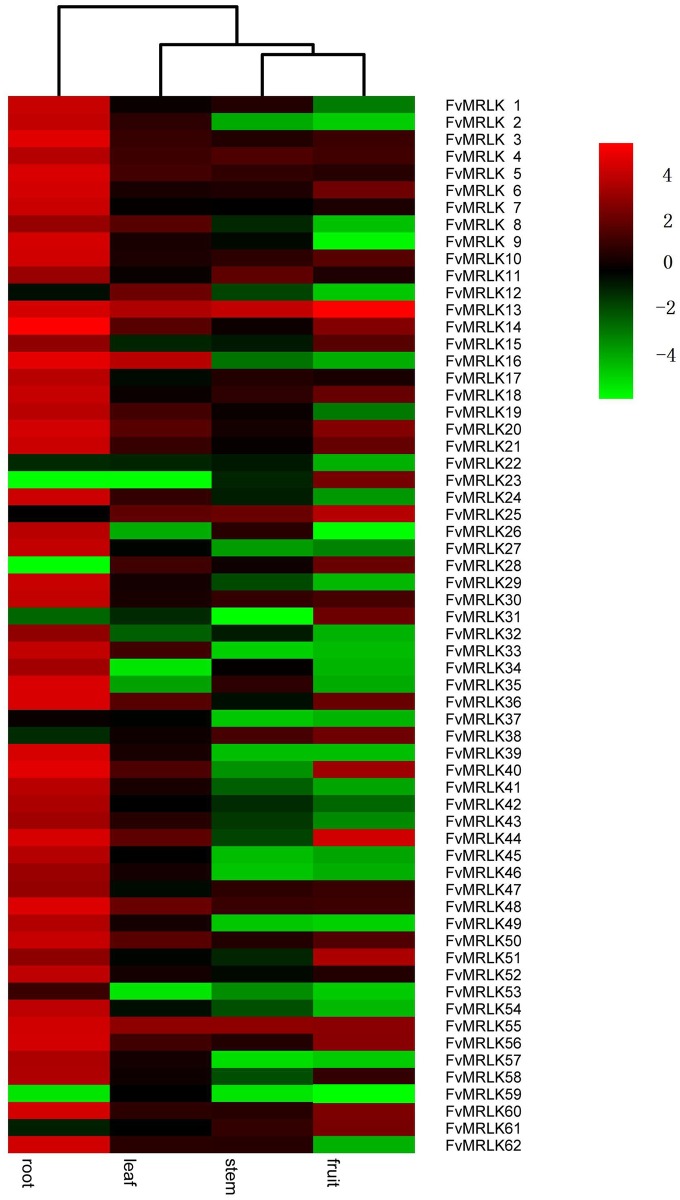
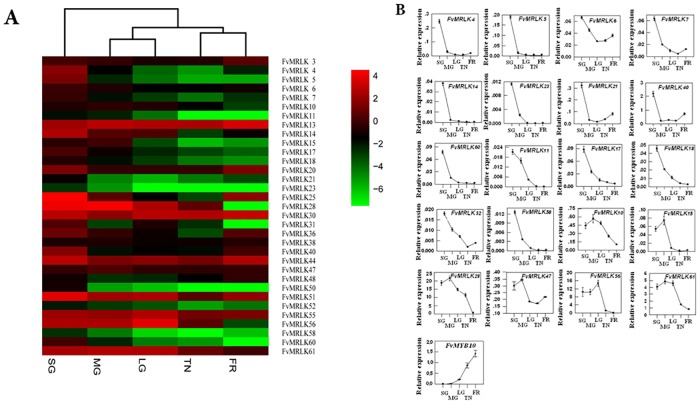
The expression profiles of the FvMRLKs varied in different organs or tissues of strawberry. As shown in Fig 5, high levels of FvMRLK gene expression were found in roots. The transcript levels of more than 80% of the genes were much higher in roots than in other organs, suggesting that FvMRLKs is a regulator of root growth and development. Although the expression levels of most of the genes were lower in fruits than in roots, 33 members genes were found to be expressed in fruits (Fig 6a). More than 60% of these 33 genes were expressed at high levels only in the early stages of fruit development and exhibited dramatic decreases in expression as fruit development progressed (Fig 6b). While the expression of FvMRLK4, 5, 7, 14, 21, 23, 40 and 60 declined sharply in the earliest developmental stage (SG) [42], the expression of FvMRLK6, 11,17,18, 52 and 58 dropped dramatically at the MG stage and the expression of FvMRLK10, 15,28,27,47,56 and 61 dropped dramatically at the LG stage. This result suggests that many FvMRLKs play important roles in the regulation of strawberry fruit growth and development and that there may be synergistic effects between different FvMRLKs that participate in these processes.
Fig 5. Expression profiles of the strawberry FvMRLK genes in different tissues.
The heat map was generated based on the mean values of the qRT-PCR data from different samples. Red and green represent higher and lower expression, respectively.
Fig 6. Changes in the expression profiles of FvMRLK genes during strawberry fruit growth and development.
(a) Heat map showing the expression profiles of the FvMRLK genes. The heat map was generated based on the mean values of the qRT-PCR data of the different samples. Red and green colors represent higher or lower expression. (b) Expression of the FvMRLKs decreased during strawberry fruit growth and development. The data are the mean of 3 replicates ±SE.
Expression of FvMRLKs in response to environmental and internal cues
Strawberry fruit development and ripening are regulated by environmental cues, such as temperature and drought stress, and internal signals, such as auxin (IAA), abscisic acid (ABA), and sucrose. To establish whether FvMRLKs are indeed involved in strawberry fruit development and ripening, we examined the expression of selected FvMRLKs in response to IAA, ABA, and sucrose treatment as well as to temperature and dehydration stresses. As shown in Fig 7a, the expression of nearly all of the FvMRLK genes examined were sensitive to low temperature. The gene expression of some FvMRLKs increased more than 25-fold in response to low temperature treatment. Some FvMRLKs were also sensitive to the dehydration treatment, but the dehydration-induced changes in gene expression were generally smaller than those following low temperature treatment. As a negative regulator, IAA plays critical roles in the regulation of strawberry fruit development and ripening, whereas both ABA and sucrose have been demonstrated to be positive regulators of strawberry fruit ripening [43]. Consistent with this, IAA treatment was found to result in a dramatic increase in gene expression for most of the FvMRLK genes examined. By contrast, ABA and sucrose treatments resulted in a decrease in expression of some of the FvMRLK genes examined (Fig 7b). Collectively, these results suggest that FvMRLKs play important roles, and moreover, most of them function as negative regulators in the regulation of strawberry fruit development and ripening.
Fig 7. Expression of the FvMRLKs in response to environmental cues and internal signals that play critical roles in the regulation of strawberry fruit development and ripening.
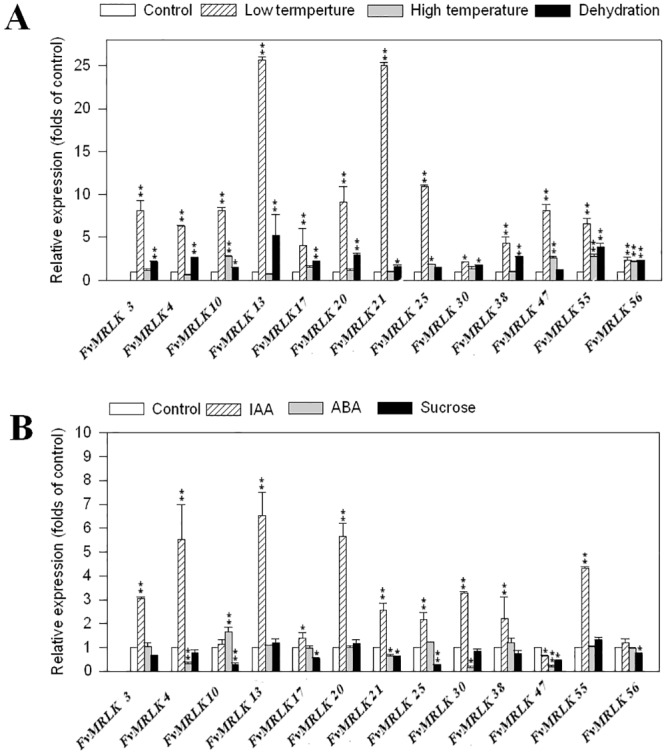
(a) Effect of environmental cues on the expression of FvMRLKs; (b) effect of internal signals on the expression of FvMRLKs. Gene expression was determined by qRT-PCR. The data are the mean of 3 replicates ±SE.
Discussion
The malectin and malectin-like domain containing RLKs family in relation to the CrRLK1L gene family
In Arabidopsis, CrRLK1Ls are a unique subfamily of RLKs comprised of 17 members. Given that CrRLK1Ls were found to contain malectin or malectin-like domains in their extracellular sequence [31], the CrRLK1Ls family has been commonly thought to be the same in terminology with the family of proteins containing malectin and malectin-like domain. In the present study, to compare FvMRLKs with the MRLKs in Arabidopsis, we searched the Arabidopsis genmone for the identification of the proteins containing the malectin and malectin-like domain. The results indicated that the Arabidopsis genome harbors 35 proteins containing malectin or malectin-like domains (S3 Table), which is greater than the 17 Arabidopsis CrRLK1L genes. Since the number of the malectin and malectin-like domain-containing proteins is much larger than that of the CrRLK1L family, it is clearly not reasonable to regard the CrRLK1L family as the family of proteins containing malectin and malectin-like domain. Because of this, in the present study, we categorized the malectin and malectin-like domain containing proteins into a unique family, and thus named as the MRLK gene family.
Phylogenetic characteristics of FvMRLKs
Strawberry genome size varies by species. With 34,809 genes resulted from gene prediction modeling, the genome of diploid strawberry, F. vesca, is about twice size of the Arabidosis genome (240 Mb for F. vesca versus 125 Mb for Arabidopsis)[44]. In the present study, we identified 62 FvMRLKs in the strawberry genome database and 35 MRLKs members in the Arabidopsis genmone. Based on a phylogenetic tree generated by an alignment of the MRLK protein sequences from strawberry and Arabidopsis, we classified the 62 FvMRLKs into six major groups (Fig 3). Members within the same group shared a similar gene structure, length, and amino acid motif composition, indicating their close evolutionary relationship. FvMRLK21, 22, 23, 24, and 25, which were similar in motif type and the distribution and contained tandem duplications, were clustered on chromosome 3. The change in motif composition from group I to group VI may reflect the evolutionary process of the FvMRLK gene family. Tandem duplications and segmental/whole-genome duplications represent two of the major mechanisms for the expansion of the RLK gene family in plants [25].The existence of ten clustered FvMRLK genes indicated that gene duplications had occurred. Notably, group III was strawberry specific; no members from Arabidopsis were classified into this group, suggesting that this group of genes may have special roles in the regulation of growth and development in strawberry. As shown in Fig 5, among the 62 FvMRLK genes, 33 were found to be expressed primarily in fruits, and these fruit-expressed genes included some homologs of FER, HERK1/2, and THE1 (i.e.,FvMRLK10, FvMRLK30, and FvMRLK47). Except for FvMRLK7 and FvMRLK40, nine genes from group III were not expressed in the fruits, possibly implying a special role of group III genes in the regulation of vegetative growth and development in strawberry.
Universal roles of FvMRLKs in the regulation of plant growth and development
Ripening of fleshy fruits is a complex process that involves dramatic changes in color, firmness, flavor, and aroma [43, 45]. Strawberry is a model plant for the research of the non-climacteric fruits. Limited information is available on the mechanisms controlling fruit growth and development, especially for the non-climacteric fruits. A decrease in fruit firmness is a critical event in fruit ripening. Fruit firmness is essentially controlled by cell wall metabolism and the decrease in fruit firmness primarily results from the breakdown of cellulose and pectin. The CrRLK1L protein kinases have been shown to function in cell wall metabolism [46–48]. Given the close relationship between cell wall metabolism and fruit ripening and the relationship between CrRLK1Ls and cell metabolism, CrRLK1Ls may be important regulators of fruit development and ripening. Consist with this inference, in the present study, we had found that many MRLK genes were specifically and highly expressed in strawberry fruits (Fig 5). Interestingly, among the MRLKs expressed in the fruits, the expression profiles of more than two-thirds of the MRLKs were found to be tightly correlated with the progress of strawberry fruit development and ripening (Fig 6). It has been well established that strawberry fruit development and ripening is regulated by both internal and environmental cues [49]. We examined the expression of MRLKs in response to these cues, such as IAA, ABA, sucrose, temperature and dehydration stresses. As shown in Fig 7A, most of the genes examined were sensitive to low temperature treatment, for example, in response to low temperate treatment, the expression of FvMRLK13 and FvMRLK21 increased to more than 25 folds compared with the control. The expression of FvMRLKs were also found to be strongly regulated by the internal cues. Furthermore, whereas treatments of ABA and sucrose, two positive signals controlling strawberry fruit ripening [42,49,50], caused an increase in the expression of some genes, the treatment of IAA [51], negative signal of fruit ripening, contrarily caused a dramatic increase in the gene expression (Fig 7B). Collectively, these data implies a potential role of MRLKs in the regulation of strawberry fruit development and ripening.
FER, HERK1/2, ANX1/2, and THE1 are several representative proteins that have been functionally elucidated in the CrRLK1L gene family [31]. FER together with its two closest homologs ANX1/2, controls polarized cell growth, thereby playing a critical role in the regulation of pollen tube and root hair elongation [52, 53]. THE1 functions to mediate the responses of plant cells to the perturbation of cellulose synthesis [24]. Acting redundantly with THE1, HERK1/2 also functions to regulate cell elongation. FER, HERK1/2, ANX1/2, and THE1 may have important roles in universal biological processes. For example, FER might be implicated in ethylene biosynthesis [16], starch accumulation[17], seed development[18],vegetative growth[21, 23], mechanical signal transduction[54], and pathogen resistance[19, 20]. Given the importance of FER, HERK1/2, ANX1/2, and THE1, identification of their homologs in the strawberry genome is important for understanding the roles of FvMRLKs in regulating strawberry plant growth and development. As shown in Fig 3, phylogenetic analysis showed that the homologs of AXN1/2, FvMRLK33, and FvMRLK34, and the homologs of FER and FvMRLK47, are in group I. Three homologs of THE1, FvMRLK10, FvMRLK26, and FvMRLK58, and one homolog of HERK1, are in group II, implying the importance of members of group I and group II in the regulation of strawberry growth and development.
Analysis of the expression profiles of the FvMRLK genes indicated that more than 80% of the FvMRLKs were expressed in roots with transcript levels much higher than in other tissues, suggesting that FvMRLKs play a major role in the regulation of root growth and development (Fig 5). Further analysis showed that among the 62 FvMRLK genes, only 33 were expressed in the early stages of fruit development. Interestingly, among these 33 genes, the transcript levels of more than 20 genes were found to be dramatically decreased at advanced stages of fruit development. Strawberry fruit ripening is sensitive to both environmental and internal cues. Low temperature inhibits ripening, whereas high temperature promotes it. IAA, ABA, and sucrose are key signals controlling strawberry fruit ripening; thus, we examined the responses of some FvMRLKs to these compounds. As expected, the expression of many FvMRLKs was found to be sensitive to the environmental cues and the internal signals. These observations suggest that FvMRLKs play important roles in the regulation of strawberry fruit development and ripening.
Specific expression of a gene in a tissue or cell may imply that the gene plays a specific role in that tissue or cell. To identify the FvMRLKs implicated in strawberry fruit development and ripening, we hoped to identify FvMRLKs that were only expressed in fruits but not in other tissues. However, nearly all of the FvMRLKs that were expressed in fruits were also found to be expressed in other organs or tissues, especially in roots, although only about half of the FvMRLKs were expressed in the fruits. Assuming that some FvMRLKs are critical regulators of fruit development and ripening, these FvMRLKs may play different roles in different tissues or cells. This was demonstrated in a recent study. For example, OST1 is a key protein controlling stomatal movement and the ortholog of OST1 also plays a critical role in the regulation of strawberry fruit ripening. Similarly, while FER was found to play a critical role in the regulation of pollen tube growth and sperm release, it also plays a critical role in the regulation of root hair elongation as well as many other biological processes as described above. Based on the expression profile of the FvMRLK genes, further investigation is needed to establish how different FvMRLKs coordinately regulate a common biological process, and by contrast, how individual FvMRLK genes regulate distinct biological processes. Collectively, the present study provides valuable information about FvMRLKs in strawberry, and thereby casts light on the mechanisms underlying the regulation of fruit development and ripening in this model non-climacteric fruit.
Conclusions
Sixty-two FvMRLKs were characterized in the strawberry genome, and classified these genes into six subfamilies with distinct malectin domains in the extracellular regions of the encoded proteins. More than 80% of the FvMRLKs were expressed in various tissues, with higher levels in roots than in other organs. Thirty-three FvMRLKs were expressed in fruits during the early stages of development, and over 60% of these exhibited dramatic decreases in expression during fruit growth and development. Moreover, the expression of some FvMRLKs was sensitive to both environmental and internal cues that play critical roles in regulating strawberry fruit development and ripening.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
This work was supported by the National Natural Science Foundation (31471851; 31672133; 31572104), the Scientific Research Program of Beijing Municipal Commission of Education (PXM2016-014207-000008; CEFF-PXM2016-014207-000038) and Fok Ying-Tong Education Foundation (151027).
Abbreviations
- SG
small green stage
- MG
medium green stage
- LG
large green stage
- TN
turn stage
- FR
full red stage
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Natural Science Foundation (31471851; 31672133; 31572104), the Scientific Research Program of Beijing Municipal Commission of Education (PXM2016-014207-000008; CEFF-PXM2016-014207-000038) and Fok Ying-Tong Education Foundation (151027). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Shiu S H, Bleecker AB. Plant receptor-like kinase gene family: diversity, function, and signaling. Sci STKE. 2001;(113): re22 10.1126/stke.2001.113.re22 [DOI] [PubMed] [Google Scholar]
- 2.Torii KU. Leucine-rich repeat receptor kinases in plants: structure, function, and signal transduction pathways. Int Rev Cytol. 2004; 234:1–46. 10.1016/S0074-7696(04)34001-5 [DOI] [PubMed] [Google Scholar]
- 3.Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci U S A. 2001; 98 (19):10763–10768. 10.1073/pnas.181141598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shiu SH, Bleecker AB. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003;132 (2):530–543. 10.1104/pp.103.021964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schulze-Muth P, Irmler S, Schroder G, Schroder J. Novel type of receptor-like protein kinase from a higher plant (Catharanthus roseus). cDNA, gene, intramolecular autophosphorylation, and identification of a threonine important for auto- and substrate phosphorylation. J Biol Chem. 1996; 271 (43):26684–26689. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, Zafian P, Choudhary M, Lawton M. The PR5K receptor protein kinase from Arabidopsis thaliana is structurally related to a family of plant defense proteins. Proc Natl Acad Sci U S A. 1996; 93 (6): 2598–2602. 10.1073/pnas.93.6.2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung AY, Wu HM. THESEUS 1, FERONIA and relatives: a family of cell wall-sensing receptor kinases? Curr Opin Plant Biol. 2011; 14 (6):632–641. 10.1016/j.pbi.2011.09.001 [DOI] [PubMed] [Google Scholar]
- 8.Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, et al. The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science. 2007; 317 (5838): 656–660. 10.1126/science.1143562 [DOI] [PubMed] [Google Scholar]
- 9.Huck N. The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development. 2003; 130 (10): 2149–2159. 10.1242/dev.00458 [DOI] [PubMed] [Google Scholar]
- 10.Duan Q, Kita D, Johnson EA, Aggarwal M, Gates L, Wu HM, et al. Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nat Commun. 2014; 5: 3129 10.1038/ncomms4129 [DOI] [PubMed] [Google Scholar]
- 11.Ngo QA, Vogler H, Lituiev DS, Nestorova A, Grossniklaus U. A calcium dialog mediated by the FERONIA signal transduction pathway controls plant sperm delivery. Dev Cell. 2014; 29 (4):491–500. 10.1016/j.devcel.2014.04.008 [DOI] [PubMed] [Google Scholar]
- 12.Kessler SA, Lindner H, Jones DS, Grossniklaus U. Functional analysis of related CrRLK1L receptor-like kinases in pollen tube reception. EMBO Rep. 2015; 16 (1):107–115. 10.15252/embr.201438801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu F, Qian L, Nibau C, Duan Q, Kita D, Levasseur K, et al. FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proc Natl Acad Sci U S A. 2012; 109 (36): 14693–14698. 10.1073/pnas.1212547109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duan Q, Kita D, Li C, Cheung AY, Wu HM. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci U S A. 2010; 107 (41):17821–17826. 10.1073/pnas.1005366107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang GQ, Li E, Ge FR, Li S, Wang Q, Zhang CQ, et al. Arabidopsis RopGEF4 and RopGEF10 are important for FERONIA-mediated developmental but not environmental regulation of root hair growth. New Phytol. 2013; 200 (4):1089–1101. 10.1111/nph.12432 [DOI] [PubMed] [Google Scholar]
- 16.Mao D, Yu F, Li J, Van de Poel B, Tan D, Li J, et al. FERONIA receptor kinase interacts with S-adenosylmethionine synthetase and suppresses S-adenosylmethionine production and ethylene biosynthesis in Arabidopsis. Plant Cell Environ. 2015; 38 (12): 2566–2574. 10.1111/pce.12570 [DOI] [PubMed] [Google Scholar]
- 17.Yang T, Wang L, Li C, Liu Y, Zhu S, Qi Y, et al. Receptor protein kinase FERONIA controls leaf starch accumulation by interacting with glyceraldehyde-3-phosphate dehydrogenase. Biochem Biophys Res Commun. 2015; 465 (1):77–82. 10.1016/j.bbrc.2015.07.132 [DOI] [PubMed] [Google Scholar]
- 18.Yu F, Li J, Huang Y, Liu L, Li D, Chen L, et al. FERONIA receptor kinase controls seed size in Arabidopsis thaliana. Mol Plant .2014; 7 (5):920–922. 10.1093/mp/ssu010 [DOI] [PubMed] [Google Scholar]
- 19.Keinath NF, Kierszniowska S, Lorek J, Bourdais G, Kessler SA, Shimosato-Asano H, et al. PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J Biol Chem. 2010; 285 (50):39140–39149. 10.1074/jbc.M110.160531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kessler SA, Shimosato-Asano H, Keinath NF, Wuest SE, Ingram G, Panstruga R, et al. Conserved molecular components for pollen tube reception and fungal invasion. Science. 2010; 330 (6006):968–971. 10.1126/science.1195211 [DOI] [PubMed] [Google Scholar]
- 21.Guo H, Li L, Ye H, Yu X, Algreen A, Yin Y. Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2009; 106 (18):7648–7653. 10.1073/pnas.0812346106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo H, Ye H, Li L, Yin Y. A family of receptor-like kinases are regulated by BES1 and involved in plant growth in Arabidopsis thaliana. Plant Signal Behav. 2009; 4 (8):784–786. 10.1073/pnas.0812346106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deslauriers SD, Larsen PB. FERONIA is a key modulator of brassinosteroid and ethylene responsiveness in Arabidopsis hypocotyls. Mol Plant. 2010; 3 (3):626–640. 10.1093/mp/ssq015 [DOI] [PubMed] [Google Scholar]
- 24.Hematy K, Sado PE, Van Tuinen A, Rochange S, Desnos T, Balzergue S, et al. A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr Biol. 2007; 17 (11): 922–931. 10.1016/j.cub.2007.05.018 [DOI] [PubMed] [Google Scholar]
- 25.Hematy K, Hofte H. Novel receptor kinases involved in growth regulation. Curr Opin Plant Biol. 2008; 11 (3):321–328. 10.1016/j.pbi.2008.02.008 [DOI] [PubMed] [Google Scholar]
- 26.Boisson-Dernier A, Kessler SA, Grossniklaus U. The walls have ears: the role of plant CrRLK1Ls in sensing and transducing extracellular signals. J Exp Bot. 2011; 62 (5):1581–1591. 10.1093/jxb/erq445 [DOI] [PubMed] [Google Scholar]
- 27.Boisson-Dernier A, Roy S, Kritsas K, Grobei MA, Jaciubek M, Schroeder JI, et al. Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development. 2009; 136 (19):3279–3288. 10.1242/dev.040071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schallus T, Jaeckh C, Feher K, Palma AS, Liu Y, Simpson JC, et al. Malectin: a novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Mol Biol Cell. 2008; 19 (8): 3404–3414. 10.1091/mbc.E08-04-0354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schallus T, Feher K, Sternberg U, Rybin V, Muhle-Goll C. Analysis of the specific interactions between the lectin domain of malectin and diglucosides. Glycobiology. 2010; 20 (8):1010–1020. 10.1093/glycob/cwq059 [DOI] [PubMed] [Google Scholar]
- 30.Takeda K, Qin SY, Matsumoto N, Yamamoto K. Association of malectin with ribophorin I is crucial for attenuation of misfolded glycoprotein secretion. Biochem Biophys Res Commun. 2014; 454 (3):436–440. 10.1016/j.bbrc.2014.10.102 [DOI] [PubMed] [Google Scholar]
- 31.Lindner H, Muller LM, Boisson-Dernier A, Grossniklaus U. CrRLK1L receptor-like kinases: not just another brick in the wall. Curr Opin Plant Biol.2012; 15 (6):659–669. 10.1016/j.pbi.2012.07.003 [DOI] [PubMed] [Google Scholar]
- 32.Nguyen QN, Lee YS, Cho LH, Jeong HJ, An G, Jung KH. Genome-wide identification and analysis of Catharanthus roseus RLK1-like kinases in rice. Planta. 2015; 241 (3):603–613. 10.1007/s00425-014-2203-2 [DOI] [PubMed] [Google Scholar]
- 33.Niu E, Cai C, Zheng Y, Shang X, Fang L, Guo W. Genome-wide analysis of CrRLK1L gene family in Gossypium and identification of candidate CrRLK1L genes related to fiber development. Mol Genet Genomics. 2016; 10.1007/s00438-016-1169-0 [DOI] [PubMed] [Google Scholar]
- 34.Petersen TN, Brunak S, Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011; 8(10):785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 35.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007; 23 (21):2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 36.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28 (10): 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voorrips RE. MapChart: Software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93(1), 77–78. 10.1093/jhered/93.1.77 [DOI] [PubMed] [Google Scholar]
- 38.Rambaldi D, Ciccarelli FD. Fancy Gene: dynamic visualization of gene structures and protein domain architectures on genomic loci. Bioinformatics. 2009; 25 (17):2281–2282. 10.1093/bioinformatics/btp381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37 (Web Server issue):W202–208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holub EB. The arms race is ancient history in Arabidopsis, the wildflower. Nat Rev Genet. 2001; 2:516–527. 10.1038/35080508 [DOI] [PubMed] [Google Scholar]
- 41.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004; 4, 10 10.1186/1471-2229-4-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jia H, Wang Y, Sun M, Li B, Han Y, Zhao Y, et al. Sucrose functions as a signal involved in the regulation of strawberry fruit development and ripening. New Phytol. 2013; 198 (2):453–65. 10.1111/nph.12176 [DOI] [PubMed] [Google Scholar]
- 43.Seymour GB, Østergaard L, Chapman NH, Knapp S, Martin C. Fruit development and ripening. Annu Rev Plant Biol. 2013; 64: 219–241. 10.1146/annurev-arplant-050312-120057 [DOI] [PubMed] [Google Scholar]
- 44.Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, et al. The genome of woodland strawberry (Fragaria vesca). Nat Genet. 2011; 43 (2), 109–16. 10.1038/ng.740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giovannoni J. MOLECULAR BIOLOGY OF FRUIT MATURATION AND RIPENING. Annu Rev Plant Physiol Plant Mol Biol. 2001; 52:725–749. 10.1146/annurev.arplant.52.1.725 [DOI] [PubMed] [Google Scholar]
- 46.Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science. 2014; 343 (6169): 408–411. 10.1126/science.1244454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li S, Zhang Y. To grow or not to grow: FERONIA has her say. Mol Plant. 2014;7 (8): 1261–1263. 10.1093/mp/ssu031 [DOI] [PubMed] [Google Scholar]
- 48.Wolf S, Hofte H. Growth Control: A Saga of Cell Walls, ROS, and Peptide Receptors. Plant Cell. 2014; 26 (5):1848–1856. 10.1105/tpc.114.125518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han Y, Dang R, Li J, Jiang J, Zhang N, Jia M, et al. SUCROSE NONFERMENTING1-RELATED PROTEIN KINASE2.6, an ortholog of OPEN STOMATA1, is a negative regulator of strawberry fruit development and ripening. Plant Physiol. 2015;167 (3):915–930. 10.1104/pp.114.251314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aharoni A, Keizer LC, Van Den Broeck HC, Blanco-Portales R, Munoz-Blanco J, Bois G, et al. Novel insight into vascular, stress, and auxin-dependent and -independent gene expression programs in strawberry, a non-climacteric fruit. Plant Physiol. 2002; 129 (3):1019–1031. 10.1104/pp.003558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Given NK, Venis MA, Gierson D.Hormonal regulation of ripening in the strawberry, a non-climacteric fruit. Planta.1988; 174(3):402–406. 10.1007/BF00959527 [DOI] [PubMed] [Google Scholar]
- 52.Miyazaki S, Murata T, Sakurai-Ozato N, Kubo M, Demura T, Fukuda H, et al. ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Curr Biol. 2009; 19 (15):1327–1331. 10.1016/j.cub.2009.06.064 [DOI] [PubMed] [Google Scholar]
- 53.Kanaoka MM, Torii KU. FERONIA as an upstream receptor kinase for polar cell growth in plants. Proc Natl Acad Sci U S A. 2010; 107 (41):17461–17462. 10.1073/pnas.1013090107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shih HW, Miller ND, Dai C, Spalding EP, Monshausen GB. The receptor-like kinase FERONIA is required for mechanical signal transduction in Arabidopsis seedlings. Curr Biol. 2014; 24 (16):1887–1892. 10.1016/j.cub.2014.06.064 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.